Abstract
It has been estimated that worldwide up to 10% of all human cancers are the result of viral infection, with 7.2% of all cancers in the developed world have a viral aetiology. In contrast, 22.9% of infections in the developing world are the result of viral infections. This number increases to 30% in Sub-Saharan Africa. The ability of viral infections to induce the transformation of normal cells into cancerous cells is well documented. These viruses are mainly Hepatitis B and C viruses, Epstein Barr virus, Human papillomavirus and Human Cytomegalovirus. They can induce the transformation of normal cells into cancer cells and this may be the underlying cause of carcinogenesis in many different types of cancer. These include liver cancer, lymphoma, nasopharyngeal cancer, cervical cancer, gastric cancer and even glioblastoma. Long non-coding RNAs (LncRNAs) can function by regulating the expression of their target genes by controlling the stability of the target mRNAs or by blocking translation of the target mRNA. They can control transcription by regulating the recruitment of transcription factors or chromatin modification complexes. Finally, lncRNAs can control the phosphorylation, acetylation, and ubiquitination of proteins at the post-translation level. Thus, altering protein localisation, function, folding, stability and ultimately expression. In addition to these functions, lncRNA also regulate alternate pre-mRNA splicing in ways that contribute to the formation of tumours. This mainly involves the interaction of lncRNAs with splicing factors, which alters their activity and function. The ability of lncRNAs to regulate the stability, expression and function of tumour suppressor proteins is important in the development and progression of cancers. LncRNAs also regulate viral replication and latency, leading to carcinogenesis. These factors all make lncRNAs ideal targets for the development of biomarker arrays that can be based on secreted lncRNAs leading to the development of affordable non-invasive biomarker tests for the stage specific diagnosis of tumours. These lncRNAs can also serve as targets for the development of new anticancer drug treatments.
Keywords: Non-coding RNAs, hepatitis C virus, hepatitis B virus, epstein barr virus, human papilloma virus, hepatocellular carcinoma, nasopharyngeal cancer, gastric cancer, head and neck cancer, lymphoma, glioma
Introduction
In 1909, Dr. Francis Peyton Rous extracted tumour cells from a chicken and grafted these cells into other chickens. These chickens were then found to also develop tumours [1]. The transforming factor was eventually identified as the RNA virus, the Rous sarcoma virus. This discovery led to Dr. Rous receiving a Nobel prize for medicine in 1966 [2]. It is estimated that up to 10% of all human cancers are caused by infection with oncogenic viruses [3]. The number of cancer cases that are attributable to viral infections is much higher in developing nations where 22.9% of all cancers are the result of infection with oncogenic viruses, compared to 7.2% in the developed world [4]. However, in general, only a relatively small percentage of individuals infected with an oncovirus develop cancer. An indication of the number of cancer cases caused by oncoviruses contribute to the total number of cancer cases is given in Figure 1. This percentage is even higher in Sub-Saharan Africa where up to 30% of all cancers are due to infections with oncogenic viruses [5]. These include the avian Rous sarcoma virus, cottontail rabbit papillomavirus, mouse mammary tumour virus [4], adenovirus and simian virus 40 (SV40), Epstein-Barr virus (EBV), human T-cell lymphoma virus-1 (HTLV-1), high risk human papillomaviruses (HPVs), Kaposi’s sarcoma-associated herpes virus (KSHV), Merkel cell polyomavirus (MCV) [6]. Some of these viruses as well as details concerning their taxonomy, the nature of their genetic material, the oncogenes they express and their related cancers are given in Table 1. The way in which these viruses are able to initiate cancer development was first discovered by Harold Varmus and Michael Bishop in 1976. They identified the viral oncogene v-src. Further characterisation of other oncogenes led to the discovery of p53, which is an antigen-associated protein for viral oncogenes such as SV40 [7]. Viral oncogenes manipulate the infected cells’ machinery in order for the virus to replicate and survive in the host. The expressed proteins then lead to the transformation of the cells. Alternate mRNA splicing is required for the effective expression of viral oncogenes. This process allows for the generation of a more diversified viral transcriptome [8]. In addition to viral oncogenes that code for proteins, these viruses encode multiple non-coding RNAs (ncRNAs).
Figure 1.
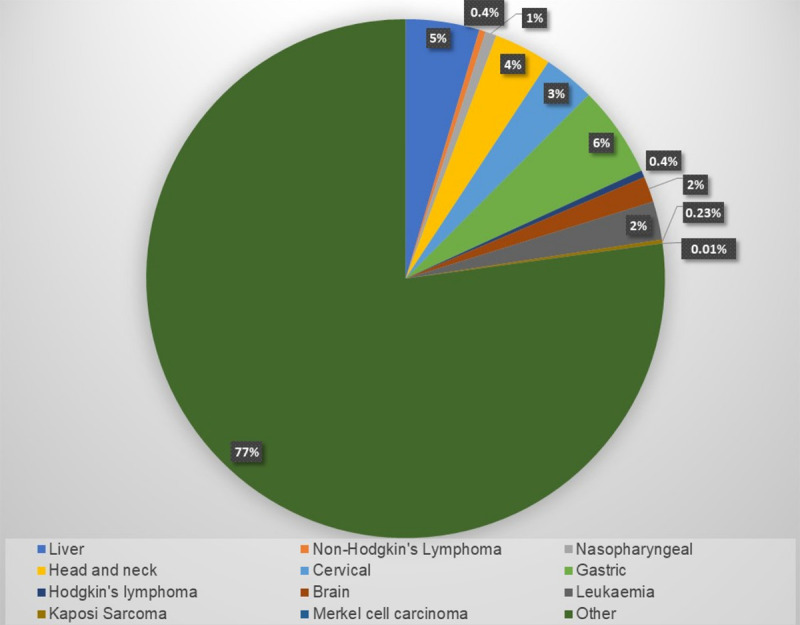
Prevalence of different cancers with a viral aetiology. The cancers that can potentially be caused by viral infection, accounted for approximately 23% of all cancer cases in 2018.
Table 1.
Oncoviruses and their associate cancers
| Virus | Taxonomy | DNA/RNA | Oncogenes | Tumour type |
|---|---|---|---|---|
| Adenovirus 12, 18 | Adenoviridae | DNA | E1A, E1B | Solid tumours in rodents |
| BK virus | Papovaviridae | DNA | T antigens | Solid tumours in primates and rodents |
| EBV | Herpesviridae | DNA | LMP-1, BARF-1 | Burkitt’s lymphoma Hodgkin’s lymphoma, stomach cancer nasopharyngeal carcinoma |
| HBV | Hepadriaviridae | DNA | HBx | Hepatocellular carcinoma |
| Hepatitis C | Flavaviridae | RNA | - | Hepatocellular carcinoma |
| HPV 16, 18, 21, 45 | Papillomaviridae | DNA | E6, E7 | Cervical, anal, oral cancers |
| hTCLV1 | Retroviridae | RNA | Tax | Adult T-cell leukoma lymphoma |
| KHSV | Herpesviridae | DNA | vGPCR | Kaposi’s sarcoma |
| Merkel cell polyomaviridea | Papovaviridae | DNA | T antigen | Merkel cell carcinoma |
| Human cytomegalovirus (HCMV) | Cytomegaloviridae | Herpesviridae | DNA | Liver cancer |
NcRNAs are functional RNA molecules that are not translated into proteins, but are transcribed from DNA, and these include housekeeping (transfer RNAs, ribosomal RNAs, small nuclear RNAs, small nucleolar RNAs, telomerase RNAs) and regulatory noncoding RNAs. These regulatory non-coding RNAs include both small noncoding and long noncoding RNAs. One of the common types of noncoding RNAs are long non-coding RNAs (lncRNAs). LncRNAs are non-coding transcripts that are more than 200 nucleotides long and Next-generation sequencing (NGS) technology has revealed that lncRNAs are involved in differentiation, proliferation, and cell death [9,10]. As a result, these molecules play a role in cancer development and progression [11]. Viral infections lead to changes in the host transcriptome including the expression of lncRNAs. Infected cells may also express viral lncRNAs [12].
Long non-coding RNAs
The majority of lncRNAs are transcribed by RNA polymerase II, and they are 5’-capped, spliced, and polyadenylated to form a structure similar to mRNA. Multiple large-scale projects have been undertaken to identify lncRNAs and have resulted in the identification over 20,000 lncRNAs [13]. LncRNAs function by interacting with either DNA or RNA or proteins, influencing the formation and function of the secondary and tertiary structures of these molecules. When they are localised within the nucleus, they can guide chromatin-modifying-complexes or transcription factors. In the cytosol, they can control the stability of mRNA or compete with endogenous mRNA for access to the protein expression machinery [14].
