Abstract
INTRODUCTION:
Histone modification activity of the polycomb repressive complexes 1 and 2 (PRC1 and PRC2) is critical for the establishment and maintenance of gene expression patterns and, thus, to the maintenance of cell identity. Distinct classes of cofactor proteins are known to regulate the functional activity and interplay of these two complexes, but we presently lack a comprehensive, mechanistic understanding of this process. Furthermore, PRC2 cofactors like AEBP2 and JARID2 also play a role in mediating the cross-talk between different histone posttranslational modifications and PRC2 recruitment and activity—a function that is important for the regulated control of gene expression.
RATIONALE:
PRC1 is an E3 ubiquitin ligase responsible for the monoubiquitination of histone H2A (H2AK119ub1), a histone mark recognized by PRC2 and linked to its genomic recruitment. We used cryo-electron microscopy (cryo-EM) and biochemical activity assays to probe the role played by PRC2 cofactors JARID2 and AEBP2 in the recognition of H2AK119ub1 and the regulation of PRC2 activity. We extended our cryo-EM and biochemical activity analysis to examine the possible role played by JARID2 and AEBP2 in the cross-talk between the histone H3K4me3, H3K36me3 modifications linked to transcriptionally active regions, and PRC2 activity.
RESULTS:
We find that JARID2 recognizes both the ubiquitin moiety in H2AK119ub1 and the conserved histone H2A-H2B acidic patch. We also observe that the tandem zinc fingers of AEBP2 interact with ubiquitin and the histone H2A-H2B surface on the other side of the nucleosome. Biochemical assays show a secondary activation of PRC2 by JARID2 and AEBP2 on H2AK119ub1-containing nucleosomes besides the primary EED-mediated allosteric activation of PRC2 by methylated JARID2. Furthermore, we also find that the joint presence of JARID2 and AEBP2 partially reduces the inhibition of PRC2 methyltransferase activity by the transcriptionally active histone posttranslational modifications H3K4me3 and H3K36me3. Cryo-EM visualization of PRC2 that contains JARID2 and AEBP2 interacting with a H3K4me3-containing nucleosome shows the coexistence of states in which the histone H3 tail is either absent or engaged and reaching the catalytic site in PRC2, which provides a physical basis for the partial activity of the complex on H3K4me3-containing nucleosomes.
CONCLUSION:
Our studies indicate that cofactors JARID2 and AEBP2 play a crucial role in both the recruitment and activation of PRC2 through their recognition of H2AK119ub1, which is generated by PRC1. Additionally, our work suggests that JARID2 and AEBP2 are likely to play a key role in regulating PRC2 activity on genomic regions with active transcription marks. The examination of the genomic distribution in embryonic stem cells of PRC2 core proteins together with JARID2 and AEBP2 will be important to further define their role in the tight regulation of PRC2 activity.
Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) cooperate to determine cell identity by epigenetic gene expression regulation. However, the mechanism of PRC2 recruitment by means of recognition of PRC1-mediated H2AK119ub1 remains poorly understood. Our PRC2 cryo-electron microscopy structure with cofactors JARID2 and AEBP2 bound to a H2AK119ub1-containing nucleosome reveals a bridge helix in EZH2 that connects the SET domain, H3 tail, and nucleosomal DNA. JARID2 and AEBP2 each interact with one ubiquitin and the H2A-H2B surface. JARID2 stimulates PRC2 through interactions with both the polycomb protein EED and the H2AK119-ubiquitin, whereas AEBP2 has an additional scaffolding role. The presence of these cofactors partially overcomes the inhibitory effect that H3K4me3 and H3K36me3 exert on core PRC2 (in the absence of cofactors). Our results support a key role for JARID2 and AEBP2 in the cross-talk between histone modifications and PRC2 activity.
Graphical Abstract
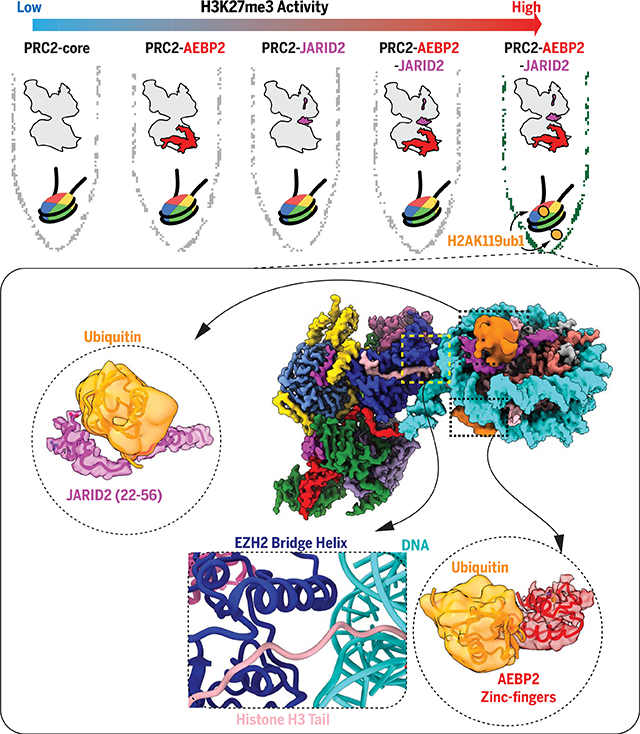
Regulation of PRC2 methyltransferase activity by cofactors JARID2 and AEBP2 and by histone posttranslational modification.
Although core PRC2 is a weak enzyme, it is allosterically activated by JARID2 and AEBP2. The presence of monoubiquitinated histone H2A, the product of PRC1 activity, is recognized by both JARID2 and AEBP2 through interactions that likely mediate recruitment of PRC2 to polycomb sites in the genome and further activate the methyltransferase activity of PRC2 on K27 of histone H3.
Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) are chromatin modifiers critical for lineage commitment during embryonic development and for the maintenance of cell type identity after differentiation (1, 2). PRC1 is an E3 ubiquitin ligase that catalyzes the monoubiquitination of histone H2A at lysine 119 (H2AK119ub1) through its RING1A or RING1B subunits (3, 4). The core of PRC2 (PRC2-core) contains four proteins (EZH2, EED, SUZ12, and RBAP46 or RBAP48) and catalyzes the mono-, di-, and trimethylation of histone H3 at lysine 27 [H3K27me1, H3K27me2, and H3K27me3 (H3K27me1/me2/me3)] (5). Accessory subunits, also referred to as cofactors, associate with the PRC2-core to constitute PRC2 variants with distinct biological functions (6). Two mutually exclusive forms of PRC2 have been reported, sometimes referred to as PRC2.2 (containing JARID2 and AEBP2) and PRC2.1 (containing PHF1, MTF2, PHF19, or EPOP or PALI1) (4, 6). PRC2 cofactors have been proposed to have roles in both the initial recruitment of the complex to chromatin and the allosteric regulation of its histone methyltransferase (HMTase) activity through interactions with long noncoding RNAs (lncRNAs), GC-rich DNA, and the histone mark H2AK119ub1 (7–9). Recent studies have suggested a highly regulated hierarchical recruitment model involving a complex cross-talk between the activities of the different PRC1 and PRC2 complexes in the establishment of polycomb heterochromatin domains (3, 10). The loss of H2AK119ub1 on polycomb target genes has been observed to result in a corresponding reduction in H3K27me3 deposition, which suggests a strong connection between PRC1 and PRC2 catalytic activities (11, 12). However, the specific role played by PRC1-mediated H2AK119ub1 in the recruitment of PRC2 has remained elusive, and there have been conflicting observations reporting both inhibition and activation of PRC2 by this histone mark (13, 14). H2AK119ub1 nucleosomes are recognized by PRC2 that contains AEBP2 and JARID2, which are thought to be involved in both the H2AK119ub1-dependent recruitment and the regulation of HMTase activity (13). Recent studies have shown that this recruitment is mediated by a putative ubiquitin interaction motif (UIM) located near the N terminus of JARID2 (7). Despite these biochemical insights, the mechanistic details of H2AK119ub1 recognition by PRC2 remain unknown. In addition to their role in H2AK119ub1 recognition, PRC2-core together with AEBP2 and JARID2 colocalize to CpG-rich promoter regions with H3K4me3 and to bivalent domains containing both H3K4me3 and H3K27me3 in embryonic stem cells (15–19). It has been accepted in the field that active transcription marks (H3K4me3 and H3K36me3) inhibit PRC2 HMTase activity; however, the effect of cofactors AEBP2 and JARID2 in the H3K4me3- and H3K36me3-mediated inhibition remains poorly understood (20, 21). Using cryo-electron microscopy (cryo-EM) and biochemical assays, we provide mechanistic insight into how the cofactors AEBP2 and JARID2 specifically recognize H2AK119ub1 and how they affect PRC2 HMTase activity in the context of H2AK119ub1-, H3K4me3-, or H3K36me3-containing nucleosomes.
