Abstract
Long non-coding RNAs (lncRNAs) are increasingly recognized as promising targets in cancer treatment. However, compared to targeting the ordinary protein-coding genes, suppressing non-coding RNAs expressed in cancer cells has been a more challenging task. The major hurdles lay on the requirement of a tumor-specific delivery system for the designated inhibitor to suppress the target transcripts within the cellular compartment. EGFR is a cancer driver gene which is frequently associated with the triple-negative phenotype of breast cancer. Prior studies have shown that expression of the tumor-promoting lncRNA HOTAIR (HOX antisense intergenic RNA) is positively regulated by the epithelial growth factor receptor (EGFR) in triple-negative breast cancer (TNBC), and consistently the expression of both genes is closely correlated in breast cancer. Here we show that a chimeric aptamer recognizing the epithelial growth factor receptor (EGFR) coupled with a siRNA against HOTAIR (EGFR aptamer-coupled siHOTAIR) preferentially and effectively down-regulated HOTAIR in EGFR-expressing cancer cells. Functionally, the EGFR aptamer-coupled siHOTAIR more potently inhibited the growth, migration, and invasion of EGFR-expressing TNBC cells as well as cells with reconstituted EGFR compared to cancer cells with low EGFR expression. Our results demonstrate a novel strategy of targeting cancer progression by aptamer-directed delivery of anti-lncRNA RNA interference that can be applicable to other cellular contexts and cancer types.
Keywords: EGFR, receptor tyrosine kinase, long non-coding RNA, HOTAIR, aptamer, triple-negative breast cancer, TNBC, siRNA
Introduction
Triple-negative breast cancer (TNBC), a major breast cancer subtype accounting for 10%-20% of breast cancer, is characterized by the lack of estrogen receptor, progesterone receptor, and the receptor tyrosine-protein kinase erbB-2 (ErbB2/HER2) [1,2]. TNBC is characterized by promoted tumor growth and treatment resistance, and is often accompanied by enhanced aggressiveness. Due to the lack of conventional therapeutic targets, TNBC is the only subtype of breast cancer for which no FDA-approved targeted therapies are available. Currently chemotherapy is the main therapeutic option for TNBC, but resistance to the treatment frequently occurs in relapsed advanced tumors. Blockade of immune checkpoint is emerging as a promising intervention for certain TNBC. However, the response of patients with metastatic TNBC to PD-1/PD-L1 blockade has been low, with response rates around 5% [3-5]. Thus, developing effective therapeutic approaches to TNBC remains an unmet medical need with significant clinical demands.
Expression of the receptor tyrosine kinase EGFR occurs in about 50% of TNBC tumors and therefore has been viewed as a promising target of TNBC [6]. Activation of EGFR has profound effect in cancer biology with multiple pathways such as MAPK, PI3K/AKT, and Wnt/β-catenin as the downstream effectors of activated EGFR for tumor promotion [6-8]. Mounting evidence derived from the clinic and in vitro studies supports that inhibition of EGFR promotes cell death, suppresses cancer cell growth, and enhances tumor progression, indicating a critical role in breast cancer biology [6,9,10]. However, the outcome of clinical trials for the therapeutic efficacy of targeting EGFR as single therapy in breast cancer has been disappointing due to intrinsic insensitivity or acquired resistance [11,12], suggesting that targeting EGFR alone is unlikely to have major therapeutic impact in advanced breast cancer.
We previously showed that in TNBC cells EGFR activation induced nuclear translocation of the oncogenic transcription factor β-catenin which in turn induced gene expression of the lncRNA HOTAIR through two LIF1/TCF4-binding sites [13]. HOTAIR is a 2.3-kb transcript transcriptionally derived from the intergenic region of the HOXC homeotic gene cluster [14], and was the first lncRNA identified with tumor-promoting function whose expression is correlated with poor prognosis in breast cancer [15,16]. HOTAIR forms a platform assembling epigenetic moderators to regulate gene expression. In addition, it has been reported that HOTAIR can regulate miRNA to promote cancer cell growth and migration [17,18]. Expression of HOTAIR enhanced the growth and metastasis of xenograft tumors of mammary fat pad [15]. We have identified the chondroitin sulfate sulfotransferase CHST15 as one of the HOTAIR-regulated genes [19]. CHST15 modified the context of glycan modifications of cell surface proteins by increasing the contents of the chondroitin 4,6-disulfate which in turn promote cell migration, invasion, and tumor growth. Importantly, downregulation of HOTAIR resulted in loss of sensitivity of TNBC to EGFR inhibitors [13]. Thus, HOTAIR as well as the biological functions regulated by HOTAIR are critical tumor-promoting mechanism of the EGFR cascade in TNBC cells, suggesting that the HOTAIR lncRNA is a promising target in EGFR-expressing TNBC cells.
