Abstract
Background
Analyses for the presence of SARS-CoV‑2 in the tissues of COVID-19 patients is important in order to improve our understanding of the disease pathophysiology for interpretation of diagnostic histopathological findings in autopsies, biopsies, or surgical specimens and to assess the potential for occupational infectious hazard.
Material and methods
In this review we identified 136 published studies in PubMed’s curated literature database LitCovid on SARS-CoV‑2 detection methods in tissues and evaluated them regarding sources of error, specificity, and sensitivity of the methods, taking into account our own experience.
Results
Currently, no sufficiently specific histomorphological alterations or diagnostic features for COVID-19 are known. Therefore, three approaches for SARS-CoV‑2 detection are used: RNA, proteins/antigens, or morphological detection by electron microscopy. In the preanalytical phase, the dominant source of error is tissue quality, especially the different intervals between sample collection and processing or fixation (and its duration) and specifically the interval between death and sample collection in autopsies. However, this information is found in less than half of the studies (e.g., in only 42% of autopsy studies). Our own experience and first studies prove the significantly higher sensitivity and specificity of RNA-based detection methods compared to antigen or protein detection by immunohistochemistry or immunofluorescence. Detection by electron microscopy is time consuming and difficult to interpret.
Conclusions
Different methods are available for the detection of SARS-CoV‑2 in tissue. Currently, RNA detection by RT-PCR is the method of choice. However, extensive validation studies and method harmonization are not available and are absolutely necessary.
Supplementary Information
The online version of this article (10.1007/s00292-021-00920-1) contains supplementary material, which is available to authorized users.
Keywords: COVID-19, Electron microscopy, Fluorescence in situ hybridization, Reverse transcriptase polymerase chain reaction, Preanalytical phase
Abstract
Hintergrund
Die Analyse von SARS-CoV‑2 in Geweben von COVID-19-Patienten ist wichtig für ein besseres Verständnis der Pathophysiologie der Krankheit, die Interpretation der diagnostischen histopathologischen Befunde in Autopsien, Biopsien und Resektaten oder um ein potenzielles berufsbedingtes Infektionsrisiko einzuschätzen.
Material und Methoden
In dieser Übersichtsarbeit haben wir 136 publizierte Studien zu Detektionsmethoden von SARS-CoV‑2 in Gewebe in der kuratierten Literaturdatenbank LitCovid von PubMed identifiziert und bezüglich Fehlerquellen, Spezifität und Sensitivität der Methoden unter Berücksichtigung eigener Erfahrungen ausgewertet.
Ergebnisse
Es gibt keine ausreichend spezifischen histomorphologischen Veränderungen bzw. diagnostischen Merkmale von COVID-19. Daher werden 3 Ansätze zum SARS-CoV-2-Nachweis genutzt: Nachweis von RNA, Proteinen/Antigenen oder morphologischer Nachweis mittels Elektronenmikroskopie. In der präanalytischen Phase liegt die dominante Fehlerquelle in der Gewebequalität, insbesondere den unterschiedlichen Intervallen zwischen Probenentnahme und -aufarbeitung, sowie spezifisch in Autopsien im Intervall zwischen Tod und Probenentnahme. Diese Angaben finden sich in weniger als der Hälfte der Studien (z. B. nur in 42 % der Autopsiestudien). Eigene Erfahrungen und erste Studien belegen die deutlich höhere Sensitivität und Spezifität von RNA-basierten Nachweismethoden gegenüber Antigen- bzw. Proteinnachweis mittels Immunhistochemie oder Immunfluoreszenz. Der Nachweis mittels Elektronenmikroskopie ist zeitintensiv und die Interpretation schwierig.
Schlussfolgerungen
Es stehen verschiedene Methoden zum Nachweis von SARS-CoV‑2 im Gewebe zur Verfügung. Derzeit ist der RNA-Nachweis mittels RT-PCR die Methode der Wahl. Notwendige, umfangreiche Validationsstudien und Methodenharmonisierung sind derzeit noch nicht verfügbar.
Schlüsselwörter: COVID-19, Elektronenmikroskopie, Fluoreszenz-in-situ-Hybridisierung, Reverse-Transcriptase-Polymerase-Kettenreaktion, Präanalytische Phase
Several methods are available for SARS-CoV‑2 detection: electron microscopy, antigen detection by immunohistochemistry and immunofluorescence, and nucleic acid detection by in-situ hybridization and reverse transcriptase polymerase chain reaction (RT-PCR). Due to various factors in the preanalytical, analytical, and postanalytical phase, virus detection in pathology material presents specific diagnostic challenge. This article gives an overview of several SARS-CoV‑2 detection methods and discusses existing data on specific sources of error, validity, and robustness of these methods.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of the pandemic COVID-19 disease, is a novel pathogen that enters the human body via the upper respiratory tract and spreads from there to the lower respiratory tract. Therefore, reverse transcriptase polymerase chain reaction (RT-PCR) or, increasingly, rapid antigen detection tests, from nasopharyngeal swabs, bronchoalveolar lavage, or other lower respiratory tract or lung material are used in the clinical diagnosis of SARS-CoV‑2 infection in patients. These methods are now relatively well established and validated. In contrast, detection methods in tissue are much less well studied and validated. Virus detection in tissue is important for a better understanding of COVID-19 pathophysiology and interpretation of histopathological findings as well as for assessment of a potential occupational infection risk.
