Abstract
Hepatoblastoma is a rare childhood liver cancer without known explicit etiology. Base excision repair (BER) pathway genes have been implicated in the pathophysiology of cancer, yet the role of BER pathway gene single nucleotide polymorphisms (SNPs) on hepatoblastoma risk still awaits to be explored. This study aims to determine whether hepatoblastoma risk be modulated by polymorphisms in the BER pathway genes based on genotyped data from 313 cases and 1446 controls. We applied TaqMan assay to genotype these included samples. We comprehensively genotyped 20 SNPs across six genes of BER, and estimated odds ratio (ORs), 95% confidence intervals (CIs), and P-values of the selected SNPs’ contribution to the risk of hepatoblastoma using logistic regression models. Only SNP rs293795 in the hOGG1 gene could significantly enhance hepatoblastoma risk under recessive model (adjusted OR=3.78, 95% CI=1.01-14.17, P=0.047). Stratified analysis revealed that rs159153 TC/CC genotype decreased hepatoblastoma risk in male subgroup. Moreover, rs293795 GG and 1-3 risk genotypes could increase hepatoblastoma risk in clinical stages I+II and male subgroups, respectively. False-positive report probability validated the reliability of the significant results. Our findings provide some clues of a potential risk effect of BER pathway gene hOGG1 SNPs on hepatoblastoma. Further investigation is warranted to confirm these findings and to better elucidate the biological pathways involved.
Keywords: Hepatoblastoma, BER, polymorphism, susceptibility
Introduction
Hepatoblastoma is a rare malignant neoplasm that originated from undifferentiated liver cells during embryonic development [1,2]. The incidence of hepatoblastoma is about 1/1.5*106~1/1.0*106, 90% of which occurs under the age of 5 years [3]. Though rare in incidence, hepatoblastoma takes up about 80% of primary liver malignancies in children [4]. Surgery with complete resection is the most effective cure option for hepatoblastoma. However, a large portion of hepatoblastoma children failed to accept this surgery [5,6]. Preoperative chemotherapy for children with hepatoblastoma can greatly increase the 5-year overall survival rate to 70%~90% [7-9]. Therefore, early screening and timely treatment of hepatoblastoma are particularly important.
Unlike adult hepatocellular carcinoma, there is no significant correlation between hepatoblastoma development and hepatitis b virus, chronic hepatitis, or cirrhosis [10,11]. According to relevant reports, the causes of hepatoblastoma include preterm birth, parental tobacco use, familial adenomatous polyposis, trisomy 18, FGFR3 mutations, low birth weight, and Beckwith-Wiedemann syndrome [12-17]. However, so far, no clear exposures can lead to the occurrence of hepatoblastoma. In addition, even if parents are exposed to the same environmental factors, only a very small number of offspring eventually develop hepatoblastoma. Increasing evidence suggests that genetic predisposition may play an important role in the occurrence of hepatoblastoma. To date, only a handful of case-control studies have analyzed the effects of single nucleotide polymorphisms (SNPs) on the risk of hepatoblastoma, with sample sizes of less than 100 [18,19]. Our research group also conducted several epidemiological investigations of hepatoblastoma [20-22]. There is no doubt that more characteristics of genetic variation in hepatoblastoma susceptibility will contribute to understanding the etiology of hepatoblastoma.
The human genome was continuously exposed to exogenous (ionizing radiation chemicals, ultraviolet light) and endogenous (metabolic by-products, intracellular hydrolysis) DNA damages [23,24]. If not repaired accurately, DNA damages may cause genomic instability and eventually impact tumor susceptibility [25]. DNA repair systems inherently exist in preserving the integrity of genome [26]. Base excision repair (BER) pathway, a primary DNA repair system, is responsible for repairing base lesions and AP sites [27,28]. BER pathway generally consists of human 8-oxoguanine DNA glycosylase (hOGG1), poly(ADP) ribose polymerase 1 (PARP1), apurinic/apyrimidinic endonuclease (APE1/APEX1), flap endonuclease 1 (FEN1), DNA ligase III (LIG3), and x-ray repair cross-complementing group 1 (XRCC1). In repairing DNA damage, the BER process may be generally divided into four steps: recognize and excise the damaged base, incise the DNA backbone, fill the nucleotide gap, and seal the remaining gap. Considerable evidence suggests the implication of abnormal expression of BER pathway proteins in multiple diseases including cancers [29]. Many BER pathway gene polymorphisms have been reported to contribute to risk of cancer [30]. Further molecular mechanism analysis showed that SNPs in the BER pathway genes may change protein dynamics, thereby limiting DNA repair ability and ultimately promoting the occurrence and development of cancer [31,32].
