Abstract
Memory B cells play a fundamental role in host defenses against viruses, but to date, their role has been relatively unsettled in the context of SARS-CoV-2. We report here a longitudinal single-cell and repertoire profiling of the B cell response up to 6 months in mild and severe COVID-19 patients. Distinct SARS-CoV-2 spike-specific activated B cell clones fueled an early antibody-secreting cell burst as well as a durable synchronous germinal center response. While highly mutated memory B cells, including pre-existing cross-reactive seasonal Betacoronavirus-specific clones, were recruited early in the response, neutralizing SARS-CoV-2 RBD-specific clones accumulated with time and largely contributed to the late, remarkably stable, memory B cell pool. Highlighting germinal center maturation, these cells displayed clear accumulation of somatic mutations in their variable region genes over time. Overall, these findings demonstrate that an antigen-driven activation persisted and matured up to 6 months after SARS-CoV-2 infection and may provide long-term protection.
Keywords: COVID-19, germinal center, somatic hypermutation, extrafollicular response, plasma cells, neutralizing antibodies, spike protein, RBD, OC43, HKU1
Graphical abstract
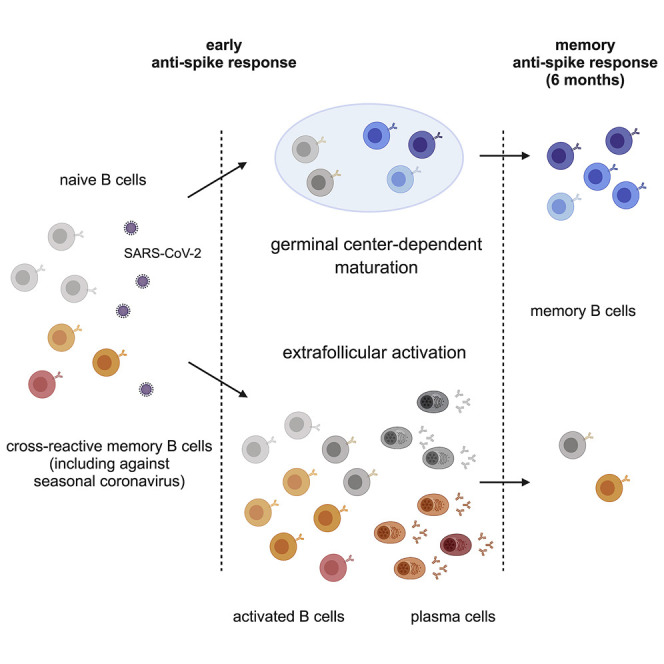
The B cell response against SARS-CoV-2 shows a temporal shift from an extrafollicular reaction that includes memory B cells against seasonal coronaviruses toward a germinal center-dependent memory response encoding (spike-specific) neutralizing antibodies.
Introduction
The new emerging coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected 88 million people and killed over 1.9 million individuals worldwide since the beginning of the pandemic. Understanding the mechanisms underlying the establishment of protective immune memory in recovering individuals is a major concern for public health and for anticipating vaccination outcomes.
In response to viral infection, virus-specific T and B cells are activated, expand, and differentiate into effector cells (Wherry and Ahmed, 2004). At the early phase of coronavirus disease 2019 (COVID-19), most infected individuals generate antibodies targeting the viral nucleocapsid (N) and the spike (S) proteins of SARS-CoV-2, including S receptor binding domain (RBD)-specific antibodies with strong neutralizing potential (Wajnberg et al., 2020). COVID-19 patients also develop potent early CD8+ and CD4+ T cell-specific responses (Braun et al., 2020; Ferretti et al., 2020; Grifoni et al., 2020, 2020; Le Bert et al., 2020; Meckiff et al., 2020; Peng et al., 2020; Sekine et al., 2020; Swadling and Maini, 2020).
Studies have documented that B cells from patients with severe COVID-19 harbored low mutations frequencies in their heavy-chain variable region (VH) genes, notably those producing anti-RBD antibodies (Nielsen et al., 2020). This suggested an active early extrafollicular response in which naive unmutated B cells first engage in cognate interactions with T cells and become fully activated to divide and differentiate into plasma cells (PCs) (Jenks et al., 2019). Along with limited somatic hypermutation (SHM) and selection, cells derived from extrafollicular responses are usually considered as short lived (MacLennan et al., 2003).
Long-term humoral immunity following infection relies on two types of cells derived from germinal center (GC) responses: long-lived plasma cells (LLPCs), which continuously secrete antibodies (Slifka et al., 1998), and memory B cells (MBCs), which can expand and differentiate into antibody-secreting cells (ASCs) upon a new antigenic challenge (Yoshida et al., 2010). MBCs can also adapt to cope with antigenic changes linked to virus mutations, as exemplified by the coevolution of neutralizing antibodies against mutating epitopes during HIV infection (Escolano et al., 2016; Liao et al., 2013; Wu et al., 2011). Assessing whether an MBC response, with the ability to rapidly produce SARS-CoV-2 neutralizing antibodies upon a new infectious challenge months or years after initial infection, is established in convalescent COVID-19 patients, thus of immediate relevance for ongoing modeling of herd immunity and the formulation of vaccine strategies.
Serological studies have so far reported contradictory results on the persistence of humoral immunity in asymptomatic, mild, and severe patients (Iyer et al., 2020; Long et al., 2020; Luchsinger et al., 2020; Pierce et al., 2020; Reynolds et al., 2020; Ripperger et al., 2020; Röltgen et al., 2020; Wajnberg et al., 2020; Weisberg et al., 2021). Furthermore, although MBCs have been observed up to 6 months after infection in an increasing number of recent publications (Chen et al., 2020; Dan et al., 2021; Gaebler et al., 2021; Hartley et al., 2020; Nguyen-Contant et al., 2020; Rodda et al., 2020; Vaisman-Mentesh et al., 2020), a recent study suggested that severe SARS-CoV-2 infection may blunt the GC response and subsequently compromise the generation of long-lived, affinity-matured MBCs (Kaneko et al., 2020). It was also suggested that the strong extrafollicular B cell activation observed after symptomatic SARS-CoV-2 infection could prevent or delay a GC-dependent immune response (Woodruff et al., 2020).
In this context, analysis of the longevity and functionality of the anti-SARS-CoV-2 MBC response becomes a major issue, notably to evaluate the protection afforded by a prior infection to subsequent viral challenges.
In the present study, we present a longitudinal deep profiling of the anti-SARS-CoV-2 MBC response in two parallel cohorts of patients with severe (S-CoV) and mild (M-CoV) COVID-19. We combined single-cell transcriptomics, single-cell culture, and immunoglobulin heavy chain (IgH) variable, diversity, and joining (VDJ) genes sequencing to track and characterize the cellular and molecular phenotype and clonal evolution of S-specific MBC clones from early time points after SARS-CoV-2 infection up to 6 months after the initial symptoms. Our results provide new insights on the origin, magnitude, and stability of the anti-SARS-CoV-2 MBC response and revealed in most patients the robust acquisition of a GC-derived immune memory.
Results
Investigating anti-SARS-CoV-2 humoral response in mild and severe COVID-19 patients
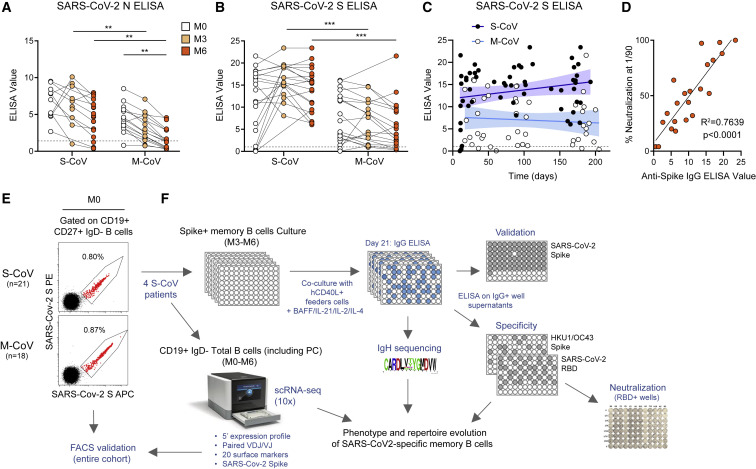
To characterize the longitudinal evolution of the humoral response against SARS-CoV-2, we set up a prospective cohort of patients with S-CoV requiring oxygen (n = 21, median age: 57 years [40-78], corresponding to WHO progression scale >4 and ≤8) and mild ambulatory forms of COVID-19 (M-CoV, n = 18, median age 35.5 years [28–64], corresponding to WHO progression scale <4; Marshall et al., 2020) (MEMO-COV, NCT04402892, see STAR methods and Table S1). Initial samples were collected from patients in median 18.8 days (±SD: 8.8) and 35.5 days (±SD: 12.8) after disease onset (M0) for S-CoV and M-CoV respectively. Two additional blood samples were collected at 3 (M3) and 6 months (M6) at a stage when all patients had fully recovered. All sera were first examined for SARS-CoV-2-specific immunoglobulin G (IgG) antibodies against the immunodominant N and S proteins using commercial ELISA. As previously observed in other studies (Ripperger et al., 2020), N-specific IgG antibodies showed a rapid decrease in both cohorts (Figure 1 A). This decrease was particularly pronounced in M-CoV patients, among which 9 out of 18 had titers below the threshold of positivity at 6 months (Figure 1A; Table S1). In contrast, S-specific IgG antibodies appeared quite stable with time and only two M-CoV patients showed anti-S antibody titers below detection threshold at the time of our last sampling (Figures 1B and 1C). Levels of S-specific IgG antibodies at 3 and 6 months, however, were significantly higher in patients who had developed a more severe form of the disease (Figures 1B and 1C). Elevated titers of anti-S IgG strongly correlated with enhanced neutralization potential of patients’ sera in vitro (Figure 1D; Figures S1 A and S1B), confirming the S trimetric glycoprotein as a primary target of the protective immune response in patients.
Figure 1.
Longitudinal characterization of the humoral response against SARS-CoV-2 in severe and mild COVID-19 patients
(A and B) Anti-SARS-CoV-2 nucleocapsid (N) (A) and spike (S) (B) serum IgG levels were measured by ELISA in 21 severe COVID-19 (S-CoV) and 18 mild COVID-19 (M-CoV) patients at M0 (white), M3 (light orange), and M6 (dark orange). The dashed line indicates the positivity threshold provided by the manufacturer.
(C) Evolution of anti-SARS-CoV-2 S serum IgG levels over time post symptom onset in S-CoV (black dots, dark blue line) and M-CoV (white dots, light blue line) patients. Continuous lines indicate linear regression, and colored area between dashed lines indicates error bands (R2 = 0.049 for S-CoV, ns and 0.0061 for M-CoV, ns, Pearson correlation).
(D) Correlation between anti-SARS-CoV-2 S serum IgG levels and in vitro neutralization potential (% neutralization achieved at a 1/90 dilution) at M6 (n = 10 S-CoV and 11 M-CoV patients). The line represents a simple linear regression (R2 and p value with Pearson correlation are shown)
(E) Representative FACS plot of His-tagged SARS-CoV-2 S staining in gated live CD19+CD38int/−CD27+IgD− B cells at M0 in two representative S-CoV (upper plot) and M-CoV patients (lower plot).
(F) Overall study design.
ANOVA and two-tailed Mann-Whitney tests (A and B). Linear regression with Pearson correlation analysis (D). ∗∗∗p < 0.001, ∗∗p < 0.01. See also Figure S1 and Table S1.
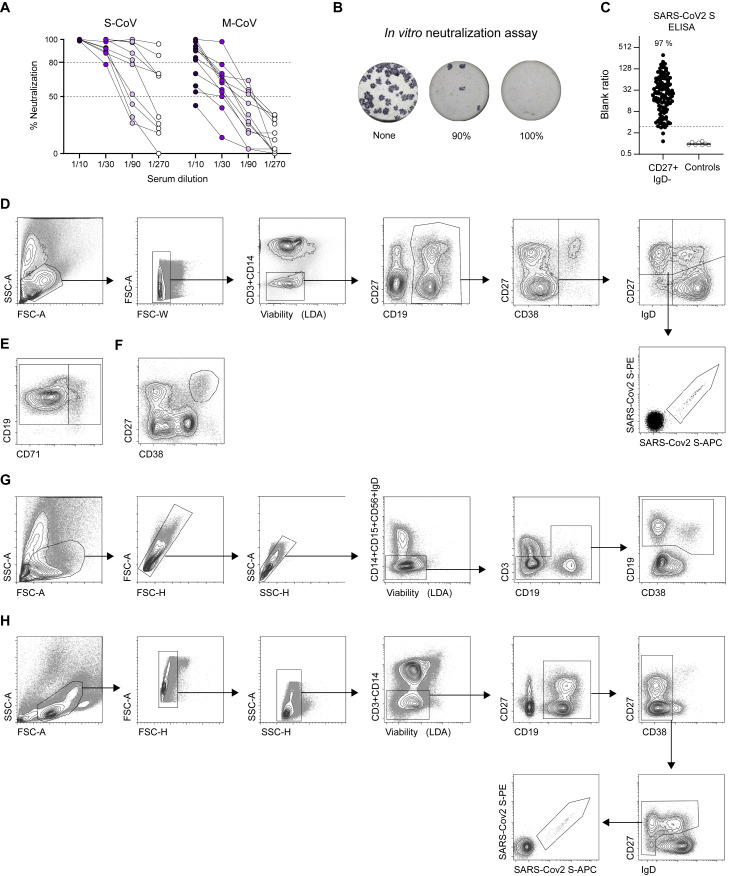
Figure S1.
