Abstract
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most lethal common cancer because of late diagnosis. Novel biomarkers for PDAC early detection are urgently needed. tRNA-derived small RNAs (tsRNAs) are novel small RNAs might serve as biomarkers for cancer diagnosis and participate in diverse physiological and pathological process. We investigated whether the expression of tsRNAs in serum could be a noninvasive method in the early detection of PDAC. Blood sample of PDAC patients and healthy controls were collected from Ruijin Hospital, Shanghai, China. Tumor and adjacent normal pancreas tissues were collected from 51 patients with PDAC undergoing therapeutic surgery. The testing cohort comprised 6 PDAC patients and 6 healthy controls and the expression of small RNAs in serum was analyzed by small RNA sequence. We verified the diagnostic performance of serum tsRNAs by qPCR in validation cohort including 110 PDAC patients and 100 healthy controls. Expression level of tsRNAs in tissue was also verified in another independent cohort including 51 tumor and 51 adjacent normal pancreas tissues. Unpaired t-test and paired t-test are used for comparing depending on whether the samples are paired. The predictive performance of tsRNAs was evaluated by Kaplan-Meier survival and receiver operating characteristic (ROC) curve. There were 45 tsRNAs expressed at remarkably higher levels, 6 tsRNAs expressed at lower levels in PDAC patients, respectively, compared with healthy volunteers. tsRNA-ValTAC-41, tsRNA-MetCAT-37 and tsRNA-ThrTGT-23 expressed significant highly (P < 0.05) in serum of PDAC patients in validation cohort. tsRNA-ValTAC-41 or tsRNA-MetCAT-37 combined with CA19-9 could increase the AUC of PDAC prediction (AUC = 0.947 and 0.949 respectively), relative to CA19-9 test alone. Besides, patients with higher serum tsRNA-ValTAC-41 level showed shorter overall survival (OS). tsRNA-ValTAC-41 also expressed at remarkably higher level in tumor tissue, and it was obviously associated with tumor staging both in serum and tissue. We provide tsRNAs profiles observed by small RNA sequencing. The diagnostic accuracy of tsRNA-ValTAC-41 and tsRNA-MetCAT-37 in serum of PDAC patients were verified. Further studies for tsRNA-ValTAC-41 are needed to confirm the findings. These tsRNAs may be promising and effective candidates in the development of highly sensitive, noninvasive biomarkers for PDAC diagnosis.
Keywords: tsRNAs, tRF-3, PDAC, diagnosis, biomarker
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers, with the 5-year survival rate less than 9% [1]. Most patients with pancreatic cancer remain asymptomatic until advanced stage [2]. Early detection is considered the most effective way to improve survival. While the only approved biomarker, serum carbohydrate antigen 19-9 (CA19-9), still have the limitation of non-specific elevated in other forms of digestive tract cancer and some non-cancerous conditions [3]. Therefore, novel non-invasion diagnostic biomarkers are urgently needed to capture the early development or the progression of the disease.
Small RNAs are short untranslated RNA molecules, including microRNAs (miRNAs), Piwi-interacting RNAs (piRNAs) and tRNA-derived small RNAs (tsRNAs). In last two decades, substantial progress has produced various evidences of the fundamental roles of small RNAs in virtually all biological pathways and may have oncogenic or tumor suppressive properties. Recently, small RNAs have been associated with cancer initiation, progression and drug response [4].
As novel small RNAs, tsRNAs generate from precursor or mature transfer RNAs (tRNAs). tsRNAs mostly produced by specific nucleases in particular cells or tissues or under certain conditions such as stress and hypoxia [5]. tsRNAs can be roughly divided into 3’ U tRFs from 3’ end, mature tRNA-derived fragments (tRFs) as well as tRNA halves (tRHs) [6]. tRFs are grouped into three subclasses: tRF-5s, tRF-3s and inter tRFs (i-tRFs). tRHs are further classified into 5’ half and 3’ half of mature tRNA [7]. tsRNAs were verified to be associated with numerous diseases, such as metabolic disorder, pathological stress injuries, neurodegenerative diseases, virus infection as well as cancer [7]. In fields related to cancer research, expression profile and biological function of tsRNAs were reported in chronic lymphocytic leukemia (CLL) [8,9], lung cancer [4,8,10], colorectal cancer [4,11], breast cancer [4,12-15], ovarian cancer [4,16,17] and prostate cancer [18,19]. These studies suggested tsRNAs might have the potential of diagnostic and therapeutic targets for cancers. However, tsRNAs have not been elucidated in PDAC. To improve the understanding of these novel small RNAs, we determined the expression profile of small RNAs in PDAC.
In this study, we focused on investigating whether tsRNAs could play a role in the diagnosis of PDAC. Based on tsRNAs sequencing analysis, we identified a group of tsRNAs that were differentially expressed in PDAC. The purpose of this study is to identify tsRNAs as biomarkers in diagnosis of PDAC as a noninvasive method, in order to improve the specificity and sensitivity of PDAC diagnosis.
Materials and methods
Patient characteristics
The PDAC patients were enrolled for testing and validation studies between 2016 and 2019, from Ruijin Hospital, Shanghai, China, as well as healthy individuals in the testing and validation sets, respectively. The study was supported by the Ethics committee of Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. All patients were validated by histopathological examination. It was conducted, according to the ethical principles of the World Medical Association Declaration of Helsinki and local legislation. All patients had confirmed informed consent to this study prior. 6 PDAC patients and 6 healthy controls were enrolled in testing set. 110 PDAC patients and 100 healthy controls were enrolled in validation set1 and another independent 51 PDAC patients were enrolled in validation set2. Detailed clinical data are summarized in Table 1.
Table 1.
Study cohort clinicopathological parameters
| Testing Set | Validation Set1 (Serum) | Validation Set2 (Tissue) | |||
|---|---|---|---|---|---|
|
|
|
|
|||
| PDAC patients (N = 6) | Healthy controls (N = 6) | PDAC patients (N = 110) | Healthy controls (N = 100) | PDAC patients (N = 51) | |
| male | 3 (50) | 5 (83) | 75 (68) | 47 (47) | 33 (64.71) |
| female | 3 (50) | 1 (17) | 35 (32) | 53 (53) | 18 (35.29) |
| 62.3 (48-70) | 33.4 (26-42) | 63.3 (48-73) | 34.19 (21-63) | 62.88 (44-85) | |
| I | 0 | Not applicable | 24 | Not applicable | 14 |
| II | 1 | 36 | 16 | ||
| III | 4 | 18 | 15 | ||
| IV | 1 | 22 | 6 | ||
| N.A. | 0 | 10 | 0 | ||
Venous blood (5 ml) samples included in this study were collected in a vacuum blood tube. After clotted for 30 min at room temperature, the blood samples were centrifuged at 4,000 rpm for 10 min at 4°C. Clear yellow supernatant was collected as serum sample. Serum samples were preserved in -80°C.
