Abstract
Cell division site positioning is precisely regulated but the underlying mechanisms are incompletely understood. In the social bacterium Myxococcus xanthus, the ~15 MDa tripartite PomX/Y/Z complex associates with and translocates across the nucleoid in a PomZ ATPase-dependent manner to directly position and stimulate formation of the cytokinetic FtsZ-ring at midcell, and then undergoes fission during division. Here, we demonstrate that PomX consists of two functionally distinct domains and has three functions. The N-terminal domain stimulates ATPase activity of the ParA/MinD ATPase PomZ. The C-terminal domain interacts with PomY and forms polymers, which serve as a scaffold for PomX/Y/Z complex formation. Moreover, the PomX/PomZ interaction is important for fission of the PomX/Y/Z complex. These observations together with previous work support that the architecturally diverse ATPase activating proteins of ParA/MinD ATPases are highly modular and use the same mechanism to activate their cognate ATPase via a short positively charged N-terminal extension.
Research organism: Other
Introduction
Accurate positioning of the cell division site ensures the formation of daughter cells of correct size and shape. In most bacteria, cell division initiates with positioning of the tubulin homolog FtsZ at the incipient division site (Du and Lutkenhaus, 2019). Subsequently, FtsZ polymerizes to form a dynamic ring-like structure, the so-called (Fts)Z-ring, which serves as a scaffold for recruitment of other components of the division machinery (Du and Lutkenhaus, 2019). Accordingly, systems that regulate division site positioning act at the level of Z-ring formation (Eswara and Ramamurthi, 2017). While the core components of the division machinery are conserved, the regulatory systems that ensure Z-ring positioning and, thus, the division site, are surprisingly diverse and also incompletely understood (Eswara and Ramamurthi, 2017). Interestingly, several of these systems have at their core a member of the ParA/MinD superfamily of P-loop ATPases that interacts with system-specific components to generate system-specific dynamic localization patterns that bring about correct Z-ring positioning (Lutkenhaus, 2012). These patterns include pole-to-pole oscillations in the Escherichia coli MinCDE system, bipolar gradient formation in the Caulobacter crescentus MipZ/ParB system, polar localization in the Bacillus subtilis MinCDJ/DivIVA system, and translocation across the nucleoid in the PomXYZ system of Myxococcus xanthus (Treuner-Lange and Søgaard-Andersen, 2014; Schumacher and Søgaard-Andersen, 2017).
ParA/MinD ATPases are key players in orchestrating the subcellular organization of bacterial cells and are not only involved in division site positioning but also in chromosome and plasmid segregation as well as positioning of other macromolecular complexes (Lutkenhaus, 2012). The function of ParA/MinD ATPases critically depends on the ATPase cycle during which they alternate between a monomeric form and a dimeric ATP-bound form (Leonard et al., 2005; Hu et al., 2003; Wu et al., 2011; Scholefield et al., 2011). Generally, ParA/MinD ATPases have a low intrinsic ATPase activity that is stimulated by a single cognate ATPase Activating Protein (AAP) (Lutkenhaus, 2012). While ParA/MinD ATPases share overall sequence conservation, the AAPs are much less conserved. The ParABS systems for chromosome and plasmid segregation and the E. coli MinCDE system have provided fundamental insights into how ParA/MinD ATPase/AAP pairs interact to establish dynamic localization patterns: The AAP ParB stimulates ATPase activity of ATP-bound ParA dimers bound non-specifically to DNA (Leonard et al., 2005; Bouet and Funnell, 1999; Hester and Lutkenhaus, 2007). In chromosome segregation systems, ParB binds to multiple parS sites at the origin of replication, forming a large complex (Lin and Grossman, 1998; Yamaichi et al., 2007; Mohl and Gober, 1997) while ATP-bound ParA dimers bind non-specifically to the nucleoid (Castaing et al., 2008; Hester and Lutkenhaus, 2007; Leonard et al., 2005; Scholefield et al., 2011). Upon replication, one of the duplicated ParB-parS complexes interacts with nucleoid-bound ParA dimers, thereby stimulating ATP hydrolysis and causing the release of ParA monomers from the nucleoid (Bouet and Funnell, 1999; Ptacin et al., 2010; Schofield et al., 2010; Vecchiarelli et al., 2013). Released ParA monomers undergo nucleotide exchange and rebind to the nucleoid (Bouet and Funnell, 1999; Ptacin et al., 2010; Schofield et al., 2010; Vecchiarelli et al., 2013). Repeated interactions between the large ParB-parS complex and nucleoid-bound ParA result in translocation of the ParB-parS complex to the opposite cell half (Vecchiarelli et al., 2014; Lim et al., 2014; Ptacin et al., 2010; Schofield et al., 2010). In the MinCDE system, the AAP MinE, which is non-homologous to ParB, stimulates ATPase activity of membrane- and ATP-bound MinD dimers (Hu and Lutkenhaus, 2001; Lackner et al., 2003; Park et al., 2011; Hu and Lutkenhaus, 2003; Szeto et al., 2002). In vivo, dimeric ATP-bound MinD forms a complex with the inhibitor of Z-ring formation MinC at the membrane (de Boer et al., 1991; Hu and Lutkenhaus, 1999; Hu and Lutkenhaus, 2003; Hu et al., 1999). Upon stimulation of MinD ATPase activity by MinE, the MinD/C complex is released from the membrane (Hu et al., 2002; Hu and Lutkenhaus, 2001; Lackner et al., 2003; Park et al., 2012). Subsequently, MinD undergoes nucleotide exchange and rebinds to the membrane together with MinC. These interactions ultimately result in the coupled pole-to-pole oscillations of the MinC/D complex and MinE (Hu and Lutkenhaus, 1999; Raskin and de Boer, 1999).
In the rod-shaped M. xanthus cells, Z-ring formation and positioning at midcell between two segregated chromosomes are stimulated by the tripartite PomX/Y/Z complex (Schumacher et al., 2017; Treuner-Lange et al., 2013; Harms et al., 2013). PomZ is a ParA/MinD ATPase while PomX and PomY separately have AAP activity and function to synergistically stimulate the low intrinsic ATPase activity of DNA- and ATP-bound dimeric PomZ (Schumacher et al., 2017; Treuner-Lange et al., 2013). PomX and PomY are non-homologous, and share homology with neither ParB nor MinE. The Pom proteins form a dynamically localized complex with an estimated size of ~15 MDa in vivo that is visible as a cluster by epifluorescence microscopy (Schumacher et al., 2017). This complex associates with the nucleoid via PomZ, and early during the cell cycle, it is positioned on the nucleoid away from midcell, that is off-center, close to the new cell pole (Schumacher et al., 2017). Subsequently, the complex translocates by biased random motion on the nucleoid to the midnucleoid, which coincides with midcell. At midnucleoid, the PomXYZ complex undergoes constrained motion and stimulates Z-ring formation. Intriguingly, during cell division, the PomX/Y/Z complex undergoes fission, with the two ‘portions’ segregating to the two daughters (Schumacher et al., 2017).
The Pom proteins interact in all three pairwise combinations in vitro and PomX is essential for cluster formation by PomY and PomZ in vivo (Schumacher et al., 2017): In vitro PomX spontaneously polymerizes in a cofactor-independent manner to form filaments, and alone forms a cluster in vivo. PomY bundles PomX filaments in vitro; in vivo PomY is recruited by PomX to form a PomX/Y complex, which is not associated with the nucleoid and stalled somewhere in cells. ATP- and DNA-bound PomZ dimers, but not monomeric PomZ, are recruited to the PomX/Y complex by interactions with PomX as well as PomY, resulting in the association of the PomX/Y/Z complex to the nucleoid. Due to the AAP activity of PomX and PomY, PomZ is rapidly turned over in the PomX/Y/Z complex and released in a monomeric form to the cytosol. Motion of the PomX/Y/Z complex depends on non-specific DNA binding and ATP hydrolysis by PomZ (Schumacher et al., 2017). Translocation to midnucleoid and constrained motion at midnucleoid arise from the continuous turnover of PomZ in the complex together with the diffusive PomZ flux on the nucleoid into the PomX/Y/Z complex (Schumacher et al., 2017). Finally, while all three Pom proteins are important for Z-ring formation at midcell, PomY and PomZ in the PomX/Y/Z complex are thought to be directly involved in recruiting FtsZ to the division site based on protein-protein interaction analyses (Schumacher et al., 2017).
The Pom system displays unusual spatiotemporal dynamics and is also unusual because it incorporates two AAPs. It remains elusive how PomX and PomY stimulate PomZ ATPase activity. With the long-term goal to understand the spatiotemporal dynamics of the Pom system, we focused on the function of PomX in vivo and in vitro. Here, we show that PomX is a triple regulator of cell division and displays three activities: The AAP activity resides in the N-terminal domain, the C-terminal domain serves as a scaffold for PomX/Y/Z complex formation, and the PomX/Z interaction is important for PomX/Y/Z complex fission during division. Moreover, our findings support the notion that that AAPs of ParA/MinD ATPases use the same mechanism to activate their cognate ATPase.
Results
PomX consists of two functional domains with distinct functions
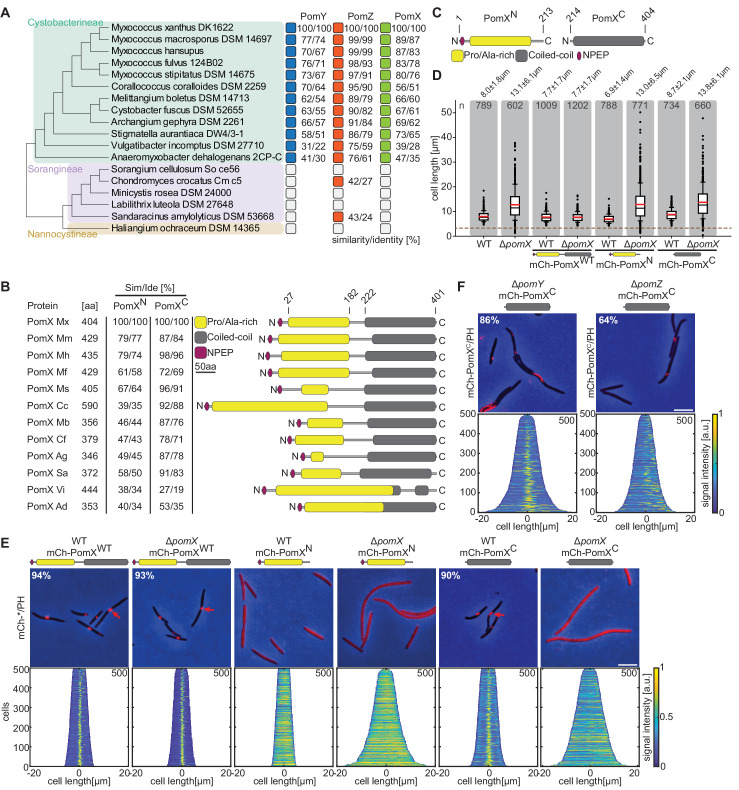
PomX and PomY co-occur with PomZ in Cystobacterineae (Schumacher et al., 2017). PomZ sequences are highly conserved, whereas PomX and PomY sequences are more divergent (Figure 1A). In M. xanthus PomX, the region from residues 27 to 182 is Ala and Pro-rich (23% Pro; 19% Ala) and the region from residues 222 to 401 contains a coiled-coil domain (Figure 1B). Although PomX homologs are of different length, their overall architecture is similar and with a high level of similarity in the C-terminal regions while the N-terminal regions vary in length and similarity (Figure 1B). To analyze how PomX functions, we divided PomX into two parts. From hereon, we refer to these two parts as the N-terminal domain (PomXN, residues 1–213) and the C-terminal domain (PomXC, residues 214–404) (Figure 1C).
Figure 1. PomX consists of two domains that are both required for function.
(A) Similarity and identity analysis of PomX, PomY, and PomZ homologs. The three Myxococcales suborders are indicated. An open box indicates that a homolog is not present. (B) Similarity and identity of PomX domains in different PomX homologs. Similarity and identity were calculated based on the domains of M. xanthus PomX shown in C. (C) PomX truncations used in this study. Numbers on top indicate the start and stop positions of the truncations relative to full-length PomXWT. (D) Cell length distribution of cells of indicated genotypes. Cells below stippled line are minicells. Numbers indicate mean cell length±STDEV. In the boxplots, boxes include the 25th and the 75th percentile, whiskers data points between the 10% and 90% percentile, outliers are shown as black dots. Black and red lines indicate the median and mean, respectively. Number of analyzed cells is indicated. In the complementation strains, pomX alleles were expressed from plasmids integrated in a single copy at the attB site. (E) Fluorescence microscopy of cells of indicated genotypes. Phase-contrast and fluorescence images of representative cells were overlayed. Numbers indicate fraction of cells with fluorescent clusters. Demographs show fluorescence signals of analyzed cells sorted according to length and with off-center signals to the right. Numbers in upper right indicate number of cells used to create demographs. Scale bar, 5 µm. (F) Fluorescence microscopy of cells of indicated genotypes. Images of representative cells and demographs were created as in (E). Scale bar, 5 µm. For experiments in D, E and F similar results were obtained in two independent experiments.
Figure 1—figure supplement 1. PomX variants accumulate in M.xanthus.
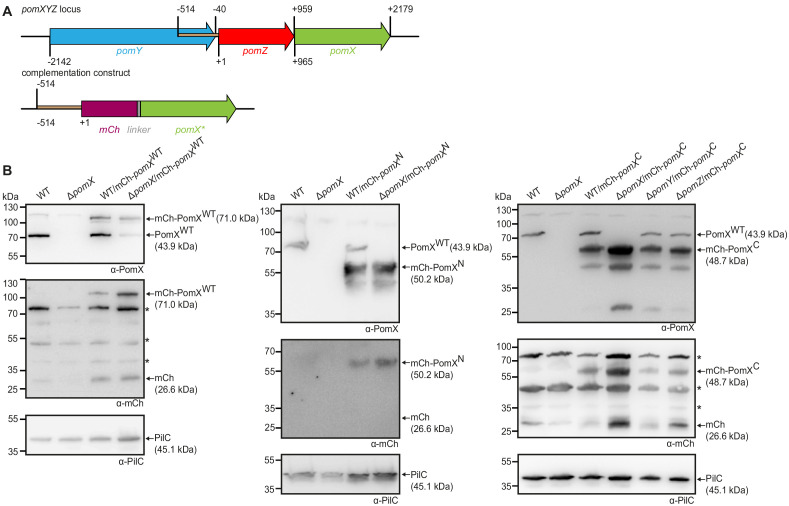
We fused the two PomX domains to mCherry (mCh) and expressed them ectopically (Figure 1—figure supplement 1A). mCh-PomXN, mCh-PomXC, and full-length mCh-PomXWT accumulated at or above PomXWT levels in ΔpomX and pomX+ cells (Figure 1—figure supplement 1B). WT M. xanthus cells have a cell length of 8.0 ± 1.8 µm (mean ± standard deviation (STDEV)), while ΔpomX cells are filamentous with a length of 13.1 ± 6.1 µm and also generate DNA-free minicells (Figure 1D). mCh-PomXWT complemented the division defect of the ΔpomX mutant in agreement with previous observations (Schumacher et al., 2017), while the truncated variants did not.
mCh-PomXWT formed a single well-defined cluster in 93% and 94% of ΔpomX and WT cells, respectively (Figure 1E). These clusters localized in the off-center position (defined as clusters outside the midcell region at 50 ± 5% of cell length) in short cells and at midcell in long cells (Figure 1E). The truncated mCh-PomX variants displayed diffuse localization in ΔpomX cells (Figure 1E). However, mCh-PomXC but not mCh-PomXN formed a single cluster in 90% of pomX+ cells and localized as mCh-PomXWT (Figure 1E). This cluster formation by mCh-PomXC was independent of PomY and PomZ (Figure 1F and Figure 1—figure supplement 1B), supporting that mCh-PomXC is integrated into the PomX/Y/Z cluster via interaction with PomX. As described (Schumacher et al., 2017), the PomX clusters were more elongated in the absence of PomY (Figure 1F). The incorporation of PomXC into the PomX/Y/Z complex interfered with neither PomX/Y/Z complex formation nor function (Figure 1D,E). We conclude that both PomX domains are essential for function and that mCh-PomXC can integrate into the PomX/Y/Z complex via interaction with PomX while mCh-PomXN cannot.