Quantification and functional assessment of the anti-SARS-CoV-2 humoral immune response in COVID-19 convalescent patients, Related to Figure 1
(A) Percentage of in vitro neutralization shown by individual sera from S-CoV (n = 10) and M-CoV (n = 12) patients at M6 at increasing dilutions.
(B) Representative wells for the neutralization assay. Blue spots represent SARS-CoV-2 positive cells.
(C) Representative results of an anti-SARS-CoV-2 S IgG ELISA on supernatants from sorted SARS-CoV-2 S-specific MBCs (dark dots) and non spike-specific MBCs (white dots), as validation of FACS SARS-CoV-2 S-staining. Lines indicate median value. Dashed line indicates the positivity threshold (≥3 x blank).
(D–H) Flow cytometric gating strategies for the analysis and sorting of major B cell populations from PBMCs of convalescent COVID-19 patients. (D) Gating strategy to analyze SARS-CoV-2 S-specific B cell population. Lymphocytes were first gated based on morphology, before exclusion of doublets, dead cells and CD3/CD14 cells. CD19+ cells were next gated before exclusion of CD38hi plasma cells. CD38int/- cells were then divided in four quadrants using CD27 and IgD. Upper left quadrant defines memory B cells (MBCs), lower left quadrant double-negative (DN), upper right quadrant CD27+IgD+ cells (MZB) and lower right quadrant naive B cells (excluding CD38hi transitional). SARS-CoV-2 S-specific B cells were then analyzed within the B cell population of interest using a double-staining strategy with anti-His antibodies of a His-tagged SARS-CoV-2 S protein. (E) Gating strategy to separate activated and classical switched B cells using CD71, within the IgD-CD27+ gate (F) Gating strategy to analyze CD27hiCD38hi plasma cells in CD3-CD14- live cells. (G) Gating strategy for sorting of CD19+IgD- cells from PBMCs for 10X Genomics single cell experiment. Lymphocytes were gated before exclusion of doublets and of CD14/CD15/CD56/IgD+ cells, before sorting of CD19+ cells. (H) Gating strategy for single-cell sorting of SARS-CoV-2 S-specific B cells for single cell culture at M3 and M6. Lymphocytes were gated before double exclusion of doublets, dead cells and CD3/CD14 cells. CD19+ cells were then gated before exclusion of CD38hi plasma cells. CD38int/- cells were further divided into four quadrants using CD27 and IgD. Double stained SARS-CoV-2 S specific cells were sorted from all three non-naive quadrants (not IgD+CD27-).
To further track the SARS-CoV-2-specific MBC response, we next implemented two complementary in-depth approaches on four S-CoV patients’ samples. First, we generated His-tagged trimeric SARS-CoV-2 S ectodomain and used successive staining with His-tagged S protein and two fluorescently labeled anti-His antibodies to stain and perform high purity single-cell sorting of SARS-CoV-2 S-specific B cells (Figure 1E). Single B cells were further cultured in an optimized in vitro culture assay (McCarthy et al., 2018) (Figure 1F). After 21 days of culture, RNA was extracted from single-cell cultures to determine their IgH VDJ sequence. Culture supernatants with IgG concentration over 1 μg/mL were additionally tested by ELISA to validate the cell specificity toward SARS-CoV-2 S protein (over 95% purity in this assay; Figure S1C), investigate its recognition of the SARS-CoV-2 S RBD and its neutralization potential, and analyze potential cross-reactivity to S proteins from seasonal coronaviruses (HCoV-HKU1/HCoV-OC43). In parallel, to get a broader view of the overall B cell response toward SARS-CoV-2 at both early (M0) and late time points (M6), we sorted CD19+IgD− B cells from the same patients (Figures S1D–S1H) and performed single-cell RNA sequencing (scRNA-seq) using the 10X Genomics technology. This allowed us to couple both 5′ scRNA-seq, V(D)J profiling, and feature barcoding of 20 known B cell surface markers, and we also developed anti-His barcoded antibodies to reveal His-tagged SARS-CoV-2 S-specific B cells.
SARS-CoV-2-responding B cells harbor phenotypically and transcriptionally an activated phenotype and a time-dependent maturation toward the MBC lineage
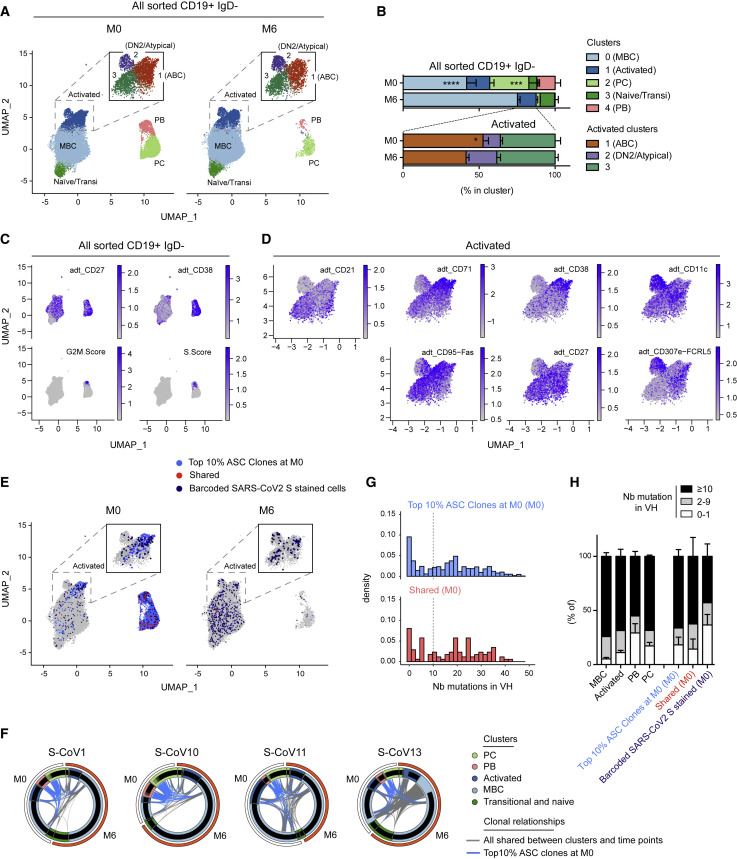
Unsupervised clustering analysis of our scRNA-seq dataset revealed that CD19+IgD− B cells could be divided in five major clusters according to their gene expression profile (Figures 2A and 2B; Figures S2 A and S2B; Table S2), a clustering which correlated nicely with the expression of barcoded surface markers included in our dataset (Figures 2C and 2D; Figure S2C). Two of these clusters corresponded to ASCs, with both a short-lived plasmablast (PB) cluster, enriched for expression of cell proliferation-related genes, and non-dividing PCs. Both PB/PC populations were strongly reduced by 6 months, confirming the resolution with time of the primary extrafollicular response in these severe COVID-19 patients (Figure 2B; Table S2). The remaining B cells were separated in three populations: a mixture of naive/transitional B cells, a resting MBC population and a CD95+ activated cluster. This activated cluster could be further subdivided in three distinct populations: CD21lowCD27+CD38+CD71+ activated B cells (ABCs), CD21lowCD27lowCD38−CD71lowCD11c+FcRL5+ cells, likely corresponding to atypical memory and/or double-negative 2 (DN2) population (Sanz et al., 2019), and CD21+CD27int/+CD38−CD71lowCD95+ cells corresponding to a cluster with intermediate characteristics between ABC and MBCs (Figures 2C and 2D; Figures S2B and S2C). The contraction of the primary response, clearly observed for ASCs, could also be seen, albeit less pronounced, for ABCs, with a parallel increase in resting MBCs. All other clusters remained stable (Figure 2B).
Figure 2.
Characterization of the B cell response against SARS-CoV-2 in acute (M0) and convalescent (M6) S-CoV patients
(A) UMAP and clustering of 41,083 B cells analyzed by scRNA-seq from four S-CoV patients at M0 (left panel) and M6 (right panel) (see Table S2). Upper square in both panels shows the results of increased clustering resolution for the “Activated” cell cluster.
(B) Relative cluster distribution at M0 and M6 for all sorted cells (top panel) and cells falling in the “Activated” cluster at initial clustering resolution. Bar indicates mean with SEM.
(C) Feature plots showing scaled normalized counts for CD27 and CD38 barcoded antibodies as well as S and G2/M signature scores in all cells.
(D) Feature plots showing scaled normalized counts for CD21, CD71, CD38, CD11c, CD95, CD27, and CD307e (FcRL5) barcoded antibodies in cells from the “Activated” cluster.
(E) UMAP of all cells at M0 or M6, with cells belonging to one of the top 10% antibody-secreting cell (ASC) clones highlighted (light blue). Cells stained by the barcoded-anti-His/His-tagged SARS-CoV-2 S protein combination (barcoded-SARS-CoV-2 S stained cells) are also highlighted (red when members of one of the top 10% ASC clones at M0 [“shared”], dark blue otherwise).
(F) Circus plots showing clonal relationships between cells from different UMAP clusters and time points. Blue lines indicate clonal relationships with one of the top 10% ASC clones at M0 and gray line all other clonal relationships.
(G) Histograms showing the number of mutations in VH genes for cells belonging to one of the top 10% ASC clones at M0 according to their barcoded-SARS-CoV-2 S staining (top = unstained; bottom = stained).
(H) Graph showing the frequencies of cells at the M0 time point harboring 0–1 (white), 2–9 (gray), or ≥10 (black) mutations in their VH genes for all sorted cells according to their cluster of origin (left) as well as for cells stained by the barcoded-SARS-CoV-2 S protein and/or members of the top 10% ASC clones at M0, as defined in (E).
Repeated measures (RM) two-way ANOVA and Sidak’s multiple comparison tests (B). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗p < 0.05. See also Figure S2 and Table S2.
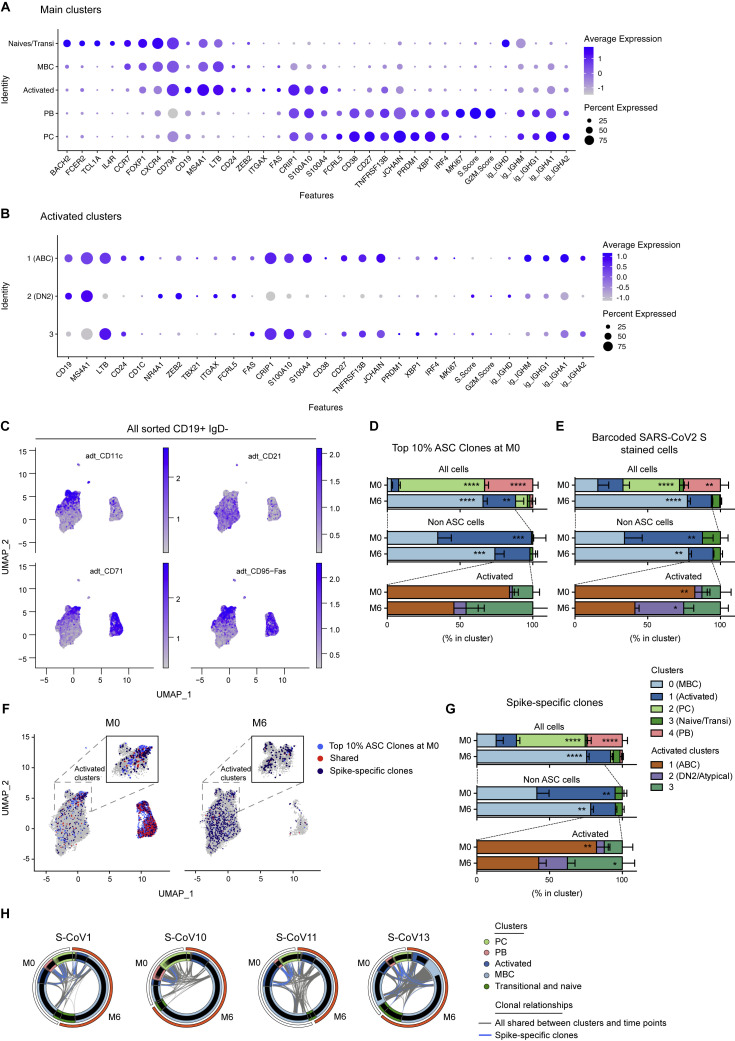
Figure S2.
Gene expression markers and cluster assignments for the main single-cell populations identified in acute and convalescent S-CoV patients, Related to Figure 2
(A and B) Dot plots showing expression of selected genes in cells from the main clusters (A) or in cells from the “Activated” cluster (B). Size of dots represents the percentage of cells in the cluster in which transcripts for that gene are detected. Dot color represents the average expression level (scaled normalized counts) of that gene in the population.
(C) Feature plots showing scaled normalized counts for CD21, CD71, CD11c, CD95 barcoded antibodies in all sorted CD19+IgD-.
(D and E) Relative cluster distribution at M0 and M6 for cells belonging to one of the top 10% ASC clones (D) or stained by the barcoded-anti-His/His-tagged SARS-CoV-2-S protein combination (barcoded-SARS-CoV-2-S) (E). Top panels represent cluster distribution for all cells, middle panels represent cluster distribution for non-ASC cells and bottom panels represent cluster distribution for cells belonging to the “Activated” cluster. Bars indicate mean with SEM.
(F) UMAP of all cells at M0 or M6, with cells belonging to one of the top 10% ASCs clones highlighted (light blue). Cells belonging to Spike-specific clones, as defined in the Methods section, are also highlighted (red when members of one of the top 10% ASC clones at M0, dark blue otherwise).
