Abstract
Transmembrane serine protease (TMPRSS2) plays an oncogenic role in prostate cancer as the fusion gene with ERG, and has also been demonstrated to be essential for the cellular entry of severe acute respiratory syndrome coronaviruses (SARS-CoV). Thus, targeting TMPRSS2 is a promising strategy for therapies against both prostate cancer and coronavirus infection. Although Nafamostat and Camostat have been identified as TMPRSS2 inhibitors, severe side effects such as cerebral hemorrhage, anaphylactoid reaction, and cardiac arrest shock greatly hamper their clinical use. Therefore, more potent and safer drugs against this serine protease should be further developed. In this study, we developed a fluorescence resonance energy transfer (FRET)-based platform for effectively screening of inhibitors against TMPRSS2 protease activity. The disruption of FRET between green and red fluorescent proteins conjugated with the substrate peptide, which corresponds to the cleavage site of SARS-CoV-2 Spike protein, was measured to determine the enzymatic activity of TMPRSS2. Through an initiate pilot screening with around 100 compounds, Flupirtine, a selective neuronal potassium channel opener, was identified as a potential TMPRSS2 inhibitor from an FDA-approved drug library by using this screening platform, and showed inhibitory effect on the TMPRSS-dependent infection of SARS-CoV-2 Spike-pseudotyped lentiviral particles. This study describes a platform proven effective for rapidly screening of TMPRSS2 inhibitors, and suggests that Flupirtine may be worthy of further consideration of repurposing to treat COVID-19 patients.
Keywords: COVID-19, FRET, SARS-CoV-2, TMPRSS2
Introduction
Transmembrane serine protease (TMPRSS2), which is also known as epitheliasin and belongs to the type II transmembrane serine proteases (TTSPs), possesses a trypsin-like endopeptidase activity and presents with a serine residue acting as a nucleophile in its catalytic active site [1]. Three highly conserved residues, Ser (S), His (H), and Asp (D), constitute the catalytic triad by working as a charge-transmit network and by forming the negatively charged pocket. This prefers to cleave amide bonds at Lys (K) or Arg (R) residues [2]. The expression of TMPRSS2 was found mostly in epithelial cells of various tissues, such as prostate, small intestine, lung and more [3]. Although the recent report proposed that TMPRSS-2 may associate with an endocytosis-independent pathway for SARS-CoV-2 virus to enter human primary lung cells [4], the physiological functions of TMPRSS2 remain incompletely defined.
The knowledge about TMPRSS2 begins from its role in progression of prostate cancer after the discovery of its oncogenic fusion genes with erythroblast transformation specific (ETS) transcription factors, such as ETS-related gene (ERG) [5]. TMPRSS2-ERG fusion, found in roughly 100,000 cases every year, is the most common molecular alteration (40-50%) in prostate cancer patients [6]. TMPRSS2 level is increased in both primary and metastatic prostate tumors and positively correlates with Gleason score [7]. Reduction of TMPRSS2 expression by gene silence and knockout strategies have been shown to decrease tumor progression in a prostate cancer-xenograft mice model [8], and the study with Transgenic Adenocarcinoma Mouse Prostate (TRAMP) models also suggests the pro-oncogenic and pro-metastatic role of TMPRSS2 in prostate cancer [9]. By screening libraries of synthetic peptides, a proteolytic cleavage sequence by TMPRSS2 was identified in pro-HGF. Activation of HGF by TMPRSS2-depedendent cleavage promoted the invasion ability of DU145 prostate cancer cells through activation of c-MET signaling, providing the mechanistic insight into the TMPRSS2-induced epithelial-mesenchymal transition phenotype in the TRAMP mouse model [9]. In addition, the proteolytic activation of matriptase by TMPRSS2 also contributes to tumor progression of prostate cancer by enhancing the cleavage of extracellular matrix nidogen-1 and laminin β1 [8]. In addition to its proteolytic activity, TMPRSS2 also increases respectively up to 6000- and 50-fold higher expressions of transcription factors ERG [6] and ETV1 [10], which play roles in development, differentiation, proliferation, and metastasis of prostate cancer [11]. These findings indicate that targeting TMPRSS2 enzymatic activity is a potential therapeutic strategy for prostate cancer [12,13].