Aptamers are short (arbitrarily defined up to ~200 nucleotides) single-stranded nuclease-stabilized oligonucleotide. Through a series of selecting cycles (namely the selection processes of systematic evolution of ligands by exponential enrichment; SELEX), the enriched aptamers specifically recognizing a molecular target can be isolated [20,21]. Successful aptamers have a dynamic three dimensional conformation which specifically recognizes the designated target in a mode similar to antibodies, with the biological advantages of high tissue penetration, low immunogenicity, and long in vivo stability [22-24]. With this strategy, a series of EGFR-targeting aptamers have been developed [25-27]. The EGFR aptamer E07 (hereafter referred to as aptEGFR) binds to EGFR with high affinity (Kd = 2.4 nM), specifically targets EGFR-expressing breast cancer cells, blocks EGFR activity in vivo, and penetrates cells with its cargo [27,28].
In the current study, we constructed a chimeric aptamer coupling E07 with a siRNA of HOTAIR in the same molecule and tested the chimera for its efficacy to deliver anti-HOTAIR RNA interference and suppression of cell growth and invasion in TNBC cells through EGFR.
Results
A synthetic DNA fragment containing the EGFR-targeting E07 aptamer, as reported by Li et al. [27], fused at the 3’ end with the antisense sequence of a HOTAIR siRNA (siHOTAIR) was produced (Supplementary Figure 1). The synthesized DNA oligomers were then amplified by PCR with a T7 promoter sequence incorporated in the 5’ primer followed by a T7-based in vitro transcription reaction. The RNA product of the EGFR aptamer coupled with siHOTAIR, hereafter termed aptEGFR-siHOTAIR. As a negative control, an aptamer in which the siHOTAIR sequence replaced with a control siRNA (aptEGFR-siCtrl) was generated.
We reasoned that the anti-EGFR aptamers should target cancer cells expressing EGFR while spare cells devoid of EGFR expression. Furthermore, the conjugated siRNA transported with the aptamer into the cells should downregulate the endogenous HOTAIR transcript. To test this hypothesis, we compared the breast cancer cell lines MDA-MB-231 and MDA-MB-468, both are EGFR-expressing cell lines, and MCF-7, which does not express EGFR. Analyses with confocal fluorescence microscopy of Cy3-labled aptEGFR-siHOTAIR and aptEGFR-siCtrl showed that the aptamers were able to enter MDA-MB-231 and MDA-MB-468 cells but not MCF-7 cells (Figure 1). The aptamer molecules in MDA-MB-231 and MDA-MB-468 cells formed punctates in the cytoplasm with partial co-localization with the multivesicular bodies (MVBs) as indicated by the endosomal marker GW182 [29], consistent with a cell-entry mechanism entailing membrane internalization. In drastic contrast, no aptamer was observed in MCF-7 cells. These results suggest that the EGFR aptamers targeted cancer cells through EGFR.
Figure 1.

Cell entry of the aptamers to EGFR-positive cancer cells. MDA-MB-231, MDA-MB-468 and MCF-7 cells cultured in regular media were treated with Cy3-labeled aptEGFR-siHOTAIR or aptEGFR-siCtrl as indicated for 4 hr. Cells were then fixed and immunostained for GW182 (FITC). The nuclei were stained with DAPI. Magnification, 200×; Scale bar, 50 µm.
Both MDA-MB-231 and MCF-7 cells express endogenous the HOTAIR lncRNA. In line with its preferential internalization in MDA-MB-231 cells, aptEGFR-siHOTAIR, but not its negative control aptEGFR-siCtrl, significantly downregulated HOTAIR expression in MDA-MB-231 cells (Figure 2A). In contrast, aptEGFR-siHOTAIR had no effect in HOTAIR expression in MCF-7 cells (Figure 2B). These results predict that treatment with aptEGFR-siHOTAIR should have cancer-inhibitory activities in MDA-MB-231 but not MCF-7 cells. Indeed, compared to treatment with aptEGFR-siCtrl, aptEGFR-siHOTAIR treatment inhibited the viability of MDA-MB-231, but not MCF-7, cells (Figure 2C).