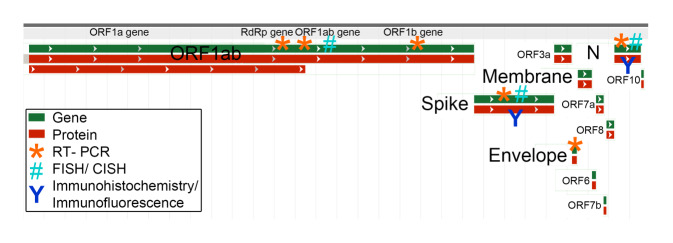
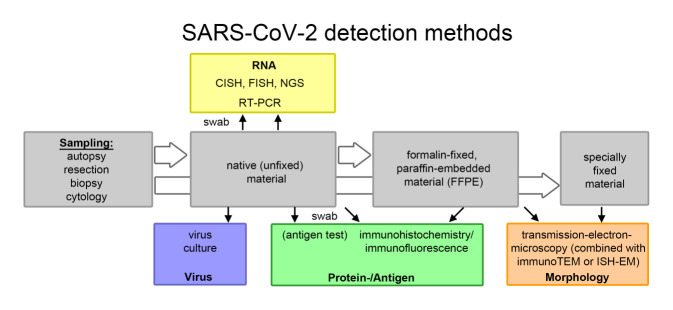
General information on SARS-CoV‑2 can be found in a separate article in this special issue [24]. SARS-CoV‑2 is a single-stranded RNA virus. The viral genome contains several genes: the E (envelope protein) gene, the M (membrane protein) gene, the N (nucleocapsid protein) gene, the RdRp (RNA-dependent RNA polymerase) gene, the S (spike protein) gene, and several ORF (open reading frame) genes encoding 10 proteins. For detection of SARS-CoV‑2, several methods can be used to detect various viral components (Fig. 1), including antigen/protein detection by immunohistochemistry/immunofluorescence (spike protein and nucleocapsid protein), RNA detection by in-situ hybridization (ORF1ab gene, spike protein gene, and nucleocapsid protein gene) or RT-PCR (mostly RdRp gene, ORF1ab gene, spike protein gene, coat protein gene, and nucleocapsid protein gene), and morphological detection of intact virus particles by electron microscopy (Fig. 2). Another option is in vitro culture of the virus—ultimately the only method that can confirm with certainty the presence of an infectious virus. This method is mainly offered in specialized virological laboratories.
Fig. 1.
SARS-CoV‑2 gene sequence (green) and protein sequence (red), with indication of currently used molecular detection methods (severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu‑1, complete genome. Figure modified from NCBI reference sequence: NC_045512.2). CISH chromogenic in situ hybridization, FISH fluorescence in situ hybridization, RT-PCR reverse transcriptase polymerase chain reaction, ORF open reading frame
Fig. 2.
SARS-CoV‑2 detection methods in pathology. CISH chromogenic in situ hybridization, FISH fluorescence in situ hybridization, NGS next-generation sequencing, RT-PCR reverse transcriptase polymerase chain reaction, ImmunoTEM immuno-transmission electron microscopy, ISH-EM in situ hybridization electron microscopy
All publications on detection methods of SARS-CoV‑2 in tissues (up until 01 November 2020) from the curated literature database LitCovid of PubMed were included (136 publications in total). Evaluated were the detection methods and the information on preanalytical factors, particularly the postmortem interval (period between death and autopsy); intraanalytical factors, particularly information on control tissue; postanalytical factors, particularly interpretation of the result; and the type of publication (original paper, case report, letter to the editor, other).
Non-method-specific factors and aspects
As a general strategy, to validate a method, detection of one or more targets by different methods or at least detection of multiple targets of the pathogen of interest should be demonstrated, and specificity and sensitivity should be analyzed with appropriate positive and negative controls. The strategy of using more than one method for virus detection was used in 22 of 62 evaluated original papers on the detection of SARS-CoV‑2 (37%, Table S1). More than one method for virus detection was used in 17 of 49 case reports evaluated (35%, Table S1), 4 of 12 letters to the editor (30%, Table S1), and 6 of 13 other publication formats (46%, e.g., technical report, Table S1). Appropriate control tissues were used for immunohistochemistry and in situ hybridization in 63% of the studies. For RT-PCR, control tissue or an RNA standard was used in 49% of studies, whereas for transmission electron microscopy, control tissue was used in only 4% of studies (Table S1) [13].
Preanalytical factors
Variability in the test material is one of the most important sources of error. Postmortem swabs of tissues or organs (nasopharynx, cornea, lung, colon, etc.), cryopreserved tissue, otherwise fixed (e.g., glutaraldehyde-fixed) tissue, or formalin-fixed paraffin-embedded (FFPE) tissue can be used as test material (Fig. 2). The timing and type of specimen collection, transport, storage, and further processing (fixation) are important. In particular, autolysis and associated degradation processes are not uncommon in autopsies, which can affect all levels of detection. The postmortem interval, which is essential for interpretation of study results, was reported in 40 of 96 published studies on autopsy material (42%). Ischemia time was not reported in any of 38 studies on non-autopsy material (skin or kidney biopsies, resected specimens, and placenta). The duration of formalin fixation, which affects RNA integrity [48], was reported in few studies and ranged from 1.5 h to 10 days (Table S1).