LncRNAs can be classed by their mode of action into four separate categories or by their genomic origin into seven categories. In terms of genomic location, the seven groups are as follows: i) Intergenic lncRNAs are coded for by intergenic regions between protein coding genes, ii) Sense or intronic lncRNAs are encoded by intronic regions in protein coding genes, iii) Antisense lncRNAs are encoded by the opposite strand, iv) Bi-directional lncRNAs are transcribed in the promoter regions of protein coding genes, v) Enhancer lncRNAs are transcribed from the enhancer regions [15], vi) Circular RNAs result from the joining of mRNAs or ncRNAs to other RNA molecules at 3’upstream or 5’downstream splice sites forming a looped circular RNA [16], and finally, vii) pseudogenes that can originate from gene duplication and may lose the ability to code for proteins due to mutations [17].
In terms of mode of action, lcRNAs can be broadly divided into four categories as follows: i) Decoy lncRNAs bind protein or RNAs, resulting in the negative regulation of protein expression, ii) These include Guide lncRNAs direct protein localisation by binding to proteins, iii) Signal lncRNAs interact with transcription factors or chromatin modifying enzymes, resulting in the regulation of transcription, and signalling pathways, iv) The final class are scaffold lncRNAs that act as an organising structure where molecules can bind and interact with each other more easily [18,19]. These broad categories group the many ways that lncRNA can control gene expression (Figure 2), through epigenetic silencing, splicing regulation, sequestering miRNAs, protein interaction, and genetic variation [18,20,21]. Since LncRNAs are involved in many important biological processes, their aberrant expressions result in the development of diseases, especially cancer (Figure 2) [22,23]. Non-coding RNAs have been shown to play an important role in tumour viruses, where non-coding RNAs might utilize non-coding RNAs to manipulate gene expression in an infected cell [24].
Figure 2.
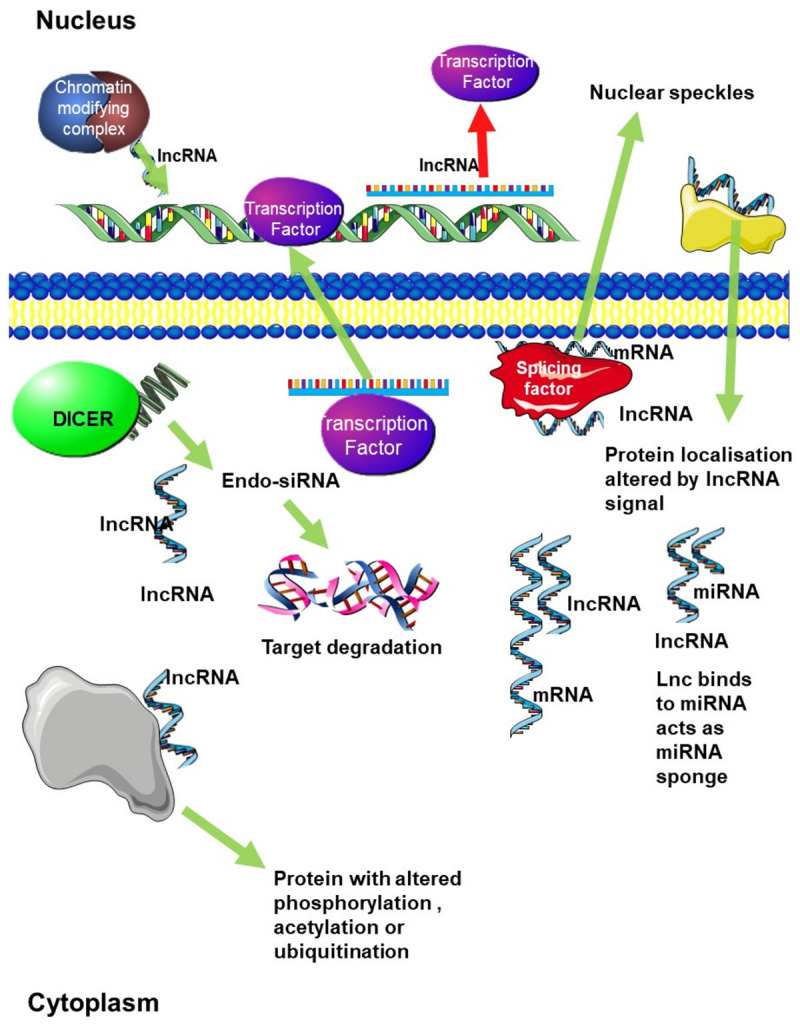
Regulation of gene expression by LncRNAs. LncRNAs can regulate gene expression in a number of ways. Firstly, through chromatin remodelling, by recruiting chromatin modifying complexes to specific genomic loci. LncRNAs can also regulate transcription by either recruiting transcription factors to their target gene promoters or by blocking transcription factors from binding to target gene promoters. Decoy lncRNAs bind miRNAs acting as a miRNA sponge sequestering them and altering protein expression. They can also be processed to produce small-interfering RNAs (siRNAs) leading to degradation of target mRNA. Finally, they can bind directly bind to mRNA and regulate mRNA stability, or competitively bind to mRNA to improve mRNA stability. Guide lncRNAs direct protein localisation. LncRNAs also bind to proteins and control protein phosphorylation, acetylation, and ubiquitination at the post-translation level. Alternative splicing can be regulated by lncRNA which interact with splicing factors to regulate splicing.
The role played by lnc RNAS in different cancers
Hepatocellular carcinoma (HCC)
The development of cancer is a multistep process resulting from chronic inflammation, DNA damage, chromosomal instability, epigenetic modifications, senescence, and telomerase reactivation. These processes can also promote carcinogenesis in hepatocytes through genetic damage and epigenetic changes [25]. The prognosis for hepatocellular carcinoma (HCC) is poor, but early detection and diagnosis does improve prognosis. However, the use of techniques such as ultrasound will not detect small liver cancers and currently, there are no reliable biomarkers to detect liver cancer [26]. During the development of HCC, aberrant lncRNAs expression can promote proliferation, apoptosis, invasion, metastasis, and angiogenesis. HCC cell division and proliferation are promoted by the lncRNA PVT1. PVT1 recruits Enhancer of zeste homolog 2 (EZH2), a histone-lysine N-methyltransferase enzyme that inhibits transcription through histone methylation and apoptosis [27]. Risk factors for HCC include infection with Hepatitis C or B virus (HCV/HBV), alcohol abuse and metabolic disease [28].
The expression of different lncRNAs in HCC depends on the stage of HCC and whether the cancer was caused by HCV infection [29]. HCV infection is a major cause HCC [30], as a result of the virus contributing to the development and progression of fibrosis [31]. HCV is a positive-strand RNA virus that does not integrate into the host cells genome. The viral genome consists of a single reading frame that codes for a large 3000 amino acid protein that is then processed into 10 smaller proteins [28]. The 10 smaller proteins include structural, core, E1, and E2, and non-structural, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B, proteins that play important roles in the viral life cycle and interact with the components of the host cell to regulate cell signalling, transcription, cell proliferation, apoptosis, vesicular trafficking, and translational regulation [32,33]. These proteins can also promote carcinogenesis in hepatocytes by inducing genetic damage and epigenetic dysregulation [25]. By comparing lncRNAs in healthy liver tissue, preneoplastic lesions and HCC, seven lncRNAs were identified with de-regulated expressions. These include LINC01419, BC014579, BC014579, AK021443, RP11-401P9.4, RP11-304 L19.5, AF070632 and CTB-167B5.2 [34]. In early HCC stages, LINC01419 transcripts are found to be highly expressed. The expression levels of this lncRNA were even higher in HCV-related HCC. The levels of both the AK021443 and AF070632 lncRNAs change as the cancer progresses. In advanced stage HCC, AK021443 levels were found to be increasing, while the expression of AF070632 decreases. LncRNAs such as AK021443, whose level decreases as HCC progresses were found to be responsible for metabolic processes not required for cancer cells. i.e. the Krebs cycle and oxidative phosphorylation, while the lncRNAs LINC01419 and AF070632 were found to be highly expressed in HCC, and are involved in the regulation of cell cycle genes [34].
Prader Willi/Angelman region RNA 5 (PAR5), and the lncRNA hypoxia-inducing factor a (aHIF) are two lncRNAs that are downregulated in HCV-related HCC. At the same time, the lncRNA human downregulated expression by HBx (hDREH) was up-regulated in HCV-related HCC [35,36]. The lncRNA, Hox transcript antisense intergenic RNA (HOTAIR) is highly expressed in HCC and is associated with elevated incidence of metastases and therefore, poor prognosis in [37]. Through the control of histone methylation, HOTAIR regulates the expression of homeobox D (HOXD), transcription factors that control developmental pathways. Increased levels of HOTAIR also lead to the overexpression of HER2 by binding to and sequestering the miRNA that controls the expression of HER2 and regulating the stability of HER2 mRNA [38]. Two other lncRNAs have been implicated in the development and progression of HCC, WRAP53 and EGOT. The expression of the tumour suppressor TP53 is controlled through the action of the lncRNA WD repeat containing antisense to TP53 (WRAP53) [36]. Finally the urothelial carcinoma associated-1 (UCA1) binds to miR-203, sequestering it, leading to increased expression of the target of this miRNA, the transcription factor Snail2 [36]. The long non-coding RNA EGOT is expressed during HCV infection and acts against the hosts’ antiviral response resulting in increased viral replication. The expression of the miR-33a-5p results in the formation of an EGOT/miR-33a-5p/HMGA2 complex and acts to inhibit EGOT function [39]. The chromatin interaction protein, High mobility group protein A2 (HMGA2), binds to the chromatin to change its structure to alter the interaction of transcription factors with DNA and changing gene transcription, thereby contributing to tumorigenesis. The expression of HMGA2 is inhibited by the miRNA 33-a-5p [40]. EGOT and miR-33a-5p share a binding site on HMGA2. EGOT thereby blocks HMGA2 activation, promoting cancer development and progression [41]. Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is another pro-oncogenic lncRNA that is highly expressed in HCV positive HCC [42].