Figure 2.
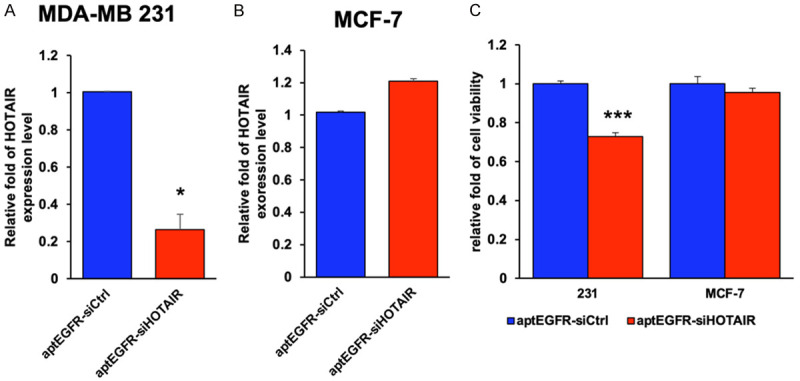
Depletion of the endogenous HOTAIR by the aptamer-delivered siHOTAIR. MDA-MB-231 and MCF-7 cells cultured in completed medium were transduced with the aptEGFR-siHOTAIR (siHOTAIR) or aptEGFR-siCtrl (siCtrl) for 72 hr. Cellular RNA of MDA-MB-231 (A) and MCF-7 (B) was extracted and the level of HOTAIR was measured by RT-PCR Endogenous actin was used as the internal control. (C) Cell viability of MDA-MB-231 and MCF-7 cells treated with the indicated aptamer was assessed by crystal violet staining with the optical density (OD) measured by absorbance at 595 nm. Bar, standard deviation. *, P < 0.05; ***, P < 0.005.
HOTAIR is well known for its function in enhancing migration and invasion of cancer cells [13,15]. Administrating aptEGFR-siHOTAIR, but not aptEGFR-siCtrl, significantly suppressed the migratory ability of MDA-MB-231 (Figure 3A). More importantly, aptEGFR-siHOTAIR, but not aptEGFR-siCtrl, suppressed the invasive activity of MDA-MB-231 cells (Figure 3B), indicating that the strategy of the chimeric aptamer is an efficient strategy to suppress invasiveness mediated by HOTAIR.
Figure 3.

Inhibition of cell migration and invasion by aptamer-delivered siHOTAIR. A. Wound healing activity of MDA-MB-231 cells treated by aptEGFR-siHOTAIR or aptEGFR-siCtrl are determined at 20 hr after wounding. B. Representative images of Boyden invasion chamber assay in which cell were treated by aptEGFR-siHOTAIR or aptEGFR-siCtrl. The data are plotted with standard deviation indicated. ***, P < 0.005. Magnification, 50×; scale bar, 500 µm.
To further test the dependence of EGFR in the biological function of the chimeric aptamer, MCF-7 cells were engineered to express ectopic EGFR or vector control alone (Figure 4A; Supplementary Figure 2). Expression of EGFR enhanced cell entry of the aptEGFR aptamer, with partial co-localization with the endosomal compartment under confocal microscopy analysis (Figure 4B). This was accompanied with reduced migratory activity of MCF-7 overexpressing EGFR, but not for the parental MCF-7 cells (Figure 4C). Importantly, introduction of EGFR enhanced invasive activity of MCF-7 cells and treatment with aptEGFR-siHOTAIR, but not aptEGFR-siCtrl, significantly suppressed invasion mediated by EGFR expression (Figure 4D).
Figure 4.
Inhibition of cell migration and invasion by aptamer-delivered siHOTAIR which specifically targeted to EGFR. (A) Ectopic EGFR expression in MCF-7. The original blots were cut according to the molecular weights of the proteins of interest. The resulted chemiluminescent images were cropped to keep only the relevant signals for space saving. (B) Confocal imaging of aptEGFR-siHOTAIR which was delivered in the presence of EGFR. Magnification, 50×; scale bar, 5 µm. Cell migration (C) and invasion (D) performed in ectopic EGFRE expression of MCF-7 cells under treatment of aptEGFR-siHOTIAR or aptEGFR-siCtrl for 72 hr, followed by the corresponding assays for the indicated times. Representative images of Boyden invasion chamber assay in which cell were treated by aptEGFR-siHOTAIR or aptEGFR-siCtrl are shown. The data are plotted with standard deviation indicated. *, P < 0.05; **, P < 0.01. Magnification, 50×; scale bar, 500 µm.
