Abstract
Immunotherapy is regarded as the most promising treatment for cancers. Various cancer immunotherapies, including adoptive cellular immunotherapy, tumor vaccines, antibodies, immune checkpoint inhibitors, and small-molecule inhibitors, have achieved certain successes. In this review, we summarize the role of macrophages in current immunotherapies and the advantages of targeting macrophages. To better understand and make better use of this type of cell, their development and differentiation characteristics, categories, typical markers, and functions were collated at the beginning of the review. Therapeutic strategies based on or combined with macrophages have the potential to improve the treatment efficacy of cancer therapies.
Subject terms: Tumour immunology, Drug development
As a type of phagocytic cell that was initially identified as clearing foreign pathogens by Elie Metchnikoff, macrophages have gradually been considered for cancer immunotherapy in recent years. In light of their positive roles in current therapeutic strategies, they have become a promising target for improved cancer treatments. To facilitate the use of macrophages in cancer immunotherapy, we summarize their related characterization and research progress in this review.
Categories and characterization of macrophages
Development and differentiation of macrophages
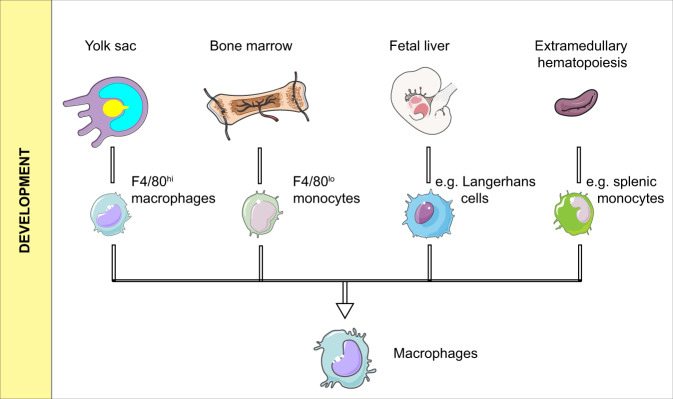
It is now widely accepted that macrophages in tissues, as well as monocytes in the peripheral blood, are classified as the mononuclear phagocytic system (MPS). This concept has developed over a long history, and its current version takes the origin, morphology, function, and kinetics of the cells into consideration.1 In MPS, macrophages originate from bone marrow stem cells, and their development goes through sequential stages as granulocyte–monocyte progenitor cells, pro-monocytes, and mature monocytes. After entering various tissues, monocytes differentiate into macrophages.2 However, in some lower multicellular organisms without circulating monocytes, such as Porifera, macrophages still exist. For patients with monocytopenia, their macrophages do not diminish correspondingly.3 These phenomena indicate that macrophages could come from other sources in addition to monocytes. This notion has been supported by additional studies. As shown in Fig. 1, based on studies from mouse models, macrophages possibly have at least four origins:4,5 (1) F4/80hi macrophages from the yolk sac that mainly reside in tissues such as the liver, spleen, lung, brain, pancreas, and kidney; (2) F4/80lo macrophages derived from bone marrow and developed through a mature stage as Ly6C+ monocytes; (3) Langerhans cells from the fetal liver (regarding Langerhans cells as macrophages but not DC cells6); (4) A few studies also have claimed that a minority of tumor-associated macrophages may come from extramedullary hematopoiesis, especially in the spleen.7,8 It has been reported that Ly6C− patrolling monocytes are mainly responsible for detecting pathogens intravascularly and maintaining vascular integrity, while Ly6C+ inflammatory monocytes are recruited to infectious sites and injuries, mediating extravascular inflammatory responses and then differentiating into macrophages.4,9 Some studies have also indicated that both Gr1+/Ly6Chigh10–12 and Gr1−/Ly6Clo monocytes have the potential to enter tissues and turn into macrophages,13 but the former are more likely to become M1 macrophages, while the latter are M2 phenotypes.14 Above all, macrophages in tissues are probably a mixture of embryo- and adult-derived cells.
