Abstract
Kawasaki disease (KD) is a paediatric vasculitis associated with coronary artery aneurysms (CAA). Genetic variants influencing susceptibility to KD have been previously identified, but no risk alleles have been validated that influence CAA formation. We conducted a genome-wide association study (GWAS) for CAA in KD patients of European descent with 200 cases and 276 controls. A second GWAS for susceptibility pooled KD cases with healthy paediatric controls from vaccine trials in the UK (n = 1609). Logistic regression mixed models were used for both GWASs. The susceptibility GWAS was meta-analysed with 400 KD cases and 6101 controls from a previous European GWAS, these results were further meta-analysed with Japanese GWASs at two putative loci. The CAA GWAS identified an intergenic region of chromosome 20q13 with multiple SNVs showing genome-wide significance. The risk allele of the most associated SNV (rs6017006) was present in 13% of cases and 4% of controls; in East Asian 1000 Genomes data, the allele was absent or rare. Susceptibility GWAS with meta-analysis with previously published European data identified two previously associated loci (ITPKC and FCGR2A). Further meta-analysis with Japanese GWAS summary data from the CASP3 and FAM167A genomic regions validated these loci in Europeans showing consistent effects of the top SNVs in both populations. We identified a novel locus for CAA in KD patients of European descent. The results suggest that different genes determine susceptibility to KD and development of CAA and future work should focus on the function of the intergenic region on chromosome 20q13.
Subject terms: Genetics research, Vasculitis, Aneurysm
Introduction
Kawasaki disease (KD) is now the leading cause of acquired heart disease in children in developed countries [1]. Although introduction of effective treatment with intravenous immunoglobulin (IVIG) reduces the incidence of coronary artery aneurysm (CAA), aneurysm formation continues to occur in a significant proportion of children. Recent reports suggest that using recommended Z score criteria for CAA, 10–30% of KD cases develop CAA [2–4]. The ongoing high rates of CAA, despite availability of treatment, have a number of possible explanations, including delayed diagnosis. However, most of the patients who develop CAA, including those who are diagnosed in the first 10 days of the illness, have evidence of coronary artery (CA) dilatation or CAA at initial presentation [3]. Others who go on to develop CAA may have inadequate response to IVIG and other agents and represent a group of patients with “resistant KD” who have a severe inflammatory process often poorly responsive to anti-inflammatory agents [5].
Attempts to identify patients at risk of CAA among patients presenting with KD have been only partially successful. Patients who have recurrent fever after IVIG (IVIG resistance) are known to have an increased risk of CAA [5]. However, clinical scoring systems that predict resistance to IVIG in Japan have been poorly predictive in other populations [6, 7]. Attempts to identify unique biomarkers that predict development of CAA have been largely unsuccessful [8, 9].
Genetic factors are now well established as playing a role in susceptibility to KD in both European descent and Asian populations [10]. Genes with validated associations with susceptibility to KD in multiple ethnic groups include FCGR2A and ITPKC [11, 12]. A genetic contribution to CAA seems likely as before effective therapy with IVIG was introduced, only 25–30% of affected children developed CAA [13].
A number of small genome-wide association studies (GWASs) and candidate gene studies have suggested a genetic contribution to CAA formation in KD but have not been validated [14–20]. Three GWASs for CAA have been published, but each had fewer than 50 cases and therefore very limited power [14–16]. In a GWAS comparing subjects who develop CAA with those who do not, we identified a new locus on chromosome 20 (Chr. 20) with genome-wide significance.
We further constructed a new KD susceptibility GWAS by pooling the newly genotyped KD cases and comparing with healthy controls. This cohort was meta-analysed with a previously reported European KD GWAS [11] and with publicly available GWAS summary data from Japanese KD susceptibility GWASs [12, 21].
Methods
Study design
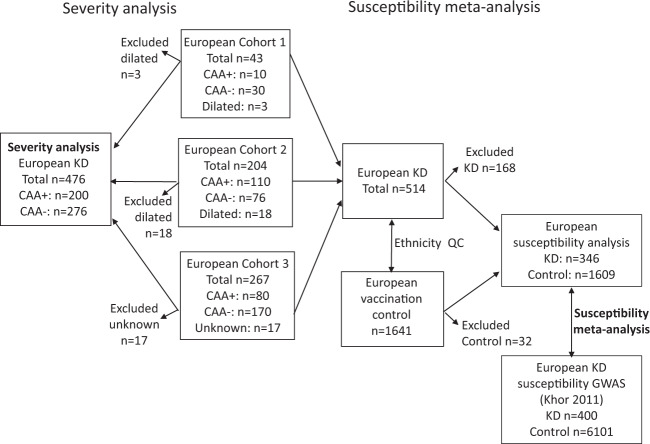
The overall study design is shown in Fig. 1.
Fig. 1. Study design.
Flow diagram describes cohorts used in the study. Cohort 1: US subjects; Cohort 2: US and Finnish subjects; Cohort 3: UK and Dutch subjects. We first performed a severity analysis comparing KD subjects who developed CAA with those who did not. We next performed a new susceptibility GWAS comparing KD subjects and healthy controls and meta-analyzed this with GWAS data from the previously reported KD GWAS of European descent subjects [11]. Arrows leaving boxes denote subjects deleted from cohorts following more stringent ethnicity quality control or deleted because of missing information on CA status. Subjects who were classified as having dilated CA were excluded from the severity analysis but included for the susceptibility analysis. CA coronary artery, CAA coronary artery aneurysm.
Participant selection
Subjects meeting the AHA KD diagnostic criteria who presented to paediatric centres in USA, UK, Holland, and Finland between 2001 and 2018 were included. CAA were identified by echocardiography at each participating centre. Subjects were categorized by echocardiography using CA Z scores (internal diameter of the right CA or left anterior descending artery normalized for body surface area) as having giant CAA (Z score ≥ 10 or absolute dimension ≥8 mm). For the US and Dutch cohorts, lesser CAA were defined as Z ≥ 2.5. Patients scored as dilated coronary arteries (Z score 2–2.5) were excluded from the CAA comparison but included in the susceptibility GWAS. In the UK, Z scores were not routinely used to assign aneurysm size prior to 2017. Therefore, CAA were described according to the Japanese Ministry of Health criteria based on absolute luminal diameters (Research Committee of the Japanese Society of Pediatric, 2014). The majority of UK patients were retrospectively recruited to the study either through local hospitals or through the UK KD Support Group, a family-based group.