(G) Relative cluster distribution at M0 and M6 for cells belonging to spike-specific clones. Top panel represent cluster distribution for all cells, middle panel represent cluster distribution for non-ASC cells and bottom panels represent cluster distribution for cells belonging to the “Activated” cluster. Bars indicate mean with SEM.
(H) Circos plots showing clonal relationships between cells from different UMAP clusters and time points. Blue lines indicate clonal relationships with one of the spike-specific clones and gray line all other clonal relationships.
RM two-way ANOVA and Sidak’s multiple comparison tests (D-E, G). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.01, ∗∗p < 0.01, ∗p < 0.05.
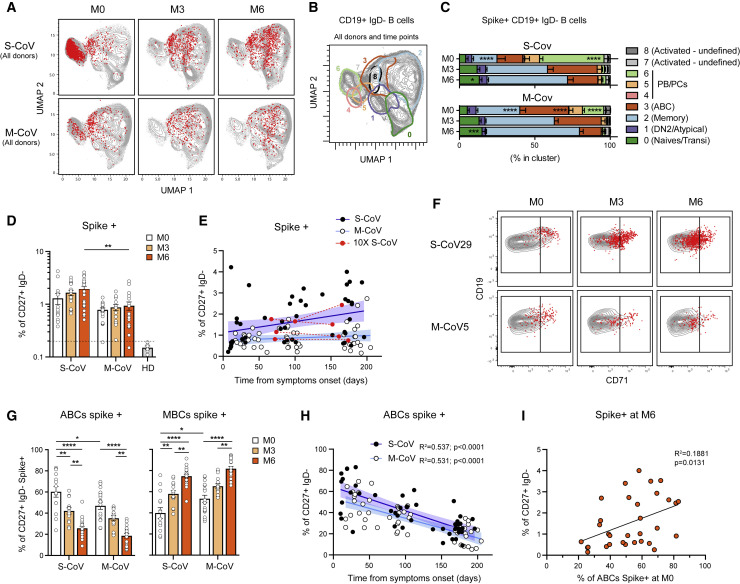
To gain insight into the specificity of this B cell response and the clonal relationship between the different B cell clusters, we first used the initial burst of ASCs seen at M0 as a proxy for the overall SARS-CoV-2-specific B cell response. We thus focused on the top 10% ASC clones, which represents 40%–55% of the PB/PC analyzed (Table S2). Analysis of these top 10% ASC clones revealed a strong link between the ABC cluster and the ASC population (Figures 2E and 2F; Figure S2D; Table S2). The bimodal distribution of SHM seen in ASCs further suggests the existence of a strong extrafollicular reaction recruiting newly engaged naive and MBCs, which fuels the primary response toward SARS-CoV-2 (Figures 2G and 2H). In line with the resolution of the immune response, most of these clones were no longer present in ASCs 6 months after SARS-CoV-2 infection, with only few cells clonally related to the original ASC burst still detectable in residual PC, while they were increased in the resting MBC compartment by that time (Figure 2E; Figure S2D). Parallel analysis of barcoded-S reads in our dataset allowed us to identify clear populations of S-specific ABCs and ASCs at M0 (Figure 2E; Figure S2E; Table S2). By 6 months, S-barcoded-specific cells mainly localized in the resting MBC cluster (Figure 2E). We performed a second analysis of S-specific cells in our dataset, identified either as barcoded-S cells or as cells clonally related to cells identified as S-specific by single-cell culture, to increase their representation (Figures S2G and S2F; description in STAR methods). We found limited clonal overlap of S-specific MBCs at M6 with the initial ASC response, suggesting that two distinct, albeit synchronous, responses take place in COVID-19 patients (Figures S2F–S2H; Table S2).
S-specific MBCs mature from ABCs and accumulate up to 6 months post-infection in COVID-19 patients
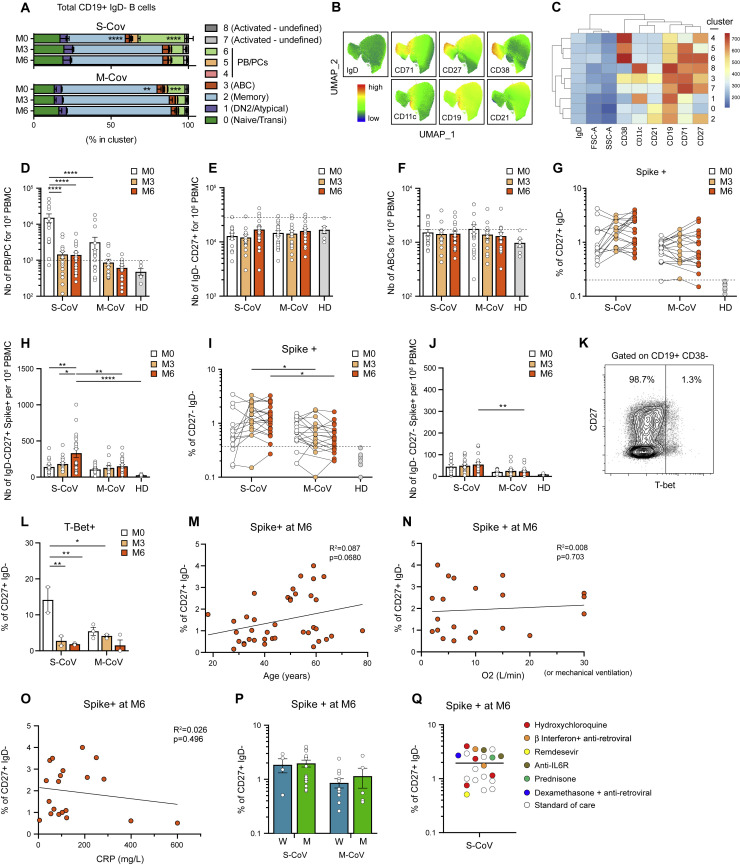
To further analyze the dynamics of the overall and SARS-CoV-2 S-specific B cell response at the level of our entire cohort, we next performed multi-parametric FACS analysis using a flow panel that included seven surface markers identified in our scRNA-seq dataset as major markers of ABCs and ASC subsets involved in the initial response (CD19, CD21, CD27, CD38, CD71, CD11c, IgD, as well as His-tagged trimeric S protein). Unsupervised analysis of CD19+IgD− switched B cell populations (Figures S3 A–S3C), concatenated from 83 samples collected at M0, M3, and M6 from patients in our cohort, confirmed our initial observation that S-specific B cells were enriched in a cluster of CD27+CD38int/+CD71+ ABCs and in ASCs at M0 (Figures 3 A–3C; Table S3). The enrichment in ASCs was most prominent in S-CoV patients and is in line with the strong ASC burst seen in these patients (Figure S3A). S-specific ASCs were marginally detectable at 6 months, at which time point S-specific B cells mostly resided in the CD21+CD27+CD38−CD71int/− resting MBC compartment in both S-CoV and M-CoV convalescent patients. Of note, some S-specific cells were still found in the CD71+ ABC cluster at M6 in both groups of patients but were much less frequent in the DN2/atypical compartment (Figure 3C).
Figure S3.
Phenotypic characterization of total and SARS-CoV-2 S-specific B cell populations in convalescent COVID-19 patients, Related to Figure 3
(A) Cluster distribution of all analyzed CD19+IgD- B cell across FlowSOM-identified clusters for S-CoV (n = 15) and M-CoV (n = 16) donors (Table S1) at all three time points (M0, M3 and M6).
(B) UMAP of concatenated down-sampled cells from all 83 samples analyzed colored according to the scaled fluorescence intensity for IgD, CD71, CD27, CD38, CD11c, CD19, CD21 expression.
(C) Heatmap representing the mean fluorescent intensity for IgD, CD71, CD27, CD38, CD11c, CD19, CD21 in each identified FlowSOM cluster.
(D–F) Absolute number of antibody secreting cells (PC+PB) (D), CD19+CD27+IgD- MBCs (E), and ABCs (F) at indicated time points in S-CoV and M-CoV patients. Bars indicate the mean with SEM. Dashed line indicates the threshold based on healthy donors (mean + 2 SD)
(G and H) Percentage (G) and absolute numbers (H) of SARS-CoV-2 spike-specific CD19+CD27+IgD- MBCs at indicated time points in S-CoV and M-CoV patients.
(I and J) Percentage (I) and absolute number (J) of SARS-CoV-2 spike-specific cells among CD27-IgD- DN cells. (G and I) Each dot represents a patient and lines connect the different time points for each patient. Dashed line indicates the threshold based on healthy donors (mean + 2 SD). (H and J) Bars indicate the mean with SEM.
(K) Representative dot plot for T-bet staining in CD19+CD38- B cells from COVID-19 patients.
(L) Percentage of intra-cellular T-Bet staining in SARS-CoV-2 spike-specific CD19+IgD-CD27+CD38- cells from S-CoV (n = 2) and M-CoV (n = 3) patients at indicated time points. Bar indicate the mean with SEM.
(M–O) Correlation between the number of SARS-CoV-2 spike-specific CD19+CD27+IgD- MBCs at M6 and age at diagnosis (M), maximum oxygen flow during hospitalization (liters/minutes, only in S-CoV) (N) and maximum CRP during hospitalization (mg/L, available only in S-CoV) (O) using linear regression.
(P) Number of spike-specific CD19+CD27+IgD- MBCs at M6 according to the initial disease severity (S-CoV and M-CoV) and the sex of the patient (Women, W, blue and Men, M, green). Bars indicate the mean with SEM.
(Q) Plot showing the number of spike-specific CD19+CD27+IgD- cells in the 21 S-CoV patients analyzed at M6 colored according to the treatment received during initial hospitalization. Standard of care included oxygen, low dose anticoagulant, antibiotic if needed and symptomatic treatments.
RM two-way ANOVA and Sidak’s multiple comparison tests (A). ANOVA and two-tailed Mann-Whitney tests (D–F, H, J, L). Linear regression with Pearson correlation analysis (M-0). ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05.
Figure 3.
Phenotypic evolution of the SARS-CoV-2 S-specific B cell response up to 6 months post-infection in M- and S-CoV patients
(A) UMAP projections of concatenated multi-parametric FACS analysis of CD19+IgD− cells from all S-CoV (n = 15) and M-CoV (n = 16) patients analyzed over time in our cohort (Table S1). His-tagged labeled SARS-CoV-2 S-specific cells are overlaid as red dots.
(B) Unsupervised clustering (FlowSOM) was performed on the concatenated FACS dataset based on IgD, CD71, CD27, CD38, CD11c, CD19, and CD21 fluorescence intensity. 95% density contours for each identified cluster are overlaid in the UMAP projection.
(C) Cluster distribution for all analyzed SARS-CoV-2 S-specific cells in identified clusters at indicated time point. Bars indicate mean with SEM.
(D) Proportion of SARS-CoV-2 S-specific CD19+IgD−CD27+CD38− MBCs at M0, M3, and M6 in S-CoV and M-CoV patients, as compared to six pre-pandemic healthy donor (HD) controls. Each dot represents one patient, and bars indicate mean with SEM. Dashed line indicates the mean + 2 SD of the detected frequency of SARS-CoV-2 S-specific MBCs in HD.
(E) Proportion of S-specific MBCs over time post symptom onset in S-CoV (black dots, dark blue line) and M-CoV (white dots, light blue line) patients. Lines indicates linear regression (R2 = 0.051, p value = 0.02 for S-CoV, and R2 = 0.018, ns, for M-CoV, Pearson correlation). Colored area between dashed lines indicates error bands. Red dots and lines represent data obtained for the four S-CoV patients analyzed as part of our scRNA-seq dataset.
(F) Representative dot plot showing CD71 and CD19 expression in CD19+IgD−CD38− B cell at indicated time points from two representative S-CoV and M-CoV patients. SARS-CoV-2 S-specific cells are overlaid as red dots. Gating strategy for ABCs and classical MBCs according to CD71 expression is further displayed.
(G) Proportion of SARS-CoV-2 S-specific cells displaying an ABC (CD19+CD27+IgD−CD71+) (left) or MBC (CD19+CD27+IgD−CD71low/int) (right) phenotype at indicated time points. Bars indicate mean with SEM.
(H) Proportion of activated MBCs over time post symptom onset in S-CoV (black dots, dark blue line) and M-CoV (white dots, light blue line) patients. Lines represents the linear regression. Colored area between dashed lines indicates error bands. R2 and p value with Pearson correlation.
(I) Correlation between the frequency of SARS-CoV-2 S-specific MBCs at M6 (expressed as % of CD27+IgD− B cells) and of ABCs at M0 in all M-CoV and S-CoV patients.
RM two-way ANOVA and Sidak’s multiple comparison tests (C). ANOVA and two-tailed Mann-Whitney tests (D and G). Linear regression with Pearson correlation analysis (I). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.01, ∗∗p < 0.01, ∗p < 0.05. See also Figure S3 and Table S3.
Based on these results, and using a more traditional gating strategy, we next focused our analysis on the switched CD27+IgD−CD38int/− MBC compartment, which includes both ABCs and resting MBCs (Figure S1D). The first overall analysis confirmed a contraction of the ASC compartment, with a stable size of both IgD−CD27+ B cells and ABCs with time (Figures S3D–S3F). In contrast to the rapid disappearance of S-specific ASCs, both the percentage and absolute number of S-specific CD27+IgD− B cells appeared stable up to 6 months in the vast majority of patients in our cohort, and even continuously increased up to that time point in a subset of convalescent S-CoV patients (Figures 3D and 3E; Figures S3G and S3H). Of note, and as previously mentioned, the double-negative population contained few S-specific cells (Figures S3I and S3J).