Discussion
Although EGFR is well known for its roles in driving cancer progression, metastasis, and resistance to treatments, targeting EGFR as single therapy has not made its way for approved breast cancer treatment due to unsatisfactory responsiveness of the patients. The observation that EGFR and c-ABL synergistically upregulate the oncogenic lncRNA HOTAIR provide an example that targeting downstream targets converging the signaling multiple pathways is a more promising strategy than targeting a single cancer driver. Alternatively, targeting key downstream targets dictated by the EGFR and c-ABL pathways can be an effective approach.
Our results showed that, compared to MCF-7 cells, MDA-MB-231 cells treated by aptEGFR-siHOTAIR preferentially downregulated HOTAIR expression with suppression of growth and invasion. The causal relationship of EGFR with the responsiveness is supported by ectopic expression of EGFR in MCF-7 cells which restored the cancer-inhibiting phenotypes by aptEGFR-siHOTAIR treatment. Loss-of-function approach by knocking down EGFR from DMA-MB-231 cells turned out to be non-feasible because of poor cell viability upon depleting EGFR in the cell line (data not shown). Overall, our finding is accountable for the scenario that the aptamer binds to the target EGFR and internalized through the endosome compartment with the cargo. It remains to be determined how the endosomal aptamer complex is released or gain access to the target RNA transcript in the cytosol. Our result showed that the internalized aptamers were colocalized with GW182, a protein enriched in the MVBs and the associated RNA-induced silencing complexes (RISCs) engaging in miRNA- and siRNA-mediated gene silencing [29].
The tumor-promoting functions of non-coding RNAs have been increasingly appreciated [30,31], and the next hurdle to be overcome is the tumor-specific targeting of these biological moieties. To our knowledge this is the first report of delivering lncRNA-targeting RNA interference through aptamers targeting a receptor tyrosine kinase on cancer cells. Given that RTKs play important roles in tumor progression and metastasis, the concept proving principles demonstrated in the current study have substantial translational implication in cancer therapy and warrants further study with systemic administration of the aptamer in animal models for which an efficient and specialized delivery system is required [32]. Specifically the delivery system for the EGFR-targeting RNA aptamer should be able to present the EGFR-targeting moiety of the aptamer for tumor homing while protects the RNA cargos from nucleolytic attacks during circulation and allows efficient endosomal escape after entering the cells. Furthermore, the presentation of the double-stranded conformation of the aptamer in the cytosol may trigger immune response by the cytosolic nucleic acid-recognizing stress-response machinery, an important issue should be factored in to the assessment of the therapeutic outcome [33]. Overall, the current study provides a proof of the concept that aptamers homing on to the preys on the cell surface can be exploited for targeting intracellular non-coding tumor-promoting genes which are otherwise spared from the conventional therapeutics. Future study is warranted to test the in vivo efficacy in suppressing tumor progression and metastasis.
Methods
Cell lines, antibodies, and reagents
The human breast cancer cell lines: MCF-7 (ATCC®HTB-22TM) and MDA-MB-231 (ATCC®HTB-26TM) were obtained from the American Type Culture Collection (ATCC). The complete medium is DMEM/F-12 supplemented with 10% fetal bovine serum (FBS), and 1% Pen-Strep human breast cancer cell lines. All cell lines were maintained at 37°C and 5% CO2. Primary antibodies used in this research include the following: mouse monoclonal anti-GW182 (ab70522, Abcam), rabbite polyclonal EGFR (ab52894, Abcam), mouse monoclonal anti-Actin (sc32251, Santa Cruz Biotechnolgy). qRT-PCR primers of human genes: HOTAIR-forward, GGTAGAAAAAGCAACCACGAAGC; HOTAIR-reverse, ACATAAACCTCTGTCTGTGAGTGCC; 18S-forward, AGGATCCATTGGAGGGCAAGT; 18S-reverse, TCCAACTACGAGCTTTTTAACTGCA; Actin-forward, CTTCCCCTCCATCGTGGG; Actin-reverse, GTGGTACGGCCAGAGGCG; GAPDH-forward, CGGAGTCAACGGATTTGGTCGTA; GAPDH-reverse, AGCCTTCTCCATGGTGGTGAAGAC.