Controls for the susceptibility analysis were 1609 healthy infants undergoing routine vaccination in the UK [22]. For validation and meta-analysis of the susceptibility GWAS, we used the genotype data from 400 KD cases and 6101 European controls described in Khor et al. [11], and summary data from KD susceptibility GWASs of individuals of Japanese ancestry [12, 21] at two putative loci.
Genotyping and statistical analysis
A detailed description of genotyping quality control (QC), imputation, statistical analysis, heritability estimation and Hi-C are in an Online Supplement Information. Briefly, after QC, multidimensional scaling (MDS) was applied (www.cog-genomics.org/plink/1.9/) to identify a genetically homogeneous group of European descent subjects for both severity and susceptibility GWAS. The genotype data were merged with the 1000 Genomes (1000G) [23] data and MDS was applied to determine the ancestry of the KD subjects. All data were imputed using the Haplotype Reference Consortium (HRC) reference panel (http://www.haplotype-reference-consortium.org) [24, 25]. To account for both ethnic diversity and cryptic relatedness, genome-wide association testing used logistic regression mixed models as implemented by the GMMAT software [26]. The top ten MDS components and two indicator variables for genotyping batch were included as fixed effects in both the CAA GWAS and new susceptibility GWAS. GWAS of the Khor KD susceptibility data used logistic regression in Plink2 (www.cog-genomics.org/plink/2.0/) with the top ten MDS components as covariates and was subsequently meta-analysed with the new GWAS. Loci were defined by ±250 kb of the top SNV in the region. Allelic heterogeneity was explored at each locus by conditional analyses in which analyses were rerun for all SNVs in loci, conditioning in the top SNV in the region.
The MAGMA software [27] was used for gene tests. Linkage disequilibrium (LD) score regression [28] and GCTA [29, 30] were used for heritability analyses. The functional impact of variants of interest were explored by querying eQTL associations in relevant tissues: specifically, whole blood, arteries and the heart using the GTEx database (https://www.gtexportal.org/home/) and the online FUMA tool (https://fuma.ctglab.nl/) [31] was used to query RegulomeDB and CADD scores.
Genome-wide summary statistics for the CAA GWAS and KD susceptibility meta-analysis are available to download from the EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/studies/GCST90013538 and https://www.ebi.ac.uk/gwas/studies/GCST90013537).
Results
Characteristics of the study population
The cohorts of CAA+ and CAA− subjects after QC and removal of non-European subjects are described in Table 1 and Fig. 1.
Table 1.
Description of KD cohorts with and without coronary artery aneurysms used in this analysis.
| Cohort | Source | Platform | CAA+: n (% male) | CAA−: n (% male) | Total: n (% male) | ||
|---|---|---|---|---|---|---|---|
| Small–medium CAA | Giant CAA | CAA+ subtotal | |||||
| 1 | US | Illumina 1.2 million SNV array | 10 (70) | 0 | 10 (70) | 30 (60) | 40 (63) |
| 2 | US | Illumina HumanCoreExome array customized | 63 (63) | 35 (60) | 98 (62) | 40 (78) | 138 (67) |
| Finland | 8 (63) | 4 (75) | 12 (67) | 36 (67) | 48 (67) | ||
| 3 | UK | Illumina HumanCoreExome array | 46 (70) | 1 (100) | 47 (70) | 122 (65) | 169 (66) |
| Holland | 21 (71) | 12 (92) | 33 (79) | 48 (46) | 81 (59) | ||
| Total | 148 (67) | 52 (69) | 200 (68) | 276 (63) | 476 (65) | ||
QC of the genomic data
Supplementary Fig. 1A shows the MDS plot of the selected subjects by cohort and CAA status. Despite the relative genetic heterogeneity, there was an even spread by CAA status. The individuals clustered to the right of this figure were recruited in Finland. Supplementary Fig. 1B shows the MDS plot of KD subjects pooled with 1000G subjects and indicates those KD subjects determined to be of European ancestry who were used for the GWAS.