Convalescent S-CoV patients showed significantly higher frequencies of S-specific MBCs at the 6-month time point of our study. Most M-CoV patients, however, still harbored a sizeable population of S-specific MBCs at 6 months post-infection (mean 0.94% ± 0.17% of MBCs) and only one (M-CoV24) out of the 16 M-CoV patients analyzed at 6 months in our study showed a frequency of S-specific switched CD27+ MBCs below that of pre-pandemic healthy donors. Even more striking was that both M-CoV patients whose serum levels of S-specific IgG had dropped below detectable levels by 6 months (correlating with an absence of in vitro neutralizing potential) still harbored a clear population of S-specific MBCs at that time point (0.41% and 0.60% of MBCs, respectively, both above the threshold of 0.20% (>2 SD of the mean value observed in healthy donors). Altogether, these results demonstrate the induction of a robust and stable S-specific MBC population in both M- and S-CoV patients. Aside from severity, none of the clinical parameters or treatment that we could monitor, notably corticosteroids, appeared to have any clear influence on the long-term establishment of these S-specific MBCs (Figures S3M–S3Q).
To ensure that such stable levels of S-specific CD27+ switched MBCs did not simply reflect a sustained immune response in these convalescent COVID-19 patients, we further used CD19 and CD71 to separate ABCs (CD19highCD71+) and resting MBCs (CD19+CD71low) among S-specific cells over time, as previously described (Ellebedy et al., 2016). Confirming our initial observation in the four S-CoV patients included in the scRNA-seq dataset (Figure 2E) and the clear enrichment of S-specific B cells in cluster 3 of our unsupervised FACS analysis (Figures 3A–3C), the majority of S-specific MBCs displayed a CD19highCD71+ ABC phenotype in the days following infection (Figure 3F). In both S-CoV and M-CoV patients, the proportion of S-specific ABCs steadily decreased over time, along with an increase of S-specific classical, resting MBCs (Figures 3G and 3H). Interestingly, ABCs were still detectable in both cohorts at 6 months (approximately 20% in both cohorts), suggesting continued antigen-driven activation, as has been described after Ebola infection (Davis et al., 2019). In S-CoV patients, a small fraction of these early responding MBCs expressed the transcription factor T-bet, but such expression appeared only transient (Figures S3K and 3L). More interestingly, we noted a significantly higher frequency of ABCs among S-specific MBCs in S-CoV as compared to M-CoV patients, as well as a large inter-patient variability for this parameter in both groups (Figure 3G). This heterogeneity could not be simply explained by inter-patient variations in the time of initial sampling. To evaluate whether early B cell activation may impact later formation of MBCs, we plotted the frequency of CD27+CD71+ ABC cells at M0 versus the frequency of S-specific MBCs at M6. Interestingly, there was a positive, albeit modest, correlation between both values, indicating that such early B cell activation does not prevent the development of B cell memory against SARS-CoV-2 (Figure 3I).
Acquisition of somatic mutations in RBD-specific Ig VH genes of MBCs
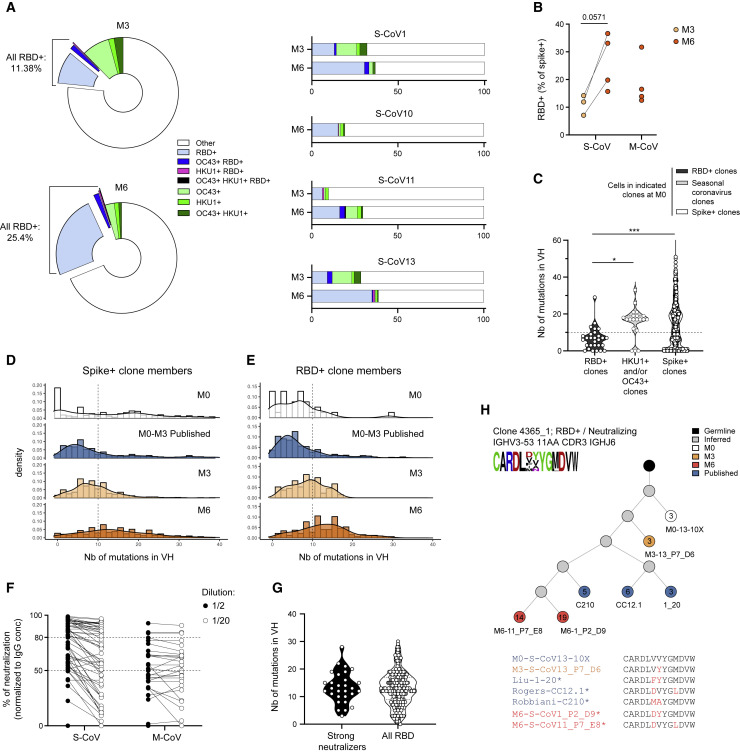
In order to better understand the origin, fine specificity, and developmental pathways involved in the generation of MBCs against SARS-CoV-2, we next performed single-cell sorting and culture of S-specific MBCs at 3 and 6 months post-infection in all four donors previously analyzed in the context of our scRNA-seq experiment (Figure 1E). A total of 2,412 cells at M3 and 2,423 cells at M6 were sorted from the four donors (Table S4), and 488 and 1,027 supernatants (respectively at M3 and M6) post single-cell culture were tested for specificity against the SARS-CoV-2 S protein, S proteins from two other related seasonal betacoronaviruses (HCoV-HKU1 and HCoV-OC43) (Huang et al., 2020), and the RBD domain of the SARS-CoV-2 S protein, which is the main target of neutralizing antibodies in COVID-19 patients (Figures 4 A; Table S4). We found that a sizeable proportion of SARS-CoV-2 S-specific MBCs also recognized the S protein of HCoV-HKU1 and HCoV-OC43, with many of these cells recognizing both, as previously reported (Shrock et al., 2020; Wec et al., 2020). The proportion of HCoV-HKU1- and HCoV-OC43-specific MBC clones, however, readily decreased between M3 and M6 (12.4% ± 4.9% to 4.8% ± 1.7% respectively), suggesting that these specificities, while actively participating in the early response, are not preferentially recruited in the later MBC compartment. In contrast, the proportion of SARS-CoV-2 RBD-specific MBC clones clearly increased between the M3 and M6 time points in all donors (11.1% ± 2.1% and 26.3% ± 5.1% respectively) (Figures 4A and 4B; Figure S4 A). SARS-CoV-2 RBD-specific clones were mostly sorted from resting CD71−CD27+ switched MBCs at these late time points (Figures S4B and S4C) and were detectable within the same proportion in M-CoV and S-Cov patients at 6 months (Figure 4B). Finally, only a minority of SARS-CoV-2 RBD-specific clones cross-reacted with HCoV-HKU1 and HCoV-OC43 S protein, as anticipated by the low similarity of the RBD domain between these different viruses (Figures 4A). Overall, these results suggested an early and rapid recruitment of cross-reactive MBCs through the extrafollicular pathway, in parallel with the establishment of a new immune response against epitopes unique to SARS-CoV-2, with a delayed and progressive output of MBCs.
Figure 4.
Maturation of the SARS-CoV-2 S-specific repertoire up to 6 months post-infection in M- and S-CoV patients
(A) Left: pie charts showing the average percentage of RBD+ and cross-reactive specificities (RBD+OC43+, RBD+HKU1+OC43+, RBD+HKU1+, OC43+, HKU1+, and OC43+HKU1+) among single-cell cultured S-specific B cells as determined by ELISA. Three and four S-CoV patients, all previously included in our scRNA-seq dataset, were analyzed at M3 and M6, respectively. Right: bar plot showing the proportion of RBD specificity and cross-reactive specificities among S-specific cells for each patient.
(B) Average percentage of RBD-specific cells among SARS-CoV-2 S-specific sorted cells from indicated S-CoV and M-CoV patient at M3 and M6.
(C) Violin plot showing the number of mutations in the Ig VH segment of cells in the original M0 10X Genomics scRNA-seq VDJ dataset found to be in clonal relationship with M3 or M6 SARS-CoV-2 S-specific sorted cells showing specificity against RBD (RBD+ clones), seasonal betacoronavirus S proteins (HKU1-CoV or OC43-CoV) or against SARS-CoV-2 S (Spike+ clones).
(D and E) Histograms showing the relative distribution of mutation numbers in the Ig VH segment from all SARS-CoV-2 S- (D) or RBD- (E) specific clone members (10X Genomics scRNA-seq dataset and single-cell heavy-chain sequencing data) at M0, M3, and M6 as well as from sequences from the literature (Brouwer et al., 2020; Kreer et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Shi et al., 2020; Wec et al., 2020; Zost et al., 2020), mainly determined between M0 and M3.
(F) Plot showing the percentage of neutralization, normalized to the IgG concentration, for single-cell culture supernatants from identified SARS-CoV-2 RBD-specific B cells at M6. Two dilutions (1/2 and 1/20) were assayed for each supernatant tested. Dashed lines indicate 80% and 50% neutralization.
(G) Violin plot representing the number of mutations in the Ig VH sequence in strong neutralizing antibodies at M6 (neutralization >80% at 1/2 dilution) versus the number of mutations in the Ig VH sequence of all cells from anti-RBD clones at M6.
(H) Evolutionary tree of an RBD-specific and neutralizing clone, built on sequences from 10X Genomics scRNA-seq, cell culture, and literature. Each circle represents a unique sequence from that clone. Circle color indicates time point of origin, and the number inside indicates the calculated number of mutations from an inferred unmutated common ancestor (“germline”). Grey indicates a theoretically inferred common precursor. ∗ indicates that the antibody associated with that sequence has been validated as neutralizing in vitro. CDR3 from all sequences in the tree are represented as a frequency plot logo (top left) as well as below the tree, where each amino acid in red indicates a change compared to the top listed CDR3.
ANOVA and two-tailed Mann-Whitney tests (B and C). ∗∗∗p < 0.01, ∗p < 0.05. See also Figure S4 and Table S4.
Figure S4.
Cross-reactivity, convergence, and accumulation of somatic mutations in anti-SARS-CoV-2 S-specific memory B cells from convalescent COVID-19 patients, Related to Figure 4
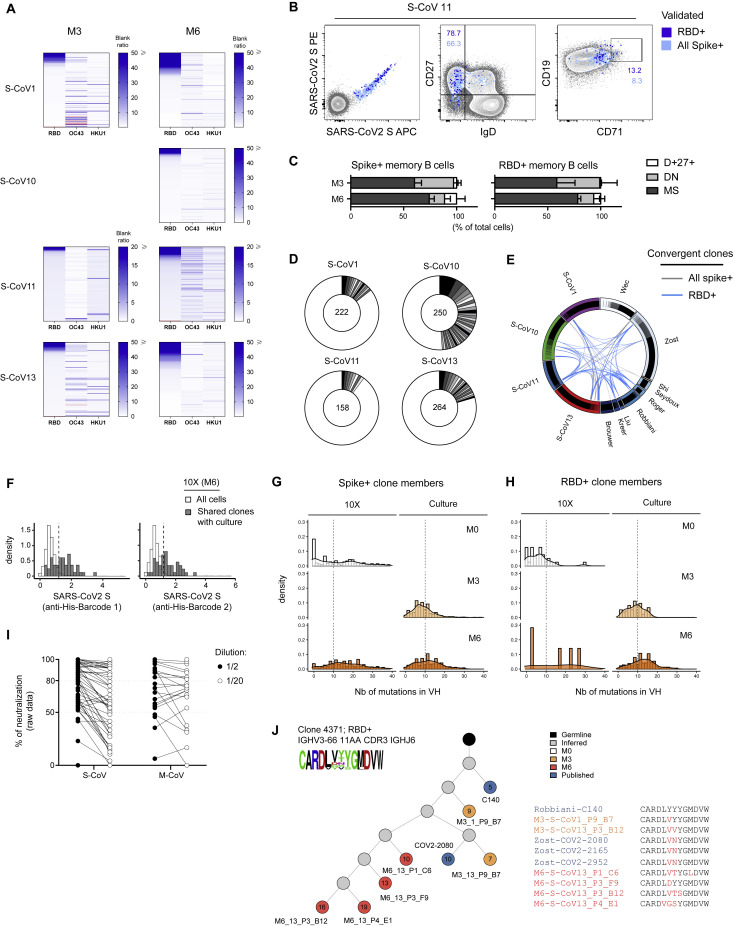
(A) Heatmaps showing the RBD, OC43 and HKU1 ELISA blank ratio for all tested SARS-CoV-2-spike-specific single cell culture supernatants at M3 and M6. Each line represents one tested supernatant. Light red lines indicate that no value is available for that supernatant.
(B) FACS plots representing index sorting data of spike-specific cells according to their specificity for SARS-CoV-2 spike or RBD.
(C) Repartition of the sorted SARS-CoV-2 spike- and RBD-specific cells at M3 and M6 between the MBCs, DN or IgD+CD27+ compartments.
(D) Pie chart representing clone size in all the sequences generated via single cell culture for each of the 4 patients previously included in our scRNaseq analysis. Total number of sequenced cells is indicated in the middle of the pie.
(E) Circus plot showing SARS-CoV-2 spike- (gray) or RBD- (light blue) specific clones shared between patients in our dataset or with sequences from the literature.
(F) Histogram showing log-normalized counts for both barcoded his-tagged SARS-CoV-2 S proteins in all cells from the M6 time point of our 10X Genomics scRNA-seq dataset (white) and cells in clonal relationship with sequence from single cell culture (dark gray)
(G and H) Histograms showing the distribution of mutations in Ig VH for SARS-CoV-2 spike- (G) and RBD- (H) specific clones at M0, M3 and M6 according to the assay of origin.