Aptamer production
Aptamer is produced by in vitro transcription. Briefly, the aptEGFR-siHOTAIR template was amplified with the E07-F and E07-HOTAIR-R primers, with the T7 promoter (5’-GATAATACGACTCACTATA-3’) incorporated in the E07 primer and the anti-sense sequence of the siHOTAIR siRNA (5’-GAACGGGAGTACAGAGAGA-3’) in the E07-HOTAIR-R primer. The PCR product was then transcribed in vitro using the Platinum II Taq DNA polymerase (Invitrogen) with which 1 μg PCR DNA template was estimated to produce about 100~150 μg of RNA product. The resulted RNA transcript contained the E07 aptEGFR aptamer linked with the anti-sense strand of the siHOTAIR siRNA. The RNA product was purified using the DirectzolTM Kit (Zymoresearch). The same approach was used to produce the control aptamer aptEGFR-siCtrl in which the siRNA was replaced with a scrambled sequence as the siCtrl (5’-GAGCGAGGGCGACTTAACCTTAGG-3’). The purified RNA (50 μM) was annealed with synthetic oligoribonucleotide of siHOTAIR or siCtrl by denaturing in annealing buffer (10 mM Tris, pH8.0, 1 mM EDTA and 50 mM NaCl) at 95°C for 5 min and let cool down to room temperature in the thermal cycler, -0.1°C/sec. For labeling, the RNA product was labeled with Cy3 by the Label IT Nucleic Acid Labeling kit (miR3600; Mirus).
The following oligonucleotides were used: aptEGFR-siHOTAIR-5’-GGCGCTCCGACCTTAGTCTCTGTGCCGCTATAATGCACGGATTTAATCGCCGTAGAAAAGCATGTCAAAGCCGGAACCGTGTAGCACAGCAGAAAAATCTCTCTGTACTCCCGTTCTT-3’ (the two underlined sequences are of aptEGFR and siHOTAIR, respectively); aptEGFR-siCtrl-5’-GGCGCTCCGACCTTAGTCTCTGTGCCGCTATAATGCACGGATTTAATCGCCGTAGAAAAGCATGTCAAAGCCGGAACCGTGTAGCACAGCAGAAAAAGAGCGAGGGCGACTTAACCTTAGGTT-3’ (the two underlined sequences are of aptEGFR and siCtrl, respectively); E07-F-GATAATACGACTCACTATAGGCGCTCCGACCTTAGTCTCTG (the T7 promoter sequence is underlined); E07-HOTAIR-R-AAGAACGGGAGTACAGAGAGATTTTTCTGCTGTGCTACACGGTTCCG (the two underlined sequences are of siHOTAIR and aptEGFR, respectively); E07-Ctrl-R-AACCTAAGGTTAAGTCGCCCTCGCTCTTTTTCTGCTGTGCTACACGGTTCCG (the two underlined sequences are of siCtrl and aptEGFR, respectively).
HOTAIR quantitation
MDA-MB-231 and MCF-7 cells were plated in 6-well plates at a cell density of ~60%. Prior to aptamer treatment, cells were incubate in 500 μl of OPTI-MEM media at 37°C for 30 min, then treated with 400 nM of annealed aptamers in 500 μl of OPTI-MEM in the presence of 1 μl of RNaseOUTTM (Invitrogen), continue incubation for 1 hr, then added with 1170 μl of DMEM/F-12 with 14% FBS and incubated further for 72 hr. Cells were then lysed and the total RNA was extracted for quantitative RT-PCR (qRT-PCR). iQTM SYBR® Green supermix (Bio-rad) was employed in qRT-PCR and a typical 2-step real-time PCR protocol was performed with an initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 10 sec and primer annealing and extension at 60°C for 30 sec.
Cell entry assay and endosome colocalization
MDA-MB-231 and MCF-7 cells were plated in 24-well plates (2.5×104 cells/well) for overnight. The cultured cells were washed and 300 μl of OPTI-MEM media was added to each well and incubated for 30 min at 37°C. 50 nM of Cy3-labeled aptamers in 300 μl of OPTI-MEM media was added and incubated at 37°C for 2 hr, followed by addition of 700 μl DMEM media with 14% and further incubation for another 2 hr. Cells were fixed with pre-warmed 4% formaldehyde in 5% acetic acid for 30 min at room temperature, permeabilized in 0.25% Triton X-100/PBS for 10 min at temperature and then subjected to immunostaining with the GW182 antibody.