PomX AAP activity resides in PomXN
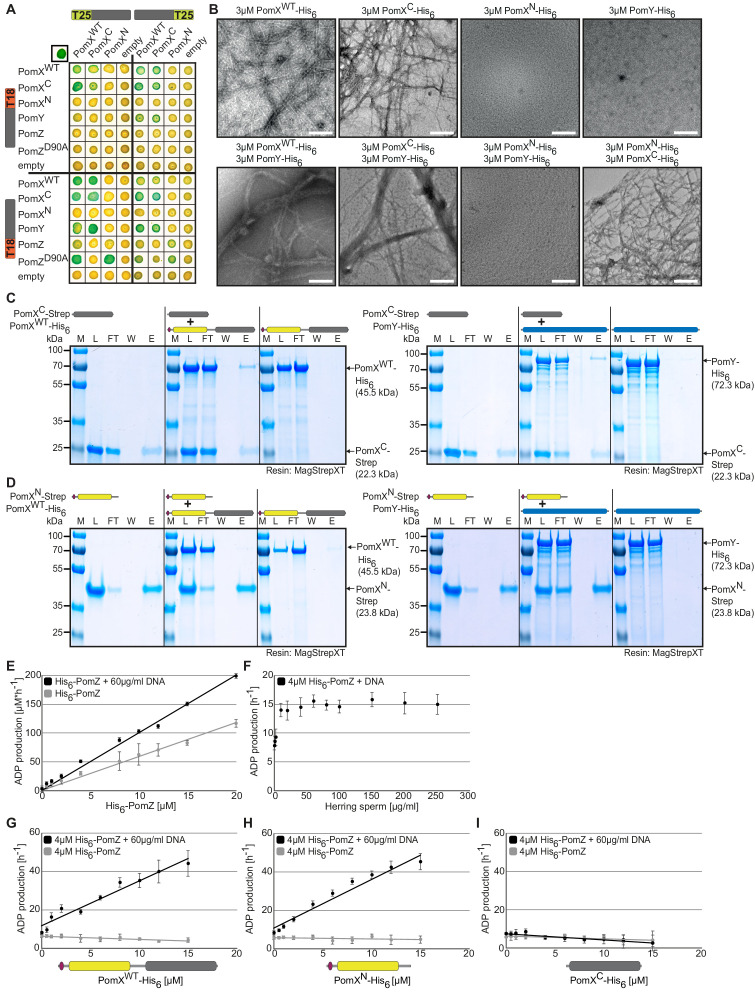
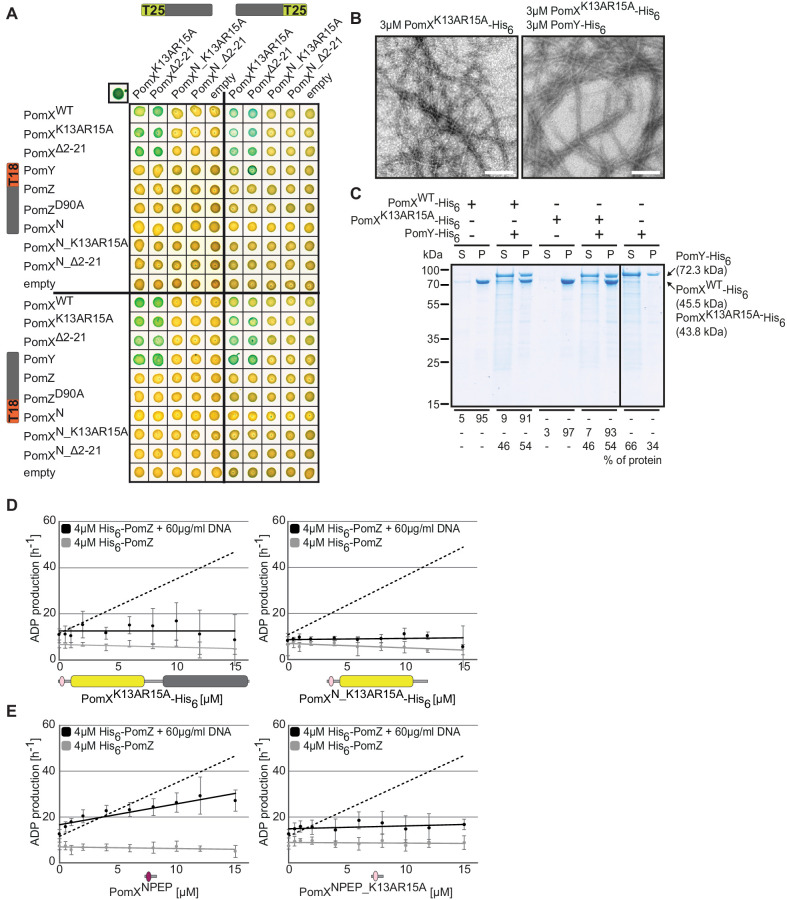
We used the bacterial adenylate cyclase two-hybrid system (BACTH) to test for protein-protein interactions involving the two PomX domains. In agreement with previous observations using purified proteins, full-length PomXWT self-interacted and interacted with PomY (Figure 2A). PomXC self-interacted and also interacted with full-length PomX and PomY, while PomXN neither self-interacted nor interacted with PomXWT, PomXC, or PomY. To test for interactions with PomZ, we used the two PomZ variants PomZWT and PomZD90A, the latter of which is locked in the DNA-binding, dimeric, ATP-bound form that interacts strongly with the PomX/PomY cluster in vivo (Schumacher et al., 2017). PomXWT and PomXN interacted with PomZWT and PomZD90A while PomXC did not; also, PomZD90A interacted more strongly with PomXN than PomZWT (Figure 2A).
Figure 2. PomXC interacts with PomX and PomY while PomXN stimulates PomZ ATPase activity.
(A) BACTH analysis of interactions between Pom proteins. The indicated protein fragments were fused to T18 and T25 as indicated. Blue colony indicates an interaction, white no interaction. Positive control in upper left corner, leucine zipper of GCN4 fused to T25 and T18. For negative controls, co-transformations with empty plasmids were performed. Images show representative results and were performed in three independent experiments. (B) TEM images of negatively stained purified proteins. Proteins were applied to the EM grids alone or after mixing in a 1:1 molar ratio as indicated before staining. Scale bar, 200 nm. Images show representative results of several independent experiments. (C, D) In vitro pull-down experiments with purified PomXC-Strep, PomXN-Strep, PomXWT-His6, and PomY-His6. Instant Blue-stained SDS-PAGE shows load (L), flow-through (FL), wash (W), and elution (E) fractions using MagStrep XT beads in pull-down experiments with 10 µM of indicated proteins alone or pre-mixed as indicated on top. Molecular size markers are shown on the left and proteins analyzed on the right together with their calculated MW. Note that PomXWT-His6 (Schumacher et al., 2017) and PomXN-Strep migrate aberrantly and according to a higher MW. All samples in a panel were analyzed on the same gel and black lines are included for clarity. Experiments were repeated in two independent experiments with similar results. (E–I) His6-PomZ ATPase activity. ADP production rate was determined in an NADH-coupled photometric microplate assay in the presence of 1 mM ATP at 32°C. DNA and PomX variants were added as indicated. Spontaneous ATP hydrolysis and NADH consumption was accounted for by subtracting the measurements in the absence of His6-PomZ. Data points show the mean±STDEV calculated from six independent measurements.
Figure 2—figure supplement 1. Purification and analysis of Pom proteins.
To test for interactions between the Pom proteins in vitro, we purified tagged variants of PomXWT, PomXN, PomXC, PomY, and PomZ (Figure 2—figure supplement 1A). We confirmed by negative stain transmission electron microscopy (TEM) that PomXWT-His6 formed filaments that were bundled by PomY-His6, while PomY-His6 on its own did not form higher order structures (Figure 2B; Schumacher et al., 2017). Consistently, when analyzed separately in high-speed centrifugation experiments, 93–95% of PomXWT-His6 and 29–35% of PomY-His6 were recovered in the pellet fraction, whereas 71–86% and 48–49%, respectively of PomXWT-His6 and PomY-His6 were in the pellet fraction when mixed in equimolar amounts (Figure 2—figure supplement 1B,C,D).
As noted for PomXWT, PomXN-His6 (molecular weight (MW) of monomer: 24.3 kDa) migrated aberrantly in SDS-PAGE (Figure 2—figure supplement 1A). By size exclusion chromatography (SEC), the majority of PomXN-His6 eluted corresponding to a globular protein with a MW of ~136 kDa and in a smaller peak corresponding to a MW of ~306 kDa (Figure 2—figure supplement 1E). PomXN-His6 neither formed higher-order structures by TEM nor in high-speed centrifugation experiments (Figure 2B and Figure 2—figure supplement 1C,D). Because PomXN does not self-interact in BACTH, migrates aberrantly by SDS-PAGE, and is rich in Ala/Pro residues and, therefore, may not have a globular conformation, it is unclear whether SEC reflects the formation of PomXN oligomers. PomXC-His6 in SDS-PAGE migrated at the expected size (MW of monomer: 22.8 kDa) (Figure 2—figure supplement 1A); however, the protein could not be analyzed by SEC because it did not enter the matrix. Accordingly, PomXC-His6 spontaneously formed filaments visible by TEM and mostly accumulated in the pellet fraction in high-speed centrifugation experiments (Figure 2B and Figure 2—figure supplement 1B,D). PomY-His6 bundled the PomXC-His6 filaments and was enriched in the pellet fraction in centrifugation assays in the presence of PomXC-His6 (Figure 2B and Figure 2—figure supplement 1B). Interactions between PomXN-His6 and PomXWT-His6, PomXC-His6 and PomY-His6 were observed by neither TEM nor high-speed centrifugation (Figure 2B and Figure 2—figure supplement 1C,D). Finally, we confirmed in pull-down experiments using truncated PomX-Strep variants (Figure 2—figure supplement 1A) that PomXWT-His6 and PomY-His6 interact with PomXC-Strep (Figure 2C) but not with PomXN-Strep (Figure 2D) and that PomXC-His6 did not interact with PomXN-Strep (Figure 2—figure supplement 1F).
To test in vitro for interactions between PomX variants and PomZ, we used PomZ ATPase activity as a readout. First, we established a base-line for these analyses. The amount of hydrolyzed ATP increased with His6-PomZ concentration in the absence of DNA (specific activity: 7 ± 1 ATP hr−1, 4 µM His6-PomZ) (Figure 2E). In the presence of increasing concentrations of non-specific herring sperm DNA, His6-PomZ ATP hydrolysis was stimulated, reaching saturation at ~40 µg/ml DNA. At this concentration, His6-PomZ ATP hydrolysis was stimulated twofold (specific activity: ~15 ATP hr−1, 4 µM His6-PomZ) (Figure 2F). In the presence of saturating concentrations of DNA (60 µg/ml), His6-PomZ ATPase activity also increased with concentration (Figure 2E). For comparison, in an average M. xanthus cell with one chromosome, the DNA concentration is ~3400 µg/ml suggesting that PomZ in vivo works under fully DNA-saturating conditions.
PomXWT-His6 only stimulated His6-PomZ ATPase activity in the presence of DNA (Figure 2G). Stimulation increased with increasing PomXWT-His6 concentrations (specific activity: 44 ± 7 ATP hr−1 at 15 µM PomXWT-His6, 4 µM His6-PomZ), and His6-PomZ ATPase activity did not reach a plateau even at the highest PomXWT-His6 concentration (Figure 2G). Importantly, and in agreement with the BACTH analysis, PomXN-His6 stimulated His6-PomZ ATP hydrolysis in the presence of DNA as efficiently as PomXWT-His6 (specific activity: 43 ± 4 ATP hr−1 at 15 µM PomXN-His6, 4 µM His6-PomZ) (Figure 2H). By contrast, PomXC-His6 did not stimulate His6-PomZ ATPase activity (Figure 2I). The observations that PomXN does not spontaneously form filaments in vitro while full-length PomX does and these two PomX variants stimulate PomZ ATPase activity with equal efficiency provide evidence that PomX filament formation is not essential for AAP activity. These observations also strongly support that the PomXN domains in a PomX filament act independently of each other to stimulate PomZ ATPase activity.
Altogether, we conclude that (1) PomX consists of two domains with distinct functions that are both required for PomX activity in vivo; (2) PomXN interacts with PomZ and harbors the entire AAP activity; (3) PomXC is required and sufficient to mediate PomX self-interaction with spontaneous filament formation in vitro and PomX-dependent cluster incorporation in vivo; (4) PomXC interacts with PomY; and, (5) the PomXN domains in a PomX filament function independently of each other to stimulate PomZ ATPase activity.
Two positively charged residues in PomXNPEP are important for division site and PomX/Y/Z cluster positioning at midcell
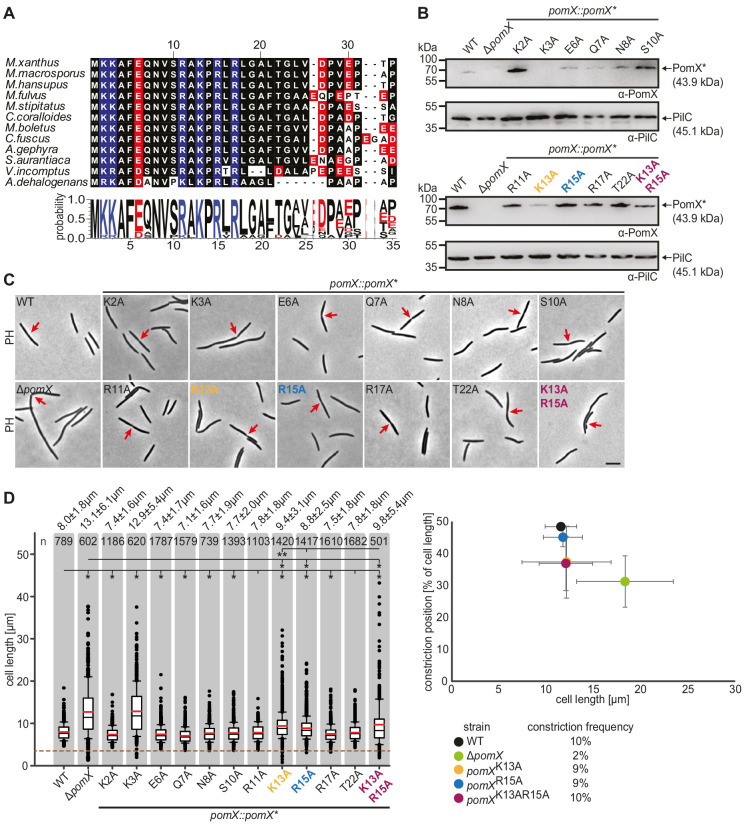
To define the PomXN region involved in AAP activity, we performed a detailed sequence analysis of the N-terminal domain of PomX homologs. This analysis revealed a stretch of highly conserved amino acids at the N-terminus (residues 1–22, from hereon PomXNPEP) that is enriched in charged amino acids, six of which are positively charged (Figures 1B and 3A and Figure 3—figure supplement 1).
