ABSTRACT
Pandemic influenza, typically caused by the reassortment of human and avian influenza viruses, can result in severe or fatal infections in humans. Timely identification of potential pandemic viruses must be a priority in influenza virus surveillance. However, the range of host species responsible for the generation of novel pandemic influenza viruses remains unclear. In this study, we conducted serological surveys for avian and human influenza virus infections in farmed mink and determined the susceptibility of mink to prevailing avian and human virus subtypes. The results showed that farmed mink were commonly infected with human (H3N2 and H1N1/pdm) and avian (H7N9, H5N6, and H9N2) influenza A viruses. Correlational analysis indicated that transmission of human influenza viruses occurred from humans to mink, and that feed source was a probable route of avian influenza virus transmission to farmed mink. Animal experiments showed that mink were susceptible and permissive to circulating avian and human influenza viruses, and that human influenza viruses (H3N2 and H1N1/pdm), but not avian viruses, were capable of aerosol transmission among mink. These results indicate that farmed mink could be highly permissive “mixing vessels” for the reassortment of circulating human and avian influenza viruses. Therefore, to reduce the risk of emergence of novel pandemic viruses, feeding mink with raw poultry by-products should not be permitted, and epidemiological surveillance of influenza viruses in mink farms should be urgently implemented.
KEYWORDS: Mink (Mustelidae), human influenza virus, avian influenza virus, surveillance, reassortment
Introduction
Influenza A virus epidemics occur regularly worldwide and are estimated to cause 3–5 million cases of severe illness, with 290,000–650,000 deaths each year [1]. Periodic pandemics can be much worse in terms of scale, morbidity and mortality. Historical pandemic influenza viruses, such as the 1957 H2N2, 1968 H3N2, and 2009 H1N1/pdm virus, were all derived from novel reassortant viruses [2,3]. Host species that are susceptible to both human and animal influenza viruses could, therefore, serve as “mixing vessels,” as has been described for pigs [4], through co-infections to generate novel reassortant viruses with the potential to cause pandemics. Mink, a fur-bearing animal, and the related ferret are members of the family Mustelidae. The ferret has been widely accepted as an excellent animal model for understanding the virulence and transmission of influenza viruses as it shares similar lung physiology and sialic acid (SA) receptor distribution to humans [5,6]. Similar to ferrets, the SA receptors SA-α-2,3-Gal and SA-α-2,6-Gal are both found in the respiratory tracts of mink [7]. Since 1984, several influenza A virus subtypes, such as avian influenza viruses (AIVs) H10N4, H5N1, and H9N2, and human influenza viruses H3N2 and H1N1/pdm [7–11], have been isolated from mink, indicating that the host species could serve as an intermediate influenza virus host in transmission between poultry and humans. However, due to a lack of systematic surveillance of influenza viruses circulating in mink, their contribution to influenza virus evolution and susceptibility to prevailing avian and human influenza viruses are unknown.
In recent years, mink farming in China has grown rapidly, with a corresponding increase in the use of animal feed, which primarily comprises raw poultry by-products. Influenza epidemiology data have indicated that there are three major subtypes of influenza viruses prevalent in poultry populations in China: H5N6, H7N9, and H9N2 [12–14]; in human populations, the predominant subtypes are H1N1/pdm and H3N2 [15]. Thus, there may be a possibility that farmed mink are regularly exposed to human and avian influenza viruses. As mink in China are not vaccinated against influenza viruses, it is relatively straightforward to perform sero-surveillance for influenza virus infections in this species. In this study, we conducted serological surveys for avian and human influenza virus infections in farmed mink and determined the susceptibility of mink to the prevailing avian and human influenza virus subtypes.
Materials and methods
Ethical compliance
All animal experiments were approved by the Beijing Association for Science and Technology (approval SYXK [Beijing] 2007-0023) and conducted in accordance with the Beijing Laboratory Animal Welfare and Ethics guidelines, as issued by the Beijing Administration Committee of Laboratory Animals, and in accordance with the China Agricultural University Institutional Animal Care and Use Committee guidelines (SKLAB-B-2010-003).