While BER pathway genes work as probably carcinogenic to humans and several epidemiological studies reported associations between these gene polymorphisms and cancers, no available reports were found on the hepatoblastoma. To elucidate these relationships, we perform a multi-center case-control study among children of Chinese ancestry.
Material and methods
Study subjects
The selection of subjects has been described previously [33-35]. Cases with hepatoblastoma were recruited from seven regional hospitals in China. Controls were randomly selected from the hospital visitors and frequency-matched to the cases by age and sex (Table 1). Controls were free of hepatoblastoma history and residing in the same region as the cases. All subjects signed their informed consent for agreeing the collection and use of blood samples in clinical research. The study was conducted after obtaining ethical approvals of hospital institutional review board.
Table 1.
Frequency distribution of selected variables in hepatoblastoma patients and cancer-free controls
| Variables | Cases (n=313) | Controls (n=1446) | P a | ||
|---|---|---|---|---|---|
|
|
|
||||
| No. | % | No. | % | ||
| Age range, month | 0.03-149.97 | 0.004-156.00 | |||
| Mean ± SD | 23.75 ± 25.93 | 25.23 ± 19.38 | 0.251b | ||
| <17 | 168 | 53.67 | 642 | 44.40 | |
| ≥17 | 145 | 46.33 | 804 | 55.60 | |
| Gender | 0.983 | ||||
| Female | 129 | 41.21 | 595 | 41.15 | |
| Male | 184 | 58.79 | 851 | 58.85 | |
| Clinical stages | |||||
| I | 97 | 30.99 | / | / | |
| II | 63 | 20.13 | / | / | |
| III | 64 | 20.45 | / | / | |
| IV | 27 | 8.63 | / | / | |
| NA | 62 | 19.81 | / | / | |
SD, standard deviation, NA, not available.
Two-sided χ2 test for distributions between hepatoblastoma cases and cancer-free controls.
T-test for age distribution between hepatoblastoma patients and cancer-free controls.
Genotyping
We first used dbSNP database for SNPs identification and then used SNPinfo software to further extract those with potential function. A total of 20 SNPs in six BER pathway genes were screened out for analysis [36]. Genomic DNA was extracted from peripheral blood samples following the protocol of QIAamp DNA Blood mini kit (QIAGEN Inc., Valencia, CA). The genotyping was running on a TaqMan platform (Applied Biosystems, Foster City, CA, USA), with details in a previous study [37]. For quality control, we took several measures in genotyping, including: 1) case and control samples were blindly genotyped by technicians, 2) both positive and negative control (water) samples were included in each 384-well plate, and 3) re-genotyping 10% randomly selected samples (100% concordant rate).
Statistical analysis
Clinical variables were analyzed using a Chi-square test (gender) or t test (age), as appropriate. To determine the associations between SNPs and hepatoblastoma risk, unconditional logistic regression models were used with adjustment for age and gender. Odds ratios (ORs) and 95% confidence intervals (CIs) generated from the models were applied to quantify the associations. False-positive report probability (FPRP) analysis was performed to assess noteworthy associations. All statistical analyses were carried out with SAS v10.0 (SAS Institute Inc., Cary, NC). All tests for statistical significance used a two-sided alpha of 0.05.
Result
Association between BER pathway gene SNPs and hepatoblastoma risk
Detailed clinical characteristics information of hepatoblastoma cases (n=313) and cancer-free controls (n=1446) was presented in our previous published studies [35,38]. A successful genotype rate of more than 95% was achieved. Relationships between polymorphisms in BER pathway genes and hepatoblastoma susceptibility are shown in Table 2. Specifically, there were 3, 3, 2, 3, 3, and 6 SNPs genotyped in the PARP1, hOGG1, FEN1, APEX1, LIG3, and XRCC1 genes, respectively. In the single locus analysis, only one BER gene SNP, hOGG1 gene rs293795, significantly impacts hepatoblastoma risk under recessive model (adjusted OR=3.78, 95% CI=1.01-14.17, P=0.047). No significant effect on risk of hepatoblastoma was observed for the rest of SNPs under dominant and recessive models (Figures 1, 2).
Table 2.
