Abstract
Background
Globally, chicken is known to be a reservoir for the spread of antimicrobial resistance genes to humans. In Nigeria, antimicrobial drugs are readily accessible for use in poultry production, either for preventive or therapeutic purposes. Extended-spectrum beta-lactamase-producing Escherichia coli (ESBL-EC) are transmissible to humans because of their zoonotic potentials. People working very closely with chickens either on farms or markets are at greater risk. The aim of this study was to investigate the prevalence and zoonotic transmission of ESBL-EC among poultry-workers, chickens, and poultry environments in Abuja, Nigeria.
Methods
We conducted a cross-sectional study among workers, chickens and poultry environment in selected farms/chicken markets in Abuja. Stool, faecal, and environmental samples were collected from apparently healthy workers, chickens, and farm/market environments from December 2018 to April 2019. Data were collected electronically using an open data kit (ODK) installed on a Smartphone. Antimicrobial resistance was determined using broth micro-dilution methods against a panel of 14 antimicrobial agents. We carried out the phenotypic and genotypic characterization of the isolates. Data were analyzed by computing frequencies, proportions and spearman’s correlation (ρ).
Results
Of 429 samples, 26.8% (n = 115) were positive for Escherichia coli (E. coli). Of the 115 E. coli isolates, 32.2% (n = 37) were confirmed ESBL producers by phenotypic characterization. Prevalence of ESBL-EC was highest among both poultry-workers (37.8%; n = 14) and chickens (37.8%; n = 14) followed by the environment (24.3%; n = 9). Both human and chicken isolates showed similar patterns of multidrug resistance to tested antimicrobials with a positive correlation (ρ = 0.91). Among ESBL producers, we observed the dissemination of blaCTX-M (10.8%; n = 4) genes. The coexistence of blaCTX-M-15 and blaTEM-1 genes was observed in 8.1% (n = 3) of the isolates, out of which (66.7%; n = 2) were chicken isolates from the farm, while a single human isolate was from the chicken market.
Conclusions
ESBL-EC isolates were prevalent amongst apparently healthy individuals, chickens and the poultry farm/market environment in Abuja. It is important to educate healthcare workers that people in proximity with poultry are a high-risk group for faecal carriage of ESBL-EC, hence pose a higher risk to the general population for the spread of antimicrobial resistance.
Keywords: Escherichia coli, Antimicrobial resistance, Chicken, Extended-spectrum β-lactamases (ESBL), Nigeria
Background
Antimicrobial resistance (AMR) in recent times has been a topical issue and gained global attention owing to the emergence of multi-drug (MDR) resistant organisms such as antimicrobial-resistant Escherichia coli [1–3]. Drug-resistant infections in humans and food animals is increasingly a global public health issue requiring measures worldwide [2]. The World Health Organization (WHO) has stated that AMR is a bigger crisis than HIV-AIDS [3]. In 2016, official studies conducted in the UK stated that AMR accounted for about 700,000 deaths annually. It has also been estimated that by 2050 the number of deaths attributable to AMR will increase to 10 million annually if not tackled now, with 40% of these deaths occurring in Africa, second only to Asia [3].
AMR can lead to more deaths since available antimicrobials are no longer effective for the treatment of common infections in humans and animals [4]. It is characterized by the spread of AMR genes and treatment is very expensive. In the animal population, AMR is facilitated by several factors such as inappropriate medication/ route of administration, non-observance of drug withdrawal periods, poor biosecurity measures and poor surveillance amongst others [5]. According to the WHO, increased threat of antibiotic resistance is a direct result of overuse and misuse of antibiotics in animals and humans [6]. The “Path of Resistance” begins by administering antimicrobials to food-animals such as chickens to keep them healthy. These antimicrobials protect the chickens against known bacterial infections. The acquired resistance of the bacteria, however, resists the antimicrobials making them ineffective. Humans become infected by resistant bacteria via various sources such as contact through the food chain, contaminated environment, occupational exposure [6, 7].
Extended-spectrum beta-lactamase-producing E. coli (ESBL–EC), which are zoonotic in nature, is one of the commonest resistant pathogens responsible for human infections. Our recently published work showed that occupational exposure for over 10 years to poultry on farms and in live bird markets was a risk factor for acquiring multidrug-resistance (MDR) E. coli [7]. It has been documented that drug resistance among E. coli isolates has increased globally mainly as a result of the high prevalence of ESBL producing bacteria [8, 9]. This high prevalence of ESBL-EC has resulted from growing reservoirs in food animals such as chickens and the use of antimicrobials [8, 9]. Studies have shown that ESBL genes which were previously found on chromosomes, but now carried on plasmids are derivatives of plasmid-mediated β-lactamases like blaTEM as well as environmentally derived types like blaCTX-M [9, 10].
It has been documented that ESBL-EC are resistant to several antibiotics especially penicillins and cephalosporins but remain susceptible to cephamycins and carbapenems [11]. Animal food sources such as chickens have been reported as potential reservoirs for the spread of ESBL-EC to humans in close proximity or via the food chain [9, 12, 13]. The mechanism of spread of antibiotic resistance from animals to humans and vice versa remains controversial. Studies have reported that E. coli which can be isolated from water, food and farm animals is associated with antimicrobial resistance [14].
A systematic review of all AMR studies done in Nigeria as at 2017 showed that these studies are either in human and animal populations or focused on animals and the food production environment [7, 15]. However, there is paucity of data on ESBL-EC in humans, animals and the food production environments. This is one of the gaps identified in 2017 when Nigeria conducted a situation analysis on AMR in humans, food animals and the environment in response to the 68th World Health Assembly resolution 68.7 [7, 16].
We hypothesized that chickens harbouring ESBL-EC can become potential sources of transmission of resistant genes to humans exposed to chickens based on their occupation as well as to the chicken market or farm environments. To better understand the association between human ESBL-EC isolates and the potential poultry/environmental sources, we investigated the occurrence of ESBL-EC among humans handling chickens, the chickens themselves and selected poultry farms/market environments in Abuja, Nigeria using disc diffusion and whole-genome sequencing. This will generate baseline data for the implementation of the AMR National Action Plan in Nigeria using a One Health approach.