Tumor and normal tissue samples from patients with PDAC were taken in the middle of cancer and adjacent normal pancreas tissue. Tissues were fresh frozen and stored at -80°C until use. All samples were reevaluated by a pathologist to confirm stage and grade. Samples were classified according to 8th edition of AJCC staging [20].
Small RNA sequencing
Total RNA was extracted for small RNA sequencing using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Each RNA sample was measured by a Nanodrop instrument (Thermo Fisher Scientific, Inc.) for the quality and concentration. The integrity of RNA was assessed using agarose gel (2%) electrophoresis. The libraries were prepared using the NEB Next Multiplex Small RNA Library Prep Kit (New England BioLabs, Ipswich, MA). A linker primer was added to both ends of the RNA fragment, and the complementary DNA (cDNA) constructs was created by PCR. The PCR production was separated by gel electrophoresis, and the 135-170 nt fragments were recycled. The small RNA library was quantified by Qubit 3.0 (Invitrogen), and the insert fragment size of the library was determined by Agilent 2200 Bioanalyzer (Agilent, Santa Clara, CA, USA). These libraries were sequenced using Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA).
Bioinformatics analysis of tsRNAs-seq data
The short reads (< 15 nt) and low-quality reads were filtered out from the raw sequencing data. To identify tsRNAs, all clean reads were aligned to the miRBase database (http://www.mirbase.org/), piwi interacting RNA (piRNA) database, NCBI, Genomic tRNA database (http://gtrnadb.ucsc.edu/), tRFdb (http://genome.bioch.virginia.edu/trfdb/) and MintBase to identify known and novel tsRNAs. The tsRNAs with fold change ≥ 2 and P ≤ 0.05 were selected as significantly differentially expressed (DE) tsRNAs. For the target genes predication of DE tsRNAs, the miRanda (http://www.microrna.org/microrna/home.do) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid) was used for analysis on line. The significantly target genes were estimated by cutoff criteria (score ≥ 150 and energy < -20 for MiRanda; Energy < -25 for RNAhybrid). These target genes were mapped to Gene Ontology (GO) database and Kyoto encyclopedia of genes and genomes (KEGG) database to obtain functional annotation. Then, the candidate tsRNAs interactions network was constructed through cytoscape 2.8.3 for functional analysis.
RNA isolation from serum and tissue for validation
Total serum RNA was isolated using the miRNeasy Serum/Plasma Kit (Qiagen, catalogue number 217184) per the manufacturer’s protocol. Total tissue RNA was extracted using the mirVana™ miRNA Isolation Kit (Invitrogen, AM1561), according to the kit protocol.
Residual DNA was eliminated by deoxyribonuclease using the DNA-free™ DNA Removal Kit (Invitrogen, AM1907). RNA quantity was measured using Epoch 2 microplate spectrophotometer (BioTek).
tsRNAs quantification
In order to measure tsRNAs levels in each sample, tsRNAs were amplified selectively without full-length tRNA quantification by existing protocol [21,22]. The serum or tissue RNA was poly-A tailed using E. coli Poly(A) Polymerase (NEB, M0276) for 30 minutes at 37°C followed by 10 minutes at 65°C. Upon annealing a linker-oligo (dT) primer (ATGCCATAATACGACTCACTATAGGGAGAAGTACTTTTTTTTTTTTTTT), the poly-A tailed RNA population was subjected to first strand cDNA synthesis using the Superscript III First-Strand Synthesis kit (Invitrogen, 18080051) in accordance with the manufacturer’s instructions. A linker-specific primer (TAATACGACTCACTATAGGGAGA) and tsRNA specific primers (tsRNA-MetCAT-37: GAAGGGTATAACCAACATTTTCAAAA; tsRNA-ValTAC-41: GTCAAGTTAAGTTGAAATCTCCTAAGTGTAAG; tsRNA-ThrTGT-23: GAGGCCCCAGCGAGAATTGAA) were then used for quantitative PCR. Quantitative PCR was done using SYBR GreenRealtime PCR Master Mix (TOYOBO, QPK-201) according to the manufacturer’s instructions. The expression of tsRNAs was calculated using the formula 2-ΔCq*10000 and RNA-U6B (specific primers: RNU6B-F: GCTTCGGCAGCACATATACTAAAAT; RNU6B-R: CGCTTCACGAATTTGCGTGTCAT) was used as endogenous reference genes.
Statistical analysis
Unpair-t test was used to compared serum or tissue tsRNAs level in cohort. Pair-t test was used in expression level in paired tissue. ROC analysis was done to determine the diagnostic sensitivity and specificity of tsRNA expression in serum. Patients were then classified into low- and high-expression level groups based on the optimal cut-off value determined by X-Tile software [23]. Kaplan-Meier (K-M) survival curves were used to analyze the survival time of patients. All statistical tests were performed with IBM SPSS Statistics (IBM, Armonk, NY, United States), version 22. Graphics were draw by GraphPad Prism 7. P value < 0.05 were considered statistically significant.
Results
Small RNA expression profiling of PDAC and healthy controls
To comprehensively profile the serum small RNAs, samples from 6 PDAC patients and 6 controls were collected. Small RNA-seq library was prepared for high-throughput sequencing as well. After validated the quality of raw sequencing data, the different expression of miRNAs, tsRNAs and piRNAs were analyzed respectively (Table 2; Supplementary Figure 1). There was slightly different trend in length of miRNAs between PDAC patients and healthy controls (Supplementary Figure 2).
Table 2.