Mink blood sampling and influenza virus antibody detection
Blood samples of mink were collected at slaughter, mostly from February to March and August to October according to animal slaughter plans. From September 2016 to March 2019, a total of 2455 blood samples were collected from 34 mink farms in northern China, of which 1772 were located in Shandong province (which produces more than 50% of the mink in China), and 683 were from Hebei province. For each group of 500–3000 mink, blood samples were collected on the same day from 20–150 mink (a 5% sample). This provided a 95% confidence level, that is, if the minimum prevalence of seropositive mink was 10%, at least one seropositive mink would be identified. Detailed production information was collected from each group tested which included age, feed type, location, and group size. Haemagglutination inhibition (HI) assays were performed as previously described [16]. Based on the World Health Organization (WHO) guideline for vaccine evaluation, HI antibody titres of ≥40 would be considered positive. Virus neutralization (VN) assay was used to confirm positive HI results. VN antibody titres of ≥10 were considered positive. Mink sera were considered seropositive only if they were both HI- and VN-positive (HI+/VN+). From 2016 to 2019, H1N1/pdm and H3N2 influenza viruses were prevalent in human populations in northern China [15]. The AIV subtypes H5N6, H7N9, and H9N2 are endemic in domestic poultry [12,13]. Thus, human H1N1/pdm and H3N2 viruses, and avian H5N6, H7N9, and H9N2 viruses were chosen as diagnostic antigens for the HI and VN assays. Taking into consideration antigenic drift of influenza viruses over time, A/Hebei/1104/2016 (H1N1/pdm), A/Beijing/1230/2016 (H3N2), A/duck/Eastern China/S0711/2014 (H5N6), A/chicken/Henan/KF/2014 (H7N9), and A/chicken/Shandong/1/2016 (H9N2) viruses were used to detect seroconversion in samples from 2016 to 2017; A/Beijing/0212/2018 (H1N1/pdm), A/Tianjin/0122/2018 (H3N2), A/chicken/Northern China/F0130c/2018 (H5N6), A/chicken/Hebei/0417/2018 (H7N9), and A/chicken/Shandong/0322/2018 (H9N2) were used on sera from 2018 to 2019.
Mink influenza virus infection and transmission
Four-month-old female mink, weighing 1000–1200 g, were purchased from a farm in Yantai, Shandong province. All were seronegative for currently circulating influenza viruses (H1, H3, H5, H7, and H9). Three mink in each group were anaesthetized with ketamine (20 mg/kg) and xylazine (1 mg/kg), and inoculated intranasally with 106 50% tissue culture infective dose (TCID50) of test virus in a 1 mL volume (500 μL per nostril). The animals were subsequently euthanised at 4 days post-infection (dpi), and nasal turbinate, trachea, lung, liver, kidney, spleen, intestine, and brain samples were collected for virus titration in Madin–Darby canine kidney (MDCK) cells. Nasal turbinate, trachea, and lung tissues were also used for histopathological analysis and immuno-detection of viral nucleoprotein. In virus transmission, groups of three mink were anaesthetized and inoculated intranasally with 106 TCID 50 of virus. The following day, a respiratory-droplet contact mink was housed in a wire frame cage 5 cm from the infected donor mink. To monitor virus shedding, nasal washes were collected from all animals every other day for 12 days and titrated for virus in MDCK cells. Sera were collected from both the inoculated and contact animals at 14 dpi. Seroconversion was analysed using HI assays. Clinical signs, temperature and body weight were recorded daily for all animals. A/Beijing/0212/2018 (H1N1/pdm), A/Tianjin/0122/2018 (H3N2), A/mink/Northern China/F0130 m/2018 (H5N6, a highly pathogenic AIV [HPAIV]), A/chicken/Hebei/0417/2018 (H7N9, a low pathogenicity avian influenza virus [LPAIV]), and A/chicken/Shandong/0322/2018 (H9N2) viruses were used for mink infection. All experiments with H5 and H7 subtype viruses were performed in biosafety level 3 containments.
Statistical analyses
The relationship between seroprevalence of human influenza viruses in mink and human infection cases was evaluated using Spearman rank correlation coefficient (rho, ρ). The degree of correlation was considered weak if ρ was <0.4, moderate if ρ was ≥0.4 and <0.6, and strong if ρ was ≥0.6. Human case data were obtained from the weekly influenza surveillance reports of the Chinese Centre for Disease Control and Prevention (CDC) from 2016 to 2019 [15]. Pearson’s chi-squared test or Fisher's exact test was used to assess significant differences in seroprevalence between the groups with different feed types. For all analyses, a P-value of <.05 was considered to be statistically significant. All analyses were performed using SPSS software version 19.0 (IBM Corporation, Armonk, NY, USA).