Fig. 1.
Development of macrophages
Wherever the macrophages originated from, the macrophage colony-stimulating factor 1 receptor (CSF1R) is a key receptor that induces their differentiation. CSF1 and IL-34 are two ligands of CSF1R. These two factors function in different ways. It has been reported that macrophages in the liver, spleen, or bone marrow are typically regulated by CSF1-mediated signals,15 while IL-34 dominates the development of macrophages in the brain.16 Given the importance of CSF1R, its inhibitors are often used in scientific studies to deplete macrophages. In addition, the lack of Sfpi1, which is a pioneering transcriptional regulator in myeloid lineage development, could result in a total depletion of CD11b+F4/80+ macrophages.17 An expression disparity of Sfpi1 determines the differentiation of Ly6Chi monocytes into iNOS+ macrophages or monocyte-derived dendritic cells.18 Id3 is indispensable for liver macrophages.19 PPARγ maintains the anti-inflammatory function of alveolar macrophages.20 Gata6 controls the proliferative renewal of peritoneal macrophages.21 LXR deficiency could cause a failure in the generation of splenic marginal zone macrophages and metallophilic macrophages.22 Epigenetic changes drive the differentiation of monocytes into macrophages.23 More factors participating in the differentiation of macrophages have been described in previous reviews.4,24,25
Categories
Macrophages are widely distributed in various tissues. According to their histological locations, macrophages residing in specific tissues can be categorized into Kupffer cells in the liver, microglial cells in the brain, osteoclasts in the osseous tissue, alveolar macrophages in the lung, mesangial cells in the kidney, subcapsular sinus macrophages in lymph, and so on.26,27 A summary of the ontogeny, functions, and markers of macrophages in different tissues is listed in Table 1. It has been shown that macrophages from different tissues possess diverse expression profiles for transcripts and proteins, which can have a profound impact on their phenotypes and functions.28,29
Table 1.
Ontogeny, functions and identifying markers of different macrophages
| Tissue | Macrophage | Ontogeny | Function | Identifying markers | Refs. |
|---|---|---|---|---|---|
| Liver | Kupffer cells | Yolk sac derived | Clearance of bacteria, aged erythrocytes, and cellular debris from the blood; regulation of the immune response; involvement in liver injury repair |
F4/80hi CD11blo Siglec-1+ CD68+ Galectin-3+ CD80lo/− PPARδ+ Ly6C− CX3CR1− Clec4F+ TIM-4+ |
27,62,223–225 |
| Monocyte-derived liver macrophage (MoMFs) | Monocyte derived | Rapid accumulation and involvement in immune responses after organ damage |
F4/80+ CD11bhi MHC-II+ CCR2lo (transferring from CCR2hi) CD64+ CX3CR1hi (transferring from CX3CR1lo) |
226–229 | |
| Liver capsular macrophages | Monocyte derived | Sensing bacteria reaching the hepatic capsule; inhibition of the hepatic spread of peritoneal pathogens; recruiting neutrophils |
F4/80+ MHC-II+ CD11b+ CD64+ CD103− CX3CR1+ TIM-4− CD207+ |
230 | |
| Lung | Alveolar macrophages | Yolk sac and fetal liver progenitors | Immune surveillance; phagocytosis of inhaled particles |
F4/80lo CD11blo CD11chi CD14lo CD200Rhi DEC205inter MHC-IIlo CD68+ Siglec F+ MARCO+ CD206+ Dectin-1+ Galectin-3+ Mertk+ CD64+ Siglec-1+ |