(I) Plot showing the raw percentage of neutralization for single cell culture supernatants from identified SARS-CoV-2 RBD-specific B cells at M6. Two dilutions (1/2 and 1/20) were assayed for each supernatant tested. Dashed lines indicate 80% and 50% neutralization (see Figure 4F).
(J) Evolutionary tree of a convergent RBD-specific and neutralizing clone, built on sequences from 10X Genomics scRNA-seq and cell culture from two patients and from the literature. Each circle represents a unique sequence from that clone. Circle color indicates time-point of origin and the number inside indicates the calculated number of mutations from an inferred unmutated common ancestor (germline). Grey indicates a theoretically inferred common progenitor. CDR3 from all sequences in the tree are represented as a frequency plot logo (top left) as well as below the tree, where each amino-acid in red indicates a change compared to the first listed CDR3 sequence.
To evaluate whether such MBC output corresponds to an ongoing GC response, we next evaluated the occurrence and evolution of somatic mutations in S-specific MBCs. Heavy-chain VH sequencing from SARS-CoV-2 S- or RBD-specific wells (Table S4) in our in vitro culture assay, at both M3 and M6 (Figure S4D), allowed us to identify clonal relationships with sequences obtained in our scRNA-seq dataset at both M0 and M6. Mutations in VH sequences of SARS-CoV-2 S-specific clones at M0 displayed a bimodal distribution with both near germline and highly mutated sequences. In line with an initial recruitment of cross-reactive MBCs into the anti-SARS-CoV-2 early humoral response, HCoV-HKU1- and HCoV-OC43-specific clones contained sequences that were already highly mutated at M0 (Figure 4C). In contrast, SARS-CoV-2 RBD-specific clones displayed low mutation numbers in VH sequences, as previously described in numerous studies during the early stages of this pandemic (Brouwer et al., 2020; Kreer et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Wec et al., 2020; Zost et al., 2020). Both early SARS-CoV-2 S- and RBD-specific Ig VH sequences showed high levels of convergence between donors in our dataset but also with published SARS-CoV-2 S- and RBD-specific antibody sequences (43/874 sequences in clonal relationship with cells from our dataset) (Figure S4E). Although near germline sequences were initially present in S-specific clones, and even predominated in those with SARS-CoV-2 RBD specificity in both our M0 dataset and in the literature, longitudinal tracking of clonal relationships at 3 and 6 months revealed a progressive acquisition of mutations in VH sequences over time (Figures 4D–4E; Figures S4G and S4H). Most importantly, over 85% of SARS-CoV-2 RBD-specific culture supernatants at M6 showed intermediate to strong neutralizing potential against SARS-CoV-2 in vitro in S-CoV patients and over 50% in M-CoV patients (Figure 4F; Figure S4I; Table S4). Progressive accumulation of mutations in VH sequences was not associated with a loss of neutralization potential (Figure 4G; Table S4). In contrast, a strong neutralizing capacity was identified along lineage trees including previously described near-germline anti-SARS-CoV-2 RBD antibodies and the mutated versions that we identified at M6 in this work. (Figures 4H and S4J). Dynamics of such clones and emergence of highly mutated neutralizing sequences, along with the robustness of the memory response observed in S-CoV and M-CoV patients, demonstrate that MBCs bearing unique specificities and neutralizing potential against SARS-CoV-2 can expand and mature over time through an ongoing GC response.
Discussion
The long-term stability of humoral immune memory following viral infection is provided by separate pools of protective LLPCs and MBCs that can last for a lifetime (Amanna et al., 2007; Crotty et al., 2003; Mamani-Matsuda et al., 2008). Upon reinfection, long-lived MBCs generate a new wave of short-lived PCs and LLPCs while preserving the pool of MBCs. In this study, we focused on SARS-CoV-2-infected patients with mild or severe disease and investigated the establishment of humoral memory and how this memory relates to the primary B cell response that emerges at the early stage of infection.
Experimental infections of volunteers with betacoronaviruses responsible for common colds and longitudinal serological studies have suggested the absence of durable immunity against these seasonal infections (Sariol and Perlman, 2020). Furthermore, early reports have shown potentially defective GC formation in S-CoV patients in the intensive care unit (Kaneko et al., 2020) and predominantly near-germline anti-SARS-CoV-2 neutralizing antibodies (Ju et al., 2020; Nielsen et al., 2020; Robbiani et al., 2020). Such observations suggested a strong bias of the anti-SARS-CoV-2 immune response toward the extrafollicular pathway in these patients, which may preclude the formation of long-term memory.
Our study demonstrates, in contrast, a remarkable stability of the overall S-specific MBC population up to 6 months after infection, and even its expansion in severe patients, extending recent observations on memory persistence in COVID-19 (Gaebler et al., 2021; Rodda et al., 2020). This is also consistent with recent results showing in a large cohort of patients that SARS-CoV-2 antigen-specific MBCs persist up to 8 months after infection (Dan et al., 2021).
Tracking of S-specific B cells provided a more complex picture than previously anticipated, characterized by two synchronous responses with distinct dynamics throughout the extrafollicular reaction. Indeed, near-germline B cell clones recognizing the SARS-CoV-2 S protein were mobilized but also pre-existing highly mutated MBCs specific for the S protein of other seasonal betacoronaviruses.
The longitudinal VDJ sequencing of S-specific MBCs revealed that neutralizing SARS-CoV-2 RBD-specific clones, including convergent antibody rearrangements across donors, accumulated with time and acquired somatic mutations in their VH genes, the hallmark of a GC-dependent immune response. Such GC imprinting is of major importance because it usually predicts the generation of long-lived MBCs (Mesin et al., 2016). Along this line, a recent report observed the presence of viral proteins in the intestine of patients several months after the initial diagnostic, which could sustain such B cell response (Gaebler et al., 2021).
Cross-reactivity against the S domain of betacoronaviruses has already been documented in serological studies showing reactivity to conserved regions of the S protein (Aydillo et al., 2020; Wang et al., 2020), by the presence of pre-existing antibodies recognizing conserved epitopes in the S2 protein of SARS-CoV-2 in uninfected individuals (Ng et al., 2020), and by cross-reactivity of sorted S-specific B cells with seasonal coronavirus (Song et al., 2020; Wec et al., 2020). Such cross-reactivity also exists for T cells (Braun et al., 2020; Grifoni et al., 2020; Le Bert et al., 2020), but its protective potential remains debated (Ng et al., 2020). The proportion of cross-reactive clones, however, decreased from 3 to 6 months, suggesting that, after an initial activation, these cross-reactive clones were not positively selected in the memory pool.
As observed previously by others for antibody titers (Ripperger et al., 2020), the overall strength and stability of the MBC response was positively correlated with the initial severity of COVID-19-associated pathologies in convalescent patients. Interestingly, the magnitude of the initial SARS-CoV-2-specific ABC response was not predictive of an impaired B cell memory, displaying in contrast a positive, albeit modest, correlation with the frequency of S-specific MBCs at 6 months. ABCs in COVID-19 patients have been previously reported with various phenotypes (Juno et al., 2020; Mathew et al., 2020; Oliviero et al., 2020; Woodruff et al., 2020), including a double-negative CD11c-positive population (DN2). One study has also suggested the presence of SARS-CoV-2 RBD-specific DN2 cells as a correlate of critical illness (Kaneko et al., 2020). We found here that most S-responding B cells harbored an activated phenotype that notably differed from DN2 but was similar to the one described after Ebola or influenza virus infection (Ellebedy et al., 2016). These SARS-CoV-2 S-specific ABCs were in a clonal relationship with expanded cells from the initial burst of ASCs and also with resting MBCs that persisted at 6 months. Given the limited clonal overlap between the early ASC burst and the later mutated MBC response, it is tempting to suggest that a distinct part of ABCs is dedicated to fueling the GC response in these patients while another part contributes to the ASC burst. Of note, the ABC population remains detectable up to 6 months post-infection in convalescent patients, with some, albeit few, S-specific cells, suggesting, as described for Ebola (Davis et al., 2019), an antigen-driven activation that persisted for months after infection.
Collectively, taking into account this report and previous studies, the COVID-19 infection seems in most non-critical cases to induce both an immediate, possibly protective antibody response, together with the ongoing maturation of MBCs, which should give rise to neutralizing ASCs upon reinfection. While this overall picture is positive, notably in the context of vaccine campaigns to come, the long-term duration of the protective MBC response remains an open question.
Limitations of study
The relatively small size of the cohort does not allow us to identify patients’ characteristics that would predict the maintenance of the immune memory. Furthermore, patients with S- and M-CoV were not matched for age and comorbidities, thus differences between these two groups must be interpreted with caution. Finally, this study did not investigate whether MBCs are generated after asymptomatic COVID-19. Thus, larger cohorts of individuals with a wider spectrum of disease severity would be mandatory in the future to provide a comprehensive picture of B cell memory against SARS-CoV-2.
STAR★METHODS
Key resources table
| REAGENT OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibody | ||
| CD3 | Biolegend | UCHT1; Cat#300425; RRID: AB_830754 |
| CD14 | BD Bioscience | M ϕP9; Cat#561709; RRID: AB_1645464 |
| CD19 | BD Bioscience | HIB19; Cat# 562321; RRID: AB_11154408 |
| CD38 | BD Bioscience | HIT2; Cat# 551400; RRID AB_394184 |
| CD27 | Biolegend | M-T271; Cat# 356417; RRID: AB_2562598 |
| CD11c | BD Bioscience | S-HCL-3; Cat#744436; RRID: AB_2742232 |
| IgD | Life technologies | Polyclonal; Cat# H15501; RRID: AB_2536563 |
| His Tag | Biolegend | J095G46; Cat# 362605; RRID:AB_2563634 |
| His Tag | Biolegend | J095G46; Cat# 362603; RRID:AB_2715818 |
| CD71 | Biolegend | CY1G4; Cat# 334111; RRID:AB_2563118 |
| CD21 | BD Bioscience | B-ly4; Cat#563163; RRID:AB_2741028 |
| CD14 | BD Bioscience | M5E2; Cat#555398; RRID:AB_396590 |
| CD15 | BD Bioscience | HI98; Cat#562371; RRID:AB_395802 |
| CD56 | BD Bioscience | B159; Cat# 556647; RRID:AB_395906 |
| IgD | BD Bioscience | IA6-2; Cat# 555779; RRID:AB_396114 |
| CD3 | BD Bioscience | UCHT1; Cat#561027; RRID:AB_10561682 |
| T-Bet | Biolegend | 4B10; Cat#644823; RRID:AB_2561760 |
| CD38 | Biolegend | HIT2; Cat#303509; RRID:AB_314361 |
| CD38 (Totalseq-C) | Biolegend | HB-7; Cat# 356637; RRID:AB_2820007 |
| CD138 (Totalseq-C) | Biolegend | MI15; Cat#356539; RRID:AB_2810567 |
| CD27 (Totalseq-C) | Biolegend | O323; Cat#302853; RRID:AB_2800747 |
| CD11c (Totalseq-C) | Biolegend | S-HCL-3; Cat#371521; RRID:AB_2801018 |
| CD21 (Totalseq-C) | Biolegend | Bu32; Cat#354923; RRID:AB_2800953 |
| CD71 (Totalseq-C) | Biolegend | CY1G4; Cat#334125; RRID:AB_2800885 |
| CXCR4 (Totalseq-C) | Biolegend | 12G5; Cat#306533; RRID:AB_2800791 |
| CXCR3 (Totalseq-C) | Biolegend | G025H7; Cat#353747; RRID:AB_2800949 |
| CD69 (Totalseq-C) | Biolegend | FN50; Cat#310951; RRID:AB_2800810 |
| HLA-DR (Totalseq-C) | Biolegend | L243; Cat#307663; RRID:AB_2800795 |
| CD307d (FCRL4; Totalseq-C) | Biolegend | 413D12; Cat#340213; RRID:AB_2832666 |
| CD307e (FCRL5; Totalseq-C) | Biolegend | 509f6; Cat#340309; RRID:AB_2819969 |
| CD95 (Fas; Totalseq-C) | Biolegend | DX2; Cat#305651; RRID:AB_2800787 |
| Integrin B7 (Totalseq-C) | Biolegend | FIB504; Cat#321229; RRID:AB_2810481 |
| CD49d (Integrin alpha 4; Totalseq-C) | Biolegend | 9F10; Cat#304345; RRID:AB_2814137 |
| CD103 (Totalseq-C) | Biolegend | Ber-ACT8; Cat#350233; RRID: AB_2800933 |
| LAG-3 (Totalseq-C) | Biolegend | 11C3C65; Cat#369335; RRID:AB_2814327 |
| TIM3 (Totalseq-C) | Biolegend | F38-2E2; Cat#345049; RRID:AB_2800925 |
| CD305 (LAIR1; Totalseq-C) | Biolegend | NKTA255; Cat#342807; RRID:AB_2832668 |
| NKG2D (Totalseq-C) | Biolegend | 1D11; Cat#320837; RRID:AB_2800844 |
| CD73 (Totalseq-C) | Biolegend | AD2; Cat#344031; RRID:AB_2800916 |
| His Tag (Totalseq-C) | Biolegend and custom conjugation | J095G46; Cat#362601; RRID: AB_2721736 |
| His Tag (Totalseq-C) | Biolegend and custom conjugation | J095G46; Cat#362601; RRID: AB_2721736 |
| N SARS-CoV-2 (Rabbit polyclonal) | Institut Pasteur, N. Escriou | N/A |
| S SARS-CoV-2 (Human monoclonal) | Institut Pasteur, H. Mouquet | N/A |
| Biological samples | ||
| Cryopreserved PBMCs from S-Cov and M-Cov patients | Henri Mondor Hospital, Assistance Publique des Hôpitaux de Paris | N/A |
| Cryopreserved PBMCs from pre-pandemic subjects | French Blood Agency (EFS) | N/A |
| SARS-CoV-2 virus (BetaCoV/France/IDF0372/2020) | Institut Pasteur, CNR Respiratory Viruses (S.Van der Werf) | N/A |
| Chemical, peptides, and recombinant proteins | ||
| SARS-CoV-2 Spike | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| SARS-CoV-2 RBD | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| HKU1 Spike | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| OC43 Spike | Institut Pasteur, Virologie Structurale (F. Rey) | N/A |
| Live dead aqua | Life technologies | Cat#L34957 |
| Recombinant human IL-2 | PeproTech | Cat#200-02 |
| Recombinant human IL-4 | PeproTech | Cat#200-04 |
| Recombinant human IL-21 | PeproTech | Cat#210-21 |
| Recombinant human BAFF | PeproTech | Cat#310-13 |
| Oligonucleotide Conjugation Kit | Abcam | Cat#ab218260 |
| Deposited Data | ||
| Single-cell RNAseq data | This study | ArrayExpress: E-MTAB-9995 |
| Software and algorithms | ||
| Kaluza v2.1 | Beckman Coulter | https://www.beckman.fr |
| Flowjo v10.7.1 | FlowJo, LLC | https://www.flowjo.com |
| GraphPad Prism v8 | GraphPad | https://www.graphpad.com |
| Codon Code Aligner v9 | Codon Code Corporation | https://www.codoncode.com/ |
| Python v3.7.6 | Python Software Foundation | https://www.python.org/ |
| R v4.0.2 | R Foundation | https://www.r-project.org |
| RStudio v1.3.1056 | RStudio | https://rstudio.com |
| IgBLASTn v1.16.0 | NCBI | https://www.ncbi.nlm.nih.gov/igblast/ |
| Docker desktop v2.5.0.0 | Docker, Inc | https://www.docker.com/products/docker-desktop |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Matthieu Mahévas (matthieu.mahevas@aphp.fr).