In addition to the oncogenic roles in prostate cancer, TMPRSS2 also mediates the cellular entry of coronaviruses. The coronavirus disease 2019 (COVID-19) outbreak, caused by infection with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has led to enormous health and economic challenges to the entire world. There is no effective vaccine or approved antiviral drug for infected patients at present except for remdesivir that was approved for the emergency use [14]; repurposing existing drugs is, thus, an urgent strategy to offer rapid path for COVI-19 treatment. The interaction of SARS-CoV-2 and host cells relies on the binding of Spike (S) protein with angiotensin-converting enzyme 2 (ACE2) receptor on the plasma membrane of human cells. The human serine protease TMPRSS2 and endocytosis are required for the viral entry of SARS-CoV2 into the cells. TMPRSS2 is widely expressed in human respiratory tract, and exerts its enzymatic activity to cleave S protein of SARS-CoV-2 [15] into S1 and S2, which is a shared mechanism for various coronaviruses to facilitate viral entry into cells [16,17]. S1 subunit binds to ACE2 on host cells, and the S2 subunit facilitates the membrane fusion between virus and host cells. The cleavage activation of S protein by TMPRSS2 is critical for the virus invasion and virulence [18]. TMPRSS-2 has also recently been found to possess an endocytosis-independent pathway for SARS-CoV-2 virus to enter human lung primary cells [4]. While multiple mutated versions of SARS-CoV-2 have been reported [19-21], raising complication for vaccine development in the future. TMPRSS-2 is a human serine protease which is required for coronavirus infection into human cells, thus, drugs to inhibit TMPRSS-2 protease activity should be able to inhibit infection of SARS-CoV family (including mutated SARS-CoV-2) coronavirus into human cells. The anticoagulants Nafamostat and Camostat mesylate, which are approved in Japan for the therapy of acute pancreatitis and disseminated intravascular coagulation currently, have been identified as a TMPRSS2 inhibitors [22]. These drugs potently inhibits SARS-CoV-2 S protein-mediated fusion and SARS-CoV-2 infection in vitro [23], and Nafamostat mesylate therapy in combination with the antiviral agent favipiravir also showed the low mortality rate (9%; 1/11 cases) of COVID19 patients who required invasive mechanical ventilation in the intensive care unit [24]. However, the association between some server adverse effects, including cerebral hemorrhage [25], anaphylactoid reaction [26,27], and cardiac arrest shock [28] during treatment with Nafamostat has been proposed. Thus, more potent and safer drugs against this serine protease should be further developed.
As TMPRSS2 is a promising therapeutic target for prostate cancer and COVID-19 treatments, repurposing FDA-approved agents on the catalytic domain of TMPRSS2 could show therapeutic valuable. Since no experimental structure for TMPRSS2 has been documented, comparative modeling strategies using domains with highly conserved structures were employed to establish structural models for TMPRSS2 serine protease domain [2,29]. However, the accuracy of this in silico prediction for TMPRSS2 inhibitor is a major concern [30,31]. To overcome this limitation, we developed a FRET-based assay aiming at detecting TMPRSS2 activity, and Flupirtine has been identified through initial screening with 100 compounds from an FDA-approved compound library.