Severity GWAS
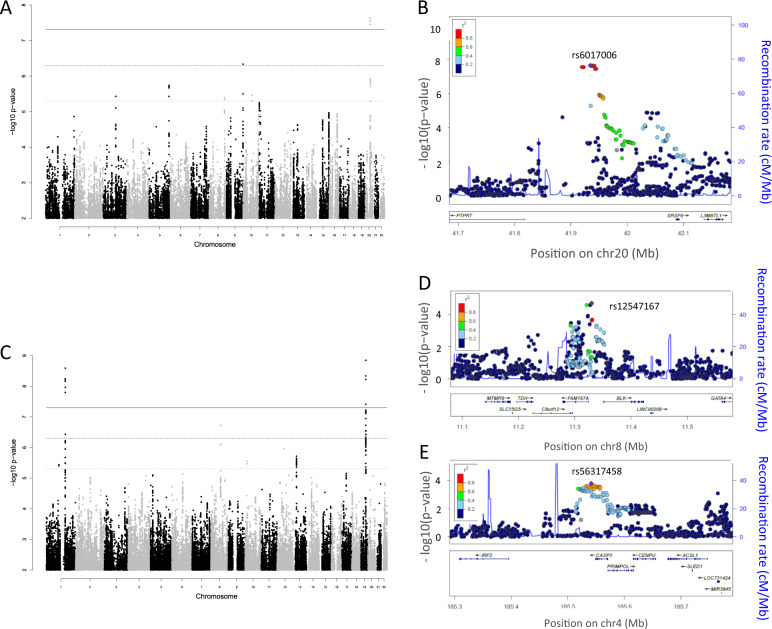
After QC, 4,873,589 genotyped and imputed SNVs remained for the severity GWAS (CAA+ vs CAA−) and the resultant Manhattan plot is shown in Fig. 2A. The variance inflation factor (1.001) indicated that population stratification had been adequately accounted for (Supplementary Fig. 2). A single region on Chr. 20 spanning 41.9198–41.9449 Mb was identified for which 15 SNVs showed genome-wide significant differences between KD subjects with and without CAA (p < 5 × 10−8) (Table 2 and Supplementary Table 1). The top associated SNV rs6017006 had a p value of 2.3 × 10−8 and odds ratio for the minor risk allele A (vs G) of 5.0 (95% CI 2.8–9.0). The regional association plot for this association is shown in Fig. 2B. The imputation quality score R2 for rs6017006 was 0.996, 0.923 and 0.954 in Cohorts 1, 2 and 3 (see Fig. 1 and Supplementary Methods), respectively. The frequency of the A allele of rs6017006 was 13 and 4% in subjects with and without CAA, respectively (Table 2). Amongst East Asian populations sampled by the 1000G project, the risk allele A is only observed in the Dai population of western China. In the other global populations covered by 1000G, the risk allele frequency is 1% in Africans, 5% in European descent, 3% in Americans and 3% in South Asians. The OR for giant CAA (giant CAA vs CAA−) was 4.0 (95% CI 1.5–10.7) comparable to the whole CAA+ group (see above). To ensure this association was not driven by population stratification arising from the cluster of Finnish samples, the analysis was rerun with these samples removed (11 cases, 35 controls), this gave a p value for rs6017006 of 3.5 × 10−9, OR = 6.1 (95% CI 3.2–11.6).
Fig. 2. Severity and susceptibility GWAS results.
A Manhattan plot for CAA GWAS (severity GWAS) showing genome-wide significant locus in intergenic region, Chr. 20 and five regions with association p < 5 × 10−6 (UNC5B, MAN1B1 and DOCK2 and two in intergenic regions). B Regional association plot of Chr. 20 intergenic SNV. C Manhattan plot for susceptibility GWAS showing confirmation of genome-wide significant SNVs in IPTKC and FCGR2A. D Regional association plots for previously published variants (FAM167A-BLK). E Regional plot of the CASP3 region associated with susceptibility. CAA coronary artery aneurysm.
Table 2.
Top variants in loci associated with CAA formation with p < 5 × 10−6.
| SNV | Chr | Positiona | Gene region | Allele | Alt. allele frequencies | ||||
|---|---|---|---|---|---|---|---|---|---|
| Ref | Alt | CAA (+) | CAA (−) | OR (95% CI) | p | ||||
| rs6017006 | 20 | 41934620 | Intergenic | G | A | 0.13 | 0.04 | 5.0 (2.8–9.0) | 2.3 × 10−8 |
| rs7871579 | 9 | 139990530 | MAN1B1 intron | T | C | 0.26 | 0.15 | 2.8 (1.8–4.2) | 4.6 × 10−7 |
| rs2449565 | 5 | 169135662 | DOCK2 intron | A | G | 0.75 | 0.61 | 2.3 (1.6–3.3) | 1.9 × 10−6 |
| rs10762437 | 10 | 73058469 | UNC5B intron | A | G | 0.25 | 0.16 | 2.5 (1.7–3.8) | 3.5 × 10−6 |
| rs35932034 | 3 | 106262305 | Intergenic | T | C | 0.03 | 0.11 | 0.2 (0.09–0.4) | 3.7 × 10−6 |
| rs1989051 | 8 | 129294118 | Intergenic | T | C | 0.69 | 0.57 | 2.2 (1.6–3.1) | 4.0 × 10−6 |
ORs were calculated using a Wald test, and p values were calculated using a Score test. All effects are relative to the alternate allele, ref/alt alleles are defined by the HRC reference panel.
Chr. chromosome, CAA coronary artery aneurysm, OR odds ratio.
aGRCh37/hg19 position.
In addition to this region on Chr. 20, we identified five other loci (top SNV) showing suggestive significance (p < 5 × 10−6). (Table 2 and Supplementary Fig. 3A–E). These included a Chr. 9 region containing MANB1, a Chr. 5 region containing DOCK2, a Chr. 10 region containing UNC5B and intergenic regions on Chr. 3 and 8. The regional association plots for the six loci indicated that allelic heterogeneity is unlikely, this was confirmed by conditional analyses, which resulted in no SNVs reaching suggestive significance.
Supplementary Table 2 shows the top 50 genes from the MAGMA test, the most significant gene was CDH3 with p = 8.3 × 10−6. The only SNV to show significant eQTL association (p < 5 × 10−5) was rs7871579 in MAN1B1, which is significantly associated with the expression of MAN1B1-AS1 in arteries (aorta, tibial), heart (coronary, left ventricle and atrial appendage) and whole blood. UAP1L1, SAPCD2, DPP7 and NPDC1 also had significant eQTL associations with rs7871579 (Supplementary Table 3). None of the other variants had an eQTL association at p < 5 × 10−5. Two intergenic SNVs on Chr. 20 and one intronic SNV on DOCK2, which were in LD with rs6017006 on Chr. 20 and rs2449565 on DOCK2, respectively, had high CADD deleterious scores. However, the RegulomeDB score and chromatin state for biological evidence to be a regulatory element were not strong for these SNVs (Supplementary Table 4).
Exploring the function of the Chr. 20 lead SNV
To explore how an intergenic region might affect the expression of genes involved in CAA formation, we analysed published data using chromosomal conformational capture (3C) methodology on a genome-wide scale (Hi-C analysis) [32]. Conformational capture methods identify genomic regions that interact with each other in three-dimensional space and that might affect gene function or expression of genes that are not in linear proximity with the region of interest. Hi-C did not identify any interactions within 1 Mb of the identified SNV. However, there were interactions identified at greater distance (Supplementary Table 5). While long range interactions are less likely to be reproducible, PLCB1, the most significant interaction, was implicated as a risk locus for CAA in a Taiwanese study [33].