Engagement of PRC2 with a ubiquitinated nucleosome and the bridge helix
We reconstituted the complete human PRC2 with cofactors AEBP2 and JARID2 (amino acids 1 to 450) [referred to hereafter as PRC2-AJ1–450 (Fig. 1A)] with a H2AK119ub1-containing nucleosome (Ncl-ub) (Fig. 1B), and we obtained the cryo-EM structure (3.5 Å) of a 1:1 Ncl-ub-PRC2-AJ1–450 complex. The overall architecture (Fig. 1C and movie S1) agrees very well with our previous cryo-EM structure of PRC2 containing AEBP2 and JARID2 (22). The engagement of the nucleosome also agrees with what we had visualized for the substrate nucleosome in the context of the five-subunit PRC2-AEBP2 complex bound to a dinucleosome (23). Improvements in cryo-EM sample preparation—that overcame the need for cross-linking and stabilized the structure from the deleterious effect of the air-water interface—resulted in an intact PRC2 bound to the nucleosome, which allowed us to structurally model several previously unmodeled regions of the complex (figs. S1 to S4). Our study provides the structures of JARID2 and AEBP2 segments recognizing the ubiquitinated nucleosome, regions of EZH2 linking the SANT2 and the SET domains, a near-complete model for SUZ12, and previously unobserved segments of AEBP2 (Fig. 1C, bottom). In addition to this 1:1 stoichiometry, our cryo-EM data processing also identified a small subpopulation with two PRC2-AJ1–450, each engaging one of the two histone H3 tails, on the opposite sides of the nucleosome (fig. S5). The two PRC2 complexes in that arrangement do not visibly interact with each other and, therefore, likely represent independent binding events. The interaction of cofactors JARID2 and AEBP2 with the ubiquitinated nucleosome is indistinguishable—at the resolution of the reconstruction—from that seen in the 1:1 stoichiometry complex (fig. S5).
Fig. 1. Cryo-EM structure of PRC2-AEBP2-JARID2 bound to a H2AK119ub1 nucleosome.
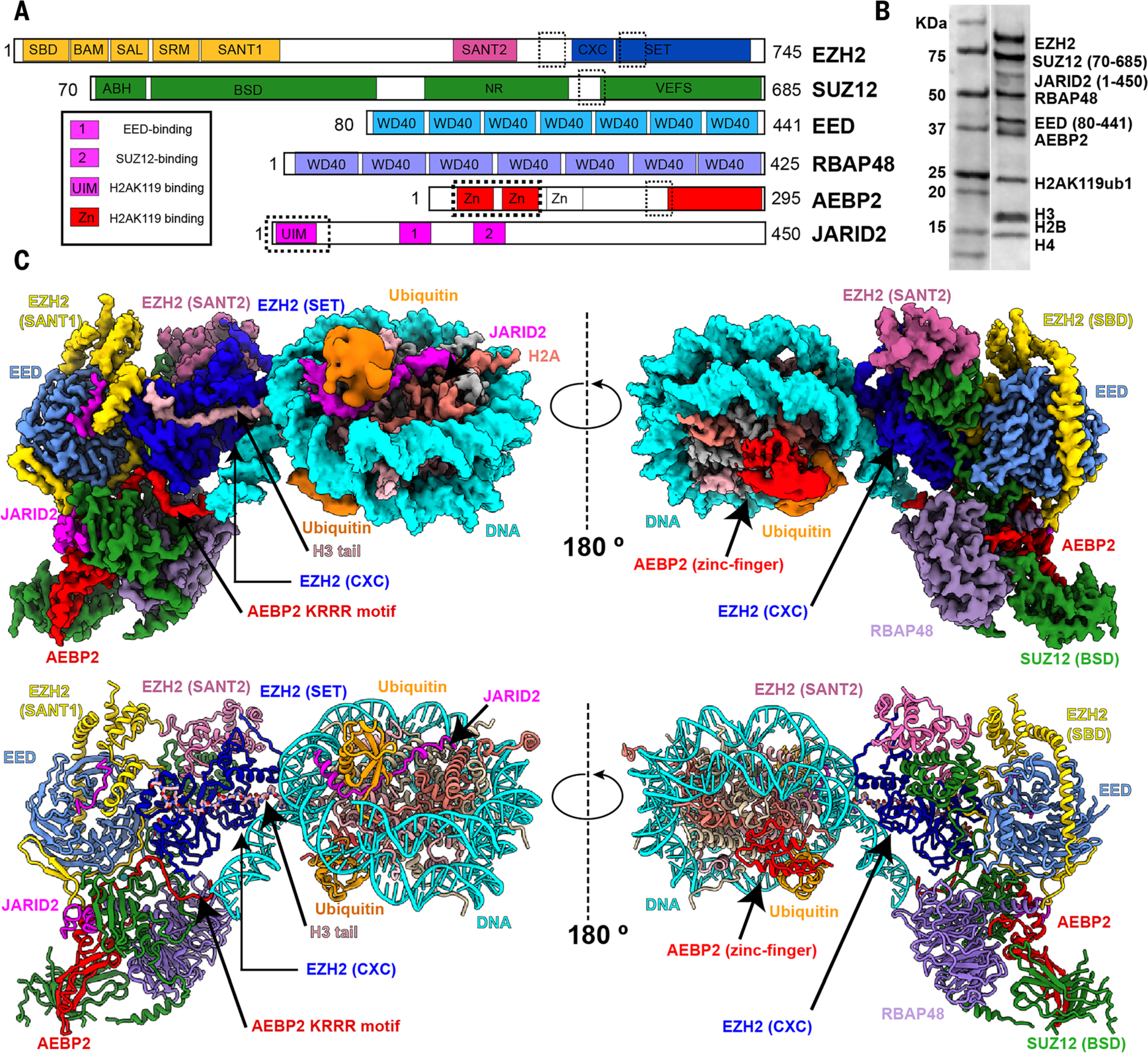
(A) Schematic representation of the proteins in the PRC2-AEBP2-JARID2 complex. Newly modeled regions in EZH2, JARID2, and AEBP2 that contribute to the interaction with the nucleosome are marked by dashed boxes. (B) Coomassie-stained gel showing all of the PRC2-core subunits, cofactors JARID2 and AEBP2, and histone proteins in the sample used for structural studies. (C) (Top) Cryo-EM density map for PRC2-AJ1–450 bound to an H2AK119ub1-containing nucleosome. (Bottom) Atomic model of PRC2-AJ1–450 bound to an H2AK119ub1-containing nucleosome, with EZH2 (SANT1) highlighted in gold, EZH2 (SANT2) in hot pink, EZH2 (SET) in blue, EED in light blue, RBAP48 in light purple, SUZ12 in green, JARID2 in magenta, AEBP2 in red, ubiquitin in orange, histone H3 in light pink, H2A in salmon, H4 and H2B in khaki, and nucleosome DNA in cyan. The image shown is a composite map where the PRC2 and nucleosome core correspond to the local resolution-filtered map from multibody refinement, whereas the ubiquitin, JARID2 UIM, and AEBP2 zinc finger densities correspond to the local resolution-filtered map from the consensus refinement. This color coding also applies to movie S1.
Our structure shows that the SET and CXC domains of EZH2 bind the H2AK119ub1 nucleosome, which contains unmodified histone H3K27, with the same interaction interface previously shown for the substrate nucleosome in our cryo-EM reconstruction of PRC2 bound to a dinucleosome (23) (Fig. 1C and fig. S6). The nucleosomal DNA is further contacted by the KRRKLKNKRRR motif (amino acids 171 to 181) of AEBP2, in agreement with previous studies that have suggested its potential role in DNA interaction (17, 24) (fig. S6). Cryo-EM analysis of the conformational heterogeneity in the sample revealed that these two PRC2-nucleosome interfaces are flexible (fig. S6). In the present structure, we have identified and modeled a region of EZH2 rich in lysines and arginines (amino acids 487 to 513) that interacts extensively with the nucleosomal DNA. The region includes lysines K509 and K510 within the IQLKK motif of EZH2, which had been predicted to be disordered and had been shown to be modified during PRC2 automethylation (25, 26). Mass spectrometry analysis of our PRC2-AJ1–450 sample confirms the presence of dimethylation of both K509 and K510. When bound to substrate nucleosome, the amino-acids-497-to-511 segment in EZH2 forms an alpha helix that is sandwiched between the nucleosomal DNA, the EZH2 (SET) domain, and the H3 tail, which we therefore call the EZH2 bridge helix (Fig. 2A). Its tripartite interaction results in the stabilization of this EZH2 region that has otherwise been unstructured or highly flexible in previous structural studies of PRC2 lacking nucleosome substrate (22, 27, 28). The residues in the EZH2 bridge helix that interact with the histone H3 tail, the nucleosome DNA, and the automethylated lysines K509 and K510 are highly conserved from plants to humans (fig. S7). The resolution of the map allowed us to build a large region of the histone H3 tail (amino acids 21 to 40) de novo and to describe a network of van der Waals and hydrogen bonding interactions that guide the H3 tail into the catalytic site of EZH2 (Fig. 2B). Notably, cancer mutations frequently observed in the EZH2 (SET) and EZH2 (CXC) domains map to regions that interact with the histone H3 tail and DNA (Fig. 2C). Our findings extend our current knowledge of substrate nucleosome engagement, providing an atomic model of DNA and histone tail contacts by EZH2.
Fig. 2. Interaction of EZH2 with the histone H3 tail and nucleosomal DNA.
