Introduction
There is a near-consensus view that severe acute respiratory syndrome coronavirus (SARS-CoV-2), the causative agent of COVID-19, has a natural zoonotic origin; however, several characteristics of SARS-CoV-2 taken together are not easily explained by a natural zoonotic origin hypothesis. These include a low rate of evolution in the early phase of transmission; the lack of evidence for recombination events; a high pre-existing binding to human angiotensin-converting enzyme 2 (ACE2); a novel furin cleavage site (FCS) insert; a flat ganglioside-binding domain (GBD) of the spike protein which conflicts with host evasion survival patterns exhibited by other coronaviruses; and high human and mouse peptide mimicry. Initial assumptions against a laboratory origin by contrast have remained unsubstantiated. Furthermore, over a year after the initial outbreak in Wuhan, there is still no clear evidence of zoonotic transfer from a bat or intermediate species. Given the immense social and economic impact of this pandemic, identifying the true origin of SARS-CoV-2 is fundamental to preventing future outbreaks. The search for SARS-CoV-2′s origin should include an open and unbiased inquiry into a possible laboratory origin.
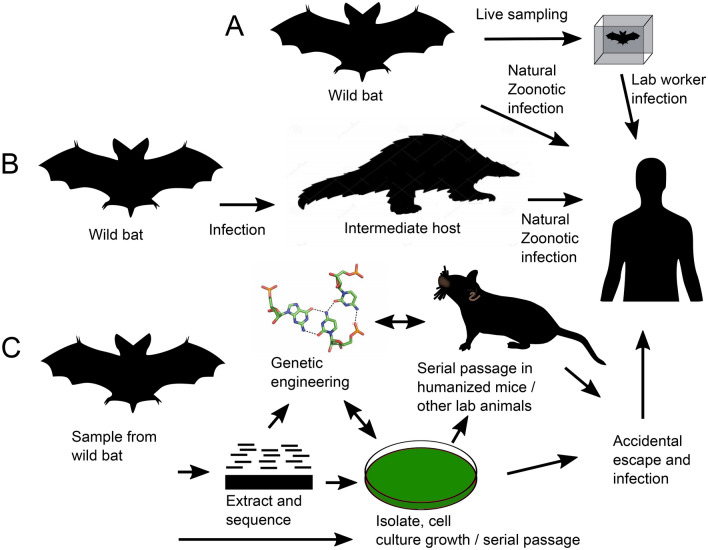
SARS-CoV-2 is a novel Betacoronavirus of lineage B (subgenus Sarbecovirus) and the causative agent of COVID-19, the first detected cases of which were identified in Wuhan in December 2019 (Huang et al. 2020a). The near-consensus view of the origin of SARS-CoV-2 is a natural zoonosis (Zhu et al. 2020; Wu et al. 2020b; Zhou et al. 2020b). Bats are thought to be the natural reservoir for SARS-related coronaviruses (SARS-r CoVs) (Li et al. 2005; Wang et al. 2006) and have been identified as the ancestral source from which severe acute respiratory syndrome coronavirus (SARS-CoV) evolved (Janies et al. 2008; Sheahan et al. 2008). While several intermediate host species have been proposed as the zoonotic source for SARS-CoV-2 (Xiao et al. 2020; Lam et al. 2020; Zhang et al. 2020b; Zhou and Shi 2021), the source of direct bat to human or intermediate animal to human zoonotic transmission of SARS-CoV-2 has not been established. An alternative hypothesis, that SARS-CoV-2 leaked from a laboratory, has been widely dismissed (Rasmussen 2021), yet very few papers counter this theory with data analysis (Andersen et al. 2020; Liu et al. 2020b; Graham and Baric 2020) (Fig. 1).
Fig. 1.
Possible routes of SARS-CoV-2′s transmission to humans. a Direct infection from bats to humans either natural or due to virus sampling. b Infection of humans via an intermediate host such as pangolins or other mammals. c Laboratory hypothesis: sampling from wild bats followed by different laboratory steps such as RNA extraction and sequencing, virus isolation or synthesis from a given sequence, growth in cell culture and infection assays, genetic engineering, passage in humanized mice or other animal models. Human infection may be caused by accidental escape of the virus from the laboratory environment
Here we address the main arguments in support of a natural origin of SARS-CoV-2 and outline the various points which support the alternative, that a laboratory origin is still a valid possibility that should not be discounted. To help prevent future viral pandemics, it is of pivotal importance to identify the source of the virus, and this is only possible with an unbiased analysis of all data available. We couple this work with calls from recent opinion pieces and comparative studies questioning a zoonotic origin (Sousa 2020; Sirotkin and Sirotkin 2020; Relman 2020; Segreto and Deigin 2020; Butler 2020; Sallard et al. 2021) via a review of the latest literature and propose an alternative to the natural zoonosis hypothesis.
Early outbreak and the search of an intermediate host
The earliest detected cases of COVID-19 were located in Wuhan, China. The Huanan seafood market in Wuhan had at first been posited as a possible location of initial zoonotic transfer from wild animals to humans (Huang et al. 2020a, b). However, three of the four patients with the earliest recorded onset of COVID-19 symptoms had no association with the seafood market (Huang et al. 2020a), and the ancestral T8782 and C28144 genotype was not associated with the seafood market (Chen et al. 2021). Phyloepidemiologic analysis of early cases also discounted this theory (Yu et al. 2020).
Although the exact zoonotic agent of the original severe acute respiratory syndrome coronavirus (SARS-CoV) has not been identified, Chiroptera are considered to be the natural reservoir of SARS-r CoVs (Li et al. 2005). Initially, palm civets and raccoon dogs were proposed as zoonotic agents (Chinese SARS Molecular Epidemiology Consortium 2004) or intermediate hosts (Tang et al. 2006), but it remains possible that they were infected by humans (Janies et al. 2008).
For SARS-CoV-2, several authors have proposed pangolins, Manis javanica, as an intermediate host due to the similarity of the receptor binding domain (RBD) for pangolin coronavirus to SARS-CoV-2 RBD (Xiao et al. 2020; Lam et al. 2020; Zhang et al. 2020b). Pangolins are, however, unlikely to be the intermediate host for SARS-CoV-2. Although two pangolin coronaviruses (Xiao et al. 2020; Liu et al. 2020a) exhibited strong binding to human ACE2 (hACE2), binding to pangolin ACE2 was approximately tenfold weaker and binding to bat Rhinolophus ferremequinum ACE2 was very weak, with similar relative binding relationships exhibited by SARS-CoV-2 (Wrobel et al. 2021). This indicates that neither pangolin coronavirus had adapted well to pangolins and that more research is required to validate the viability of coronaviruses to spread naturally between pangolins. Because of a 10–15% divergence throughout the entire spike protein with the exclusion of the N-terminal domain, Boni et al. (2020) concluded that SARS-CoV-2 is unlikely to be a recombinant of an ancestor of pangolin coronavirus and the closest SARS-CoV-2 relative, RaTG13.
All published pangolin coronavirus genome sequences with a nearly identical spike RBD to SARS-CoV-2 were sourced from a single batch of smuggled pangolins (Chan and Zhan 2020), raising the question whether pangolins may have been infected from another host species or from humans during trafficking (Choo et al. 2020; Wenzel 2020). Unlike other species demonstrated to be vectors for coronaviruses, pangolins are not trafficked together live caged in large groups for extended periods of time, making this an unlikely scenario for viral enhancement. Also, pangolins are critically endangered (Choo et al. 2020), exhibit a solitary nature, potentially have limited infection resistance (Choo et al. 2016), and a recent screening of 334 pangolins revealed a lack of coronavirus infections of pangolins in the wild (Lee et al. 2020). Finally, the discovery of synthetic DNA sequences in pangolin coronavirus metagenomic raw sequence reads by Zhang (2020) and the interpretation that the pangolin coronavirus genomes were generated from a synthetic construct, requires further investigation.
After outbreaks reported in several mink farms in Europe (Hammer et al. 2021) and the USA (Zhou and Shi 2021) from a human source with back-transmission to humans, minks, Neovison vison, have been suggested to be a potential intermediate host for SARS-CoV-2 (Zhou and Shi 2021). Studies on ferrets, closely related mustelids to minks, have demonstrated that SARS-CoV-2 binds to ferret ACE2 albeit at a lower efficiency than to human ACE2 (Huang et al. 2020b; Conceicao et al. 2020). Furthermore, SARS-CoV-2 infection and transmission among ferrets has been experimentally determined (Kim et al. 2020b; Schlottau et al. 2020; Richard et al. 2020b) with the animals exhibiting mild effects and limited or no detectable lower respiratory tract involvement (Kim et al. 2020b; Shi et al. 2020). However, several observations indicate mustelids to be an unlikely intermediary host: no outbreaks in mink farms are known in China or surrounding countries; mink farms in China are located in north-eastern regions, while the closest related coronavirus to SARS-CoV-2, RaTG13, was sampled by the Wuhan Institute of Virology (WIV) from bats in Yunnan in south-western China (Zhou et al. 2020c); that ferrets are not affected by the severe disease characteristics exhibited by humans (Johansen et al. 2020) indicates that for ferrets to be an intermediate host, significant adaptation of SARS-CoV-2 to humans would have occurred after zoonotic transfer from mustelids, which has not been observed. Finally, the identification of several SARS-CoV-2′s variants in minks, some of which characterized by mutations previously unseen in humans (Schlottau et al. 2020; Hammer et al. 2021), suggests that minks are not likely to be a natural reservoir for the virus but rather new hosts which require adaptation.