Intraanalytical factors
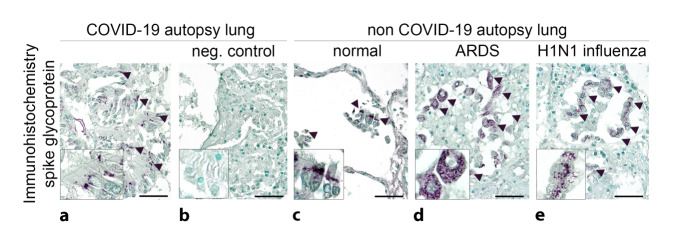
The lack of appropriate positive and negative control tissue can be another source of error. All pattern or image recognition-based detection methods, including histomorphology and ultrastructural morphology, immunohistology, immunofluorescence, and in situ hybridization, can only be interpreted in the context of positive and negative controls, especially if the antibodies or probes are not validated by the manufacturer for the particular use. An ideal positive control would theoretically be tissue comparable to the investigated material, with a confirmed viral presence and optimal preprocessing, e.g., postmortem biopsies of confirmed COVID-19 cases within a few hours of death [8]. However, such tissue is usually not available. An alternative positive control are SARS-CoV-2-infected cells from cell culture, which can be embedded in paraffin as a pellet [31]. These are currently not commercially available. To detect a nonspecific signal in SARS-CoV-2-negative tissues, comparable COVID-19-negative autopsy tissues or non-autopsy tissues should be examined as a negative control (Fig. 3). The result of the above methods is usually qualitative or semiquantitative. Quantitative analysis usually requires digitization of the images. In the case of RT-PCR, standardization with a specific in vitro-transcribed RNA quantification standard is possible [11]. Another intraanalytical source of error lies in the different detection kits, some of which may show significantly different performance, e.g., regarding sensitivity (own unpublished results).
Fig. 3.
SARS-CoV‑2 detection methods in lung tissue. a Monoclonal anti-SARS spike glycoprotein antibody (mouse, monoclonal, Abcam, Cambridge, UK, Ab272420, 1:100) with apparent specific cytoplasmic granular staining pattern (a: arrowheads) in bronchial epithelia in autopsy lung tissue from a COVID-19-positive patient (SARS-CoV‑2 E [envelope protein] gene, RdRp [RNA-dependent RNA polymerase] gene, and N [nucleocapsid protein] gene positive in reverse transcriptase polymerase chain reaction [RT-PCR], disease duration 38 days). The regular negative control shows the lack of nonspecific binding of the secondary antibody (b: biotinylated horse anti-mouse, 1:300; scale bar = 40 µm). c–e Nonspecific binding of the primary antibody with similar granular staining pattern in bronchial epithelial cells and macrophages in autopsy lung tissue: c without any lung disease, d in acute respiratory distress syndrome (ARDS), and e in H1N1 influenza (scale bar = 40 µm)
Postanalytical factors
Uncritical interpretation of an apparently specific morphology or signal may lead to a false-positive result (Fig. 3). Validation of a positive result using at least one additional method was reported in only 31 of 136 publications (23%, Table S1), with half of these publications not reporting comparable tissues for both methods or, in the case of RT-PCR, the use of a standard as a control. A statement on smoking history, duration of clinical symptoms until sampling of the test material, and type and duration of treatment (and ventilation) can help to interpret the results in a clinical context.
SARS-CoV-2 detection methods
Detection methods for SARS-CoV‑2 can be morphological (ultrastructural morphology), protein/antigen based (immunohistology, immunofluorescence), or RNA based (in situ hybridization, PCR; Fig. 2). Other methods such as viral genomics or in vitro culture of infectious virus from tissue have been applied only sporadically in pathology material.
There are currently no studies comparing the specificity and sensitivity of fresh and FFPE tissues, and most current studies were performed on FFPE material. The significance of the discrepancy between often inconspicuous histomorphology (e.g., extrapulmonary endothelial cells) and positive viral detection by immunohistochemistry [5, 19] or RT-PCR [41] remains unclear.
Morphology
Morphology of organ damage
Specific histomorphological changes that would allow COVID-19 diagnosis or specific SARS-CoV-2-induced viropathic morphological changes have not been reported to date [26, 32, 41, 42]. At present, it seems unlikely that such changes could be identified. In the lung, the picture of diffuse alveolar damage with pneumocyte proliferation, edema, hyaline membranes, and squamous metaplasia in the early phase of infection (< 10 days) and fibrosis with multinucleated (CD68-positive) giant cells in the late phase (> 10 days) has been described. The histopathological appearance described for SARS and SARS-CoV‑2 infection seems to be identical [13, 37, 44]. Frequent detection of thromboemboli and microthrombi has been described as a striking finding in COVID-19 autopsies [1, 12, 18, 25, 30, 41, 43, 51], but these are also observed in diffuse alveolar damage of other etiologies, albeit less frequently [4]. A possible overinterpretation of postmortem clots as intravital thrombi has been discussed [38, 45]. For organ-specific effects of SARS-CoV‑2, the authors refer the reader to the relevant articles in this issue [2, 3].