Materials and methods
Cloning, expression and purification of TMPRSS2 catalytic domain and fluorescent protein substrate of TMPRSS2
The gene encoding the catalytic domain of human TMPRSS2 (residues 256-492, UniProt accession: O15393) with preferential codon usage in Escherichia coli was synthesized and subcloned into pET21 or pSol-MBP vector (Lucigen) to generate C-terminal His6-tagged or N-terminal MBP-fusion protein. The designed fluorescent protein substrate of TMPRSS2 is made up of the cleavage site from SARS-CoV-2 spike protein (675QTQTNSPRRARSVAS689) inserted between mNeonGreen (GenBank: AGG56535.1) and mRuby3 (GenBank: ATE88097.1). The gene fragment containing this designed GFP-RFP pair was synthesized and constructed into a pET16b plasmid. This plasmid was transformed into E. coli BL21 (DE3) followed by the growth in LB broth at 37°C. Overexpression of MBP-tagged TMPRSS2256-492 was induced by the addition of L-rhamnose at a final concentration of 0.2% when the culture reach OD600 of 0.6. The cultures were incubated for further 20 hours at 16°C and harvested by centrifugation at 4500 g. The cells were resuspended in buffer containing Tris (50 mM at pH 8.0), NaCl (500 mM), glycerol (10%), imidazole (5 mM), lysed by sonication, and centrifuged at 28,000 g, 4°C for 1 hour. The supernatant was then loaded onto a Ni-column and purified by ÄKTA™ go chromatography system (Cytiva) according to manufacturer instructions. Overexpression of the designed GFP-RFP protein substrate was induced by the addition of 0.5 mM IPTG and continue shaking at 20°C for 18-20 hours. The purification procedure of the designed GFP-RFP protein substrate was the same as the recombinant TMPRSS2 mentioned above. The fractions containing TMPRSS2 and its protein substrate were concentrated using an Amicon® Ultra-15 (Millipore) and further purified by size-exclusion chromatography (HiLoad Superdex 200, GE Healthcare). The prepared protein samples were stored at -80°C until use.
Fluorescence resonance energy transfer (FRET)-based enzyme activity assay
To access the protease activity of the recombinant human TMPRSS2, the reaction mixture containing 25 μM MBP-tagged TMPRSS2256-492 and 60 μM tested compound in 25 mM Tris 8.0, 200 mM NaCl, 5% glycerol was pre-incubated at room temperature for 30 min in 96-well black Optiplate. The designed fluorescent protein substrate was then added to start the reaction. The fluorescent signals were obtained by detecting the cleaved products (excitation: 506 nm/emission: 536 nm) using a microplate reader. All the data were normalized to 1% DMSO control and repeated at least twice.
Cell culture
HEK-293T, VeroE6, TMPRSS2-expressing VeroE6 cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1x GlutaMAX, and 1% penicillin/streptomycin, and were incubated at 37°C and 5% CO2.
Transient transfection and drugs treatment
Approximately 3×105 of 293T cells (ATCC) were transfected with p-CMV5-TMPRSS2 by using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s protocol. After 6 hours of incubation, cells were treated with Flupirtine in dose-dependent manner for 2 days and total lysates were prepared for Western blot analysis.
Western blot analysis
Total lysates were extracted with lysis buffer (SDS (10%) and urea (5 μM)), analyzed by SDS-PAGE, and then transferred to PVDF membranes. After blocking with milk (5%), membranes were incubated with primary antibodies against TMPRSS2 (Santa Cruz) or actin (Invitrogen) for overnight at 4°C followed by incubation with HRP-conjugated secondary antibodies for 1 hour at room temperature. The immunoreactive signals were detected by using enhanced chemiluminescence with ECL reagent (Bio-Red).
Viral pseudo-particles (Vpp) infection and its inhibition
VeroE6 and VeroE6-TMPRSS2 cells (10,000 cells/well) were seeded in 96-well plates, and were pre-treated with different concentrations (1 μM, 5 μM, 25 μM) of Flupirtine or DMSO (vehicle control) for 1 h at 37°C followed by infection with the Vpp harboring SARS-2-S and luciferase reporter (purchased from National RNAi Core Facility (NRC), Academia Sinica) and centrifugation at 1250 g for 30 min. After incubation for 24 hours, the cell viability was measured by using Cell Counting Kit-8 (CCK-8) assay (Dojindo Laboratories). Then, each sample mixed with an equal volume of ready-touse luciferase substrate Bright-Glo Luciferase Assay System (Promega), and luminescence was measured immediately by the GloMax Navigator System (Promega). Raw luminescence values (indicating luciferase activity) were recorded as counts per second, and the relative light unit (RLU) was normalized with cell viability. The effect of Flupirtine on the relative infection efficiencies were calculated with control group set as 100%.
Statistical analysis
The difference between the experimental and control groups was analyzed with Student’s t-test. The significance is defined if the P-value is <0.05.