The differently expressed serum tsRNAs in PDAC, compared with healthy controls
| AccID | Database_ID | Fragment sequence, 5’-3’ | length, nt | |||
|
| ||||||
| tsrna-18478 | tRF-23-YOY9Q867D2 | TTCAATTCTCGCTGGGGCCTCCA | 23 | |||
| tsrna-26246 | tRF-25-OB9ZFH690M | GAAAATGTTTAGACGGGCTCACATC | 25 | |||
| tsrna-19142 | tRF-30-V47P596VW631 | TAGTGGTTAGGATTCGGCGCTCTCACCGCC | 30 | |||
| tsrna-09704 | tRF-25-Q99P9P9NH5 | GCTTCTGTAGTGTAGTGGTTATCAC | 25 | |||
| tsrna-18943 | tRF-21-V29K9UV30 | TAGGATGGGGTGTGATAGGTG | 21 | |||
| tsrna-05695 | tRF-47-FBVWNEB01XNYH2SBUL4 | AGAAATATGTCTGATAAAAGAGTTACTTTGATAGAGTAAATAATAGG | 47 | |||
| tsrna-01888 | tRF-20-18YKISQI | AGTGGTTAGGATTCGGCGCT | 20 | |||
| tsrna-14173 | tRF-42-WLV47PU9XW983FPD3 | TCGTATAGTGGTTAGTACTCTGCGTTGTGGCCGCAGCAACCT | 42 | |||
| tsrna-13349 | tRF-35-86V8WPMN1E8Y7Z | TCCCATATGGTCTAGCGGTTAGGATTCCTGGTTTT | 35 | |||
| tsrna-02508 | tRF-38-2YU04DYJIO3ZU30F | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAA | 38 | |||
| tsrna-09141 | tRF-34-DWY2MJ8F81KL0U | AATGTTTAGACGGGCTCACATCACCCCATAAACA | 34 | |||
| tsrna-09134 | tRF-25-DWY2MJ8F81 | AATGTTTAGACGGGCTCACATCACC | 25 | |||
| tsrna-12286 | tRF-24-7XO8Q6J61J | GTTTAGACGGGCTCACATCACCCC | 24 | |||
| tsrna-25751 | tRF-27-N3WB884U1D2 | CTGGTTCGAATCCGGCTCGAAGGACCA | 27 | |||
| tsrna-25935 | tRF-41-N5EX62Z6EXEY0VWUD | CTTACACTTAGGAGATTTCAACTTAACTTGACCGCTCTGAC | 41 | |||
| tsrna-16124 | tRF-18-YRRHQFD2 | TTCCCGGGCGGCGCACCA | 18 | |||
| tsrna-09136 | tRF-27-DWY2MJ8F81J | AATGTTTAGACGGGCTCACATCACCCC | 27 | |||
| tsrna-07409 | tRF-25-INVDRI2Q2R | ATGTTTAGACGGGCTCACATCACCC | 25 | |||
| tsrna-10865 | tRF-25-7343RX6NMH | GTGGTCTAGTGGTTAGGATTCGGCG | 25 | |||
| tsrna-07191 | tRF-32-IK9NJ4S2I7L7M | ATGGGTGGTTCAGTGGTAGAATTCTCGCCTGC | 32 | |||
| tsrna-21275 | tRF-25-7JPJ60MV9J | GTGGCGCAGCGGAAGCGTGCTGGGC | 25 | |||
| tsrna-12357 | tRF-28-79MP9PMNH5DE | GTTTCCGTAGTGTAGCGGTTATCACATT | 28 | |||
| tsrna-06963 | tRF-17-H9R8B7J | ATCTCGGTGGAACCTCC | 17 | |||
| tsrna-25171 | tRF-34-4R94SX73V2Y81W | CTCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCT | 34 | |||
| tsrna-03139 | tRF-30-JQJYSWRYVMMV | CATATCATTGGTCGTGGTTGTAGTCCGTGC | 30 | |||
| tsrna-16151 | tRF-26-YSV4VH7Q2QE | TTCCGTAGTGTAGCGGTTATCACATT | 26 | |||
| tsrna-02481 | tRF-43-2YBE7XO8Q6J6K6UDV | CACTGAAAATGTTTAGACGGGCTCACATCACCCCATAAACACC | 43 | |||
| tsrna-23729 | tRF-34-RKVP4P9L5FZUHM | GGGGGTATAGCTCAGTGGTAGAGCATTTGACTGC | 34 | |||
| tsrna-02513 | tRF-46-2YU04DYJIO3ZU3U0IO0 | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAAAGATTAAG | 46 | |||
| tsrna-03857 | tRF-37-KSBE78YLKZKWE52 | CCCCGAAAATGTTGGTTATACCCTTCCCGTACTACCA | 37 | |||
| tsrna-02512 | tRF-45-2YU04DYJIO3ZU3U0IO | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAAAGATTAA | 45 | |||
| tsrna-10930 | tRF-25-73V2Y8L981 | GTGGTTAGGATTCGGCGCTCTCACC | 25 | |||
| tsrna-04539 | tRF-29-387SFRJ4O1E2 | CCTGGGTTCGAGCCCCAGTGGAACCACCA | 29 | |||
| tsrna-07404 | tRF-20-INVDRI2Q | ATGTTTAGACGGGCTCACAT | 20 | |||
| tsrna-14691 | tRF-25-P4R8YP9LON | GCATGGGTGGTTCAGTGGTAGAATT | 25 | |||
| tsrna-02515 | tRF-50-2YU04DYJIO3ZU3U0IO5B | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAAAGATTAAGAGAA | 50 | |||
| tsrna-13738 | tRF-22-WBKY4VXV2 | TCGAACCCTGCTCGCTGCGCCA | 22 | |||
| tsrna-09139 | tRF-31-DWY2MJ8F81KLB | AATGTTTAGACGGGCTCACATCACCCCATAA | 31 | |||
| tsrna-14300 | tRF-24-WP9N1EWJI5 | TCTAGTGGTTAGGATTCGGCGCTC | 24 | |||
| tsrna-15779 | tRF-27-Q6S8V0J8O9Q | GCTCAGTCGGTAGAGCATGGGACTCTT | 27 | |||
| tsrna-02514 | tRF-47-2YU04DYJIO3ZU3U0IOL | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAAAGATTAAGA | 47 | |||
| tsrna-02510 | tRF-41-2YU04DYJIO3ZU3U0B | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAAAGA | 41 | |||
| tsrna-25233 | tRF-16-489B3RB | CTCGGTGGAACCTCCA | 16 | |||
| tsrna-09129 | tRF-19-DWY2MJJ1 | AATGTTTAGACGGGCTCAC | 19 | |||
| tsrna-11835 | tRF-24-7SFRJ4O1E2 | GTTCGAGCCCCAGTGGAACCACCA | 24 | |||
| tsrna-10864 | tRF-24-7343RX6N19 | GTGGTCTAGTGGTTAGGATTCGGC | 24 | |||
| tsrna-26278 | tRF-36-O045DBNIB9I1KQ0 | GAAAGCTCACAAGAACTGCTAACTCATGCCCCCATG | 36 | |||
| tsrna-02509 | tRF-40-2YU04DYJIO3ZU3U0 | CACTGTAAAGCTAACTTAGCATTAACCTTTTAAGTTAAAG | 40 | |||
| tsrna-01896 | tRF-29-18YKISQI451V | AGTGGTTAGGATTCGGCGCTCTCACCGCC | 29 | |||