It has also been hypothesized that frozen food might be a possible transmission vector for SARS-CoV-2 (Sun et al. 2021; Han et al. 2020) and even conjectured as a cause for the initial outbreak in Wuhan. Prolonged persistence of the virus at low temperatures, its detection on the outside of packages of frozen food and few clusters of cases in China, which have been proposed to have been caused by food delivery from other countries, have been used to support this hypothesis. However, data on infectivity under these conditions are lacking and an earlier outbreak in the countries where the food originated must have occurred for the food to be contaminated, yet no outbreak of SARS-CoV-2 anywhere in the world prior to the late 2019 outbreak in Wuhan is known.
To summarize, pangolins and mustelids are unlikely to be intermediate species through which SARS-CoV-2 was transferred to humans, while SARS-CoV-2-contaminated imported frozen food is an exceedingly unlikely source of the initial outbreak in Wuhan.
Evolutionary adaptation and recombination
Unlike SARS-CoV in its early and middle phases (Chinese SARS Molecular Epidemiology Consortium 2004; Sheahan et al. 2008; Janies et al. 2008) or the evolution of Middle East respiratory syndrome-related coronavirus (MERS-CoV) (Lau et al. 2017; Forni et al. 2017), SARS-CoV-2 exhibits limited diversity across its genomes (Dearlove et al. 2020; van Dorp et al. 2020; Zhan et al. 2020; Jia et al. 2020). A very recent emergence of SARS-CoV-2 into the human population has been proposed based on the sampling of eight nearly identical complete genomes in December 2019 (Lu et al. 2020). From earliest strains in Wuhan in 2019, SARS-CoV-2 resembled SARS-CoV in the late phase of its 2003 epidemic after SARS-CoV had developed several advantageous adaptations for human transmission (Zhan et al. 2020).
While there is no record of a process of early evolutionary adaptation, SARS-CoV-2′s receptor binding domain (RBD) appears to be highly optimized for binding to human ACE2 (Fig. 2) (Delgrado Blanco et al. 2020; Damas et al. 2020). In this respect, 43% of modelled mutations destabilize the binding energy of the SARS-CoV-2 spike protein RBD to human ACE2, while just 1% of the mutations stabilize it (Delgrado Blanco et al. 2020). Substitution of any of the eight SARS-CoV-2 RBD residues proximal to the human ACE2 binding interface with the residues found in the RaTG13 RBD were shown to be detrimental to human ACE2 binding (Conceicao et al. 2020). Furthermore, Piplani et al. (2020) in a study of 13 animal species including pangolin and bat Rhinolophus sinicus found that the SARS-CoV-2 spike protein had the highest overall binding energy for human ACE2.
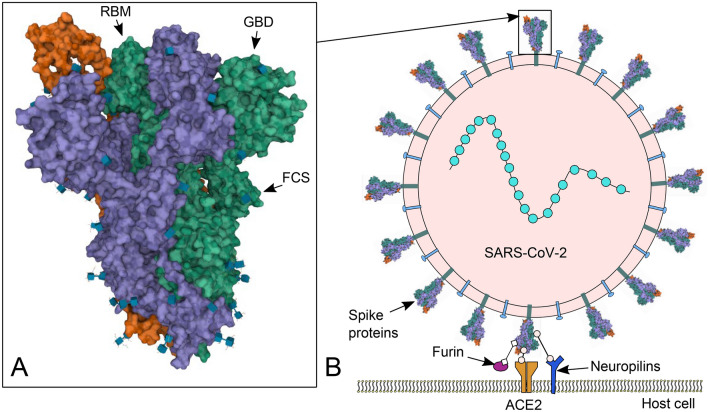
Fig. 2.
Severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) spike and cell binding. a Protein Data Bank (RCSB PDB) 3D structure 6VYB (Walls et al. 2020) image of SARS-CoV-2 spike ectodomain structure (open state) (https://www.rcsb.org/3d-view/6vyb) (Sehnal et al. 2018). Locations of the receptor binding motif (RBM), ganglioside-binding domain (GBD) and furin cleavage site (FCS) indicated by arrows. b Cartoon representation of SARS-CoV-2 binding to a human cell via spike receptor binding domain (RBD) to human angiotensin-converting enzyme 2 (ACE2) and alternate entry route via cell surface neuropilins. Spike priming/activation by furin. Box at the top of the image shows the indicative position of a
Because bats are considered to be natural reservoirs of SARS-r CoVs (Li et al. 2005), SARS-CoV-2 to bat ACE2 binding ability is expected to be high. However, tested bat species are poorly infected by SARS-CoV-2, and they are therefore unlikely to be the direct source for human infection. SARS-CoV-2 does not replicate in R. sinicus kidney or lung cells (Chu et al. 2020), binds poorly to R. sinicus ACE2 (Tang et al. 2020; Li et al. 2020a; Piplani et al. 2020), and exhibits no binding to R. ferrumequinum ACE2 (Tang et al. 2020). In addition, in-silico modelling of the binding affinity for 37 bat species (Damas et al. 2020) showed that eight species exhibited very low binding affinity (including R. ferrumequinum), and the other 29 exhibited low (including R. pearsonii and R. sinicus) binding ability. Although the host of RaTG13, R. affinis, was not modelled by Damas et al. (2020), these results are perplexing as it indicates a significant and unexplained evolutionary distance between SARS-CoV-2 and bats. Curiously, RaTG13 also exhibits poor binding to R. sinicus ACE2 (Li et al. 2020a), R. pusillus ACE2 (Chu et al. 2020), R. ferremequinum (Wrobel et al. 2021) and human ACE2 (Wrobel et al. 2020, 2021). The combination of high human adaptation and poor bat susceptibility from the first sampled strains of SARS-CoV-2 differs greatly from the evolution of MERS-CoV and SARS-CoV.
Recombination processes have been proposed by several authors as a mechanism by which SARS-CoV-2 may have evolved. Interestingly, there is no evidence of recombination events in studies of SARS-CoV-2 by Richard et al. (2020a) (6546 genome sequences as of September 2020) or Bobay et al. (2020) (218 sequences as of August 2020). This is in contrast with MERS-CoV, where despite a much smaller sample size, recombination events were detected. Furthermore, there is also no indication of recombination between the subgenus Sarbecorvirus and other Betacoronavirus subgena or species of the Alpha, Gamma or Deltacoronavirus genera. Indeed, in the subgenera of Betacoronaviruses: Embecovirus, Merbecovirus and Sarbecovirus, gene exchange is restricted to members of the same subgroup (Bobay et al. 2020). The hypothesis that the receptor binding domain (RBD) of the SARS-CoV-2 spike protein arose via a recent recombination with a pangolin-hosted coronavirus RBD (Andersen et al. 2020; Xiao et al. 2020; Li et al. 2020b; Lam et al. 2020; Zhang et al. 2020b) is not likely (Bobay et al. 2020; Paraskevis et al. 2020), and poor taxon sampling by Zhang et al. (2020b), Lam et al. (2020), and Xiao et al. (2020) is discussed by Wenzel (2020). Although earlier recombination and mutations have been proposed (Bobay et al. 2020; Wang et al. 2021; Patiño-Galindo et al. 2020), given that Sarbecoviruses have not been shown to recombine with other coronavirus genera, or other Betacoronavirus subgena, the acquisition of an RBD or a novel furin cleavage site insert by SARS-CoV-2 (Tang et al. 2021) is not likely to have happened through this natural mechanism. The hypothesis of Gallaher (2020) that SARS-CoV-2′s furin cleavage site might have resulted from a recombination event of a RaTG13-like coronavirus and HKU-9, which is a lineage D Betacoronavirus, is also unlikely to be valid, especially in light of RaTG13 being hosted by mircobats (Rhinolophus genus) and HKU-9 by megabats (Rousettus genus).
Overall, SARS-CoV-2 was remarkably well adapted to humans from its first appearance, yet poorly adapted to bat infection, the natural reservoirs for SARS-r-CoVs, with little evidence for gaining its human adaptation through natural recombination.
Furin cleavage site
SARS-CoV-2 is the only Sarbecovirus to contain a furin cleavage site (FCS) (Coutard et al. 2020). Indeed, no coronavirus with a spike protein sequence homology of greater than 40% to SARS-CoV-2 has a FCS (Wu et al. 2020a). The multibasic FCS (Fig. 3) (‘RRAR↓,’ the arrow indicates site of proteolytic cleavage) in SARS-CoV-2 plays a key role in its pathogenesis (Johnson et al. 2020; Hoffman et al. 2020; Shokeen et al. 2020; Qiao and Olvera de la Cruz 2020; Lau et al. 2020; Shang et al. 2020) and enhances its human pathogenicity over a minimal FCS ‘RXXR↓’ (Thomas 2002). It is also unusual, diverging from the canonical ‘RX[K/R]R’ motif (Tang et al. 2021). The presence of an arginine at the third position P3 before the FCS increases the efficiency of the FCS tenfold (Henrich et al. 2003). Its presence is also rare, occurring in only 5 out of 132 known FCSs (Lemmin et al. 2020). The ‘RRAR’ motif conforms to the ‘[R/K]XX[R/K]’ ‘C-end rule,’ creating a binding site for cell surface neuropilin (NRP1 and NRP2) receptors (Teesalu et al. 2009), which are more widely expressed than ACE2. NRP1 has been demonstrated as an alternate route for virus entry (Cantuti-Castelvetri et al. 2020; Daly et al. 2020).