Results
Seroprevalence of antibodies against influenza viruses in farmed mink
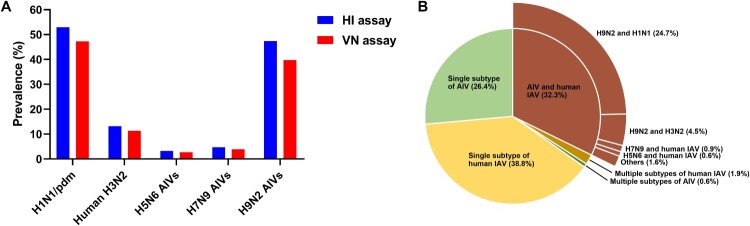
The overall seroprevalence of human and avian influenza viruses in the collected mink samples was 76.8% (1,885/2,455 [positive for both HI and VN assays, HI+/VN+]), of which 47.3% (1,162/2,455) and 11.4% (279/2,455) were positive for H1N1/pdm and H3N2 influenza viruses, respectively; and 2.8% (68/2,455), 3.9% (96/2,455), and 39.7% (975/2,455) were positive for avian viruses H5N6, H7N9, and H9N2, respectively (Figure 1(A) and Figure 2).
Figure 1.
Seroprevalence of influenza viruses in farmed mink in northern China. (A) Seroprevalence of influenza viruses in mink by combined HI and VN assays. A total of 2455 serum samples were assayed by HI (titres of ≥40 were considered positive). All HI positive samples were confirmed by VN assays (titres of ≥10 were considered positive). (B) Breakdown of positive samples by virus subtypes. Details of the number of positive samples are shown in Table S1. Note: HI: haemagglutination inhibition; VN: virus neutralization.
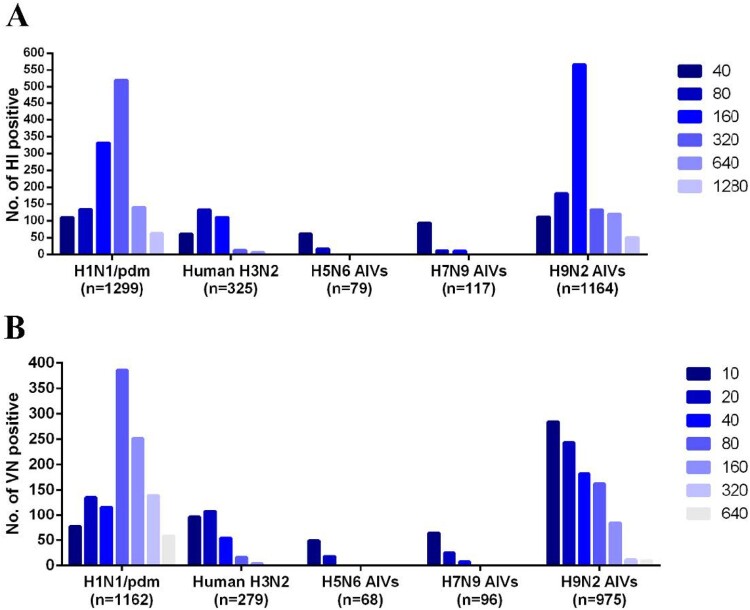
Figure 2.
Antibody titres in serum samples from farmed mink that were positive for influenza viruses. (A) HI titres in samples positive for human H1N1/pdm and H3N2 viruses, or avian H5N6, H7N9 and H9N2 viruses from a total of 2455 samples (titres of ≥40 were considered positive). (B) VN titres in samples positive for human H1N1/pdm and H3N2 viruses, or avian H5N6, H7N9, or H9N2 viruses from a total of 2108 HI-positive samples (titres of ≥10 were considered positive). The number of positive serum samples for each virus is indicated.
Further analysis of co-infection prevalence in the 1885 HI+/VN+ samples showed that 34.7% (655/1885) of the samples were seropositive for multiple influenza virus subtypes, of which 32.3% (608/1885) contained both avian and human influenza antibodies (Figure 1(B) and Supplementary Figure 1). Specifically, the seroprevalence in mink for avian H9N2 and human H1N1/pdm viruses was 26.2% (494/1885), seroprevalence for avian H5N6 and human influenza viruses was 0.8% (15/1885), and seroprevalence for avian H7N9 and human influenza viruses was 1.2% (23/1885) (Supplementary Table 1).