Wound healing assay
Wound healing assays was performed with two methods: a) Cells treated with either aptEGFR-siCtrl or aptEGFR-siHOTAIR were plated in 96-well plates (2×104 cells/well in MCF-7 while 1×104 cells/well in MDA-MB-231). The cell monolayer was scratched using a yellow pipette tip. The closure (migration distance) was assessed by photographing at 3, 6, 9, 12 and 24 hr, and measured by ImageJ. b) Cell migration was also measured by the IncuCyte ZOOM 96-well Scratch Migration assay protocol according to the manufacturer’s instructions (Essen BioScience). A scratch wound was created in each well using the IncuCyte WoundMarker. Cells treated with either aptEGFR-siCtrl or aptEGFR-siHOTAIR were plated in 96-well plates. The wound healing process was monitored continuously in the IncuCyte Live-cell Imaging System. Images were captured every 2 hr for 24 hr.
Invasion assay
Cell invasion assays were performed in Transwell chamber inserts (Corning) which were coated with a thin layer of Matrigel (Corning). 5×104 cells/well were suspended in serum-free medium and plated into each insert with the lower chambers filled with 500 μl complete medium. After incubation for 24 hr or 48 hr, invading cells on the side of the Transwell facing the lower chamber were stained with crystal violet and quantitated by measuring the absorbance at OD 595 nm after solubilized in isopropanol.
Acknowledgements
The authors thank Pheruza Tarapore for technical assistance in aptamer production. This study was supported in part by the National Health Research Institute NHRI-EX106-10602BI (to S.-C.W), Ministry of Health and Welfare Cancer Research Center of Excellence grants MOHW109-TDU-B-212-134024 and MOHW109-TDU-B-212-010001 (to S.-C.W.), and the CMUH grant DMR-CELL-17025 (to S.-C.W.). The work was also financially supported in part by the Drug Development Center, China Medical University from the Ministry of Education in Taiwan (S.-C.W.). Experiments and data analysis were performed in part through the use of the Medical Research Core Facilities Center, Office of Research & Development at China medical University, Taichung, Taiwan.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.O’Brien KM, Cole SR, Tse CK, Perou CM, Carey LA, Foulkes WD, Dressler LG, Geradts J, Millikan RC. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina Breast Cancer Study. Clin Cancer Res. 2010;16:6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pal SK, Childs BH, Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res Treat. 2011;125:627–636. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams S, Schmid P, Rugo HS, Winer EP, Loirat D, Awada A, Cescon DW, Iwata H, Campone M, Nanda R, Hui R, Curigliano G, Toppmeyer D, O’Shaughnessy J, Loi S, Paluch-Shimon S, Tan AR, Card D, Zhao J, Karantza V, Cortés J. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol. 2019;30:397–404. doi: 10.1093/annonc/mdy517. [DOI] [PubMed] [Google Scholar]
- 4.Dirix LY, Takacs I, Jerusalem G, Nikolinakos P, Arkenau HT, Forero-Torres A, Boccia R, Lippman ME, Somer R, Smakal M, Emens LA, Hrinczenko B, Edenfield W, Gurtler J, von Heydebreck A, Grote HJ, Chin K, Hamilton EP. Avelumab, an anti-PD-L1 antibody, in patients with locally advanced or metastatic breast cancer: a phase 1b JAVELIN Solid Tumor study. Breast Cancer Res Treat. 2018;167:671–686. doi: 10.1007/s10549-017-4537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voorwerk L, Slagter M, Horlings HM, Sikorska K, van de Vijver KK, de Maaker M, Nederlof I, Kluin RJC, Warren S, Ong S, Wiersma TG, Russell NS, Lalezari F, Schouten PC, Bakker NAM, Ketelaars SLC, Peters D, Lange CAH, van Werkhoven E, van Tinteren H, Mandjes IAM, Kemper I, Onderwater S, Chalabi M, Wilgenhof S, Haanen JBAG, Salgado R, de Visser KE, Sonke GS, Wessels LFA, Linn SC, Schumacher TN, Blank CU, Kok M. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat Med. 2019;25:920–928. doi: 10.1038/s41591-019-0432-4. [DOI] [PubMed] [Google Scholar]
- 6.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi G, Ueno N. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchholz TA, Tu X, Ang KK, Esteva FJ, Kuerer HM, Pusztai L, Cristofanilli M, Singletary SE, Hortobagyi GN, Sahin AA. Epidermal growth factor receptor expression correlates with poor survival in patients who have breast carcinoma treated with doxorubicin-based neoadjuvant chemotherapy. Cancer. 2005;104:676–681. doi: 10.1002/cncr.21217. [DOI] [PubMed] [Google Scholar]