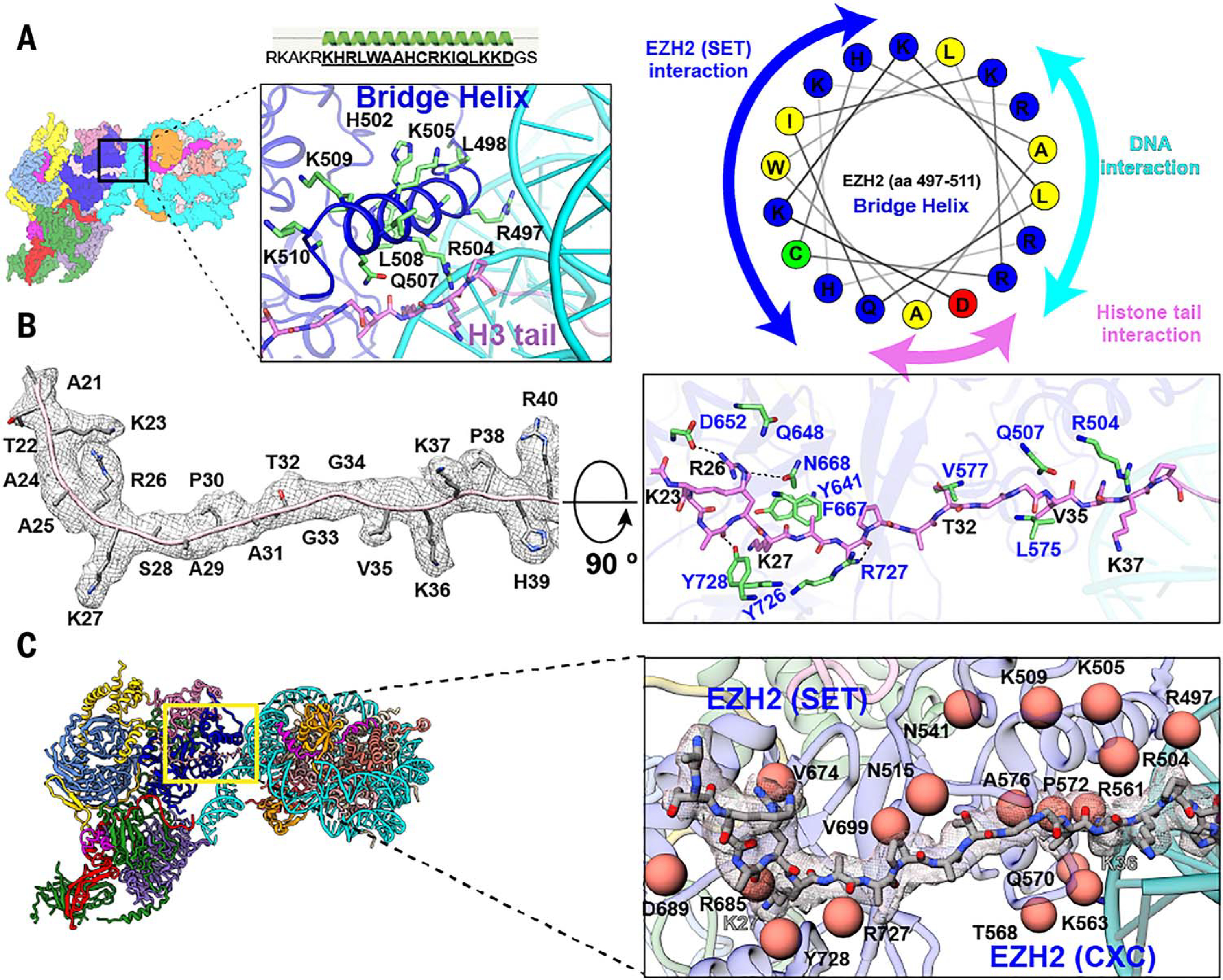
(A) (Top) Cartoon representation of the newly defined EZH2 bridge helix (amino acids 497 to 513) that interacts with nucleosomal DNA and the H3 tail. (Left) Cryo-EM structure of the PRC2-AEBP2-JARID2 complex bound to an H2AK119ub1-containing nucleosome. The zoom out shows the bridge helix, with residues interacting with the EZH2 (SET) domain, nucleosomal DNA, and the histone tail depicted in stick representation. (Right) Helix wheel diagram for the bridge helix that shows the distribution of positively charged residues on the nucleosomal DNA interacting face (cyan), positive and hydrophobic residues on the EZH2 (SET) interacting face (blue), and the residues interacting with the backbone of the H3 tail (pink). aa, amino acids. (B) (Left) Density map of the histone H3 tail (amino acids 21 to 40) (contour level: 0.024) with the corresponding atomic model. Clear density for residues R26 and K27, which have been previously observed, as well as density for K23, K36, K37, and V35 allow us to define the full extent of interaction between EZH2 (SET) and the histone H3 tail. (Right) Extensive electrostatic and van der Waals interactions between the residues in EZH2 (SET) (blue; shown in green stick representation) and the histone H3 tail (black; shown in pink stick representation) guide the H3 tail into the catalytic site. (C) Close-up view of the interaction of EZH2 (SET and CXC) with the histone H3 tail (stick representation with corresponding cryo-EM density in transparency), which shows residues involved in either direct or indirect interaction with the histone H3 tail and nucleosome DNA that are also frequently found mutated in cancers as orange spheres [COSMIC database (57)]. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
JARID2 and AEBP2 independently bind ubiquitin in H2AK119ub1-containing nucleosomes
Proximal to the EZH2 (SET) domain, an N-terminal segment of JARID2 (amino acids 22 to 56) is sandwiched between one of the two ubiquitin molecules and the H2A-H2B acidic patch (Fig. 3A). The JARID2 segment containing amino acids 24 to 39, which corresponds to the reported UIM, interacts extensively with both the ubiquitin and nucleosomal DNA. The ubiquitin conjugated to H2AK119C is oriented such that the JARID2 UIM interacts with the hydrophobic patch on ubiquitin that includes I44, which had previously been shown to be important for JARID2 interaction with ubiquitin (7) (fig. S7). Additionally, the JARID2 segment, containing amino acids 40 to 56 rich in positively charged arginine and lysine residues, interacts with the H2A-H2B acidic patch (Fig. 3A). A cryo-EM reconstruction of PRC2-AEBP2-JARID2 (amino acids 106 to 450) (referred to as PRC2-AJ106–450) that lacks amino acids 22 to 56 of JARID2 also lacks the density we assigned to the JARID2 UIM when bound to Ncl-ub, which confirms our model (fig. S8). In the absence of amino acids 22 to 56 of JARID2, the ubiquitin conjugated to H2AK119C is conformationally flexible and therefore becomes invisible in our cryo-EM reconstructions (fig. S8).
Fig. 3. JARID2 and AEBP2 interact with ubiquitin and the H2A-H2B surface.
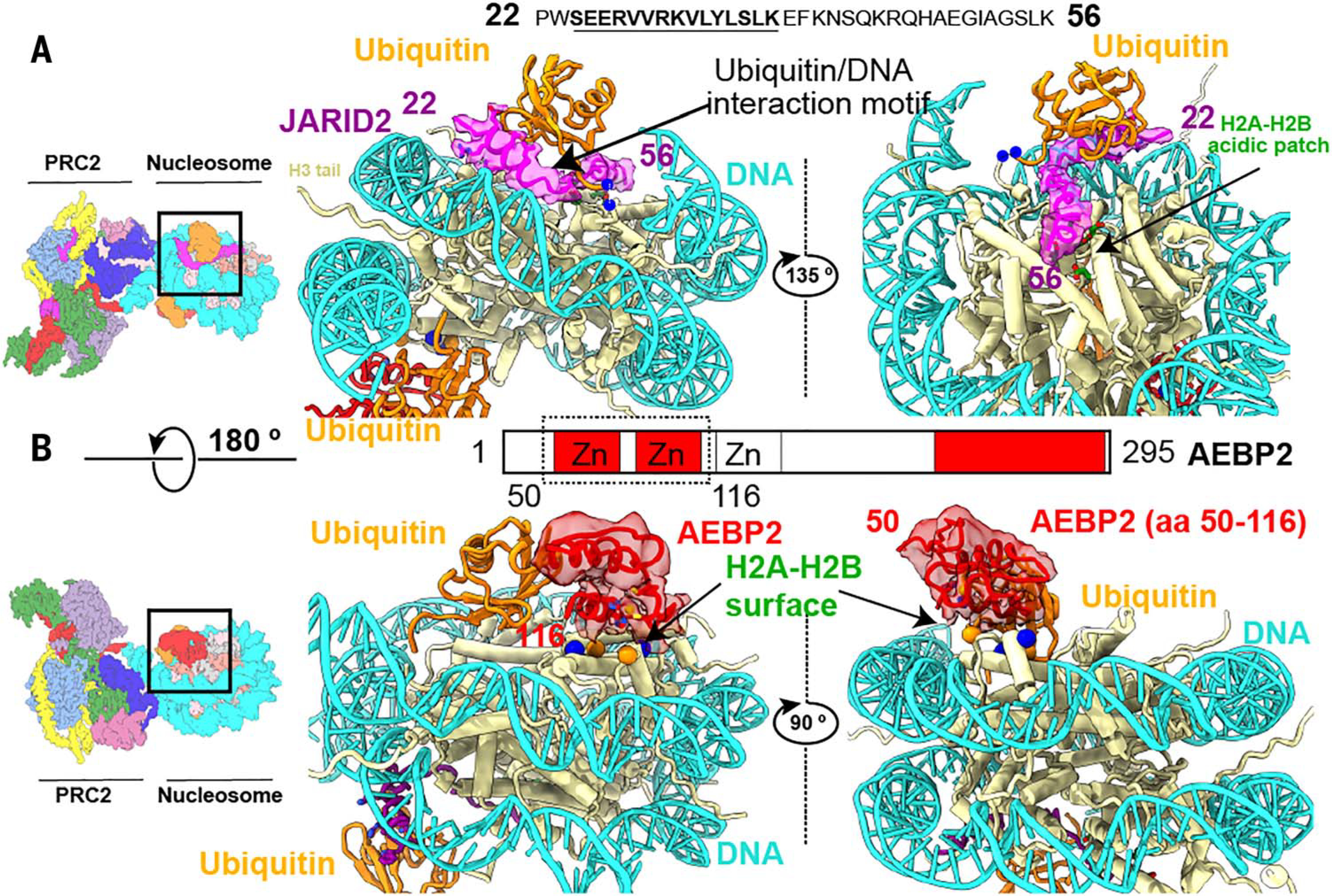
(A) Cryo-EM density (contour level: 0.007) and model of JARID2 (amino acids 22 to 56) (magenta) showing how this segment of JARID2 is sandwiched between ubiquitin and the histone surface. The JARID2 UIM (amino acids 24 to 41) forms an alpha helix that interacts with both ubiquitin and nucleosomal DNA. JARID2 (amino acids 42 to 56), which contains positively charged arginine and lysine residues, sits on top of the conserved H2A-H2B acidic patch. The blue spheres with dashed lines connecting them indicate the H2AK119C-G76C conjugation. (B) The tandem C2H2 zinc fingers of AEBP2 (amino acids 50 to 116) interact with ubiquitin, the H2A-H2B surface, and nucleosomal DNA. The second AEBP2 zinc finger (amino acids 85 to 116) interacts extensively with an H2A-H2B surface that contains a mixture of positively and negatively charged residues (blue and orange balls, respectively).