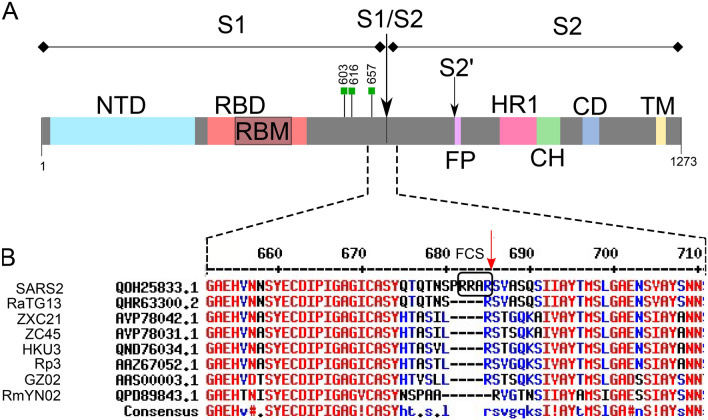
Fig. 3.
Severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) spike protein showing subunits and domains as well as local sequence alignments with other Betacoronaviruses. a Spike domains: N-terminal domain (NTD), receptor binding domain (RBD), receptor binding motif (RBM), fusion peptide (FP), hetapad repeat 1 (HR1), central helix (CH), connector domain (CD), transmembrane domain (TM), multibasic furin cleavage site (FCS) indicated by black box. The S1↓S2 and S2′ cleavage sites are indicated by arrows. Green boxes indicate location of N-glycosylated residues proximal to FCS. b MultAlin alignments using SARS-CoV-2 spike protein sequence numbering reference (Corpet 1988) (http://www.sacs.ucsf.edu/cgi-bin/multalin.py). Definition and accession numbers as follows: SARS2: SARS-CoV-2 Wuhan-Hu-1 (QOH25833.1); RaTG13 (QHR63300.2); ZXC21: bat-SL-CoVZXC21 (AVP78042.1); ZC45: bat-SL-CoVZC45 (AVP78031.1); HKU3: Bat SARS coronavirus HKU3 (QND76034.1); Rp3: Rp3/2004 (AAZ67052.1); GZ02: SARS coronavirus GZ02 (AAS00003.1); RmYN02: Bat coronavirus RmYN02 (QPD89843.1). SARS-CoV-2 referenced sequence indexes are shown
Because of the insertion of the furin cleavage site (FCS), not only furin, but also several other proteolytic enzymes are able to activate SARS-CoV-2′s spike protein (Jaimes et al. 2020). The proline residue at position P5 (5th residue prior to the FCS) is rare and only appears in 5 out of 132 known FCSs (Lemmin et al. 2020). Proline has a restricted phi angular range in peptide bond formation (Morgan and Rubenstein, 2013) which imposes conformal restraints on the peptide chain and results in the separation of the cleavage site from other structural elements, facilitating exposure to host proteases (Lemmin et al. 2020). In comparison with 132 known FCSs in FurinDB (Tian et al. 2011) (http://www.nuolan.net/substrates.html), SARS-CoV-2′s FCS exhibits several intriguing features. The P11-P1 ‘QTQTNSPRRAR’ motif is homologous to neurotoxins from Ophiophagus and Bungarus genera and neurotoxin-like regions from Rabies lyssavirus strains and may act as a superantigenic fragment (Cheng et al. 2020). The ‘XXRR[A/S/C/G/T/V/I/L]R↓’ motif is shared by only two other FCSs; the ‘XPXRXX↓’ motif also only occurs in one other FCS in FurinDB. The ‘XXRRAR↓XX’ was found to be only shared by the bacterial toxin proaerolysin (in FurinDB) (Abrami et al. 1998) and Alphacoronavirus AcCoV-JC34 (Ge et al. 2017).
Another unique feature of SARS-CoV-2 when compared to related coronaviruses is a longer loop containing the S1/S2 cleavage site (Lemmin et al. 2020): it is at least four amino acids longer around the site containing the FCS than any other known Sarbecovirus. The combination of FCS and extended loop length facilitate SARS-CoV-2 activation by transmembrane serine protease TMPRSS13, as well as TMPRSS2, albeit at one-third the effectiveness of TMPRSS2 (Laporte et al. 2020). Mutants with a deleted ‘PRRA’ insert or a shortened FCS loop with deleted preceding amino acids ‘QTQTN’ abrogated the effectiveness of both TMPRSS2 and TMPRSS13 for facilitating cleavage (Laporte et al. 2020).
Moreover, the FCS in SARS-CoV-2 is coded by rare codons, leaving it out of frame with the rest of the sequence, thus violating the rules of the copy choice recombination mechanisms that postulate in-frame insertions. Additionally, its insertion causes a peculiar split of one of the codons, serine (TCA) when compared with the close relatives MP789 and RaTG13 (Segreto and Deigin 2020). The recent acquisition of the FCS by SARS-CoV-2 via a natural insert was proposed by Wu and Zhao (2021) on the basis of the existence of FCS in other, more distant Betacoronaviruses with different loop positions to SARS-CoV-2 and the existence of a partial natural insert in the same region in RmYN02 (Zhou et al. 2020a). The reliability of the conclusions of Zhou et al. (2020a) has been questioned by Deigin and Segreto (2020), who particularly challenge the claim that RmYN02 has an insertion around the site of the FCS insertion in SARS-CoV-2 and instead point to a two amino acid deletion in RmYN02 at that locus. Therefore, RmYN02 should not be used as evidence of the natural origin of SARS-CoV-2′s FCS until its claimed insertion is properly validated.
In several viruses, low affinity attachment to heparan sulphate (HS) improves the chances of binding to a more specific entry receptor by increasing viral concentration at the cell surface (Schneider-Schaulies 2000; Zhu et al. 2011, Cagno et al. 2019). The binding to HS or allied polysaccharide heparin by SARS-CoV-2 has been demonstrated by several studies (Mycroft-West et al. 2020; Kim et al. 2020a; Zhang et al. 2020a; Clausen et al. 2020; Tiwari et al. 2020; Kwon et al. 2020), and heparin-binding affinity in SARS-CoV-2 is much higher than in SARS-CoV or MERS-CoV (Kim et al. 2020a). We note that the SARS-CoV-2 FCS ‘PRRAR↓S’ motif in its uncleaved state is consistent with the heparan sulphate binding region motif ‘XBBXBX’ (where B is a basic and X is a hydropathic residue), one of two consensus motifs determined by Cardin and Weintraub (1989) by comparing several potential heparin-binding sites in selected proteins. This particular site in the FCS was demonstrated to have the highest heparan sulphate binding affinity among the three glycosaminoglycan binding motifs identified in the spike protein of SARS-CoV-2 by Kim et al. (2020a). Cell culture adaptation to ‘Cardin-Weintraub’ motifs has been demonstrated in multiple cell passage studies (de Haan et al. 2005, 2008; Millet et al. 2020), and it should be considered as a possible reason for the strong heparan sulphate binding affinity identified in SARS-CoV-2.
Indeed, while there is no significant O-glycosylation on the spike protein in human cells (Wang et al. 2020a), the use of insect cell culture and baculovirus display system, where glycoprotein sialylation is not a major biochemical process (Marchal et al. 2001), could allow O-glycosyltransferases access to the furin cleavage site (FCS). While in human cells we interpret this region to be shielded by N-linked complex large glycans N616 and N657 and mixed Oligomannose/complex type glycans at site N603 (Casalino et al. 2020, Sun et al. 2020, Watanabe et al. 2020), in insect cells we predict that O-glycosylation on S685 would prevent cleavage by furin and preferentially bind heparan sulphate (HS). Repeated passage through an insect cell culture from an inserted more potent artificial FCS motif could then lead to the generation of a cell culture adaptive O-glycosylated ‘RRAR’ signature.
A minimal FCS could potentially have evolved via a single point mutation T678R (Li et al. 2015), which is evolutionarily more parsimonious than a complete 12nt insertion of ‘PRRA’. A multi basic cleavage site is also plausible with an additional mutation N679R. We note that deletions but not insertions frequently happen at the S1/S2 junction of SARS-CoV-2 during serial cell passage (Peacock et al. 2020) and have also been detected in strains isolated from hamsters and humans (Lau et al. 2020; Liu et al. 2020c). The acquisition of the FCS via a natural insert, when a FCS could have evolved far more easily though point mutation, we believe is highly unlikely.
Because the presence and coding sequence of a FCS is important for pathogenesis, host range, and cell tropism (Nagai et al. 1993; Millet et al. 2015), the addition of a FCS into viruses has been an active area of gain-of-function research. A FCS can be easily inserted using seamless technology (Yount et al. 2002; Sirotkin and Sirotkin 2020) without any need for cell passage, as previously performed in experiments on virulence and host tropism (Cheng et al. 2019). Insertions to change the properties of SARS-r CoV viruses are documented by Ren et al. (2008) and Wang et al. (2008). Considering that natural mutations have a very low probability to result in a stretch of 12 amino acids coding for an optimized FCS without any known intermediate form in Sarbecovirus, an artificial insertion of the FCS in SARS-CoV-2 may provide a more parsimonious explanation for its presence than natural evolution.
In summary, the FCS confers SARS-CoV-2 enhanced human pathogenicity and has never been identified in another Sarbecovirus. At the same time, FCSs have been routinely inserted into coronaviruses in gain-of-function experiments, and we provide a hypothesis through which the specific amino acid sequence of SARS-CoV-2′s FCS may have been generated through cell culture.