27,223,231–233 |
| Interstitial macrophages | Fetal liver and bone marrow-derived monocytes | Immune surveillance |
F4/80+ CD11b+ CD11c+ CD68+ MHC-II+ CD24− CD86+ Ly6C− Siglec F- CD64+ |
233–236 | |
| Central nervous system | Microglial cells | Yolk sac derived | Functioning as immune surveillance; promote neuronal survival and remove dead neurons; synaptic remodeling |
F4/80+ CD11b+ CD45lo CX3CR1hi Iba-1+ P2RY12+ |
26,27,236,237 |
| Perivascular macrophages | Yolk sac or fetal liver progenitors |
Blood–brain barrier integrity; phagocytosis; antigen presentation; lymphatic clearance |
CD45hi CD11b+ F4/80hi CX3CR1hi Iba-1hi P2RY12− CD163+ CD206+ Lyve-1+ |
237–248 | |
| Meningeal macrophages | Yolk sac derived | Immune surveillance |
F4/80+ CD11b+ CD45hi CX3CR1hi Iba-1+ CD209b+ Chnrb4+ |
27,237,249 | |
| Bone | Osteoclast | Monocyte derived | Resorption of organic matter and minerals from the bone matrix |
Calcitonin receptor+ Calcr+ RANKL+ |
26,27,250–252 |
| Bone marrow macrophages | Yolk sac derived or fetal liver-derived monocytes | Promoting erythropoiesis; maintenance of the hematopoietic stem cells niche |
Siglec-1+ ER-HR3+ F4/80+ Tartrate-resistant acid phosphatase (TRAP)− |
250,253 | |
| Spleen | Marginal zone macrophages | Bone marrow-derived monocytes | Clearance of pathogens present in the circulation; retention of marginal zone B cells |
CD68+ Dectin-2+ F4/80lo LXRα+ MARCO+ TIM-4+ SIGN-R1+ |
22,254–256 |
| Marginal metallophilic macrophages | Bone marrow-derived monocytes | Clearance of pathogens present in the circulation |
CD68 + F4/80lo LXRα+ Siglec-1+ |
257 | |
| White pulp (tingible body) macrophages | Not clear | Clearance of apoptotic B cells |
CD68 + MFG-E8+ Mertk+ TIM-4+ CD36+ |
257–259 | |
| Red pulp macrophages | Yolk sac-derived or fetal liver-derived monocytes | Clearance of effete red blood cells; immunosurveillance; detoxification; iron recycling; antigen delivery to DCs |
F4/80hi CD11blo Siglec-1lo CD68+ MHC-IIlo CSF1R+ SIRPα+ Siglec F− CD163+ Dectin-2+ VCAM1+ Spi-C+ Heme Oxigenase+ Ferroportin+ |
223,255,259–261 | |
| Kidney | Mesangial cell | Monocyte derived | Intraglomerular mesangial cells; regulation of glomerular filtration; mesangial matrix formation; phagocytosis; monitoring of glucose concentrations |
F4/80+ CD11blo CD103− CX3CR1+ SIRPα+ Siglec F- |
223 |
| Lymph node | Subcapsular sinus macrophages | Yolk sac-derived or bone marrow-derived monocyte | Limiting the systemic dissemination of pathogens and bacterial infections; promote the presentation of antigens |
F4/80lo MARCO+ Siglec-1hi CD11bhi Ligands for the cysteine-rich domain of the mannose receptor+ |
223,262,263 |
| Medullary macrophages | Bone marrow-derived monocytes | Highly phagocytic and rapidly clear pathogens |
CD11b+ Siglec-1+ F4/80+ MARCO+ SIGN-R1+ |
223,263,264 | |
| Serosal Tissues | Pleural macrophages | Bone marrow-derived monocytes | Immune surveillance |
CD11bhi F4/80hi Siglec F− RELMα+ TIM-4+ |
265–267 |
| Large peritoneal macrophages | Yolk sac-derived or fetal liver-derived monocytes | Regulation of IgA production in the gut by peritoneal B1 cells |
CD11bhi CD11clo SIGN-R1− F4/80hi GATA-6+ MHC-IIlo/− CD62L– TIM-4+ |
268–270 | |
| Small peritoneal macrophages | Bone marrow-derived monocytes | Immune surveillance |