Methods
Study design and sample collection
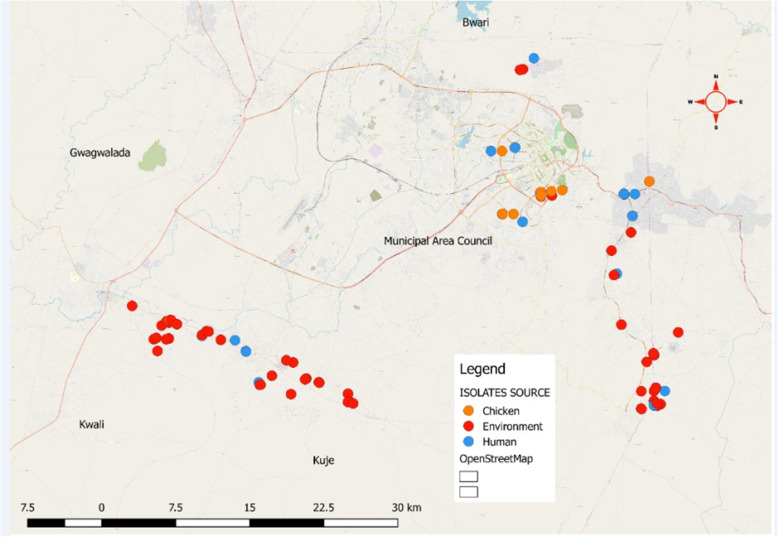
This cross-sectional study was conducted in Abuja, North Central Nigeria (Fig. 1). Fifty-two commercial poultry farms and eight live bird markets willingly participated in this study. Sampling took place from December 2018 to April 2019. We collected freshly passed stool samples from randomly selected apparently healthy consenting participants who either worked in poultry farms or chicken markets. The stool samples were collected using sterile stool containers. Fresh faecal samples from chickens that had not contacted the soil were randomly collected using a sterile spoon and stool container. Environmental samples such as litter and water samples were randomly collected from different locations on the poultry farms and live bird markets. We randomly collected 30 g of litter samples from 12 different spots in the poultry houses and 100 mL of water from each study site using sterile containers. All samples were transported in cool boxes to the National Reference Laboratory, Nigeria Centre for Disease Control Gaduwa, Abuja and processed within 3 hours of sample collection for the presence of E. coli as previously described [7]. A total of 429 samples were collected for this study, comprising 122 human stool samples, 111 chicken samples, and 196 environmental samples. Some aspects of the study in humans focusing on the risk factors for acquiring multidrug-resistant E. coli has already been published in recent times [7].
Fig. 1.
Map of Abuja showing the distribution of ESBL-EC human, chicken and environmental isolates from farms and chicken markets. This map highlights five out of six area councils in Abuja, Federal Capital Territory namely: Bwari, Kwali, Kuje, Gwagwalada and Municipal area councils. Our study was done at two area councils (Kuje and Municipal area councils) with the highest volume of poultry production in Abuja. Each dot represents an ESBL-EC isolate obtained either from humans, chickens or the environment (Source of the Map is the Federal Ministry of Agriculture and Rural Development, Abuja, Nigeria)
Microbiological analysis
Isolation and identification of E. coli isolates
Briefly, about one gram of human stool sample, one gram of poultry faeces and 30 g of litter samples were inoculated respectively in enrichment broth (buffered peptone water) in a 1:10 sample to broth ratio and incubated at 37 °C for 24 h. Subsequently, a 10ul loop-full of overnight culture from enrichment broth was plated onto MacConkey lactose agar and incubated at 37 °C for 24 h. Suspected E. coli colonies, usually pink to red were picked and further streaked on Eosin Methylene Blue (EMB) agar. Colonies suggestive of E. coli were then sub-cultured onto Tryptic Soy Agar (TSA) plates and incubated for 24 h at 37 °C under aerobic conditions for the isolation of pure cultures as previously described [7]. All the E. coli isolates were further tested for indole, methyl red, Voges–Proskauer, and citrate utilization biochemical tests [7]. Isolates that were presumptive for E. coli in the screening tests were later subjected to further testing using the commercially available biochemical test strip, Microbact GNB 24E (Oxoid, UK), for confirmation according to the Manufacturer’s instructions.
Isolation of E. coli from water samples
For the isolation of E. coli from poultry farm/live bird market water samples; the Membrane filtration technique was used for the isolation and the identification of E. coli from water samples. Single sterile 0.45 μm pores filter disks were placed in a filtration unit to filter each 100 ml of the water sample. The filter membranes were then placed on EMB agar plates and incubated at 37 °C for 24 h. All the E. coli isolates were further tested as stated above.
Antimicrobial susceptibility testing
The minimum inhibitory concentrations (MIC) of ESBL-EC isolates were determined by broth microdilution assay methods using the Gram-negative Sensititre™ (CMV3AGNF) plate (Trek Diagnostic Systems, OH) against a panel of 14 antimicrobial agents. Of these, five were β-lactam antimicrobials comprising ampicillin, amoxicillin/clavulanic acid, ceftriaxone, ceftiofur, and cefoxitin while the rest were non-β-lactam antimicrobials (streptomycin, gentamicin, azithromycin, ciprofloxacin, chloramphenicol, trimethoprim/sulfamethoxazole, tetracycline, sulfisoxazole, and nalidixic acid. Briefly, three distinct colonies were picked from the overnight culture on TSA plates and suspended into 4 ml of sterile deionized water. Next, this was adjusted to a 0.5 McFarland standard after which 10 μl of suspension was mixed with Mueller-Hinton broth. Next, 50 μl of the suspension was inoculated to each well of a Sensititre™ plate using the Sensititre™ auto-inoculator (Trek Diagnostic Systems, OH) and incubated at 37 °C for 24 h. Plates were read using Sensititre™ ARIS automated system which interprets isolates based on the MIC as susceptible, intermediate or resistant using the guidelines of Clinical and Laboratory Standards Institute (CLSI) M100 28th Edition [17]. E. coli ATCC25922 was used for internal quality control and categorized ESBL-EC isolates with intermediate MIC levels as resistant [17]. We defined multidrug resistance (MDR) as resistance to three or more classes of antimicrobials.