Susceptibility GWAS
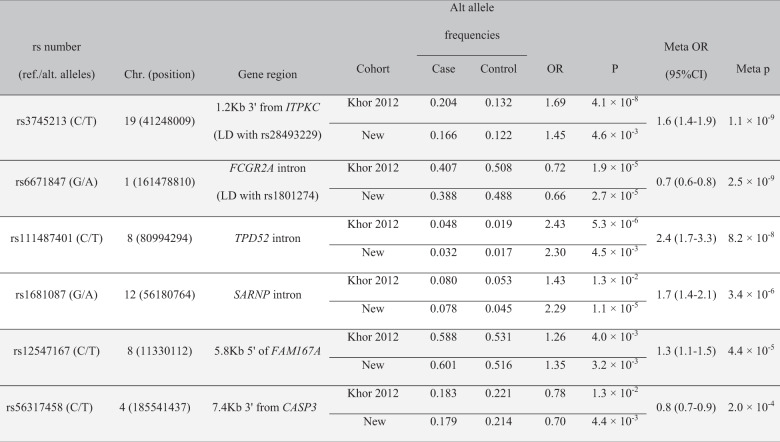
There were 5,784,964 genotyped and imputed SNVs passing QC in the new and Khor cohorts. The Manhattan plot for the KD susceptibility meta-analysis of these GWAS cohorts is shown in (Fig. 2C) and quantile–quantile plots for the individual GWASs and meta-analyses are shown in Supplementary Fig. 4A–C. The analysis identified two loci (top SNV ±250 kb) meeting genome-wide significance and two new loci with suggestive significance (p < 5 × 10−6). Table 3 shows results for the top SNV at each locus. Regional association plots for the loci in Table 3 are shown in Supplementary Fig. 5. Conditional analyses resulted in no SNVs reaching suggestive significance suggesting allelic heterogeneity was unlikely.
Table 3.
Variants associated with susceptibility to KD in order of descending significance.
All effects are relative to the alternate allele, ref/alt alleles are defined by the HRC reference panel. Shaded genes: previously reported in other ethnic groups. Highlighted variants validate previously published variants influencing susceptibility to KD in Asian and European descent cohorts.
New newly analysed European descent cohort in this paper, Chr. chromosome, position: GRCh37/hg19, Meta meta-analysis, OR odds ratio.
The most significant association in ITPKC (p = 1.1 × 10−9 for rs3745213) is in high LD with the previously reported variant, rs28493229, replicating findings from previous European and Asian genetic studies [11, 34, 35] (Supplementary Fig. 5A). The associated region in FCGR2A (p = 2.5 × 10−9 for rs6671847) is also in LD with rs1801274 (R2 = 0.89, D′ = 1) that has been associated with KD susceptibility in previous studies [11, 36] (Supplementary Fig. 5B).
The two additional SNVs showing suggestive evidence for association with KD susceptibility in our European meta-analysis were an intronic variant in TPD52 (p = 8.2 × 10−8, rs111487401) on Chr. 8 and an intronic variant in SARNP on Chr.12. (p = 3.4 × 10−6, rs1681087) (Supplementary Fig. 5C, D). Additional three SNVs showed suggestive evidence in the primary GWAS (score test followed by weighted z meta-analysis). However, these SNVs had p > 5 × 10−6 with Wald test and inverse-variance meta-analysis (Supplementary Table 6).
Supplementary Table 7 shows the top 50 genes associated with KD susceptibility from the MAGMA test using the meta-analysis results, the most significant gene was CACNA2D3 with p = 2.4 × 10−6.
There were no significant eQTL associations for the SNVs in the two novel loci: rs111487401 in TPD52 on Chr. 8 and rs1681087 in SARNP on Chr.12 (Supplementary Table 3). However, these two SNVs were in LD with SNVs that had RegulomeDB score and chromatin state with biological evidence to be a regulatory element but their CADD scores were low (Supplementary Table 4).
Analyses of previously associated loci
Querying of the NHGRI EBI GWAS Catalog for KD susceptibility associations identified eight SNVs (with non-ambiguous alleles) with a p value of <5 × 10−8 located in six loci (Supplementary Table 8). In addition to ITPKC and FCGR2A, two other loci showed nominal levels of significance in our European meta-analysis: rs2736340 in the BLK-FAM167A locus on Chr. 8 [37] (European meta p value = 3.3 × 10−3, GWAS of Chinese subjects p = 9 × 10−10) and rs2130392 in CASP3-CENPU locus on Chr. 4 [12] (European meta p value = 0.011, GWAS of Japanese subjects p = 3 × 10−8). The loci that did not replicate were CD40 on Chr. 20 (rs1569723 [37]) and HLA-DQB2, HLA-DOB on Chr. 6 (rs2857151 [12]). Supplementary Table 9 shows our meta-analysis results for the other SNVs from the EBI GWAS Catalog with suggestive evidence for association with KD susceptibility.
Summary level data of CASP3 and BLK/FAM167A susceptibility loci were available from two GWASs of Japanese subjects [12, 21] and were meta-analysed with our European meta-analysis results. Results from meta-analysing with Japanese summary data from the CASP3 locus [12, 21] are shown in Supplementary Table 10. The most significantly associated SNV in the region was rs2720378 (European meta p = 2.5 × 10−3, Japanese p = 3.5 × 10−9, global meta p = 1.2 × 10−10, global meta OR = 0.741 (95% CI: 0.676, 0.812)). A test for heterogeneity of effects between studies showed no differences in effects between populations (p = 0.2). Regional association plots for the Japanese GWAS and the meta-analysis are shown in Supplementary Fig. 6A, B. The top SNV in the region in our study (rs56317458, Table 3 and Supplementary Fig. 5E) was not included in the Japanese study.