CD11blo CD11C− SIGN-R1+ F4/80lo MHC-IIhi CD62L+ TIM-4- |
265,269,270 | |
| Skin | Langerhans cells | Yolk sac-derived or fetal liver-derived monocytes | Interaction with T lymphocytes; immune surveillance |
CD11b + CD11c+ F4/80+ Id2+ Langerin+ RUNX3 + |
27,271,272 |
| Dermal macrophages | Bone marrow-derived monocytes | Immune surveillance |
CD11b+ CD11clo CD301+ Dectin-1+ Dectin-2+ F4/80+ CD64hi Mertk+ MHC-IIlo CD206+ Siglec-1hi |
27,223,255,273 | |
| Adipose Tissue | Adipose tissue-associated macrophages | Not clear | Adipogenesis; adaptive thermogenesis; regulation of insulin sensitivity and glucose tolerance |
CD45+ F4/80+ PPARγ+ |
274,275 |
| Gastrointestinal Tract | Intestinal lamina propria macrophages | Bone marrow-derived monocytes | Maintenance of intestinal homeostasis; recognition and removal of intestinal pathogens; maintenance of gut epithelial integrity |
CD11b+ CD11c+ CX3CR1hi F4/80+ CD64+ MHC-IIhi |
27,276 |
| Blood | Ly6Clo monocytes | Bone marrow-derived monocytes | Immune surveillance; maintenance of vascular integrity |
CD11bhi CD43+ CX3CR1+ F4/80+ Ly6Clo CSF1R+ NR4A1+ |
27,277,278 |
| Tumor | Tumor-associated macrophage | Yolk sac derived or monocyte derived | Promote tumor growth; inhibit tumoricidal immune response; initiate angiogenesis; activate matrix remodeling; aid invasion and intravasation |
Murine: Ly6C+ MHC-II+ CX3CR1+ CCR2+ CD62L+ TIE2+ Human: CD14+ CD312+ CSF1R+ CD16+ |
42,279–281 |
Based on phenotypes and functions, macrophages can be typically divided into M1 (proinflammatory, classically activated macrophages) and M2 (anti-inflammatory, alternatively activated macrophages) types (Fig. 2). In brief, M1 macrophages can be induced by IFN-γ, lipopolysaccharide (LPS), TNF-α or granulocyte–macrophage colony-stimulating factor (GM-CSF), followed by activation of Toll-like receptor signaling pathways. They play a positive role in the removal of pathogens and tumor cells. On the one hand, M1 macrophages express high levels of antigen-presenting MHC complexes, which accelerate the activation of adaptive immune responses. On the other hand, they act directly on target cells by generating nitric oxide, reactive oxygen species, and reactive nitrogen species. Moreover, M1 macrophages promote inflammatory responses by secreting cytokines such as TNF-α, IL-1α, IL-1β, IL-6, IL-12, IL-18, and IL-23.30,31 Excessive M1 macrophage-mediated responses may lead to tissue damage, which is the main cause of atherosclerosis and other chronic inflammation.32–34 M2 macrophages can be induced by cytokines, such as IL-4, IL-13, glucocorticoids, M-CSF/CSF1, IL-10, IL-33, IL-21, and TGF-β.31,35–37 Accompanied by increased production of polyamines and ornithine through the arginase pathway, high secretion of IL-10, PGE2, TGF-β, but low IL-12, they are major participants in the clearance of parasites and homeostasis, such as tissue remodeling and regeneration, wound healing and anti-inflammation.38,39 When M2 macrophages develop further, they are refined into M2a, M2b, M2c, and M2d subgroups.40 Their specific characterizations have been reviewed by Abbas Shapouri Moghaddam et al.41 Macrophages have strong plasticity. It has been shown that different phenotypes could possibly transform mutually under certain conditions.
Fig. 2.