| tsrna-24916 | tRF-38-RXPIN24YDRFU8U0E | GGTTAGCACTCTGGACTCTGAATCCAGCGATCCGAGTT | 38 | |||
| tsrna-26483 | tRF-20-OMMIL0O6 | GAATCCGGCTCGAAGGACCA | 20 | |||
|
| ||||||
| Type | Anticodons | Style | PDAC_log Value | CONTROL_log Value | Log2FC | P value |
|
| ||||||
| tRF-3 | ThrTGT | up | 1.661561937 | -0.025742273 | 1.6873042 | 0.0007625 |
| i-tRF | PheGAA | up | 1.85372168 | 0.25685016 | 1.5968715 | 0.0014351 |
| i-tRF | GluCTC | up | 1.74613625 | 0.233768415 | 1.5123678 | 0.0025247 |
| tRF-5 | ValCAC | up | 2.016917869 | 0.522544893 | 1.494373 | 0.0028382 |
| tRF-5 | GlnTTG | up | 2.330731335 | 0.904535166 | 1.4261962 | 0.0043764 |
| tRF-5 | IleGAT | up | 1.051827825 | -0.348957883 | 1.4007857 | 0.0051215 |
| i-tRF | GluCTC | up | 1.100540288 | -0.270007709 | 1.370548 | 0.0061568 |
| i-tRF | HisGTG | up | 0.833999022 | -0.528906011 | 1.362905 | 0.0064468 |
| 5’-half | GluTTC | up | 0.724293908 | -0.607371448 | 1.3316654 | 0.0077645 |
| tRF-5 | LysTTT | up | 5.995384675 | 4.677263566 | 1.3181211 | 0.0084075 |
| i-tRF | PheGAA | up | 1.640360473 | 0.324212163 | 1.3161483 | 0.0085051 |
| i-tRF | PheGAA | up | 1.56317462 | 0.279689088 | 1.2834855 | 0.0102743 |
| i-tRF | PheGAA | up | 0.807589717 | -0.472889518 | 1.2804792 | 0.0104526 |
| tRF-3 | TyrGTA | down | -0.360340284 | 0.91847101 | -1.2788113 | 0.0105528 |
| tRF-3 | ValTAC | up | 1.09329157 | -0.183463465 | 1.276755 | 0.0106774 |
| tRF-3 | GlyCCC | up | 0.769550591 | -0.504691303 | 1.2742419 | 0.0108314 |
| i-tRF | PheGAA | up | 1.722985181 | 0.448880293 | 1.2741049 | 0.0108399 |
| i-tRF | PheGAA | up | 1.546748141 | 0.281568863 | 1.2651793 | 0.0114036 |
| i-tRF | GluCTC | up | 1.000015661 | -0.263967867 | 1.2639835 | 0.011481 |
| i-tRF | GlyGCC | up | 0.671082865 | -0.58569284 | 1.2567757 | 0.011958 |
| i-tRF | MetCAT | up | 0.760216887 | -0.488405465 | 1.2486224 | 0.0125187 |
| tRF-5 | ValCAC | up | 1.201057567 | -0.040164054 | 1.2412216 | 0.0130477 |
| tRF-3 | GlnCTG | up | 0.819184775 | -0.416000576 | 1.2351854 | 0.0134938 |
| 5’-half | GluCTC | up | 1.885542245 | 0.654592062 | 1.2309502 | 0.0138148 |
| i-tRF | GluTTC | down | -0.037738799 | 1.193017171 | -1.230756 | 0.0138297 |
| i-tRF | ValCAC | up | 0.820516116 | -0.404013916 | 1.22453 | 0.0143143 |
| tRF-3 | PheGAA | up | 2.234063666 | 1.012092738 | 1.2219709 | 0.0145178 |
| 5’-half | CysGCA | up | 1.029605896 | -0.191991858 | 1.2215978 | 0.0145477 |
| tRF-5 | LysTTT | up | 5.928537502 | 4.707122075 | 1.2214154 | 0.0145623 |
| tRF-3 | MetCAT | up | 1.521204137 | 0.304749721 | 1.2164544 | 0.0149653 |
| tRF-5 | LysTTT | up | 5.893796539 | 4.679316868 | 1.2144797 | 0.0151284 |
| i-tRF | GluCTC | up | 0.807945558 | -0.39532262 | 1.2032682 | 0.0160845 |
| tRF-3 | ValTAC | down | -0.484189819 | 0.710548394 | -1.1947382 | 0.0168472 |
| i-tRF | PheGAA | up | 1.617883879 | 0.43139674 | 1.1864871 | 0.0176151 |
| tRF-5 | GlyGCC | up | 0.690073474 | -0.494950751 | 1.1850242 | 0.0177544 |
| tRF-5 | LysTTT | up | 5.980603203 | 4.798564875 | 1.1820383 | 0.0180418 |
| tRF-3 | SerTGA | down | -0.395284112 | 0.784779646 | -1.1800638 | 0.0182341 |
| i-tRF | PheGAA | up | 1.350256124 | 0.189856357 | 1.1603998 | 0.0202491 |
| i-tRF | GluCTC | up | 1.501502967 | 0.350484913 | 1.1510181 | 0.0212773 |
| i-tRF | LysCTT | up | 0.71532284 | -0.431133899 | 1.1464567 | 0.0217935 |
| tRF-5 | LysTTT | up | 5.963670269 | 4.817448041 | 1.1462222 | 0.0218204 |
| tRF-5 | LysTTT | up | 5.94475542 | 4.803929443 | 1.140826 | 0.0224458 |
| tRF-3 | GlnCTG | up | 1.738047624 | 0.598514194 | 1.1395334 | 0.0225979 |
| i-tRF | PheGAA | up | 0.972545362 | -0.16338534 | 1.1359307 | 0.0230266 |
| tRF-3 | ValTAC | up | 0.738153603 | -0.39532262 | 1.1334762 | 0.0233227 |
| i-tRF | GluCTC | up | 0.830508 | -0.291686318 | 1.1221943 | 0.0247269 |
| i-tRF | SerGCT | up | 0.842881081 | -0.27418423 | 1.1170653 | 0.0253892 |
| tRF-5 | LysTTT | up | 5.899380184 | 4.786552441 | 1.1128277 | 0.0259479 |
| i-tRF | GluCTC | up | 1.456334408 | 0.349984204 | 1.1063502 | 0.0268226 |
| i-tRF | GlnCTG | down | -0.389138911 | 0.705702356 | -1.0948413 | 0.0284398 |
| tRF-3 | TyrGTA | down | 0.066874169 | 1.151799777 | -1.0849256 | 0.0298999 |
Corresponding indolent PDAC vs normal control fold changes are reported.