Phenotypic and genotypic detection of ESBLs
Detection of ESBL phenotype by disk diffusion test
All the E. coli isolates were screened for the production of extended-spectrum beta-lactamase (ESBLs) by using the disk diffusion test as described by CLSI guidelines (M100 28th Edition) [17]. From the pure cultures of bacteria grown overnight on TSA plate supplemented with sheep blood, a suspension matching 0.5 McFarland standard (1.5 × 108 CFU/ml) was prepared in normal saline. A sterile cotton swab was used to spread the bacteria on Mueller Hinton agar in order to obtain a lawn culture. After allowing the plate to dry, disks of ceftazidime (30 μg) (CAZ), ceftazidime + clavulanic acid (30/10 μg) (CAC), cefotaxime (30 μg) (CTX), cefotaxime + clavulanic acid (30/10 μg) (CEC) were placed on the surface and the plates were incubated in ambient air at 37 °C for 16–18 h. Following growth, the diameter of the zones around the disks was measured and recorded. An increase in the zone diameter by ≥5 mm around the disks containing cephalosporin with clavulanic over the disks containing cephalosporin alone indicated ESBL production according to CLSI guidelines (M100 28th Edition). E. coli ATCC 25922 and K. pneumoniae ATCC 700603 were used as negative and positive controls respectively [17].
Whole genome sequencing (WGS) of E. coli isolates
All E. coli isolates from humans, poultry and environmental samples were subjected to whole-genome sequencing. Briefly, all E. coli isolates (n = 110) were cultured overnight at 37 °C on Luria-Bertani (LB) agar for 24 h. DNA was extracted using Whole Genome DNA Isolation for Gram-Negative Bacteria protocol (Lucigen MasterPure™ Gram Positive DNA Purification Kit) following the manufacturer’s instructions. Next, DNA concentrations were quantified using the Qubit 4.0 Fluorometer assay kit (Thermo Fisher Scientific, MA). After DNA quantification, libraries for each E. coli isolates were prepared for WGS using a Nextera XT kit. Briefly, 0.3 ng/μL of DNA from each isolate was processed using a Nextera XT DNA Sample Prep Kit (Illumina Inc., San Diego, CA), pooled together, and sequenced on an Illumina Miseq platform using a 2 × 250 paired-end approach (Illumina Inc., San Diego, CA). Raw sequencing reads were demultiplexed and converted to fastq files using CLC Genomics workbench version 9.4 (Qiagen bioinformatics, Valencia, CA). The DNA sequences for each isolate were transferred to the National Center for Biotechnology (NCBI) database after which each isolate was given an accession number. In silico prediction of antimicrobial resistance was conducted by comparing the DNA sequence for each isolate against several genetic analysis databases such as ResFinder (version 3.2, database date: 2019-07-02, cge.cbs.dtu.dk), Antibiotic Resistance Gene-ANNOTation (ARG-ANNOT), ABRICATE and the Comprehensive Antibiotic Resistance Database (CARD) to identify resistance genes [18]. For each isolate, we used between 90 and 100% identity to match individual genes to an annotated resistance gene. However, the genes included in the final profile were decided using the output from ResFinder version 3.2 [19].
Multi-locus sequence typing (MLST) of MDR E. coli isolates
We performed in silico MLST-analyses using previously described schemes by Achtman [20] which considered allelic variation amongst seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) to assign sequence types (STs). WGS data were used to generate the E. coli MLST assignment for each isolate that perfectly matched the alleles in the MLST database [20]. Isolates with 100% match against known MLST alleles were assigned STs however those without perfect matches were usually identified as non-conclusive or unknown. Some isolates which were matched with MLST alleles of unknown ST in the MLST database were assigned as potential new type [21].
Data collection and analyses
Data were collected electronically using open data kit (ODK) installed on a smartphone. Data were analyzed by computing frequencies, proportions and spearman’s correlation (ρ). Out of 110 E. coli isolates sequenced from our study, the accession numbers for 108 paired-end reads have been deposited by the Thakur Molecular Epidemiology Laboratory, NC State University (GenomeTrakr Project) in the National Center for Biotechnology Information under the Bio project ID number PRJNA293225. The remaining two isolates have accession obtained from the DNA Data Bank of Japan (DDBJ) [22, 23]. The additional data file for this study contains a list of accession numbers for individual Sequence Read Archive (SRA) for the ESBL-EC isolates.
Results
Prevalence of E. coli in humans, chickens and poultry farm/market environment
Overall, 429 samples comprising human stool (n = 122), chicken faeces (n = 111), litter (n = 131) and water samples (n = 65) were collected from 52 poultry farms and 8 chicken markets in Kuje and Municipal Area councils of Abuja, Nigeria (Fig. 1). The sample size from each farm or market varied depending on the actual size in terms of the number of chickens reared or sold and the willingness of the owners to participate in the study.
The overall prevalence of E. coli from all sources was 26.8% (n = 115) out of which 61% (n = 70) were obtained from poultry farms and 39.1% (n = 45) from chicken markets. Out of a total of 70, E. coli isolates from the poultry farms, 38.6% (n = 27) were of human origin; 31.4% (n = 22) of poultry origin, while 30% (n = 21) were from the poultry farm environment (litter and water samples). Forty-five E. coli isolates were obtained from the chicken markets out of which 46.7% (n = 21) were human isolates, 33.3% (n = 15) were chicken isolates and 20% (n = 9) were from the chicken market environment (Fig. 2).
Fig. 2.
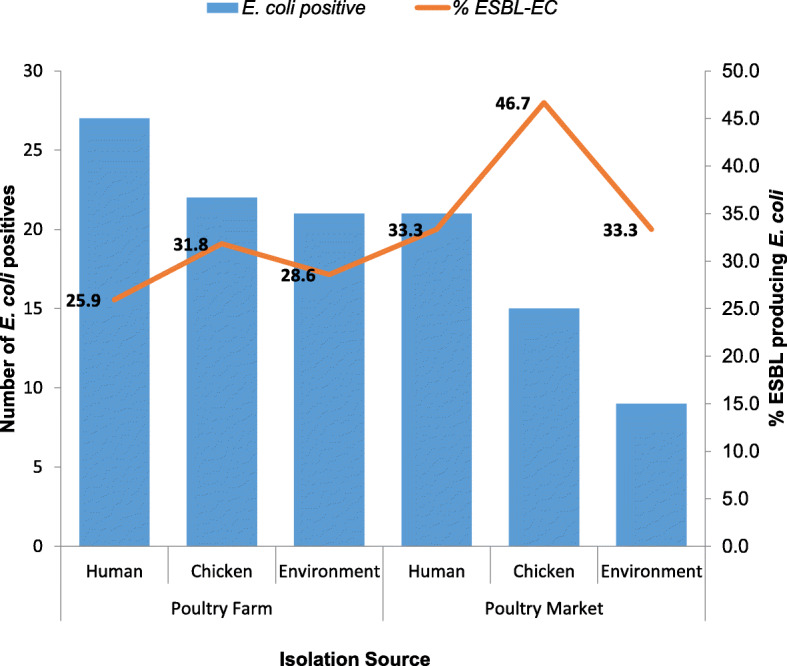
Proportion of ESBL-EC among positive E. coli isolates from humans, chickens, and poultry environment in Abuja, Nigeria, 2019. The number of E. coli positive isolates are plotted as bars on the primary axis while the proportion of these positive isolates which are ESBL-EC in percentages are plotted as a line graph on the secondary axis. The various data points on the line graph are also displayed on the chart
Detection of ESBL-EC isolates
Of the 115 E. coli isolates, 32.2% (n = 37) were confirmed ESBL producers by phenotypic characterization. Prevalence of ESBL-EC was highest among both poultry-workers 37.8% (n = 14) and chickens 37.8% (n = 14) followed by the poultry farm/market environment 24.3% (n = 9). Two (22.2%) of the environmental isolates obtained from litter and water sources were from the same chicken market. More than half (54%) of these isolates were from samples obtained from the poultry farm. The human and chicken isolates from the farm and chicken markets had similar ESBL-EC prevalence; however, the environmental isolates from the farm had doubled the prevalence of that observed in the chicken markets (Fig. 3).