Categories of macrophages
Tumor-associated macrophages (TAMs) generally represent a major component of myeloid cells present in tumors. For some solid tumors, TAMs can arise from several origins: as residual macrophages derived from the yolk sac, infiltrating macrophages as the major replenishment recruited from bone marrow/Ly6C+-circulating monocytes, and a minority from the spleen.8,42–47 It has been demonstrated that TAMs with different origins may act differently than anti-macrophage oncotherapies.43 In most established tumors, TAMs tend to be considered M2-skewed macrophages because they possess the majority of the representative properties of M2 macrophages, usually including but not limited to high expression levels of arginase-1, mannose receptor, and a low MHC class II complex.48 Transcriptome profile analysis revealed that TAMs are more similar to fetal macrophages but not inflammatory macrophages.41 However, as macrophages are plastic, there is also evidence suggesting that TAMs actually have both M1 and M2 expression patterns or expression patterns distinct from those of M1 and M2 macrophages.49 Since 90–95% of neoplasms are closely associated with a chronic inflammatory status, it has been suggested that M1 macrophages may induce tumor initiation by creating a mutagenic microenvironment, while M2 macrophages promote malignancy progression.36,50 It is also believed that TAMs may exert both tumor-promoting and tumor-inhibiting functions,51,52 which make TAMs a potential target for cancer therapies.
Typical markers
To be distinguished from other immunocytes, macrophages can be characterized by phagocytosis and the expression of CD11b, F4/80, and CSF1R in mice or CD79, CD163, CD16, CD312, and CD115 in humans.41 Specifically, to present antigens and activate adaptive immune responses, M1 macrophages often express high levels of MHC class II molecules and costimulatory molecules, such as CD40, CD80, and CD86, while M2 macrophages contain upregulated levels of endocytosis-related receptors, such as the human scavenger receptors CD163 and Stabilin-1 and C-type lectin receptors, including CD206, CD301, detin-1 and CD209.31 In addition to the proinflammatory or anti-inflammatory cytokines mentioned above, polarized macrophages generate different types of chemokines. CXCL9, CXCL10, CXCL11, and CCL5 are usually secreted by M1 macrophages to recruit Th1, Th17, and cytotoxic T cells, while CCL2, CCL17, CCL18, CCL22, and CCL24 are produced by M2 macrophages in most cases.31,38,40
Basic functions of macrophages
One of the basic functions of macrophages is phagocytosis. Through phagocytosis, macrophages can clear erythrocytes, apoptotic cells, and effete cells to maintain homeostasis. Neutropenia and splenomegaly may occur when neutrophils and erythrocytes in the spleen and liver cannot be phagocytized.53 This type of clearance process is independent of immune responses and is regarded as the fundamental function of macrophages.54 When pathogens or aberrant cells, such as tumor cells, are recognized by macrophages, they can be phagocytized and processed into antigen peptides. Macrophages present these peptides to MHC class II molecules on their surface and stimulate T-cell proliferation and activation with the synergistic effect of costimulatory molecules.55,56 It has been reported that adult macrophages are primarily responsible for host defense, while fetal macrophages are involved in tissue remodeling.40 Macrophages play an important role in the development and homeostasis. For example, microglia are required in almost every precise developmental stage of the central nervous system.57 Cardiac macrophages help maintain homeostasis in the steady-state heart by facilitating myocardial conduction.58 CCR2− macrophages are instrumental in cardiac recovery, coronary development, and postnatal coronary growth.59,60 Impaired activation or depletion of Kupffer cells leads to hepatic steatosis and insulin resistance.61–63 Defects in perivascular macrophages can give rise to the unsuccessful establishment of the blood–brain barrier.64 It is well known that macrophages are related to many diseases. Here, we will focus on its role in tumors in the following sections.