Binding domains, peptide mimicry
A ‘ganglioside-binding domain’ (GBD) in the N-terminal domain (NTD) of SARS-CoV-2 (Fig. 2a) (Pirone et al. 2020) is characterized by a large flat interface enriched in aromatic and basic amino acid residues (Fantini et al. 2020a) and contains one of three inserts in the NTD of SARS-CoV-2 identified by Zhou P. et al. (2020b). The GBD proffers SARS-CoV-2 with an additional receptor/attachment ability to sialic acid-containing glycoproteins, in addition to the primary ACE2 receptor, as well as heparan-sulphate (Clausen et al. 2020), neuropilins (Cantuti-Castelvetri et al. 2020; Daly et al. 2020) and L-SIGN/DC-SIGN (Chiodo et al. 2020; Gao et al. 2020; Soh et al. 2020; Thépaut et al. 2020), constituting enhanced receptor pathways compared with SARS-CoV. The importance of the GBD in SARS-CoV-2 infectivity was indicated by Chi et al. (2020) and McCallum et al. (2021) who identify potent binding antibodies which provide strong neutralizing activity against SARS-CoV-2 by binding to residues in this domain. Fantini et al. (2020b) discuss the flat structural topography of the GBD which proffers improved functional interaction and because of this attribute and sequence peculiarities in the spike protein, raise questions concerning the proximal origin of SARS-CoV-2. The flat topography of the GBD was also observed by Seyran et al. 2021 as anomalous compared with other human coronaviruses, which typically exhibit hidden sugar-binding site localization as an evolutionary measure to evade host immune surveillance, termed the ‘Canyon Hypothesis’ (Rossman 1989; Chen and Li 2013; Li 2015).
Another curious feature of SARS-CoV-2 is its binding efficiency to human ACE2, being much more effective than SARS-CoV. Khatri et al. (2020) measured a large interaction surface with high binding affinity between SARS-CoV-2 and ACE2 as > 15-fold stronger than between SARS-CoV and ACE2. This is supported by Wrapp et al. (2020) who find ~ tenfold to 20-fold higher binding efficiency. The increased SARS-CoV-2 to ACE2 binding efficiency has been proposed to be due to a larger hydrophobic interaction surface for SARS-CoV-2 over SARS-CoV (Gussow et al. 2020; Wan et al. 2020; Lai et al. 2020; Khatri et al. 2020; Brielle et al. 2020) with an increased number of interacting residues (Brielle et al. 2020; Wang et al. 2020b) and extra charge interaction (Sørensen et al. 2020; Gussow et al. 2020; Wang et al. 2020c). Closer interaction distances between the N-terminal end of ACE2 and the central region of the receptor binding motif (RBM) for SARS-CoV-2 over SARS-CoV (Wang et al. 2020c) also facilitates coupling. These modifications indicate a more highly adapted ability for SARS-CoV-2 to bind to ACE2 than seen for SARS-CoV. While SARS-CoV to human ACE2 affinity relied on five key residues all of which exhibited natural mutation in the early stages of adaptation to a new host (Wan et al. 2020), SARS-CoV-2 displays from even the very first isolates, a more optimized configuration without any evidence of early natural mutations (Zhan et al. 2020).
Other indications of significant human adaptation are seen in peptide mimicry by SARS-CoV-2. Eight and 9-mer peptide mimicry between SARS-CoV-2 and the human reference genome was analysed by Venkatakrishnan et al. (2020) who found unique mimicry of four major histocompatibility complex binding peptides not shared by SARS-CoV, MERS or other human coronaviruses. Mimicry of these peptides which are expressed in the lung, oesophagus, arteries, heart, pancreas, and macrophages is potentially associated with autoinflammation in some COVID-19-infected patients (Venkatakrishnan et al. 2020). Sørensen et al. (2020) confirm that the SARS-CoV-2 spike protein is remarkably well adapted to humans, with a 78.4% similarity to 6-mer human epitopes. This finding is consistent with work by Kanduc and Shoenfeld (2020) who observe an anomalously high 6 and 7-mer peptide sharing between SARS-CoV-2's spike glycoprotein and human and Mus musculus proteins. Interestingly, mouse ACE2 does not effectively bind to the SARS-CoV-2′s spike protein (Li et al. 2020a; Tang et al. 2020; Praharaj et al. 2020; Damas et al. 2020). Extensive passage in mice with humanized lungs and immune systems (Cockrell et al. 2018; Wahl et al. 2019) would explain such an improbable peptide sharing. Indeed, Friend and Stebbing (2021) propose knockout mice with human ACE2 receptors may be the intermediate animal host for SARS-CoV-2.
In summary, the flat ganglioside-binding region of SARS-CoV-2 does not fit the ‘Canyon Hypothesis’ whereby a virus structurally hides residues involved in host receptor recognition from the host's immune system, while peptide mimicry of proteins in major functional human organs as well as mouse proteins by SARS-CoV-2 could be explained by passage in transgenic humanized mice.
O-linked glycans
Theoretical predictions for O-linked glycans in the SARS-CoV-2 spike protein have been used as evidence of a ‘mucin-like domain’ that might be involved in immunoevasion in an animal host by shielding epitopes or key residues on the SARS-CoV-2 spike protein (Andersen et al. 2020) and hence supporting the argument for natural evolution of SARS-CoV-2.
O-glycosylation and/or N-linked sulphated glycans on full length SARS-CoV-2 spike protein constructs (Zhao et al. 2020; Watanabe et al. 2020; Klein and Zaia 2020; Sanda et al. 2021) and subunits (Shajahan et al. 2020) have been reported by several groups, albeit at relatively low levels of site occupation. Wang et al. (2020a), however, in a comprehensive, high-fidelity mass spectrometric approach based on glycan reporter signature ions-triggered electron-transfer/ higher-energy collisional dissociation (EThcD) mass spectrometry, did not observe any detectable occupied O-glycosylation sites. The use of EThcD allowed the sites of glycosylation to be unambiguously determined with a greater proportion of fragment ions observed (Riley et al. 2020). This method provides an increased degree of confidence in the results over conventional collision-induced dissociation, higher-energy collisional dissociation (HCD) (Watanabe et al. 2020; Zhang et al. 2020c; Shajahan et al. 2020; Klein and Zaia 2020), stepped collision energy HCD (Zhao et al. 2020), or HCD fragmentation and modulated normalized collision energy (Sanda et al. 2021) methods.
Furthermore, glycan sequons can actually arise in vitro in the presence of antibodies, as was recently observed during serial passaging of SARS-CoV-2 (Andreano et al. 2020) and may also arise in the laboratory during in vivo passaging of viruses in, for example, humanized mice.
Critically, contrary to the Andersen et al. (2020) supposition, there is no O-linked glycosylation on the neighbouring residues of the S1/S2 junction or at a significant level anywhere along the spike protein. No interaction of SARS-CoV-2 with a host immune system based on O-linked glycans can be claimed, and hence, this mechanism does not support the argument for natural evolution of SARS-CoV-2.
Reverse-genetic systems and virus backbone
The observation that SARS-CoV-2 was not derived from a previously used virus backbone was used as an argument by Andersen et al. (2020) and Liu et al. (2020b) as evidence against a laboratory origin hypothesis. In contrast, the Betacoronavirus RaTG13 was fully sequenced in 2018 (Zhou et al. 2020c) but only published after the identification of SARS-CoV-2 (Zhou et al. 2020b) and more unpublished sequences existed in a WIV database that was deleted after the beginning of the pandemic (Segreto and Deigin 2020). SARS-CoV-2 could have been engineered using one of the over 1500 strains openly collected by institutions associated with WIV (Sirotkin and Sirotkin 2020), a completely undocumented backbone, or one of several fairly well-correlated bat coronaviruses could have been used in combination with directed evolution, a widely used technique for introducing mutations and selection to achieve proteins with desired properties (Badran and Liu 2015; Standage-Beier and Wang 2017; Simon et al. 2019). Specifically, this technique has been used for engineering novel virus variants (Excoffon et al. 2009; Lin et al. 2012; Meister et al. 2019). Furthermore, novel yet undocumented reverse-genetic systems could also have potentially been used. Indeed, multiple groups have developed SARS-CoV-2 reverse genetics systems for SARS-CoV-2 research in short periods of time (Hou et al. 2020; Torii et al. 2020; Thi Nhu Thao et al. 2020). Additionally, seamless ‘No See’em’ technology pioneered nearly 20 years ago allows reverse engineering to be used without leaving any traces (Yount et al. 2002).
We disagree with the hypothesis by Andersen et al. (2020) that the high-affinity binding solution of SARS-CoV-2′s RBD to human ACE2, which differs from the optimal binding solution modelled for SARS-CoV (Wan et al. 2020), provides strong evidence that SARS-CoV-2 could not have been engineered in a laboratory. Computational prediction is not necessary for generating novel human pathogenic viruses. Culturing and adapting coronaviruses and influenza A virus to different cell lines, including human airway epithelial cells, has been conducted in various laboratories (Tse et al. 2014; Menachery et al. 2015; Zeng et al. 2016; Jiang et al. 2020); furthermore, experimental creation of chimeric viruses by directed engineering as discussed above does not require prior modelling.
Overall, the observations that SARS-CoV-2 is not derived by previously published backbones or using known reverse-genetic systems cannot be used as a strong argument against its possible laboratory origin. The same applies to its high-affinity binding to human ACE2 which differs from the one modelled for SARS-CoV.