Fig. 3.
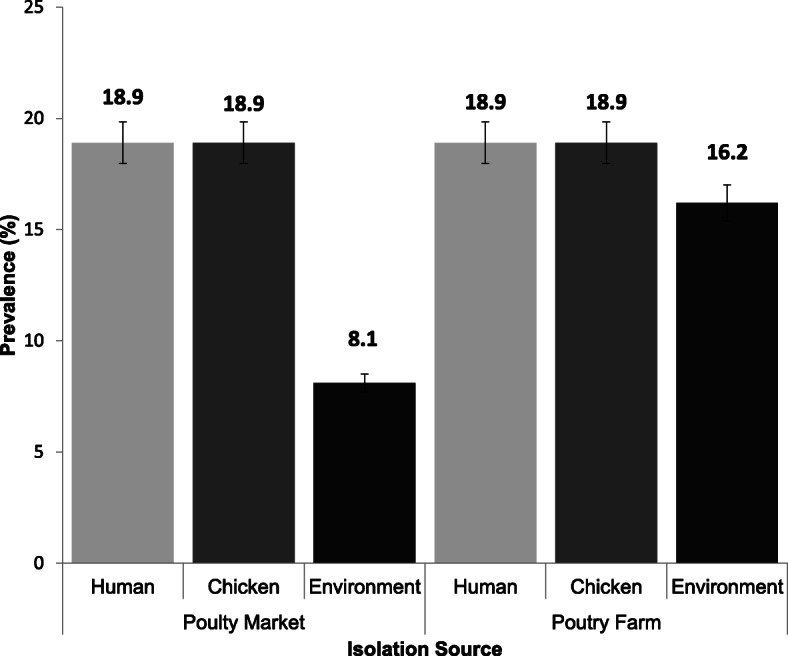
Prevalence of ESBL-EC isolated from humans, chickens and poultry environment, Abuja, Nigeria, 2019. Bars represent the proportion of ESBL-EC isolates from each isolation source with 95% confidence intervals. Error bars represent Standard Error of the mean prevalence. Data were obtained from two sources: poultry farms and poultry markets
Antimicrobial susceptibility testing of ESBL-EC isolates
Of 37 ESBL-EC isolates, 94.6% (n = 35) were multidrug-resistant (MDR). Out of these, 37.1% (n = 13) were human isolates, 37.1% (n = 13) were chicken isolates while 25.7% (n = 9) were environmental isolates (Table 1). Overall, the frequency of resistance in ESBL-EC to the antibiotics tested were tetracycline (97.3%), ampicillin (97.3%), sulfamethoxazole + trimethoprim (83.7%), sulfisoxazole (81.1%), Cefoxitin (78.4%), streptomycin (75.7%), gentamicin (45.9%), nalidixic acid (45.9%), azithromycin (35.1%), chloramphenicol (32.4%), and ciprofloxacin (27.0%).
Table 1.
Antimicrobial resistance profiles of ESBL-EC isolates from Humans, chickens and farm/market environments in Abuja - Nigeria, 2019
| Drug Class | Drug | MIC Resistance breakpoint μg/mL | Humans, n = 14 (%) |
Chickens n = 14 (%) |
Environment n = 9 (%) |
|---|---|---|---|---|---|
| Tetracyclines | Tetracyclines (TET) | ≥ 16 | 14 (100) | 13 (92.9) | 9 (100) |
| Folate Pathway antagonists | Sulfamethoxazole Trimethoprim (SXT) | ≥ 4/76 | 12 (85.7) | 10 (71.4) | 9 (100) |
| Sulfisoxazole (FIS) | ≥512 | 12 (85.7) | 9 (64.3) | 9 (100) | |
| Penicillins | Ampicillin (AMP) | ≥ 32 | 13 (92.9) | 14 (100) | 9 (100) |
| Quinolones | Nalidixic acid (NAL) | ≥ 32 | 5 (35.7) | 7 (50.00) | 5 (55.6) |
| Ciprofloxacin (CIP) | ≥ 4 | 3 (21.4) | 5 (35.7) | 2 (22.2) | |
| Aminoglycosides | Streptomycin (STR) | ≥ 32 | 10 (71.4) | 10 (71.4) | 8 (88.9) |
| Gentamicin (GEN) | ≥ 16 | 5 (35.7) | 7 (50.00) | 5 (55.6) | |
| Phenicols | Chloramphenicol (CHL) | ≥ 32 | 3 (21.4) | 7 (50.00) | 2 (22.2) |
| Macrolide Antibiotics | Azithromycin (AZI) | ≥ 32 | 4 (28.6) | 4 (28.6) | 5 (55.6) |
| B-lactam inhibitors | Amoxicillin-clavulanate (AMC) | ≥ 32/16 | 1 (7.14) | 0 (0) | 0 (0) |
| Cephem | Ceftriaxone (CRO) | ≥ 4 | 2 (14.3) | 2 (14.3) | 0 (0) |
| Cefoxitin (FOX) | ≥ 32 | 14 (100) | 14 (100) | 1 (11.1) | |
| Ceftiofur (XNL) | ≥ 8 | 2 (14.3) | 2 (14.3) | 1 (11.1) | |
| Resistance to 3 or more classes of antibiotics | MDR | n/a | 13 (92.9) | 13 (92.9) | 9 (100) |
ESBL level genes in humans, chickens and poultry environment
Both human and chicken isolates showed similar patterns of multidrug resistance to tested antimicrobials with a positive correlation (ρ = 0.91). Among ESBL producers, we observed the dissemination of blaCTX-M 10.8% (n = 4) genes out of which 50% (n = 2) were chicken isolates from different chicken farms while 50% (n = 2) were human isolates from the same chicken market. Of the blaCTX-M genes detected, 75% (n = 3) were of the subtype blaCTX-M-15 while 25% (n = 1) was of the subtype blaCTX-M-65. Both human isolates and one chicken isolate were of the blaCTX-M-15 subtype.