In addition to HCV, HBV is another aetiological factor in liver cancer development. The lncRNA, highly up regulated in Liver Cancer (HULC) is expressed at high levels in HCC and activates HBV replication. The lncRNA achieves this by decreasing the expression of the cell cycle regulation gene, APOBEC3B, and by HULC up-regulating microRNA-539. HULC also increases the stability of HBV covalently closed circular DNA through this inhibition of APOBEC3B [43]. The HBV X protein alters the expression of lncRNAs, which can assist in the development and progression of HCC. One of these deregulated lncRNAs is the Semaphorin 6Aeantisense RNA 1 (SEMA6A-AS1). In HBV positive HCC, the levels of SEMA6A-AS1 are reduced and this reduced expression of SEMA6A-AS1 inversely correlates with tumour stage, metastasis, and mortality. This low level of SEMA6A-AS1 expression is also a good indicator of poor treatment outcome in HBV-associated HCC [44].
Lymphoma
EBV infection of human primary resting B lymphocytes (RBLs) results in these cells expressing viral latency genes, which alter gene expression. This can result in the development of cancer through the transformation of RBLs to lymphoblastoid cells. Once an individual is infected with EBV, the virus is present in a latent form in B lymphocytes for the rest of the individual’s life. The presence of latent viral infection in the lymphocytes can lead to the development of cancer such as Burkitt’s lymphoma and Hodgkin’s lymphoma [45]. During this transformation, the expression of a number of lncRNAs changes such as CYTOR and NORAD that play an important role in cell growth. The increased expression of these RNAs is directly associated with cell transformation [46]. NORAD is also involved in stimulating DNA replication and repair by binding to proteins involved in this process [47]. The lncRNAs 7SL, H19, and H19-AS play a role in the development of many cancers including lymphomas [48]. Infected lymphocytes release nine lncRNAs into the extracellular spaces through the use of exosomes. These include 7SL, H19, H19 upstream conserved 1&2, H19 antisense, HAR1B, HOXA6as, NDM29, SNHG5, and Tsix. Of these, only H19 and H19-AS were up-regulated [49].
In addition to altering the expression of host lncRNA, EBV encodes its own ncRNAs that can influence the development and progression of cancer, named EBV-encoded small RNAs (EBERs). Some of these RNAs accomplish this by binding to host proteins, forming ribonucleoprotein (RNP).
Nasopharyngeal carcinoma
EBV infection is a common risk factor for many different types of lymphoid cancer. This includes nasopharyngeal carcinoma (NPC). The viral proteins associated with the development of NPC are encoded for by the viral RNA transcript, Bam-HI A rightward transcripts (BARTs). This transcript also codes for viral microRNAs and lncRNAs. These lncRNAs affect the expression of cell adhesion, oxidoreductase activity, inflammation, and immunity genes [50]. One of these genes, IKAROS family zinc finger 3 (IKZF3/Aiolos), plays a role in lymphocyte development and cell attachment. BART lncRNA induces the expression of this gene. It is also likely that the expression of BART lncRNA interacts with the hosts’ chromatin remodelling machinery to control and maintain EBV latency [50].
Genome wide association studies and NGS have revealed that the expression of 2192 lncRNAs change in the development and progression of NPC. This results in the upregulation of 62 genes that are controlled through these lncRNAs [51]. These include CD44 (Hyaluronan/CD44) and interleukin 1 receptor associated kinase 1 (IRAK1). IRAK is involved in EBV infection while CD44 is involved in cancer progression [51]. LncRNAs that were downregulated in NPC include NR2F2 antisense RNA1 (NR2F21), lncRNA-family with sequence similarity 95 member C (FAM95C), and long intergenic nonprotein coding RNA 1106 (LINC01106). Those that were upregulated include lncRNA-CH507-513H4.6, lncRNA-THAP9 antisense RNA 1 (THAP9-AS1), lncRNACH507-513H4.3 and lncRNA-RP4-794H19.1 [51].
EBV expresses many miRNAs, which can interact with host lncRNAs to regulate their function. NPC associated gene 7 (NAG7) is a lncRNA that acts as a tumour suppressor gene by inhibiting cell cycle progression and promoting apoptosis. EBV expresses a ncRNA called EBER1, which negatively regulates NAG7 promoting metastasis [52]. EBV-miR-BART6-3p binds to lncRNA LOC553103 leading to a decrease in the expression of this, tumour suppressor, lncRNA [53].
All these previously discussed lncRNAs act as tumour suppressors and their expression negatively correlates with NPC progression. However, some circulating lncRNAs found in the blood of NPC patients are associated with a poor prognosis. These include the lncRNAs MALAT1, APAF1-AS1, and AL359062 [54].
Head and neck cancer
Head and neck squamous cell carcinoma (HNSCC) are the sixth most common cancer worldwide. It has a poor prognosis and does not respond well to treatment, being associated with high relapse rates. Recent studies have reported that ncRNAs might play critical role in the development and progression of HNSCC [55]. Other than tobacco and alcohol, some of the biggest risk factors for the development of this cancer is HPV infection [56]. HPV-related HNSCC has a different molecular basis underlying the progression of the disease compared to non-HPV related HNSCC and therefore, has different treatment outcomes as well.
A comparison between HPV positive and HPV negative HNSCC revealed that there are 132 lncRNAs that appeared in the HPV positive group and not the HPV negative group [57]. The four lncRNAs HOTAIR, PROM1, CCAT1, and MUC19 are found in HPV positive HNSCC and are associated with a decreased number of myeloid-derived suppressor cells (MDSCs). These immune cells are normally found in high numbers in individuals suffering from infection or diagnosed with cancer. They play a role in tumour immunity suppression and have been observed to be elevated in HNSCC [58], with even higher numbers of MDSCs in HPV positive HNSCC. This implies that one of the ways in which HPV can promote the formation of HNSCC is by manipulating the immune system [59]. Since the four lncRNAs mentioned above seem to inhibit this process, they may be targets for the HPV E7 oncoprotein. The main function of the E7 protein in HPV infection is to prevent cell death [57].
Cervical cancer
Cervical cancer is the third most common cancer worldwide and the fourth leading cause of cancer-associated mortality in women. The most important risk factor for cervical cancer is infection with HPV. This infection is possibly the initial cause of carcinogenesis in cervical cancer. The ability of HPV to induce cervical cancer relies on the viral oncoproteins E6 and E7 (Figure 3) to inactivate tumour suppressor proteins such as TP53and Rb. The expression of the viral proteins inhibits the tumour suppressor proteins such as TP53 by forming a complex with the E3 ubiquitin ligase UBE3A. This E6/E7/UBE3A complex ubiquitinates TP53, leading to its proteasomal degradation of TP53 [60,61]. The expression of HPV-16 E6 also leads to changes in the expression of lncRNAs. The lncRNA, Damage-induced (DINO), is expressed in response to TP53 signalling and DNA damage. DINO functions to stabilise TP53 and is able to inhibit the viral protein induced degradation of TP53 by inhibiting the formation of the E6/E7/UBE3A complex. This stabilisation of TP53 results in an increase in the transcription of the targets of TP53 (Figure 4). In HPV positive cervical cancer, the expression of DINO is downregulated. The degradation of TP53 resulting from the complex formed by the E3 ligase and viral protein is thought to be the result of the decreased expression of DINO [62].
Figure 3.
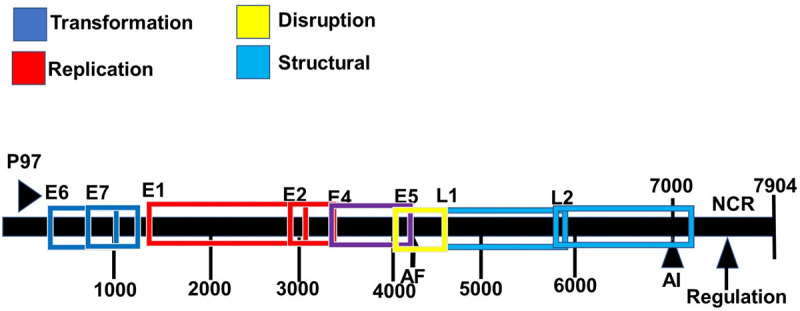
Genomic organization of the human papillomavirus type 16. ORFs deduced from the DNA sequence are designated E1 to E7, and L1 and L2. The non-coding region (NCR, also known as a long control region) is also shown. AE and AL indicate early and late polyadenylation sites. The functions of these genes are given by the color coded boxes.