Conclusion
More than a year after the initial documented cases in Wuhan, the source of SARS-CoV-2 has yet to be identified, and the search for a direct or intermediate host in nature has been so far unsuccessful. The low binding affinity of SARS-CoV-2 to bat ACE2 studied to date does not support Chiroptera as a direct zoonotic agent. Furthermore, the reliance on pangolin coronavirus receptor binding domain (RBD) similarity to SARS-CoV-2 as evidence for natural zoonotic spillover is flawed, as pangolins are unlikely to play a role in SARS-CoV-2′s origin and recombination is not supported by recent analysis. At the same time, genomic analyses pointed out that SARS-CoV-2 exhibits multiple peculiar characteristics not found in other Sarbecoviruses. A novel multibasic furin cleavage site (FCS) confers numerous pathogenetically advantageous capabilities, the existence of which is difficult to explain though natural evolution; SARS-CoV-2 to human ACE2 binding is far stronger than SARS-CoV, yet there is no indication of amount of evolutionary adaptation that SARS-CoV or MERS-CoV underwent. The flat topography of the ganglioside-binding domain (GBD) in the N-terminal domain (NTD) of SARS-CoV-2 does not conform with typical host evasion evolutionary measures exhibited by other human coronaviruses. The combination of binding strength, human and mouse peptide mimicry, as well as high adaptation for human infection and transmission from the earliest strains might suggest the use of humanized mice for the development of SARS-CoV-2 in a laboratory environment. The application of mouse strains expressing human ACE2 for SARS-CoV-related research is well documented (Ren et al. 2008; Hou et al. 2010; Menachery et al. 2015; Cockrell et al. 2018; Jiang et al. 2020). Additionally, culturing and adapting coronaviruses to different cell lines, including human airway epithelial cells, has been experimentally conducted in various laboratories (Tse et al. 2014; Menachery et al. 2015; Zeng et al. 2016; Jiang et al. 2020). While a natural origin is still possible and the search for a potential host in nature should continue, the amount of peculiar genetic features identified in SARS-CoV-2′s genome does not rule out a possible gain-of-function origin, which should be therefore discussed in an open scientific debate.
Acknowledgements
We are grateful to the Decentralised Radical Autonomous Search Team Investigating COVID‐19 (DRASTIC) Twitter group for their investigative work in uncovering a significant number of previously unpublished facts about SARS‐CoV‐2 and its relative strains.
Abbreviations
- ACE2
Angiotensin-converting enzyme 2
- FCS
Furin cleavage site
- GBD
Ganglioside-binding domain
- HS
Heparan sulphate
- MERS-CoV
Middle East respiratory syndrome-related coronavirus
- NTD
N-terminal domain
- RBD
Receptor binding domain
- RBM
Receptor binding motif
- SARS-CoV
Severe acute respiratory syndrome coronavirus
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus type 2
- SARS-r CoV
SARS-related coronavirus
- TMPRSS
Transmembrane serine protease
- WIV
Wuhan Institute of Virology
Declarations
Conflict of interest
The authors declare no competing financial interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abrami L, Fivaz M, Decroly E, Seidah NG, Jean F, Thomas G, Leppla SH, Buckley JT, van der Goot FG. The pore-forming toxin proaerolysin is activated by furin. J Biol Chem. 1998;273(49):32656–32661. doi: 10.1074/jbc.273.49.32656. [DOI] [PubMed] [Google Scholar]
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, Dal Monego S, Pantano E, Manganaro N, Manenti A, Manna R (2020) SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. BioRxiv. 10.1101/2020.12.28.424451 [DOI] [PMC free article] [PubMed]
- Badran AH, Liu DR. In vivo continuous directed evolution. Curr Opin Chem Biol. 2015;24:1–10. doi: 10.1016/j.cbpa.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay L, O’Donnell AC, Ochman H. Recombination events are concentrated in the spike protein region of Betacoronaviruses. PLOS Genet. 2020;16(12):e1009272. doi: 10.1371/journal.pgen.1009272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni MF, Lemey P, Jiang X, Lam T-YT, Perry BW, Castoe TA, Rambaut A, Robertson DL (2020) Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 5(11):1408–1417. 10.1038/s41564-020-0771-4 [DOI] [PubMed]
- Brielle ES, Schneidman-Duhovny D, Linial M. The SARS-CoV-2 Exerts a Distinctive Strategy for Interacting with the ACE2 Human Receptor. Viruses. 2020;12(5):497. doi: 10.3390/v12050497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler CD (2020) Plagues, pandemics, health security, and the war on nature. J Hum Secur 16(1). 10.12924/johs2020.16010053
- Cagno T, Jones T. Heparan sulfate proteoglycans and viral attachment: true receptors or adaptation bias? Viruses. 2019;11(7):596. doi: 10.3390/v11070596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L, Ojha R, Pedro LD, Djannatian M, Franz J. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin AD, Weintraub HJ (1989) Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler Off J Am Hear Assoc Inc 9(1): 21–32. 10.1161/01.ATV.9.1.21 [DOI] [PubMed]
- Casalino L, Gaieb Z, Goldsmith JA, Goldsmith JA, Hjorth CK, Dommer AC, Harbison AM, Fogarty CA, Barros EP, Taylor BC, McLellan JS, Fadda E. Beyond Shielding: The Roles of Glycans in the SARS-CoV-2 Spike Protein. ACS Cent Sci. 2020;6(10):1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YA, Zhan SH (2020) Single source of pangolin CoVs with a near identical Spike RBD to SARS-CoV-2. BioRxiv. 10.1101/2020.07.07.184374.
- Chen L, Li F. Structural analysis of the evolutionary origins of influenza virus hemagglutinin and other viral lectins. J Virol. 2013;87(7):4118–4120. doi: 10.1128/JVI.03476-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li S, Wu W, Geng S, Mao M (2021) Distinct mutations and lineages of SARS-CoV-2 virus in the early phase of COVID-19 pandemic and subsequent global expansion. BioRxiv. 10.1101/2021.01.05.425339 [DOI] [PMC free article] [PubMed]
- Cheng J, Zhao Y, Xu G, Zhang K, Jia W, Sun Y, Zhoa J, Xue J, Hu Y, Zhang G. The S2 Subunit of QX-type infectious bronchitis coronavirus spike protein is an essential determinant of neurotropism. Viruses. 2019;11(10):972. doi: 10.3390/v11100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng MH, Zhang S, Porritt RA, Noval Rivas M, Paschold L, Willscher E, Binder M, Arditi M, Bahar I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc Natl Acad Sci. 2020;117(41):25254–25262. doi: 10.1073/pnas.2010722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium (2004) Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 303(5664):1666–1669. 10.1126/science.1092002 [DOI] [PubMed]
- Chiodo F, Bruijns S, Rodriguez E, Li RJE, Molinaro A, Silipo A, Di Lorenzo F, Garcia-Rivera D, Valdes-Balbin Y, Verez-Bencomo V, van Kooyk Y (2020) Novel ACE2-independent carbohydrate-binding of SARS-CoV-2 spike protein to host lectins and lung microbiota. BioRxiv. 10.1101/2020.05.13.092478
- Choo SW, Rayko M, Tan TK, Hari R, Komissarov A, Wee WY, Yurchenko AA, Kliver S, Tamazian G, Antunes A, Wilson RK (2016) Pangolin genomes and the evolution of mammalian scales and immunity. Genome Res 26(10):1312–1322. 10.1101/gr.203521.115 [DOI] [PMC free article] [PubMed]
- Choo SW, Zhou J, Tian X, Zhang S, Qiang S, O'Brien SJ, Tan KY, Platto S, Koepfli KP, Antunes A, Sitam FT (2020) Are pangolins scapegoats of the COVID-19 outbreak-CoV transmission and pathology evidence? Conserv Lett 13(6):1–12. 10.1111/conl.12754
- Chu H, Chan JF, Yuen TT, Shuai H, Yuan S, Wang Y, Hu B, Yip CC, Tsang JO, Huang X, Chai Y (2020) Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe 1(1):14–23. 10.1016/S2666-5247(20)30004-5 [DOI] [PMC free article] [PubMed]
- Clausen TM, Sandoval DR, Spliid CB, Pihl J, Perrett HR, Painter CD, Narayanan A, Majowicz SA, Kwong EM, McVicar RN, Thacker BE (2020) SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183(4):1043–1057.e15. 10.1016/j.cell.2020.09.033 [DOI] [PMC free article] [PubMed]
- Cockrell AS, Leist SR, Douglas MG, Baric RS. Modeling pathogenesis of emergent and pre-emergent human coronaviruses in mice. Mamm Genome. 2018;29(7–8):367–383. doi: 10.1007/s00335-018-9760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceicao C, Thakur N, Human S, Kelly JT, Logan L, Bialy D, Bhat S, Stevenson-Leggett P, Zagrajek AK, Hollinghurst P, Varga M (2020) The SARS-CoV-2 Spike protein has a broad tropism for mammalian ACE2 proteins. PLOS Biol 18(12):e3001016. 10.1371/journal.pbio.3001016 [DOI] [PMC free article] [PubMed]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176(January):104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly JL, Simonetti B, Klein K, Chen KE, Williamson MK, Antón-Plágaro C, Shoemark DK, Simón-Gracia L, Bauer M, Hollandi R, Greber UF (2020) Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 370(6518):861–865. 10.1126/science.abd3072 [DOI] [PMC free article] [PubMed]
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli KP, Pfenning AR, Zhao H, Genereux DP (2020) Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci 117(36):22311–22322. 10.1073/pnas.2010146117 [DOI] [PMC free article] [PubMed]
- Dearlove B, Lewitus E, Bai H, Li Y, Reeves DB, Joyce MG, Scott PT, Amare MF, Vasan S, Michael NL, Modjarrad K (2020) A SARS-CoV-2 vaccine candidate would likely match all currently circulating variants. Proc Natl Acad Sci 117(38):23652–23662. 10.1073/pnas.2008281117 [DOI] [PMC free article] [PubMed]