Electron microscopy-based detection of virus particles
As with other viruses, the detection of intact virus particles is possible ultrastructurally using transmission electron microscopy (EM). Advantages of this method include morphological localization in specific cells and subcellular compartments. Another advantage is the detection of intact virus particles as opposed to the detection of structural components such as proteins and RNA using other methods. To date, attempted SARS-CoV‑2 virus detection has been published in 46 studies. Positive detection of virus or “virus-like particles” was reported in 24 and 11 studies, respectively (Table S1). A negative result was found in 11 studies (Table S1). Glutaraldehyde at a concentration of 1.5–4% was generally used as a fixative; in individual studies, tissue was primarily fixed in formaldehyde for up to 10 days and later transferred to glutaraldehyde [17, 23]. In only two studies were infected cells from cell culture used as positive controls or COVID-19-negative tissue from autopsies used as a negative control [14, 22]. The postmortem interval or ischemia time was not reported in the majority of studies (Table S1).
Interpretation of ultrastructural SARS-CoV‑2 detection is difficult because several cellular components show virus-like size and morphology, which can easily be confused with viruses, especially by non-experts. In particular, these include clathrin-coated vesicles, multilamellar bodies, and rough endoplasmic reticulum [14, 16, 36]. In infected cells from cell culture with high virus load, ultrastructural detection is usually possible [14, 49], whereas virus detection on autopsy material is extremely difficult, time consuming, and successful only in exceptional cases and in the hands of designated experts [22]. Besides, the magnifications required for ultrastructural detection of viruses are not routinely used, even in centers performing diagnostic EM [22]. A more detailed discussion of SARS-CoV‑2 detection by EM has recently been published elsewhere [22].
Protein/antigen detection
Protein/antigen detection is a very well established and validated method in pathology for the diagnosis of viral and non-viral diseases. In the case of SARS-CoV‑2, a distinctive signal in immunohistochemistry or immunofluorescence should usually be interpreted with caution (Fig. 3), especially in the absence of positive and negative controls. The recently introduced and available rapid antigen tests might also find their utility in pathology diagnostics. In particular, they could provide rapid information on possible SARS-CoV‑2 infection in autopsies or frozen section/biobank by swab testing. There is currently no published work on this, and it is unclear whether the sensitivity and specificity might be sufficient. Other novel methods for protein detection, such as mass spectrometry, have not been used as virus detection methods in pathology material.
Immunohistochemistry
The potential advantages of immunohistochemistry are the possible localizability of the signal in specific cells and correlation with pathological changes of the respective tissue, as well as the very wide availability of these methods in pathology labs. One source of error in the detection of SARS-CoV‑2 by immunohistochemistry is a nonspecific signal that can only be identified when positive controls (e.g., infected cells from cell culture) and negative controls (e.g., tissue from non-COVID-19 autopsies) are used (Fig. 3; [35]). The commercially available antibodies against SARS-CoV‑2 proteins have not been tested by the manufacturers for specificity and sensitivity. Omitting the primary antibody is not suitable as a sole negative control [20]. Information on control tissues is found in 22 of 40 studies (55%). Appropriate negative controls to detect false-positive signals (e.g., lung tissue from non-COVID-19 autopsies) were used in 13 of 31 studies that reported positive detection of SARS-CoV‑2 (42%). The positive signal in these studies was found in glandular cells of the nasopharyngeal mucosa, pneumocytes (diffuse cytoplasmic, weakly extracellular), multinucleated giant cells, respiratory epithelial cells, peribronchial glands, alveolar macrophages, hyaline membranes (diffuse, strongly positive), endothelial cells (granular cytoplasmic), syncytiotrophoblast and cytotrophoblast of the placenta, renal tubular epithelial cells, glomerular endothelial cells, and eccrine glands of the skin (granular cytoplasmic). In 9 studies, SARS-CoV‑2 detection by immunohistochemistry was negative (Table S1). One study showed a weak nonspecific signal throughout the renal parenchyma [29]. Fifteen different antibodies against the spike and nucleocapsid proteins have been published (Fig. 1 and Table S1). The specificity of the staining of two commonly used antibodies has been questioned in various commentaries [5, 29, 33, 47].
The sensitivity and specificity of detection of SARS-CoV‑2 by immunohistochemistry compared to in situ hybridization and RT-PCR were assessed in a study of eight COVID-19 autopsies and non-COVID-19 autopsies as negative controls. When compared to RT-PCR results, the study showed a sensitivity of 86.7% and specificity of 100% for in situ hybridization, and a sensitivity of 85.7% and specificity of 53.3% for immunohistochemistry, with low to moderate interobserver variability. Detection of SARS-CoV‑2 by immunohistochemistry was successful only in the lung, while no virus could be detected by any of the methods in the heart, liver, kidney, small intestine, skin, adipose tissue, and bone marrow [34].