Association between incidence of human influenza virus infection in humans and corresponding seroprevalence in farmed mink
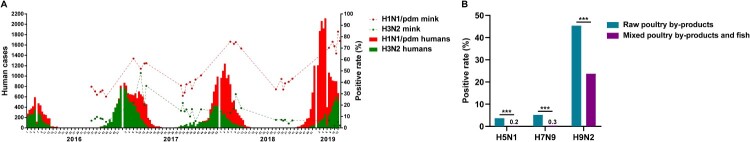
Analysis of monthly seroprevalence data for all viruses found that the seroprevalence of human influenza virus in mink correlated with circulating virus levels in humans; it was high from February to March and relatively low from August to October (Supplementary Table 2). We further analysed the correlations of the incidence of human influenza virus infections between mink and human populations. Cumulative human cases of H1N1/pdm virus infections had increased annually from 2016 (Figure 3(A)). Seroprevalence of H1N1/pdm virus in mink followed the same rising trend, peaking at 72.0% in 2019 (Figure 3(A)), and was positively correlated with the incidence of human infection (P = .005, ρ = .746) (Table 1). Human cases of human H3N2 virus infections peaked in 2017 followed by subsequent decline. Similarly, the seroprevalence of H3N2 virus in mink was at its highest in 2017 (at 20.2%), which was followed by decline. There was no significant correlation between the seroprevalence of H3N2 virus antibodies in mink and incidence of H3N2 virus infections in humans (Table 1). However, given that influenza seroconversion in mink would lag behind those in human populations, and if this lag period of one or two months is taken into account, the correlation between the two host populations for H3N2 virus infection was statistically significant (Table 1).
Figure 3.
Correlational analysis of mink infection with human and avian influenza viruses. (A) Statistical analysis of the number of human seasonal influenza A (H1N1/pdm and H3N2) cases and the seroprevalence of influenza virus infection in farmed mink in northern China from 2016 to 2019. Coloured columns indicate weekly number of human cases of influenza virus infection from the influenza surveillance reports of China CDC from 2016 to 2019. Dashed lines show the seroprevalence of H1N1/pdm and H3N2 viruses in different mink farms. (B) Comparison of the seroprevalence values according to the types of food used to feed mink in farms. ***P < .001, between farms feeding mink with poultry by-products only and those feeding mink with both poultry by-products and fish.
Table 1.
Correlations of seroprevalence of human influenza viruses between mink and human populations.
| Number of months lagged | H1N1/pdm | H3N2 | ||
|---|---|---|---|---|
| rhoa | P valuea | rho | P value | |
| 0 | 0.746 | .005 | 0.445 | .147 |
| 1 | 0.739 | .006 | 0.616 | .033 |
| 2 | 0.789 | .002 | 0.674 | .016 |
Note: Statistically significant differences (P < .05) in italics.
Spearman correlation test.
Association between feed type and avian influenza virus infection in farmed mink
Previous research found that influenza virus could be transmitted from swine to mink following feeding animals with a ration composed of uncooked meat by-products of swine [11]. Here, we examined whether feed was a possible route for AIV transmission to farmed mink. Mink are usually fed with raw poultry by-products (offal and carcasses of poultry) from two months old. For better reproduction performance, some mink farms use fish products supplemented with 20–30% poultry by-products. The majority of farms investigated (24/34) used only poultry by-products, and the remaining 10 farms used both poultry by-products and fish (mixed-feed farms) (Supplementary Table 3). In farms that used only poultry by-products, the seroprevalence values of avian H5N6, H7N9, and H9N2 viruses were 3.7%, 5.2%, and 45.4%, respectively. These were significantly higher (P < .001) than the seroprevalence values in the mixed-feed farms, which were 0.2%, 0.3%, and 23.7% for H5N6, H7N9, and H9N2 viruses, respectively (Figure 3(B)).