Resistance determinants detected in ESBL-EC isolates
This study identified more than 30 different resistance determinants from 37 ESBL-EC isolates (Table 2). Aminoglycosides accounted for the majority of these resistance determinants with about 10 different variants (aadA1, aadA2, aadA5, aac(3)-IIa, aac(3)-IId, aac(3)-Ib, aac(6)-Ib-cr, aph(3)-Ia, aph(3)-Ib, aph(6)-Id) detected. More than half of the ESBL-EC isolates (25) exhibited aph(3)-Ib gene which is a metabolic enzyme conferring aminoglycoside resistance. It is important to note that we detected aac(6)-Ib-cr gene which is responsible for the reduction in the activity of ciprofloxacin in one ESBL-EC isolate. We also observed the aac(3)-IId gene which is responsible for conferring resistance to gentamicin among ten ESBL-EC isolates. Among the aminoglycoside resistance genes in high prevalence was aph(6)-Id which is a plasmid-encoded gene. This was followed by β-lactam resistance genes which were of six different types (blaTEM-1, blaTEM-20, blaOXA-1, blaOXA-129, blaCTX-M-15, blaCTX-M-65) out of which blaCTX-M type was classical of the ESBL producing E. coli. Resistance to fluoroquinolones, one of the WHO listed critically important antimicrobials, were also detected with five different variants (qnrB1, qnrB19, qnrS1, qnrS2, aac(6)-Ib-cr) and this is usually associated with mutations in the gyrA and parC genes. Other resistance determinants that we observed included phenicol resistance (cmlA1, catA1, catA2, catB3, floR), rifampicin resistance (ARR-3), sulphonamide resistance (sul1, sul2, sul3), tetracycline resistance (tetA, tetB) and trimethoprim resistance (dfrA1, dfrA12, dfrA14, dfrA17, dfrA21). Our study detected one plasmid-mediated colistin resistance gene (mcr-1.1).
Table 2.
Resistance determinants detected in ESBL-EC isolates from humans, chickens and farm/market environment in Abuja - Nigeria, 2019
| Antibiotic classa | Resistance determinants of ESBL-EC isolates (no of isolates)b |
|---|---|
| Aminoglycoside | aadA1 (6), aadA2 (4), aadA5 (3), aac(3)-IIa (4), aac(3)-IId (11), aac(3)-Ib (1), aac(6)-Ib-cr (1), aph(3)-Ia (6), aph(3)-Ib (25), aph(6)-Id (22) |
| β-lactamases | blaTEM-1 (33), blaTEM-20 (1), blaOXA-1 (1), blaOXA-129 (1), blaCTX-M-15 (3), blaCTX-M-65 (1) |
| Colistin | mcr-1.1 (1) |
| Macrolide | mdfA (34), mphA (11), mphB (1) |
| Phenicol | cmlA1 (2), catA1 (4), catA2 (1), catB3 (1), floR (6) |
| Rifampicin | ARR-3 (1) |
| Quinolone | qnrB1 (1), qnrB19 (6), qnrS1 (24), qnrS2 (2), aac(6)-Ib-cr (1) |
| Sulphonamide | sul1 (7), sul2 (30), sul3 (3) |
| Tetracycline | tetA (31), tetB (3) |
| Trimethoprim | dfrA1 (3), dfrA12 (3), dfrA14 (23), dfrA17 (4), dfrA21 (1) |
aDrugs corresponding to each antibiotic class used in our study are as follows: aminoglycosides, streptomycin, gentamicin; beta-lactams, ampicillin, ceftriaxone, cefotaxime, ceftazidime, ceftiofur, cefoxitin, amoxicillin-clavulanic acid, and cefpodoxime; quinolones, nalidixic acid, and ciprofloxacin; phenicols, chloramphenicol; sulfonamides and trimethoprim, trimethoprim-sulfamethoxazole, Sulfisoxazole; tetracyclines, tetracycline; macrolides, azithromycin.
bThe number of isolates carrying each resistance determinant are presented in parentheses
Multi-locus sequence determination of ESBL-EC isolates
The 37 ESBL-EC isolates belonged to 27 different sequence types (ST), out of which one was non-conclusive and two were novel. ST-155 (13.5%; n = 5), ST-10 (10.8%; n = 4), ST-48 (8.1%; n = 3) and ST-1196 (5.4%; n = 2) were the most commonly observed STs in silico analysis of ESBL-EC (Fig. 4).
Fig. 4.