- Deigin Y, Segreto R (2020) The bat coronavirus RmYN02 is characterized by a 6-nucleotide deletion at the S1/S2 junction, and its claimed PAA insertion is highly doubtful. arXiv. http://arxiv.org/abs/2012.00627
- Delgado Blanco J, Hernandez-Alias X, Cianferoni D, Serrano L. In silico mutagenesis of human ACE2 with S protein and translational efficiency explain SARS-CoV-2 infectivity in different species. PLOS Comput Biol. 2020;16(12):e1008450. doi: 10.1371/journal.pcbi.1008450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CAM, Li Z, te Lintelo E, Bosch BJ, Haijema BJ, Rottier PJM. Murine coronavirus with an extended host range uses heparan sulfate as an entry receptor. J Virol. 2005;79(22):14451–14456. doi: 10.1128/JVI.79.22.14451-14456.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan CAM, Haijema BJ, Schellen P, et al. Cleavage of Group 1 Coronavirus Spike Proteins: How Furin Cleavage Is Traded Off Against Heparan Sulfate Binding Upon Cell Culture Adaptation. J Virol. 2008;82(12):6078–6083. doi: 10.1128/JVI.00074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJDA, Koerber JT, Dickey DD, et al. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc Natl Acad Sci. 2009;106(10):3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J, Di Scala C, Chahinian H, Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection (2020a) Int J Antimicrob Agents. 2020;55(5):105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J, Chahinian H, Yahi N (2020b) Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem Biophys Res Commun. 10.1016/j.bbrc.2020.10.015 [DOI] [PMC free article] [PubMed]
- Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25(1):35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend T, Stebbing J (2021) What is the intermediate host species of SARS-CoV-2? Future Virol. 10.2217/fvl-2020-0390
- Gallaher WR. A palindromic RNA sequence as a common breakpoint contributor to copy-choice recombination in SARS-COV-2. Arch Virol. 2020;165(10):2341–2348. doi: 10.1007/s00705-020-04750-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Zeng J, Jia N, Stavenhagen K, Matsumoto Y, Zhang H, Li J, Hume AJ, Mühlberger E, van Die I, Kwan (2020) SARS-CoV-2 spike protein interacts with multiple innate immune receptors. BioRxiv. 10.1101/2020.07.29.227462
- Ge X-Y, Yang W-H, Zhou J-H, et al. Detection of alpha- and betacoronaviruses in rodents from Yunnan, China. Virol J. 2017;14(1):98. doi: 10.1186/s12985-017-0766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RL, Baric RS. SARS-CoV-2: combating coronavirus emergence. Immunity. 2020;52(5):734–736. doi: 10.1016/j.immuni.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussow AB, Auslander N, Faure G, Wolf YI, Zhang F, Koonin EV. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci. 2020;117(26):15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer AS, Quaade ML, Rasmussen TB, Fonager J, Rasmussen M, Mundbjerg K, Lohse L, Strandbygaard B, Jørgensen CS, Alfaro-Núñez A, Rosenstierne MW (2021) SARS-CoV-2 transmission between Mink (Neovison vison) and Humans, Denmark. Emerg Infect Dis 27(2):547–551. 10.3201/eid2702.203794 [DOI] [PMC free article] [PubMed]
- Han J, Zhang X, He S, Jia P. Can the coronavirus disease be transmitted from food? A review of evidence, risks, policies and knowledge gaps. Environ Chem Lett. 2020;19(1):5–16. doi: 10.1007/s10311-020-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich S, Cameron A, Bourenkov GP, et al. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Mol Biol. 2003;10(7):520–526. doi: 10.1038/nsb941. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Kleine-Weber H, Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78(4):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Peng C, Yu M, et al. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch Virol. 2010;155(10):1563–1569. doi: 10.1007/s00705-010-0729-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon III KH, Kato T, Lee RE, Yount BL, Mascenik TM, Chen G (2020) SARS-CoV-2 Reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 182(2):429–446.e14. 10.1016/j.cell.2020.05.042 [DOI] [PMC free article] [PubMed]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zhang C, Pearce R, Omenn GS, Zhang Y. Identifying the Zoonotic Origin of SARS-CoV-2 by Modeling the Binding Affinity between the Spike Receptor-Binding Domain and Host ACE2. J Proteome Res. 2020;19(12):4844–4856. doi: 10.1021/acs.jproteome.0c00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaimes JA, Millet JK, Whittaker GR. Proteolytic Cleavage of the SARS-CoV-2 Spike Protein and the Role of the Novel S1/S2 Site. Science. 2020;23(6):101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janies D, Habib F, Alexandrov B, Hill A, Pol D (2008) Evolution of genomes, host shifts and the geographic spread of SARS-CoV and related coronaviruses. Cladistics 24:111–130. 10.1111/j.1096-0031.2008.00199.x [DOI] [PMC free article] [PubMed]
- Jia Y, Shen G, Nguyen S, Zhang Y, Huang K-S, Ho H-Y, Hor W-S, Yang CH, Li C, Wang WL (2020) Analysis of the mutation dynamics of SARS-CoV-2 reveals the spread history and emergence of RBD mutant with lower ACE2 binding affinity. BioRxiv. 10.1101/2020.04.09.034942
- Jiang R-D, Liu M-Q, Chen Y, Shan C, Zhou Y-W, Shen X-R, Li Q, Zhang L, Zhu Y, Si HR, Wang Q (2020) Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182(1):50–58.e8. 10.1016/j.cell.2020.05.027 [DOI] [PMC free article] [PubMed]
- Johansen MD, Irving A, Montagutelli X, et al. Animal and translational models of SARS-CoV-2 infection and COVID-19. Mucosal Immunol. 2020;13(6):877–891. doi: 10.1038/s41385-020-00340-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Xie X, Kalveram B, Lokugamage KG, Muruato A, Zou J, Zhang X, Juelich T, Smith JK, Zhang L, Schindewolf C (2020) Furin cleavage site is key to SARS-CoV-2 pathogenesis. BioRxiv. 10.1101/2020.08.26.268854
- Kanduc D, Shoenfeld Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: implications for the vaccine. Immunol Res. 2020;68(5):310–313. doi: 10.1007/s12026-020-09152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri I, Staal FJT, van Dongen JJM. Blocking of the high-affinity interaction-synapse between SARS-CoV-2 spike and human ACE2 proteins likely requires multiple high-affinity antibodies: an immune perspective. Front Immunol. 2020;11(September):1–9. doi: 10.3389/fimmu.2020.570018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Jin W, Sood A, et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antiviral Res. 2020;181(January):104873. doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-I, Kim S-G, Kim S-M, Kim E-H, Park S-J, Yu K-M, Chang J-H, Kim EJ, Lee S, Casel MA, Um J (2020b) Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 27(5):704–709.e2. 10.1016/j.chom.2020.03.023 [DOI] [PMC free article] [PubMed]
- Klein JA, Zaia J (2020) Assignment of coronavirus spike protein site-specific glycosylation using GlycReSoft. BioRxiv. 10.1101/2020.05.31.125302
- Kwon PS, Oh H, Kwon S-J, Linhardt RJ, Dordick JS. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6(1):50. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai HTT, Nguyen LH, Kranjc A, Nguyen TT, Nguyen-Manh D (2020) Elucidating the differences in the molecular mechanism of receptor binding between 2019-nCoV and the SARS-CoV viruses using computational tools. BioRxiv. 10.1101/2020.04.21.053009
- Lam TTY, Jia N, Zhang YW, Shum MHH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ (2020) Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285. 10.1038/s41586-020-2169-0 [DOI] [PubMed]
- Laporte M, Stevaert A, Raeymaekers V, Van Berwaer, R, Martens K, Pöhlmann S, Naesens L (2020) The SARS-CoV-2 and other human coronavirus spike proteins are fine-tuned towards temperature and proteases of the human airways. BioRxiv. 10.1101/2020.11.09.374603 [DOI] [PMC free article] [PubMed]
- Lau S, Wong A, Lau T, Woo P. Molecular Evolution of MERS Coronavirus: Dromedaries as a Recent Intermediate Host or Long-Time Animal Reservoir? Int J Mol Sci. 2017;18(10):2138. doi: 10.3390/ijms18102138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S-Y, Wang P, Mok BW-Y, Jinxia Zhang A, Chu H, Lee AC-Y, Deng S, Chen P, Chan KH, Song W, Chen Z (2020) Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect 9(1):837–842. 10.1080/22221751.2020.1756700 [DOI] [PMC free article] [PubMed]
- Lee J, Hughes T, Lee M-H, Field H, Rovie-Ryan JJ, Sitam FT, Sipangkui S, Nathan SK, Ramirez D, Kumar SV, Lasimbang H (2020) No evidence of coronaviruses or other potentially zoonotic viruses in Sunda pangolins (Manis javanica) entering the wildlife trade via Malaysia. EcoHealth 17(3):406–418. 10.1007/s10393-020-01503-x [DOI] [PMC free article] [PubMed]
- Lemmin T, Kalbermatter D, Harder D, Plattet P, Fotiadis D. Structures and dynamics of the novel S1/S2 protease cleavage site loop of the SARS-CoV-2 spike glycoprotein. J Struct Biol X. 2020;4:100038. doi: 10.1016/j.yjsbx.2020.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J (2005) Bats are natural reservoirs of SARS-like coronaviruses. Science 310(5748):676–679. 10.1126/science.1118391 [DOI] [PubMed]
- Li W, Wicht O, van Kuppeveld FJM, He Q, Rottier PJM, Bosch B-J (2015) A single point mutation creating a furin cleavage site in the spike protein renders porcine epidemic diarrhea coronavirus trypsin independent for cell entry and fusion. J Virol 89(15):8077–8081. 10.1128/JVI.00356-15 [DOI] [PMC free article] [PubMed]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol. 2015;89(4):1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang H, Tang X, Fang S, Ma D, Du C, Wang Y, Pan H, Yao W, Zhang R, Zou X (2020a) SARS-CoV-2 and three related coronaviruses utilize multiple ACE2 orthologs and are potently blocked by an improved ACE2-Ig. J Virol 94(22):1–14. 10.1128/JVI.01283-20 [DOI] [PMC free article] [PubMed]