Results
To establish the FRET-based TMPRSS2 enzymatic activity assay, the catalytic domain (256-492) of the TMPRSS2 was cloned into pET21 (pET21-TMPRSS2256-492; 26 kDa) and expressed in E. coli. The protein was not soluble and was obtained in the form of inclusion bodies, which were separated from cell pellet (Figure 1A). To improve the solubility of this target protein, we adopted a fusion partner strategy by tagging maltose Binding Protein (MBP) to TMPRSS2256-492 (MBP-TMPRSS2256-492; 63 kDa) [32]. As shown in Figure 1B, the induced MBP-tagged TMPRSS2 was obtained in soluble lysates although the majority remains detected in insoluble pellet.
Figure 1.
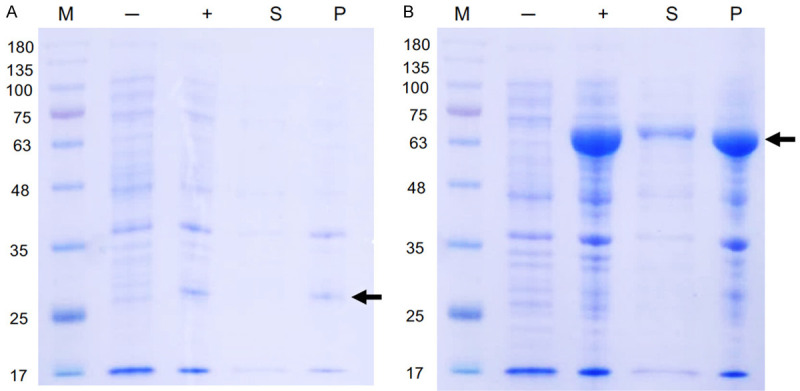
Evaluation of different expression strategy for TMPRSS2 serine protease domain. Small-scale expression of (A) pET21-TMPRSS2256-492 (B) MBP-TMPRSS2256-492 by E. coli BL21 (DE3). M, marker; -, no induction; +, IPTG induction; S, soluble fraction, P, pellet.
By taking advantage of the known sequence of protein cleavage site by TMPRSS2 [15], we have developed an assay for TMPRSS2 enzymatic activity which utilizes a FRET-based screening platform. Green fluorescent protein (GFP) and red fluorescent protein (RFP), the donor fluorophore and acceptor fluorophore, were respectively fused with amino- and carboxy ends of a peptide substrate (QTQTNSPRRARSVASO) for TMPRSS2 (Figure 2A). Before the cleavage by TMPRSS2, GFP donor fluorophore non-radiatively transfers its excitation energy to the neighboring RFP, thereby causing the emission of red fluorescence from this acceptor fluorophore. Green-red FRET pairs showed several advantages, including less autofluorescence, less phototoxicity, and greater spectra separation [33]. While the cleavage of substrate protein by TMPRSS2, the GFP fluorophore was spatially separated from the RFP fluorophore, thus leading to disassembling the substrate and restoring GFP fluorescence. The fluorescence protein substrate for TMPRSS2, GFP-QTQTNSPRRARSVASO-RFP, was synthesized and purified with immobilized metal affinity chromatography (Figure 2B). The purified GFP/RFP fluorescent substrate protein was detected approximately at 57 kDa in SDS-PAGE, which is consistent with its theoretical M.W. (Figure 2C).
Figure 2.

Establishment of FRET-based TMPRSS2 enzyme activity assay. A. Schematic presentation of FRET-based enzyme activity assay of TMPRSS2. B. Immobilized metal affinity chromatography of the fluorescent protein substrate of TMPRSS2 (GFP-QTQTNSPRRARSVAS-RFP). C. SDS-PAGE of the purified fluorescent protein substrate. The estimated MW is consistent with its theoretical MW of 57.2 kDa.