Figure 3. PomXN harbors a conserved N-terminal peptide crucial for cell division site positioning at midcell.
(A) Multiple sequence alignment of the conserved PomX N-terminus. Black background indicates similar amino acids. Positively and negatively charged residues are indicated in blue and red, respectively. Weblogo consensus sequence is shown below. (B) Western blot analysis of accumulation of PomX variants. Protein from the same number of cells was loaded per lane. Molecular mass markers are indicated on the left. PilC was used as a loading control. (C) Phase-contrast microscopy of strains of indicated genotypes. Representative cells are shown. Red arrows indicate cell division constrictions. Scale bar, 5 µm. (D) Analysis of cell length distribution and cell division constrictions of cells of indicated genotypes. Left panel, boxplot is as in Figure 1D. Number of cells analyzed is indicated at the top. *p<0.001; **p<0.05 in Mann-Whitney test. Right panel, cell division position in % of cell length is plotted as a function of cell length. Dots represent mean ± STDEV. Numbers below indicate cell division constriction frequency. In B, C, and D, similar results were obtained in two independent experiments.
Figure 3—figure supplement 1. PomX homologs are highly conserved.
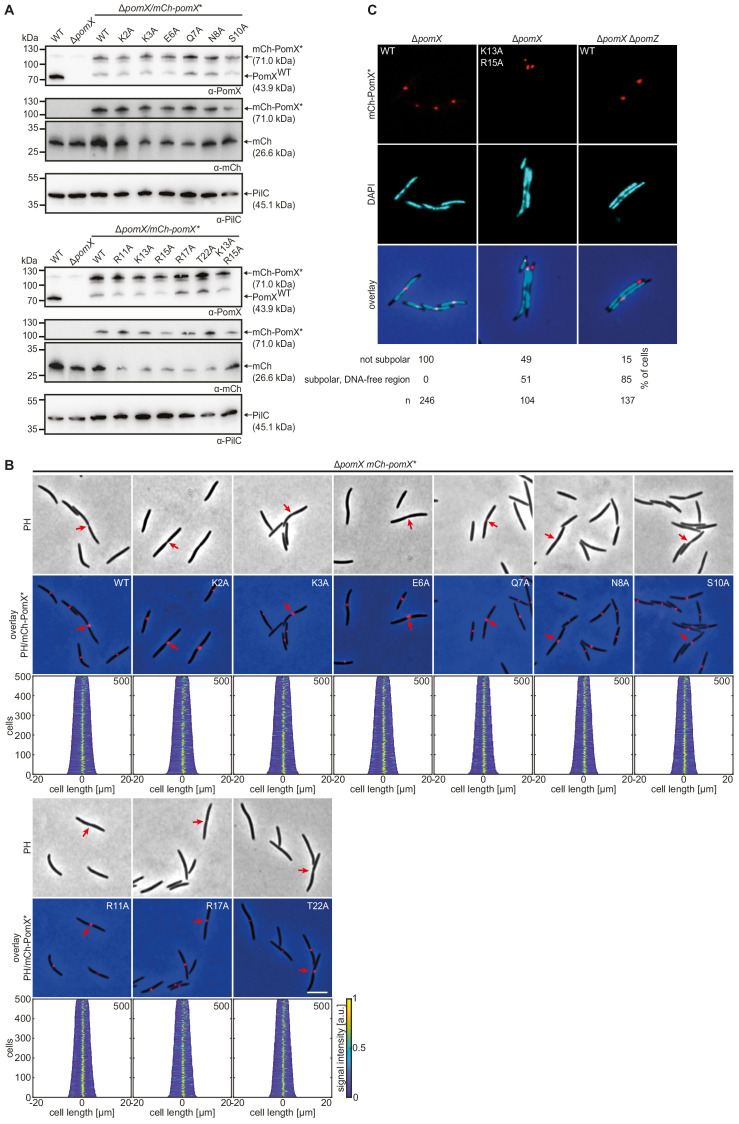
To probe the role of PomXNPEP, we generated PomX variants lacking residues 2–21 (PomXΔ2-21) with and without mCh; however, none of these variants accumulated in M. xanthus. Therefore, we performed Ala scanning of PomXNPEP in which all charged or hydrophilic residues were replaced by Ala. Then the mutant alleles replaced the pomXWT allele at the pomX locus. All 11 PomX variants except for PomXK3A accumulated similarly to or at slightly lower or higher levels than PomXWT (Figure 3B). Most strains had a cell length similar to WT and cell division constrictions at midcell (Figure 3C,D). As expected, pomXK3A cells were similar to ΔpomX cells (but see also details below about mCh-PomXK3A). More importantly, the pomXK13A and pomXR15A mutants generated filamentous cells and minicells, but the filamentous cells were shorter than ΔpomX cells (Figure 3D left panel). pomXK13A and pomXR15A cells had a cell division constriction frequency similar to WT (Figure 3D right panel), but these constrictions were mostly not at midcell (Figure 3C,D right panel). The pomXK13AR15A double mutant had a more pronounced filamentation phenotype than the single mutants, formed minicells, had a constriction frequency similar to WT, and mostly with the constrictions away from midcell (Figure 3C,D). PomXK13AR15A accumulated similarly to PomXWT (Figure 3B). We conclude that Lys13 and Arg15 in PomXNPEP are important for PomX function. Because the PomXK13A, PomXR15A, and PomXK13AR15A variants caused defects distinct from the ΔpomX mutant, i.e., cells are shorter, and with more constrictions, we conclude that substitution of Lys13 and/or Arg15 does not result in a complete loss of PomX function (see Discussion).
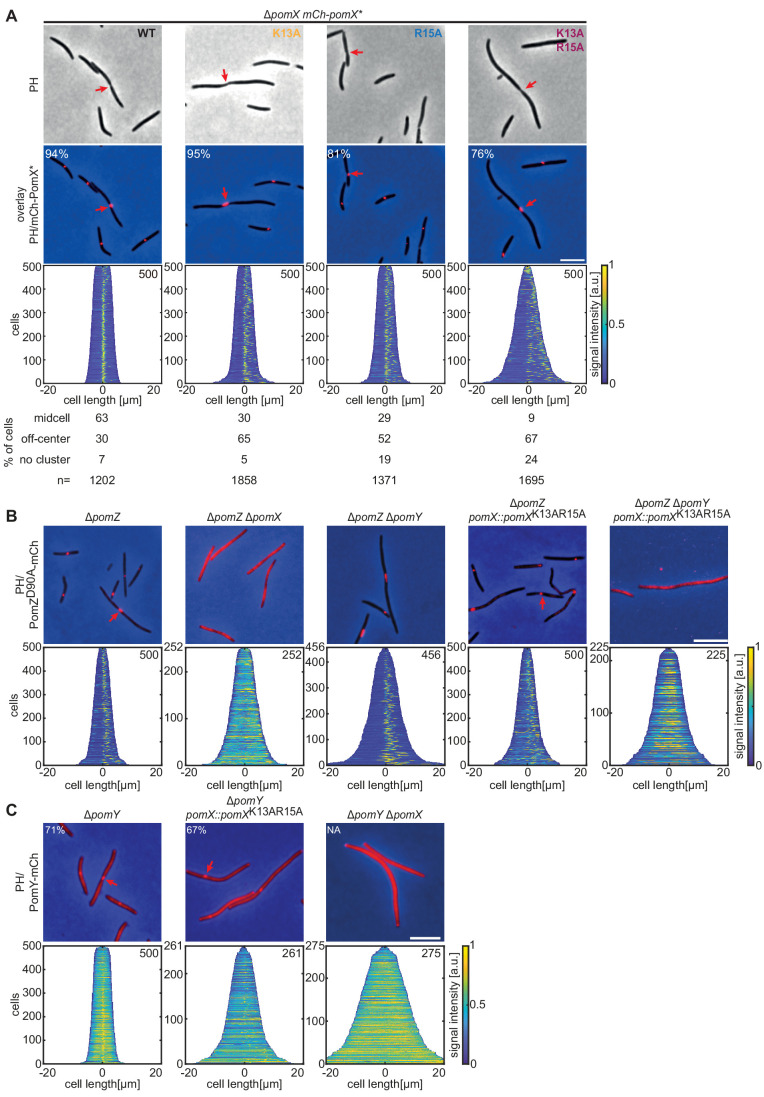
Next, we determined the localization of the PomXNPEP variants with Ala substitutions using mCh fusion proteins ectopically expressed in the ΔpomX mutant. All 11 fusion proteins with a single substitution as well as mCh-PomXK13AR15A accumulated at the same level as mCh-PomXWT in M. xanthus (Figure 4—figure supplement 1A). Most strains including the one expressing mCh-PomXK3A (cell length: 7.9 ± 1.8 µm) had a cell length similar to WT (cell length: 7.7 ± 1.7 µm) and cell division constrictions at midcell (Figure 4—figure supplement 1B) demonstrating that Lys3 is not important for PomX function. More importantly, mCh-PomXK13A, mCh-PomXR15A, and mCh-PomXK13AR15A formed clusters in vivo; however, these were mostly not at midcell and in the case of mCh-PomXK13AR15A ~50% localized in the DNA-free subpolar regions while this localization pattern was not observed for mCh-PomXWT (Figure 4A and Figure 4—figure supplement 1C). The clusters formed by these three mCh-PomX variants, similarly to those of mCh-PomXWT, colocalized with cell division constrictions. Overall, these observations demonstrate that mCh-PomXK13A, mCh-PomXR15A, and mCh-PomXK13AR15A are functional in forming clusters, defining the division site, and stimulating division but cannot correctly position the division site at midcell. All other PomX variants with substitutions in PomXNPEP, including mCh-PomXK3A, localized as mCh-PomXWT (Figure 4—figure supplement 1B).
Figure 4. PomXK13AR15A forms clusters and interacts with PomY but not with PomZ in vivo.
(A-C) Fluorescence microscopy of cells of indicated genotypes. Phase-contrast (PH) images and/or overlays of fluorescence images and PH of representative cells. Red arrows indicate division constrictions. Scale bar, 5 µm. In A, numbers in overlays indicate fraction of cells with a cluster and numbers below indicate localization patterns in % and number of cells analyzed. Demographs are as in Figure 1E. Similar results were observed in two independent experiments.
Figure 4—figure supplement 1. The PomXK13AR15A variant is impaired in function.
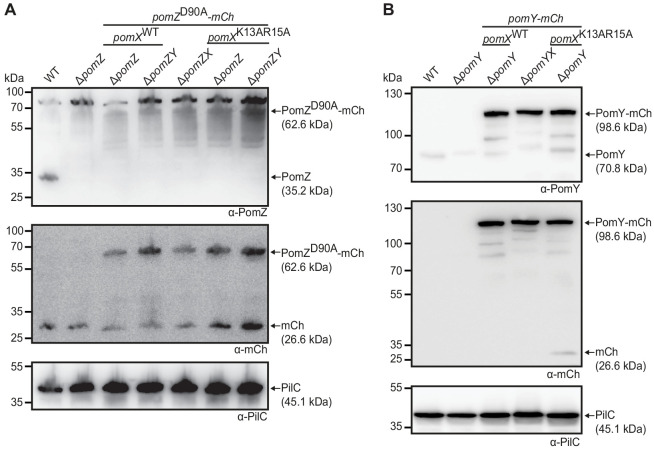
Figure 4—figure supplement 2. Western blot analysis of PomY-mCh and PomZD90A-mCh accumulation.
Because recruitment of the PomX/Y cluster to the nucleoid and PomX/Y/Z cluster localization at midcell depend on PomZ, these observations pointed in the direction that the PomZ and PomX interaction involves Lys13 and Arg15. To test for an interaction between PomZ and PomXNPEP variants in vivo, we explored PomXK13AR15A in more detail and took advantage of the ATP-locked PomZD90A variant. In the presence of PomXWT, PomX/Y/ZD90A clusters are randomly positioned on the nucleoid and rarely at midcell due to lack of PomZ ATPase activity (Schumacher et al., 2017; Figure 4B). We confirmed that in the absence of PomX, PomZD90A-mCh did not form clusters and instead colocalized with the nucleoid, while PomZD90A-mCh still formed clusters in the absence of PomY (Figure 4B and Figure 4—figure supplement 2A). In the presence of PomXK13AR15A, PomZD90A-mCh formed clusters (Figure 4B). Because PomZ also interacts with PomY, we speculated that this cluster incorporation resulted from PomY recruiting PomZD90A-mCh. Indeed, upon additional deletion of pomY, PomZD90A-mCh no longer formed clusters and colocalized with the nucleoid when PomXK13AR15A was the only source of PomX (Figure 4B).
An active PomY-mCh fusion did not form clusters in the absence of PomX but formed clusters in the presence of PomXK13AR15A (Figure 4C and Figure 4—figure supplement 2B). As expected, in the presence of PomXK13AR15A, the PomY-mCh clusters generally localized away from midcell as opposed to clusters in the presence of PomXWT.
Altogether, these observations support that PomXNPEP is required for the interaction between PomX and PomZ but for neither PomX self-interaction nor PomX interaction with PomY. Moreover, they support that the PomX/Y/Z clusters formed in the presence of PomXK13AR15A are proficient in stimulating cell division but deficient in efficiently localizing to midcell, consistent with the PomX/PomZ interaction being perturbed. Finally, they support that PomZ in its ATP-bound dimeric form can be recruited independently by PomX and PomY to the PomX/Y complex.
PomXNPEP is required and sufficient for stimulation of PomZ ATP hydrolysis
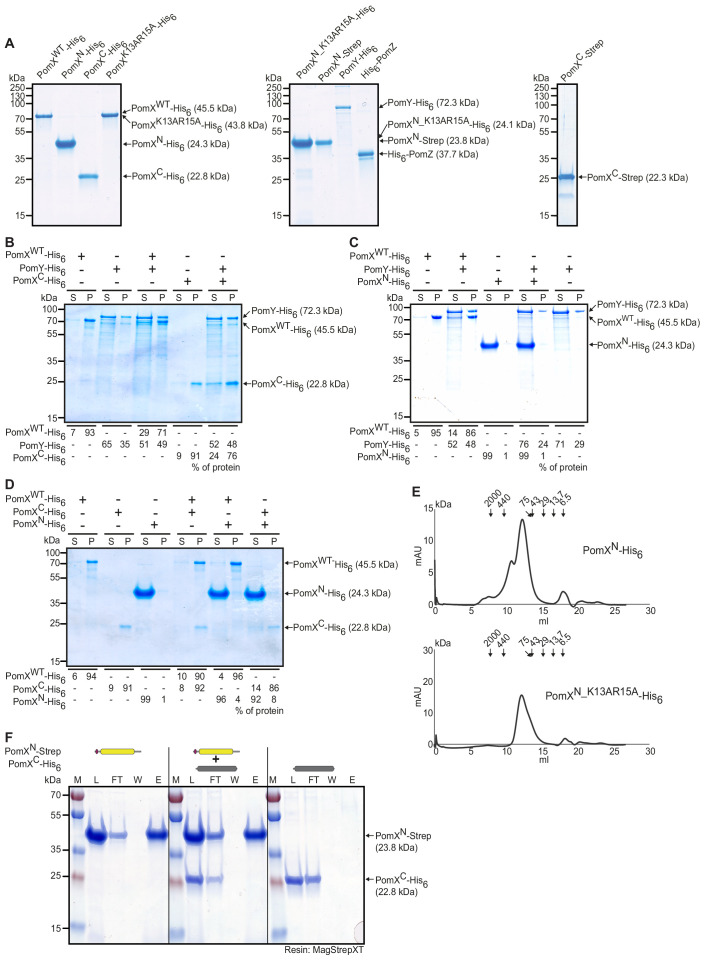
We generated PomX constructs for BACTH analysis that were either truncated for PomXNPEP (PomXΔ2-21 and PomXN_Δ2-21) or contained substitutions in PomXNPEP (PomXK13AR15A and PomXN_K13AR15A). PomXK13AR15A and PomXΔ2-21 self-interacted, and also interacted with PomXWT and PomY (Figure 5A). Consistently, in vitro PomXK13AR15A-His6 formed filaments that were bundled by PomY-His6, accumulated in the pellet fraction after high-speed centrifugation, and brought PomY-His6 to the pellet fraction similarly to PomXWT-His6 (Figure 5B,C). As expected, in the BACTH neither PomXN_ Δ2-21 nor PomXN_K13AR15A interacted with PomXWT and PomY (Figure 5A). Importantly, all four PomXNPEP variants (PomXΔ2-21, PomXN_Δ2-21, PomXK13AR15A and PomXN_K13AR15A) were dramatically reduced in interaction with PomZ and PomZD90A (Figure 5A; see also Figure 2A). Altogether, these findings further support that PomXNPEP is specifically important for the PomX/PomZ interaction and not for PomX/PomX and PomX/PomY interactions.