On the opposite, distal face of the nucleosome, the first two of the three tandem C2H2 zinc fingers of AEBP2 (amino acids 50 to 116) interact with ubiquitin and with an H2A-H2B surface containing both acidic and basic residues (Fig. 3B). The interaction surface between the H2A-H2B and the AEBP2 zinc fingers is similar to that previously observed for the zinc finger of the DUB (deubiquitinase) domain in the yeast cofactor SAGA interacting with the histone surface (29, 30) (fig. S7). The cryo-EM reconstruction of PRC2-AJ1–450 bound to an unmodified nucleosome (lacking H2AK119ub1) shows no density for either the JARID2 amino acids 22 to 56 or the AEBP2 tandem zinc finger regions, which indicates that the interaction of these motifs with the H2A-H2B surface is weak in the absence of ubiquitin (fig. S8). This conclusion was further confirmed by cryo-EM reconstructions of PRC2-AEBP2 bound to an unmodified nucleosome (fig. S8). In summary, our cryo-EM structures show that both JARID2 and AEBP2 mediate the recognition of H2AK119ub1, providing additional anchoring interactions with the nucleosome besides those of EZH2 with nucleosomal DNA and the H3 tail, and thus supporting a mechanism of JARID2- and AEBP2-dependent recruitment of PRC2 to PRC1-modified nucleosomes.
Effect of cofactors on PRC2 activity on ubiquitinated nucleosomes
Given both the role of JARID2 and AEBP2 in H2AK119ub1 recognition and recruitment to chromatin and the conflicting data on the effect of this modification on PRC2 HMTase activity, we decided to compare the enzymatic activity of PRC2 complexes in the presence or absence of these cofactors. We carried out activity assays for five different PRC2 complexes—PRC2-core, PRC2-AEBP2 (PRC2-A), PRC2-JARID2 (amino acids 106 to 450) (PRC2-J106–450), PRC2-AJ1–450, and PRC2-AEBP2ΔZn-JARID2 (amino acids 1 to 450; lacking the zinc fingers in AEBP2) (PRC2-AΔZn-J1–450)—on either unmodified or H2AK119ub1-containing mononucleosomes (Fig. 4, A and B, and figs. S9 and S10). We performed our HMTase activity assays probing both for methyltransferase activity on the basis of total S-adenosyl homocysteine (SAH) production (thus quantifying H3K27me1, H3K27me2, and H3K27me3 cumulatively), and for H3K27me3 specifically using Western blot analysis. In agreement with previous results, our analysis of HMTase activity shows that PRC2 containing both AEBP2 and JARID2 is a much more active methyltransferase compared with either PRC2-core or PRC2 containing only AEBP2, both of which exhibit weak HMTase activity (31) (Fig. 4, A and B). As expected, the PRC2-core does not distinguish the ubiquitinated from the unmodified nucleosome. Despite the interaction of the AEBP2 tandem zinc fingers with both the H2AK119ub1 and H2A-H2B surfaces, the HMTase activity of PRC2-AEBP2 on unmodified nucleosomes was only slightly lower (based on total methylation) than that on Ncl-ub. A PRC2-J106–450, containing the JARID2-K116me3 stimulatory region but lacking the H2AK119ub1-recognizing UIM, displayed enhanced but otherwise similar HMTase activity on either unmodified nucleosomes or H2AK119ub1-containing nucleosomes, as expected on the basis of our structure. The inclusion of the larger JARID2 version (amino acids 1 to 450), containing the H2AK119ub1-recognizing UIM in addition to the stimulatory JARID2-K116me3 segment, results in a further stimulation of PRC2 HMTase activity on H2AK119ub1-containing nucleosomes compared with unmodified nucleosomes (based on both total methylation and specific trimethylation). The deletion of the AEBP2 tandem zinc fingers results in reduction of the additional stimulatory effect observed for H2AK119ub1-containing nucleosomes compared with unmodified nucleosomes (Fig. 4, A and B). Taken together, our results suggest that the primary allosteric activation of PRC2 by JARID2 involves the binding of JARID2 K116me3 to EED that results in the stabilization of the EZH2 (SET) catalytic site, whereas there is a secondary, modest contribution resulting from the interaction of both JARID2 and AEBP2 with H2AK119ub1 and core histones on opposite faces of the nucleosome. Our structure suggests that both JARID2 and AEBP2 may play a role during the recruitment of PRC2 by means of H2AK119ub1 recognition. The increase in HMTase activity of PRC2-AJ1–450 on H2AK119ub1-containing nucleosomes could be because of an increased residence time of PRC2 resulting from additional anchoring interactions of the cofactors with the ubiquitin and the histone core.
Fig. 4. HMTase activity of different PRC2 complexes on modified nucleosomes.
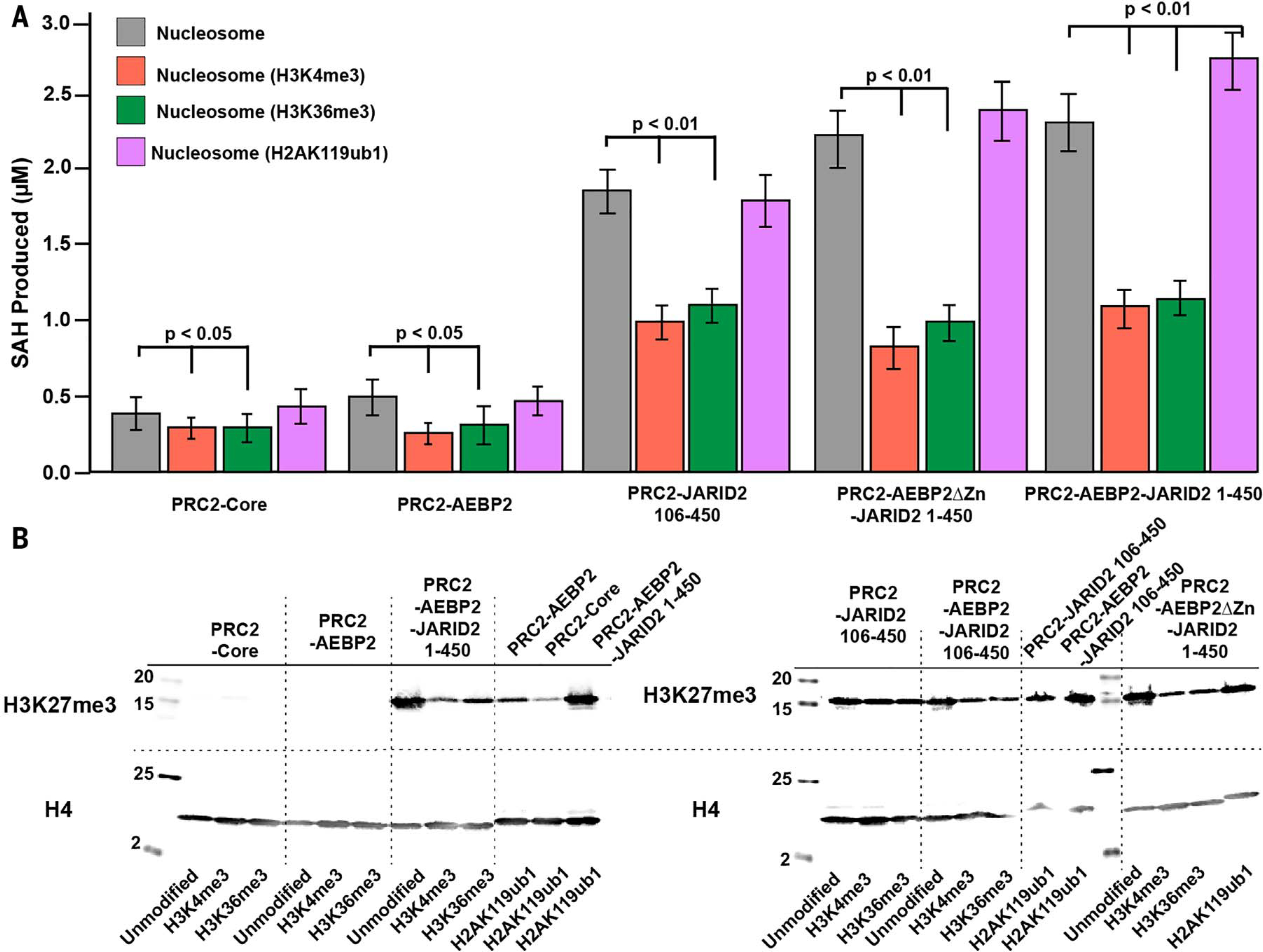
(A) Bar graph showing the comparison of end-point, cumulative HMTase activity (H3K27me1/me2/me3) of PRC2-core, PRC2-AEBP2, PRC2-JARID2 (amino acids 106 to 450), PRC2-AEBP2ΔZn-JARID2 (amino acids 1 to 450), and PRC2-AEBP2-JARID2 (amino acids 1 to 450) on unmodified nucleosomes (gray), H3K4me3-containing (orange), H3K36me3-containing (green), and H2AK119ub1-containing nucleosomes (magenta). The differences in end-point measurements that are statistically significant are indicated by the P values. (B) Western blot analysis comparing the H3K27me3 production for unmodified nucleosomes versus the H3K4me3-containing, H3K36me3-containing, and H2AK119ub1-containing nucleosomes.