Relationship between polymorphisms in base excision repair pathway genes and hepatoblastoma susceptibility in Chinese children
| Gene | Polymorphism | Allele | Cases | Controls | AOR (95% CI)a | P a | AOR (95% CI)b | P b | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||||
| W | M | WW | WM | MM | WW | WM | MM | ||||||
| PARP1 | rs1136410 | A | G | 101 | 157 | 49 | 501 | 665 | 279 | 1.08 (0.83-1.40) | 0.561 | 0.79 (0.56-1.10) | 0.157 |
| PARP1 | rs2666428 | T | C | 185 | 111 | 11 | 911 | 492 | 42 | 1.12 (0.87-1.45) | 0.365 | 1.22 (0.62-2.41) | 0.560 |
| PARP1 | rs8679 | A | G | 272 | 33 | 2 | 1279 | 162 | 4 | 0.99 (0.67-1.46) | 0.971 | 2.38 (0.43-13.07) | 0.318 |
| hOGG1 | rs1052133 | G | C | 103 | 153 | 55 | 449 | 749 | 247 | 0.91 (0.70-1.18) | 0.484 | 1.05 (0.76-1.44) | 0.789 |
| hOGG1 | rs159153 | T | C | 254 | 52 | 5 | 1139 | 292 | 14 | 0.84 (0.61-1.14) | 0.263 | 1.68 (0.60-4.71) | 0.323 |
| hOGG1 | rs293795 | A | G | 273 | 34 | 4 | 1289 | 151 | 5 | 1.14 (0.78-1.67) | 0.492 | 3.78 (1.01-14.17) | 0.049 |
| FEN1 | rs174538 | A | G | 95 | 142 | 76 | 491 | 635 | 319 | 1.18 (0.90-1.53) | 0.230 | 1.13 (0.85-1.51) | 0.402 |
| FEN1 | rs4246215 | T | G | 94 | 142 | 77 | 490 | 641 | 314 | 1.19 (0.91-1.55) | 0.198 | 1.17 (0.88-1.56) | 0.271 |
| APEX1 | rs1130409 | T | G | 111 | 147 | 54 | 481 | 704 | 260 | 0.90 (0.70-1.16) | 0.416 | 0.96 (0.69-1.32) | 0.784 |
| APEX1 | rs1760944 | T | G | 110 | 156 | 46 | 487 | 687 | 271 | 0.93 (0.72-1.20) | 0.582 | 0.75 (0.53-1.05) | 0.097 |
| APEX1 | rs3136817 | T | C | 251 | 57 | 4 | 1176 | 253 | 16 | 1.06 (0.78-1.45) | 0.701 | 1.18 (0.39-3.55) | 0.773 |
| LIG3 | rs1052536 | C | T | 147 | 127 | 36 | 680 | 634 | 132 | 0.98 (0.77-1.26) | 0.897 | 1.31 (0.88-1.93) | 0.182 |
| LIG3 | rs3744356 | C | T | 301 | 7 | 2 | 1398 | 48 | 0 | 0.88 (0.43-1.81) | 0.729 | / | / |
| LIG3 | rs4796030 | A | C | 92 | 142 | 76 | 426 | 720 | 300 | 0.99 (0.75-1.29) | 0.922 | 1.25 (0.93-1.66) | 0.137 |
| XRCC1 | rs1799782 | G | A | 143 | 140 | 27 | 710 | 611 | 125 | 1.13 (0.88-1.44) | 0.347 | 1.00 (0.65-1.55) | 0.997 |
| XRCC1 | rs25487 | C | T | 169 | 116 | 25 | 803 | 529 | 114 | 1.04 (0.82-1.34) | 0.732 | 1.02 (0.65-1.60) | 0.939 |
| XRCC1 | rs25489 | C | T | 253 | 51 | 6 | 1169 | 250 | 27 | 0.95 (0.69-1.30) | 0.753 | 1.07 (0.44-2.62) | 0.886 |
| XRCC1 | rs2682585 | G | A | 234 | 69 | 7 | 1126 | 292 | 28 | 1.15 (0.87-1.54) | 0.328 | 1.19 (0.52-2.76) | 0.681 |
| XRCC1 | rs3810378 | G | C | 165 | 122 | 23 | 777 | 541 | 128 | 1.02 (0.80-1.30) | 0.878 | 0.82 (0.51-1.30) | 0.387 |
| XRCC1 | rs915927 | T | C | 236 | 72 | 2 | 1125 | 297 | 24 | 1.11 (0.83-1.49) | 0.470 | 0.39 (0.09-1.65) | 0.200 |
AOR, adjusted odds ratio; CI, confidence interval, HWE, Hardy-Weinberg equilibrium.
Adjusted for age and sex for dominant model (MM/WM vs. WW).
Adjusted for age and sex for recessive model (MM vs. WW/WM).
Figure 1.