Susceptibility of mink to circulating avian and human influenza viruses
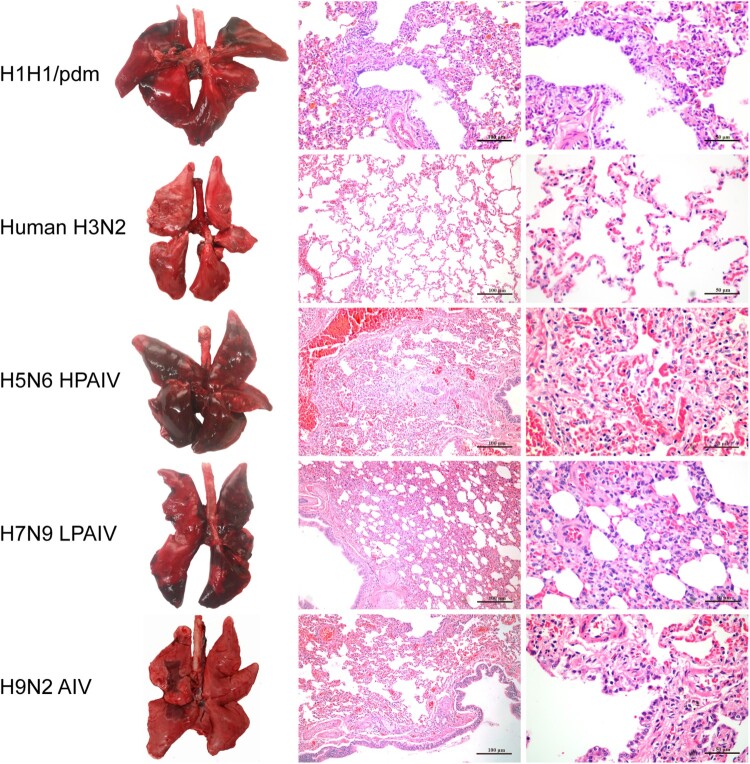
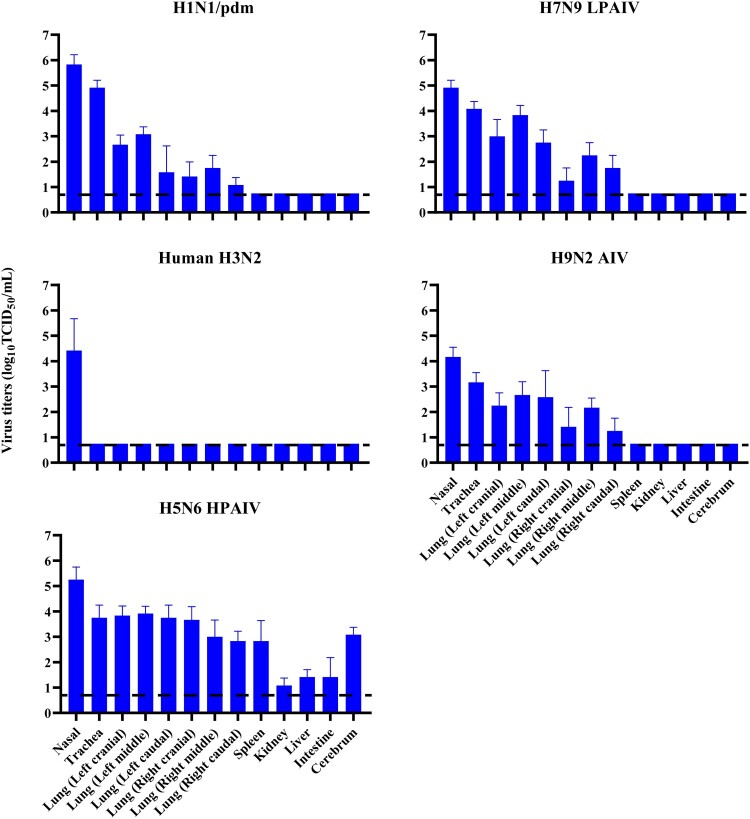
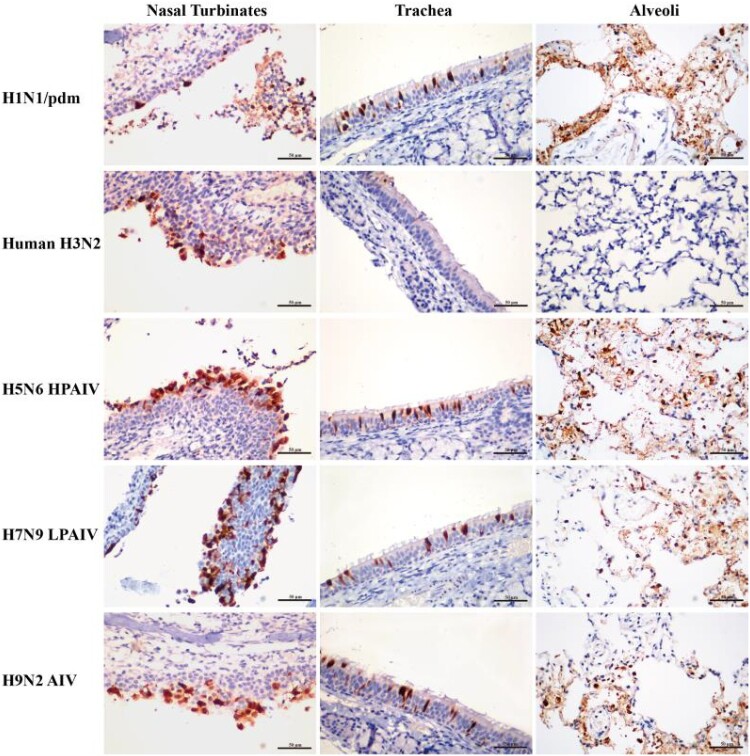
To determine host susceptibility, mink were intranasally infected with human (H1N1/pdm and H3N2) and avian (H5N6, H7N9, and H9N2) influenza viruses. Clinically, mink infected with human H3N2 virus were asymptomatic, while those infected with H1N1/pdm, H7N9, and H9N2 influenza viruses exhibited transient signs of pyrexia (Supplementary Figure 2(A)), sneezing and nasal discharge. H5N6 virus was the most pathogenic, and infected mink displayed severe pyrexia and weight loss (>15%), which led to the euthanasia of all three mink at 10 dpi (Supplementary Figure 2(B)). No apparent abnormality was found in the lungs of mink with H3N2 virus infection; however, multifocal areas of pulmonary consolidation were present in the lungs of mink infected with H1N1/pdm, H7N9, and H9N2 viruses (Figure 4). Consistent with the clinical severity, H5N6 virus infections caused the most severe gross and histopathological changes in the lungs (Figure 4). Virus shedding up to 6 dpi was evident from infected mink for all avian and human influenza viruses, with a virus titre range of 103.5–105.5 TCID50/mL based on nasal washes. With avian H5N6 and H9N2 viruses, shedding continued to be detected at 8 dpi (Supplementary Table 4). Virus spread to other tissues was consistent with the severity of infection. Human H3N2 virus was detected only in the nasal turbinate, whereas H1N1/pdm, H7N9, and H9N2 viruses were detected throughout the respiratory tract (Figures 5 and 6). The systemic spread of the H5N6 virus was evident by its presence in all organs examined (Figures 5 and 6).