JARID2 partially overcomes the inhibitory effect of H3K4me3 and H3K36me3
In view of the involvement of cofactors in the cross-talk between H2AK119ub1 and PRC2 HMTase activity, we asked how the presence of JARID2 and/or AEBP2 could also have an effect on HMTase activity in the presence of the active transcription marks H3K4me3 and H3K36me3. To our knowledge, the in vitro effect of these modifications on PRC2 HMTase activity has been reported only for the PRC2-core complex lacking cofactors. Again, we performed HMTase activity assays probing for total methyltransferase activity and for H3K27me3 specifically using five different PRC2 complexes: PRC2-core, PRC2-A, PRC2-J106–450, PRC2-AJ1–450, and PRC2-AΔZn-J1–450. We found that in both types of assays, the HMTase activities of PRC2-core and PRC2-AEBP2 complexes are weak in the presence of unmodified nucleosomes and for nucleosomes containing either H3K4me3 or H3K36me3, which is in agreement with previous studies (20, 32) (Fig. 4, A and B, and figs. S9 and S10). Although PRC2 complexes containing both AEBP2 and JARID2 (amino acids 1 to 450) displayed some inhibition for cumulative activity (H3K27me1/me2/me3) on nucleosomes containing H3K4me3 or H3K36me3, they still retained substantial activity on both (on the basis of cumulative methylation and H3K27me3 Western blot analysis). HMTase activity assays with human nucleosomes containing H3K4me3 or H3K36me3 generated by means of native ligation mirror the observations for Xenopus nucleosomes containing H3K4me3 or H3K36me3 generated as a methyl-lysine analog (33) (figs. S9 and S10). This activity of PRC2-AJ1–450 is further confirmed by the detection of peptides containing H3K27me1/me2/me3 by mass spectrometry analysis of HMTase reactions of this complex with either H3K4me3 or H3K36me3 nucleosomes. Thus, our data show that in the presence of both AEBP2 and JARID2, there is still an inhibitory effect of the two marks but that the allosteric activation via JARID2-K116me3 results in activity that is substantially increased with respect to that without JARID2. We propose that complete PRC2 inhibition at actively transcribing regions may require a combination of H3K4me3- or H3K36me3- and RNA-mediated effects or perhaps the eviction of PRC2 containing JARID2 and/or AEBP2 (9, 20, 34–36). To gain a mechanistic understanding of the inhibition observed for H3K36me3-containing nucleosomes, we carried out structural modeling of the H3K36me3 within our cryo-EM structure of PRC2-AJ1–450 bound to a H2AK119ub1-containing nucleosome. Modeling suggests that although there is limited space for the bulky trimethyl group, it could still be accommodated by means of alternative side chain conformations (Fig. 5). In addition to possible steric effects of accommodating H3K36me3, the loss of electrostatic interaction between the H3K36 and the phosphate backbone of nucleosomal DNA may also contribute to the inhibition of HMTase activity by H3K36me3. These conclusions agree with those recently reached by Müller and co-workers from the analysis of H3 tail binding in their structure of PRC2-PHF1 bound to a dinucleosome (37).
Fig. 5. Location of H3K36 at the PRC2-nucleosomal DNA interface.
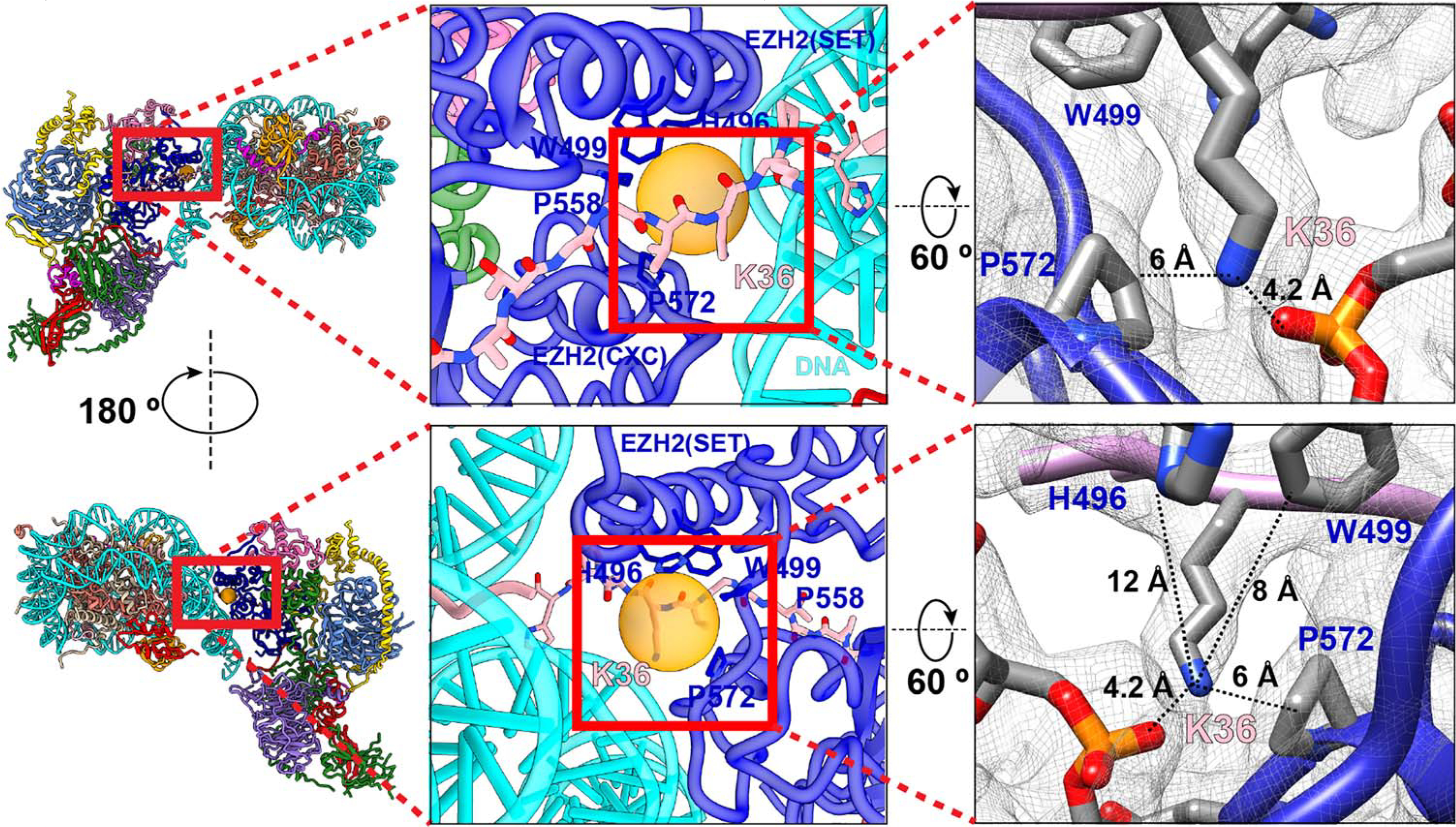
Close-up view of the location of histone H3K36 and its interactions with nucleosomal DNA and PRC2. Bulky residues in PRC2 close to H3K36 are shown in stick representation, with the orange sphere indicating the potential space available for alternative conformations of the H3K36 side chain. On the right is the cryo-EM density (contour level: 0.024) with the atomic model shown, illustrating the distances between the H3K36 terminal amine group and the nearest bulky residues.
To investigate the effect of H3K4me3 on the engagement of PRC2 with nucleosomes containing this modification, we used cryo-EM to visualize PRC2-AJ1–450 bound to a human H3K4me3-containing nucleosome. Our analysis shows the presence of two distinct states in this sample: one in which the H3 tail is engaged with the SET domain, as seen for the unmodified tail, and one in which the H3 tail is not engaged (figs. S11 and S12). In both cases, our cryo-EM reconstructions (~7-Å resolution) show that the interaction of PRC2 with this H3K4me3-modified nucleosome is indistinguishable from that seen with the H2AK119ub1-containing nucleosome (fig. S12). In addition to the presence or absence of an engaged H3 tail, we noticed differences in the region of PRC2 proximal to EED and the SANT1 domain, where the allosteric activator segments (JARID2 K116me3 or H3K27me3) bind (figs. S11 and S12). We hypothesize that a less-stable H3 tail engagement with the active site in the presence of H3K4me3, as indicated by the coexistence of the two states we observe by cryo-EM, could contribute to the H3K4me3-mediated inhibition of PRC2 HMTase activity. Because PRC2 associates with different cofactors in vivo, more-detailed structural and biochemical analyses of PRC2 in complex with cofactors and bound to H3K4me3- or H3K36me3-containing nucleosomes will be necessary to better understand the inhibitory effect of these marks on PRC2 activity and the reduction of that inhibition by cofactors.
Conclusion
Our structural work has identified a key region—the EZH2 bridge helix—for the binding of PRC2 to nucleosomes and its engagement of the H3 tail. It also defines the details of how PRC2 cofactors JARID2 and AEBP2 recognize nucleosomes containing H2AK119ub1 and thus provides a mechanistic explanation for the recruitment of PRC2 to PRC1-modified genomic regions. Our activity assays indicate that H2AK119ub1 results in a modest increase in HMTase activity in the presence of both cofactors and that their presence reduces the inhibitory effect of H3K4me3 or H3K36me3. We speculate that the reduced inhibition in the presence of cofactors could be relevant in the context of establishing and maintaining bivalent domains containing both H3K4me3 and H3K27me3 marks in embryonic stem cells and during imprinting (38). Taken together, our results emphasize the extent to which cofactors like AEBP2 and JARID2 can serve as regulators of PRC2 activity in different chromatin environments. Additional studies will be required to shed further light onto the complex regulatory mechanisms of PRC2 activity.