In serum of PDAC patients, miRNAs were most abundant (97.59%), tsRNAs were next to miRNAs, accounting for 2.18% of total small RNAs (Figure 1A), and tsRNAs significantly increased in PDAC samples compared to healthy controls (Figure 1B). There were 6 types of tsRNAs in PDAC and healthy controls, including 3’-half, 5’-half, i-tRF, tRF-1, tRF-3 and tRF-5 (Figure 1C), i-tRF, tRF-3 and tRF-5 are the three most abundant tsRNAs in serum. We found 51 differentially expressed tsRNAs among these, 45 tsRNAs were upregulated and 6 were downregulated in PDAC (defined as log2-fold expression difference > 2 and P value < 0.05) (Table 2; Supplementary Figure 3). The differential tsRNAs expression was shown in a heatmap (Figure 1D).
Figure 1.
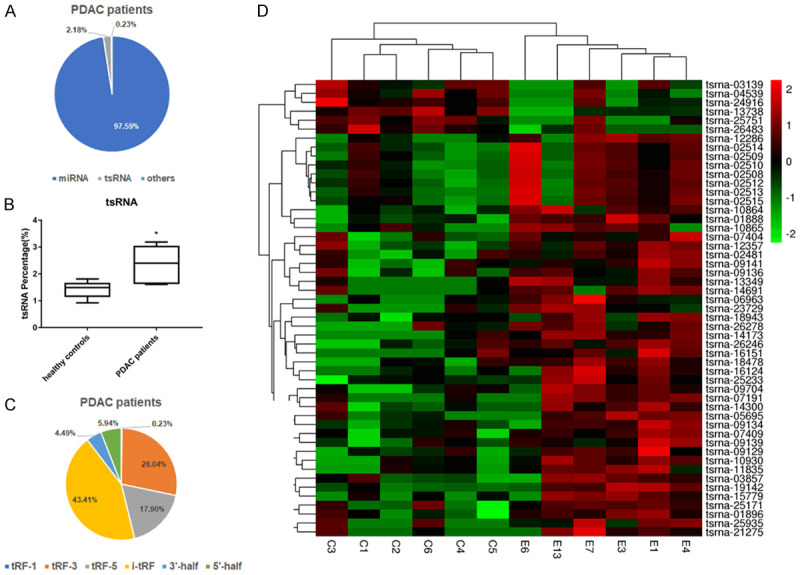
Differential expression analysis of serum tsRNAs in PDAC patients, compared with healthy controls. A. The types of small RNAs in serum from PDAC patients. B. The proportion of tsRNAs in small RNA have statistical significance between PDAC and health controls. C. The types of tsRNAs in serum from PDAC patients. D. Heatmap depicting the expression of the 51 differentially expressed tsRNAs across all 12 samples of PDAC and healthy controls. C1, C2, C3, C4, C5, C6 refer to samples from healthy normal people (n = 6); E1, E3, E4, E6, E7, E13 refer to samples from PDAC patients (n = 6).
Relative levels of tsRNAs normalized to RNU6 as reference gene were increased in serum of patients with PDAC vs healthy controls
The tsRNAs-seq data was studied by analyzing their levels using quantitative PCR in a validation set, which included 110 PDAC patients and 100 healthy controls. Based on the evaluation by qPCR in PDAC patients and healthy controls, 3 candidate tRF-3s, tsRNA-MetCAT-37, tsRNA-ValTAC-41, tsRNA-ThrTGT-23 were identified, 2-ΔCq*10000 value of the tsRNAs after normalizing to RNBU6 was evaluated. The expression level of tsRNA-MetCAT-37, tsRNA-ValTAC-41 and tsRNA-ThrTGT-23 were significant upregulated in serum of PDAC patients (P = 0.0004, 0.0019 and 0.0038) (Figure 2).
Figure 2.
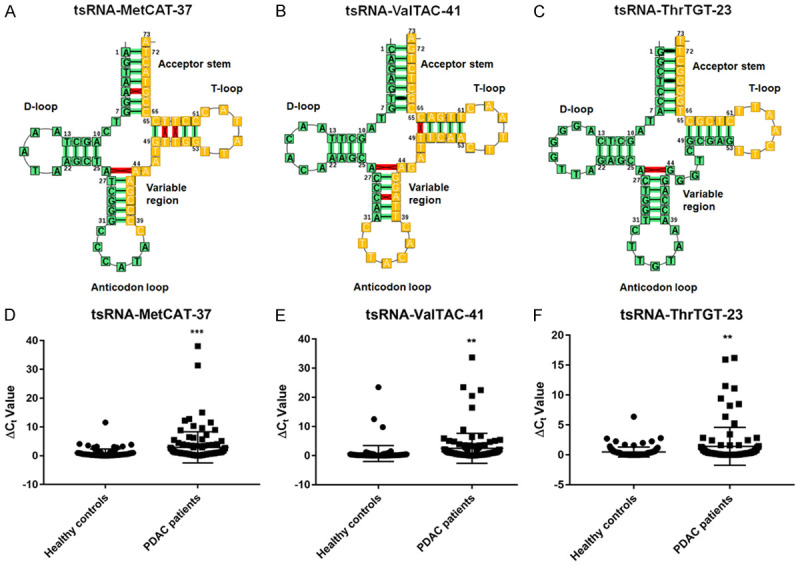
Structure and qPCR data of select tsRNAs in the validation set. The structure of tsRNA-MetCAT-37 (A), tsRNA-ValTAC-41 (B), and tsRNA-ThrTGT-23 (C). tRNA covariance model fold borrowed and modified from F. Jühling, M. Mörl, R. K. Hartmann, M. Sprinzl, P. F. Stadler, and J. Pütz. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res., 2009, Vol. 37, (Database issue): D159-D162. The relative expression level of tsRNA-MetCAT-37 (D), tsRNA-ValTAC-41 (E), and tsRNA-ThrTGT-23 (F) between PDAC 110 patients and 100 healthy controls. Unpaired-t test was used to test for statistical differences. One numeric difference in ΔCt represents a 2-fold difference in the amount of validated tsRNAs. (P value of student t-test: **P ≤ 0.01, ***P ≤ 0.001).
ROC analyses indicated that tsRNA-MetCAT-37 and tsRNA-ValTAC-41 had diagnostic potential
110 PDAC patients and 100 healthy controls were then identified for testing the accuracy of diagnosis. tsRNA-MetCAT-37 and tsRNA-ValTAC-41 showed statistically significant curve and the AUC were 0.687 and 0.793 respectively (Figure 3).
Figure 3.