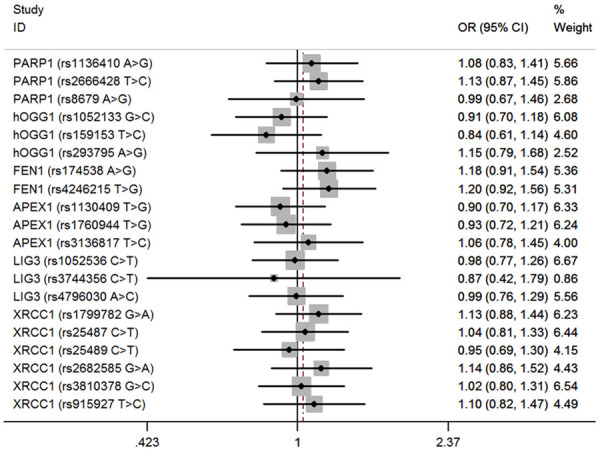
Forest plot for the association between BER gene SNPs and hepatoblastoma susceptibility under the recessive model (MM vs. WW/WM). For each SNP, the estimates of OR and its 95% CI are plotted with a box and a horizontal line.
Figure 2.
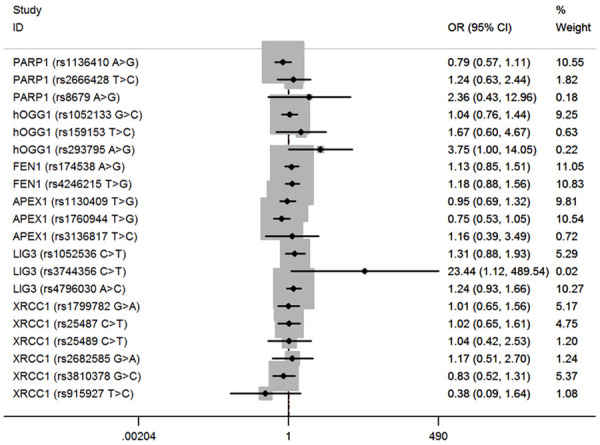
Forest plot for the association between BER gene SNPs and hepatoblastoma susceptibility under the dominant model (MM/WM vs. WW). For each SNP, the estimates of OR and its 95% CI are plotted with a box and a horizontal line.
Stratification analysis
We next carried out stratification analysis in the strata of age, gender, and clinical stage (Table 3). Regarding SNP rs159153, TC/CC genotype was significantly associated with decreased hepatoblastoma risk in male (adjusted OR=0.60, 95% CI=0.39-0.93, P=0.022). Compared with the AA/AG genotype, the rs293795 GG genotype increased hepatoblastoma risk in children with clinical stages I+II tumor (adjusted OR=5.67, 95% CI=1.34-24.05, P=0.019). We further set rs1052133 GG, rs159153 CC, and rs293795 AG/GG genotypes as risk genotypes. After combining the risk genotypes, we observed that patients with 1-3 risk genotypes were more likely to develop hepatoblastoma in male (adjusted OR=1.39, 95% CI=1.01-1.92, P=0.045).
Table 3.
Stratification analysis for the association between hOGG1 genotypes and hepatoblastoma susceptibility in Chinese children
| Variables | rs159153 (case/control) | AOR (95% CI)a | P a | rs293795 (case/control) | AOR (95% CI)a | P a | Risk genotypesb (case/control) | AOR (95% CI)a | P a | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||
| TT | TC/CC | AA/AG | GG | 0 | 1-3 | |||||||
| Age, month | ||||||||||||
| <17 | 135/516 | 31/126 | 0.93 (0.60-1.44) | 0.739 | 164/641 | 2/1 | 7.62 (0.69-84.76) | 0.099 | 87/372 | 79/270 | 1.28 (0.91-1.81) | 0.160 |
| ≥17 | 119/623 | 26/180 | 0.76 (0.48-1.19) | 0.231 | 143/799 | 2/4 | 2.88 (0.52-15.89) | 0.226 | 83/469 | 62/334 | 1.06 (0.74-1.51) | 0.764 |
| Sex | ||||||||||||
| Female | 99/484 | 29/110 | 1.29 (0.81-2.05) | 0.282 | 127/593 | 1/1 | 4.67 (0.29-75.10) | 0.277 | 75/332 | 53/262 | 0.89 (0.60-1.32) | 0.563 |
| Male | 155/655 | 28/196 | 0.60 (0.39-0.93) | 0.022 | 180/847 | 3/4 | 3.57 (0.79-16.13) | 0.098 | 95/509 | 88/342 | 1.39 (1.01-1.92) | 0.045 |
| Clinical stages | ||||||||||||
| I+II | 134/1139 | 26/306 | 0.73 (0.47-1.13) | 0.154 | 157/1440 | 3/5 | 5.67 (1.34-24.05) | 0.019 | 92/841 | 68/604 | 1.02 (0.73-1.42) | 0.902 |
| III+IV | 76/1139 | 14/306 | 0.68 (0.38-1.22) | 0.197 | 90/1440 | 0/5 | / | / | 46/841 | 44/604 | 1.34 (0.87-2.05) | 0.180 |
AOR, adjusted odds ratio; CI, confidence interval.