A potential source of a possible false-positive interpretation of a nonspecific staining pattern may also be placental tissue from COVID-19-positive mothers or when placental tissue is used as a positive control, as both placental endothelial cells and syncytiotrophoblast may show a distinctive but false-positive signal [21]. In one study, no cross-reactivity of a SARS-CoV nucleocapsid protein antibody was found with influenza A (H1N1), influenza B, respiratory syncytial virus, parainfluenza virus type 3, human coronavirus (HCoV) 229E, or MERS-CoV in PCR-validated control tissues [33]. The antibodies usually stain for both SARS and SARS-CoV‑2, but this should not cause diagnostic difficulties. The cross-reactivity of most antibodies with other viruses, particularly with other coronaviruses, remains unclear.
In summary, based on current data and compared to RNA-based methods, immunohistochemistry is not recommended for the detection of SARS-CoV‑2.
Immunofluorescence
Compared to immunohistochemistry, immunofluorescence has been used less frequently for the detection of SARS-CoV‑2 proteins/antigens [9, 28, 31, 40], possibly because in FFPE tissue, stronger autofluorescence is a confounding intraanalytical factor and frozen tissues remain potentially infectious. Furthermore, the exact localization of the positive signal and the recognition of possible morphological correlates of viral infection are hampered by the lack of overview staining, particularly in the presence of additional autolytic changes in autopsy material.
RNA detection
Viral RNA detection has been established and validated as a standard method for the diagnosis of COVID-19 swabs, similarly to the RNA-based detection of SARS in 2003. Predominantly, genomic viral RNA is detected, although detection of viral transcripts or transcriptome is also possible [39]. Various genes can be used (Fig. 1 and Table S1). The E (envelope protein) gene is a highly conserved gene that is identical in SARS and SARS-CoV‑2 (pan-sarbecovirus gene). Therefore, one protocol recommends using E (envelope protein) gene amplification as a pretest for the detection of SARS virus and, in the case of a positive result, subsequently amplifying one or two other genes from SARS-CoV‑2 as a confirmatory test (N [nucleocapsid protein] gene, RdRp [RNA-dependent RNA polymerase] gene) [10]. The S (spike protein) gene has more frequent mutations due to high selection pressure, such that using the S gene alone is not recommended. However, it can be used as a third target molecule to detect some mutations [27]. For robust detection, at least two targets should be amplified using the most sensitive and regionally established assay, as recommended by the World Health Organization (WHO) and the Robert Koch Institute in Germany [50]. In addition to all controls recommended in the respective kit, using a positive control, e.g., from a confirmed COVID-19 case, is recommended. A separate negative control does not seem to be necessary.
RT-PCR of (postmortem) swabs
A possible strategy to detect SARS-CoV‑2 RNA in tissues is by swabbing tissues or organs (e.g., nasopharynx, cornea, lung, trachea, colon) during autopsy (or in frozen section or biobank). Positive detection of SARS-CoV‑2 RNA 12 days postmortem has been described (Table S1). RT-PCR from swabs is very widely established and available, offering a method of choice for labs that do not perform RNA-based testing. The correlation between intra- and postmortem detection of viral RNA from swab material has not been described so far. In a study comparing detection of SARS-CoV‑2 from corneal swabs with postmortem nasopharyngeal swab and intravital detection, corneal swab did not detect a single COVID-19 case, thus rendering it not useful as a screening method before corneal transplantation. Similarly, postmortem nasopharyngeal swab was also of rather limited use [15].
When a standard is used, the viral RNA copy number can be quantified as the viral load. Due to the often unclear or highly variable preanalytical conditions, accurate quantification is not always sufficiently precise. For diagnostic purposes, qualitative findings, i.e., positive vs. negative, are sufficient in most applications or cases. Thus, an RNA quantification standard is not mandatory. For research purposes and for some specific questions, it may be useful, e.g., to distinguish between a high and a low viral load. For example, in autopsy cases, lung tissues show a high viral load and other organs show a low viral load. Of 56 publications evaluated that used RT-PCR for virus detection from postmortem swabs or tissue, at least two different target molecules were amplified in 21 cases (37.5%), three in nine cases (16%), and seven or eight in one case each (Table S1).
RT-PCR in tissue
RT-PCR for detection of viral RNA is the most frequently published method for virus detection in tissue, mainly in autopsies (45 publications). In 40 studies, SARS-CoV‑2 RNA was detected in tissues, whereas five studies did not detect SARS-CoV‑2 RNA using RT-PCR (Table S1). The postmortem interval was reported in the majority of publications. Possible sources of preanalytical error in RT-PCR include RNA degradation during the postmortem interval or ischemia time and RNA fragmentation during formalin fixation. Correlations between quantitative RNA detection in vivo and quantitative RNA detection postmortem do not exist to date. A postanalytical confounding factor of RT-PCR might be the limited comparability of results between two sites due to different PCR devices, kits with different types and numbers of target genes (Fig. 1), and different numbers of PCR cycles (cycle threshold, Ct value) determining a positive result as a cut-off value. Comparisons between the different methods and approaches , as well as comparisons between different laboratories, are not yet available. In the majority of publications, either the Ct value between < 25 cycles and ≤ 45 cycles or the quantitative viral load after standardization is given. A disadvantage of RT-PCR is the lack of a possibility to assign the detected viral RNA to specific cells.