Figure 5. PomX AAP activity resides in PomXNPEP.
(A) BACTH analysis of interactions between Pom proteins and PomX variants. Experiments were performed in parallel with those in Figure 2A. For presentation purposes, the results for PomXWT and PomXN T25 fusion proteins and their interaction with PomZ and PomZD90A T18 fusion proteins were not included but are included in Figure 2A. Images show representative results and similar results were obtained in three independent experiments. (B) TEM images of negatively stained purified proteins. Experiments were done as in Figure 2B. Scale bar, 200 nm. Images show representative results of several independent experiments. (C) Sedimentation assays with indicated purified proteins. Proteins were analyzed at a concentration of 3 µM alone or in combination. After high-speed centrifugation, proteins in the supernatant (S) and pellet (P) fractions were separated by SDS-PAGE and stained with Instant Blue. Molecular size markers are shown on the left and analyzed proteins on the right including their calculated MW. Numbers below indicate % of proteins in different fractions. Similar results were obtained in two independent experiments. All samples were analyzed on the same gel; the black line indicates that lanes were removed for presentation purposes. (D, E) His6-PomZ ATPase activity. Experiments were done and analyzed as in Figure 2E–I in the presence or absence of DNA and the indicated proteins and peptides. Data points show the mean±STDEV calculated from six independent measurements. In (D), stippled lines indicate the regression of the ADP production rate in the presence of PomXWT-His6 (left, Figure 2G) and PomXN-His6 (right, Figure 2H). In E, the stippled line indicates the regression of the ADP production rate in the presence of PomXWT-His6.
Next, we tested whether PomXNPEP is important for PomX AAP activity. Because His6-tagged truncated PomXNPEP variants could not be overexpressed in E. coli, we focused on the PomXK13AR15A-His6 and PomXN_K13RAR15A-His6 variants (Figure 2—figure supplement 1A). PomXN_K13AR15A-His6 (calculated MW: 24.1 kDa) behaved similarly to PomXN-His6 in SEC and eluted as a single peak corresponding to a globular protein of ~136 kDa (Figure 2—figure supplement 1E). Remarkably, neither PomXK13AR15A-His6 nor PomXN_K13AR15A-His6 stimulated PomZ ATPase activity (Figure 5D). Consequently, we tested whether a peptide consisting of the 22 PomXNPEP residues alone is sufficient to stimulate His6-PomZ ATPase activity. The PomXNPEP peptide alone stimulated His6-PomZ ATPase activity in the presence of DNA (specific activity: 27 ± 5 ATP hr−1 at 15 µM PomXNPEP, 4 µM His6-PomZ) while a PomXNPEP peptide with the K13AR15A substitutions did not (Figure 5E). We conclude that PomXNPEP is required and sufficient for stimulation of PomZ ATPase activity by PomX.
The PomX/PomZ interaction is important for PomX/Y/Z cluster fission during division
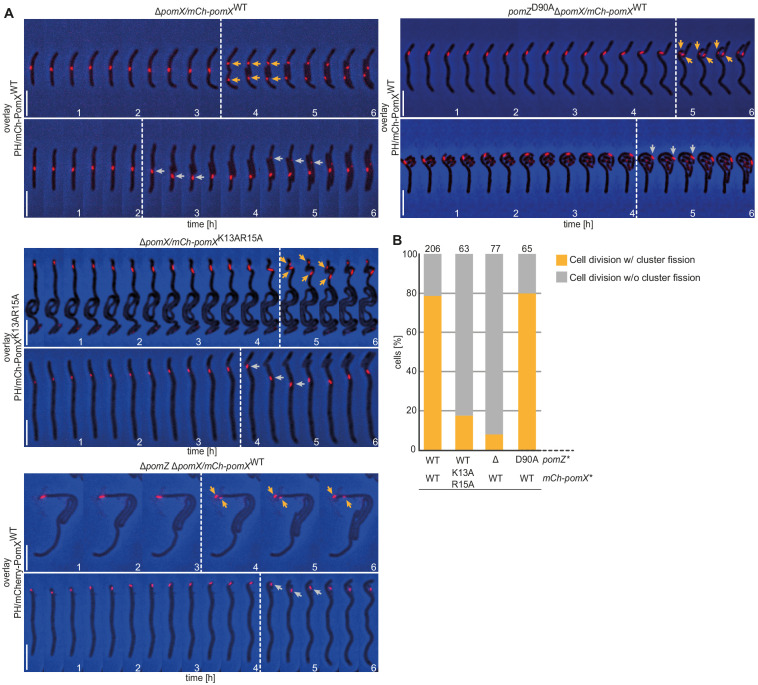
Twenty-four percent of cells containing mCh-PomXK13AR15A lacked a visible cluster compared to only 6% in the presence of mCh-PomXWT (Figure 4A). Tagged and untagged PomXK13AR15A accumulate at the same level as tagged and untagged PomXWT (Figure 3B and Figure 4—figure supplement 1A), suggesting that this difference in cluster formation is not caused by differences in gene expression or protein stability. We, therefore, investigated whether mCh-PomXK13AR15A causes a cluster fission defect during cell division.
In the presence of mCh-PomXWT, ~80% of divisions are accompanied by symmetric or asymmetric cluster fission, with each portion of a divided cluster segregating to a daughter (Figure 6A,B). In the remaining ~20%, cluster splitting did not visibly occur, the undivided cluster segregated to one of the daughters, and ‘empty’ daughter cells eventually regenerated a cluster that was visible after ~2 hr (Figure 6A). mCh-PomXK13AR15A clusters showed the same three patterns during division. However, cluster fission and segregation to daughters occurred in only ~20% of cells (Figure 6A,B).
Figure 6. The PomX/PomZ interaction is important for cluster fission during division.
(A) Fluorescence time-lapse microscopy of mCh-PomX variants in cells of indicated genotypes. Overlays of representative mCh images and PH are shown in 20 min intervals. Stippled lines indicate cell division events. Orange and gray arrows mark mCh-PomX clusters in daughter cells after cell division with cluster fission and without cluster fission, respectively. Scale bar, 5 µm. (B) Quantification of cluster fission during cell division in cells of indicated genotypes. Cell division events were divided into those with (orange) and without (gray) cluster fission. Number of analyzed cell divisions is shown on top. The same results were obtained in two independent experiments.
Because PomXK13AR15A has reduced PomZ AAP activity, we tested cluster fission in ΔpomZ cells and in cells containing PomZD90A. In ΔpomZ cells, division occurs at a reduced frequency but still over the PomX/Y cluster (Schumacher et al., 2017). In these cells, mCh-PomXWT clusters rarely underwent fission and mostly segregated into one daughter (Figure 6A,B). In cells with PomZD90A, divisions also occurred over the cluster, and ~80% of clusters underwent fission during division and segregated to both daughters (Figure 6A,B). Altogether, these observations suggest that the interaction between PomZ and PomX is important for PomX/Y/Z cluster fission, while ATP hydrolysis by PomZ is not.
Discussion
In the present study, we used in vivo and in vitro approaches to functionally dissect the cell division regulatory protein PomX. We demonstrate that PomX is composed of two domains and has three functions in vivo. The N-terminal PomXN domain contains the PomZ AAP activity; the C-terminal PomXC domain is essential for PomX self-interaction, for the interaction with PomY, and functions as a scaffold for PomX/Y/Z cluster formation; and, the PomX/Z interaction, but not PomZ ATPase activity, is important for PomX/Y/Z cluster fission during division.
PomXN, which is Ala/Pro-rich and, therefore, likely unstructured, interacts with PomZ and activates ATPase activity of DNA-bound PomZ in vitro as efficiently as PomXWT. Moreover, a peptide comprising the N-terminal 22 residues of PomX (PomXNPEP) is sufficient to activate PomZ ATPase activity. In PomXNPEP, two positively charged residues (Lys13 and Arg15) are essential for AAP activity. Of note, the AAP activity of PomXNPEP was lower than that of PomXN. Similarly, in the case of the ParB homologs Spo0J of Thermus thermophilus and SopB of plasmid F of E. coli, as well as MinE of Neisseria gonorrhoeae, peptides comprising the N-terminal 20, 52, and 22 residues, respectively are sufficient for AAP activity (Ah-Seng et al., 2009; Ghasriani et al., 2010; Leonard et al., 2005). In the case of the shorter Spo0J and MinE peptides, the AAP activity was also lower than for the full-length proteins (Ghasriani et al., 2010; Leonard et al., 2005), while the longer SopB peptide was as efficient as full-length SopB (Ah-Seng et al., 2009). Because BACTH analyses did not reveal an interaction between PomZ and PomX variants lacking PomXNPEP or containing the K13AR15A substitutions, we speculate that the lower AAP activity of PomXNPEP indicates that the context of PomXNPEP might be important for its AAP activity; however, we cannot rule out that interactions between PomZ and PomXN beyond NPEP may also be important for PomZ ATPase activation.
PomXC with its predicted coiled-coil domain self-interacts and also interacts with PomXWT and PomY. Specifically, PomXC, similarly to PomXWT, formed filaments in vitro that were bundled by PomY. In vivo mCh-tagged PomXC integrated into clusters containing PomXWT but alone was not sufficient to form a cluster. By contrast, PomXN did not form or integrate into a cluster under any conditions tested; moreover, PomXN interacted with neither PomXWT nor PomY in BACTH or in vitro. Because PomXWT alone can form a cluster and is essential for PomX/Y/Z cluster formation in vivo, these observations support a model whereby PomXWT serves as a scaffold protein for cluster formation in vivo and in which PomXC has a key role in this scaffolding function by self-interacting and interacting with PomY. The observation that neither PomXN nor PomXC alone is sufficient to form a cluster in vivo suggests that these two domains may also interact. Our experiments suggest that these interactions are of low affinity because there were not detected by any of the methods used here (BACTH, TEM, high-speed centrifugation, and pull-down experiments). Alternatively, the conformation of the two separated domains could be different from that in PomXWT, and, therefore, no interactions were detected. Altogether, we suggest that PomXWT monomers in vivo interact via their coiled-coil domain in PomXC to spontaneously form a polymeric structure that is stabilized or modified by PomXN; this polymeric structure, in turn, serves as a scaffold to recruit PomY resulting in formation of the PomX/Y complex. PomZ in its ATP-bound dimeric form is recruited to this PomX/Y complex and associates it to the nucleoid. Because PomXWT and the likely unstructured PomXN stimulate PomZ ATPase activity to the same extent, these observations also strongly support that PomXWT monomers in the polymeric structure in vitro and cluster in vivo stimulate PomZ ATPase activity independently of each other. In vitro, purified PomXWT spontaneously polymerizes under all conditions tested. We speculate that this spontaneous polymerization ensures that one, and only one, PomX scaffold is formed per cell in vivo, thus guaranteeing that only one PomX/Y/Z complex is formed per cell as would be required for a complex that defines the site of cell division.
By analyzing PomZ variants, we previously showed that PomZ ATPase activity is essential for PomX/Y/Z cluster translocation and cluster localization to midcell. Because the PomX variants with reduced AAP activity resulted in the formation of PomX/Y/Z clusters that were typically not at midcell, we conclude that the low intrinsic PomZ ATPase activity is not sufficient to fuel translocation of the PomX/Y/Z cluster to midcell and that this translocation is fueled by PomX, and likely also by PomY, stimulated ATP hydrolysis by PomZ. In the PomX AAP mutants, constrictions were formed at WT frequencies over the PomX/Y/Z cluster. Because these clusters were typically not at midcell, these PomX variants resulted in divisions away from midcell giving rise to filamentous cells and minicells. Thus, PomX AAP mutants are fully competent in stimulating cell division but deficient in positioning the PomX/Y/Z cluster at midcell. These observations also support that PomX-stimulated PomZ ATPase activity is not important for stimulation of Z-ring formation. In cells lacking the PomX protein, PomY and PomZ do not form clusters and cell divisions occur at a low frequency (Schumacher et al., 2017); by contrast, the PomX AAP mutants still support PomX/Y/Z cluster formation and cell division. Thus, PomX AAP mutants retain partial PomX activity. Importantly, in mutants lacking PomZ, the PomX/Y cluster is also mostly away from midcell, and Z-rings and constrictions are formed over the cluster away from midcell, but at a much-reduced frequency compared to WT. Thus, PomX AAP mutants and a mutant lacking PomZ phenocopy each other with respect to PomX/Y/Z cluster positioning but not with respect to constriction frequency. We suggest that this difference is the result of different cluster compositions, that is the Pom clusters formed in the AAP mutants contain PomZ as well as PomY, which both interact with FtsZ (Schumacher et al., 2017), while the Pom clusters formed in the absence of PomZ only contain PomX and PomY. Thus, the division defects in Δpom mutants are a consequence of reduced formation and mislocalization of division constrictions, while the PomX AAP mutants are only deficient in cell division localization.
DNA binding by PomZ is important for the low intrinsic as well as PomX-stimulated ATPase activity. This is similar to what has been described for several DNA-binding ParA ATPases and their partner AAPs (Ah-Seng et al., 2009; Kiekebusch et al., 2012; Leonard et al., 2005; Lim et al., 2014; Scholefield et al., 2011; Schumacher et al., 2017). Similarly, MinE-dependent stimulation of ATP hydrolysis by a MinD variant that does not bind the membrane is reduced (Hu and Lutkenhaus, 2003). However, it remains unknown how DNA or membrane binding makes these ATPases competent for ATPase activity. Likewise, the precise molecular mechanism by which AAPs stimulate ATPase activity of their cognate ParA/MinD family ATPase remains unknown. However, in the case of the E. coli MinD/MinE system, it has been suggested that MinD ATPase activation by MinE involves the asymmetric interaction of the N-terminus of a MinE monomer with the ATP-bound MinD dimer (Park et al., 2012). Our data support that activation of many ParA/MinD family ATPases by their partner AAP may involve a shared mechanism: In agreement with the findings here that PomXNPEP is enriched in positively charged residues and that Lys13 and Arg15 are essential for PomX AAP activity, it has previously been noted that ParB-type AAPs, MinE-type AAPs and the non-homologous AAP ParG of plasmid TP228 that functions together with the ParA ATPase ParF contain a stretch of N-terminal residues rich in positively charged amino acids (Barillà et al., 2007; Leonard et al., 2005; Ah-Seng et al., 2009; Ghasriani et al., 2010; Figure 7). For several of these AAPs, it has been shown that one or more of the positively charged residues are important for AAP activity (Figure 7; Leonard et al., 2005; Scholefield et al., 2011; Barillà et al., 2007; Ah-Seng et al., 2009; Park et al., 2012; Hu and Lutkenhaus, 2001; Ghasriani et al., 2010). Also, as noted above, in the case of Spo0J of T. thermophilus, plasmid F SopB, and MinE of N. gonorrhoeae, 20, 52, and 22 N-terminal residues are sufficient for AAP activity. Thus, PomX is the fourth type of ParA/MinD AAP that displays this characteristic N-terminus and in which positively charged residues are important for AAP activity. Intriguingly, TlpT, which is the suggested AAP of the ParA-like ATPase PpfA involved in translocation and positioning of the large cytoplasmic chemoreceptor cluster in Rhodobacter sphaeroides (Roberts et al., 2012), and McdB, which is the AAP of the ParA-like ATPase McdA important for carboxysome translocation and positioning in Synechococcus elongatus (MacCready et al., 2018) both have an N-terminal extension enriched in positively charged residues (Figure 7). Altogether, these findings lend further support to the notion that many AAPs of ParA/MinD ATPases use the same mechanism to stimulate ATPase activity (Leonard et al., 2005; Park et al., 2011; Zhang and Schumacher, 2017; Barillà et al., 2007). Moreover, they support that ParA/MinD AAPs display remarkable plasticity and modularity in which a stretch of N-terminal amino acids enriched in positively charged residues grafted onto an interaction domain, which is involved in protein-protein, protein-DNA, or protein-membrane interaction, can generate an AAP. Interestingly, PomY, the second AAP of PomZ (Schumacher et al., 2017), does not have an N-terminus enriched in positively charged residues suggesting that its mode of action could be different from that of previously described AAPs.