To assess the effectiveness of the established FRET-based TMPRSS2 enzymatic activity assay, we next tested the inhibitory efficacy of Nafamostat and Camostat, which are trypsin-like serine proteases inhibitors and have been shown to repress cellular entry of SARS-CoV2 by targeting TMPRSS [22], on the enzymatic activity of TMPRSS2. Both Nafamostat and Camostat reduced the enzymatic activity of TMPRSS2 by nearly 50% as compared to vehicle control (Figure 3). These results validated the established FRET-based TMPRSS-2 enzymatic activity assay as an efficient platform to screen potential TMRPSS-2 inhibitors for anti-SARS-CoV-2 infection. By using this strategy, our initial screening for the first 100 compounds from the FDA-approved compound library already identified one FDA-approved drug, Flupirtine inhibiting TMPRSS-2 activity by about 50% (Figure 3). TMPRSS2 is capable of autocleavage [34], and is expressed as a full-length form at 65 kDa and a cleaved form at 26 kDa. The autocleavage of TMPRSS2 was repressed by Flupirtine, supporting its anti-TMPRSS2 proteolytic activity (Figure 4). In addition, treatment with Flupirtine also suppressed the infection of TMPRSS2-overexpressing VeroE6 cells with SARS-CoV-2 Spike-pseudotyped lentiviral particles (Figure 5), supporting its activity to suppress virus infection is also TMPRSS2-dependent. Flupirtine, a selective neuronal potassium channel opener, functions as an indirect NMDA receptor antagonist and GABAA receptor modulator [35]. This drug has been used as an analgesic for acute pain, including back pain, migraines, oncology, postoperative care, and gynecology. Since the drug has been approved clinically for individual indications with low safety issues and side effects. And the other two known compounds targeting TRPRSS2 are known to have significant side effects and high safety concern [25-27,36,37], the identified drug, Flupirtine may have potential of repurposing to overcome the COVID-19 pandemic.
Figure 3.

Flupirtine is a TMRPSS2 inhibitor. The relative inhibitory effect of clinically approved inhibitors, Nafamostat, Camostat, and Flupirtine, on TMPRSS2 enzymatic activity in FRET-based assay.
Figure 4.

Suppression of TMPRSS2 autocleavage by Flupirtine. HEK-293T cells were transfected with TMPRSS2 followed by treatment with indicated doses of Flupirtine or Nafamostat for 24 hours. Total lysates were prepared and subjected to Western blot analysis with anti-TMPRSS2 and anti-Actin antibodies. The autocleaved form of TMPRSS2 at 26 kDa was suppressed by Flupirtine or Nafamostat and the quantitative data was normalized with the level of actin and shown in lower panel.
Figure 5.

Flupirtine suppresses TMPRSS-dependent infection of SARS-CoV-2 Spike-pseudotyped lentiviral particles. VeroE6 cells and VeroE6 cells transfected with TMPRSS-2 expressing vector were treated with Flupirtine at indicated concentrations followed by infection with SARS-CoV-2 Spike-pseudotyped lentiviral particles containing luciferase reporter. The infection efficacy was determined by the luciferase activity normalized with cell viability. *, p value <0.001 between two cell models. #, p value <0.001 compared with vehicle control of VeroE6 +, p value <0.001 compared with vehicle control of VeroE6-TMPRSS2.
Discussion
To fight with SARS-CoV2, most of therapeutic strategies were developed by aiming to target viral proteins such as Mpro and RNA-dependent RNA polymerase [38] include Remdesirvir, GC376, and tafenoquine [39-41]. However, the viral evolution and genome variability of RNA viruses driven by the high mutation rate enable viruses to increase the infectivity or escape the attack from host immunity [20,42]. Development of antiviral agent or vaccine by targeting the viral proteins may encounter the problem of drug-resistant mutations in the target enzyme [43], driving the need to develop the alternative therapeutic strategy targeting cellular protein in host cells. In this study, we developed a FRET-based human TMPRSS2 enzymatic assay platform to screen the FDA-approved drugs repositioning as promising TMPRSS2 inhibitors for the treatment of COVID-19 disease.