Functions of macrophages in cancers
By secreting various factors and affecting other immune cells, macrophages not only play a role in chronic inflammation but also initiate, promote, or suppress the development of cancer. Ornithine, VEGF, EGF, and TGF-β are examples of tumor-promoting factors derived from macrophages, while nitric oxide generated by inducible nitric oxide synthase in macrophages can inhibit tumor growth.32,33,65,66 Macrophages have been demonstrated to be involved directly or indirectly in several key features of malignant tumors, including angiogenesis, invasiveness, metastasis, regulation of the tumor microenvironment, and therapeutic resistance (Fig. 3).
Fig. 3.
Functions of macrophages in cancers. (1) promotion of angiogenesis; (2) induction of invasiveness and metastasis; (3) regulation of the tumor microenvironment; and (4) induction of therapeutic resistance
Angiogenesis
By expressing WNT7B, WNT5A, WNT11, VEGF-C, VEGF-D, and other factors, macrophages are deeply involved in vasculogenesis and lymphogenesis.67–70 In addition, TAMs can enhance tumor hypoxia and glycolysis,71 two important causes of angiogenesis.72,73 HIF-1a is a protein induced in hypoxia conditions. It has been demonstrated that HIF-1a is an important transcriptional factor regulating the transcription of angiogenesis-associated genes, such as VEGF, bFGF, PDGF, and PGE2 in TAMs.74,75 Through the synthesis of WNT7B, macrophages also stimulate vascular endothelial cells to produce VEGF.76 Other TAM-produced proangiogenic molecules that recruit or activate endothelial cells include CXCL12, TNF-α, IL-1β, IL-8, Sema4d, adrenomedullin, and thymidine phosphorylase.41,77,78 Studies on liver diseases have revealed that in addition to producing proangiogenic molecules, macrophages can benefit the formation of complex vascular networks by interacting with the sprouting vasculature.79 Live imaging showed that macrophages drive sprouting angiogenesis via VEGF signaling and coordinate blood vessel regression in wound healing by clearing apoptotic endothelial cells.80 Preventing macrophages from entering avascular areas by blocking the Sema3A/Nrp1 signaling pathway could inhibit angiogenesis.81 It has been reported that angiogenic macrophages are similar to fetal counterparts based on their characteristic expression of TIE2.77,82 Targeting TIE2 or its ligand ANG2 inhibits angiogenesis in certain tumor models, such as those for breast and pancreatic cancers.82 Depletion of TAMs inhibits angiogenesis.74,83 A close relationship between macrophages and angiogenesis has been discussed in previous reviews.84,85
Invasiveness and metastasis
Macrophages can not only increase the density of blood vessels but also promote the invasiveness and metastasis of tumor cells. By expressing matrix metalloproteinases, cathepsin, urokinase plasminogen activator, and matrix remodeling enzymes, such as lysyl oxidase and osteonectin, macrophages dissolve the extracellular matrix to pave the path for tumor cell escape.4 TAMs upregulate cytokines, such as IL-1ra, to promote metastasis by enhancing tumor cell stemness.86 Secretion of TGF-β and growth factors, such as EGF analogs, promotes epithelial–mesenchymal transition and invasiveness of tumor cells.87–90 Exosomes released from M2 macrophages are responsible for cancer metastasis by transferring certain miRNAs into cancer cells, such as colorectal cancer and pancreatic ductal adenocarcinoma cells.91,92
In addition to macrophages in primary tumors, macrophages can also assist in tumor survival and colonization at premetastatic lesions. It has been demonstrated that macrophages are required for the early dissemination of breast cancer, and early disseminated macrophages contribute to late metastasis.93 Tumor exosomes are crucial in tumor organotropic metastasis. It has been observed that pancreatic cancer cell-derived exosomes preferentially colocalize with macrophages in liver metastasis sites.94 Exosome-educated macrophages facilitate premetastatic niche formation via secretion of TGF-β.95 In addition, the interplay between integrin a4 on macrophages and VECAM1 on tumor cells promotes cancer cell survival.96 Results from other studies support the indispensable role of monocytes/macrophages recruited to premetastatic niches in promoting circulating tumor cell survival and colonization in metastatic lesions.97,98 At lung metastasis nodules of breast cancer, CCL2 produced by tumor cells is an important chemokine for the recruitment and retention of inflammatory monocytes/macrophages.99 By recruiting Ly6C+ monocytes via CCL2, fibrocytes prepare a premetastatic niche in the lung for melanoma cells.100 After differentiating of CCR2+Ly6C+ inflammatory monocytes into Ly6C− macrophages, these cells accelerate tumor cell extravasation by generating VEGF.101