Figure 7. AAPs of MinD/ParA ATPases are diverse but share common features.
Left, domain analysis of known and predicted AAPs of ParA/MinD ATPases with key below. Sequences on the right, N-terminus of indicated proteins. Positively charged amino acids are indicated on blue, and positively charged residues experimentally demonstrated to be important for AAP activity on red. Underlined sequences indicate peptides experimentally demonstrated to have AAP activity. Spo0J of T. thermophilus and B. subtilis, and SopB of plasmid F are ParB homologs.
In addition to similarities at the biochemical level, systems incorporating a DNA-binding ParA ATPase and an AAP(s) also share similarities in their translocation. The PomX/Y complex, ParB-parS complexes, cytoplasmic chemoreceptor clusters and carboxysomes are all large structures that are translocated as cargo on the nucleoid in a mechanism that depends on an AAP(s) stimulating ATP hydrolysis by the cognate DNA-binding ParA ATPase. Importantly, the AAP(s) is an integral part of the translocated cargo. We speculate that the integration of the AAP activity into a large cargo structure provides an elegant solution to guarantee that ATPase activation is spatially restricted to the transported cargo.
In addition to serving as a scaffold for PomX/Y/Z cluster formation and as a PomZ AAP, we report that PomX has a third important function in PomX/Y/Z cluster fission during division. In the presence of PomXWT, the cluster visibly undergoes fission during ~80% of divisions in otherwise WT cells, and those cells that do not receive a cluster slowly rebuild a cluster [here; (Schumacher et al., 2017)]. As a result, most WT cells contain a visible PomX/Y/Z cluster. By contrast, a PomX AAP mutant did not support fission as efficiently as PomXWT and fewer cells contain a cluster. We speculate that the reduced frequency of cluster fission events contributes to the cell division defect in the PomX AAP mutants. In cells lacking PomZ, the PomX/Y cluster also does not split efficiently during division, while the ATP-locked dimeric PomZD90A stimulates fission as efficiently as PomZWT. In all these mutants, division occurs over the Pom cluster. Additionally, in cells treated with cephalexin, which blocks cell division, cluster fission also does not occur (Schumacher et al., 2017; Treuner-Lange et al., 2013). Because cephalexin-treated cells still segregate their chromosomes (Schumacher et al., 2017; Treuner-Lange et al., 2013), these observations support that cell division is essential for fission, chromosome segregation is not sufficient, and the PomX/Z interaction is important while PomZ ATPase activity is not. This is in contrast to the SopB/SopA system for plasmid F segregation. For this system, it has been shown that SopB-stimulated ATPase activity of SopA is important to split plasmid clusters (Ah-Seng et al., 2013) suggesting that the splitting of plasmid clusters and PomX/Y/Z cluster fission may occur by different mechanisms. In the future, it will be interesting to establish the mechanism for PomX/Y/Z cluster fission in detail.
Materials and methods
M. xanthus and E. coli strains and growth
Strains, plasmids, and primers are listed in the Key Resources Table. M. xanthus strains are derivatives of DK1622 WT (Kaiser, 1979). M. xanthus strains were cultivated in 1% CTT medium (1% casitone, 10 mM Tris-HCl pH 7.6, 1 mM KPO4 pH 7.6, 8 mM MgSO4) or on 1% CTT 1.5% agar plates (Hodgkin and Kaiser, 1977). Kanamycin, oxytetracycline, and gentamycin were added at concentrations of 50 µg/ml, 10 µg/ml, and 10 µg/ml, respectively. Growth was measured as an increase in optical density (OD) at 550 nm. M. xanthus cells were transformed by electroporation. In-frame deletions were generated as described (Shi et al., 2008). Plasmids were integrated by site-specific recombination at the Mx8 attB locus or by homologous recombination at the endogenous site. All plasmids were verified by sequencing. All strains were verified by PCR. Non-motile strains were generated to allow time-lapse microscopy for several hours by deletion of mglA (Miertzschke et al., 2011; Schumacher and Søgaard-Andersen, 2018). E. coli strains were grown in LB or 2xYT medium in the presence of relevant antibiotics or on LB plates containing 1.5% agar (Sambrook and Russell, 2001). Plasmids were propagated in E. coli NEB Turbo cells (New England Biolabs) (F' proA+B+ lacIq ∆lacZM15/fhuA2 ∆(lac-proAB) glnV galK16 galE15 R(zgb-210::Tn10) TetS endA1 thi-1 ∆(hsdS-mcrB)5). Growth of E. coli was measured as an increase in OD at 600 nm.
Fluorescence microscopy and live cell imaging
Fluorescence microscopy was performed as described (Schumacher et al., 2017). Briefly, exponentially growing cells were transferred to slides with a thin 1.0% agarose pad (SeaKem LE agarose, Cambrex) with TPM buffer (10 mM Tris-HCl pH 7.6, 1 mM KH2PO4 pH 7.6, 8 mM MgSO4), covered with a coverslip and imaged using a temperature-controlled Leica DMi6000B inverted microscope with an HCX PL FLUOTAR objective at 32°C. Phase-contrast and fluorescence images were recorded with a Hamamatsu ORCA-flash 4.0 sCMOS camera using the LASX software (Leica Microsystems). Time-lapse microscopy was performed as described (Schumacher and Søgaard-Andersen, 2018). Briefly, cells were transferred to a coverslip mounted on a metallic microscopy slide and covered with a pre-warmed 1% agarose pad supplemented with 0.2% casitone in TPM buffer. Slides were covered with parafilm to retain humidity of the agarose. Live-cell imaging was performed at 32°C. For DNA staining, cells were incubated with 1 mg/ml 2-(4-Amidinophenyl)−6-indolcarbamidine-dihydrochloride (DAPI) for 10 min at 32°C before microscopy. Image processing was performed with Metamorph_v 7.5 (Molecular Devices). For image analysis, cellular outlines were obtained from phase-contrast images using Oufti and manually corrected if necessary (Paintdakhi et al., 2016). Fluorescence microscopy image analysis was performed with a custom-made Matlab script (Matlab R2018a, MathWorks) available at https://github.com/SBergeler/ImageAnalysisMyxo copy archived at 500 cells from a dataset (unless otherwise stated) and calculating the length of the cells' centerlines (using meshes obtained from Oufti). Next, the fluorescence intensity of each cell was corrected by the background fluorescence locally around each cell. Cells were ordered by length and oriented according to the cell segment with the brightest intensity. The brightest 3% of the intensity values of all cells were set to the maximal intensity value, which is scaled to one, to be able to visualize fluorescence intensity variations inside the cells despite very bright clusters.
Bacterial two hybrid assay
BACTH experiments were performed as described (Karimova et al., 1998). Relevant genes were cloned into the appropriate vectors to construct N-terminal and C-terminal fusions with the 25 kDa N-terminal or the 18 kDa C-terminal adenylate cyclase fragments of B. pertussis. Restoration of cAMP production was observed by the formation of blue color on LB agar supplemented with 80 µg/ml 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) and 0.25 mM isopropyl-β-D-thiogalactoside (IPTG). As positive control, the leucine zipper from GCN4 was fused to the T18 and the T25 fragment. As a negative control, plasmids were co-transformed that only expressed the T18 or T25 fragment. All tested interactions were spotted on the same LB agar plates with positive control and all corresponding negative controls.
Western blot analysis
Western blot analyses were performed as described (Sambrook and Russell, 2001) with rabbit polyclonal α-PomX (1:15000), α-PomY (1:15000), α-PomZ (1:10000) (Schumacher et al., 2017), α-PilC (1:3000) (Bulyha et al., 2009), or α-mCh (1:10000; Biovision) primary antibodies together with horseradish-conjugated goat α-rabbit immunoglobulin G (Sigma-Aldrich) as secondary antibody (1:25000). Blots were developed using Luminata Forte Western HRP Substrate (Millipore) and visualized using a LAS-4000 luminescent image analyzer (Fujifilm).
Protein purification
His6-PomZ was purified from E. coli as described (Treuner-Lange et al., 2013). PomXWT-His6 and PomY-His6 were purified as described (Schumacher et al., 2017) using plasmids pEMR3 and pEMR1, respectively. PomXK13AR15A-His6 was purified as PomX-His6. Briefly, plasmid pSH58 was propagated in E. coli NiCo21(DE3) cells (NEB), grown in LB medium with 50 µg/ml kanamycin at 30°C to an OD600 of 0.6–0.7. Protein expression was induced with 0.4 mM IPTG for 16 hr at 18°C. Cells were harvested by centrifugation at 6000 g for 20 min at 4°C. Cells were washed with lysis buffer 1 (50mM NaH2PO4; 300 mM NaCl; 10 mM imidazole; pH 8.0 (adjusted with NaOH)) and lysed in 50 ml lysis buffer 2 (lysis buffer 1 with 0.1 mM EDTA; 1 mM β-mercaptoethanol; 100 mg/ml phenylmethylsulfonyl fluoride (PMSF); 1× complete protease inhibitor (Roche Diagnostics GmbH); 10 U/ml DNase 1) by sonication in three rounds of sonication for 5 min with a Branson Sonifier (Duty cycle 4; output control 40%) (Heinemann) on ice. Cell debris was removed by centrifugation at 4700 g for 45 min at 4°C. PomXK13AR15A-His6 was affinity purified with Protino Ni-NTA resin (Macherey-Nagel) from a batch, equilibrated in lysis buffer 1. PomXK13AR15A-His6 was eluted from the resin by washing 1× with 5 ml elution buffer 1 (lysis buffer 1 with 50 mM imidazole) and 3 x with 5 ml elution buffer 2 (lysis buffer 1 with 250 mM imidazole). Purified PomXK13AR15A-His6 was dialyzed 4× against 2 l dialysis buffer (50 mM Hepes/NaOH pH 7.2; 50 mM KCl; 0.1 mM EDTA; 1 mM β-mercaptoethanol; 10% (v/v) glycerol). Proteins were frozen in liquid nitrogen and stored at −80°C until used.
To purify PomXN-His6 plasmid pAH157 was propagated in E. coli NiCo21(DE3) cells, grown in 2×YT medium with 50 µg/ml kanamycin and 0.5% glucose at 30°C to an OD600 of 0.6–0.7. Protein expression was induced with 0.4 mM IPTG for 16 hr at 18°C. Cells were harvested and lysed as described for PomXK13AR15A. PomXN-His6 was affinity purified with Protino Ni-NTA resin (Macherey-Nagel) from a batch, equilibrated in lysis buffer 1. Contaminating proteins were eluted from the resin by washing 6× with 40 ml wash buffer 1 (lysis buffer 1 with 20 mM imidazole), 1× with 40 ml wash buffer 2 (lysis buffer 1 with 50 mM imidazole). PomXN-His6 was eluted from the resin by washing with 1 × 10 ml elution buffer 1 (lysis buffer 1 with 100 mM imidazole), 1× with 10 ml elution buffer 2 (lysis buffer 1 with 150 mM imidazole) and 1× with 10 ml elution buffer 3 (lysis buffer 1 with 200 mM imidazole). The elution fractions were pooled and loaded onto a HiLoad 16/600 Superdex 200 pg gel filtration column (GE Healthcare) that was equilibrated with dialysis buffer. Elution fractions were pooled. Proteins were frozen in liquid nitrogen and stored at −80°C until used.
For purification of PomXN_K13AR15A-His6 plasmid pAH165 was propagated in E. coli Rosetta2(DE3) cells, grown in 2×YT medium with 50 µg/ml kanamycin, 30 µg/ml chloramphenicol and 0.5% glucose at 30°C to an OD600 of 0.6–0.7. Protein expression was induced with 0.5 mM IPTG for 16 hr at 18°C. Cells were harvested and lysed as described for PomXK13AR15A. PomXN_K13AR15A-His6 was purified from cleared lysates with a 5 ml HiTrap Chelating HP column loaded with NiSO4 and equilibrated with lysis buffer 1. The column was washed with 20 column volumes (CVs) lysis buffer 1. The protein was eluted with elution buffer (50 mM NaH2PO4; 300 mM NaCl; 500 mM imidazole; pH 8.0 [adjusted with NaOH]) with a gradient of 20 CV. Fractions containing PomXN_K13AR15A-His6 were pooled and concentrated with an Amicon Ultra-15 centrifugation filter device with a cutoff of 3 kDa and loaded onto a HiLoad 16/600 Superdex 200 pg gel filtration column (GE Healthcare) that was equilibrated with dialysis buffer. Elution fractions were pooled. Proteins were frozen in liquid nitrogen and stored at −80°C until used.
To purify PomXC-His6 plasmid pAH152 was propagated in E. coli Rosetta2(DE3) cells, grown in 2×YT medium with 50 µg/ml kanamycin, 30 µg/ml chloramphenicol and 0.5% glucose at 37°C to an OD600 of 0.6–0.7. Protein expression was induced with 0.5 mM IPTG for 4 hr at 37°C. Cells were harvested and lysed as described for PomXK13AR15A. PomXC-His6 was affinity purified with Protino Ni-NTA resin (Macherey-Nagel) from a batch, equilibrated with lysis buffer 1. Contaminating proteins were eluted from the resin by washing 6× with 40 ml wash buffer 1 (lysis buffer 1 with 20 mM imidazole), 1× with 40 ml wash buffer 2 (lysis buffer 1 with 50 mM imidazole) and 2× with 40 ml wash buffer 3 (lysis buffer 1 with 100 mM imidazole) and 1× with 40 ml wash buffer 4 (lysis buffer 1 with 150 mM imidazole). PomXC-His6 was eluted from the resin with 3 × 10 ml elution buffer (lysis buffer 1 with 250 mM imidazole). The elution fractions were pooled and dialyzed against 4 × 2 l of dialysis buffer at 4°C. Proteins were frozen in liquid nitrogen and stored at −80°C until used.