ACE2 and TMPRSS2, the receptor and co-factor of SARS-CoV-2 respectively, are expressed in lung secretory cells [44], and are two key cellular proteins for the entry of coronaviruses [22]. Preventing the interaction of human ACE2 and coronaviruses by blocking the receptor-binding domain (RBD) of the viral S-protein has been proposed as a potential therapeutic strategy for COVID-19 disease [45,46]. However, ACE2 is a key component of the renin-angiotensin-aldosterone system (RAAS), and enhancement of ACE2 expression due to RAAS blockade of ACE inhibitors has been demonstrated with experimental evidence. It raises the concerns about the deleterious effects in patients with underlying cardiovascular disease [47]. Since TMPRSS2 is another critical cellular protein for the entry of coronaviruses [22] and the tumor progression of prostate cancer [8], targeting TMPRSS2 activity would be another idea therapeutic strategy for theses disease. To establish a rapid and high throughput screen system, we adopted in vitro protease enzymatic activity assay by using the recombinant and purified TMPRSS2 catalytic domain as the enzyme and the fluorescence protein-conjugated peptide as the substrate. As a transmembrane protein, however, bacterial-expressed TMPRSS2 catalytic domain is poorly soluble and exists in the inclusion body [48]. To improve its solubility, this catalytic domain was fused with MBP (Figure 1), which is one of the most effective solubility enhancers [49] without affecting the enzymatic activity of TMPRSS2. Recently, an additional protease Furin was also demonstrated to mediate the priming of S protein of SARA-CoV-2 but not SARS-CoV [50]. By employing the same principle, specific inhibitors against Furin could also be identified in the FRET-based protease activity assay.
By using this screening system, Flupirtine, an FDA-approved drug used for as an analgesic for the moderate-to-severe cases of acute pain, was identified as a potential TMPRSS2 inhibitor. Although several computer-aided screening systems have been proposed for potential TMPRSS2 inhibitors [2,29,51], Flupirtine was not identified in these virtual screening platforms probably due to the use of comparative modeling, but not experimental, structure of TMPRSS22 serine protease domain in these systems. Our FRET-based TMPRSS2 enzymatic assay provides a rapid and high throughput screening platform for the identification of selective and potent TMPRSS2 inhibitors for both COVID19 and prostate cancer therapies and will be a power strategy to identify potential inhibitors through high throughput screening.
Acknowledgements
This work was supported by grants from the Ministry of Science and Technology of Taiwan (grant no. MOST 108-2311-B-241-001, to Y. Chen and MOST 109-2327-B-039-003 to M.-C. Hung), from China Medical University (grant no. CMU106-ASIA-18, to W.-C. Huang), and from China Medical University Hospital (grant no. DMR107-023, to C.-H. Chen). This work was also financially supported by the “Drug Development Center, China Medical University” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Disclosure of conflict of interest
None.
References
- 1.Patel S. A critical review on serine protease: key immune manipulator and pathology mediator. Allergol Immunopathol (Madr) 2017;45:579–591. doi: 10.1016/j.aller.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singh N, Decroly E, Khatib AM, Villoutreix BO. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur J Pharm Sci. 2020;153:105495. doi: 10.1016/j.ejps.2020.105495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacquinet E, Rao NV, Rao GV, Zhengming W, Albertine KH, Hoidal JR. Cloning and characterization of the cDNA and gene for human epitheliasin. Eur J Biochem. 2001;268:2687–2699. doi: 10.1046/j.1432-1327.2001.02165.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann M, Mosbauer K, Hofmann-Winkler H, Kaul A, Kleine-Weber H, Kruger N, Gassen NC, Muller MA, Drosten C, Pohlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- 5.Clark JP, Cooper CS. ETS gene fusions in prostate cancer. Nat Rev Urol. 2009;6:429–439. doi: 10.1038/nrurol.2009.127. [DOI] [PubMed] [Google Scholar]