Materials availability
No unique materials were generated for this study.
Data and code availability
The accession number for all the single cell RNA sequencing data reported in this paper is ArrayExpress: E-MTAB-9995. Single cell culture VDJ sequencing data reported in Figure 4 are directly included in this study as part of Table S4.
Experimental model and subject details
Study participants
In total 39 patients with COVID-19 were recruited. We included 21 COVID-19 patients requiring oxygen (S-CoV) and 18 healthcare workers, with a mild COVID-19 disease (M-CoV). SARS-CoV-2 infection was defined as confirmed reverse transcriptase polymerase chain reaction (RT-PCR) on nasal swab or clinical presentation associated with typical aspect on CT-scan and/or serological evidence. Samples were collected in mean 18.8 (±SD: 8.8 days) days after disease onset in S-CoV, and in mean 35.5 days (±SD: 12.8 days) for M-CoV. Samples were additionally collected 3 months (mean ± SD: 89.8 ± 15.8 days for S-CoV and 94.2 ± 8.2 for M-CoV), and 6 months (mean ± SD: 170.5 ± 12.9 days for S-CoV and 184.6 ± 10.2 for M-CoV) after onset. Clinical and biological characteristics of these patients are summarized in Table S1. B cells from four S-CoV patients were further analyzed by scRNaseq, these four patients being selected for the absence of specific treatment (corticosteroids, IL6/IL6R inhibitors) that could interfere with the B cell response. Patients were recruited at Henri Mondor University Hospital (AP-HP), between March and May 2020. Samples from healthy donors were obtained at EFS Henri-Mondor and frozen before 2019. MEMO-COV-2 study (NCT04402892) was approved by the ethical committee Ile-de-France VI (Number: 40-20 HPS), and was performed in accordance with the French law. Written informed consent was obtained from all participants.
Method details
SARS-CoV-2 viral detection by RT-PCR
Nasopharyngeal (NP) swabs in transport media and plasma samples were held at 4°C (or −80°C if > 12 h) prior to testing on the Alinity M SARS CoV-2 Amp kit and/or Cepheid Xpert Xpress SARS-CoV-2 kit. The GeneXpert ®Dx System (Cepheid) and Alinity M (Abbott) performed automated specimen processing and real-time RT-PCR analysis. The Cepheid Xpert Xpress SARS-CoV-2 kit detects N and E genes of the SARS CoV-2 genome; Alinity M SARS CoV-2 Amp kit detects RdRp and N genes.
Anti-S and anti-N commercial assays
Serological assays were performed for IgG anti-N, IgG anti-S and total antibodies (ab) anti-S detection. Serum samples were processed for anti-Nucloprotein (N) detection on Abbott SARS-CoV-2 IgG chemiluminescent microparticle immunoassay following the manufacturer’s instructions. Serum samples were further analyzed using the VITROS Immunodiagnostic Products, for IgG anti-spike (S) SARS-CoV-2 detection and for total Ab anti-spike (S) detection (Ortho Clinical Diagnostics). All assays were performed by trained laboratory technicians according to the respective manufacturer’s standard procedures. Qualitative results and index values reported by the instruments were used in analysis.
Recombinant protein purification
Construct design
The human coronavirus Spike (S) proteins were expressed as ectodomains that were stabilized to preserve their trimeric prefusion conformation. The SARS-CoV-2 spike protein (residues 1-1208) was stabilized by introducing six proline substitutions (F817P, A892P, A899P, A942P, K986P, V987P), a GSAS substitution at the furin cleavage site (residues 682–685) and a C-terminal Foldon trimerization motif (Hsieh et al., 2020), followed by Hisx8 and Strep tags. This construct was cloned with its endogenous signal peptide in pcDNA3.1(+). The OC43 S ectodomain (residues 15-1263) was cloned in the pCAGGS vector with a CD5 N-terminal signal peptide and a C-terminal GCN4 trimerization motif, a thrombin cleavage site and a single Strep-tag. Additionally, it contains mutations to abolish the furin cleavage (754-RRSRG-758 to 754-GGSGG-758) at the S1-S2 junction (Tortorici et al., 2019). The HKU1-CoV S protein (residues 14-1276) was stabilized by a double-proline mutation (N1067P, L1068P), the substitution of the RRSRG motif by GGSGG (residues 754-758) to avoid a potential S1/S2 furin cleavage, and a C-terminal Foldon motif. This construct was cloned in pcDNA3.1(+) with an IgK signal peptide and a thrombin cleavage site at the C terminus that was followed by a Hisx8 tag.
The SARS-CoV-2 Receptor Binding Domain (RBD) was cloned in pcDNA3.1(+) encompassing residues 331-528 from the Spike ectodomain, and it was flanked by an N-terminal IgK signal peptide and a C-terminal Thrombin cleavage site followed by a Hisx8-tag.
Protein expression and purification
The plasmids coding for the recombinant proteins were transiently transfected in Expi293F™ cells (Thermo Fischer) using FectroPRO® DNA transfection reagent (Polyplus), according to the manufacturer’s instructions. The cells were incubated at 37°C for 5 days and then the culture was centrifuged and the supernatant was concentrated. The proteins were purified from the supernatant by affinity and size-exclusion chromatography (SEC). The first purification step was performed using StrepTactin columns (IBA) (SARS-CoV-2 S, OC43 S) or His-Trap™ Excel columns (GE Healthcare) (HKU1 S, SARS-CoV-2 RBD). The different S ectodomains were further purified using a Superose6 10/300 colum (GE Healthcare) equilibrated in PBS, while a Superdex200 10/300 column was used for SARS-CoV-2 RBD.
Flow cytometry and cell sorting
PBMCs were isolated from venous blood samples via standard density gradient centrifugation and used after cryopreservation at −150°C. Cells were thawed using RPMI-1640 (GIBCO) 10% FBS, washed twice and incubated with 10 μg of the SARS-CoV-2 His-tagged spike protein in 100 μL of PBS (GIBCO) 2% FBS during 20 min on ice. Cells were washed and resuspended in the same conditions, then the fluorochrome-conjugated antibody cocktail including the 2 anti-His antibodieswas added at pre-titrated concentrations for 20 min at 4°C and viable cells were identified using a LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific) incubated with conjugated antibodies (see key resources table). If a permeabilization was needed (T-Bet staining), cells were washed and resuspended in 250 μL of Fix/perm (eBioscience) for 25 min, then washed with appropriate buffer before adding the conjugated antibody on the cell pellet for a further 25 min at 4°C. Samples were acquired using a LSR Fortessa SORP (BD Biosciences). For cell sorting, cells were stained using the same protocol and then sorted in 96 plates using an ultra-purity mode on a MA900 cell sorter (SONY), or Aria III (BD Biosciences). Data were analyzed with FlowJo or Kaluza softwares. Detailed gating strategies for individual markers are depicted in Figure S1. Control samples included PMBC collected from heathy donors before SARS-CoV-2 pandemic and incubation of S-CoV and M-CoV samples with anti-His antibodies but no His-tagged spike protein to determine the labeling background.
For UMAP generation and visualization (Figures 3A–3C), data from all 83 samples from patients with complete panel acquisition at M0, M3 and M6 (Table S1) in our dataset were individually down-sampled to 3000 cells each using the Downsample (v3.3) plugin in FlowJO. All samples were subsequently concatenated and FlowJO UMAP (v3.1) plugin was used to calculate the UMAP coordinates for the resulting 249,000 cells (with 30 neighbors, metric = euclidian and minimum distance = 0.5 as default parameters) (Figure 3A). The FlowSOM plugin was used in parallel on the same downsampled dataset to create a self-organizing map (using n = 9 clusters as default parameter) (Figure 3B). Upsampling was then performed via the FlowSOM plugin by applying this self-organizing map to the initial FCS files from all 83 samples to calculate both total and spike-specific memory B cell repartition in identified clusters on all collected cells for each donor (Figure 3C; Figure S3A; Table S3). Both UMAP and FlowSOM plugin were run taking into account fluorescent intensities from the following parameters: FSC-A, SSC-A, CD19, CD21, CD11c, CD71, CD38, CD27 and IgD. Contour plots (equal probability contouring, with levels set to 5% of gated populations) for each identified cluster were further overlaid on UMAP projection in FlowJO. For visualization purposes, only the outermost density representing 95% of the total gated cells was kept for the final figure (Figure 3B), all other levels were removed in Adobe Illustrator.
Single-cell culture
Single cell culture was performed as previously described (Crickx et al., 2019): single B cells were sorted in 96-well plates containing MS40 cells expressing CD40L (kind gift from G. Kelsoe). Cells were co-cultured at 37°C with 5% CO2 during 21 or 25 days in RPMI-1640 (Invitrogen) supplemented with 10% HyClone FBS (Thermo Scientific), 55 μM 2-mercaptoethanol, 10 mM HEPES, 1 mM sodium pyruvate, 100 units/mL penicillin, 100 μg/mL streptomycin, and MEM non-essential amino acids (all Invitrogen), with the addition of recombinant human BAFF (10 ng/mL), IL2 (50 ng/mL), IL4 (10 ng/mL), and IL21 (10 ng/mL; all Peprotech). Part of the supernatant was carefully removed at days 4, 8, 12, 15 and 18 and the same amount of fresh medium with cytokines was added to the cultures. After 21 days of single cell culture, supernatants were harvested and stored at −20°C. Cell pellets were placed on ice and gently washed with PBS (GIBCO) before being resuspended in 50 μL of RLT buffer (QIAGEN) supplemented with 10% 2-mercaptoethanol and subsequently stored at −80°C until further processing.
ELISA
IgG and SARS-CoV-2, HKU1-CoV and OC43-CoV spike-specific IgG from culture supernatants were detected by home-made ELISA. Briefly, 96 well ELISA plates (Thermo Fisher) were coated with either goat anti-human Ig (10 μg/mL, Invitrogen) or recombinant SARS-CoV-2, HKU1 or OC43 spike or SARS-CoV-2 RBD protein (2.5 μg/mL each) in sodium carbonate during 1 h at 37°C. After plate blocking, cell culture supernatants were added for 1 h, then ELISA were developed using HRP-goat anti-human IgG (1 μg/mL, Immunotech) and TMB substrate (Eurobio). OD450 and OD620 were measured and Ab-reactivity was calculated after subtraction of blank wells. Supernatants whose ratio of OD450-OD620 over control well (consisting of supernatant from wells that contained single-cell sorted spike-negative memory B cells from the same single cell culture assay) was over 3 were considered as positive for RBD ELISA. A cut-off at a ratio of 5 was chosen for HKU1 and OC43 spike ELISA. PBS was used to define background OD450-OD620.
Single-cell IgH sequencing
Clones whose culture had proven successful (IgG concentration ≥1 μg/mL at day 21-25) were selected and extracted using the NucleoSpin96 RNA extraction kit (Macherey-Nagel) according to the manufacturer’s instruction. A reverse transcription step was then performed using the SuperScript IV enzyme (ThermoFisher) in a 14 μL final volume (42°C 10 min, 25°C 10 min, 50°C 60 min, 94°C 5 min) with 4 μl of RNA and random hexamers (GE Healthcare). A PCR was further performed based on the protocol established by Tiller et al. (Tiller et al., 2008). Briefly, 3.5 μL of cDNA was used as template and amplified in a total volume of 40 μL with a mix of forward L-VH primers (Table S4) and reverse Cγ primer and using the HotStar® Taq DNA polymerase (QIAGEN) and 50 cycles of PCR (94°C 30 s, 58°C 30 s, 72°C 60 s). PCR products were sequenced with the reverse primer CHG-D1 and read on ABI PRISM 3130XL genetic analyzer (Applied Biosystems). Sequence quality was verified with the CodonCode Aligner software (CodonCode Corporation) and data were analyzed with the IMGT/HighV-QUEST web portal (from The International Immunogenetics Information System) or in parallel with the VDJ sequences generated as part of our scRNA-seq dataset (see below).