For purification of PomXN-Strep, plasmid pDS232 was propagated in E. coli Rosetta2(DE3) cells, grown in LB medium with 50 µg/ml kanamycin, 30 µg/ml chloramphenicol and 0.5% glucose at 32°C to an OD600 of 0.6–0.7. Protein expression was induced with 0.5 mM IPTG for 2 hr at 32°C. Cells were harvested and lysed as described for PomXK13AR15A-His6 but in StrepTag lysis buffer (100 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT)). PomXN-Strep was purified from cleared lysates with a 5 ml StrepTrap HP column equilibrated with StrepTag lysis buffer. The column was washed with 20 CV StrepTag lysis buffer. The protein was eluted with StrepTag elution buffer (StrepTag lysis buffer with 2.5 mM D-desthiobiotin). Fractions containing PomXN-Strep were pooled and dialyzed against 2 × 3 l of dialysis buffer at 4°C. Proteins were frozen in liquid nitrogen and stored at −80°C until used.
For purification of PomXC-Strep, plasmid pDS333 was propagated in E. coli Rosetta2(DE3) cells, grown in LB medium with 50 µg/ml kanamycin, 30 µg/ml chloramphenicol and 0.5% glucose at 32°C to an OD600 of 0.6–0.7. Protein expression was induced with 1 mM IPTG for 18 hr at 18°C. Cells were harvested and lysed as described for PomXN-Strep. PomXC-Strep was purified from a batch using 2 ml Strep-TactinXT 4Flow resin (iba), equilibrated with StrepTag lysis buffer. The resin was incubated with the cleared lysate for 1 hr at 4°C on a rotary shaker. Contaminating proteins were eluted from the resin by washing 5× with 10 ml StrepTag lysis buffer. The protein was eluted with 1× BXT buffer (100 mM Tris-HCl pH8.0, 150 mM NaCl, 1 mM EDTA, 50 mM biotin) (iba). Fractions containing PomXC-Strep were pooled and dialyzed against 2 × 5 l of dialysis buffer at 4°C. Proteins were frozen in liquid nitrogen and stored at −80°C until used.
Protein sedimentation assay
Before sedimentation experiments, a clearing spin was performed for all proteins to be analyzed at 20,000 g for 10 min at 4°C. Proteins at a final concentration of 3 µM in a total volume of 50 µl were mixed and incubated for 1 hr at 32°C in a buffer (50 mM Hepes/NaOH, pH 7.2, 50 mM KCl, 1 mM β-mercaptoethanol, 10 mM MgCl2). Samples were separated into soluble and insoluble fractions by high-speed centrifugation (160,000 g, 60 min, 25°C). Insoluble and soluble fractions were separated, and volumes adjusted with 1× SDS sample buffer. Fractions were separated by SDS-PAGE and stained with Instant Blue (expedion) for 10 min.
In vitro pull-down experiments
Ten µM protein alone or pre-mixed as indicated were incubated for 1 hr at 32°C in reaction buffer (50 mM HEPES/NaOH pH 7.2, 50 mM KCl, 10 mM MgCl2) in a total volume of 200 µl and applied to 20 µl 5% (v/v) MagStrepXT beads (iba) for 30 min. Magnetic beads were washed 10× with 200 µl reaction buffer. Proteins were eluted with 200 µl 1× BXT buffer (100 mM Tris-HCl pH8.0, 150 mM NaCl, 1 mM EDTA, 50 mM biotin) (iba). 10 µl per sample were separated by SDS-PAGE.
Negative stain transmission electron microscopy
To fix and stain protein samples for negative stain TEM, 10 µl of a protein sample of interest (protein concentration before application onto the EM grid 3 µM) was applied onto an EM grid (Plano) and incubated for 1 min at 25°C. Residual liquid was blotted through the grid by applying the grid's unused side on Whatman paper. The grid was washed twice with double-distilled H2O. For staining, 10 µl of 1% uranyl acetate solution was applied onto the grid for 1 min and blotted through with a Whatman paper. If protein mixtures were applied to the EM grid, proteins of interest at a concentration of 3 µM were pre-mixed in a low-binding microtube (Sarstedt) and incubated for 10 min at 25°C before application onto the EM grid. Finished grids were stored in a grid holder for several months at room temperature. Electron microscopy was performed with a CM120 electron microscope (FEI) at 120kV.
ATPase assay
ATP hydrolysis was determined using a 96-well NADH-coupled enzymatic assay (Kiianitsa et al., 2003) with modifications. Protein concentration was determined using Protein Assay Dye Reagent Concentrate (BioRad). Assays were performed in reaction buffer (50 mM HEPES/NaOH pH 7.2, 50 mM KCl, 10 mM MgCl2) with 0.5 mM nicotinamide adenine dinucleotide (NADH) and 2 mM phosphoenolpyruvate and 3 µl of a pyruvate kinase/lactate dehydrogenase mix (PYK/LDH; Sigma). PomXNPEP and PomXNPEP_K13AR15A peptides (MKKAFEQNVSRAKPRLRLGALT and MKKAFEQNVSRAAPALRLGALT) were purchased from Thermo Scientific. If appropriate, herring sperm DNA was added at a concentration of 60 µg/ml unless otherwise stated. Buffer was pre-mixed with proteins in low-binding microtubes (Sarstedt) on ice. To correct for glycerol in the assays, dialysis buffer was added if necessary. A total of 100 µl mixtures were transferred into transparent UV-STAR µCLEAR 96-well microplates (Greiner bio-one). The reaction was started by the addition of 1 mM ATP. Measurements were performed in an infinite M200PRO (Tecan) for 2 hr in 30 s intervals at 32°C shaking at 340 nm wavelength. To account for background by spontaneous ATP hydrolysis and UV-induced NADH decomposition, all assays were performed without the addition of His6-PomZ and measurements were subtracted. The light path was determined experimentally with known NADH concentrations to be 0.248 cm. The extinction coefficient of NADH ε340 = 6220 M−1cm−1 was used.
Analytical size-exclusion chromatography
Experiments were carried out in dialysis buffer (50 mM Hepes/NaOH pH 7.2; 50 mM KCl; 0.1 mM EDTA; 1 mM β-mercaptoethanol; 10% (v/v) glycerol). PomXN-His6 and PomXN_K13AR15A-His6 were applied onto a Superdex 200 10/300 GL gel filtration column equilibrated with dialysis buffer. Blue dextran (2000 kDa), ferritin (440 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), RNAse A (13.7 kDa), and aprotinin (6.5 kDa) were used as standards with the same buffer conditions to calibrate the column.
Bioinformatics
Gene and protein sequences of PomX, PomY, and PomZ were obtained from NCBI. PomX homologs were identified in a best-best hit reciprocal BlastP analysis from fully-sequenced genomes of Myxobacteria (Altschul et al., 1990). The similarity and identity of proteins were calculated from pairwise sequence alignments with EMBOSS Needle (Madeira et al., 2019). Domain analyses were performed with SMART (Letunic and Bork, 2018), PROSITE and Pfam (El-Gebali et al., 2019). Multiple sequence alignments were created with MUSCLE (Madeira et al., 2019) and further edited with Bioedit (https://bioedit.software.informer.com/7.2/). Consensus sequences of multiple sequence protein alignments were created with Weblogo 3 (Crooks et al., 2004). Proteins used: PomXMx (ABF89666; MXAN_0636), PomXMm (ATB45064; MYMAC_000648), PomXMh (AKQ69458; A176_006370), PomXMf (AKF79435; MFUL124B02_03625), PomXMs (AGC41991; MYSTI_00641), PomXCc (AFE03552; COCOR_00544), PomXMb (ATB30647; MEBOL_004108), PomXCf (ATB35470; CYFUS_000883), PomXAg (AKI99966; AA314_01593), PomXSa (ADO75429; STAUR_7674), PomXVi (AKU92216; AKJ08_2603), PomXAd (ABC83600; Adeh_3834). Spo0J Tt (AAS81946.1), Spo0J Bs (P26497.2) ParG TP228 Ec (WP_139578510.1), SopB F Ec (BAA97917.1), MinE Ng (AAK30127.1), MinE Ec (EFB7450413.1), TlpT Rs (ABA79218.1), McdB Se (ABB57864.1).
Statistics
The mean and standard deviation (STDEV) were calculated with Excel 2016. Localization patterns from fluorescence microscopy data were quantified based on the indicated n-value per strain. Boxplots were generated with SigmaPlot 14.0 (Systat). Statistical analysis was performed with SigmaPlot 14.0. All data sets were tested for normality using a Shapiro-Wilk test. For data with a non-normal distribution, a Mann-Whitney test was applied to test for significant differences.
Acknowledgements
We thank Sabrina Huneke-Vogt for assistance with plasmid constructions, Manon Wigbers for help with the image analysis script, and Anke Treuner-Lange for many helpful discussions.
Appendix 1
Appendix 1—key resources table.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. xanthus) | pomX | NCBI |
mxan_0636
new locus tag MXAN_RS03090 |
|
| Gene (M. xanthus) | pomY | NCBI |
mxan_0634
new locus tag MXAN_RS03080 |
|
| Gene (M. xanthus) | pomZ | NCBI |
mxan_0635
new locus tag MXAN_RS03085 |
|
| Strain, strain background (E. coli) | Arctic Express DE3 RP | Agilent Technologies | E. coli B F– ompT hsdS(rB– mB–) dcm+TetR gal λ(DE3) endA Hte [cpn10 cpn60 GentR] | Used for protein expression |
| Strain, strain background (E. coli) | Rosetta 2 DE3 | Novagen | F- ompT hsdSB(rB- mB-) gal dcm (DE3) pRARE2 (CamR) | Used for protein expression |
| Strain, strain background (E. coli) | NiCo21(DE3) | New England Biolabs | can::CBD fhuA2 [lon] ompT gal (λ DE3) [dcm] arnA::CBD slyD::CBD glmS6Ala ∆hsdS λ DE3 = λ sBamHIo ∆EcoRI-B int::(lacI::PlacUV5::T7 gene1) i21 ∆nin5 | Used for protein expression |
| Strain, strain background (E. coli) | NEB Turbo | New England Biolabs | F' proA+B+ lacIq(∆lacZM15/fhuA2 ∆(lac-proAB) glnV galK16 galE15 R(zgb-210::Tn10) TetS endA1 thi-1 ∆(hsdS-mcrB)5) | Used for cloning |
| Strain, strain background (M. xanthus) | DK1622 | DOI: 10.1073/pnas.76.11.5952 | Wildtype | |
| Strain, strain background (M. xanthus) | SA3108 | DOI: 10.1111/mmi.12094 | ΔpomZ | Strain with an in-frame deletion in pomZ |
| Strain, strain background (M. xanthus) | SA3146 | DOI: 10.1111/mmi.12094 | ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a ΔpomZ background |
| Strain, strain background (M. xanthus) | SA4223 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomX | Strain with an in-frame deletion in pomX |
| Strain, strain background (M. xanthus) | SA4252 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a ΔpomX background |
| Strain, strain background (M. xanthus) | SA4703 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomY | Strain with an in-frame deletion in pomY |
| Strain, strain background (M. xanthus) | SA4712 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔpomY; attB::PpilA pomY-mCh (pDS7) | Strain expressing PomY-mCh in a ΔpomY background |
| Strain, strain background (M. xanthus) | SA4797 | https://doi.org/10.1016/j.devcel.2017.04.011 | ΔmglA; ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a non-motile ΔpomX background |
| Strain, strain background (M. xanthus) | SA4297 | this study | Wild-type; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in WT background |
| Strain, strain background (M. xanthus) | SA6100 | this study | pomX::pomXK13AR15A | Strain with a replacement of pomX with the pomXK13AR15A allele |
| Strain, strain background (M. xanthus) | SA7014 | this study | ΔpomX; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomX and pomZ deletion background |
| Strain, strain background (M. xanthus) | SA7061 | this study | ΔmglA; ΔpomZ; ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a non-motile pomX and pomZ deletion background |
| Strain, strain background (M. xanthus) | SA7063 | this study | ΔpomZ; ΔpomX; attB::Pmxan0635 mCh-pomX (pAH53) | Strain expressing mCh-PomX in a pomX and pomZ deletion background |
| Strain, strain background (M. xanthus) | SA8240 | this study | pomX::pomXK13AR15A; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomZ deletion background with pomXK13AR15A mutation. |
| Strain, strain background (M. xanthus) | SA8250 | this study | pomX::pomXK13AR15A; ΔpomY; attB::PpilA pomY-mCh (pDS7) | Strain expressing PomZD90A-mCh in a pomXK13AR15A background |
| Strain, strain background (M. xanthus) | SA8268 | this study | pomX::pomXK13AR15A; ΔpomY; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomZ and pomY deletion background with pomXK13AR15A mutation |
| Strain, strain background (M. xanthus) | SA9700 | this study | pomX::pomXE6A | Strain with a replacement of pomX with the pomXE6A allele |
| Strain, strain background (M. xanthus) | SA9701 | this study | pomX::pomXQ7A | Strain with a replacement of pomX with the pomXQ7A allele |
| Strain, strain background (M. xanthus) | SA9702 | this study | pomX::pomXN8A | Strain with a replacement of pomX with the pomXN8A allele |
| Strain, strain background (M. xanthus) | SA9714 | this study | pomX::pomXR11A | Strain with a replacement of pomX with the pomXR11A allele |
| Strain, strain background (M. xanthus) | SA9715 | this study | pomX::pomXK3A | Strain with a replacement of pomX with the pomXK3A allele |
| Strain, strain background (M. xanthus) | SA9716 | this study | pomX::pomXR17A | Strain with a replacement of pomX with the pomXR17A allele |
| Strain, strain background (M. xanthus) | SA9717 | this study | pomX::pomXT22A | Strain with a replacement of pomX with the pomXT22A allele |
| Strain, strain background (M. xanthus) | SA9718 | this study | pomX::pomXK2A | Strain with a replacement of pomX with the pomXK2A allele |
| Strain, strain background (M. xanthus) | SA9719 | this study | pomX::pomXR15A | Strain with a replacement of pomX with the pomXR15A allele |
| Strain, strain background (M. xanthus) | SA9720 | this study | ΔpomY; ΔpomZ; attB::Pmxan0635 pomZD90A-mCh (pKA43) | Strain expressing PomZD90A-mCh in a pomZ and pomY deletion background |
| Strain, strain background (M. xanthus) | SA9721 | this study | ΔpomX; ΔpomY; attB::PpilA pomY-mCh (pDS7) | Strain expressing PomY-mCh in a pomY and pomX deletion background |
| Strain, strain background (M. xanthus) | SA9726 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXN (pDS252) | Strain expressing mCh-PomXN in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9727 | this study | Wild-type; attB::Pmxan0635 mCh-pomXN (pDS252) | Strain expressing mCh-PomXN in a WT background |
| Strain, strain background (M. xanthus) | SA9731 | this study | pomX::pomXK13A | Strain with a replacement of pomX with the pomXK13A allele |
| Strain, strain background (M. xanthus) | SA9732 | this study | pomX::pomXS10A | Strain with a replacement of pomX with the pomXS10A allele |
| Strain, strain background (M. xanthus) | SA9739 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXQ7A (pDS317) | Strain expressing mCh-PomXQ7A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9740 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXN8A (pDS318) | Strain expressing mCh-PomXN8A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9741 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXR17A (pDS323) | Strain expressing mCh-PomXR17A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9742 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXT22A (pDS324) | Strain expressing mCh-PomXT22A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9743 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXS10A (pDS319) | Strain expressing mCh-PomXS10A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9744 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXR11A (pDS320) | Strain expressing mCh-PomXR11A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9747 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK13A (pDS321) | Strain expressing mCh-PomXK13A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9748 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXR15A (pDS322) | Strain expressing mCh-PomXR15A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9749 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK2A (pDS314) | Strain expressing mCh-PomXK2A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9750 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK3A (pDS315) | Strain expressing mCh-PomXK3A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9751 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXE6A (pDS316) | Strain expressing mCh-PomXE6A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9752 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXK13AR15A (pDS325) | Strain expressing mCh-PomXK13AR15A in a pomX deletion background |