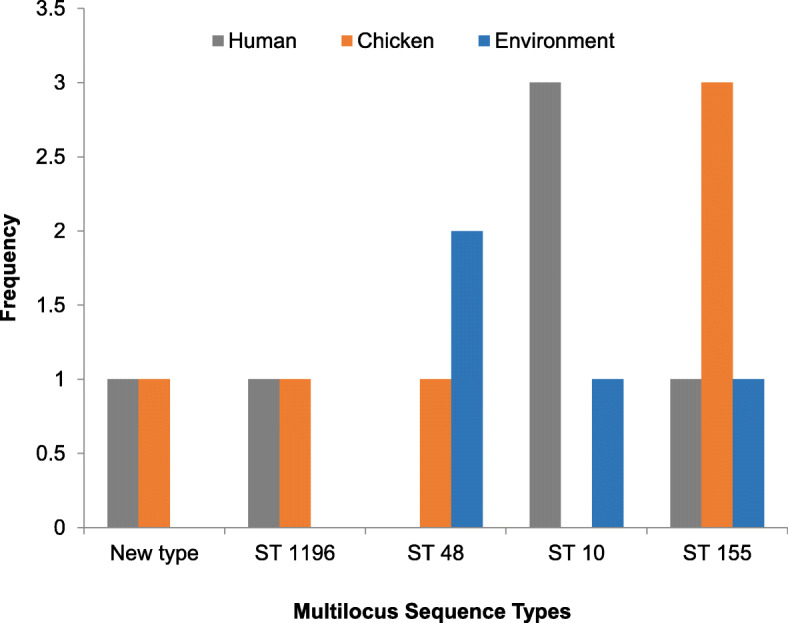
Multilocus Sequence Types for ESBL-EC isolated from humans, chickens and poultry environment, Abuja, Nigeria, 2019. Each bar represents the various ESBL-EC sequence types for isolates obtained from humans, chickens and chicken farm/market environments
In the ST-10 group, the most represented isolate was from humans handling chickens on the farms and in the chicken market (3/4), while the least common ST in this group was from the chicken market environment (1/4). This was followed by the ST-155 group majority of which were from the chicken market (3/5 from chickens, 1/5 from human and 1/5 from the poultry market environment). The most common ST observed in the poultry farm environment was ST-48 (2/3). The in silico β-lactamase analysis of the ESBL-EC showed that the most detected gene was blaCTX-M (10.8%). All of the blaCTX-M isolates belonged to four different sequence types; being a novel ST, a non-conclusive ST, ST-10, and ST-2179.
Discussion
Globally, studies have documented that E. coli isolated from food-producing animals particularly chickens are usually resistant to β-lactam antimicrobial agents [7, 24, 25]. It has also been reported that chickens are an important source of ESBL-EC transmission to humans although this has not been clearly demonstrated [1, 13, 26].
Our study attempted to investigate the prevalence of ESBL-EC among people who handle chickens either on the farm or market as well as in the various environment where the chickens are reared or sold. Our findings showed that antimicrobial-resistant E. coli, particularly ESBL-EC, are present in the poultry farm environment, as well as the market environment, where these chickens are sold acting as a reservoir of antimicrobial-resistant bacteria and eventually pose as a health risk to humans working in such environment. This study observed that the prevalence of ESBL-EC was 37.8%, 37.8% and 24.3% in humans, chickens and poultry environment, respectively. Our study prevalence of ESBL-EC in humans, animals, and the environment was higher than what was observed in another similar study [2, 12]. Similar studies conducted elsewhere reported a much higher prevalence of ESBL-EC in chickens and apparently healthy individuals [11, 12]. Our study prevalence of ESBL antimicrobial resistance in human and chicken isolates was similar however, a much lower prevalence was observed in the environmental isolates. A possible explanation may be as a result of easy access to antimicrobials for both human and veterinary use as opposed to the developed economies where these are strictly prescription-only medicines [15, 27, 28].
Tetracycline, sulphonamides and aminoglycosides class of antimicrobials accounted for the majority of the resistance determinants observed in this study. This is similar to the findings of other studies that reported high tetracycline and sulphonamides resistance on poultry farms [12, 15, 27, 28]. The high prevalence of AMR genes belonging to these classes of antimicrobials observed in this study is most likely because these antimicrobials are commonly used in chicken farms in Nigeria either for prophylaxis or therapeutic purposes [15, 27, 28].
Our study showed that among the ESBL-EC isolates, one of the most prevalent resistance genes observed were tetA and tetB that are responsible for resistance against tetracycline. This finding was not surprising as tetracycline is one of the most abused antibiotics in poultry production in Nigeria [27, 29, 30].
In our study, blaCTX-M was the only ESBL gene type observed among human and chicken isolates and this is consistent with findings from other studies [13, 31, 32]. The blaCTX-M-15 genes detected in this study were observed in human isolates from the same chicken market however our study cannot clearly demonstrate that this occurred as a result of horizontal gene transfer from one chicken seller to the other. The chicken isolates had two different CTX-M subtypes: CTX-M-15 and CTX-M-65. These chicken isolates where CTX-M genes were observed were also sourced from two different poultry farms. Interestingly, the CTX-M-15 subtype from one chicken isolate was similar to that of the human isolates. It is important to note that chickens sold in the chicken markets in Abuja are usually sourced from various farms, thus providing an enabling environment for the spread of antimicrobial resistance genes.
Although studies have provided some evidence that food animals are critical in the transfer of antimicrobial-resistant E. coli to the human population, our study cannot clearly demonstrate that the resistance observed in the apparently healthy humans were from the chickens or the farm/market environment. In contrast to our findings, a study done in Ghana demonstrated that four human isolates and broiler isolates were closely related suggesting a possibility of spread of resistance between the two populations [13]. The possible explanation for the disparity in our findings when compared with the documented evidence by the Ghana study was that the isolates in the Ghana study were obtained from sick children while our source was apparently healthy individuals.
The genetic relatedness of the ESBL-EC isolates in our study was determined using multilocus sequence typing (MLST) of whole-genome sequences. Our findings showed the genetic diversity of ESBL-EC as we observed 27 different sequence types (STs) and this is consistent with findings from other studies [12, 13, 31, 32]. However, the most common STs were ST-10, ST-48 and ST-155. In our study, ST-10 was observed in humans and chicken farm/market environment but not in chickens although previous studies have reported ST-10 as one of the most common in human and chicken populations [13, 32]. Our findings showed that the ESBL-EC strains from the humans, chickens, and chicken market environments had identical ST-155, thus suggesting that a possible transmission between these hosts and the environment may have occurred. This finding demonstrates that the co-colonization of antimicrobial-resistant E. coli from a shared source is also possible. There is documentary evidence that E. coli ST-155 represents an important zoonotic strain responsible for the transmission of ESBL-EC to humans [33, 34]. It is also important to note that ST-155 has been detected in E. coli isolates from water samples [34] and this is in agreement with findings from our study where this ST was detected in isolates from environmental samples. A possible explanation for our findings is that there is a possibility for the circulation of host-adapted lineages of ESBL-EC.
To the best of our knowledge, this study is the first in Nigeria to report plasmid-mediated colistin resistance gene (mcr-1.1) in the chicken market environment. The mcr-1.1 positive E. coli strain was of ST-10 and isolated from poultry water at the chicken market. This finding is rather not surprising as colistin, which is considered an antimicrobial agent of last resort is often used for therapeutic purposes in poultry production in Nigeria [7, 27]. Our results are in agreement with documentary evidence that ST-10 is the most common ST globally carrying the mcr-1 gene as well as the dominant ST in both animal and water samples [35]. This finding has highlighted the potential for the mcr-1 gene to spread from the poultry farm to the chicken market environment, hence should be considered a potential public health risk especially to the chicken sellers.