Materials and methods
Protein purification
PRC2 constructs containing full-length EZH2, EED (amino acids 80 to 441), SUZ12 (amino acids 70 to 685), full-length RBAP48, the embryonic isoform of AEBP2, either full length or missing the tandem zinc fingers (AEBP2ΔZn), and different JARID2 constructs were expressed and purified as previously described (22). PRC2-core components together with cofactors strep-tagged AEBP2 and FLAG-tagged JARID2 were assembled into a multibac construct for HighFive insect cell MacroBac baculovirus expression system (39) The cells were transfected with baculovirus at 28°C for 66 hours and frozen in liquid nitrogen until use. PRC2 constructs were purified by resuspending cell pellets in 25 mM HEPES pH 7.9, 250 mM NaCl, 2 mM MgCl2,1 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1% NP40, 10% glycerol supplemented with 10 μM leupeptin, 0.2 mM PMSF, DNAse, and protease inhibitor cocktail (Roche). All purification steps were performed at 4°C. After cell lysis, debris were removed by centrifugation at 15,000 rpm for 35 min. The supernatant was incubated with Step-tactin superflow plus resin (Qiagen) overnight (12 hours), and resin was washed with low-salt buffer A (25 mM HEPES pH 7.09, 250 mM NaCl, 2 mM MgCl2,0.01% NP40, 1 mM TCEP, 10% glycerol, 10 μM leupeptin, 0.2 mM PMSF, and protease inhibitor cocktail). The resin was further washed with high-salt buffer (buffer A with 1 M NaCl). Subsequently, the resin was washed with buffer A without NP40 and eluted with 10 mM Desthiobiotin. The elutes were pooled and incubated with anti-Flag M2 agarose affinity gel and washed with buffer A with 1 M NaCl. PRC2 complexes were eluted in buffer A (without NP40) with 150 μg/ml 3XFLAG peptide and subjected to overnight TEV cleavage to remove all affinity tags. The complexes were further purified over Superose 6 increase (GE Healthcare) equilibrated with 25 mM HEPES pH 7.9, 150 mM NaCl, 2 mM MgCl2, 10% glycerol, and 1 mM TCEP. Protein was flash frozen in liquid nitrogen as single-use aliquots and stored at −80°C.
Nucleosome reconstitution
Xenopus histones (H2A, H2B, H3, and H4) were expressed and purified as described previously (40). All of the nucleosomal DNA used in this study contained a CpG Island sequence and a 5′ biotin tag and was assembled by large scale polymerase chain reaction, purified over an anion exchange column, and further purified by ethanol precipitation. The 227-base pair (bp) nucleosome DNA sequence used for all the studies with Xenopus nucleosomes was 5′ (biotin)CACGCGACTGTGTGCCCGTCAGACGCgtgccgaggccgctcaattggtcgtagacagctctagcaccgcttaaacgcacgtacgcgctgtcccccgcgttttaaccgccaaggggattactccctagtctccaggcacgtgtcagatatatacatcctgtatgcatgcatatcattcgatcggagctcccgatcgatgc - 3′. The 601-nucleosome positioning sequence is underlined, and the CG-rich sequence used is indicated in uppercase. For unmodified nucleosomes, equimolar amounts of all histones were dialyzed, and the octamer was purified over Superdex 200 (GE Healthcare). The DNA and octamer were mixed in a 1:1.1 ratio and purified over a BioRad prep cell after overnight gradient salt dialysis, as described previously (40). For modified nucleosomes, MLA analogs of H3K4me3 and H3K36me3 were first generated before octamer reconstitution and nucleosome generation (33). For the generation of ubiquitinated nucleosomes, first equimolar amounts of H2A (K119C) were cross-linked with 6xHis-ubiquitin (G76C) using dichloroacetone and purified under denaturing conditions over a nickel column, as described previously (41). The purified H2A-ub was dialyzed into water and lyophilized for storage. For H2A-ub-containing octamer reconstitution, roughly equimolar amounts of H2A-ub were mixed with H2B under denaturing conditions, and the H2Aub-H2B dimer was purified similarly to the octamer as described above. The H3-H4 tetramer was reconstituted, and the purified H3-H4 tetramer, H2A-ub-H2B dimer, and nucleosomal DNA were mixed in 1.2:2.4:1 ratio and purified using a BioRad Prep cell after overnight salt dialysis. Nucleosomes were stored on ice for up to 1 month. All human nucleosomes were purchased from Epicypher with the 5′ biotinylated 187-bp DNA sequence containing the 601-nucleosome positioning sequence: 5′(BioTEG)GGACCCTATACGCGGCCGCCCtggagaatcccggtctgcaggccgctcaattggtcgtagacagctctacgtggcgaatttgcgtgcatgcgcctgtcccccgcgttttaaccgccaaggggattactccctagtctccaggcacgtgtcagatatatacatcctgtGCCGGTCGCGAACAGCGACC 3′. All recombinant Xenopus nucleosomes containing H3K4me3 or H3K36me3 were generated as MLA analogs, whereas all human nucleosomes purchased from Epicypher contained native ligated H3K4me3 or H3K36me3.
Kinetic assays
For PRC2 activity assays on different modified mononucleosomes, approximately twofold dilutions of nucleosomes at concentrations from 1 μM to 40 nM were used, together with 20 μM S-adenosyl methionine (SAM). The reaction, carried out at room temperature (RT), was initiated by the addition of PRC2 complexes to a final concentration of 40 nM. Ten microliters of the reaction was taken at 0.5-min, 1-min, and 2-min time points and quenched immediately by addition of trifluoroacetic acid at a final concentration of 0.1%. The quenched reactions were then transferred to a 384-well, white round-bottom microplate to profile for methyltransferase activity (cumulative mono-, di-, and trimethylation) using the MTase-Glo assay (Promega), which detects the production of SAH as a luminescent indicator. The raw luminescence values were correlated to SAH concentration using standard curves after accounting for basal luminescence of PRC2, nucleosome, and buffer without SAM. The initial rates were determined by linear regression and kinetic parameters determined by fitting the initial rates. All reactions were performed in duplicate, and the entire experiment was repeated twice for each PRC2-nucleosome combination. Concentrations lower than 40 nM PRC2 complex could not be used because of PRC2 dissociation (on the basis of negative stain EM analysis). Michaelis-Menten fits of the rate of reaction as a function of substrate concentration for Vmax (maximal velocity achieved by the system at saturating substrate concentrations) and KM (Michaelis constant), were carried out using in-house scripts. Notably, PRC2 is an allosteric enzyme and thus precise interpretation of Michaelis-Menten parameters is not suitable. However, qualitative comparison of the largely similar KM values for the different PRC2 complexes with different nucleosome substrates suggests that the overall binding affinity is similar, as observed previously (9, 20).
Endpoint methyltransferase measurements were performed under similar concentrations, with a 10-μl reaction containing 40 nM PRC2, 1 μM nucleosome, and 40 μM SAM, incubated for 2 hours at RT. The reactions were quenched by addition of trifluoroacetic acid to a final concentration of 0.1%. This was used for luminescent detection using MTase-Glo assay (Promega), as above. Each reaction was done in triplicate, and the whole experiment was repeated twice. The statistical significance of the differences in measurements was obtained using unpaired t test followed by P value estimation.
Western blots
PRC2 complex (200 nM) was incubated together with 400 nM nucleosomes and 40 μM SAM in HMTase buffer containing 25 mM HEPES pH 7.9, 50 mM NaCl, 2 mM MgCl2, 0.5 mM EDTA, and 1 mM dithiothreitol (DTT) at RT for 90 min. The reactions were then run on a 4–20% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gel for 30 min at 150 V. Amersham Hybond Nitrocellulose membrane was used for the transfer. Transfers were performed in a 4°C cold room in an ice bath at 90 V for 10 min followed by 60 V for 30 min. Primary antibodies from Cell Signaling Technology for H3K27me3 (rabbit monoclonal) and H4 (mouse monoclonal) were used. Secondary antibodies from ECL Plex containing Cy5 (goat-anti-rabbit) or Cy3 (goat-anti-mouse) were used to detect H3K27me3 and H4, respectively. This Cell Signaling antibody was highly specific for H3K27me3 on either Xenopus H3 or human H3.2 or H3.3 containing H3K27me3, with no cross reactivity detected toward H3K4me3 or H3K36me3. The repetition of Western blot assay (fig. S9D) was performed with 200 nM PRC2 complex, 1 μM nucleosome substrates, and 80 μM SAM.
Cryo-EM sample preparation
Streptavidin monolayer affinity grids were used for cryo-EM grid preparation. Quantifoil Au 2/2 Streptavidin grids were made in-house using procedures described previously (42). PRC2-nucleosome complexes were assembled by incubating 200 nM nucleosome with 600 nM PRC2 complex and 80 μM SAH in 25 mM HEPES pH 7.9, 40 mM KCl, 1 mM TCEP, and 1 mM MgCl2 for 30 min at RT. Three microliters of the complex was then incubated with streptavidin monolayer affinity grids (prewashed in the buffer above) and incubated for 3 to 8 min in a humidified chamber. The grid was washed twice with 10 μl of wash buffer containing 25 mM HEPES pH 7.9,40 mM KCl, 1 mM MgCl2, 1 mM TCEP, 4% Trehalose, and 0.01% NP40 or 0.05% beta-octaglucoside. After the washes, the buffer was wicked away using Whatman filter paper, and 2 μl of the wash buffer described above was added immediately. The grid was then transferred to the FEI Mark IV Vitrobot and manually blotted for 2 s at 5°C and 100% humidity and then plunged into liquid ethane.