Tissue-resident macrophages have also been demonstrated to promote or restrict metastasis. Alveolar macrophages promote hepatocellular carcinoma lung metastasis by producing an inflammatory mediator, leukotriene B4.102 By suppressing Th1 responses, alveolar macrophages facilitate breast tumor cells to metastasize.103 Kupffer cells engulf cancer cells in a Dectin-2-dependent manner to limit liver metastasis.104
Effects of macrophages on tumor microenvironment
Many factors, such as CSF1, VEGF-A, CXCL12, ANG2, CCL5, and CCL2, in solid tumors, can recruit angiogenic macrophages.77,101,105–108 This enrichment allows macrophages to play a major role in the construction of the tumor immune microenvironment. Granulin generated by TAMs can induce fibrosis, which prevents T cells from infiltrating.109,110 Attenuation of the TAM antigen presentation ability results in a reduction in T-cell activation and proliferation.40 Exosomes consisting of various miRNAs derived from TAMs orchestrate an immunosuppressive tumor microenvironment by causing Treg/Th17 imbalance.111 It has been summarized that tumor-associated macrophages support a suppressive tumor microenvironment in three ways:112 (1) by consuming the metabolites, (e.g., L-arginine, which is essential for T-cell activation, can be metabolized by TAMs with high expression of ARG1.) (2) by producing the cytokines and chemokines, IL-10, TGF-β, and PGE2, which are primarily secreted by TAMs, to inhibit the activation and function of various immune cells, including cytotoxic T cells, but induce and maintain regulatory T cells, (3) by expressing inhibitory molecules. TAMs elicit immune suppression through the expression of inhibitory receptors or immune checkpoint ligands (e.g., MHC-I molecules, PD-L1, PD-L2, CD80, CD86, B7-H4 and VISTA). These molecules deliver an inhibitory signal to ligand- or receptor-expressing immune cells.
Therapeutic resistance
Macrophages are also an important cell-extrinsic factor that mediates the resistance of tumor cells to chemotherapy or radiotherapy. By expressing IL-6, TNF-α, cathepsin B and S, or inducing other cells to produce IL-6, macrophages activate STAT3 in tumor cells, which enhances the proliferation and survival of malignant cells under treatment with several chemotherapeutics.113 The epithelial to mesenchymal transition, which can be elicited by macrophages, has been demonstrated to be another mechanism behind chemoresistance.114–116 Exosomal miR-223 from macrophages has been reported to cause a chemoresistant phenotype after being delivered into epithelial ovarian cancer cells.117 miR-21 derived from macrophages is responsible for cisplatin resistance in gastric cancer cells.118 Macrophages exacerbate fatty acid beta-oxidation of gastric cancer cells by generating growth differentiation factor 15 so that the cancer cells are more resistant to 5-fluorouracil treatment.119 Metabolites, including deoxycytidine, from macrophages, weakened the therapeutic effect of gemcitabine in pancreatic ductal adenocarcinoma.120 Murine pancreatic ductal adenocarcinoma models showed an enhanced therapeutic response toward gemcitabine after depleting macrophages with liposomal clodronate.121 As summarized by Marek Nowak et al., TAMs contribute to chemoresistance by inducing prosurvival and antiapoptotic signals in cancer cells, as well as their protumoral polarization.122
It has been reported that irradiation promotes the accumulation and M2 polarization of macrophages.123 Heparin-binding epidermal growth factor, which is primarily secreted by macrophages, could reduce the radiosensitivity of head and neck cancer cells by activating the nonhomologous end-joining pathway.124 TNF-α has a radioprotective function in a TAM-produced VEGF-dependent manner.125 Carcinoembryonic antigen has been identified as a radioresistance marker in colorectal cancer because it induces M2 polarization of macrophages.126 Inhibition of differentiation of M2 macrophages showed enhanced responses to radiotherapy in breast cancer.127 Of note, dying cancer cells after treatment with chemotherapeutics or radiation might also initiate antitumor immune responses. Whether the function of macrophages leads to sensitization or resistance to traditional therapy is complex.128,129 Better understanding of the mechanisms can improve the efficacy of traditional oncotherapy.