Conclusions
ESBL-EC isolates were prevalent amongst apparently healthy individuals, chickens and the poultry farm/market environment in Abuja. Among ESBL-EC, the highest resistance was observed to tetracycline, sulphonamides, and aminoglycosides which are classes of antimicrobials commonly used on chicken farms for therapeutic purposes in Abuja. Our study detected ESBL genes, blaCTX-M in human isolates from the same chicken market however, the chicken isolates with blaCTX-M genes were sourced from two different poultry farms. ST155 was the only ST detected in humans, chickens and chicken farm environments in this study. The possibility of the spread of ESBL-EC from other environmental sources on the chicken farm/market to either humans or chickens may have been due to poor biosecurity measures observed on the farms or markets where samples were collected. It is therefore important to educate healthcare workers that people in proximity with poultry are a high-risk group for faecal carriage of ESBL-EC, hence pose a higher risk to the general population for the spread of AMR. It is also important to educate farmers on the need to observe biosecurity measures on chicken farms and markets.
Supplementary information
Acknowledgments
The authors would like to appreciate Dr. Chikwe Ihekweazu, Director General, Nigeria Center for Disease Control, Abuja for providing laboratory space and support towards this research at the National Reference Laboratory. The authors acknowledge the intellectual contributions of Dr. Adebayo Adedeji, Mrs. Mba Nwando, Miss Eme Ekeng, Mr. Akinpelu Afolabi, Mr. Chris Chukwu and Mr. Sunday Thomas Sola of the National Reference Laboratory, Gaduwa, Abuja, towards the success of this research. The authors will like to appreciate the efforts of members of the Thakur Molecular laboratory, North Carolina State University; Erin Harrell, Nigatu Atlaw, Ayanna Glaize, Morgan Young, Luke Raymond, Bryson Staley and Steven Branz towards the success of the research. The whole genome sequencing was completed by the FDA GenomeTrakr program funded grant 1U18FD00678801.
Abbreviations
- AMR
Antimicrobial resistance
- ARG-ANNOT
Antibiotic Resistance Gene-ANNOTation,
- CARD
Comprehensive Antibiotic Resistance Database
- CLSI
Clinical and Laboratory Standards Institute
- DDBJ
DNA Data Bank of Japan
- ESBL
Extended-Spectrum Beta-Lactamase
- ESBL-EC
Extended-Spectrum Beta-Lactamase producing E. coli
- LB
Luria-Bertani
- MDR
Multi-drug Resistance
- MIC
Minimum Inhibitory Concentration
- MLST
Multi-locus Sequence Typing
- NCBI
National Center for Biotechnology Information
- ODK
Open data kit
- SRA
Sequence Read Archive
- ST
Sequence Type
- TSA
Tryptic Soy Agar
- WGS
Whole-genome Sequencing
- WHO
World Health Organization
Authors’ contributions
MA was the principal investigator, designed data collection tools, collected data, isolated the organism, performed antibiotic sensitivity testing on the isolates, analyzed and interpreted the data and wrote the first draft of the manuscript. JK made substantial contributions to conception and design. JK, LH, and ST supervised the laboratory aspect of the research. LH generated the whole genome sequence profile for all the isolates. MA and DH performed bioinformatics analysis. JK, EO, RSH, and ST revised the article critically for important intellectual content. All authors read and approved the final manuscript.
Funding
The author(s) did not receive any specific funding for this work.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. All data generated or analyzed during this study are also included in this published article [and its supplementary information files].
Ethics approval and consent to participate
For this study, we obtained ethics approval from the Scientific and Ethical committee of the FCT Health Research Ethics Committee in July 2018 (Approval Number: FHREC/2018/01/84/16–07-18). We also obtained permission from the owners of each poultry farm and management of chicken markets prior to commencement of study. The confidentiality of the information obtained was assured. An informed consent form clearly stating the purpose and benefits of the study was given to all study participants willing to participate in the study and duly signed prior to sample collection.
Ethics approval for this study was also obtained from the Scientific and Ethical Committee of the Ahmadu Bello University Committee on Animal Use and Care (Approval Number: ABUCAUC/2020/35). An informed consent form was given to the owners of farm and chicken market authority which was duly signed prior to sample collection.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s42522-020-00014-7.
References
- 1.Gbonon M, Carole V, Kouadio GN, Baguy OM, Djénéba OG, Ayayi A, et al. Antimicrobial resistance profile and molecular characterization of extended-spectrum Beta- lactamase genes in Enterobacteria isolated from human. Anim Environ. 2018;10(1):1–9. 10.9734/JAMB/2018/3995.
- 2.Pormohammad A, Nasiri MJ, Azimi T. Prevalence of antibiotic resistance in escherichia coli strains simultaneously isolated from humans, animals, food, and the environment: A systematic review and meta-analysis. Infection Drug Resistance. 2019;12:1181–1197. doi: 10.2147/IDR.S201324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Humphreys G, Fleck F. United nations meeting on antimicrobial resistance. Bull World Health Organ. 2016;94:638–639. doi: 10.2471/BLT.16.020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antimicrobial resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. Accessed 10 Dec 2019.
- 5.Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24:718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stop using antibiotics in healthy animals to prevent the spread of antibiotic resistance. https://www.who.int/news-room/detail/07-11-2017-stop-using-antibiotics-in-healthy-animals-to-prevent-the-spread-of-antibiotic-resistance. Accessed 10 Dec 2019.