| Strain, strain background (M. xanthus) | SA9753 | this study | ΔmglA; ΔpomX; attB::Pmxan0635 mCh-pomXK13AR15A (pDS325) | Strain expressing mCh-PomXK13AR15A in a non-motile pomX deletion background |
| Strain, strain background (M. xanthus) | SA9754 | this study | ΔmglA; ΔpomZ; ΔpomX; Pmxan0635 mCh-pomX (pAH53); mxan18-19::Pmxan0635 pomZD90A (pDS80) | Strain expressing mCh-PomX in a non-motile pomX and pomZ deletion background that expresses PomZD90A. |
| Strain, strain background (M. xanthus) | SA9755 | this study | Wild-type; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a WT background |
| Strain, strain background (M. xanthus) | SA9756 | this study | ΔpomY; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a pomY deletion background |
| Strain, strain background (M. xanthus) | SA9757 | this study | ΔpomZ; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a pomZ deletion background |
| Strain, strain background (M. xanthus) | SA9762 | this study | ΔpomX; attB::Pmxan0635 mCh-pomXC (pDS329) | Strain expressing mCh-PomXC in a pomX deletion background |
| Recombinant DNA reagent | pAH27 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomX, KmR | |
| Recombinant DNA reagent | pAH53 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Pmxan0635 mCh-pomX, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS1 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomY, KmR | |
| Recombinant DNA reagent | pDS7 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | PpilA pomY-mCh, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS16 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomY and pomZ, KmR | |
| Recombinant DNA reagent | pDS80 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Pmxan0635 pomZD90A, mxan_18–19 intergenic region, TcR | |
| Recombinant DNA reagent | pEMR3 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Overexpression of PomX-His6, KmR | |
| Recombinant DNA reagent | pKA1 (plasmid) |
DOI: 10.1111/mmi.12094 | Construct for in-frame deletion of pomZ, KmR | |
| Recombinant DNA reagent | pKA3 (plasmid) |
DOI: 10.1111/mmi.12094 | Overexpression of His6-PomZ, KmR | |
| Recombinant DNA reagent | pKA43 (plasmid) |
DOI: 10.1111/mmi.12094 | Pmxan0635 pomZD90A-mCh, Mx8 attB, TcR | |
| Recombinant DNA reagent | pMAT12 (plasmid) |
https://doi.org/10.1016/j.devcel.2017.04.011 | Construct for in-frame deletion of pomZ and pomX, KmR | |
| Recombinant DNA reagent | pAH152 (plasmid) |
this study | Overexpression of PomXC-His6, KmR | |
| Recombinant DNA reagent | pSL16 (plasmid) |
DOI: 10.1038/emboj.2011.291 | Construct for in-frame deletion of mglA, KmR | |
| Recombinant DNA reagent | pUT18 (plasmid) |
https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pUT18C (plasmid) |
https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pKT25 (plasmid) |
https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pKNT25 (plasmid) |
https://doi.org/10.1073/pnas.95.10.5752 | BACTH plasmid | |
| Recombinant DNA reagent | pAH154 (plasmid) |
this study | Pmxan0635 mCh-pomXN, Mx8 attB, KmR | |
| Recombinant DNA reagent | pAH157 (plasmid) |
this study | Overexpression of PomXN-His6, KmR | |
| Recombinant DNA reagent | pAH165 (plasmid) |
this study | Overexpression of PomXN_K13AR15A-His6, KmR | |
| Recombinant DNA reagent | pDS100 (plasmid) |
this study | BACTH plasmid for pomZ (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS103 (plasmid) |
this study | BACTH plasmid for pomX (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS106 (plasmid) |
this study | BACTH plasmid for pomX (pKT25), KmR | |
| Recombinant DNA reagent | pDS109 (plasmid) |
this study | BACTH plasmid for pomZ (pUT18), AmpR | |
| Recombinant DNA reagent | pDS110 (plasmid) |
this study | BACTH plasmid for pomX (pUT18), AmpR | |
| Recombinant DNA reagent | pDS114 (plasmid) |
this study | BACTH plasmid for pomX (pKNT25) KmR | |
| Recombinant DNA reagent | pDS115 (plasmid) |
this study | BACTH plasmid for pomZD90A (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS117 (plasmid) |
this study | BACTH plasmid for pomZD90A (pUT18), AmpR | |
| Recombinant DNA reagent | pDS120 (plasmid) |
this study | BACTH plasmid for pomY (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS122 (plasmid) |
this study | BACTH plasmid for pomY (pUT18), AmpR | |
| Recombinant DNA reagent | pDS184 (plasmid) |
this study | BACTH plasmid for pomXΔ2-21 (pUT18), AmpR | |
| Recombinant DNA reagent | pDS185 (plasmid) |
this study | BACTH plasmid for pomXΔ2-21 (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS186 (plasmid) |
this study | BACTH plasmid for pomXΔ2-21 (pKT25), KmR | |
| Recombinant DNA reagent | pDS187 (plasmid) |
this study | BACTH plasmid for pomXΔ2-21 (pKNT25) KmR | |
| Recombinant DNA reagent | pDS188 (plasmid) |
this study | BACTH plasmid for pomXC (pUT18), AmpR | |
| Recombinant DNA reagent | pDS189 (plasmid) |
this study | BACTH plasmid for pomXC (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS190 (plasmid) |
this study | BACTH plasmid for pomXC (pKT25), KmR | |
| Recombinant DNA reagent | pDS191 (plasmid) |
this study | BACTH plasmid for pomXC (pKNT25) KmR | |
| Recombinant DNA reagent | pDS192 (plasmid) |
this study | BACTH plasmid for pomXN (pUT18), AmpR | |
| Recombinant DNA reagent | pDS193 (plasmid) |
this study | BACTH plasmid for pomXN (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS194 (plasmid) |
this study | BACTH plasmid for pomXN (pKT25), KmR | |
| Recombinant DNA reagent | pDS195 (plasmid) |
this study | BACTH plasmid for pomXN (pKNT25) KmR | |
| Recombinant DNA reagent | pDS232 (plasmid) |
this study | Overexpression of PomXN-Strep, KmR | |
| Recombinant DNA reagent | pDS252 (plasmid) |
this study | Pmxan0635 mCh-pomXN, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS253 (plasmid) |
this study | BACTH plasmid for pomXN_K13AR15A (pUT18), AmpR | |
| Recombinant DNA reagent | pDS254 (plasmid) |
this study | BACTH plasmid for pomXN_K13AR15A (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS255 (plasmid) |
this study | BACTH plasmid for pomXN_K13AR15A (pKT25), KmR | |
| Recombinant DNA reagent | pDS256 (plasmid) |
this study | BACTH plasmid for pomXN_K13AR15A (pKNT25) KmR | |
| Recombinant DNA reagent | pDS257 (plasmid) |
this study | BACTH plasmid for pomXN_Δ2-21 (pUT18), AmpR | |
| Recombinant DNA reagent | pDS258 (plasmid) |
this study | BACTH plasmid for pomXN_Δ2-21 (pUT18C), AmpR | |
| Recombinant DNA reagent | pDS259 (plasmid) |
this study | BACTH plasmid for pomXN_Δ2-21 (pKT25), KmR | |
| Recombinant DNA reagent | pDS260 (plasmid) |
this study | BACTH plasmid for pomXN_Δ2-21 (pKNT25) KmR | |
| Recombinant DNA reagent | pDS303 (plasmid) |
this study | nat. site codon exchange for pomXK2A, KmR | |
| Recombinant DNA reagent | pDS304 (plasmid) |
this study | nat. site codon exchange for pomXK3A, KmR | |
| Recombinant DNA reagent | pDS305 (plasmid) |
this study | nat. site codon exchange for pomXE6A, KmR | |
| Recombinant DNA reagent | pDS306 (plasmid) |
this study | nat. site codon exchange for pomXQ7A, KmR | |
| Recombinant DNA reagent | pDS307 (plasmid) |
this study | nat. site codon exchange for pomXN8A, KmR | |
| Recombinant DNA reagent | pDS308 (plasmid) |
this study | nat. site codon exchange for pomXS10A, KmR | |
| Recombinant DNA reagent | pDS309 (plasmid) |
this study | nat. site codon exchange for pomXR11A, KmR | |
| Recombinant DNA reagent | pDS310 (plasmid) |
this study | nat. site codon exchange for pomXK13A, KmR | |
| Recombinant DNA reagent | pDS311 (plasmid) |
this study | nat. site codon exchange for pomXR15A, KmR | |
| Recombinant DNA reagent | pDS312 (plasmid) |
this study | nat. site codon exchange for pomXR17A, KmR | |
| Recombinant DNA reagent | pDS313 (plasmid) |
this study | nat. site codon exchange for pomXT22A, KmR | |
| Recombinant DNA reagent | pDS314 (plasmid) |
this study | Pmxan0635 mCh-pomXK2A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS315 (plasmid) |
this study | Pmxan0635 mCh-pomXK3A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS316 (plasmid) |
this study | Pmxan0635 mCh-pomXE6A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS317 (plasmid) |
this study | Pmxan0635 mCh-pomXQ7A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS318 (plasmid) |
this study | Pmxan0635 mCh-pomXN8A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS319 (plasmid) |
this study | Pmxan0635 mCh-pomXS10A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS320 (plasmid) |
this study | Pmxan0635 mCh-pomXR11A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS321 (plasmid) |
this study | Pmxan0635 mCh-pomXK13A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS322 (plasmid) |
this study | Pmxan0635 mCh-pomXR15A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS323 (plasmid) |
this study | Pmxan0635 mCh-pomXR17A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS324 (plasmid) |
this study | Pmxan0635 mCh-pomXT22A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS325 (plasmid) |
this study | Pmxan0635 mCh-pomXK13AR15A, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS329 (plasmid) |
this study | Pmxan0635 mCh-pomXC, Mx8 attB, KmR | |
| Recombinant DNA reagent | pDS333 (plasmid) |
this study | Overexpression of PomXC-Strep, KmR | |
| Recombinant DNA reagent | pEMR1 (plasmid) |
this study | Overexpression of PomY-His6, KmR | |
| Recombinant DNA reagent | pSH1 (plasmid) |
this study | nat. site codon exchange for pomXK13AR15A, KmR | |
| Recombinant DNA reagent | pSH36 (plasmid) |
this study | BACTH plasmid for pomXK13AR15A (pKNT25) KmR | |
| Recombinant DNA reagent | pSH37 (plasmid) |
this study | BACTH plasmid for pomXK13AR15A (pKT25), KmR | |
| Recombinant DNA reagent | pSH38 (plasmid) |
this study | BACTH plasmid for pomXK13AR15A (pUT18), AmpR | |
| Recombinant DNA reagent | pSH39 (plasmid) |
this study | BACTH plasmid for pomXK13AR15A (pUT18C), AmpR | |
| Recombinant DNA reagent | pSH58 (plasmid) |
this study | Overexpression of PomXK13AR15A-His6, KmR | |
| Sequence-based reagent | pomX BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGAAGAAAGCCTTTGAAC-3’ |
| Sequence-based reagent | pomX BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGGCGCACCGTGGCCTGAC-3’ |
| Sequence-based reagent | pomY BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGGTGAGCGACGAGCGTCCG-3’ |
| Sequence-based reagent | pomY BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGAGCGGCGAAGTATTTGTG-3’ |
| Sequence-based reagent | pomZ BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGGAAGCGCCGACGTAC-3’ |
| Sequence-based reagent | pomZ BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGGCCGGCCTGCTGGGTGCC-3’ |
| Sequence-based reagent | pomXΔ2–21 BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGACGGGCCTCGTCGACCCC-3’ |
| Sequence-based reagent | pomXC BTH fwd XbaI | this study | PCR primer | 5’-GCGTCTAGAGATGGCCACCGTGGCGGAGGCG-3’ |
| Sequence-based reagent | pomXN BTH rev KpnI | this study | PCR primer | 5’-GCGGGTACCCGGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | 0636 up fwd | this study | PCR primer | 5’-GCGGGATCCGTCACCCCAAGCCATTC-3’ |
| Sequence-based reagent | PomX K2A rev native | this study | PCR primer | 5’-CAAAGGCTTTCGCCATGGTTCTCAG-3’ |
| Sequence-based reagent | PomX K2A fwd native | this study | PCR primer | 5’-CTGAGAACCATGGCGAAAGCCTTTG-3’ |
| Sequence-based reagent | 0636 HindIII rev stop | this study | PCR primer | 5’-GCGAAGCTTTCAGCGCACCGTGGCCTGAC-3’ |
| Sequence-based reagent | PomX K3A rev native | this study | PCR primer | 5’-CTGTTCAAAGGCCGCCTTCATGGTTC-3’ |
| Sequence-based reagent | PomX K3A fwd native | this study | PCR primer | 5’-GAACCATGAAGGCGGCCTTTGAACAG-3’ |
| Sequence-based reagent | PomX E6A rev | this study | PCR primer | 5’-GGACACGTTCTGCGCAAAGGCTTTCTT-3’ |
| Sequence-based reagent | PomX E6A fwd | this study | PCR primer | 5’-AAGAAAGCCTTTGCGCAGAACGTGTCC-3’ |
| Sequence-based reagent | PomX Q7A rev | this study | PCR primer | 5’-GCGGGACACGTTCGCTTCAAAGGCTTT-3’ |
| Sequence-based reagent | PomX Q7A fwd | this study | PCR primer | 5’-AAAGCCTTTGAAGCGAACGTGTCCCGC-3’ |
| Sequence-based reagent | PomX N8A rev | this study | PCR primer | 5’-GGCGCGGGACACCGCCTGTTCAAAGGC-3’ |
| Sequence-based reagent | PomX N8A fwd | this study | PCR primer | 5’-GCCTTTGAACAGGCGGTGTCCCGCGCC-3’ |
| Sequence-based reagent | PomX S10A rev | this study | PCR primer | 5’-CGGCTTGGCGCGCGCCACGTTCTGTTC-3’ |
| Sequence-based reagent | PomX S10A fwd | this study | PCR primer | 5’-GAACAGAACGTGGCGCGCGCCAAGCCG-3’ |
| Sequence-based reagent | PomX R11A rev | this study | PCR primer | 5’-GCGCGGCTTGGCCGCGGACACGTTCTG-3’ |
| Sequence-based reagent | PomX R11A fwd | this study | PCR primer | 5’-CAGAACGTGTCCGCGGCCAAGCCGCGC-3’ |
| Sequence-based reagent | PomX K13A rev | this study | PCR primer | 5’-GCGGAGGCGCGGCGCGGCGCGGGACAC-3’ |
| Sequence-based reagent | PomX K13A fwd | this study | PCR primer | 5’-GTGTCCCGCGCCGCGCCGCGCCTCCGC-3’ |
| Sequence-based reagent | PomX R15A rev | this study | PCR primer | 5’-GCCCAGGCGGAGCGCCGGCTTGGCGCG-3’ |
| Sequence-based reagent | PomX R15A fwd | this study | PCR primer | 5’-CGCGCCAAGCCGGCGCTCCGCCTGGGC-3’ |
| Sequence-based reagent | PomX R17A rev | this study | PCR primer | 5’-CAGCGCGCCCAGCGCGAGGCGCGGCTT-3’ |
| Sequence-based reagent | PomX R17A fwd | this study | PCR primer | 5’-AAGCCGCGCCTCGCGCTGGGCGCGCTG-3’ |
| Sequence-based reagent | PomX T22A rev | this study | PCR primer | 5’-GTCGACGAGGCCCGCCAGCGCGCCCAG-3’ |
| Sequence-based reagent | PomX T22A fwd | this study | PCR primer | 5’-CTGGGCGCGCTGGCGGGCCTCGTCGAC-3’ |
| Sequence-based reagent | PomX K13AR15A rev | this study | PCR primer | 5’-CAGAACGTGTCCCGCGCCGCGCCGGCCCTCCGCCTGGGCGCGCTG-3’ |
| Sequence-based reagent | PomX K13AR15A fwd | this study | PCR primer | 5’-CAGCGCGCCCAGGCGGAGGGCCGGCGCGGCGCGGGACACCTTCTG-3’ |
| Sequence-based reagent | mCherry XbaI fwd | this study | PCR primer | 5’-GCGTCTAGAGTGAGCAAGGGCGAGGAG-3’ |
| Sequence-based reagent | PomX K2A rev | this study | PCR primer | 5’-TTCAAAGGCTTTCGCCATGGCTCCGCC-3’ |