Adjusted for age and sex, omitting the corresponding stratify factor.
Risk genotypes were carriers with rs1052133 GG, rs159153 CC and rs293795 AG/GG genotypes.
False-positive report probability (FPRP) analysis
FPRP analysis was conducted to confirm the significant findings (Table 4). The threshold for FPRP was preset as 0.2. At the prior probability level of 0.25, findings for male in rs159153 TC/CC vs. TT and male in risk genotypes 1-3 vs. 0 remained noteworthy.
Table 4.
False-positive report probability analysis for significant findings
| Genotype | OR (95% CI) | P a | Statistical powerb | Prior probability | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||||
| rs293795 A>G | ||||||||
| GG vs. AA/GA | 3.752 (1.002-14.055) | 0.0497 | 0.092 | 0.619 | 0.830 | 0.982 | 0.998 | 1.000 |
| Stage I/II | 5.503 (1.303-23.246) | 0.0204 | 0.046 | 0.570 | 0.799 | 0.978 | 0.998 | 1.000 |
| rs159153 TC/CC vs. TT | ||||||||
| Male | 0.604 (0.392-0.931) | 0.0223 | 0.330 | 0.169 | 0.378 | 0.870 | 0.985 | 0.999 |
| Risk genotypes 1-3 vs. 0 | ||||||||
| Male | 1.379 (1.000-1.900) | 0.0497 | 0.699 | 0.176 | 0.390 | 0.876 | 0.986 | 0.999 |
OR, odds ratio; CI, confidence interval.
Chi-square test was used to calculate the genotype frequency distributions.
Statistical power was calculated using the number of observations in each subgroup and the corresponding ORs and P values in this table.
Discussion
The current knowledge of genetic predisposition to hepatoblastoma is incomplete. Challenge remains to fully unearth the full spectrum of hepatoblastoma susceptibility variations. In this study, we set as a pioneer to comprehensively genotype 20 SNPs of the critical genes in BER pathway. We here obtained a significant hepatoblastoma risk-associated SNP rs293795 of the hOGG1 gene. The findings of our research may contribute to the identification of individuals susceptible to hepatoblastoma for tailored early detection or other preventive interventions.
Intensive investigations have been performed regarding the impact of BER pathway gene SNPs on susceptibility of cancer. Using the Spanish sample, Jonine D. Figueroa et al. [39] comprehensively determined the relationship between 43 candidate SNPs in 12 BER genes (XRCC1, hOGG1, LIG1, MUTYH, PARP1, PARP3, PARP4, POLB, APEX1, POLD1, PCNA, and LIG3) and the risk of bladder cancer. They detected that POLB rs3136717 and PARP1 rs1136410 significantly predispose to bladder cancer, whereas hOGG1 rs125701 protects from getting bladder cancer. Our research team also focuses on the role of BER gene polymorphisms on cancer risk. We have observed significant associations between hOGG1 rs1052133, FEN1 rs4246215, FEN1 rs174538 polymorphisms and susceptibility of Wilms tumor in the Chinese population [30]. More recently, of the 20 SNPs in BER pathway genes genotyped, only FEN1 gene rs174538 could impact the risk of neuroblastoma [36]. So far, the role of BER pathway gene SNPs in hepatoblastoma has not yet been illustrated. Given the specific role of BER pathway gene SNPs in specific cancer, it is necessary to carry out another study regarding the hepatoblastoma.
The current analysis revealed that among the 20 SNPs analyzed, only hOGG1 gene rs293795 significantly predisposed to hepatoblastoma. Further stratification analysis did reveal some significant relationships among hOGG1 gene SNPs with hepatoblastoma risk under some subgroups. FPRP analysis further validated the strength of the significant findings. The current negative results were plausible as most of the SNPs are only with small to moderate impact on the risk of cancer. hOGG1 is a multifunctional DNA glycosylase that plays a major role in the repair of DNA oxidative damage [40]. hOGG1 could specifically recognize the 8-OH-dG damage and then efficiently catalyze and excise the damage [41]. hOGG1 gene is located on chromosome 3p25 and consists of eight exons. hOGG1 is a polygenetic gene that has been reported to be greatly involved in multiple cancers [42,43]. Moreover, SNPs in hOGG1 gene are also reported in cancer etiology. Mohammed Alanazi et al. [44] found that hOGG1 gene rs293795 did not show any association with breast cancer. Qin et al. [45] also failed to detect a relationship between hOGG1 gene rs293795 and risk of non-small cell lung cancer. A similar negative result was also obtained in our previous study regarding neuroblastoma [46]. The conflicting role of the same rs293795 on different cancers indicating that the same SNP may exert a different role in different cancers.