- 6.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 7.Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R, Nelson PS. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J Pathol. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- 8.Ko CJ, Huang CC, Lin HY, Juan CP, Lan SW, Shyu HY, Wu SR, Hsiao PW, Huang HP, Shun CT, Lee MS. Androgen-induced TMPRSS2 activates matriptase and promotes extracellular matrix degradation, prostate cancer cell invasion, tumor growth, and metastasis. Cancer Res. 2015;75:2949–2960. doi: 10.1158/0008-5472.CAN-14-3297. [DOI] [PubMed] [Google Scholar]
- 9.Lucas JM, Heinlein C, Kim T, Hernandez SA, Malik MS, True LD, Morrissey C, Corey E, Montgomery B, Mostaghel E, Clegg N, Coleman I, Brown CM, Schneider EL, Craik C, Simon JA, Bedalov A, Nelson PS. The androgen-regulated protease TMPRSS2 activates a proteolytic cascade involving components of the tumor microenvironment and promotes prostate cancer metastasis. Cancer Discov. 2014;4:1310–1325. doi: 10.1158/2159-8290.CD-13-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vitari AC, Leong KG, Newton K, Yee C, O’Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, Mohan S, Pandita A, Hongo JA, Arnott D, Wertz IE, Gao WQ, French DM, Dixit VM. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 11.Shaikhibrahim Z, Wernert N. ETS transcription factors and prostate cancer: the role of the family prototype ETS-1 (review) Int J Oncol. 2012;40:1748–1754. doi: 10.3892/ijo.2012.1380. [DOI] [PubMed] [Google Scholar]
- 12.Martin CE, List K. Cell surface-anchored serine proteases in cancer progression and metastasis. Cancer Metastasis Rev. 2019;38:357–387. doi: 10.1007/s10555-019-09811-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arora K, Barbieri CE. Molecular subtypes of prostate cancer. Curr Oncol Rep. 2018;20:58. doi: 10.1007/s11912-018-0707-9. [DOI] [PubMed] [Google Scholar]
- 14.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh MD, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC ACTT-1 Study Group Members. Remdesivir for the treatment of covid-19 - preliminary report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Kleine-Weber H, Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784. e775. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia S, Liu Q, Wang Q, Sun Z, Su S, Du L, Ying T, Lu L, Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniloski Z, Guo X, Sanjana NE. The D614G mutation in SARS-CoV-2 spike increases transduction of multiple human cell types. bioRxiv. 2020:2020.06.14.151357. [Google Scholar]
- 20.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI Sheffield COVID-19 Genomics Group. McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. e819. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, Liu H, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Zhang L, Li X, Huang W, Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto M, Kiso M, Sakai-Tagawa Y, Iwatsuki-Horimoto K, Imai M, Takeda M, Kinoshita N, Ohmagari N, Gohda J, Semba K, Matsuda Z, Kawaguchi Y, Kawaoka Y, Inoue JI. The anticoagulant nafamostat potently inhibits SARS-CoV-2 S protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses. 2020;12:629. doi: 10.3390/v12060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doi K, Ikeda M, Hayase N, Moriya K, Morimura N COVID-UTH Study Group. Nafamostat mesylate treatment in combination with favipiravir for patients critically ill with Covid-19: a case series. Crit Care. 2020;24:392. doi: 10.1186/s13054-020-03078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hifumi T, Isokawa S, Otani N, Ishimatsu S. Adverse events associated with nafamostat mesylate and favipiravir treatment in COVID-19 patients. Crit Care. 2020;24:497. doi: 10.1186/s13054-020-03227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shindo M, Ookawara S, Kitano T, Ishii H, Miyazawa H, Ito K, Ueda Y, Hirai K, Hoshino T, Morishita Y. Sustained severe intestinal edema after nafamostat mesilate-associated anaphylactic reaction during hemodialysis. Nefrologia. 2019;39:202–204. doi: 10.1016/j.nefro.2018.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Higuchi N, Yamazaki H, Kikuchi H, Gejyo F. Anaphylactoid reaction induced by a protease inhibitor, nafamostat mesilate, following nine administrations in a hemodialysis patient. Nephron. 2000;86:400–401. doi: 10.1159/000045822. [DOI] [PubMed] [Google Scholar]