Figure 4.
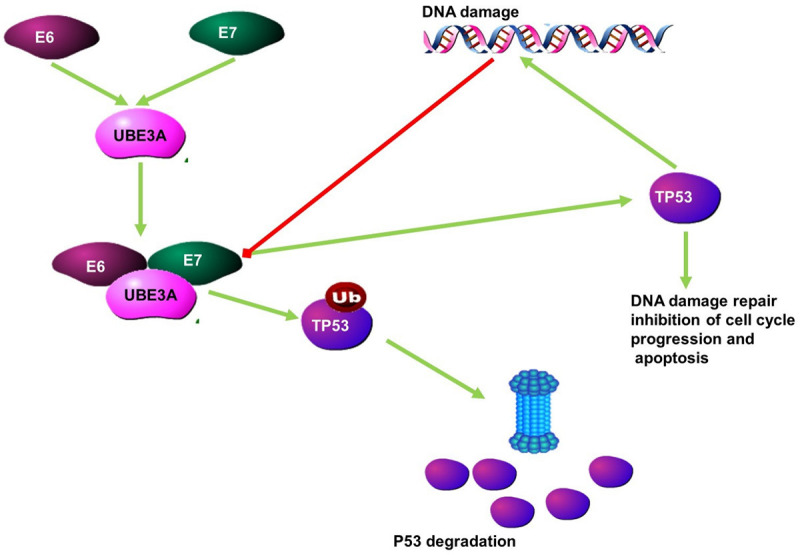
The function of the lncRNA DINO as anti-oncogenic. Following DNA damage, the lncRNA Damage-induced (DINO), is expressed. DINO stabilises TP53 and disrupts the E6/E7/ubiquitin ligase complex which ubiquitinates TP53, leading to its degradation.
Some of the other lncRNAs whose expression is disrupted by HPV infection include GAS5, H19, and FAM83H-AS1 [63]. FAM83H-AS1 expression was increased following E6 expression and led to increased cellular proliferation and decreased apoptosis. Increased levels of FAM83H-AS1 is associated with a poor prognosis [63]. NGS analysis identified 19 lncRNAs, 3 novel lncRNAs were found to be differentially expressed in three cervical cancer patients infected with HPV16. All of these lncRNAs play roles in the carcinogenesis and development of cervical cancer [64].
Gastric cancer
Gastric cancer is most common in South America (ASR 13.8) and Asia (ASR 9.5) [65]. The mortality rate amongst gastric cancer patients is high. Risk factors for gastric cancer include H. pylori infection, which leads to the formation of ulcers, gut inflammation, pernicious anaemia and the presence of polyps in the stomach [66]. Infection with EBV is also thought to be related to the development and progression of gastric cancer in up to 10% of gastric cancer patients [67]. Gastric cancer associated with EBV infection is known as EBV-associated gastric carcinoma (EBVaGC). EBV infection leads to changes in lncRNA expression, that may contribute to the development and progression of gastric cancer.
The expression of the lncRNA Small nucleolar RNA host gene 8 (SNHG8) is increased in gastric cancer cells infected with EBV [68]. The level of expression is a good indicator of tumour stage with increased expression indicating a more advanced stage of cancer progression. This lncRNA influences the expression of multiple genes such as TRIM28 (a mediator of gene silencing), EIF4A2 (RNA helicase involved in mRNA translation), NAP1L1 (promoter of cell proliferation), PLD3 (Regulator inflammatory cytokine responses), RPL18A (a ribosome unit), and TRPM7 (a regulator of apoptosis and necrosis) [68]. The downregulation of this lncRNA inhibits cell growth and causes cell cycle arrest and increases apoptosis [69]. RNAs that regulate other RNAs by competing for binding to miRNAs that target these other RNAs, are known as competing endogenous RNAs (ceRNAs). Two lncRNAs were identified as playing a role in the EBV associated gastric cancer by acting as ceRNAs. These lncRNAs, RP5-1039K5.19 and TP73-AS1, regulate signalling pathways [70]. The pro-oncogene Guanine-nucleotide-binding protein beta polypeptide 2-like promotes cell growth and metastasis. The expression of this protein is promoted by the lncRNA OR3A4 in virus-associated gastric cancer [71].
Brain cancers
Malignant brain tumours can be classified according to the tissue or cell type that gives rise to them (Figure 5). Gliomas start in the glial cells of the brain or spine. Malignant gliomas are highly invasive and have a high mortality rate [72]. Risk factors that contribute to the development of gliomas include exposure to ionizing radiation, polymorphisms in genes involved In DNA repair and cell cycle regulation as well as infection by retroviruses such as cytomegalovirus reviewed in [73]. Human cytomegalovirus (HCMV) is a member of the herpes virus family that is associated with the development of gliomas. The virus expresses more than 200 proteins, which can be divided into three groups based on the stage during which they are expressed. These three categories are immediate early (IE), early (E), and late (L) [74]. HCMV genomic sequences as well as HCMV proteins have been isolated from gliomas. A high HCMV viral load in brain cancer patients is associated with lower survival times and poor treatment outcomes [75].
Figure 5.
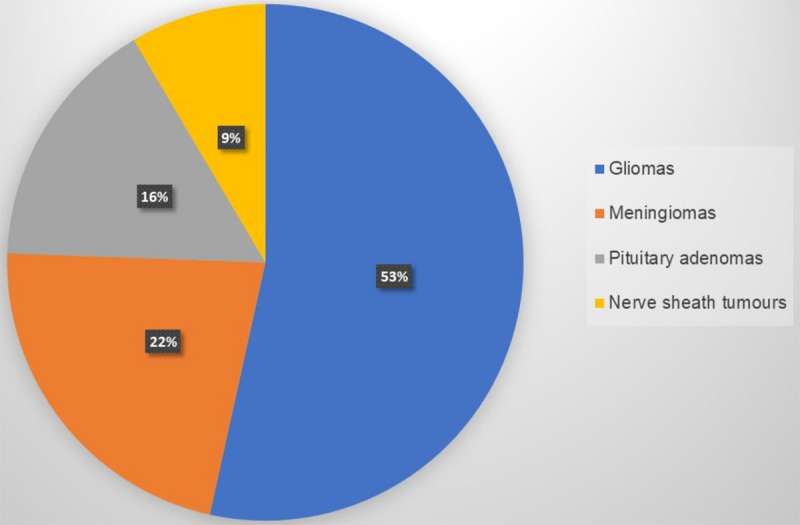
Types of brain cancer by incidence. Malignant brain tumours are named after the tissue from which they develop. Gliomas are the most common type of malignant brain tumour.
Latent infection also involved the expression of 646 lncRNAs (208 known lncRNAs and 438 novel lncRNAs) and decreased expression of 424 (140 known and 284 novel) [76]. Whole transcriptome sequencing of HCMV infected and uninfected CD14 (+) and CD34 (+) cells, led to the identification of two lncRNAs in the infected cells. These lncRNAs, named RNA4.9 and RNA2.7, enable the virus to exist in a latent state within these cells. It is now known that RNA4.9 is involved in the repression of host gene transcription [77] and is involved in promoting viral replication [78]. Another lncRNA, RNA1.2, is involved in the inhibition of the hosts’ immune response to the virus. RNA1.2 prevents the expression of IL-6 by inhibiting the NF-κB pathway [78].
The expression of lncRNAs also differ between glioma tissues and normal brain. The lncRNA FGD5-AS1 was overexpressed in glioma cells and is associated with the viability, migration, and invasion of glioma cells [79]. The expression of the lncRNA colon cancer-associated transcript-1 (CCAT1) is higher than in normal tissue. It functions as a competitor for the binding of miR-181b to target proteins such as fibroblast growth factor receptor 3 (FGFR3) and platelet-derived growth factor receptor (PDGFR). This leads to the decreased expression of these proteins. In this way, CCAT1 increases the expression of these growth factors leading to cancer progression [80]. The levels of the lncRNA LOC728196 is higher in glioma cells than in normal nerve tissue, where it prevents the inhibition of transcription factor 7 (TCF7). This transcription factor is involved in excessive activation of the Wnt/β-catenin signalling pathway [81].
The immune response to glioma and viral infection can be modulated by the lncRNA, LINC00152. The exact mechanism behind this is unknown. Another nine lncRNAs are also related to the immune response surrounding glioma cells. These include Phosphoglucomutases 5-antisense RNA 1 (PGM5-AS1), ST20-antisense RNA 1 (ST20-AS1), ankyrin repeat and PH domain 2-antisense RNA 1 (AGAP2-AS1), MIR155 host gene (MIR155HG), SNHG8, LINC00937, TUG1, MAPK activated protein kinase 5-antisense RNA 1 (MAPKAPK5-AS1) and HLA complex group 18 (HCG18). Additionally, the lncRNA TUG1 is also increased in glioma cells [82,83].