- 7.Aworh MK, Kwaga J, Okolocha E, Mba N, Thakur S. Prevalence and risk factors for multi-drug resistant Escherichia coli among poultry workers in the Federal Capital Territory, Abuja, Nigeria. PLoS One. 2019;14:e0225379. doi: 10.1371/journal.pone.0225379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veenemans J, Overdevest IT, Snelders E, Willemsen I, Hendriks Y, Adesokan A, et al. Next-generation sequencing for typing and detection of resistance genes: performance of a new commercial method during an outbreak of extended-spectrum-beta-lactamase-producing Escherichia coli. J Clin Microbiol. 2014;52:2454–2460. doi: 10.1128/JCM.00313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, the Netherlands. Emerg Infect Dis. 2011;17:1216–1222. doi: 10.3201/eid1707.110209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oduro-Mensah D, Obeng-Nkrumah N, Bonney EY, Oduro-Mensah E, Twum-Danso K, Osei YD, et al. Genetic characterization of TEM-type ESBL-associated antibacterial resistance in Enterobacteriaceae in a tertiary hospital in Ghana. Ann Clin Microbiol Antimicrob. 2016;15:29. doi: 10.1186/s12941-016-0144-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleem R, Ejaz H, A. Zafar SY. Phenotypic characterization of extended-spectrum-beta - lactamase producing E. Coli from healthy individuals, patients, sewage sludge, cattle, chickens and raw meat. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nguyen VT, Jamrozy D, Matamoros S, Carrique-Mas JJ, Ho HM, Thai QH, et al. Limited contribution of non-intensive chicken farming to ESBL-producing Escherichia coli colonization in humans in Vietnam: an epidemiological and genomic analysis. J Antimicrob Chemother. 2019;74:561–570. doi: 10.1093/jac/dky506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falgenhauer L, Imirzalioglu C, Oppong K, Akenten CW, Hogan B, Krumkamp R, et al. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol. 2019;9. 10.3389/fmicb.2018.03358. [DOI] [PMC free article] [PubMed]
- 14.Ghosh P, Mahanti A, Samanta I, Joar SN, Batabyal K, Dey S, et al. Occurrence of extended-spectrum cephalosporinase producing Escherichia coli in kuroiler birds. Veterinarski Arhiv. 2017;87(6):745–757. doi: 10.24099/vet.arhiv.160719a. [DOI] [Google Scholar]
- 15.Oloso N, Fagbo S, Garbati M, Olonitola S, Awosanya E, Aworh M, et al. Antimicrobial resistance in food animals and the environment in Nigeria: A review. Int J Environ Res Public Health. 2018;15:1284. doi: 10.3390/ijerph15061284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nigeria Centre for Disease Control . Antimicrobial use and resistance in Nigeria. 2017. pp. 1–158. [Google Scholar]
- 17.CLSI . M100 performance standards for antimicrobial susceptibility testing A CLSI supplement for global application. 2018. [Google Scholar]
- 18.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, et al. ARG-annot, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F. M., Larsen M. V. Identification of acquired antimicrobial resistance genes. Journal of Antimicrobial Chemotherapy. 2012;67(11):2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, et al. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.BioSample for BioProject (Select 293225) - BioSample - NCBI. https://www.ncbi.nlm.nih.gov/biosample?Db=biosample&DbFrom=bioproject&Cmd=Link&LinkName=bioproject_biosample&LinkReadableName=BioSample&ordinalpos=1&IdsFromResult=293225. Accessed 16 Feb 2020.
- 23.Escherichia coli (ID 601337) - BioProject - NCBI. https://www.ncbi.nlm.nih.gov/bioproject/601337. Accessed 16 Feb 2020.
- 24.Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: A developing country-perspective. Front Microbiol. 2016;7:1881. 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed]
- 25.Sharma C, Rokana N, Chandra M, Singh BP, Gulhane RD, Gill JPS, et al. Antimicrobial resistance: its surveillance, impact, and alternative management strategies in dairy animals. Frontiers Vet Sci. 2018;4:237. 10.3389/fvets.2017.00237. [DOI] [PMC free article] [PubMed]
- 26.Chang Q, Wang W, Regev-Yochay G, Lipsitch M, Hanage WP. Antibiotics in agriculture and the risk to human health: how worried should we be? Evol Appl. 2015;8:240–247. doi: 10.1111/eva.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oluwasile B, Agbaje M, Ojo O, Dipeolu M. Antibiotic usage pattern in selected poultry farms in Ogun state. Sokoto J Vet Sci. 2014;12:45. doi: 10.4314/sokjvs.v12i1.7. [DOI] [Google Scholar]
- 28.Agyare Christian, Etsiapa Boamah Vivian, Ngofi Zumbi Crystal, Boateng Osei Frank. Antimicrobial Resistance - A Global Threat. 2019. Antibiotic Use in Poultry Production and Its Effects on Bacterial Resistance. [Google Scholar]
- 29.Baba Galad H, Ahmad Geid Y, Usman Sham B, Ibrahim Ab H, Ibrahim B, Waziri A. Survey of antimicrobial residue in table eggs among layer poultry farmers in Maiduguri Metropolis, Borno state. Asian J Anim Vet Adv. 2018;13:101–108. doi: 10.3923/ajava.2018.101.108. [DOI] [Google Scholar]
- 30.Adebowale OO, Adeyemo OK, Awoyomi O, Dada R, Adebowale O. Antibiotic use and practices in commercial poultry laying hens in Ogun state Nigeria. Rev d’élevage médecine vétérinaire des pays Trop. 2016;69:41. doi: 10.19182/remvt.31170. [DOI] [Google Scholar]
- 31.Hussain A, Shaik S, Ranjan A, Suresh A, Sarker N, Semmler T, et al. Genomic and functional characterization of poultry Escherichia coli from India revealed diverse extended-Spectrum β-lactamase-producing lineages with shared virulence profiles. Front Microbiol. 2019;10:2766. doi: 10.3389/fmicb.2019.02766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Day MJ, Hopkins KL, Wareham DW, Toleman MA, Elviss N, Randall L, et al. Extended-spectrum Î2-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. 2019. [DOI] [PubMed] [Google Scholar]
- 33.Salim A, Babu P, Mohan K, Moorthy M, Raj D, Kallampillil Thirumeni S, et al. Draft genome sequence of an Escherichia coli sequence type 155 strain isolated from sewage in Kerala, India. Microbiol Resour Announc. 2019;8:e01707–18. 10.1128/%20MRA.01707-18. [DOI] [PMC free article] [PubMed]
- 34.Gomi R, Matsuda T, Matsumura Y, et al. Whole-genome analysis of antimicrobial-resistant and Extraintestinal pathogenic Escherichia coli in river water. Appl Environ Microbiol. 2017;83(5):e02703–e02716. doi: 10.1128/AEM.02703-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elbediwi, Li, Paudyal, Pan, Li, Xie, Rajkovic, Feng, Fang, Rankin, Yue Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980–2018) Microorganisms. 2019;7(10):461. doi: 10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. All data generated or analyzed during this study are also included in this published article [and its supplementary information files].