The results from meta-analysing with Japanese summary data from the FAM167A-BLK locus [12] are shown in Supplementary Table 11. The most significant SNV in the meta-analysis was rs35393613 (European meta p = 6.9 × 10−4, OR = 1.3, Japanese p = 3.6 × 10−12, OR = 1.8, global meta p = 5.2 × 10−13, global meta OR = 1.51 (95% CI: (1.35, 1.688)) [12]. There was evidence for a difference in effect sizes between populations (p = 0.02), this could be explained by differences in LD between rs35393613 and the causal variant in the European and Japanese populations. Regional association plots for the Japanese GWAS and the meta-analysis are shown in Supplementary Fig. 7A, B. The top SNV in our analysis was not included in the Japanese data (rs12547167, see Table 3 and Supplementary Fig. 5F).
Finally, in Supplementary Table 12, we report results from our study of SNVs reported in the first European GWAS for KD susceptibility [38]. None of the 37 overlapping SNPs were significant at p < 0.05.
Cross-phenotype associations
Supplementary Table 13 shows comparison between studies of SNVs associated in the susceptibility and severity GWASs. Only rs10762437 (UNC5B intron) shows nominal cross-phenotype association. The top SNV rs12547167 at the FAM167A locus did not pass QC in the CAA GWAS, the SNV reported has R2 = 0.6 with it. Results are not shown for the MAN1B or TPD52 as neither passed QC in the Khor GWAS.
Heritability
Estimates of the heritability of CAA risk gave inconclusive results with very wide confidence intervals using both GCTA and LD score. This is likely because of the relatively small sample size of this GWAS. Assuming the prevalence of KD in individuals of European descent to be 2 in 10,000 [3], LD score estimated heritability on the liability scale in our population of European descent to be 0.119 (95% CI: 0.046, 0.191). The heritability in other ethnic groups could be quite different because of differences in genetic and/or environmental contributions to KD susceptibility. Assuming the odds ratios and minor allele frequencies shown in Table 3, the explained heritability on the liability scale for the top SNVs in ITPKC, FCGR2A, FAM167A and CASP3 were 0.0026. 0.0033, 0.0019 and 0.0009, respectively, yielding a total of 0.008. These results suggest that there are many other causative variants contributing to risk of KD and that risk is likely the result of combined effect of multiple variants in each individual.
Discussion
Our new GWAS pooling cases from UK, Holland, USA and Finland identified a region of Chr. 20 significantly associated with CAA. Previous GWAS and candidate gene studies have implicated a number of genes or gene regions in CAA development but these reports have been in cohorts with relatively small numbers of CAA cases and therefore limited power to identify significant associations [14–20].
We have identified a novel association between CAA formation and a SNV rs6017006 located in an intergenic region of chromosome 20 which is in LD with rs6030760 (19 kb 5′ upstream of rs6017006, R2 = 1) that alters the binding motif for CCCTC-binding factors (CTCF). CTCF and cohesin generate the unknotted loop of DNA where enhancer–promoter interactions occur during cellular differentiation [39]. To understand the functional significance of this variant in the context of the 3D genome, we performed Hi-C analysis using published data in human B‐lymphoblastoid cell lines as they provide the deepest coverage [32]. This analysis identified an association with PLCB1, which has been implicated in CAA formation in a Taiwanese GWAS [17]. However, as the association is 35 Mb distant from the identified SNV, there is less confidence in the robustness of this association [40–42]. Further studies are needed to evaluate the biological mechanisms by which the associated region influences development of CAA. It is noteworthy that the Chr. 20 variant does not influence susceptibility to KD, only CAA. Furthermore, comparison of SNVs associated with CAA and those associated with susceptibility showed no cross-phenotype associations, suggesting that children who develop CAA are a genetic subset of those who are susceptible.
Suggestive association was found between risk of CAA and five other loci (p < 5 × 10−6). One of these, UNC5B, is expressed on endothelial progenitor cells and is a receptor for netrin 4 with potential biological relevance to KD and CAA. Expression of UNC5B was shown to be essential for netrin 4-mediated neovascularization following ischaemic injury [43]. Another variant at rs7871579 is located in an intron of MAN1B1 (mannosidase alpha class 1B member 1), 9 Kb 5′ of MAN1B1-AS1 and 11.5 Kb 3′ of UAP1L1. This locus is reported as a quantitative trait locus (eQTL) with MAN1B1-AS1 in multiple tissues/cells including blood, heart, CA and fibroblasts (p = 2.3 × 10−6–5.3 × 10−21) and with UAP1L1 in blood (p = 5.1 × 10−17) (Supplementary Table 3). MAN1B1-AS1 is a long non-coding gene that may regulate expression of MAN1B1, which is the enzyme that removes the terminal mannose residue from the middle branch of N-glycans [44]. MAN1B1 also has a non-enzymatic function to prevent secretion of abnormally folded proteins [45]. Therefore, patients with MAN1B1 deficiency have accumulation of hybrid-type glycans and some high mannose structures in serum IgG and other glycoproteins [46]. Previous data indicate that endogenous IgG molecules from KD patients have more fucose, galactose and sialylated N-glycans, compared to healthy children [47]. UAP1L1 is also a glycosylation enzyme [48] and may influence T-cell development [49]. The variant on Chr. 5 (rs2449565) is located in an intron of DOCK2 that influences activation, migration and proliferation of T cells, B cells, NK cells, dendritic cells and neutrophils [50]. A PheWAS using the GWAS Atlas (https://atlas.ctglab.nl) of the SNVs associated with CAA development and susceptibility (Tables 2 and 3) revealed no cross-phenotype associations at p < 5 × 10−5 except rs6671847 in FCGR2A, which was in LD with the previously reported exonic non-synonymous SNV rs1801274 (Supplementary Table 15).