Our study has weaknesses that should be considered. One limitation is the possibility of selection bias of subjects, as all the subjects were hospital based. Another limitation is the lack of incorporation of genetic-environmental interaction analysis, as hepatoblastoma is a complex disease not just caused by genetic aberrance. Moreover, cautions should be taken when interpreting the conclusion here to other ethnicities, since only Chinese population was analyzed. What’s more, though as a multi-center study with moderate sample size, for subgroups the sample size is still limited. Statistical conclusion of these stratification analyses will be impaired to some extent at the present time. Of note, the exact functional role and molecule mechanisms of hOGG1 gene in hepatoblastoma await to be explored.
Conclusion
In summary, the current study was the first case-control investigation reporting the role of BER pathway gene SNPs on risk of hepatoblastoma in Chinese ancestry children. Our findings provide suggestions of hOGG1 genetic association for hepatoblastoma in Chinese ancestry children. Further genetic studies leveraging larger sample sizes are warranted to refine this association and reveal the underlying biology of hepatoblastoma.
Acknowledgements
This study was funded by grants from the Special Financial Grant from the China Postdoctoral Science Foundation (No. 2020T130132), National Natural Science Foundation of China (No: 81560262, 81960294) and Guangdong Provincial Key Laboratory of Research in Structural Birth Defect Disease (No: 2019B030301004).
Disclosure of conflict of interest
None.
References
- 1.Sharma D, Subbarao G, Saxena R. Hepatoblastoma. Semin Diagn Pathol. 2017;34:192–200. doi: 10.1053/j.semdp.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Haas JE, Muczynski KA, Krailo M, Ablin A, Land V, Vietti TJ, Hammond GD. Histopathology and prognosis in childhood hepatoblastoma and hepatocarcinoma. Cancer. 1989;64:1082–1095. doi: 10.1002/1097-0142(19890901)64:5<1082::aid-cncr2820640520>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 3.Ranganathan S, Lopez-Terrada D, Alaggio R. Hepatoblastoma and pediatric hepatocellular carcinoma: an update. Pediatr Dev Pathol. 2020;23:79–95. doi: 10.1177/1093526619875228. [DOI] [PubMed] [Google Scholar]
- 4.Feng J, Polychronidis G, Heger U, Frongia G, Mehrabi A, Hoffmann K. Incidence trends and survival prediction of hepatoblastoma in children: a population-based study. Cancer Commun (Lond) 2019;39:62. doi: 10.1186/s40880-019-0411-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spector LG, Birch J. The epidemiology of hepatoblastoma. Pediatr Blood Cancer. 2012;59:776–779. doi: 10.1002/pbc.24215. [DOI] [PubMed] [Google Scholar]
- 6.Meyers RL, Maibach R, Hiyama E, Haberle B, Krailo M, Rangaswami A, Aronson DC, Malogolowkin MH, Perilongo G, von Schweinitz D, Ansari M, Lopez-Terrada D, Tanaka Y, Alaggio R, Leuschner I, Hishiki T, Schmid I, Watanabe K, Yoshimura K, Feng Y, Rinaldi E, Saraceno D, Derosa M, Czauderna P. Risk-stratified staging in paediatric hepatoblastoma: a unified analysis from the children’s hepatic tumors international collaboration. Lancet Oncol. 2017;18:122–131. doi: 10.1016/S1470-2045(16)30598-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang T, Whitlock RS, Vasudevan SA. Surgical management of hepatoblastoma and recent advances. Cancers (Basel) 2019;11:1944. doi: 10.3390/cancers11121944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YT, Chang J, Yao YM, Li YN, Zhong XD, Liu ZL. Novel treatment of refractory/recurrent pulmonary hepatoblastoma. Pediatr Int. 2019;62:324–329. doi: 10.1111/ped.14134. [DOI] [PubMed] [Google Scholar]
- 9.Almstedt E, Elgendy R, Hekmati N, Rosen E, Warn C, Olsen TK, Dyberg C, Doroszko M, Larsson I, Sundstrom A, Arsenian Henriksson M, Pahlman S, Bexell D, Vanlandewijck M, Kogner P, Jornsten R, Krona C, Nelander S. Integrative discovery of treatments for high-risk neuroblastoma. Nat Commun. 2020;11:71. doi: 10.1038/s41467-019-13817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czauderna P, Lopez-Terrada D, Hiyama E, Haberle B, Malogolowkin MH, Meyers RL. Hepatoblastoma state of the art: pathology, genetics, risk stratification, and chemotherapy. Curr Opin Pediatr. 2014;26:19–28. doi: 10.1097/MOP.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 11.Wiwanitkit V. Hepatitis virus B is not a risk factor in hepatoblastoma patients. Asian Pac J Cancer Prev. 2005;6:213–214. [PubMed] [Google Scholar]