Oligonucleotide conjugation of anti-His antibody
Unconjugated anti-His antibodies were purchased from Biolegend and custom oligonucleotides were ordered from Integrated DNA Technologies, following the 10X Genomics protocol available at https://support.10xgenomics.com/single-cell-gene-expression/overview/doc/demonstrated-protocol-cell-surface-protein-labeling-for-single-cell-rna-sequencing-protocols and the barcode whitelist. Sequences of the 2 HPLC purified barcoded oligonucleotides were: /5AmMC12/CGGAGATGTGTATAAGAGACAGNNNNNNNNNNTGCATAGCCTGTGGANNNNNNNNNCCCATATAAGAAA and /5AmMC12/CGGAGATGTGTATAAGAGACAGNNNNNNNNNNCTCTCCAATGTACTCNNNNNNNNNCCCATATAAGAAA. Oligonucleotide antibody conjugation was performed using the Oligonucleotide Conjugation Kit from Abcam, according to the manufacturer instructions using an oligonucleotide/antibody ratio of 5:1.
Single-cell RNA-seq library preparation and sequencing
Peripheral (CD3-CD14-CD15-CD56-CD19+IgD-) B cells were FACS-sorted (MA900, Sony) in PBS/0.08% FCS from 4 patients (S-CoV) at baseline (M0) and 6 months (M6). 5x104 to 10x105 cells were obtained for each subset. The scRNA-seq libraries were generated using Chromium Next GEM Single Cell V(D)J Reagent Kit v.1.1 with Feature Barcoding (10X Genomics) according to the manufacturer’s protocol. Gene expression (mRNA), ADT and VDJ BCR libraries were constructed. Briefly, cells were counted and up to 20 000 cells were loaded in the 10X Genomics Chromium Controller to generate single-cell gel-beads in emulsion. After reverse transcription, gel-beads in emulsion were disrupted. Barcoded complementary DNA was isolated and amplified by PCR. Following fragmentation, end repair and A-tailing, sample indexes were added during index PCR. The purified libraries were sequenced on a Novaseq S2 flowcell (Illumina) with 26 cycles of read 1, 8 cycles of i7 index and 91 cycles of read 2, targeting a median depth of 50000 reads per cell for gene expression and 5000 reads per cell for each other two libraries (BCR VDJ and ADT Feature barcoding).
Single-cell gene expression analysis
Paired-end FASTQ reads for all three libraries were demultiplexed and aligned against the GRCh38 human reference genome (GENCODE v32/Ensembl 98; July 2020) using 10X Genomics’ Cell Ranger v4.0.0 pipeline. Outputs of Cell Ranger were directly loaded into Seurat v3.2.2 (Stuart et al., 2019) for further QC steps and analysis. Following manual inspection of cell quality, only genes detected in at least 10 cells and cells with more than 500 unique genes detected and less than 25% of UMI counts mapped to mitochondrial genes were kept. Reads mapping to the immunoglobulin gene locus were further stored in a separate assay at this step to avoid unwanted clustering based solely on differential isotype expression and cells with exactly one heavy chain sequence were retained for final analysis. UMI counts were then log-normalized. The top 2,000 highly variable genes were identified using the FindVariableFeatures() function in Seurat and the default vst method. Normalized counts were then scaled and centered using the ScaleData() function, removing unwanted variation related to the percentage of mitochondrial UMI counts at that step. After principal component analysis, potential donor and sort-specific batch effects were removed using the Harmony algorithm (Korsunsky et al., 2019). The first 30 corrected PCA dimensions were then used to construct a knn graph (k = 20 neighbors) and perform graph-based clustering (Louvain) with a resolution parameter of 0.2 as well as compute the UMAP coordinates for each cell. Further separation of the “Activated” cell cluster was performed using a resolution parameter of 0.4. Two small clusters with T cell (0.2%) and monocyte (0.1%) signatures were removed from further analysis as clear doublets, as well as a separate cluster of poor quality cells (2.1% of all cells), grouped solely based on high percentages of mitochondrial UMI count. G2M and S cell cycle signatures were calculated using the CellCycleScoring() function and the associated gene lists in Seurat (G2M scoring: HMGB2, CDK1, NUSAP1, UBE2C, BIRC5, TPX2, TOP2A, NDC80, CKS2, NUF2, CKS1B, MKI67, TMPO, CENPF, TACC3, FAM64A, SMC4, CCNB2, CKAP2L, CKAP2, AURKB, BUB1, KIF11, ANP32E, TUBB4B, GTSE1, KIF20B, HJURP, CDCA3, HN1, CDC20, TTK, CDC25C, KIF2C, RANGAP1, NCAPD2, DLGAP5, CDCA2, CDCA8, ECT2, KIF23, HMMR, AURKA, PSRC1, ANLN, LBR, CKAP5, CENPE, CTCF, NEK2, G2E3, GAS2L3, CBX5, CENPA; S scoring: MCM5, PCNA, TYMS, FEN1, MCM2, MCM4, RRM1, UNG, GINS2, MCM6, CDCA7, DTL, PRIM1, UHRF1, MLF1IP, HELLS, RFC2, RPA2, NASP, RAD51AP1, GMNN, WDR76, SLBP, CCNE2, UBR7, POLD3, MSH2, ATAD2, RAD51, RRM2, CDC45, CDC6, EXO1, TIPIN, DSCC1, BLM, CASP8AP2, USP1, CLSPN, POLA1, CHAF1B, BRIP1, E2F8).
Computational analyses of VDJ sequences
Processed FASTA sequences from cultured single-cell heavy chain sequencing, 10X Genomics single-cell RNA sequencing and 874 published SARS-CoV-2 RBD and/or S-specific antibodies (Brouwer et al., 2020; Kreer et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Wec et al., 2020; Zost et al., 2020) were annotated using Igblast v1.16.0 against the human IMGT reference database. Non-productively rearranged sequences were removed at that step as well as sequences from cells that did not pass the initial QC cut-offs from our scRNA-seq analysis. Cases of 10X Genomics barcodes with two or more consensus heavy chain sequences for which more than ten UMI were detected were generally flagged as potential doublets for removal from our scRNA-seq analysis. Similarly, cases where no clear heavy chains could be attributed (none above 10 UMIs) were also flagged for removal. Two exceptions were made: 1/ in cases of identical CDR3s but differing isotypes (c_call), in which case the isotype switched sequence was kept and UMI counts from both contigs were aggregated; and 2/ in cases when one the heavy chains was clearly over represented at the UMI level and the second most represented sequences did not exceed ten UMIs, in which case the most represented sequence was kept.
Clonal cluster assignment (DefineClones.py) and germline reconstruction (CreateGermlines.py) was performed using the Immcantation/Change-O toolkit (Gupta et al., 2015) on all heavy chain V sequences. Sequences that had the same V-gene, same J-gene, including ambiguous assignments, and same CDR3 length with maximal length normalized nucleotide hamming distance of 0.15 were considered as potentially belonging to the same clonal group. Mutation frequencies in V genes were then calculated using the calcObservedMutations() function from Immcantation/SHazaM v1.0.2 R package. For the analysis of the initial ASC response in our 10X Genomics dataset (Figures 2E and 2F), clonal assignments were further corrected using available light chain information (light_cluster.py script from Immcantation). Further clonal analyses were implemented in R.
Based on light chain-corrected clonal affectation, clones were ranked based on the total number of ASC (PC + PB) from the M0 time point of our analysis found in each clone for each individual donor. Clones with equal number of ASC were further ranked based on their total number of Activated and MBC cells from the M0 time point of our analysis. The top ten percent highest ranked clones were further labeled as Top 10% ASC clones (Figures 2E and 2F; Figure S2D). Based on heavy-chain only clonal affectation, clones were defined as SARS-CoV-2 S specific (Spike-specific clones) if they contained 1 or more validated single-cell culture sequence or if more than ten percent of the cells from that clone were positively stained by our barcoded His-tagged S protein in our scRNaseq dataset. All Spike-specific clones described in Figure 4 and Figures S2F and S2G were manually curated based on available light chain information (10X Genomics and published antibodies only) and CDR3 sequences. Cut-off for positive staining with barcoded His-tagged S protein in our scRNaseq dataset were set at log normalized count values of 1.2 for both barcodes. Cells in clonal relationship with single-cell culture sequences at the M6 time point served as reference (Figure S4F). Clones were defined as SARS-CoV-2 RBD or HKU1/OC43 spike-specific (Figures 4C–4G) if they contained 1 or more validated single-cell culture sequence with a positive ELISA against one of these proteins. Graphics were obtained using the ggplot2 v3.3.2 and circlize v0.4.10 packages. Phylogenetic trees were generated using the Immcantation/IgPhyML toolkit (Immcantation/suite v4.0.0 docker image) and further visualized in R using the Alakazam v1.0.2 and igraph v1.2.6 packages.
Virus neutralization assay
Virus neutralization was evaluated by a focus reduction neutralization test (FRNT). Vero E6 cells were seeded at 2x104 cells/well in a 96-well plate 24 h before the assay. Two hundred focus-forming units (ffu) of Sars-CoV-2 virus (BetaCoV/France/IDF0372/2020 strain, a kind gift from the National Reference Centre for Respiratory Viruses at Institut Pasteur, Paris, originally supplied through the European Virus Archive goes Global platform) were pre-incubated with serial dilutions of heat-inactivated sera or supernatant from B cell clones for 1 h at 37°C before infection of cells. After 2 h infection, the virus/antibody mix was removed and foci were left to develop in presence of 1.5% methylcellulose for 2 days. Cells were then fixed with 4% formaldehyde and foci were revealed using either a rabbit anti-SARS-CoV-2 N or a human anti-SARS-CoV-2 S (H2-162) antibody and matching secondary HRP-conjugated secondary antibodies. Foci were visualized by diaminobenzidine (DAB) staining and counted using an Immunospot S6 Analyzer (Cellular Technology Limited CTL). Purified IgG from healthy donors were used as negative controls (TEM10 at 100 μg/mL; TEM11 at 90 μg/mL; TEM13 at 130 μg/mL) and purified human monoclonal C3-235 as positive control (IC50 0.3 nM). Percentage of virus neutralization was calculated as (100 - (#foci Ab / #foci control) ∗100). The total amount of IgG present in the supernatants from B cells varied between S-CoV (12 μg/mL ± 7.6) and M-CoV patients (48 μg/mL ± 26). Neutralization values were normalized to the total IgG concentration present in the supernatant. As the neutralization activity may not linearly correlate with the amount of IgG, such normalization may lead to an underestimate of the neutralizing potency of B cell supernatants from M-CoV patients. Variation of the assay in weak neutralizers (i.e., < 50%) was due to intrinsic variation in counting.
Quantification and statistical analysis
Repeated Measures (RM) two-way ANOVA, Sidak’s multiple comparison tests, Kruskal-Wallis test and Mann-Whitney test were used to compare continuous variables as appropriate (indicated in Figures). A P-value ≤0.05 was considered statistically significant. Statistical analyses involved use of GraphPad Prism 8.0 (La Jolla, CA, USA).
Additional resources
ClinicalTrials.gov Identifier: MEMO-CoV2, NCT04402892.
Acknowledgments
We thank Garnett Kelsoe for providing us with the human cell culture system, together with invaluable advice. We thank the Prince of Monaco and the Government Council for free donation. We also thank Sébastien Storck, Sandra Weller, Cécile Masson, and Florence Canoui for their advice and support; we thank the physicians Constance Guillaud, Frédéric Schlemmer, Giovanna Melica, Elena Fois, Henri Guillet, Nicolas De Prost, Pascal Lim, Sébastien Gallien, and Bertrand Godeau, whose patients were included in this study. This work was supported by a grant from the Agence Nationale de la Recherche and the Fondation pour la Recherche Médicale (ANR, MEMO-COV-2), by the Fondation Princesse Grace, and, in part by an ERC Advanced Investigator Grant (B-response). Assistance Publique – Hôpitaux de Paris (AP-HP, Département de la Recherche Clinique et du Développement) was the promotor and the sponsor of MEMO-COV-2. A.S. was supported by a Poste d’Accueil from Inserm, A.R. by an Année Recherche from AP-HP and by a SNFMI fellowship.
Author contributions
Conceptualization, P.C., A.S., S. Fillatreau, J.-C.W., C.-A.R., and M. Mahévas; data curation, P.C. and A.S; formal analysis, A.S., P.C., A.R., I.A., A.V., M.B.-A., S.H., A.B.-F., F.A.R., S. Fourati, M. Michel, J.M., M. Ménager, I.F., A.M, E.C., L.L., F.N.-P., M.L., J.M., and G.B.-S.; funding acquisition, S. Fillatreau., J.-C.W., C.-A.R., and M. Mahévas; investigation, A.S., P.C., A.R., I.A., and A.V.; methodology, A.S., J.-C.W., C.-A.R., P.C., and M. Mahévas; project administration, P.C., S.B., S. Fillatreau., J.-C.W., C.-A.R., and M. Mahévas; resources, J.-M.P., F.A.R., S. Fillatreau, E.C., C.-A.R., and M. Mahévas; software, P.C.; supervision, P.C., J.-C.W., C.-A.R., and M. Mahévas; validation, A.R., I.A., and A.V.; visualization, P.C., A.S., I.A., A.R., A.V., and M. Mahévas; writing – original draft, P.C., A.S., J.-C.W., C.-A.R., and M. Mahévas; writing – review & editing, all authors.