- 10.Ueno NT, Zhang D. Targeting EGFR in triple negative breast cancer. J Cancer. 2011;2:324–328. doi: 10.7150/jca.2.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX, Liu MC, Storniolo AM, Rimawi MF, Forero-Torres A, Wolff AC, Hobday TJ, Ivanova A, Chiu WK, Ferraro M, Burrows E, Bernard PS, Hoadley KA, Perou CM, Winer EP. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J. Clin. Oncol. 2012;30:2615–2623. doi: 10.1200/JCO.2010.34.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gutteridge E, Agrawal A, Nicholson R, Leung Cheung K, Robertson J, Gee J. The effects of gefitinib in tamoxifen-resistant and hormone-insensitive breast cancer: a phase II study. Int J Cancer. 2010;126:1806–1816. doi: 10.1002/ijc.24884. [DOI] [PubMed] [Google Scholar]
- 13.Wang YL, Overstreet AM, Chen MS, Wang J, Zhao HJ, Ho PC, Smith M, Wang SC. Combined inhibition of EGFR and c-ABL suppresses the growth of triple-negative breast cancer growth through inhibition of HOTAIR. Oncotarget. 2015;6:11150–11161. doi: 10.18632/oncotarget.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang MC, Ni JJ, Cui WY, Wang BY, Zhuo W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am J Cancer Res. 2019;9:1354–1366. [PMC free article] [PubMed] [Google Scholar]
- 17.Di W, Li Q, Shen W, Guo H, Zhao S. The long non-coding RNA HOTAIR promotes thyroid cancer cell growth, invasion and migration through the miR-1-CCND2 axis. Am J Cancer Res. 2017;7:1298–1309. [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Chen X, Xu T, Xia R, Han L, Chen W, De W, Shu Y. MiR-326 regulates cell proliferation and migration in lung cancer by targeting phox2a and is regulated by HOTAIR. Am J Cancer Res. 2016;6:173–186. [PMC free article] [PubMed] [Google Scholar]
- 19.Liu LC, Wang YL, Lin PL, Zhang X, Cheng WC, Liu SH, Chen CJ, Hung Y, Jan CI, Chang LC, Qi X, Hsieh-Wilson LC, Wang SC. Long noncoding RNA HOTAIR promotes invasion of breast cancer cells through chondroitin sulfotransferase CHST15. Int J Cancer. 2019;145:2478–2487. doi: 10.1002/ijc.32319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 21.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 22.Famulok M, Hartig JS, Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- 23.Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerchia L, de Franciscis V. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010;28:517–525. doi: 10.1016/j.tibtech.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Esposito CL, Passaro D, Longobardo I, Condorelli G, Marotta P, Affuso A, de Franciscis V, Cerchia L. A Neutralizing RNA Aptamer against EGFR causes selective apoptotic cell death. PLoS One. 2011;6:e24071. doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Larson T, Nguyen HH, Sokolov KV, Ellington AD. Directed evolution of gold nanoparticle delivery to cells. Chem Commun (Camb) 2010;46:392–394. doi: 10.1039/b920865h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of cell proliferation by an Anti-EGFR aptamer. PLoS One. 2011;6:e20299. doi: 10.1371/journal.pone.0020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu HY, Yu X, Liu H, Wu D, She JX. Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Sci Rep. 2016;6:30346. doi: 10.1038/srep30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siomi H, Siomi MC. RISC hitches onto endosome trafficking. Nat Cell Biol. 2009;11:1049–1051. doi: 10.1038/ncb0909-1049. [DOI] [PubMed] [Google Scholar]
- 30.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. [Google Scholar]
- 31.Wang K, Chang H. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johannes L, Lucchino M. Current challenges in delivery and cytosolic translocation of therapeutic RNAs. Nucleic Acid Therapeutics. 2018;28:178–193. doi: 10.1089/nat.2017.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitehead KA, Dahlman JE, Langer RS, Anderson DG. Silencing or stimulation? siRNA delivery and the immune system. Ann Rev Chem Biomol Eng. 2011;2:77–96. doi: 10.1146/annurev-chembioeng-061010-114133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