- 12.Hadzic N, Cho SJ, Finegold MJ. Hepatoblastoma surveillance in infants born with very low birth weight: has the time come? J Pediatr. 2020;216:248–249. doi: 10.1016/j.jpeds.2019.09.020. [DOI] [PubMed] [Google Scholar]
- 13.Wang TY, Han YL, Gao YJ, Xu M, Gu S, Yin MZ, Zhong YM, Hu WT, Pan C, Tang JY. Retrospective analysis of childhood hepatoblastoma in a single centre in China. Clin Oncol (R Coll Radiol) 2019;31:471–478. doi: 10.1016/j.clon.2019.03.044. [DOI] [PubMed] [Google Scholar]
- 14.Sorahan T, Lancashire RJ. Parental cigarette smoking and childhood risks of hepatoblastoma: OSCC data. Br J Cancer. 2004;90:1016–1018. doi: 10.1038/sj.bjc.6601651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baynam GS, Goldblatt J. A child with an FGFR3 mutation, a laterality disorder and an hepatoblastoma: novel associations and possible gene-environment interactions. Twin Res Hum Genet. 2010;13:297–300. doi: 10.1375/twin.13.4.297. [DOI] [PubMed] [Google Scholar]
- 16.Oue T, Kubota A, Okuyama H, Kawahara H, Nara K, Kawa K, Kitajima H. Hepatoblastoma in children of extremely low birth weight: a report from a single perinatal center. J Pediatr Surg. 2003;38:134–137. doi: 10.1053/jpsu.2003.50027. [DOI] [PubMed] [Google Scholar]
- 17.Valentin LI, Perez L, Masand P. Hepatoblastoma associated with trisomy 18. J Pediatr Genet. 2015;4:204–206. doi: 10.1055/s-0035-1565265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pakakasama S, Chen TT, Frawley W, Muller C, Douglass EC, Tomlinson GE. Myeloperoxidase promotor polymorphism and risk of hepatoblastoma. Int J Cancer. 2003;106:205–207. doi: 10.1002/ijc.11191. [DOI] [PubMed] [Google Scholar]
- 19.Pakakasama S, Chen TT, Frawley W, Muller CY, Douglass EC, Lee R, Pollock BH, Tomlinson GE. CCND1 polymorphism and age of onset of hepatoblastoma. Oncogene. 2004;23:4789–4792. doi: 10.1038/sj.onc.1207499. [DOI] [PubMed] [Google Scholar]
- 20.Yang T, Li J, Wen Y, Tan T, Yang J, Pan J, Hu C, Yao Y, Zhang J, Xin Y, Li S, Xia H, He J, Zou Y. LINC00673 rs11655237 C>T polymorphism impacts hepatoblastoma susceptibility in Chinese children. Front Genet. 2019;10:506. doi: 10.3389/fgene.2019.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li L, Zhuo Z, Yang Z, Zhu J, He X, Yang Z, Zhang J, Xin Y, He J, Zhang T. HMGA2 polymorphisms and hepatoblastoma susceptibility: a five-center case-control study. Pharmgenomics Pers Med. 2020;13:51–57. doi: 10.2147/PGPM.S241100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Deng Y, Zhang K, Bai Y, Zhu J, Zhang J, Xin Y, Li L, He J, Wang W. LIN28B gene polymorphisms modify hepatoblastoma susceptibility in Chinese children. J Cancer. 2020;11:3512–3518. doi: 10.7150/jca.42798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andor N, Maley CC, Ji HP. Genomic instability in cancer: teetering on the limit of tolerance. Cancer Res. 2017;77:2179–2185. doi: 10.1158/0008-5472.CAN-16-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 27.Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen. 2017;58:235–263. doi: 10.1002/em.22087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst) 2014;19:14–26. doi: 10.1016/j.dnarep.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsden CG, Dragon JA, Wallace SS, Sweasy JB. Base excision repair variants in cancer. Methods Enzymol. 2017;591:119–157. doi: 10.1016/bs.mie.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Jia W, Wu C, Fu W, Xia H, Liu G, He J. Base excision repair gene polymorphisms and wilms tumor susceptibility. EBioMedicine. 2018;33:88–93. doi: 10.1016/j.ebiom.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokhansanj BA, Wilson DM 3rd. Estimating the effect of human base excision repair protein variants on the repair of oxidative DNA base damage. Cancer Epidemiol Biomarkers Prev. 2006;15:1000–1008. doi: 10.1158/1055-9965.EPI-05-0817. [DOI] [PubMed] [Google Scholar]