Our new KD susceptibility GWAS and meta-analysis with published GWASs have confirmed associations with ITPKC and FCGR2A variants with KD susceptibility [11, 12]. Nominal association was also observed for previously published variants in regions near FAM167A-BLK (p = 4.4 × 10−5) and CASP3 (p = 2.0 × 10−4). Meta-analysis with published data from Japan at these loci showed significance for the same SNVs across European and Japanese populations with consistent effects observed in both populations. The multi-ethnic meta-analyses of the CASP3 and FAM167A-BLK loci of Asian and European populations narrowed the association peaks and hence have better localized the causal variants in these regions. For both loci, the top SNV in the analysis of the European data was not included in the Japanese data. However, the data support the hypothesis that the same causal variants at these loci are acting in both populations. In contrast, we could not replicate the genome-wide significant associations observed at the CD40 and HLA-DQB2-HLA-DOB loci in East Asian populations in our European data.
Two new susceptibility loci with p < 5 × 10−6 were identified in our meta-analysis. The first, rs111487401, is in TPD52, which is highly expressed in mature B cells and binds to annexin V1 in a calcium-dependent manner [51]. This gene may be involved in the secretory functions of mature plasma cells. Polyclonal B cell activation is a prominent feature of KD and secretion of immunoglobulin molecules may be influenced by this protein. The second nominally associated variant, SARNP, is expressed in the heart and is involved in mRNA splicing and export. It is induced by thrombopoietin [52], which is highly expressed in acute KD [53]. These variants will require further replication.
The finding of an intergenic region on Chr. 20 that is significantly associated with development of CAA, but has no apparent influence on susceptibility, supports the hypothesis that different genes determine susceptibility to KD and development of CAA. There are likely other variants that contribute to CAA that can only be studied by comparing KD subjects with and without CAA. The functional significance of the intergenic locus on Chr. 20 should be a focus of future studies.
Limitations to our study include the lack of coverage of the X and Y chromosomes. The fact that males develop aneurysms more often than females suggests that risk alleles may reside on the sex chromosomes and should be investigated in future studies. This study focused only on risk alleles for CAA in a European descent cohort and future studies should address other ethnic groups. Although this is the largest GWAS focused on risk of CAA, the sample size was smaller than that in many GWAS. Given that only 14% of the subjects with CAA carry the risk allele on Chr. 20, it is clear that other loci that influence CAA remain to be discovered.
Finally, our findings may have relevance for the newly emerged childhood inflammatory multisystem syndrome temporally associated with exposure to SARS-CoV-2 (PIMS-TS, also called MIS-C) [54]. In countries experiencing large numbers of patients with COVID-19, there has been an increase in children meeting diagnostic criteria for KD, of whom 10–20% develop CAA [55, 56]. The genes and regions identified in our study as conferring risk of KD and CAA warrant exploration in the patients with PIMS-TS.
In summary, we identified a novel locus for CAA in KD patients using a robust dataset of European descent subjects. Comparing our pooled KD subjects with healthy controls confirmed several previously identified loci that contribute to KD susceptibility, but demonstrated that the CAA locus was not implicated in host susceptibility. These results suggest that different genes determine susceptibility to KD and development of CAA and future work should focus on the function of the intergenic region on chromosome 20q13.
Supplementary information
Acknowledgements
Recruitment from the UK Kawasaki disease family support group was greatly assisted by Sue Davidson and at St Mary’s Hospital by Sobia Mustafa. The authors would like to thank all the parents and children who participated in the study.
Funding
This work was supported in part by a grant from the Gordon and Marilyn Macklin Foundation to Burns and a grant from the Kawasaki Disease Foundation to Burns, the UK National Institute for Health Research Imperial Biomedical Research Centre at Imperial College (WMNP_P69099 [JAH and ML]), the Javon Charitable Trust (ML) and the Children of St Mary’s Intensive Care Kawasaki Disease Research Fund (JAH and ML). This project has received funding from the European Union’s Seventh Framework programme under grant agreement no. 279185 (EUCLIDS), which funded the genotyping of the vaccine cohort, collected in the Oxford Vaccine Centre Biobank.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Ethics approval
Recruitment of subjects at each participating centre was with parental consent and child assent as appropriate. The study was approved by the local research ethics board for each country and participating centre. (UCSD: IRB # 170790; Ethics Board of the Amsterdam University Medical Center: NL41023.018.12; Coordinating ethics committee of Helsinki University Hospital, #447/EO/2006; UK patients: EUCLIDS 11/LO/1982; UK Kawasaki Genetics 13/LO/0026; IRIS 09/H0712/58.)
Footnotes
Members of the The International Kawasaki Disease Genetics Consortium, UK Kawasaki Disease Genetics Consortium and EUCLIDS Consortium are listed in Supplementary Information.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Clive Hoggart, Chisato Shimizu, Jane C. Burns, Michael Levin
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-021-00838-5.