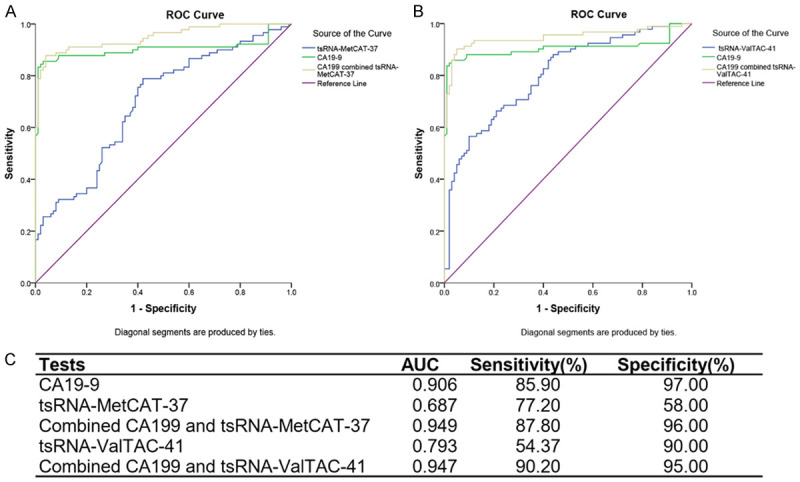
ROC analyses of PDAC prediction based on serum tsRNAs and CA19-9. (A, B) ROC curve analysis of ΔCt summation of serum CA19-9 combined with tsRNA-MetCAT-37 (A) and tsRNA-ValTAC-41 (B) of 110 PDAC patients and 100 healthy controls. (C) AUC, sensitivity and specificity of serum CA19-9, tsRNA-MetCAT-37 and tsRNA-ValTAC-41.
ROC curve of current diagnostic biomarker CA19-9 was also tested in our patients. As expected, CA19-9 for PDAC diagnosis showed an AUC value of 0.906, with a sensitivity of 85.9% and specificity of 97.0%. In order to improve the accuracy of CA19-9 alone, combing of serum tsRNAs with CA19-9 was employed. tsRNA-MetCAT-37 or tsRNA-ValTAC-41 combined with CA19-9 respectively could increase the AUC of PDAC prediction, compared with CA19-9 alone, the AUC value increased to 0.949 and 0.947 at the sensitivity of 87.8% and 90.2%, respectively (Figure 3).
tsRNA-ValTAC-41 has a potential biological function in PDAC
Moreover, patients with low serum tsRNA-ValTAC-41 level had a significantly longer OS than those with high level (LogRank P < 0.0001) (Figure 4A). Objective to investigate the relationship between tsRNAs and severity of PDAC, correlative study of the serum tsRNAs and AJCC stage was analyzed. It showed that tsRNA-ValTAC-41 in locally advanced and metastatic PDAC were significantly up-regulated compared to those in early stage (stages III and IV vs I and II, P = 0.0033) (Figure 4B). Further analysis showed that tsRNA-ValTAC-41 expressed obviously higher in patients of M1 stage compared to M0 (P = 0.0189) and especially up-regulated in patients with liver metastasis (P = 0.0189) (Supplementary Figure 3).
Figure 4.
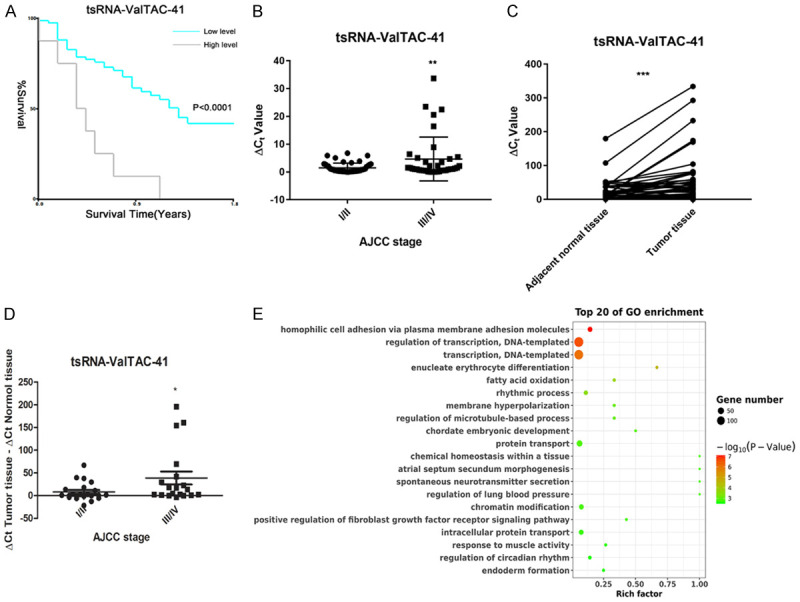
Kaplan-Meier curves, expression level and bioinformatic analyze of tsRNA-ValTAC-41 in serum and tissue of PDAC patients. A. Kaplan-Meier curves estimating the longer overall survival (OS) in patients with lower tsRNA-ValTAC-41 serum level. B. tsRNA-ValTAC-41 expression in serum was identically associated with AJCC grade I/II vs grade III/IV. C. The expression level of tsRNA-ValTAC-41 in tumor tissues and adjacent normal tissues. D. tsRNA-ValTAC-41 expression in tissue of PDAC patients was also associated with AJCC grade I/II vs grade III/IV. (P value of student t-test: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). E. GO enrichment analyzing in target genes of tsRNA-ValTAC-41.
Due to the better efficiency of tsRNA-ValTAC-41 in both diagnosis and prognosis prediction, we focus on tsRNA-ValTAC-41 for in-depth biological function research. In order to investigate the specific regulatory role of tsRNAs in tumors, another independent cohort of 51 patients were included to validate the expression level of tsRNA-ValTAC-41 in tumor tissue and adjacent normal tissue. tsRNA-ValTAC-41 showed higher expression in tumor tissue comparing to normal adjacent pancreas. Consistently, expression level of tsRNA-ValTAC-41 in tissue is also higher in locally advanced and metastatic PDAC patients (Figure 4C, stages III and IV vs I and II, P = 0.0326).
To further explore the mechanism of tsRNA-ValTAC-41 affecting tumor metastasis, bioinformatics analysis was performed to predict the target genes as well as their biological functions. GO functional enrichment analysis of the target genes indicated that tsRNA-ValTAC-41 (Figure 4D and Supplementary Figure 4) may have the potential role in cell adhesion, regulation of transcription and transcription.
Discussion
Despite the emergent improvement in diagnosis and therapy area of cancer, PDAC still remains one of the most deadly cancer with the 5-year survival less than 9% [24]. Incidence rates continue to increase in this disease in recent decades [24]. PDAC showed early recurrence and metastasis, as well as resistance to chemotherapy and radiotherapy. Surgery remains the only option to cure this disease, while only 15% of patients attend the opportunity of surgery because most patients are diagnosed in advanced stage [25]. Consider the special characteristics of PDAC, the early diagnosis is most urgently needed.