- 32.Tudek B. Base excision repair modulation as a risk factor for human cancers. Mol Aspects Med. 2007;28:258–275. doi: 10.1016/j.mam.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Chen H, Li Y, Li L, Zhu J, Yang Z, Zhang J, Li S, Xin Y, Xia H, He J. YTHDC1 gene polymorphisms and hepatoblastoma susceptibility in Chinese children: a seven-center case-control study. J Gene Med. 2020;22:e3249. doi: 10.1002/jgm.3249. [DOI] [PubMed] [Google Scholar]
- 34.Luo Z, Li G, Wang M, Zhu J, Yang Z, Li Y, Zhang J, Xin Y, Li S, Li L, Zhuo Z, He J. YTHDF1 rs6090311 A>G polymorphism reduces hepatoblastoma risk: evidence from a seven-center case-control study. J Cancer. 2020;11:5129–5134. doi: 10.7150/jca.46120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhuo ZJ, Hua RX, Chen Z, Zhu J, Wang M, Yang Z, Zhang J, Li Y, Li L, Li S, Xin Y, Xia H, He J. WTAP gene variants confer hepatoblastoma susceptibility: a seven-center case-control study. Mol Ther Oncolytics. 2020;18:118–125. doi: 10.1016/j.omto.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhuo Z, Zhou C, Fang Y, Zhu J, Lu H, Zhou H, Wu H, Wang Y, He J. Correlation between the genetic variants of base excision repair (BER) pathway genes and neuroblastoma susceptibility in eastern Chinese children. Cancer Commun (Lond) 2020;40:641–646. doi: 10.1002/cac2.12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhuo ZJ, Liu W, Zhang J, Zhu J, Zhang R, Tang J, Yang T, Zou Y, He J, Xia H. Functional polymorphisms at ERCC1/XPF genes confer neuroblastoma risk in Chinese children. EBioMedicine. 2018;30:113–119. doi: 10.1016/j.ebiom.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu P, Zhuo ZJ, Zhu J, Yang Z, Xin Y, Li S, Li L, Li Y, Wang H, He J. Association of TP53 rs1042522 C>G and miR-34b/c rs4938723 T>C polymorphisms with hepatoblastoma susceptibility: a seven-center case-control study. J Gene Med. 2020;22:e3182. doi: 10.1002/jgm.3182. [DOI] [PubMed] [Google Scholar]
- 39.Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S, Welch R, Dosemeci M, Tardon A, Serra C, Carrato A, Garcia-Closas R, Castano-Vinyals G, Rothman N, Garcia-Closas M. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007;121:233–242. doi: 10.1007/s00439-006-0294-y. [DOI] [PubMed] [Google Scholar]
- 40.Shinmura K, Yokota J. The OGG1 gene encodes a repair enzyme for oxidatively damaged DNA and is involved in human carcinogenesis. Antioxid Redox Signal. 2001;3:597–609. doi: 10.1089/15230860152542952. [DOI] [PubMed] [Google Scholar]
- 41.Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuzefovych LV, Kahn AG, Schuler MA, Eide L, Arora R, Wilson GL, Tan M, Rachek LI. Mitochondrial DNA Repair through OGG1 activity attenuates breast cancer progression and metastasis. Cancer Res. 2016;76:30–34. doi: 10.1158/0008-5472.CAN-15-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kondo S, Toyokuni S, Tanaka T, Hiai H, Onodera H, Kasai H, Imamura M. Overexpression of the hOGG1 gene and high 8-hydroxy-2’-deoxyguanosine (8-OHdG) lyase activity in human colorectal carcinoma: regulation mechanism of the 8-OHdG level in DNA. Clin Cancer Res. 2000;6:1394–1400. [PubMed] [Google Scholar]
- 44.Alanazi M, Pathan AAK, Shaik JP, Alhadheq A, Khan Z, Khan W, Al Naeem A, Parine NR. The hOGG1 Ser326Cys gene polymorphism and breast cancer risk in saudi population. Pathol Oncol Res. 2017;23:525–535. doi: 10.1007/s12253-016-0146-6. [DOI] [PubMed] [Google Scholar]
- 45.Qin H, Zhu J, Zeng Y, Du W, Shen D, Lei Z, Qian Q, Huang JA, Liu Z. Aberrant promoter methylation of hOGG1 may be associated with increased risk of non-small cell lung cancer. Oncotarget. 2017;8:8330–8341. doi: 10.18632/oncotarget.14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YZ, Zhuo ZJ, Fang Y, Li L, Zhang J, He J, Wu XM. Functional polymorphisms in hOGG1 gene and neuroblastoma risk in Chinese children. J Cancer. 2018;9:4521–4526. doi: 10.7150/jca.27983. [DOI] [PMC free article] [PubMed] [Google Scholar]