Alternate splicing and lncRNA in various cancers
Alternate splicing is the mechanism whereby the pre-mRNA encoded by a single gene is processed into multiple transcripts. These can give rise to multiple protein isoforms which can have slightly different functions or can act in a completely antagonistic manner to the other protein isoforms. The splicing process is controlled by splicing factor proteins. These proteins are RNA binding proteins (RBPs) that interact with RNA through one or more RNA binding domains, these include domains such as the RNA recognition motif (RRM), the hnRNP K homology domain (KH) or the DEAD box helicase domain [84]. There are two major families of splicing factors, the serine arginine splicing factors (SR proteins) and the heterogeneous nuclear ribonucleoproteins (hnRNPs). The splicing of mRNA can be affected through the function of lncRNAs in one of two ways. The lncRNAs can bind to splicing factors and either influence the expression of specific isoforms or by forming nuclear paraspeckles to regulate gene expression.
The lncRNA, lncRNA-p21, is transcribed from the p21 locus. Like p21 the lncRNA is involved in regulating cell proliferation, apoptosis and DNA damage response [85]. Through its binding to hnRNPK, lncRNA-p21 is able to promote TP53-mediated expression of p21 and other YP53 targets by promoting the expression of TP53 transcripts [86]. SR proteins are phosphorylated/dephosphorylated in order to properly splice pre-mRNA. Hyperphosphorylation of SR proteins changes the binding affinity to target pre-mRNA and guiding the selection of splice sites [87]. The phosphorylation state also influences the movement of the SR protein. LncRNA regulates the phosphorylation state of the SR proteins and therefore regulates alternative splicing. Partially dephosphorylated SR proteins support the first steps of the transesterification reactions. Additionally, intranuclear trafficking of SR proteins between nuclear speckles and transcription sites is dependent on their phosphorylation status [88]. The lncRNA MALAT1 was found to to regulate pre-mRNA alternate splicing by interacting with SR-splicing factors. It regulates the phosphorylation of these splicing factors, thereby changing the subcellular distribution of these SR proteins. This results in a change in the set of pre-mRNAs that the SR protein is able to interact with and regulate the splicing of [89].
LncRNAs such as MALAT1 are also able to alter splicing by hijacking splicing factors leading to the selection of specific isoforms. For example, Polypyrimidine tract binding protein (PTBP2) is a proto-oncogene whose splicing is controlled by the splicing factor, SFPQ. SFPQ achieves this by binding to PTBP2 mRNA and altering the splicing of PTBP2 to favor an anti-tumour isoform. MALAT1 binds SFPQ and disrupts the PTBP2/SFPQ splicing complex from forming. This prevents SFPQ from altering the splicing of PTBP2 to fvpur anti tumourogenic isoforms (Figure 7) [90].
Figure 7.
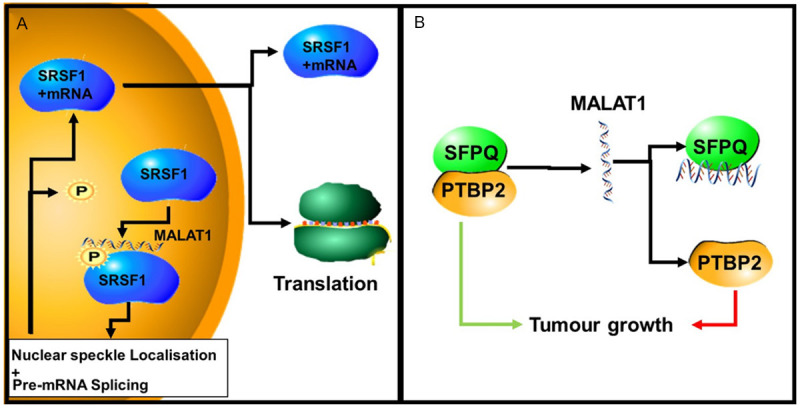
The role of MALAT1 in the regulation of alternative splicing. A. MALAT1 controls the phosphorylation of SRSF1 proteins in the nucleus, including the MALAT1-interacting SRSF1. Phosphorylated SRSF1 is accumulated in nuclear speckles (NS). Dephosphorylated SRSF1 promotes its interaction with mRNAs and transport into the cytoplasm. Here, the spliced mRNA is either translated by the ribosome or degradation by the non-sense mediated decay (NMD) machinery. B. MALAT1 binds to (hijacks) SFPQ, disrupting the SFPQ/PTBP2 complex. This results in the pro-oncogenic isoform of PTBP2, promoting tumour growth.
LncRNAs interact with RNA-binding proteins to form paraspeckles. These loci are distributed throughout the cell and are involved in gene expression. The lncRNA Nuclear Enriched Abundant Transcript 1 (NEAT1) has at least five known splice variants and is an essential component of these speckle. Of these five, the short variant is NEAT1-1 (3.7-kb) and the long variant is NEAT1-2 (23.7-kb) [91]. NEAT1 is also involved in the sequestration of related proteins. It is known to regulate the splicing of PPARγ mRNA that results in the production of the two isoforms, PPARγ1 and PPARγ2. These proteins are involved in the differentiation of adipocytes. SRp40 is involved in PPARγ splicing. SRp40 associates with NEAT1, which leads to the alteration in the splicing isoform of PPARt favoured by SRp40. Different levels of NEAT1 and SRp40 have different effects on the levels of these isoforms. With the level of NEAT1 expression changing the relative levels of PPARγ isoforms (Figure 6) [92].
Figure 6.
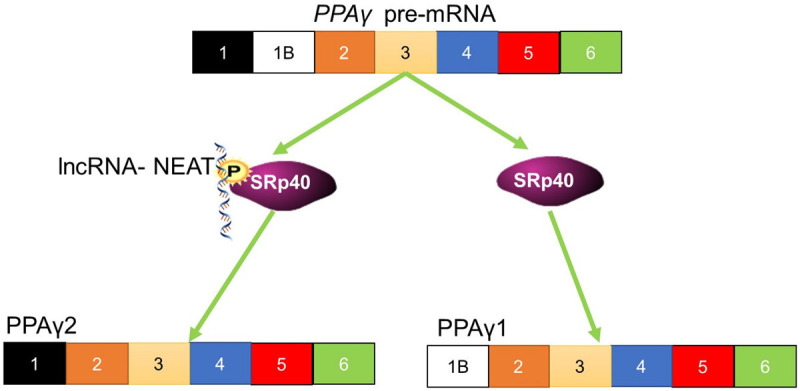
The function of NEAT1 in the splicing of PPARγ. NEAT1 interacts with and phosphorylates the splicing factor, SRp40. This splicing factor controls the pre-mRNA splicing of PPARγ. Phosphorylation of the splicing factor leads to changes in the isoform that results from pre-mRNA splicing.
LncRNAs in cancer treatment
LncRNAs as biomarkers
Currently, the vast majority of diagnostic tests rely on the symptoms and detecting physiological changes as cancer progresses. The adoption of biochemical markers, known as biomarkers, that are related to the development and progression of the disease offers distinct advantages. These include price, speed, and decreased invasiveness. For instance, HCC is normally only diagnosed following the onset of symptoms, at which point the cancer is advanced and treatment options are limited. These advanced stages are also associated with a poor prognosis. The large number of lncRNAs whose expression is altered during viral associated carcinogenesis (Table 2) makes them an attractive choice for biomarkers, which can be used to screen individuals in a regular non-invasive screening program. The selection of specific lncRNAs may be useful for diagnosing HCV or HBV specific HCC, which may influence treatment choices. Two lncRNAs that may be used for this purpose in HCV-related HCC include UCA1 and WRAP53. Not only can these biomarkers be used to diagnose HCV-related HCC, but in the case of UCA, the expression level is also associated with the size of the tumour and its ability to invade other tissues and therefore, tumour stage [29]. This makes UCA an ideal lncRNA biomarker, since it is capable to diagnose and stage the cancer. The ideal lncRNAs to be used as a biomarker would be a panel of differentially expressed circulating lncRNAs [93].
Table 2.