- 28.Kim HS, Lee KE, Oh JH, Jung CS, Choi D, Kim Y, Jeon JS, Han DC, Noh H. Cardiac arrest caused by nafamostat mesilate. Kidney Res Clin Pract. 2016;35:187–189. doi: 10.1016/j.krcp.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hutter MC. The current limits in virtual screening and property prediction. Future Med Chem. 2018;10:1623–1635. doi: 10.4155/fmc-2017-0303. [DOI] [PubMed] [Google Scholar]
- 31.Heo L, Feig M. Experimental accuracy in protein structure refinement via molecular dynamics simulations. Proc Natl Acad Sci U S A. 2018;115:13276–13281. doi: 10.1073/pnas.1811364115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pennati A, Deng J, Galipeau J. Maltose-binding protein fusion allows for high level bacterial expression and purification of bioactive mammalian cytokine derivatives. PLoS One. 2014;9:e106724. doi: 10.1371/journal.pone.0106724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bajar BT, Wang ES, Zhang S, Lin MZ, Chu J. A guide to fluorescent protein FRET pairs. Sensors (Basel) 2016;16:1488. doi: 10.3390/s16091488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- 35.Szelenyi I. Flupirtine, a re-discovered drug, revisited. Inflamm Res. 2013;62:251–258. doi: 10.1007/s00011-013-0592-5. [DOI] [PubMed] [Google Scholar]
- 36.Uno Y. Camostat mesilate therapy for COVID-19. Intern Emerg Med. 2020;15:1577–1578. doi: 10.1007/s11739-020-02345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim JY, Kim JB, Choo SJ, Chung CH, Lee JW, Jung SH. Anticoagulation during extracorporeal membrane oxygenation; nafamostat mesilate versus heparin. Ann Thorac Surg. 2016;102:534–539. doi: 10.1016/j.athoracsur.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 38.Uddin M, Mustafa F, Rizvi TA, Loney T, Suwaidi HA, Al-Marzouqi AHH, Eldin AK, Alsabeeha N, Adrian TE, Stefanini C, Nowotny N, Alsheikh-Ali A, Senok AC. SARS-CoV-2/COVID-19: viral genomics, epidemiology, vaccines, and therapeutic interventions. Viruses. 2020;12:526. doi: 10.3390/v12050526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang YC, Yang WH, Yang CS, Hou MH, Tsai CL, Chou YZ, Hung MC, Chen Y. Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug. Am J Cancer Res. 2020;10:2535–2545. [PMC free article] [PubMed] [Google Scholar]
- 40.Huang ST, Lai HC, Lin YC, Huang WT, Hung HH, Ou SC, Lin HJ, Hung MC. Principles and treatment strategies for the use of Chinese herbal medicine in patients at different stages of coronavirus infection. Am J Cancer Res. 2020;10:2010–2031. [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Yang WH, Huang LM, Wang YC, Yang CS, Liu YL, Huo MH, Tsai CL, Chou YZ, Huang BY, Hung CF, Hung YL, Chen JS, Chiang YP, Cho DY, Jeng LB, Tsai CH, Hung MC. Inhibition of severe acute respiratory syndrome coronavirus 2 main protease by tafenoquine in vitro. Biorxiv. 2020 [Google Scholar]
- 42.Pachetti M, Marini B, Benedetti F, Giudici F, Mauro E, Storici P, Masciovecchio C, Angeletti S, Ciccozzi M, Gallo RC, Zella D, Ippodrino R. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:179. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, Sheng J, Quan L, Xia Z, Tan W, Cheng G, Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lukassen S, Chua RL, Trefzer T, Kahn NC, Schneider MA, Muley T, Winter H, Meister M, Veith C, Boots AW, Hennig BP, Kreuter M, Conrad C, Eils R. SARS-CoV-2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.20105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.South AM, Diz DI, Chappell MC. COVID-19, ACE2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020;318:H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer D, Sielaff F, Hammami M, Bottcher-Friebertshauser E, Garten W, Steinmetzer T. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem J. 2013;452:331–343. doi: 10.1042/BJ20130101. [DOI] [PubMed] [Google Scholar]
- 49.Fox JD, Routzahn KM, Bucher MH, Waugh DS. Maltodextrin-binding proteins from diverse bacteria and archaea are potent solubility enhancers. FEBS Lett. 2003;537:53–57. doi: 10.1016/s0014-5793(03)00070-x. [DOI] [PubMed] [Google Scholar]
- 50.Xia S, Lan Q, Su S, Wang X, Xu W, Liu Z, Zhu Y, Wang Q, Lu L, Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Target Ther. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Idris MO, Yekeen AA, Alakanse OS, Durojaye OA. Computer-aided screening for potential TMPRSS2 inhibitors: a combination of pharmacophore modeling, molecular docking and molecular dynamics simulation approaches. J Biomol Struct Dyn. 2020:1–19. doi: 10.1080/07391102.2020.1792346. [DOI] [PMC free article] [PubMed] [Google Scholar]