FISH/CISH
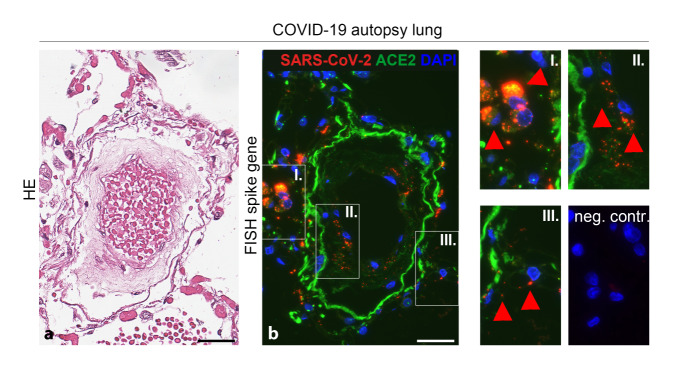
In situ hybridization (ISH) has been used in 30 of 136 studies to date. Chromogen in situ hybridization (CISH) was used in the majority of studies, but fluorescent ISH (FISH) can also be used (Fig. 4). Sixteen studies reported positive detection of SARS-CoV‑2 RNA using ISH. Fourteen studies found no SARS-CoV‑2 RNA. A comparison of immunohistochemistry and ISH with RT-PCR for virus detection showed high specificity (100%) and sensitivity (86.7%) of ISH and moderate to near-perfect interobserver variability [34]. A comparison between immunohistochemistry and ISH for the detection of SARS-CoV‑2 in the kidney, placenta, and lung tissues from COVID-19 patients and COVID-10 autopsy cases showed 100% agreement between the results of both methods [6].
Fig. 4.
SARS-CoV‑2 RNA detection by fluorescence in situ hybridization (FISH) in lung tissue from a SARS-CoV-2-positive patient (measurement bar = 50 µm), a HE lung and pulmonary vessel, b consecutive sectional step to a with positive RNA detection for SARS-CoV‑2 (red) in macrophages (I.), endothelia (II.), and capillary endothelia (III.) in the absence of signal in the negative control (neg. contr.)
Other methods
Virus RNA sequencing, next-generation sequencing (NGS) [34, 43, 45], nested PCR [46], and proteomics [39] have been applied only in single studies. Sequencing of the virus is important for genealogical determination of virus origin and detection of mutations.
The only reliable method for detection of infectious virus—inoculation of cells in cell culture with swabs from autopsy tissues—has also rarely been performed. This is the only method for detection of infectious virus and at the same time can be used as source material for virus detection by transmission electron microscopy or other methods [7, 49]. However, a disadvantage of this detection method is the need for biological safety level 3 or 4 laboratories and appropriately trained staff, which are mainly available in specialized virology laboratories.
Conclusion
The detection of SARS-CoV‑2 in tissue is possible using various methods, each of which has different advantages, disadvantages, and indications. The detection methods and particularly the interpretation of the findings are not always easy due to different factors in the preanalytical, intraanalytical, and postanalytical phases, especially in the case of negative results. Due to the best performance, RNA-based detection in FFPE material is currently seen as the method of choice. These methods have been established and validated within the framework of the German Registry for COVID-19 Autopsies (DeRegCOVID; www.DeRegCOVID.ukaachen.de) and are offered by the Institute of Pathology at the University Hospital Aachen for all interested centers. Nevertheless, more extensive validation studies, as well as interlaboratory tests for the detection methods, are necessary.
Practical conclusion
- There are several methods for the detection of SARS-CoV-2:
-
MorphologySpecific histomorphological detection is currently not possible/not known.Morphological detection by electron microscopy has so far only been successful in the hands of experts, under optimal conditions, and with comparatively intensive effort, and is currently not recommended for diagnostics.
-
Antigen/proteinCurrent data show lower specificity and sensitivity of immunohistochemistry compared to RNA-based methods; thus, sole use of immunohistochemistry for diagnostics is currently not recommended.
-
RNADue to its sensitivity and specificity, RT-PCR-based SARS-CoV‑2 detection is recommended as the method of choice in tissue diagnostics.In situ hybridization is not recommended over RT-PCR due to the more complex methodology for primary diagnostics, but this method has important applications for many research questions.
-
- For interpretation of the results and for future studies on detection methods, different factors have to be taken into account, in particular
- disease duration,
- postmortem interval (the period between death and autopsy),
- duration and type of specimen fixation,
- specification and type of positive and negative controls (if possible from the same material).
Currently, there is a lack of larger and multicenter validation studies or interlaboratory comparisons for the SARS-CoV‑2 tissue detection methods. These would be highly desirable.