LncRNAs in different cancers and their associated viral infections
| LncRNA name | Changes in cancer type | Virus | Refs |
|---|---|---|---|
| Enhancer of zeste homolog 2 (EZH2) | HCC | HCV | [27] |
| LINC01419 | early HCC stages | HCV | [34] |
| BC014579 | HCV | [34] | |
| BC014579 | HCV | [34] | |
| AK021443 | Increasing in late HCC | HCV | [34] |
| RP11-401P9.4 | HCV | [34] | |
| RP11-304 L19.5 | HCV | [34] | |
| AF070632 | Decrease in late stage HCC | HCV | [34] |
| CTB-167B5.2 | HCV | [34] | |
| hypoxia-inducing factor aHIF | Downregulated in HCC | HCV | [35,36] |
| Prader Willi/Angelman region RNA 5 (PAR5) | Downregulated in HCC | HCV | [35,36] |
| human downregulated expression by HBx (hDREH) | Up regulated in HCC | HCV | [35,36] |
| Hox transcript antisense intergenic RNA (HOTAIR) | Highly expressed in HCC | HCV | [38] |
| WD repeat containing antisense to TP53 (WRAP53) | Highly expressed in HCC | HBV | [43] |
| urothelial carcinoma associated 1 (UCA1) | Highly expressed in HCC | HBV | [43] |
| NR2F21 | Downregulated in NPC | EBV | [51] |
| FAM95C | Downregulated in NPC | EBV | [51] |
| LINC01106 | Downregulated in NPC | EBV | [51] |
| CH507-513H4.6 | Upregulated in NPC | EBV | [51] |
| HAP9-AS1 | Upregulated in NPC | EBV | [51] |
| CH507-513H4.3 | Upregulated in NPC | EBV | [51] |
| RP4-794H19.1 | Upregulated in NPC | EBV | [51] |
| LINC00312 (NAG7) | Downregulated in NPC | EBV | [52] |
| LOC553103 | Downregulated in NPC | EBV | [53] |
| APAF1-AS1 | Upregulated in NPC | EBV | [54] |
| AL359062 | Upregulated in NPC | EBV | [54] |
| HOTAIR | Upregulated in HNSSC | HPV | [58] |
| PROM1 | Upregulated in HNSSC | HPV | [58] |
| CCAT1 | Upregulated in HNSSC | HPV | [58] |
| MUC19 | Upregulated in HNSSC | HPV | [58] |
| DINO | Downregulated in cervical cancer | HPV | [62] |
| GAS5 | Altered in cervical caner | HPV | [63] |
| H19 | Altered in cervical caner | HPV | [63] |
| FAM83H-AS1 | Increased in cervical caner | HPV | [63] |
| CYTOR | Upregulated in various lymphomas | EBV | [46] |
| NORAD | Upregulated in various lymphomas | EBV | [46] |
| 7SL | Downregulated in Various cancers | EBV | [48] |
| H19 | Upregulated in Various cancers | EBV | [48] |
| H19-AS | Upregulated in Various cancers | EBV | [48] |
| NDM29 | Downregulated in lymphoma | EBV | [49] |
| HOXA6as | Downregulated in lymphoma | EBV | [49] |
| HAR1B | Downregulated in lymphoma | EBV | [49] |
| Tsix | Downregulated in lymphoma | EBV | [49] |
| SNHG8 | Upregulated in EBVaGC | EBV | [68] |
| RP5-1039 K5.19 | Upregulated in EBVaGC | EBV | [68] |
| TP73-AS1 | Upregulated in EBVaGC | EBV | [68] |
| MALAT1 | Upregulated in NPC | EBV | [54] |
| Upregulated in diffuse large B cell lymphoma | EBV | [56] |
Apart from using lncRNAs to diagnose and stage cancer, lncRNAs can also be used as a prognostic marker to predict patient survival. A circulating lncRNA that shows promise as a prognostic biomarker for HCC is the lncRNA Small Nucleolar RNA Host Gene 15 (SNHG15). Increased expression of this lncRNA results in decreased HCC patient survival [94], making it an ideal prognostic biomarker. Many of the lncRNAs that were previously discussed as useful diagnostic biomarkers would also be useful prognostic biomarkers. Other lncRNAs that may prove useful are the lncRNAs, MALAT1 and HEIH that have also been studied for their potential use as diagnostic biomarkers for HCV-related HCC. HEIH expression can be monitored in the blood [95]. Many alternatively expressed lncRNAs in EBV-associated lymphoma are exported from the cell. The release of some of these lncRNAs from the cells using exosomes is important for diagnostic purposes. These secreted lncRNAs allow for detection of many of these lncRNAs using blood tests [49].
LncRNAs as drug targets
The importance of lncRNAs in cancer development and progression makes them the ideal targets for the development of new treatments. By inhibiting the activity of these lncRNAs through the use of antagonists can inhibit the pro-carcinogenic activity of these lncRNA. This has been shown to be an effective potential strategy in multiple cancers such as HCC [96]. There are numerous methods whereby lncRNA can be therapeutically targeted. One of the methods that can be used to interfere with the activity of lncRNAs is through the use of small interfering RNAs to inhibit the expression of these pro-oncogenic lncRNAs. By silencing lncRNA using RNAi to knockdown expression of the target lncRNA, positive anti-cancer effects have been observed. Examples of silencing the expression of target lncRNAs include the lncRNA Linc00974, which is highly expressed in HCC [97,98]. As a result, targeting Lnc00974 with siRNA resulted in the inhibition of cell proliferation and invasion, and increased apoptosis in the tumour [98]. Another technique is to target the regulatory element of the lncRNA [97].
In order to effectively use these as an anticancer treatment, first the intracellular localization of cancer-related lncRNAs should be inhibited [99]. One of these solutions is the use of antisense oligonucleotides (ASOs). These can enter the nucleus and induce RNase H-dependent degradation or sterically block lncRNA activity. ASOs can also be modified to increase their stability. The lncRNA MALAT1 has been experimentally targeted using ASO to decrease its levels. This was performed in a mouse model where it reduced the growth and metastasis of tumours [100]. The delivery of these ASOs is complicated as they have low levels of intracellular uptake and low stability in the body. New techniques of delivering these ASOs include the development of biocompatible nanoparticles [101]. Another technique involves blocking the binding of lncRNAs to protein targets and this applies to the lncRNAs that interact with chromatin remodelling complexes, transcription factors or those that act as protein chaperones by binding to protein targets [101].
Conclusions
Perturbations to the gene regulation network is an important cause for the development and progression of cancer. For a long time, studies have focused on changes in gene control that occur at the transcription and expression levels, neglecting the control of gene expression that occurs at the post-transcriptional level. This includes changes in mRNA splicing, and the control of gene expression by ncRNAs, such as lncRNAs and miRNAs. Unlike splicing and miRNAs, lncRNAs interact with DNA, RNA, and protein, meaning they act to regulate gene expression at the level of transcription, post transcription and expression.
The role of lncRNAs in enhancing viral infection-mediated carcinogenesis process provides an important insight into the ability of infection by these viruses to act as causative agents in the development of these cancers. This becomes especially important in cancers such as cervical cancer, where viral infection by HPV is the most important risk factor in cervical carcinoma. The ability of these viral infections to alter the expression patterns of lncRNA and facilitate tumourigenesis has led to them being considered as probable biomarkers for the future diagnosis or prognosis of these cancers. Since lncRNAs expression is altered at multiple stages of cancer development and with cancer progression means that specific patterns of lncRNA expression can be used to not only diagnose cancer, but stratify patients based on the stage of cancer and their need for treatment. The identification of changes in the expression of lncRNAs, has been advanced by technologies such as NGS and bioinformatics. These advances not only make the study of lncRNA expression changes easier but would help in the establishment of specific lncRNAS as biomarkers.
Acknowledgements
The authors would like to thank the South African Medical Research Council (SAMRC) for funding this research.
Disclosure of conflict of interest
None.