Figure 4.
Gross pathology and histopathology of the lungs of mink infected with human (H1N1/pdm and H3N2) and avian (H5N6, H7N9, and H9N2) influenza viruses at 4 dpi. Lungs of mink with H1N1/pdm, H9N2, and H7N9 influenza virus infections showed mild to moderate pulmonary consolidation with varying degrees of bronchitis. Few remarkable pathological findings were obtained from the lungs of mink infected with the H3N2 virus. In contrast, the H5N6 influenza virus caused severe lesions with extensive pulmonary consolidation, peribronchiolitis and bronchopneumonia characterised by oedema and infiltration of inflammatory cells. Images shown are representative of three mink from three independent experiments.
Figure 5.
Virus replication in mink infected with avian and human influenza viruses. Mink were inoculated intranasally with 106 TCID50 of indicated viruses. Three mink per group were euthanised at 4 dpi, and samples of nasal turbinate, trachea, lung, spleen, liver, kidney, intestine and cerebrum were collected from each mink for virus titration by TCID50 assays on MDCK cells. Systemic spread of H5N6 virus was detected in all organs examined. All values shown are means ± SD from three independent experiments for each sample. Dashed lines indicate the lower limits of detection.
Figure 6.
Detection of viral nucleoprotein in the respiratory tract of mink infected with influenza viruses at 4 dpi. Tissue sections of nasal turbinate, trachea and lung (alveoli) were immunostained for viral nucleoprotein (red-brown staining). Extensive distribution of H1N1/pdm, H5N6, H7N9, and H9N2 influenza virus were evident in the nasal turbinate, trachea, and lung. In contrast, the H3N2 virus was mainly restricted to the nasal turbinate.