Accumulating evidence shows that small RNAs are cell-specific and tumor-specific [26], and may be used as diagnostic markers. The expression levels of miR-16, miR-21, miR-210, miR-155, miR-20a, miR-25 and miR-196a in the plasma of patients with PDAC were higher than those of the normal controls [27]. Further study verified the diagnostic sensitivity and accuracy improvement when miR-16, miR-155 and miR-25 were combined with CA19-9, respectively [28,29]. Besides the increased application to body fluids analyses of RNA-seq in recent decades, most available publications are focused on miRNAs, while the research of tsRNAs is still novel field. tRNAs are subjected to fragmentation as tsRNAs and latter are further divided into 3’ U tRFs, tRFs and itRHs. Most of tsRNAs are produced as a result of oncogenic stress such as hypoxia, which coincides with the hypoxic microenvironment of PDAC. This indicated us that there may be suitable conditions for the production of tsRNAs in this disease. Rather than randomly degraded tRNA fragments, recent studies have verified the biological function of several tsRNAs in multiple malignant tumors, including lung carcinoma [4], breast cancer [12], colorectal cancer [11] and chronic lymphocytic leukemia [30]. tsRNAs take part in cellular processes including cell proliferation [10,31], metastasis [32] and apoptosis [32,33]. Diagnostic potential of tsRNAs was also reported in prostate cancer [34], liver cancer [6] and clear cell renal cell carcinoma [21]. Although the function of tsRNAs remains largely unknown in cancer process, but they may be suitable as cancer biomarkers [35].
We firstly described the composition and expression profile of tsRNAs in serum of PDAC patients by small RNA sequencing. Most abundant small RNAs in serum were miRNAs, yet tsRNAs ranked second. The percentage of tsRNAs increased in PDAC patient cohort. 51 differentially expressed tsRNAs among this profile, 45 tsRNAs were upregulated and 6 were downregulated in PDAC. Higher expression of tsRNA-MetCAT-37, tsRNA-ValTAC-41 and tsRNA-ThrTGT-23 were verified in serum of 110 PDAC patients compared to 100 controls by qPCR. CA19-9 is routinely detected in the diagnosis of PDAC patients in clinical process, while the low specificity in non-malignant still limited the accuracy of diagnosis [4]. Combination of other high specificity biomarker could improve the diagnostic accuracy of PDAC [34,35]. Therefore, the diagnostic value of CA19-9 and selected tsRNAs were validated in this study. In the combined CA19-9 and tsRNA-MetCAT-37 or tsRNA-ValTAC-41 respectively, the AUC was obviously increased. tsRNA-MetCAT-37 or tsRNA-ValTAC-41 may be potential biomarkers in PDAC, especially with CA19-9.
Ideal biomarkers can not only be used for screening and diagnosis, but in many cases, they can also be the starting of understanding cancer biological pathway as well as the regulatory mechanisms. Similar to miRNAs, tsRNAs also have possible biological roles in PDAC apart from their use as diagnosis biomarkers. tsRNA-ValTAC-41 showed excellent prognostic value. Besides, the expression level of tsRNA-ValTAC-41 significantly increased in tumor tissue comparing to adjacent normal tissue, which indicated its potential functional role in cancer process. Adverse AJCC stage was associated with tsRNA-ValTAC-41 expression both in serum and tissue, and high expression of tsRNA-ValTAC-41 in serum was related to distant metastasis, especially liver metastasis. Bioinformatics analysis showed tsRNA-ValTAC-41 was in the process of adhesion, regulation of transcription and transcription. It suggested tsRNA-ValTAC-41 may take part in tumor progression and metastasis in PDAC. Further functional validation is needed in these field. This breakthrough study can be achieved by translating newly acquired tsRNAs knowledge into clinical practice of PDAC.
Interestingly, tsRNA-MetCAT-37, tsRNA-ValTAC-41 and tsRNA-ThrTGT-23 are all 3’-tRF. tRF-3, derived from the 3’end of mature tRNA, is produced by cleavage of ANG, Dicer, or members of ribonuclease A superfamily at the T-loop. Therefore, the tRF-3 tail contains a CCA structure specific for the 3’end of the mature tRNA, approximately 18-22 nt in length [36].
The tRF-3 was identified in human mature B cells [31], breast cancer [37], and from expression screening in HeLa and HCT-16 cells [38]. The functional studies showed that to tRF-3 could repress mRNA transcripts, suppress cell proliferation as well as modulate DNA damage response [39]. tRF-3 in breast cancer cells correlated with cell invasiveness and migration [37]. The role of tRF-3 in HeLa and HCT-16 cells in suppressing tumor growth suggested that it could be a potential new target for cancer therapy [33]. What’s the role of tRF-3, especially tsRNA-ValTAC-41 in PDAC, further studies of tsRNA-ValTAC-41 in PDAC should be the next aim.