Cryo-EM data collection and processing
All of the datasets were collected in super-resolution counting mode with image shift and active beam-tilt correction. All but one of the datasets were collected on either a Titan Krios or Talos Arctica equipped with Gatan K3 direct detector. One of the datasets was collected on a Titan Krios equipped with a Gatan K2 detector. All data were acquired as dark-subtracted, non-gain corrected movies, and gain correction was applied during motion correction using MotionCor2 (43). Data acquisition was performed using SerialEM with custom macros for automated data collection. A GIF quantum energy filter was used for collection with a 20-eV slit width. The total dose for all of the datasets was 50 electrons per square angstrom (e−/Å2). All other parameters for data collection can be found in table S1.
Negative stain on the PRC2-nucleosome complexes was performed by applying 3 to 4 μl of 50 nM nucleosome plus 150 nM PRC2 on glow-discharged 400 mesh C-flat continuous carbon grids (Protochips) and incubated for <20 s before staining. Negative stain screening to gauge the intactness of PRC2 was performed by diluting the PRC2 complexes to the appropriate concentration and immediately applying 3 to 4 μl to the glow-discharged grids. All negative stain samples were visualized on a Tecnai F20 microscope operated at 120 kV, with a Gatan Ultrascan 4000 camera, at a nominal magnification of 80,000× (1.5 Å per pixel), using a dose of 20 e−/Å2.
All data processing was done within RELION (44). The movie frames were aligned using MotionCor2 (43) and CTF parameters were fit using GCTF (42). The summed aligned movies were used to substract the background streptavidin lattice using in-house scripts (45). These subtracted, summed aligned movies were used in RELION version 3.0 and 3.1 for processing. Several algorithms were tried for particle picking including logpicker, RELION autopick based on two-dimensional (2D) templates obtained from a combination of logpicker or manual picking, EMAN neural net picking, and Topaz neural net picking (46, 47). Topaz was eventually used because it resulted in the cleanest picks and, more importantly, particles with different orientations. To pick particles within Topaz, ~200 to 500 unambiguous particles were carefully picked manually and used to train a neural net model. This neural net model was subsequently applied to the entire dataset. Topaz neural net models were generated for each dataset individually. Multibody refinement and particle subtraction were performed using the latest build of RELION 3.1. For all datasets, initial models were generated within RELION and used as reference for the first round of 3D classification. Subsequent processing steps used good classes from the initial 3D classification as a reference. After the initial 3D classification, 2D classification was performed for all the datasets to clean up the particle stack. Subsequently, the good particles were subjected to refinement and further 3D classification followed by downstream processing. For the PRC2 bound to Ncl-ub, the final set of good particles from both datasets—one collected with a Gatan K3 detector and the other collected with a Gatan K2 detector—was combined and refined, followed by 3D classification and subsequent final MultiBody refinement in RELION. The combined data, however, did not result in an improvement in resolution or in the map quality, likely because of both the fewer number of particles and the overall higher defocus of the Gatan K2 dataset compared with the Gatan K3 dataset. Soft-edged masks used during multibody refinement were generated within RELION at a contour level of 0.006 and extended by 9 Å with a soft edge of 13 Å. Soft-edged masks used for the 2:1 PRC2: nucleosome were generated within RELION at a contour level of 0.009 and extended by 9 Å with a soft edge of 13 Å. Local resolution estimation was performed within RELION using the same soft, spherical masks used during refinement.
Model building
All models were built using the cryo-EM maps of PRC2-AJ1–450 bound to Ncl-ub. The coordinates of PRC2-AEBP2-JARID2 (amino acids 106 to 450) were used as a starting model from which all the coordinates were adjusted and rebuilt in the new map using COOT (22, 48). Regions corresponding to EZH2, AEBP2, and SUZ12 were built de novo into the EM density in COOT (48). Regions of JARID2 assigned after comparison with cryo-EM maps of PRC2-AJ1.450 bound to unmodified nucleosomes and PRC2-AEBP2 bound to H2AK119ub1 ubiquitinated nucleosomes were built using Protein Data Bank ID 3AIQ as a starting model and adjusted in COOT (48, 49). The register of the JARID2 UIM region was set using the bulky Arg, Lys, or Trp side chains. The tandem zinc fingers of AEBP2 were built by first docking each zinc finger structure individually into the cryo-EM density and then adjusting it in COOT (48, 50). The resolution of the zinc finger proximal to the nucleosome allowed us to set the helix register using bulky His, Trp, Tyr, or Lys side chains and by the zinc coordinating side chains Cys and His. The model prediction for AEBP2 tandem zinc finger density using the BALBES approach as part of the CCP4 suite also returned a tandem zinc finger fold (51).
The nucleosome was built by first docking the atomic model with the 601-nucleosome positioning sequence into the EM density, and all the histones and DNA were manually adjusted and extended in COOT (48, 52). The histone H3 tail reaching into the EZH2 catalytic site was built de novo in COOT (48). The density of histone H2A reaching into the ubiquitin density was used as a marker for rigid body docking of the structure of ubiquitin, such that G76C of ubiquitin would be ~3 Å from H2AK119C. Similarly, for the ubiquitin close to the AEBP2 density on the nucleosome, the density of the H2A tail and the density of the C-terminal tail of ubiquitin were used as markers to rigid body dock the ubiquitin (29, 53).
The model for the second PRC2 bound to Ncl-ub was obtained by rigid-body docking the model for PRC2 obtained in this study, and the histone H3 tail was modeled as a Poly-Ala chain in the catalytic site of the second PRC2.
The model for the monomeric PRC2-Ncl-ub structure was subjected to global refinement and minimization in real space using PHENIX (54). These were then subjected to manual inspection and adjustment in COOT (48) followed by refinement again in PHENIX (54). Model overfitting was also evaluated in PHENIX.
Mass spectrometry
Sample preparation was performed as described previously (22). Briefly, 50 μg of each PRC2 complex was cross-linked with either 0.5 mM final concentration of isotope-labeled di-Succinimidyl Suberate (DSS-d0 or DSS-d12) (Creative Molecules Inc.) in a final volume of 37 μl of 25 mM HEPES pH 7.9,150 mM NaCl, 1 mM TCEP, and 5% glycerol and then incubated at 37°C for 30 min at 500 rpm on a Thermomixer (Eppendorf). All reactions were quenched with 50 mM final concentration of ammonium bicarbonate (NH4HCO3) and evaporated to dryness in a vacuum centrifuge. The dried pellets were dissolved in 50 μl of 8 M urea, reduced with 2.5 mM TCEP for 30 min at 37°C and, alkylated with 5 mM iodoacetamide (Sigma-Aldrich) for 30 min at RT in the dark. Digestion was carried out after diluting urea to 5 M with 50 mM NH4HCO3 and adding 1% (w/w) LysC protease (Wako Chemicals) for 2 hours at 37°C, subsequently diluting to 1 M urea with 50 mM NH4HCO3, and finally adding 2% (w/w) trypsin (Promega) for 14 hours at 37°C. Protein digestion was stopped by acidification with 1% (v/v) formic acid. Digested peptides were purified using Sep-Pak C18 cartridges (Waters) according to the manufacturer’s protocol. Cross-linked peptides were enriched by peptide size exclusion chromatography (SEC) as previously described (55). SEC fractions were then reconstituted in 5% acetonitrile and 0.1% formic acid and analyzed in duplicates on an high-performance liquid chromatography (HPLC) system (Thermo Easy-nLC 1200) coupled to a mass spectrometer (Thermo Orbitrap Fusion Lumos). Analytes were separated on an Acclaim PepMap rapid separation liquid chromatography (RSLC) column (25 cm by 75 μm, 2-μm particle size, Thermo Scientific) over a 60-min gradient from 7 to 35% acetonitrile at a flow rate of 300 nl/min. The mass spectrometer was operated in data-dependent acquisition (DDA) mode with MS acquisition in the Orbitrap analyzer at 120,000 resolution and tandem mass spectrometry (MS/MS) acquisition in the linear ion trap at normal resolution after collision-induced dissociation. DDA was set up to select precursors from an MS1 full scan with a charge state of +3 or higher within a 3-s cycle between MS1 scans and a dynamic exclusion of 30 s.
Raw files were loaded in MaxQuant (v1.5.2.8) (56) and searched against a Swiss-Prot canonical human proteome database (including common contaminant proteins), with precursor and fragment tolerances of 20 parts per million (ppm) and 0.5 Da, respectively. LFQ (label-free quantitation) mode was activated. The search was performed in target-decoy mode, including reverse decoys and including carbamidomethylation (C) as fixed modification and the following variable modifications: mono-, dimethyl (KR), trimethyl (K), oxidation (M), and phosphorylation (STY). Results were filtered at 1% peptide and protein level false discovery rate. Peptide intensities for EZH2 peptides were corrected by the abundance of EZH2 protein in each condition with a custom R script.
Mass spectrometry analysis for the detection of H3K27me1/me2/me3 in HMTase reactions with H3K4me3- or H3K36me3-containing nucleosomes was carried out by performing insolution tryptic digest of 10- to 20-μg reaction using the rapid tryptic digest kit (Promega) and following manufacturer’s instructions. Briefly, each reaction mixture was diluted 1:2 with liquid chromatography-mass spectrometry grade water, followed by 4× dilution with rapid digest buffer and then followed by the addition of reducing agent TCEP at a final concentration of 2 mM. Each reaction was then incubated at 42°C for 1 hour. The reaction mixture was cooled to RT, and freshly prepared iodoacetamide was added at a final concentration of 6 mM and incubated in the dark for 1 hour. The reaction mixtures were each digested with 2 μg of trypsin by incubation at 60°C for 2 hours followed by 37°C overnight, and they were then injected into the mass spectrometer for analysis.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Chintangal and P. Tobias for computational support, D. Reif for his contributions during the early stages of the project, R. M. Glaeser and B. G. Han for valuable advice concerning streptavidin grid preparation, B. Greber for advice during model building, and J. Müller and his laboratory members for insightful discussions.