| Sequence-based reagent | PomX K2A fwd | this study | PCR primer | 5’-GGCGGAGCCATGGCGAAAGCCTTTGAA-3’ |
| Sequence-based reagent | KA348 | this study | PCR primer | 5’-GCCAAGCTTTCAGCGCACCGTGGCCTG-3’ |
| Sequence-based reagent | PomX K3A fwd | this study | PCR primer | 5’-GGAGCCATGAAGGCGGCCTTTGAACAG-3’ |
| Sequence-based reagent | PomX K3A rev | this study | PCR primer | 5’-CTGTTCAAAGGCCGCCTTCATGGCTCC-3’ |
| Sequence-based reagent | AH142 | this study | PCR primer | 5’-GGAATTCCATATGGCCACCGTGGCGGAGGCG-3’ |
| Sequence-based reagent | KA346 | this study | PCR primer | 5’-GCCAAGCTTGCGCACCGTGGCCTGACTC-3’ |
| Sequence-based reagent | AH143 | this study | PCR primer | 5’-CCCAAGCTTGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | NdeI PomX fwd | this study | PCR primer | 5’-GGAATTCCATATGAAGAAAGCCTTTGAACAG-3’ |
| Sequence-based reagent | AH144 | this study | PCR primer | 5’-GCCAAGCTTTCAGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | KA384 | this study | PCR primer | 5’-GCGGGATCCGGCGGAGCCATGAAGAAAGCCTTTGAACAG-3’ |
| Sequence-based reagent | DS276 | this study | PCR primer | 5’-GCGAAGCTTACTTCTCGAACTGTGGGTGACTCCAGCGCACCGTGGCCTGAC-3’ |
| Sequence-based reagent | DS277 | this study | PCR primer | 5’-GCGCCATGGCCACCGTGGCGGAGGCG-3’ |
| Sequence-based reagent | PomX BspHI fwd | this study | PCR primer | 5’-GCGTCATGAAGAAAGCCTTTGAACAGAACG-3’ |
| Sequence-based reagent | PomXN rev strep-tag | this study | PCR primer | 5’-GCGAAGCTTACTTCTCGAACTGTGGGTGACTCCAGGGCAGCGGCTCCGGGCG-3’ |
| Sequence-based reagent | NdeI-PomY fwd | this study | PCR primer | 5’-GGAATTCCATATGAGCGACGAGCGTCCGGAC-3’ |
| Sequence-based reagent | PomY C-term his rev | this study | PCR primer | 5’-CGGAAGCTTAGCGGCGAAGTATTTGTGC-3’ |
| Sequence-based reagent | AH141 | this study | PCR primer | 5’-GCGGGATCCGGCGGAGCCGCCACCGTGGCGGAGGCG-3’ |
| Antibody | α-PomX (rabbit, polyclonal) | https://doi.org/10.1016/j.devcel.2017.04.011 | Western Blot (1:15000) | |
| Antibody | α-PomY (rabbit, polyclonal) | https://doi.org/10.1016/j.devcel.2017.04.011 | Western Blot (1:15000) | |
| Antibody | α-PomZ (rabbit, polyclonal) | DOI: 10.1111/mmi.12094 | Western Blot (1:10000) | |
| Antibody | α-PilC (rabbit, polyclonal) | DOI:10.1111/j.1365–2958.2009.06891.x | Western Blot (1:3000) | |
| Antibody | α-mCherry (rabbit, polyclonal) | Biovision | Cat# 5993 | Western Blot (1:10000) |
| Antibody | horseradish-conjugated α-rabbit immunoglobulin G (goat,polyclonal) |
Sigma-Aldrich | Cat# A0545-1ML | Western Blot (1:25000) |
| Peptide, recombinant protein | PomXNPEP | Thermo Scientific | MKKAFEQNVSRAKPRLRLGALT | |
| Peptide, recombinant protein | PomXNPEPK13AR15A | Thermo Scientific | MKKAFEQNVSRAAPALRLGALT | |
| Software, algorithm | Metamorph_v 7.5 | Molecular Devices | ||
| Software, algorithm | Oufti | DOI: 10.1111/mmi.13264 | http://www.oufti.org/ | |
| Software, algorithm | Matlab R2018a | MathWorks | ||
| Commercial assay or kit | Luminata Forte | Fisher scientific | Cat# 10394675 |
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Lotte Søgaard-Andersen, Email: sogaard@mpi-marburg.mpg.de.
Gisela Storz, National Institute of Child Health and Human Development, United States.
Gisela Storz, National Institute of Child Health and Human Development, United States.
Funding Information
This paper was supported by the following grants:
Deutsche Forschungsgemeinschaft TRR 174 to Erwin Frey, Lotte Søgaard-Andersen.
Max Planck Society to Lotte Søgaard-Andersen.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Resources, Data curation, Formal analysis, Validation, Investigation, Visualization, Methodology, Writing - original draft, Project administration, Writing - review and editing.
Data curation, Validation, Investigation.
Software, Methodology, Writing - review and editing.
Supervision, Funding acquisition, Writing - review and editing.
Conceptualization, Supervision, Funding acquisition, Writing - original draft, Project administration, Writing - review and editing.
Additional files
Data availability
All data supporting this study are available within the article and supporting files. Source data files have been provided for Figures 1, 2, 3, 4, 5 and 6.
References
- Ah-Seng Y, Lopez F, Pasta F, Lane D, Bouet J-Y. Dual role of DNA in regulating ATP hydrolysis by the SopA partition protein. Journal of Biological Chemistry. 2009;284:30067–30075. doi: 10.1074/jbc.M109.044800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ah-Seng Y, Rech J, Lane D, Bouet JY. Defining the role of ATP hydrolysis in Mitotic segregation of bacterial plasmids. PLOS Genetics. 2013;9:e1003956. doi: 10.1371/journal.pgen.1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barillà D, Carmelo E, Hayes F. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. PNAS. 2007;104:1811–1816. doi: 10.1073/pnas.0607216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouet JY, Funnell BE. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. The EMBO Journal. 1999;18:1415–1424. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyha I, Schmidt C, Lenz P, Jakovljevic V, Höne A, Maier B, Hoppert M, Søgaard-Andersen L. Regulation of the type IV pili molecular machine by dynamic localization of two motor proteins. Molecular Microbiology. 2009;74:691–706. doi: 10.1111/j.1365-2958.2009.06891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaing JP, Bouet JY, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Molecular Microbiology. 2008;70:1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer PA, Crossley RE, Hand AR, Rothfield LI. The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. The EMBO Journal. 1991;10:4371–4380. doi: 10.1002/j.1460-2075.1991.tb05015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Lutkenhaus J. At the heart of bacterial cytokinesis: the Z ring. Trends in Microbiology. 2019;27:781–791. doi: 10.1016/j.tim.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. The pfam protein families database in 2019. Nucleic Acids Research. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswara PJ, Ramamurthi KS. Bacterial cell division: nonmodels poised to take the spotlight. Annual Review of Microbiology. 2017;71:393–411. doi: 10.1146/annurev-micro-102215-095657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghasriani H, Ducat T, Hart CT, Hafizi F, Chang N, Al-Baldawi A, Ayed SH, Lundström P, Dillon JA, Goto NK. Appropriation of the MinD protein-interaction motif by the dimeric interface of the bacterial cell division regulator MinE. PNAS. 2010;107:18416–18421. doi: 10.1073/pnas.1007141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Treuner-Lange A, Schumacher D, Søgaard-Andersen L. Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLOS Genetics. 2013;9:e1003802. doi: 10.1371/journal.pgen.1003802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester CM, Lutkenhaus J. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. PNAS. 2007;104:20326–20331. doi: 10.1073/pnas.0705196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. PNAS. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. PNAS. 1999;96:14819–14824. doi: 10.1073/pnas.96.26.14819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gogol EP, Lutkenhaus J. Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. PNAS. 2002;99:6761–6766. doi: 10.1073/pnas.102059099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Saez C, Lutkenhaus J. Recruitment of MinC, an inhibitor of Z-ring formation, to the membrane in Escherichia coli: role of MinD and MinE. Journal of Bacteriology. 2003;185:196–203. doi: 10.1128/JB.185.1.196-203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in Escherichia coli involves rapid pole to pole oscillation of the division inhibitor MinC under the control of MinD and MinE. Molecular Microbiology. 1999;34:82–90. doi: 10.1046/j.1365-2958.1999.01575.x. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Molecular Cell. 2001;7:1337–1343. doi: 10.1016/s1097-2765(01)00273-8. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J. A conserved sequence at the C-terminus of MinD is required for binding to the membrane and targeting MinC to the septum. Molecular Microbiology. 2003;47:345–355. doi: 10.1046/j.1365-2958.2003.03321.x. [DOI] [PubMed] [Google Scholar]
- Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. PNAS. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. PNAS. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekebusch D, Michie KA, Essen LO, Löwe J, Thanbichler M. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Molecular Cell. 2012;46:245–259. doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiianitsa K, Solinger JA, Heyer WD. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Analytical Biochemistry. 2003;321:266–271. doi: 10.1016/S0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- Lackner LL, Raskin DM, de Boer PA. ATP-dependent interactions between Escherichia coli min proteins and the phospholipid membrane in vitro. Journal of Bacteriology. 2003;185:735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard TA, Butler PJ, Löwe J. Bacterial chromosome segregation: structure and DNA binding of the soj dimer--a conserved biological switch. The EMBO Journal. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Research. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim HC, Surovtsev IV, Beltran BG, Huang F, Bewersdorf J, Jacobs-Wagner C. Evidence for a DNA-relay mechanism in ParABS-mediated chromosome segregation. eLife. 2014;3:e02758. doi: 10.7554/eLife.02758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. The ParA/MinD family puts things in their place. Trends in Microbiology. 2012;20:411–418. doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCready JS, Hakim P, Young EJ, Hu L, Liu J, Osteryoung KW, Vecchiarelli AG, Ducat DC. Protein gradients on the nucleoid position the carbon-fixing organelles of cyanobacteria. eLife. 2018;7:e39723. doi: 10.7554/eLife.39723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miertzschke M, Koerner C, Vetter IR, Keilberg D, Hot E, Leonardy S, Søgaard-Andersen L, Wittinghofer A. Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity. The EMBO Journal. 2011;30:4185–4197. doi: 10.1038/emboj.2011.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/S0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- Paintdakhi A, Parry B, Campos M, Irnov I, Elf J, Surovtsev I, Jacobs-Wagner C. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Molecular Microbiology. 2016;99:767–777. doi: 10.1111/mmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KT, Wu W, Battaile KP, Lovell S, Holyoak T, Lutkenhaus J. The min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146:396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K-T, Wu W, Lovell S, Lutkenhaus J. Mechanism of the asymmetric activation of the MinD ATPase by MinE. Molecular Microbiology. 2012;85:271–281. doi: 10.1111/j.1365-2958.2012.08110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L. A spindle-like apparatus guides bacterial chromosome segregation. Nature Cell Biology. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA. MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. Journal of Bacteriology. 1999;181:6419–6424. doi: 10.1128/JB.181.20.6419-6424.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MAJ, Wadhams GH, Hadfield KA, Tickner S, Armitage JP. ParA-like protein uses nonspecific chromosomal DNA binding to partition protein complexes. PNAS. 2012;109:6698–6703. doi: 10.1073/pnas.1114000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. The EMBO Journal. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholefield G, Whiting R, Errington J, Murray H. Spo0J regulates the oligomeric state of soj to trigger its switch from an activator to an inhibitor of DNA replication initiation. Molecular Microbiology. 2011;79:1089–1100. doi: 10.1111/j.1365-2958.2010.07507.x. [DOI] [PubMed] [Google Scholar]
- Schumacher D, Bergeler S, Harms A, Vonck J, Huneke-Vogt S, Frey E, Søgaard-Andersen L. The PomXYZ proteins Self-Organize on the bacterial nucleoid to stimulate cell division. Developmental Cell. 2017;41:299–314. doi: 10.1016/j.devcel.2017.04.011. [DOI] [PubMed] [Google Scholar]
- Schumacher D, Søgaard-Andersen L. Regulation of cell polarity in motility and cell division in Myxococcus xanthus. Annual Review of Microbiology. 2017;71:61–78. doi: 10.1146/annurev-micro-102215-095415. [DOI] [PubMed] [Google Scholar]
- Schumacher D, Søgaard-Andersen L. Fluorescence Live-cell imaging of the complete vegetative cell cycle of the Slow-growing social bacterium Myxococcus xanthus. Journal of Visualized Experiments. 2018;136:57860. doi: 10.3791/57860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Wegener-Feldbrügge S, Huntley S, Hamann N, Hedderich R, Søgaard-Andersen L. Bioinformatics and experimental analysis of proteins of two-component systems in Myxococcus xanthus. Journal of Bacteriology. 2008;190:613–624. doi: 10.1128/JB.01502-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto TH, Rowland SL, Rothfield LI, King GF. Membrane localization of MinD is mediated by a C-terminal motif that is conserved across Eubacteria, archaea, and chloroplasts. PNAS. 2002;99:15693–15698. doi: 10.1073/pnas.232590599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treuner-Lange A, Aguiluz K, van der Does C, Gómez-Santos N, Harms A, Schumacher D, Lenz P, Hoppert M, Kahnt J, Muñoz-Dorado J, Søgaard-Andersen L. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Molecular Microbiology. 2013;87:235–253. doi: 10.1111/mmi.12094. [DOI] [PubMed] [Google Scholar]
- Treuner-Lange A, Søgaard-Andersen L. Regulation of cell polarity in Bacteria. Journal of Cell Biology. 2014;206:7–17. doi: 10.1083/jcb.201403136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Hwang LC, Mizuuchi K. Cell-free study of F plasmid partition provides evidence for cargo transport by a diffusion-ratchet mechanism. PNAS. 2013;110:E1390–E1397. doi: 10.1073/pnas.1302745110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchiarelli AG, Neuman KC, Mizuuchi K. A propagating ATPase gradient drives transport of surface-confined cellular cargo. PNAS. 2014;111:4880–4885. doi: 10.1073/pnas.1401025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Park K-T, Holyoak T, Lutkenhaus J. Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC. Molecular Microbiology. 2011;79:1515–1528. doi: 10.1111/j.1365-2958.2010.07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaichi Y, Fogel MA, McLeod SM, Hui MP, Waldor MK. Distinct centromere-like parS sites on the two chromosomes of Vibrio spp. Journal of Bacteriology. 2007;189:5314–5324. doi: 10.1128/JB.00416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Schumacher MA. Structures of partition protein ParA with nonspecific DNA and ParB effector reveal molecular insights into principles governing Walker-box DNA segregation. Genes & Development. 2017;31:481–492. doi: 10.1101/gad.296319.117. [DOI] [PMC free article] [PubMed] [Google Scholar]