References
- 1.McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American Heart Association. Circulation. 2017;135:e927–99. doi: 10.1161/CIR.0000000000000484. [DOI] [PubMed] [Google Scholar]
- 2.Ogata S, Tremoulet AH, Sato Y, Ueda K, Shimizu C, Sun X, et al. Coronary artery outcomes among children with Kawasaki disease in the United States and Japan. Int J Cardiol. 2013;168:3825–8. doi: 10.1016/j.ijcard.2013.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skochko SM, Jain S, Sun X, Sivilay N, Kanegaye JT, Pancheri J, et al. Kawasaki disease outcomes and response to therapy in a multiethnic community: a 10-year experience. J Pediatr. 2018;203:408–15 e3. doi: 10.1016/j.jpeds.2018.07.090. [DOI] [PubMed] [Google Scholar]
- 4.Tulloh RMR, Mayon-White R, Harnden A, Ramanan AV, Tizard EJ, Shingadia D, et al. Kawasaki disease: a prospective population survey in the UK and Ireland from 2013 to 2015. Arch Dis Child. 2019;104:640–6. doi: 10.1136/archdischild-2018-315087. [DOI] [PubMed] [Google Scholar]
- 5.Tremoulet AH, Best BM, Song S, Wang S, Corinaldesi E, Eichenfield JR, et al. Resistance to intravenous immunoglobulin in children with Kawasaki disease. J Pediatr. 2008;153:117–21. doi: 10.1016/j.jpeds.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies S, Sutton N, Blackstock S, Gormley S, Hoggart CJ, Levin M, et al. Predicting IVIG resistance in UK Kawasaki disease. Arch Dis Child. 2015;100:366–8. doi: 10.1136/archdischild-2014-307397. [DOI] [PubMed] [Google Scholar]
- 7.Sleeper LA, Minich LL, McCrindle BM, Li JS, Mason W, Colan SD, et al. Evaluation of Kawasaki disease risk-scoring systems for intravenous immunoglobulin resistance. J Pediatr. 2011;158:831–5 e3. doi: 10.1016/j.jpeds.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adjagba PM, Desjardins L, Fournier A, Spigelblatt L, Montigny M, Dahdah N. N-terminal pro-brain natriuretic peptide in acute Kawasaki disease correlates with coronary artery involvement. Cardiol Young. 2015;25:1311–8. doi: 10.1017/S1047951114002431. [DOI] [PubMed] [Google Scholar]
- 9.Sato YZ, Molkara DP, Daniels LB, Tremoulet AH, Shimizu C, Kanegaye JT, et al. Cardiovascular biomarkers in acute Kawasaki disease. Int J Cardiol. 2013;164:58–63. doi: 10.1016/j.ijcard.2011.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Onouchi Y. The genetics of Kawasaki disease. Int J Rheum Dis. 2018;21:26–30. doi: 10.1111/1756-185X.13218. [DOI] [PubMed] [Google Scholar]
- 11.Khor CC, Davila S, Breunis WB, Lee YC, Shimizu C, Wright VJ, et al. Genome-wide association study identifies FCGR2A as a susceptibility locus for Kawasaki disease. Nat Genet. 2011;43:1241–6. doi: 10.1038/ng.981. [DOI] [PubMed] [Google Scholar]
- 12.Onouchi Y, Ozaki K, Burns JC, Shimizu C, Terai M, Hamada H, et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet. 2012;44:517–21. doi: 10.1038/ng.2220. [DOI] [PubMed] [Google Scholar]
- 13.Newburger JW, Takahashi M, Burns JC, Beiser AS, Chung KJ, Duffy CE, et al. The treatment of Kawasaki syndrome with intravenous gamma globulin. N Engl J Med. 1986;315:341–7. doi: 10.1056/NEJM198608073150601. [DOI] [PubMed] [Google Scholar]
- 14.Kim JJ, Park YM, Yoon D, Lee KY, Seob Song M, Doo, et al. Identification of KCNN2 as a susceptibility locus for coronary artery aneurysms in Kawasaki disease using genome-wide association analysis. J Hum Genet. 2013;58:521–5. doi: 10.1038/jhg.2013.43. [DOI] [PubMed] [Google Scholar]
- 15.Kuo HC, Li SC, Guo MM, Huang YH, Yu HR, Huang FC, et al. Genome-wide association study identifies novel susceptibility genes associated with coronary artery aneurysm formation in Kawasaki disease. PLoS ONE. 2016;11:e0154943. doi: 10.1371/journal.pone.0154943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon YC, Kim JJ, Yu JJ, Yun SW, Yoon KL, Lee KY, et al. Identification of the TIFAB gene as a susceptibility locus for coronary artery aneurysm in patients with Kawasaki disease. Pediatr Cardiol. 2019;40:483–8. doi: 10.1007/s00246-018-1992-7. [DOI] [PubMed] [Google Scholar]
- 17.Lin YJ, Chang JS, Liu X, Tsang H, Chien WK, Chen JH, et al. Genetic variants in PLCB4/PLCB1 as susceptibility loci for coronary artery aneurysm formation in Kawasaki disease in Han Chinese in Taiwan. Sci Rep. 2015;5:14762. doi: 10.1038/srep14762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onouchi Y, Onoue S, Tamari M, Wakui K, Fukushima Y, Yashiro M, et al. CD40 ligand gene and Kawasaki disease. Eur J Hum Genet. 2004;12:1062–8. doi: 10.1038/sj.ejhg.5201266. [DOI] [PubMed] [Google Scholar]
- 19.Shimizu C, Eleftherohorinou H, Wright VJ, Kim J, Alphonse MP, Perry JC, et al. Genetic variation in the SLC8A1 calcium signaling pathway is associated with susceptibility to Kawasaki disease and coronary artery abnormalities. Circ Cardiovasc Genet. 2016;9:559–68. doi: 10.1161/CIRCGENETICS.116.001533. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu C, Matsubara T, Onouchi Y, Jain S, Sun S, Nievergelt CM, et al. Matrix metalloproteinase haplotypes associated with coronary artery aneurysm formation in patients with Kawasaki disease. J Hum Genet. 2010;55:779–84. doi: 10.1038/jhg.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onouchi Y, Ozaki K, Buns JC, Shimizu C, Hamada H, Honda T, et al. Common variants in CASP3 confer susceptibility to Kawasaki disease. Hum Mol Genet. 2010;19:2898–906. doi: 10.1093/hmg/ddq176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Connor D, Png E, Khor CC, Snape MD, Hill AVS, van der Klis F, et al. Common genetic variations associated with the persistence of immunity following childhood immunization. Cell Rep. 2019;27:3241–53 e4. doi: 10.1016/j.celrep.2019.05.053. [DOI] [PubMed] [Google Scholar]