Funding: This work was partially funded by NIH grant GM127018 to E.N. V.K. was supported by a postdoctoral fellowship from the Helen Hay Whitney Foundation and the NIH K99/R00 Pathway to Independence Award. S.P. was supported by the Alexander von Humboldt Foundation (Germany) as a Feodor-Lynen postdoctoral fellow. M.F. was supported by an EMBO Long-Term Fellowship (ALTF-343-2013). Access to the instrumentation was supported by the following grants to R.A.: Proteomics 4D (ERC-2014-AdG 670821), ULTRA-DD (FP7-JTI 115766), and ETH Scientific Equipment. E.N. is a Howard Hughes Medical Institute investigator.
Footnotes
Competing interests: The authors declare no competing interests.
SUPPLEMENTARY MATERIALS
science.sciencemag.org/content/371/6527/eabc3393/suppl/DC1
Figs. S1 to S12
Table S1
References (58, 59)
MDAR Reproducibility Checklist
Movie S1
View/request a protocol for this paper from Bio-protocol.
SUPPLEMENTARY MATERIALS http://science.sciencemag.org/content/suppl/2021/01/19/371.6527.eabc3393.DC1
Data and materials availability:
Cryo-EM density maps and fitted models have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) for the PRC2-AEBP2-JARID2 (amino acids 1 to 450) bound to Ncl-ub [EMDB: 21707 and PDB: 6WKR (HPUB)].
REFERENCES AND NOTES
- 1.Bracken AP, Helin K, Polycomb group proteins: Navigators of lineage pathways led astray in cancer. Nat. Rev. Cancer 9, 773–784 (2009). doi: 10.1038/nrc2736 [DOI] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Cavalli G, Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136, 3531–3542 (2009). doi: 10.1242/dev.033902 [DOI] [PubMed] [Google Scholar]
- 3.Di Croce L, Helin K, Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol 20, 1147–1155 (2013). doi: 10.1038/nsmb.2669 [DOI] [PubMed] [Google Scholar]
- 4.Margueron R, Reinberg D, The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). doi: 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao R et al. , Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002). doi: 10.1126/science.1076997 [DOI] [PubMed] [Google Scholar]
- 6.Hauri S et al. , A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 17, 583–595 (2016). doi: 10.1016/j.celrep.2016.08.096 [DOI] [PubMed] [Google Scholar]
- 7.Cooper S et al. , Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun 7, 13661 (2016). doi: 10.1038/ncomms13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaneko S et al. , Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol. Cell 53, 290–300 (2014). doi: 10.1016/j.molcel.2013.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X et al. , Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol 24, 1028–1038 (2017). doi: 10.1038/nsmb.3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blackledge NP et al. , Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157, 1445–1459 (2014). doi: 10.1016/j.cell.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackledge NP et al. , PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 77, 857–874.e9 (2020). doi: 10.1016/j.molcel.2019.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamburri S et al. , Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 77, 840–856.e5 (2020). doi: 10.1016/j.molcel.2019.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalb R et al. , Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol 21, 569–571 (2014). doi: 10.1038/nsmb.2833 [DOI] [PubMed] [Google Scholar]
- 14.Whitcomb SJ et al. , Histone monoubiquitylation position determines specificity and direction of enzymatic cross-talk with histone methyltransferases Dot1L and PRC2. J. Biol. Chem 287, 23718–23725 (2012). doi: 10.1074/jbc.M112.361824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendenhall EM et al. , GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLOS Genet. 6, e1001244 (2010). doi: 10.1371/journal.pgen.1001244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mikkelsen TS et al. , Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007). doi: 10.1038/nature06008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grijzenhout A et al. , Functional analysis of AEBP2, a PRC2 Polycomb protein, reveals a Trithorax phenotype in embryonic development and in ESCs. Development 143, 2716–2723 (2016). doi: 10.1242/dev.123935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G et al. , Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24, 368–380 (2010). doi: 10.1101/gad.1886410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiden AP et al. , Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell 6, 591–602 (2010). doi: 10.1016/j.stem.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitges FW et al. , Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 (2011). doi: 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- 21.Yuan W et al. , H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem 286, 7983–7989 (2011). doi: 10.1074/jbc.M110.194027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasinath V et al. , Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 359, 940–944 (2018). doi: 10.1126/science.aar5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poepsel S, Kasinath V, Nogales E, Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Biol 25, 154–162 (2018). doi: 10.1038/s41594-018-0023-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee C-H et al. , Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol. Cell 70, 435–448.e5 (2018). doi: 10.1016/j.molcel.2018.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C-H et al. , Automethylation of PRC2 promotes H3K27 methylation and is impaired in H3K27M pediatric glioma. Genes Dev. 33, 1428–1440 (2019). doi: 10.1101/gad.328773.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X et al. , Regulation of histone methylation by automethylation of PRC2. Genes Dev. 33, 1416–1427 (2019). doi: 10.1101/gad.328849.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao L, Liu X, Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 350, aac4383 (2015). doi: 10.1126/science.aac4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Justin N et al. , Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat. Commun 7, 11316 (2016). doi: 10.1038/ncomms11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morgan MT et al. , Structural basis for histone H2B deubiquitination by the SAGA DUB module. Science 351, 725–728 (2016). doi: 10.1126/science.aac5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H et al. , Structure of the transcription coactivator SAGA. Nature 577, 717–720 (2020). doi: 10.1038/s41586-020-1933-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son J, Shen SS, Margueron R, Reinberg D, Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 27, 2663–2677 (2013). doi: 10.1101/gad.225888.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge EJ, Jani KS, Diehl KL, Müller MM, Muir TW, Nucleation and propagation of heterochromatin by the histone methyltransferase PRC2: Geometric constraints and impact of the regulatory subunit JARID2. J. Am. Chem. Soc 141, 15029–15039 (2019). doi: 10.1021/jacs.9b02321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon MD et al. , The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007). doi: 10.1016/j.cell.2006.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R, PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol 20, 1258–1264 (2013). doi: 10.1038/nsmb.2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q et al. , RNA exploits an exposed regulatory site to inhibit the enzymatic activity of PRC2. Nat. Struct. Mol. Biol 26, 237–247 (2019). doi: 10.1038/s41594-019-0197-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kia SK, Gorski MM, Giannakopoulos S, Verrijzer CP, SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol 28, 3457–3464 (2008). doi: 10.1128/MCB.02019-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finogenova K et al. , Structural basis for PRC2 decoding of active histone methylation marks H3K36me2/3. eLife 9, e61964 (2020). doi: 10.7554/eLife.61964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernstein BE et al. , A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006). doi: 10.1016/j.cell.2006.02.041 [DOI] [PubMed] [Google Scholar]
- 39.Gradia SD et al. , MacroBac: New Technologies for Robust and Efficient Large-Scale Production of Recombinant Multiprotein Complexes. Methods Enzymol. 592, 1–26 (2017). doi: 10.1016/bs.mie.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dyer PN et al. , Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2003). doi: 10.1016/S0076-6879(03)75002-2 [DOI] [PubMed] [Google Scholar]
- 41.Long L, Furgason M, Yao T, Generation of nonhydrolyzable ubiquitin-histone mimics. Methods 70, 134–138 (2014). doi: 10.1016/j.ymeth.2014.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Gctf: Real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). doi: 10.1016/j.jsb.2015.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng SQ et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheres SH, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). doi: 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han BG et al. , Long shelf-life streptavidin support-films suitable for electron microscopy of biological macromolecules. J. Struct. Biol 195, 238–244 (2016). doi: 10.1016/j.jsb.2016.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bepler T et al. , Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019). doi: 10.1038/s41592-019-0575-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang G et al. , EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). doi: 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K, Coot: Model-building tools for molecular graphics. Acta Cryst D60, 2126–2132 (2004). doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 49.Sato Y et al. , Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 28, 2461–2468 (2009). doi: 10.1038/emboj.2009.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun A et al. , Structural and biochemical insights into human zinc finger protein AEBP2 reveals interactions with RBBP4. Protein Cell 9, 738–742 (2018). doi: 10.1007/s13238-017-0483-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long F, Vagin AA, Young P, Murshudov GN, BALBES: A molecular-replacement pipeline. Acta Cryst. D64, 125–132 (2008). doi: 10.1107/S0907444907050172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worden EJ, Zhang X, Wolberger C, Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome. eLife 9, e53199 (2020). doi: 10.7554/eLife.53199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vijay-Kumar S, Bugg CE, Cook WJ, Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol 194, 531–544 (1987). doi: 10.1016/0022-2836(87)90679-6 [DOI] [PubMed] [Google Scholar]
- 54.Adams PD et al. , PHENIX: Building new software for automated crystallographic structure determination. Acta Cryst. D58, 1948–1954 (2002). doi: 10.1107/S0907444902016657 [DOI] [PubMed] [Google Scholar]
- 55.Leitner A, Walzthoeni T, Aebersold R, Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat. Protoc 9, 120–137 (2014). doi: 10.1038/nprot.2013.168 [DOI] [PubMed] [Google Scholar]
- 56.Cox J, Mann M, MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). doi: 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 57.Tate JG et al. , COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 47, D941–D947 (2019). doi: 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM density maps and fitted models have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) for the PRC2-AEBP2-JARID2 (amino acids 1 to 450) bound to Ncl-ub [EMDB: 21707 and PDB: 6WKR (HPUB)].