Transmission of human influenza viruses among mink
To assess airborne transmissibility of human and avian influenza A viruses among mink, naïve mink were placed 5 cm away, in groups of three in separate wire cages, from infected mink 24 h post-infection. Human H1N1/pdm and H3N2 viruses were readily transmitted from mink that were infected earlier, via aerosol, to naïve mink at 2 and 4 dpi, respectively, with subsequent seroconversion (Figure 7 and Supplementary Table 5). No airborne transmission to naïve mink was detected with avian H5N6, H7N9, and H9N2 viruses (Figure 7 and Supplementary Table 5).
Figure 7.
Aerosol transmission of human influenza viruses between mink. Groups of three mink were inoculated intranasally with 106 TCID50 of the viruses indicated. The following day, a respiratory-droplet contact mink was housed in a wire frame cage 5 cm from infected donor mink cage. Nasal washes for virus shedding detection were collected every two days from all animals. Virus titres were determined by TCID50 assays on MDCK cells. Each coloured bar represents the virus titre in a single animal. Horizontal dashed lines indicate the lower limits of virus detection.
Discussion
We found that farmed mink in China were commonly infected with avian and human influenza viruses, of which human H1N1/pdm and avian H9N2 influenza virus infections were the most prevalent. The high seroprevalence of combined avian and human influenza viruses suggests a strong likelihood of co-infections and thus farmed mink could serve as “mixing vessels,” as in pigs, for the generation of novel reassortant viruses. Our host susceptibility analysis found that mink are susceptible and permissive to circulating avian (H7N9, H5N6, and H9N2) and human (H3N2 and H1N1/pdm) influenza A viruses. Only the human influenza viruses, not avian viruses, were capable of aerosol transmission among mink, suggesting that mink are similar to ferrets as conducive hosts for human influenza virus replication. We previously found that the avian H9N2 virus subtype shows good gene compatibility in reassortment with the human H1N1/pdm virus, which in co-infection could generate novel reassortant viruses with high human infectivity [17]. Although the co-infection rates of avian H5 or H7 viruses with human H1N1/pdm or H3N2 viruses appear low, the public health threat from such reassortant viruses should not be ignored as their pandemic potential has been experimentally demonstrated [18–21].
Infection prevalence of human H1N1/pdm or H3N2 viruses in both human and mink populations strongly indicates human-to-mink virus transmission. Previous studies suggested that mammalian and avian influenza viruses could be effectively transmitted to mink through uncooked poultry or pork by-products [7,10,11,22–24]. Our correlational analysis supports the notion that feeding mink with raw poultry by-products is a major source of AIV transmission. Avian H9N2 influenza virus is the most dominant subtype in poultry [13,25] and was also the most common seropositive subtype found in farmed mink. Notably, since the introduction of the avian H7N9 subtype vaccine to chicken flocks in China in 2017 [26], no H7N9 seropositive mink was detected in 2019.
Recently, the rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was reported in mink farms in several countries in Europe and the Americas; millions of mink were culled to control the spread of the virus [27–29]. As mink are inherently susceptible to both influenza viruses and SARS-CoV-2, it is important to prevent mink from being infected with such viruses. Clear intervention steps that should be taken are to stop the use of raw poultry products in the prevention of influenza virus transmission to mink and to isolate all workers with respiratory symptoms from mink farms. In summary, mink as a host species is highly susceptible and permissive to circulating human and avian influenza viruses. To avoid serious public health threats from the emergence of highly pathogenic virus reassortants, basic preventative husbandry steps, such as a ban on the use of raw poultry feed, along with regular virus surveillance of mink farms should be urgently implemented.
Supplementary Material
Funding Statement
This work was supported by the National Key Research and Development Program (2016YFD0500204) and National Natural Science Foundation of China (31873022 and 31761133003).
Disclosure statement
The authors have no potential competing interests.