- Li X, Giorgi EE, Marichannegowda MH, et al. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci Adv. 2020;6(27):eabb9153. doi: 10.1126/sciadv.abb9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T-Y, Dowd KA, Manhart CJ, Nelson S, Whitehead SS, Pierson TC. A novel approach for the rapid mutagenesis and directed evolution of the structural genes of west nile virus. J Virol. 2012;86(7):3501–3512. doi: 10.1128/JVI.06435-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jiang J-Z, Wan X-F, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, Chen J (2020a) Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLOS Pathog 16(5):e1008421. 10.1371/journal.ppat.1008421 [DOI] [PMC free article] [PubMed]
- Liu SL, Saif LJ, Weiss SR, Su L. No credible evidence supporting claims of the laboratory engineering of SARS-CoV-2. Emerg Microbes Infect. 2020;9(1):505–507. doi: 10.1080/22221751.2020.1733440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zheng H, Lin H, Li M, Yuan R, Peng J, Xiong Q, Sun J, Li B, Wu J, Yi L (2020c) Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol 94(17):1–9. 10.1128/JVI.00790-20 [DOI] [PMC free article] [PubMed]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y (2020) Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed]
- McCallum M, De Marco A, Lempp F, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F, Zepeda S (2021) N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. BioRxiv. 10.1101/2021.01.14.426475 [DOI] [PMC free article] [PubMed]
- Marchal I, Jarvis DL, Cacan R, Verbert A. Glycoproteins from Insect cells: Sialylated or Not? Biol Chem. 2001;382(2):151–159. doi: 10.1515/BC.2001.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister SW, Hendrikse NM, Löfblom J. Directed evolution of the 3C protease from coxsackievirus using a novel fluorescence-assisted intracellular method. Biol Chem. 2019;400(3):405–415. doi: 10.1515/hsz-2018-0362. [DOI] [PubMed] [Google Scholar]
- Menachery VD, Yount BL, Debbink K, Agnihothram S, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH (2015) A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21(12):1508–1513. 10.1038/nm.3985 [DOI] [PMC free article] [PubMed]
- Millet JK, Whittaker GR. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet JK, Jaimes JA, Whittaker GR. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol Rev. 2020;14(October):1–16. doi: 10.1093/femsre/fuaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan AA, Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS ONE. 2013;8(1):e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mycroft-West CJ, Su D, Elli S, Li Y, Guimond SE, Miller GJ, Turnbull JE, Yates EA, Guerrini M, Fernig DG, de Lima MA (2020) The 2019 coronavirus (SARS-CoV-2) surface protein (Spike) S1 Receptor Binding Domain undergoes conformational change upon heparin binding. BioXriv. 10.1101/2020.02.29.971093
- Nagai Y. Protease-dependent virus tropism and pathogenicity. Trends Microbiol. 1993;1(3):81–87. doi: 10.1016/0966-842X(93)90112-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D, Kostaki EG, Magiorkinis G, Panayiotakopoulos G, Sourvinos G, Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect Genet Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patiño-Galindo JÁ, Filip I, AlQuraishi M, Rabadan R (2020) Recombination and lineage-specific mutations led to the emergence of SARS-CoV-2. BioRxiv. 10.1101/2020.02.10.942748 [DOI] [PMC free article] [PubMed]
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, Braga L (2020) The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. BioRxiv. 10.1101/2020.09.30.318311
- Piplani S, Singh PK, Winkler DA, Petrovsky N (2020) In silico comparison of spike protein-ACE2 binding affinities across species; significance for the possible origin of the SARS-CoV-2 virus. ArXiv. http://arxiv.org/abs/2005.06199
- Pirone L, Del Gatto A, Di Gaetano S, et al. A Multi-Targeting Approach to Fight SARS-CoV-2 Attachment. Front Mol Biosci. 2020;7(August):1–5. doi: 10.3389/fmolb.2020.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praharaj MR, Garg P, Kesarwani V, Topno NA, Khan RIN, Sharma S, Panigrahi M et al (2020) SARS-CoV-2 Spike Glycoprotein and ACE2 interaction reveals modulation of viral entry in different hosts. BioRxiv. 10.1101/2020.05.08.084327 [DOI] [PMC free article] [PubMed]
- Qiao B, de la Cruz OM. Enhanced Binding of SARS-CoV-2 Spike Protein to Receptor by Distal Polybasic Cleavage Sites. ACS Nano. 2020;14(8):10616–10623. doi: 10.1021/acsnano.0c04798. [DOI] [PubMed] [Google Scholar]
- Rasmussen AL. On the origins of SARS-CoV-2. Nat Med. 2021 doi: 10.1038/s41591-020-01205-5. [DOI] [PubMed] [Google Scholar]
- Relman DA. Opinion: To stop the next pandemic, we need to unravel the origins of COVID-19. Proc Natl Acad Sci. 2020;117(47):29246–29248. doi: 10.1073/pnas.2021133117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Qu X, Li W, Han Z, Yu M, Zhou P, Zhang S-Y, Wang LF, Deng H, Shi Z (2008) Difference in receptor usage between severe acute respiratory syndrome (SARS) coronavirus and sars-like coronavirus of bat origin. J Virol 82(4):1899–1907. 10.1128/JVI.01085-07 [DOI] [PMC free article] [PubMed]
- Richard D, Owen CJ, Dorp L Van, Balloux F (2020a) No detectable signal for ongoing genetic recombination in SARS-CoV-2. BioXriv. 10.1101/2020.12.15.422866
- Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, Fentener van Vlissingen M, Rockx B, Haagmans BL, Koopmans MP, Fouchier RA (2020b) SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun 11(1):3496. 10.1038/s41467-020-17367-2 [DOI] [PMC free article] [PubMed]
- Riley NM, Malaker SA, Driessen MD, Bertozzi CR. Optimal dissociation methods differ for N - and O -glycopeptides. J Proteome Res. 2020;19(8):3286–3301. doi: 10.1021/acs.jproteome.0c00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmann MG. The canyon hypothesis. J Biol Chem. 1989;264(25):14587–14590. doi: 10.1016/S0021-9258(18)63732-9. [DOI] [PubMed] [Google Scholar]
- Sallard E, Halloy J, Casane D, Decroly E, van Helden J (2021) Tracing the origins of SARS-COV-2 in coronavirus phylogenies: a review. Environ Chem Lett. 10.1007/s10311-020-01151-1 [DOI] [PMC free article] [PubMed]
- Sanda M, Morrison L, Goldman R. N- and O-Glycosylation of the SARS-CoV-2 spike protein. Anal Chem. 2021;93(4):2003–2009. doi: 10.1021/acs.analchem.0c03173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, Höper D, Mettenleiter TC, Balkema-Buschmann A, Harder T, Grund C (2020) SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe 1(5):e218–e225. 10.1016/s2666-5247(20)30089-6 [DOI] [PMC free article] [PubMed]
- Schneider-Schaulies J. Cellular receptors for viruses: links to tropism and pathogenesis. J Gen Virol. 2000;81(6):1413–1429. doi: 10.1099/0022-1317-81-6-1413. [DOI] [PubMed] [Google Scholar]
- Segreto R, Deigin Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin. BioEssays. 2020 doi: 10.1002/bies.202000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnal D, Rose AS, Koča J, Burley SK, Velankar (2018) Mol*: towards a common library and tools for web molecular graphics. In: MolVA '18: Proceedings of the workshop on molecular graphics and visual analysis of molecular data. 10.2312/molva.20181103
- Seyran M, Pizzol D, Adadi P, El‐Aziz TMA, Hassan SS, Soares A, Kandimalla R, Lundstrom K, Tambuwala M, Aljabali AA, Lal A (2021) Questions concerning the proximal origin of SARS-CoV-2. J Med Virol 93(3):1204–1206. 10.1002/jmv.26478 [DOI] [PMC free article] [PubMed]
- Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T, Rockx B, Donaldson E, et al. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J Virol. 2008;82(5):2274–2285. doi: 10.1128/jvi.02041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, Liu R, He X, Shuai L, Sun Z, Zhao Y (2020) Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 368(6494):1016–1020. 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed]
- Shokeen K, Pandey S, Shah M, Kumar S (2020) Insight towards the effect of the multibasic cleavage site of SARS-CoV-2 spike protein on cellular proteases. BioXriv. 10.1101/2020.04.25.061507 [DOI] [PMC free article] [PubMed]
- Simon AJ, D’Oelsnitz S, Ellington AD. Synthetic evolution. Nat Biotechnol. 2019;37(7):730–743. doi: 10.1038/s41587-019-0157-4. [DOI] [PubMed] [Google Scholar]
- Sirotkin K, Sirotkin D. Might SARS-CoV-2 Have Arisen via Serial Passage through an Animal Host or Cell Culture?: A potential explanation for much of the novel coronavirus’ distinctive genome. BioEssays. 2020;42(10):1–7. doi: 10.1002/bies.202000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh WT, Liu Y, Nakayama EE, Ono C, Torii S, Nakagami H, Matsuura Y, Shioda T, Arase H (2020) The N-terminal domain of spike glycoprotein mediates SARS-CoV-2 infection by associating with L-SIGN and DC-SIGN. BioRxiv. 10.1101/2020.11.05.369264
- Sørensen B, Susrud A, Dalgleish AG. Biovacc-19: a candidate vaccine for covid-19 (SARS-CoV-2) developed from analysis of its general method of action for infectivity. QRB Discov. 2020;1:e6. doi: 10.1017/qrd.2020.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa A (2020). SARS-Cov-2 Natural or artificial? That is the question. Clin Microbiol Res. 10.31487/j.cmr.2020.01.06
- Standage-Beier K, Wang X. Genome reprogramming for synthetic biology. Front Chem Sci Eng. 2017;11(1):37–45. doi: 10.1007/s11705-017-1618-2. [DOI] [Google Scholar]