References
- 1.Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J Exp Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pagano JS, Blaser M, Buendia MA, Damania B, Khalili K, Raab-Traub N, Roizman B. Infectious agents and cancer: criteria for a causal relation. Semin Cancer Biol. 2004;14:453–471. doi: 10.1016/j.semcancer.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 3.de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 4.Simard EP, Jemal A. Commentary: Infection-related cancers in low- and middle-income countries: challenges and opportunities. Int J Epidemiol. 2013;42:228–229. doi: 10.1093/ije/dys216. [DOI] [PubMed] [Google Scholar]
- 5.Nnaji M, Adedeji OA, Sule O. Infection-related cancers in sub-saharan Africa. In: Adedeji OA, editor. Cancer in sub-saharan africa: current practice and future. Cham: Springer International Publishing; 2017. pp. 37–52. [Google Scholar]
- 6.Ajiro M, Zheng ZM. Oncogenes and RNA splicing of human tumor viruses. Emerg Microbes Infect. 2014;3:e63. doi: 10.1038/emi.2014.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 8.Epstein MA, Henle G, Achong BG, Barr YM. Morphological and biological studies on a virus in cultured lymphoblasts from burkitt’s lymphoma. J Exp Med. 1965;121:761–770. doi: 10.1084/jem.121.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salviano-Silva A, Lobo-Alves SC, Almeida RC, Malheiros D, Petzl-Erler ML. Besides pathology: long non-coding RNA in cell and tissue homeostasis. Noncoding RNA. 2018;4:3. doi: 10.3390/ncrna4010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucafo M, De Iudicibus S, Di Silvestre A, Pelin M, Candussio L, Martelossi S, Tommasini A, Piscianz E, Ventura A, Decorti G. Long noncoding RNA GAS5: a novel marker involved in glucocorticoid response. Curr Mol Med. 2015;15:94–99. doi: 10.2174/1566524015666150114122354. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 12.Fortes P, Morris KV. Long noncoding RNAs in viral infections. Virus Res. 2016;212:1–11. doi: 10.1016/j.virusres.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 15.St Laurent G, Wahlestedt C, Kapranov P. The landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–51. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzman J. Circular RNA expression: its potential regulation and function. Trends Genet. 2016;32:309–316. doi: 10.1016/j.tig.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 19.Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 20.Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma. Carcinogenesis. 2014;35:507–514. doi: 10.1093/carcin/bgt405. [DOI] [PubMed] [Google Scholar]
- 21.Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Han L, Ma P, Liu SM, Zhou X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol. 2016;37:6847–6854. doi: 10.1007/s13277-015-4568-7. [DOI] [PubMed] [Google Scholar]
- 23.Han Y, Zhou L, Wu T, Huang Y, Cheng Z, Li X, Sun T, Zhou Y, Du Z. Downregulation of lncRNA-MALAT1 affects proliferation and the expression of stemness markers in glioma stem cell line SHG139S. Cell Mol Neurobiol. 2016;36:1097–1107. doi: 10.1007/s10571-015-0303-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng ZM. Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses. Int J Biol Sci. 2010;6:730–755. doi: 10.7150/ijbs.6.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer. 2013;13:123–135. doi: 10.1038/nrc3449. [DOI] [PubMed] [Google Scholar]
- 26.European Association for the Study of the Liver. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Guo J, Hao C, Wang C, Li L. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18:98. doi: 10.1186/s12935-018-0582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 29.Zhang H, Zhu C, Zhao Y, Li M, Wu L, Yang X, Wan X, Wang A, Zhang MQ, Sang X, Zhao H. Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma. Oncotarget. 2015;6:43770–43778. doi: 10.18632/oncotarget.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maucort-Boulch D, de Martel C, Franceschi S, Plummer M. Fraction and incidence of liver cancer attributable to hepatitis B and C viruses worldwide. Int J Cancer. 2018;142:2471–2477. doi: 10.1002/ijc.31280. [DOI] [PubMed] [Google Scholar]
- 31.Jeong SW, Jang JY, Chung RT. Hepatitis C virus and hepatocarcinogenesis. Clin Mol Hepatol. 2012;18:347–356. doi: 10.3350/cmh.2012.18.4.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chisari FV. Unscrambling hepatitis C virus-host interactions. Nature. 2005;436:930–932. doi: 10.1038/nature04076. [DOI] [PubMed] [Google Scholar]
- 33.Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Curr Opin Microbiol. 2002;5:419–427. doi: 10.1016/s1369-5274(02)00341-7. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Q, Matsuura K, Kleiner DE, Zamboni F, Alter HJ, Farci P. Analysis of long noncoding RNA expression in hepatocellular carcinoma of different viral etiology. J Transl Med. 2016;14:328. doi: 10.1186/s12967-016-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang DY, Zou XJ, Cao CH, Zhang T, Lei L, Qi XL, Liu L, Wu DH. Identification and functional characterization of long non-coding RNA MIR22HG as a tumor suppressor for hepatocellular carcinoma. Theranostics. 2018;8:3751–3765. doi: 10.7150/thno.22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AHF. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Transl Res. 2016;168:134–145. doi: 10.1016/j.trsl.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, Sugimachi K, Mimori K, Wakabayashi G, Mori M. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29:946–950. doi: 10.3892/or.2012.2219. [DOI] [PubMed] [Google Scholar]
- 38.Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu ZL, Zhou GZ, Cao G, Jin L, Xie HW, Wang CM, Lv J, De W, Wu M, Cao XF. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52:908–915. doi: 10.1002/mc.21944. [DOI] [PubMed] [Google Scholar]
- 39.Peng W, Wu J, Fan H, Lu J, Feng J. LncRNA EGOT promotes tumorigenesis via hedgehog pathway in gastric cancer. Pathol Oncol Res. 2019;25:883–887. doi: 10.1007/s12253-017-0367-3. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Sun Z, Wang Y, Hu Z, Zhou H, Zhang L, Hong B, Zhang S, Cao X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci Rep. 2016;6:23170. doi: 10.1038/srep23170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu S, Ai H, Zhang K, Yun H, Xie F. Long Non-Coding RNA EGOT promotes the malignant phenotypes of hepatocellular carcinoma cells and increases the expression of HMGA2 via down-regulating miR-33a-5p. Onco Targets Ther. 2019;12:11623–11635. doi: 10.2147/OTT.S218308. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Toraih EA, Ellawindy A, Fala SY, Al Ageeli E, Gouda NS, Fawzy MS, Hosny S. Oncogenic long noncoding RNA MALAT1 and HCV-related hepatocellular carcinoma. Biomed Pharmacother. 2018;102:653–669. doi: 10.1016/j.biopha.2018.03.105. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Feng J, Sun M, Yang G, Yuan H, Wang Y, Bu Y, Zhao M, Zhang S, Zhang X. Long non-coding RNA HULC activates HBV by modulating HBx/STAT3/miR-539/APOBEC3B signaling in HBV-related hepatocellular carcinoma. Cancer Lett. 2019;454:158–170. doi: 10.1016/j.canlet.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 44.Yu S, Li N, Wang J, Fu Y, Huang Y, Yi P, Chen R, Tang D, Hu X, Fan X. Correlation of long noncoding RNA SEMA6A-AS1 expression with clinical outcome in HBV-related hepatocellular carcinoma. Clin Ther. 2020;42:439–447. doi: 10.1016/j.clinthera.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 45.Young LS, Rickinson AB. Epstein-barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 46.Wang C, Li D, Zhang L, Jiang S, Liang J, Narita Y, Hou I, Zhong Q, Zheng Z, Xiao H, Gewurz BE, Teng M, Zhao B. RNA sequencing analyses of gene expression during epstein-barr virus infection of primary B lymphocytes. J Virol. 2019;93:e00226-19. doi: 10.1128/JVI.00226-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munschauer M, Nguyen CT, Sirokman K, Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP, Subramanian V, Chen J, Schenone M, Guttman M, Carr SA, Lander ES. The NORAD lncRNA assembles a topoisomerase complex critical for genome stability. Nature. 2018;561:132–136. doi: 10.1038/s41586-018-0453-z. [DOI] [PubMed] [Google Scholar]
- 48.Gallo A, Vella S, Miele M, Timoneri F, Di Bella M, Bosi S, Sciveres M, Conaldi PG. Global profiling of viral and cellular non-coding RNAs in Epstein-Barr virus-induced lymphoblastoid cell lines and released exosome cargos. Cancer Lett. 2017;388:334–343. doi: 10.1016/j.canlet.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, Li X, Hu J, Cao P, Yan Q, Zhang S, Dang W, Lu J. Long noncoding RNAs involvement in Epstein-Barr virus infection and tumorigenesis. Virol J. 2020;17:51–51. doi: 10.1186/s12985-020-01308-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verhoeven RJA, Tong S, Mok BW, Liu J, He S, Zong J, Chen Y, Tsao SW, Lung ML, Chen H. Epstein-barr virus BART long non-coding RNAs function as epigenetic modulators in nasopharyngeal carcinoma. Front Oncol. 2019;9:1120. doi: 10.3389/fonc.2019.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li XX, Liang XJ, Zhou LY, Liu RJ, Bi W, Zhang S, Li SS, Yang WH, Chen ZC, Yang XM, Zhang PF. Analysis of differential expressions of long non-coding RNAs in nasopharyngeal carcinoma using next-generation deep sequencing. J Cancer. 2018;9:1943–1950. doi: 10.7150/jca.23481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, Huang C, Gong Z, Zhao Y, Tang K, Li X, Fan S, Shi L, Li X, Zhang P, Zhou Y, Huang D, Liang F, Zhang X, Wu M, Cao L, Wang J, Li Y, Xiong W, Zeng Z, Li G. Expression of LINC00312, a long intergenic non-coding RNA, is negatively correlated with tumor size but positively correlated with lymph node metastasis in nasopharyngeal carcinoma. J Mol Histol. 2013;44:545–554. doi: 10.1007/s10735-013-9503-x. [DOI] [PubMed] [Google Scholar]