Supplementary Information
Supplementary table 1: Characteristics of included studies: type of article, PMID, number of patients, sample type, postmortem interval (d), detection method(s) applied
Acknowledgments
Acknowledgements
The authors would like to express their sincere gratitude for the outstanding and unique dedication and scientific work of the many involved staff members of the Institute of Pathology and LaBooratory of Nephropathology. For special support, the authors would like to thank Anna Breitbach, Eva Miriam Buhl, Roman David Bülow, Jana Baues, Louisa Böttcher, Till Braunschweig, Marc Britz, Claudio Cacchi, Edgar Dahl, Sonja Djudjaj, Christina Gianussis, Barbara Klinkhammer, Ruth Knüchel-Clarke, Patrick Kühl, Simon Otten, Tessa Schade, Sophia Villwock, Dickson Wong, and Sophie Wucherpfennig (in alphabetical order). This work was supported by the German Registry for COVID-19 Autopsies (DeRegCOVID, www.DeRegCOVID.ukaachen.de), funded by the German Federal Ministry of Health (ZMVI1-2520COR201), the German Federal Ministry of Education and Research under the University Medicine Network (DEFEAT PANDEMIcs, 01KX2021), the German Research Foundation (DFG: SFB/TRR57, SFB/TRR219, BO3755/3‑1, and BO3755/6-1), and the RWTH START program (125/17).
Declarations
Conflict of interest
S. von Stillfried and P. Boor declare that they have no competing interests.
For this article no studies with human participants or animals were performed by any of the authors. All studies performed were in accordance with the ethical standards indicated in each case.
References
- 1.Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M, Werlein C, Länger F, et al. COVID-19: Auswirkungen auf Lunge und Herz. Pathologe. 2021 doi: 10.1007/s00292-021-00918-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann K, Boor P, Wiech T, et al. Covid-19-Auswirkungen auf die Niere. Pathologe. 2021 doi: 10.1007/s00292-020-00899-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrossi AV, Farver C. The pulmonary pathology of COVID-19. Cleve Clin J Med. 2020 doi: 10.3949/ccjm.87a.ccc063. [DOI] [PubMed] [Google Scholar]
- 5.Baeck M, Hoton D, Marot L, et al. Chilblains and COVID-19: why SARS-CoV-2 endothelial infection is questioned. Br J Dermatol. 2020;183:1152–1153. doi: 10.1111/bjd.19489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Best Rocha A, Stroberg E, Barton LM, et al. Detection of SARS-CoV-2 in formalin-fixed paraffin-embedded tissue sections using commercially available reagents. Lab Invest. 2020;100:1485–1489. doi: 10.1038/s41374-020-0464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borczuk AC, Salvatore SP, Seshan SV, et al. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol. 2020;33:2156–2168. doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brook OR, Piper KG, Mercado NB, et al. Feasibility and safety of ultrasound-guided minimally invasive autopsy in COVID-19 patients. Abdom Radiol. 2020 doi: 10.1007/s00261-020-02753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 12.Edler C, Schroder AS, Aepfelbacher M, et al. Dying with SARS-CoV-2 infection-an autopsy study of the first consecutive 80 cases in Hamburg, Germany. Int J Legal Med. 2020;134:1275–1284. doi: 10.1007/s00414-020-02317-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewig S. SARS. A lesson in infection epidemiology and a masterpiece of modern infection control. Pathologe. 2003;24:335–337. doi: 10.1007/s00292-003-0637-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frelih M, Erman A, Wechtersbach K, et al. SARS-CoV-2 virions or ubiquitous cell structures? Actual dilemma in COVID-19 era. Kidney Int Rep. 2020;5:1608–1610. doi: 10.1016/j.ekir.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuest M, Boor P, Knuechel R, et al. Postmortem conjunctival and nasopharyngeal swabs in SARS-CoV-2 infected and uninfected patients. Acta Ophthalmol. 2020 doi: 10.1111/aos.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith CS, Miller SE, Martines RB, et al. Electron microscopy of SARS-CoV-2: a challenging task. Lancet. 2020;395:e99. doi: 10.1016/S0140-6736(20)31188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimes Z, Bryce C, Sordillo EM, et al. Fatal pulmonary thromboembolism in SARS-coV-2-infection. Cardiovasc Pathol. 2020;48:107227. doi: 10.1016/j.carpath.2020.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosse C, Grosse A, Salzer HJF, et al. Analysis of cardiopulmonary findings in COVID-19 fatalities: high incidence of pulmonary artery thrombi and acute suppurative bronchopneumonia. Cardiovasc Pathol. 2020;49:107263. doi: 10.1016/j.carpath.2020.107263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecht JL, Quade B, Deshpande V, et al. SARS-CoV-2 can infect the placenta and is not associated with specific placental histopathology: a series of 19 placentas from COVID-19-positive mothers. Mod Pathol. 2020;33:2092–2103. doi: 10.1038/s41379-020-0639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt SM, Baskin DG, Frevert CW, et al. Controls for immunohistochemistry: the Histochemical Society’s standards of practice for validation of immunohistochemical assays. J Histochem Cytochem. 2014;62:693–697. doi: 10.1369/0022155414545224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honig A, Rieger L, Kapp M, et al. Immunohistochemistry in human placental tissue--pitfalls of antigen detection. J Histochem Cytochem. 2005;53:1413–1420. doi: 10.1369/jhc.5A6664.2005. [DOI] [PubMed] [Google Scholar]