Declaration of interests
M. Mahévas received research funds from GlaxoSmithKline, outside of the submitted work, and personal fees from LFB and Amgen, outside of the submitted work. J.-C.W. received consulting fees from Institut Mérieux, outside of the submitted work. J.-M.P. received personal fees from Abbvie, Gilead, Merck, and Siemens Healthcare, outside of the submitted work.
Published: February 2, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2021.01.050.
Supplemental information
References
- Amanna I.J., Carlson N.E., Slifka M.K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- Aydillo T., Rombauts A., Stadlbauer D., Aslam S., Abelenda-Alonso G., Escalera A., Amanat F., Jiang K., Krammer F., Carratala J., García-Sastre A. Antibody Immunological Imprinting on COVID-19 Patients. MedRxiv. 2020 doi: 10.1101/2020.10.14.20212662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zuiani A., Fischinger S., Mullur J., Atyeo C., Travers M., Lelis F.J.N., Pullen K.M., Martin H., Tong P., et al. Quick COVID-19 Healers Sustain Anti-SARS-CoV-2 Antibody Production. Cell. 2020;183:1496–1507.e16. doi: 10.1016/j.cell.2020.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickx E., Chappert P., Weller S., Sokal A., Azzaoui I., Vandenberghe A., Bonnard G., Rossi G., Fadeev T., Storck S., et al. A reservoir of rituximab-resistant splenic memory B cells contributes to relapses after B-cell depletion therapy. bioRxiv. 2019 doi: 10.1101/833343. [DOI] [Google Scholar]
- Crotty S., Felgner P., Davies H., Glidewell J., Villarreal L., Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J. Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.W., Jackson K.J.L., McElroy A.K., Halfmann P., Huang J., Chennareddy C., Piper A.E., Leung Y., Albariño C.G., Crozier I., et al. Longitudinal Analysis of the Human B Cell Response to Ebola Virus Infection. Cell. 2019;177:1566–1582.e17. doi: 10.1016/j.cell.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellebedy A.H., Jackson K.J.L., Kissick H.T., Nakaya H.I., Davis C.W., Roskin K.M., McElroy A.K., Oshansky C.M., Elbein R., Thomas S., et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 2016;17:1226–1234. doi: 10.1038/ni.3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolano A., Steichen J.M., Dosenovic P., Kulp D.W., Golijanin J., Sok D., Freund N.T., Gitlin A.D., Oliveira T., Araki T., et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166:1445–1458.e12. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased Screens Show CD8+ T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., Ladinsky M., Cho A., Jankovic M., Schaefer-Babajew D., et al. Evolution of Antibody Immunity to SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A Sequence Homology and Bioinformatic Approach Can Predict Candidate Targets for Immune Responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N.T., Vander Heiden J.A., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S.H. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics. 2015;31:3356–3358. doi: 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley G.E., Edwards E.S.J., Aui P.M., Varese N., Stojanovic S., McMahon J., Peleg A.Y., Boo I., Drummer H.E., Hogarth P.M., et al. Rapid generation of durable B cell memory to SARS-CoV-2 spike and nucleocapsid proteins in COVID-19 and convalescence. Sci. Immunol. 2020;5:eabf8891. doi: 10.1126/sciimmunol.abf8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-L., Goldsmith J.A., Schaub J.M., DiVenere A.M., Kuo H.-C., Javanmardi K., Le K.C., Wrapp D., Lee A.G., Liu Y., et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science. 2020;369:1501–1505. doi: 10.1126/science.abd0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A.T., Garcia-Carreras B., Hitchings M.D.T., Yang B., Katzelnick L.C., Rattigan S.M., Borgert B.A., Moreno C.A., Solomon B.D., Trimmer-Smith L., et al. A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat. Commun. 2020;11:4704. doi: 10.1038/s41467-020-18450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A.S., Jones F.K., Nodoushani A., Kelly M., Becker M., Slater D., Mills R., Teng E., Kamruzzaman M., Garcia-Beltran W.F., et al. Persistence and decay of human antibody responses to the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 patients. Sci. Immunol. 2020;5:eabe0367. doi: 10.1126/sciimmunol.abe0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks S.A., Cashman K.S., Woodruff M.C., Lee F.E.-H., Sanz I. Extrafollicular responses in humans and SLE. Immunol. Rev. 2019;288:136–148. doi: 10.1111/imr.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Juno J.A., Tan H.-X., Lee W.S., Reynaldi A., Kelly H.G., Wragg K., Esterbauer R., Kent H.E., Batten C.J., Mordant F.L., et al. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nat. Med. 2020;26:1428–1434. doi: 10.1038/s41591-020-0995-0. [DOI] [PubMed] [Google Scholar]
- Kaneko N., Kuo H.-H., Boucau J., Farmer J.R., Allard-Chamard H., Mahajan V.S., Piechocka-Trocha A., Lefteri K., Osborn M., Bals J., et al. Massachusetts Consortium on Pathogen Readiness Specimen Working Group Loss of Bcl-6-Expressing T Follicular Helper Cells and Germinal Centers in COVID-19. Cell. 2020;183:143–157.e13. doi: 10.1016/j.cell.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K., et al. Longitudinal Isolation of Potent Near-Germline SARS-CoV-2-Neutralizing Antibodies from COVID-19 Patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Liao H.-X., Lynch R., Zhou T., Gao F., Alam S.M., Boyd S.D., Fire A.Z., Roskin K.M., Schramm C.A., Zhang Z., et al. NISC Comparative Sequencing Program Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Liu L., Kou G., Zheng Y., Ding Y., Ni W., Wang Q., Tan L., Wu W., Tang S., et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020;58:e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Luchsinger L.L., Ransegnola B.P., Jin D.K., Muecksch F., Weisblum Y., Bao W., George P.J., Rodriguez M., Tricoche N., Schmidt F., et al. Serological Assays Estimate Highly Variable SARS-CoV-2 Neutralizing Antibody Activity in Recovered COVID-19 Patients. J. Clin. Microbiol. 2020;58:e02005–e02020. doi: 10.1128/JCM.02005-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan I.C.M., Toellner K.-M., Cunningham A.F., Serre K., Sze D.M.-Y., Zúñiga E., Cook M.C., Vinuesa C.G. Extrafollicular antibody responses. Immunol. Rev. 2003;194:8–18. doi: 10.1034/j.1600-065x.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- Mamani-Matsuda M., Cosma A., Weller S., Faili A., Staib C., Garçon L., Hermine O., Beyne-Rauzy O., Fieschi C., Pers J.-O., et al. The human spleen is a major reservoir for long-lived vaccinia virus-specific memory B cells. Blood. 2008;111:4653–4659. doi: 10.1182/blood-2007-11-123844. [DOI] [PubMed] [Google Scholar]
- Marshall J.C., Murthy S., Diaz J., Adhikari N.K., Angus D.C., Arabi Y.M., Baillie K., Bauer M., Berry S., Blackwood B., et al. WHO Working Group on the Clinical Characterisation and Management of COVID-19 infection A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 2020;20:e192–e197. doi: 10.1016/S1473-3099(20)30483-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew D., Giles J.R., Baxter A.E., Oldridge D.A., Greenplate A.R., Wu J.E., Alanio C., Kuri-Cervantes L., Pampena M.B., D’Andrea K., et al. UPenn COVID Processing Unit Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science. 2020;369:e00461-20. doi: 10.1126/science.abc8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Watanabe A., Kuraoka M., Do K.T., McGee C.E., Sempowski G.D., Kepler T.B., Schmidt A.G., Kelsoe G., Harrison S.C. Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires. Immunity. 2018;48:174–184.e9. doi: 10.1016/j.immuni.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., et al. Imbalance of Regulatory and Cytotoxic SARS-CoV-2-Reactive CD4+ T Cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L., Ersching J., Victora G.D. Germinal center B cell dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., Ulferts R., Earl C., Wrobel A.G., Benton D.J., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Contant P., Embong A.K., Kanagaiah P., Chaves F.A., Yang H., Branche A.R., Topham D.J., Sangster M.Y. S Protein-Reactive IgG and Memory B Cell Production after Human SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit. MBio. 2020;11:e01991-20. doi: 10.1128/mBio.01991-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Röltgen K., Jean G.H., Stevens B.A., Lee J.-Y., Rustagi A., Rogers A.J., et al. Human B Cell Clonal Expansion and Convergent Antibody Responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliviero B., Varchetta S., Mele D., Mantovani S., Cerino A., Perotti C.G., Ludovisi S., Mondelli M.U. Expansion of atypical memory B cells is a prominent feature of COVID-19. Cell. Mol. Immunol. 2020;17:1101–1103. doi: 10.1038/s41423-020-00542-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Oxford Immunology Network Covid-19 Response T cell Consortium. ISARIC4C Investigators Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce C.A., Preston-Hurlburt P., Dai Y., Aschner C.B., Cheshenko N., Galen B., Garforth S.J., Herrera N.G., Jangra R.K., Morano N.C., et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Sci. Transl. Med. 2020;12:eabd5487. doi: 10.1126/scitranslmed.abd5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds C.J., Swadling L., Gibbons J.M., Pade C., Jensen M.P., Diniz M.O., Schmidt N.M., Butler D.K., Amin O.E., Bailey S.N.L., et al. Discordant neutralizing antibody and T cell responses in asymptomatic and mild SARS-CoV-2 infection. Sci. Immunol. 2020;5:eabf3698. doi: 10.1126/sciimmunol.abf3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger T.J., Uhrlaub J.L., Watanabe M., Wong R., Castaneda Y., Pizzato H.A., Thompson M.R., Bradshaw C., Weinkauf C.C., Bime C., et al. Orthogonal SARS-CoV-2 Serological Assays Enable Surveillance of Low-Prevalence Communities and Reveal Durable Humoral Immunity. Immunity. 2020;53:925–933.e4. doi: 10.1016/j.immuni.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda L.B., Netland J., Shehata L., Pruner K.B., Morawski P.A., Thouvenel C.D., Takehara K.K., Eggenberger J., Hemann E.A., Waterman H.R., et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell. 2020;184:169–183.e17. doi: 10.1016/j.cell.2020.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Powell A.E., Wirz O.F., Stevens B.A., Hogan C.A., Najeeb J., Hunter M., Wang H., Sahoo M.K., Huang C., et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci. Immunol. 2020;5:eabe0240. doi: 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz I., Wei C., Jenks S.A., Cashman K.S., Tipton C., Woodruff M.C., Hom J., Lee F.E.-H. Challenges and Opportunities for Consistent Classification of Human B Cell and Plasma Cell Populations. Front. Immunol. 2019;10:2458. doi: 10.3389/fimmu.2019.02458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariol A., Perlman S. Lessons for COVID-19 Immunity from Other Coronavirus Infections. Immunity. 2020;53:248–263. doi: 10.1016/j.immuni.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.-B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Karolinska COVID-19 Study Group Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J., et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.-H., Leng Y., Robinson M.L., Sie B.M., Li M.Z., Chen Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370:eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slifka M.K., Antia R., Whitmire J.K., Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- Song G., He W.-t., Callaghan S., Anzanello F., Huang D., Ricketts J., Torres J.L., Beutler N., Peng L., Vargas S., et al. Cross-reactive serum and memory B cell responses to spike protein in SARS-CoV-2 and endemic coronavirus infection. BioRxiv. 2020 doi: 10.1101/2020.09.22.308965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling L., Maini M.K. T cells in COVID-19 - united in diversity. Nat. Immunol. 2020;21:1307–1308. doi: 10.1038/s41590-020-0798-y. [DOI] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.-J., Bosch B.-J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman-Mentesh A., Dror Y., Tur-Kaspa R., Markovitch D., Kournos T., Dicker D., Wine Y. SARS-CoV-2 specific memory B cells frequency in recovered patient remains stable while antibodies decay over time. MedRxiv. 2020 doi: 10.1101/2020.08.23.20179796. [DOI] [Google Scholar]
- Wajnberg A., Amanat F., Firpo A., Altman D.R., Bailey M.J., Mansour M., McMahon M., Meade P., Mendu D.R., Muellers K., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370:1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., van Haperen R., Gutiérrez-Álvarez J., Li W., Okba N.M.A., Albulescu I., Widjaja I., van Dieren B., Fernandez-Delgado R., Sola I., et al. Isolation of cross-reactive monoclonal antibodies against divergent human coronaviruses that delineate a conserved and vulnerable site on the spike protein. BioRxiv. 2020 doi: 10.1101/2020.10.20.346916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D., et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.-H., Wontakal S., Szabo P.A., Wells S.B., Dogra P., Gray J., et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat. Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Ahmed R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004;78:5535–5545. doi: 10.1128/JVI.78.11.5535-5545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff M.C., Ramonell R.P., Nguyen D.C., Cashman K.S., Saini A.S., Haddad N.S., Ley A.M., Kyu S., Howell J.C., Ozturk T., et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., et al. NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Mei H., Dörner T., Hiepe F., Radbruch A., Fillatreau S., Hoyer B.F. Memory B and memory plasma cells. Immunol. Rev. 2010;237:117–139. doi: 10.1111/j.1600-065X.2010.00938.x. [DOI] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for all the single cell RNA sequencing data reported in this paper is ArrayExpress: E-MTAB-9995. Single cell culture VDJ sequencing data reported in Figure 4 are directly included in this study as part of Table S4.