- 53.He B, Li W, Wu Y, Wei F, Gong Z, Bo H, Wang Y, Li X, Xiang B, Guo C, Liao Q, Chen P, Zu X, Zhou M, Ma J, Li X, Li Y, Li G, Xiong W, Zeng Z. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He B, Zeng J, Chao W, Chen X, Huang Y, Deng K, Huang Z, Li J, Dai M, Chen S, Huang H, Dai S. Serum long non-coding RNAs MALAT1, AFAP1-AS1 and AL359062 as diagnostic and prognostic biomarkers for nasopharyngeal carcinoma. Oncotarget. 2017;8:41166–41177. doi: 10.18632/oncotarget.17083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 56.Fakhry C, Gillison ML. Clinical implications of human papillomavirus in head and neck cancers. J. Clin. Oncol. 2006;24:2606–2611. doi: 10.1200/JCO.2006.06.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma X, Sheng S, Wu J, Jiang Y, Gao X, Cen X, Wu J, Wang S, Tang Y, Tang Y, Liang X. LncRNAs as an intermediate in HPV16 promoting myeloid-derived suppressor cell recruitment of head and neck squamous cell carcinoma. Oncotarget. 2017;8:42061–42075. doi: 10.18632/oncotarget.14939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ostrand-Rosenberg S. Immune surveillance: a balance between protumor and antitumor immunity. Curr Opin Genet Dev. 2008;18:11–18. doi: 10.1016/j.gde.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sunthamala N, Pientong C, Ohno T, Zhang C, Bhingare A, Kondo Y, Azuma M, Ekalaksananan T. HPV16 E2 protein promotes innate immunity by modulating immunosuppressive status. Biochem Biophys Res Commun. 2014;446:977–982. doi: 10.1016/j.bbrc.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 60.Boyer SN, Wazer DE, Band V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996;56:4620–4624. [PubMed] [Google Scholar]
- 61.Jones DL, Münger K. Analysis of the p53-mediated G1 growth arrest pathway in cells expressing the human papillomavirus type 16 E7 oncoprotein. J Virol. 1997;71:2905–2912. doi: 10.1128/jvi.71.4.2905-2912.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma S, Munger K. Expression of the long noncoding RNA DINO in human papillomavirus-positive cervical cancer cells reactivates the dormant TP53 tumor suppressor through ATM/CHK2 signaling. mBio. 2020;11:e01190-20. doi: 10.1128/mBio.01190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barr JA, Hayes KE, Brownmiller T, Harold AD, Jagannathan R, Lockman PR, Khan S, Martinez I. Long non-coding RNA FAM83H-AS1 is regulated by human papillomavirus 16 E6 independently of p53 in cervical cancer cells. Sci Rep. 2019;9:3662. doi: 10.1038/s41598-019-40094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Zhao Y, Chen M, Cui J. Identification of novel long non-coding and circular RNAs in human papillomavirus-mediated cervical cancer. Front Microbiol. 2017;8:1720. doi: 10.3389/fmicb.2017.01720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 66.Rawla P, Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. Epstein-Barr virus (EBV)-associated gastric carcinoma. Viruses. 2012;4:3420–3439. doi: 10.3390/v4123420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yue W, Zhu M, Zuo L, Xin S, Zhang J, Liu L, Li S, Dang W, Zhang S, Xie Y, Zhu F, Lu J. Early pattern of epstein-barr virus infection in gastric epithelial cells by “Cell-in-cell”. Virol Sin. 2019;34:253–261. doi: 10.1007/s12250-019-00097-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J, Yang C, Gu Y, Li C, Zhang H, Zhang W, Wang X, Wu N, Zheng C. Knockdown of the lncRNA SNHG8 inhibits cell growth in Epstein-Barr virus-associated gastric carcinoma. Cell Mol Biol Lett. 2018;23:17. doi: 10.1186/s11658-018-0070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jing JJ, Wang ZY, Li H, Sun LP, Yuan Y. Key elements involved in Epstein-Barr virus-associated gastric cancer and their network regulation. Cancer Cell Int. 2018;18:146. doi: 10.1186/s12935-018-0637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo X, Yang Z, Zhi Q, Wang D, Guo L, Li G, Miao R, Shi Y, Kuang Y. Long noncoding RNA OR3A4 promotes metastasis and tumorigenicity in gastric cancer. Oncotarget. 2016;7:30276–30294. doi: 10.18632/oncotarget.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 73.Kofman A, Marcinkiewicz L, Dupart E, Lyshchev A, Martynov B, Ryndin A, Kotelevskaya E, Brown J, Schiff D, Abounader R. The roles of viruses in brain tumor initiation and oncomodulation. J Neurooncol. 2011;105:451–466. doi: 10.1007/s11060-011-0658-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Strååt K, Liu C, Rahbar A, Zhu Q, Liu L, Wolmer-Solberg N, Lou F, Liu Z, Shen J, Jia J, Kyo S, Björkholm M, Sjöberg J, Söderberg-Nauclér C, Xu D. Activation of telomerase by human cytomegalovirus. J Natl Cancer Inst. 2009;101:488–497. doi: 10.1093/jnci/djp031. [DOI] [PubMed] [Google Scholar]
- 75.Barami K. Oncomodulatory mechanisms of human cytomegalovirus in gliomas. J Clin Neurosci. 2010;17:819–823. doi: 10.1016/j.jocn.2009.10.040. [DOI] [PubMed] [Google Scholar]
- 76.Zhang Q, Lai MM, Lou YY, Guo BH, Wang HY, Zheng XQ. Transcriptome altered by latent human cytomegalovirus infection on THP-1 cells using RNA-seq. Gene. 2016;594:144–150. doi: 10.1016/j.gene.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rossetto CC, Tarrant-Elorza M, Pari GS. Cis and trans acting factors involved in human cytomegalovirus experimental and natural latent infection of CD14 (+) monocytes and CD34 (+) cells. PLoS Pathog. 2013;9:e1003366. doi: 10.1371/journal.ppat.1003366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tai-Schmiedel J, Karniely S, Lau B, Ezra A, Eliyahu E, Nachshon A, Kerr K, Suárez N, Schwartz M, Davison AJ, Stern-Ginossar N. Human cytomegalovirus long noncoding RNA4.9 regulates viral DNA replication. PLoS Pathog. 2020;16:e1008390. doi: 10.1371/journal.ppat.1008390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y, Xiao Y, Li GC, Gong FY, Zhang XN, Hou K. Long non-coding RNAs as epigenetic mediator and predictor of glioma progression, invasiveness, and prognosis. Semin Cancer Biol. 2020 doi: 10.1016/j.semcancer.2020.08.016. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 80.Cui B, Li B, Liu Q, Cui Y. lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem. 2017;118:4548–4557. doi: 10.1002/jcb.26116. [DOI] [PubMed] [Google Scholar]
- 81.Wallmen B, Schrempp M, Hecht A. Intrinsic properties of Tcf1 and Tcf4 splice variants determine cell-type-specific Wnt/β-catenin target gene expression. Nucleic Acids Res. 2012;40:9455–9469. doi: 10.1093/nar/gks690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Wu F, Zhao Z, Wang KY, Huang RY, Wang HY, Lan Q, Wang JF, Zhao JZ. Long noncoding RNA LINC00152 is a potential prognostic biomarker in patients with high-grade glioma. CNS Neurosci Ther. 2018;24:957–966. doi: 10.1111/cns.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z, Xi Z, Li Z, Bao M, Liu Y. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–331. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 84.Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 85.Tomita S, Abdalla MO, Fujiwara S, Yamamoto T, Iwase H, Nakao M, Saitoh N. Roles of long noncoding RNAs in chromosome domains. Wiley Interdiscip Rev RNA. 2017;8 doi: 10.1002/wrna.1384. [DOI] [PubMed] [Google Scholar]
- 86.Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA, Jacks T. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3:1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 88.Cáceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z, Sui H, Tang Y, Wang Y, Liu N, Ren J, Hou F, Li Q. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Naganuma T, Hirose T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013;10:456–461. doi: 10.4161/rna.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, Cooper DR. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 2009;150:2087–2097. doi: 10.1210/en.2008-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Plissonnier ML, Herzog K, Levrero M, Zeisel MB. Non-coding RNAs and hepatitis C virus-induced hepatocellular carcinoma. Viruses. 2018;10:591. doi: 10.3390/v10110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H, Min W, Bennett AM, Gregory RI, Ding Y, Huang Y. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang C, Yang X, Qi Q, Gao Y, Wei Q, Han S. lncRNA-HEIH in serum and exosomes as a potential biomarker in the HCV-related hepatocellular carcinoma. Cancer Biomark. 2018;21:651–659. doi: 10.3233/CBM-170727. [DOI] [PubMed] [Google Scholar]
- 96.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–446. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 97.Kumar MM, Goyal R. LncRNA as a therapeutic target for angiogenesis. Curr Top Med Chem. 2017;17:1750–1757. doi: 10.2174/156802661766616111644744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tang J, Zhuo H, Zhang X, Jiang R, Ji J, Deng L, Qian X, Zhang F, Sun B. A novel biomarker Linc00974 interacting with KRT19 promotes proliferation and metastasis in hepatocellular carcinoma. Cell Death Dis. 2014;5:e1549. doi: 10.1038/cddis.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu X, Sood AK, Dang CV, Zhang L. The role of long noncoding RNAs in cancer: the dark matter matters. Curr Opin Genet Dev. 2018;48:8–15. doi: 10.1016/j.gde.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Arun G, Diermeier S, Akerman M, Chang KC, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L, Brogi E, Egeblad M, Spector DL. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev. 2016;30:34–51. doi: 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wu SY, Lopez-Berestein G, Calin GA, Sood AK. RNAi therapies: drugging the undruggable. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008362. 240ps247. [DOI] [PMC free article] [PubMed] [Google Scholar]