- 23.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM,, 1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S, Forer L, Schonherr S, Sidore C, Locke AE, Kwong A, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–7. doi: 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48:1279–83. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Wang C, Conomos MP, Stilp AM, Li Z, Sofer T, et al. Control for population structure and relatedness for binary traits in genetic association studies via logistic mixed models. Am J Hum Genet. 2016;98:653–66. doi: 10.1016/j.ajhg.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin YJ, Chang JS, Liu X, Lin TH, Huang SM, Liao CC, et al. Sorting nexin 24 genetic variation associates with coronary artery aneurysm severity in Kawasaki disease patients. Cell Biosci. 2013;3:44. doi: 10.1186/2045-3701-3-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuo HC, Yang KD, Juo SH, Liang CD, Chen WC, Wang YS, et al. ITPKC single nucleotide polymorphism associated with the Kawasaki disease in a Taiwanese population. PLoS ONE. 2011;6:e17370. doi: 10.1371/journal.pone.0017370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Onouchi Y, Gunji T, Burns JC, Shimizu C, Newburger JW, Yashiro M, et al. ITPKC functional polymorphism associated with Kawasaki disease susceptibility and formation of coronary artery aneurysms. Nat Genet. 2008;40:35–42. doi: 10.1038/ng.2007.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwon YC, Kim JJ, Yun SW, Yu JJ, Yoon KL, Lee KY, et al. Male-specific association of the FCGR2A His167Arg polymorphism with Kawasaki disease. PLoS ONE. 2017;12:e0184248. doi: 10.1371/journal.pone.0184248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YC, Kuo HC, Chang JS, Chang LY, Huang LM, Chen MR, et al. Two new susceptibility loci for Kawasaki disease identified through genome-wide association analysis. Nat Genet. 2012;44:522–5. doi: 10.1038/ng.2227. [DOI] [PubMed] [Google Scholar]
- 38.Burgner D, Davila S, Breunis WB, Ng SB, Li Y, Bonnard C, et al. A genome-wide association study identifies novel and functionally related susceptibility loci for Kawasaki disease. PLoS Genet. 2009;5:e1000319. doi: 10.1371/journal.pgen.1000319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Tessneer KL, Li C, Gaffney PM. From association to mechanism in complex disease genetics: the role of the 3D genome. Arthritis Res Ther. 2018;20:216. doi: 10.1186/s13075-018-1721-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Javierre BM, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167:1369–84 e19. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maass PG, Rump A, Schulz H, Stricker S, Schulze L, Platzer K, et al. A misplaced lncRNA causes brachydactyly in humans. J Clin Investig. 2012;122:3990–4002. doi: 10.1172/JCI65508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47:598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- 43.Lee NG, Jeung IC, Heo SC, Song J, Kim W, Hwang B, et al. Ischemia-induced Netrin-4 promotes neovascularization through endothelial progenitor cell activation via Unc-5 Netrin receptor B. FASEB J. 2020;34:1231–46. doi: 10.1096/fj.201900866RR. [DOI] [PubMed] [Google Scholar]
- 44.Saldova R, Shehni AA, Haakensen VD, Steinfeld I, Hilliard M, Kifer I, et al. Association of N-glycosylation with breast carcinoma and systemic features using high-resolution quantitative UPLC. J Proteome Res. 2014;13:2314–27. doi: 10.1021/pr401092y. [DOI] [PubMed] [Google Scholar]
- 45.Iannotti MJ, Figard L, Sokac AM, Sifers RN. A Golgi-localized mannosidase (MAN1B1) plays a non-enzymatic gatekeeper role in protein biosynthetic quality control. J Biol Chem. 2014;289:11844–58. doi: 10.1074/jbc.M114.552091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saldova R, Stockmann H, O’Flaherty R, Lefeber DJ, Jaeken J, Rudd PM. N-glycosylation of serum IgG and total glycoproteins in MAN1B1 deficiency. J Proteome Res. 2015;14:4402–12. doi: 10.1021/acs.jproteome.5b00709. [DOI] [PubMed] [Google Scholar]
- 47.Ogata S, Shimizu C, Franco A, Touma R, Kanegaye JT, Choudhury BP, et al. Treatment response in Kawasaki disease is associated with sialylation levels of endogenous but not therapeutic intravenous immunoglobulin G. PLoS ONE. 2013;8:e81448. doi: 10.1371/journal.pone.0081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai CY, Liu H, Tin KX, Huang Y, Yeh KH, Peng HW, et al. Identification of UAP1L1 as a critical factor for protein O-GlcNAcylation and cell proliferation in human hepatoma cells. Oncogene. 2019;38:317–31. doi: 10.1038/s41388-018-0442-6. [DOI] [PubMed] [Google Scholar]
- 49.Swamy M, Pathak S, Grzes KM, Damerow S, Sinclair LV, van Aalten DM, et al. Glucose and glutamine fuel protein O-GlcNAcylation to control T cell self-renewal and malignancy. Nat Immunol. 2016;17:712–20. doi: 10.1038/ni.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y, Meng F, Wang B, He L, Liu Y, Liu Z. Dock2 in the development of inflammation and cancer. Eur J Immunol. 2018;48:915–22. doi: 10.1002/eji.201747157. [DOI] [PubMed] [Google Scholar]
- 51.Tiacci E, Orvietani PL, Bigerna B, Pucciarini A, Corthals GL, Pettirossi V, et al. Tumor protein D52 (TPD52): a novel B-cell/plasma-cell molecule with unique expression pattern and Ca(2+)-dependent association with annexin VI. Blood. 2005;105:2812–20. doi: 10.1182/blood-2004-07-2630. [DOI] [PubMed] [Google Scholar]
- 52.Kang GJ, Park MK, Byun HJ, Kim HJ, Kim EJ, Yu L, et al. SARNP, a participant in mRNA splicing and export, negatively regulates E-cadherin expression via interaction with pinin. J Cell Physiol. 2020;235:1543–55. doi: 10.1002/jcp.29073. [DOI] [PubMed] [Google Scholar]
- 53.Ishiguro A, Ishikita T, Shimbo T, Matsubara K, Baba K, Hayashi Y, et al. Elevation of serum thrombopoietin precedes thrombocytosis in Kawasaki disease. Thromb Haemost. 1998;79:1096–100. [PubMed] [Google Scholar]
- 54.Whittaker E, Bamford A, Kenny J, Kafarou M, Jones CE, Shah P, et al. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA. 2020;324:259–69. doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toubiana J, Poirault C, Corsia A, Bajolle F, Fourgeaud J, Angoulvant F, et al. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ. 2020;369:m2094. doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verdoni L, Mazza A, Gervasoni A, Martelli L, Ruggeri M, Ciuffreda M, et al. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:1771–8. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.