- 22.Hopfer H, Herzig MC, Gosert R, et al. Hunting coronavirus by transmission electron microscopy - a guide to SARS-CoV-2-associated ultrastructural pathology in COVID-19 tissues. Histopathology. 2020 doi: 10.1111/his.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosier H, Farhadian SF, Morotti RA, et al. SARS-CoV-2 infection of the placenta. J Clin Invest. 2020;130:4947–4953. doi: 10.1172/JCI139569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klingel K. Biologie und Pathologie von Corona-Viren. Pathologe. 2021 doi: 10.1007/s00292-021-00923-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kommoss FKF, Schwab C, Tavernar L, et al. The pathology of severe COVID-19-related lung damage. Dtsch Arztebl Int. 2020;117:500–506. doi: 10.3238/arztebl.2020.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konopka KE, Nguyen T, Jentzen JM, et al. Diffuse alveolar damage (DAD) resulting from coronavirus disease 2019 Infection is Morphologically Indistinguishable from Other Causes of DAD. Histopathology. 2020;77:570–578. doi: 10.1111/his.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-coV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamers MM, Beumer J, Van Der Vaart J, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen CP, Bourne TD, Wilson JD, et al. Collapsing glomerulopathy in a patient with COVID-19. Kidney Int Rep. 2020;5:935–939. doi: 10.1016/j.ekir.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lax SF, Skok K, Zechner P, et al. Pulmonary arterial thrombosis in COVID-19 with fatal outcome : results from a prospective, single-center, clinicopathologic case series. Ann Intern Med. 2020;173:350–361. doi: 10.7326/M20-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Babka AM, Kearney BJ, et al. Molecular detection of SARS-CoV-2 in formalin-fixed, paraffin-embedded specimens. JCI Insight. 2020;5(12):e139042. doi: 10.1172/jci.insight.139042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansueto G. COVID-19: Brief check through the pathologist’s eye (autopsy archive) Pathol Res Pract. 2020;216:153195. doi: 10.1016/j.prp.2020.153195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martines RB, Ritter JM, Matkovic E, et al. Pathology and pathogenesis of SARS-coV-2 associated with fatal Coronavirus disease, United States. Emerg Infect Dis. 2020;26:2005–2015. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massoth LR, Desai N, Szabolcs A, et al. Comparison of RNA in situ hybridization and Immunohistochemistry techniques for the detection and localization of SARS-coV-2 in human tissues. Am J Surg Pathol. 2021;45:14–24. doi: 10.1097/PAS.0000000000001563. [DOI] [PubMed] [Google Scholar]
- 35.Meinhardt J, Radke J, Dittmayer C, et al. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat Neurosci. 2021;24:168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- 36.Miller SE, Goldsmith CS. Caution in identifying Coronaviruses by electron microscopy. J Am Soc Nephrol. 2020;31:2223–2224. doi: 10.1681/ASN.2020050755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicholls JM, Poon LL, Lee KC, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361:1773–1778. doi: 10.1016/S0140-6736(03)13413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nichols L. Pulmonary thrombotic and thromboembolic disease in COVID-19. Ann Intern Med. 2020 doi: 10.7326/L20-1275. [DOI] [Google Scholar]
- 39.Nie X, Qian L, Sun R, et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021 doi: 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puelles VG, Lutgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-coV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Remmelink M, De Mendonca R, D’haene N, et al. Unspecific post-mortem findings despite multiorgan viral spread in COVID-19 patients. Crit Care. 2020;24:495. doi: 10.1186/s13054-020-03218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi GM, Delsante M, Pilato FP, et al. Kidney biopsy findings in a critically ill COVID-19 patient with dialysis-dependent acute kidney injury: a case against “SARS-coV-2 nephropathy”. Kidney Int Rep. 2020;5:1100–1105. doi: 10.1016/j.ekir.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sauter JL, Baine MK, Butnor KJ, et al. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology. 2020;77:915–925. doi: 10.1111/his.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaller T, Hirschbuhl K, Burkhardt K, et al. Postmortem examination of patients with COVID-19. JAMA. 2020;323:2518–2520. doi: 10.1001/jama.2020.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sekulic M, Harper H, Nezami BG, et al. Molecular detection of SARS-coV-2 infection in FFPE samples and histopathologic findings in fatal SARS-coV-2 cases. Am J Clin Pathol. 2020;154:190–200. doi: 10.1093/ajcp/aqaa091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shirato K, Nao N, Katano H, et al. Development of genetic diagnostic methods for detection for novel Coronavirus 2019(ncoV-2019) in Japan. Jpn J Infect Dis. 2020;73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 47.Smith KD, Akilesh S, Alpers CE, et al. Am I a coronavirus? Kidney Int. 2020;98:506–507. doi: 10.1016/j.kint.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Susman S, Berindan-Neagoe I, Petrushev B, et al. The role of the pathology department in the preanalytical phase of molecular analyses. Cancer Manag Res. 2018;10:745–753. doi: 10.2147/CMAR.S150851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tzankov A, Jonigk D. Unlocking the lockdown of science and demystifying COVID-19: how autopsies contribute to our understanding of a deadly pandemic. Virchows Arch. 2020;477:331–333. doi: 10.1007/s00428-020-02887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogels CBF, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wichmann D, Sperhake JP, Lutgehetmann M, et al. Autopsy findings and venous thromboembolism in patients with COVID-19: a prospective cohort study. Ann Intern Med. 2020;173:268–277. doi: 10.7326/M20-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary table 1: Characteristics of included studies: type of article, PMID, number of patients, sample type, postmortem interval (d), detection method(s) applied