Conclusion
Serum tsRNA-MetCAT-37, tsRNA-ValTAC-41 and tsRNA-ThrTGT-23 were up-regulated in PDAC patients. Patients with lower tsRNA-ValTAC-41 serum level showed longer survival time. Besides, tsRNA-ValTAC-41 was positively correlated to AJCC stage in PDAC and expressed highly in tumor tissue. Moreover, tsRNA-MetCAT-37 and tsRNA-ValTAC-41 could increase the AUC of CA19-9 in PDAC diagnosis. Thus, tsRNAs may serve as novel PDAC biomarkers as well as functional participant. Further investigations are warranted to determine the functional implications of altered expression of tsRNAs in PDAC pathogenesis and progression.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant NO. 81672325 & NO. 81802316).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/S0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 3.Crawley AS, O’Kennedy RJ. The need for effective pancreatic cancer detection and management: a biomarker-based strategy. Expert Rev Mol Diagn. 2015;15:1339–1353. doi: 10.1586/14737159.2015.1083862. [DOI] [PubMed] [Google Scholar]
- 4.Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, Farina NH, Lian JB, Tomasello L, Liu CG, Palamarchuk A, Hart JR, Bell C, Carosi M, Pescarmona E, Perracchio L, Diodoro M, Russo A, Antenucci A, Visca P, Ciardi A, Harris CC, Vogt PK, Pekarsky Y, Croce CM. tsRNA signatures in cancer. Proc Natl Acad Sci U S A. 2017;114:8071–8076. doi: 10.1073/pnas.1706908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shen Y, Yu X, Zhu L, Li T, Yan Z, Guo J. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med (Berl) 2018;96:1167–1176. doi: 10.1007/s00109-018-1693-y. [DOI] [PubMed] [Google Scholar]
- 6.Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D, Xu X, Zuo Y, Zhao Y, Wei YQ, Wei XW, Peng Y. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18:74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Liu X, Pu W, Peng Y. tRNA-derived small non-coding RNAs in human disease. Cancer Lett. 2018;419:1–7. doi: 10.1016/j.canlet.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G, Rassenti LZ, Pass HI, Kipps TJ, Liu CG, Croce CM. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113:5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veneziano D, Tomasello L, Balatti V, Palamarchuk A, Rassenti LZ, Kipps TJ, Pekarsky Y, Croce CM. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2019;116:24252–24258. doi: 10.1073/pnas.1913695116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao Y, Sun Q, Liu X, Wang P, Wu R, Ma Z. tRF-Leu-CAG promotes cell proliferation and cell cycle in non-small cell lung cancer. Chem Biol Drug Des. 2017;90:730–738. doi: 10.1111/cbdd.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W, Zhang Q, Zhang L, Xue Z, Li Y, Da Y, Yang Q, Li Z, Liu L, Qiao L, Kong Y, Yao Z, Zhao P, Li M, Zhang R. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 2017;77:3194–3206. doi: 10.1158/0008-5472.CAN-16-3146. [DOI] [PubMed] [Google Scholar]
- 12.Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telonis AG, Rigoutsos I. Race disparities in the contribution of mirna isoforms and tRNA-derived fragments to triple-negative breast cancer. Cancer Res. 2018;78:1140–1154. doi: 10.1158/0008-5472.CAN-17-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun C, Yang F, Zhang Y, Chu J, Wang J, Wang Y, Zhang Y, Li J, Li Y, Fan R, Li W, Huang X, Wu H, Fu Z, Jiang Z, Yin Y. tRNA-derived fragments as novel predictive biomarkers for trastuzumab-resistant breast cancer. Cell Physiol Biochem. 2018;49:419–431. doi: 10.1159/000492977. [DOI] [PubMed] [Google Scholar]
- 15.Mo D, Jiang P, Yang Y, Mao X, Tan X, Tang X, Wei D, Li B, Wang X, Tang L, Yan F. A tRNA fragment, 5’-tiRNA, suppresses the Wnt/β-catenin signaling pathway by targeting FZD3 in breast cancer. Cancer Lett. 2019;457:60–73. doi: 10.1016/j.canlet.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Zhou K, Diebel KW, Holy J, Skildum A, Odean E, Hicks DA, Schotl B, Abrahante JE, Spillman MA, Bemis LT. A tRNA fragment, tRF5-Glu, regulates BCAR3 expression and proliferation in ovarian cancer cells. Oncotarget. 2017;8:95377–95391. doi: 10.18632/oncotarget.20709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang M, Li F, Wang J, He W, Li Y, Li H, Wei Z, Cao Y. tRNA-derived fragment tRF-03357 promotes cell proliferation, migration and invasion in high-grade serous ovarian cancer. Onco Targets Ther. 2019;12:6371–6383. doi: 10.2147/OTT.S206861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7:24766–24777. doi: 10.18632/oncotarget.8293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chun YS, Pawlik TM, Vauthey JN. 8th edition of the AJCC cancer staging manual: pancreas and hepatobiliary cancers. Ann Surg Oncol. 2018;25:845–847. doi: 10.1245/s10434-017-6025-x. [DOI] [PubMed] [Google Scholar]
- 21.Zhao C, Tolkach Y, Schmidt D, Kristiansen G, Muller SC, Ellinger J. 5’-tRNA Halves are dysregulated in clear cell renal cell carcinoma. J Urol. 2018;199:378–383. doi: 10.1016/j.juro.2017.07.082. [DOI] [PubMed] [Google Scholar]
- 22.Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816–E3825. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 24.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 25.Butturini G, Stocken DD, Wente MN, Jeekel H, Klinkenbijl JH, Bakkevold KE, Takada T, Amano H, Dervenis C, Bassi C, Büchler MW, Neoptolemos JP Pancreatic Cancer Meta-Analysis Group. Influence of resection margins and treatment on survival in patients with pancreatic cancer: meta-analysis of randomized controlled trials. Arch Surg. 2008;143:75–83. doi: 10.1001/archsurg.2007.17. [DOI] [PubMed] [Google Scholar]
- 26.Muller S, Raulefs S, Bruns P, Afonso-Grunz F, Plotner A, Thermann R, Jager C, Schlitter AM, Kong B, Regel I, Roth WK, Rotter B, Hoffmeier K, Kahl G, Koch I, Theis FJ, Kleeff J, Winter P, Michalski CW. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng JF, Zhuang YY, Huang FT, Zhang SN. Noncoding RNAs and pancreatic cancer. World J Gastroenterol. 2016;22:801–814. doi: 10.3748/wjg.v22.i2.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao L, He SB, Li DC. Effects of miR-16 plus CA19-9 detections on pancreatic cancer diagnostic performance. Clin Lab. 2014;60:73–77. doi: 10.7754/clin.lab.2013.121210. [DOI] [PubMed] [Google Scholar]
- 29.Pan W, Tang W, Yuan W, Yu Q, Zuo W, Xu C, Ma J. Expression and clinical significance of plasma small RNA in patients with pancreatic cancer. Zhonghua Zhong Liu Za Zhi. 2014;36:351–354. [PubMed] [Google Scholar]
- 30.Balatti V, Rizzotto L, Miller C, Palamarchuk A, Fadda P, Pandolfo R, Rassenti LZ, Hertlein E, Ruppert AS, Lozanski A, Lozanski G, Kipps TJ, Byrd JC, Croce CM, Pekarsky Y. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2015;112:2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, Wan F, Wang Y, Long J, Zhu X. Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer. Cancer Manag Res. 2019;11:3945–3956. doi: 10.2147/CMAR.S199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, Zhang F, Chua MS, So S, Zhang QC, Sarnow P, Kay MA. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens-Uzunova ES, Jalava SE, Dits NF, van Leenders GJ, Moller S, Trapman J, Bangma CH, Litman T, Visakorpi T, Jenster G. Diagnostic and prognostic signatures from the small non-coding RNA transcriptome in prostate cancer. Oncogene. 2012;31:978–991. doi: 10.1038/onc.2011.304. [DOI] [PubMed] [Google Scholar]
- 35.Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297–4304. doi: 10.1016/j.febslet.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou X, Zhang ZJ, Peng YH, Yang YZ, Yun JP. miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35:469–478. doi: 10.1093/carcin/bgt330. [DOI] [PubMed] [Google Scholar]
- 38.Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, Zhang F, Chua MS, So S, Zhang QC, Sarnow P, Kay MA. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