References
- 1.Influenza (Seasonal). 2018. Available from: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal)
- 2.Neumann G, Noda T, Kawaoka Y.. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459(7249):931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawaoka Y, Krauss S, Webster RG.. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J Virol. 1989;63(11):4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma W, Kahn RE, Richt JA.. The pig as a mixing vessel for influenza viruses: human and veterinary implications. J Mol Genet Med. 2008;3(1):158–166. [PMC free article] [PubMed] [Google Scholar]
- 5.Shinya K, Ebina M, Yamada S, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440(7083):435–436. [DOI] [PubMed] [Google Scholar]
- 6.van Riel D, Munster VJ, de Wit E, et al. H5n1 virus attachment to lower respiratory tract. Science. 2006;312(5772):399–399. [DOI] [PubMed] [Google Scholar]
- 7.Peng L, Chen C, Kai-yi H, et al. Molecular characterization of H9N2 influenza virus isolated from mink and its pathogenesis in mink. Vet Microbiol. 2015;176(1–2):88–96. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Li Z, Cui Y, et al. Emergence of an Eurasian avian-like swine influenza A (H1N1) virus from mink in China. Vet Microbiol. 2020;240(108509):1–7. [DOI] [PubMed] [Google Scholar]
- 9.Englund L. Studies on influenza viruses H10N4 and H10N7 of avian origin in mink. Vet Microbiol. 2000;74(1-2):101–107. [DOI] [PubMed] [Google Scholar]
- 10.Jiang W, Wang S, Zhang C, et al. Characterization of H5N1 highly pathogenic mink influenza viruses in eastern China. Vet Microbiol. 2017;201:225–230. [DOI] [PubMed] [Google Scholar]
- 11.Gagnon CA, Spearman G, Hamel A, et al. Characterization of a Canadian mink H3N2 influenza A virus isolate genetically related to triple reassortant swine influenza virus. J Clin Microbiol. 2009;47(3):796–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bi Y, Chen Q, Wang Q, et al. Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host & Microbe. 2016;20(6):810–821. [DOI] [PubMed] [Google Scholar]
- 13.Su S, Bi Y, Wong G, et al. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol. 2015;89(17):8671–8676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pu J, Wang S, Yin Y, et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A. 2015;112(2):548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Influenza Weekly Report. Available from: http://www.chinaivdc.cn/cnic/
- 16.Eichelberger M, Golding H, Hess M, et al. FDA/NIH/WHO public workshop on immune correlates of protection against influenza A viruses in support of pandemic vaccine development, Bethesda, Maryland, US, December 10-11, 2007. Vaccine. 2008;26(34):4299–4303. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Qin K, Wang J, et al. High genetic compatibility and increased pathogenicity of reassortants derived from avian H9N2 and pandemic H1N1/2009 influenza viruses. Proc Natl Acad Sci U S A. 2011;108(10):4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhang Q, Kong H, et al. H5n1 hybrid viruses bearing 2009/H1N1 virus genes transmit in Guinea pigs by respiratory droplet. Science. 2013;340(6139):1459–1463. [DOI] [PubMed] [Google Scholar]
- 20.Li C, Hatta M, Nidom CA, et al. Reassortment between avian H5N1 and human H3N2 influenza viruses creates hybrid viruses with substantial virulence. Proc Natl Acad Sci U S A. 2010;107(10):4687–4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen L-M, Davis CT, Zhou H, et al. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 2008;4(5):e1000072–e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gholipour H, Busquets N, Fernández-Aguilar X, et al. Influenza A virus surveillance in the invasive American mink (Neovison vison) from freshwater ecosystems, northern Spain. Zoonoses Public Health. 2017;64(5):363–369. [DOI] [PubMed] [Google Scholar]
- 23.Xue R, Tian Y, Hou T, et al. H9n2 influenza virus isolated from minks has enhanced virulence in mice. Transbound Emerg Dis. 2018;65(3):904–910. [DOI] [PubMed] [Google Scholar]
- 24.Yoon KJ, Schwartz K, Sun D, et al. Naturally occurring influenza A virus subtype H1N2 infection in a midwest United States mink (mustela vison) ranch. J Vet Diagn Invest. 2012;24(2):388–391. [DOI] [PubMed] [Google Scholar]
- 25.Gu M, Xu L, Wang X, et al. Current situation of H9N2 subtype avian influenza in China. Vet Res. 2017;48(1):49–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng X, Tian G, Shi J, et al. Vaccination of poultry successfully eliminated human infection with H7N9 virus in China. Science China Life Sciences. 2018;61(12):1465–1473. [DOI] [PubMed] [Google Scholar]
- 27.Koopmans M. SARS-CoV-2 and the human-animal interface: outbreaks on mink farms. Lancet Infect Dis. 2021;21(1):18–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammer AS, Quaade ML, Rasmussen TB, et al. SARS-CoV-2 transmission between mink (Neovison vison) and humans, Denmark. Emerging Infect. Dis.. 2021;27(2):547–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371(6525):172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.