- Sun C, Cheng C, Zhao T, Chen Y, Ayaz AM. Frozen food: is it safe to eat during COVID-19 pandemic? Public Health. 2021;190(January):e26. doi: 10.1016/j.puhe.2020.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Ren K, Zhang X, Chen J, Jiang Z, Jiang J, Ji F, Ouyang X, Li L (2020) Mass spectrometry analysis of newly emerging coronavirus HCoV-19 spike protein and human ACE2 reveals camouflaging glycans and unique post-translational modifications. Engineering. 10.1016/j.eng.2020.07.014 [DOI] [PMC free article] [PubMed]
- Tang XC, Zhang JX, Zhang SY, et al. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol. 2006;80(15):7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y-D, Li Y-M, Sun J, Zhang H-L, Wang T-Y, Sun M-X, Yang Y-L, Hu X, Zhao J, Cai X (2020) Cell entry of SARS-CoV-2 conferred by angiotensin-converting enzyme 2 (ACE2) of different species. BioRxiv. 10.1101/2020.06.15.153916
- Tang T, Jaimes JA, Bidon MK, Straus MR, Daniel S, Whittaker GR (2021) Proteolytic activation of SARS-CoV-2 spike at the S1/S2 boundary: potential role of proteases beyond furin. ACS Infect Dis 7(2):264–272. 10.1021/acsinfecdis.0c00701 [DOI] [PMC free article] [PubMed]
- Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci. 2009;106(38):16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thépaut M, Luczkowiak J, Vivès C, Labiod N, Bally I, Lasala F, Grimoire Y, Fenel D, Sattin S, Thielens N, Schoehn G (2020) DC/L-SIGN recognition of spike glycoprotein promotes SARS-CoV-2 trans-infection and can be inhibited by a glycomimetic antagonist. BioRxiv. 10.1101/2020.08.09.242917 [DOI] [PMC free article] [PubMed]
- Thi Nhu Thao T, Labroussaa F, Ebert N, V’kovski P, Stalder H, Portmann H, Kelly J, Steiner S, Holwerda M, Kratzel A, Gultom M (2020) Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 582:561–565. 10.1038/s41586-020-2294-9 [DOI] [PubMed]
- Thomas G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3(10):753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Huang Q, Fang Y, Wu J. FurinDB: A database of 20-residue furin cleavage site motifs, substrates and their associated drugs. Int J Mol Sci. 2011;12(2):1060–1065. doi: 10.3390/ijms12021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari V, Tandon R, Sankaranarayanan NV, Beer JC, Kohlmeir EK, Swanson-Mungersonet M, Desai UR (2020) Preferential recognition and antagonism of SARS-CoV-2 spike glycoprotein binding to 3- O -sulfated heparan sulfate. BioRxiv. 10.1101/2020.10.08.331751
- Torii, S, Ono C, Suzuki R, Morioka Y, Anzai I, Fauzyah Y, Maeda Y, Kamitani W, Fukuhara T, Matsuura Y (2020) Establishment of a reverse genetics system for SARS-CoV-2 using circular polymerase extension reaction. BioRxiv. 10.1101/2020.09.23.309849 [DOI] [PMC free article] [PubMed]
- Tse LV, Hamilton AM, Friling T, Whittaker GR. A novel activation mechanism of avian influenza virus H9N2 by furin. J Virol. 2014;88(3):1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L, Richard D, Tan CCS, Shaw LP, Acman M, Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat Commun. 2020;11(1):5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Kayal N, Anand P, Badley AD, Church GM, Soundararajan V (2020) Benchmarking evolutionary tinkering underlying human–viral molecular mimicry shows multiple host pulmonary–arterial peptides mimicked by SARS-CoV-2. Cell Death Discov 6:96. 10.1038/s41420-020-00321-y [DOI] [PMC free article] [PubMed]
- Wahl A, De C, Abad Fernandez M, Lenarcic EM, Xu Y, Cockrell AS, Cleary RA et al (2019) Precision mouse models with expanded tropism for human pathogens. Nat Biotechnol 37(10):1163–1173. 10.1038/s41587-019-0225-9 [DOI] [PMC free article] [PubMed]
- Walls AC, Park Y, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an Analysis Based On Decade-Long Structural Studies of SARS coronavirus. J Virol. 2020;94(7):1–9. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LF, Shi Z, Zhang S, Field H, Daszak P, Eaton BT. Review of bats and SARS. Emerg Infect Dis. 2006;12(12):1834–1840. doi: 10.3201/eid1212.060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-M, Wang L-F, Shi Z-L (2008) Construction of a non-infectious SARS coronavirus replicon for application in drug screening and analysis of viral protein function. Biochem Biophys Res Commun 374(1):138–142. 10.1016/j.bbrc.2008.06.129 [DOI] [PMC free article] [PubMed]
- Wang D, Baudys J, Bundy JL, Solano M, Keppel T, Barr JR. Comprehensive analysis of the glycan complement of SARS-CoV-2 spike proteins using signature ions-triggered electron-transfer/higher-energy collisional dissociation (EThcD) mass spectrometry. Anal Chem. 2020;92(21):14730–14739. doi: 10.1021/acs.analchem.0c03301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q (2020b) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181(4):894–904. 10.1016/j.cell.2020.03.045 [DOI] [PMC free article] [PubMed]
- Wang Y, Liu M, Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci. 2020;117(25):13967–13974. doi: 10.1073/pnas.2008209117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Pipes L, Nielsen R (2021) Synonymous mutations and the molecular evolution of SARS-CoV-2 origins. Virus Evol 7:1–11. 10.1093/ve/veaa098 [DOI] [PMC free article] [PubMed]
- Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M (2020) Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369(6501):330–333. 10.1126/science.abb9983 [DOI] [PMC free article] [PubMed]
- Wenzel J. Origins of SARS-CoV-1 and SARS-CoV-2 are often poorly explored in leading publications. Cladistics. 2020;36(4):374–379. doi: 10.1111/cla.12425. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel AG, Benton DJ, Xu P, et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020;27(8):763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel AG, Benton DJ, Xu P, Rosenthal PB, Skehel JJ, Gamblin SJ. Structure and binding properties of Pangolin-CoV spike glycoprotein inform the evolution of SARS-CoV-2. Nat Commun. 2021;12(1):837. doi: 10.1038/s41467-021-21006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Zheng M, Yang Y, Gu X, Yang K, Li M, Liu Y, Zhang Q, Zhang P, Wang Y, Wang Q (2020a) Furin: a potential therapeutic target for COVID-19. Science 23(10):101642. 10.1016/j.isci.2020.101642 [DOI] [PMC free article] [PubMed]
- Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhao S. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Res. 2021;50:102115. doi: 10.1016/j.scr.2020.102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou J-J, Li N, Guo Y, Li X, Shen X, Zhang Z (2020) Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583(7815):286–289. 10.1038/s41586-020-2313-x [DOI] [PubMed]
- Yount B, Denison MR, Weiss SR, Baric RS. Systematic Assembly Of A Full-Length Infectious cDNA of mouse hepatitis virus strain A59. J Virol. 2002;76(21):11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WB, Tang GD, Zhang L, Corlett RT. Decoding evolution and transmissions of novel pneumonia coronavirus using the whole genomic data. ChinaXiv Published online. 2020 doi: 10.12074/202002.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L-P, Gao Y-T, Ge X-Y, et al. Bat severe acute respiratory syndrome-like coronavirus WIV1 encodes an extra accessory protein, ORFX, Involved in Modulation of the Host Immune Response. J Virol. 2016;90(14):6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan SH, Deverman B, Chan YA (2020) SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence? BioRxiv. 10.1101/2020.05.01.073262
- Zhang D (2020) The Pan-SL-CoV/GD sequences may be from contamination. Zenodo. 10.5281/zenodo.4450267
- Zhang Q, Chen CZ, Swaroop M, Xu M, Wang L, Lee J, Wang AQ, Pradhan M, Hagen N, Chen L, Shen M (2020a) Heparan sulfate assists SARS-CoV-2 in cell entry and can be targeted by approved drugs in vitro. Cell Discov 6(1):80. 10.1038/s41421-020-00222-5 [DOI] [PMC free article] [PubMed]
- Zhang T, Wu Q, Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak. Curr Biol. 2020;30(7):1346–1351.e2. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao W, Mao Y, Chen Y, Wang S, Zhong Y, Su T, Gong M, Du D, Lu X, Cheng J (2020c) Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol Cell Proteomics. 10.1074/mcp.RA120.002295 [DOI] [PMC free article] [PubMed]
- Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH, Brindley MA (2020) Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28(4):586–601.e6. 10.1016/j.chom.2020.08.004 [DOI] [PMC free article] [PubMed]
- Zhou P, Shi Z-L (2021) SARS-CoV-2 spillover events. Science 371(6525):120–122. 10.1126/science.abf6097 [DOI] [PubMed]
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, Hughes AC (2020a) A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/s2 cleavage site of the spike protein. Curr Biol 30(11):2196–2203.e3. 10.1016/j.cub.2020.05.023 [DOI] [PMC free article] [PubMed]
- Zhou P, Yang X-L, Wang X-G, Ben H, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang CL, Chen HD (2020b) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed]
- Zhou P, Yang X-L, Wang X-G, Ben H, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang CL, Chen HD (2020c) Addendum: A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588(7836):E6–E6. 10.1038/s41586-020-2951-z [DOI] [PMC free article] [PubMed]
- Zhu N, Zhang D, Wang W, et al. A novel Coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Li J, Liang G. How does cellular heparan sulfate function in viral pathogenicity? Biomed Environ Sci. 2011;24(1):81–87. doi: 10.3967/0895-3988.2011.01.011. [DOI] [PubMed] [Google Scholar]