Involvement of macrophages in current immunotherapy
Due to the limitations and shortages of traditional cancer treatments, immunotherapy has become the most promising cancer treatment. Various cancer immunotherapy strategies have emerged. These include adoptive cellular immunotherapy, tumor vaccines, antibodies, immune checkpoint inhibitors, and small-molecule inhibitors. Although most of these strategies are not meant to target macrophages directly or originally, macrophages contribute significantly to the final outcomes.
Immune checkpoint inhibitors
To date, many immune checkpoint blockade therapies have been reported, but the most commonly used therapies in the clinic are anti-PD-1 and anti-PD-L1 therapies. Cancer immunotherapy based on inhibiting the immune checkpoints CTLA-4 and PD-1 aim at relieving immune suppression rather than simply reinforcing immune responses. Blocking the PD-1/PD-L1 pathways with inhibitors to enhance the cytotoxic function of T cells has made certain achievements in the resolution of malignancies.130 However, even if the adaptive immune system is compromised131 or the function of T cells cannot be fully recovered by PD-1 inhibitors under specific circumstances,132 PD-1/PD-L1 antagonisms can still increase antitumor efficacy. Therefore, more immune cell types should be involved in PD-1/PD-L1 inhibitor treatment. Additional studies revealed that both PD-L1 and PD-1 are expressed in TAMs,131,133,134 promoting immune suppression and escape. PD-1+ TAM phagocytosis can be rescued by PD-1/PD-L1 blockade, which leads to a direct decrease in tumor burden.131 Furthermore, anti-PD-1 or PD-L1 immune checkpoint blockade induced an M1 macrophage polarization.135,136 M1 macrophage polarization or repolarization has been linked to an enhanced antineoplastic effect by numerous studies.137–140 Of note, macrophages might play a negative role in anti-PD-1 treatment, such as by preventing cytotoxic T cells from reaching tumor cells.141 In addition, in vivo imaging showed the transfer of an anti-PD-1 antibody from CD8+ T cells to TAMs through the binding of Fc-Fcγ receptors shortly after its administration. Blocking such binding reduced the accumulation of anti-PD-1 antibody in TAMs and prolonged its retention time in CD8+ T cells, leading to the regression of tumors.142
Along with the concept of immune checkpoints on T cells, several checkpoints that are mainly associated with macrophages have also been discovered. CD47 is a poor prognostic factor in tumor cells, and its interaction with SIRPα on macrophages helps tumor cells evade phagocytic clearance by macrophages.143,144 Blocking CD47 has resulted in macrophage-mediated tumor inhibition.145 The inhibitory receptor LILRB1 expressed on macrophages prevents tumor cells from being phagocytosed by interacting with the beta-2 microglobulin (β2M) subunit of the MHC class I complex.146 The CD24-Siglec-10 axis promotes immune evasion by downregulating macrophage phagocytosis.147 Inhibition of these immune checkpoints has significantly increased cancer immunotherapy efficacy.
Tumor vaccines
Vaccines can be divided into two categories: preventive vaccines and therapeutic vaccines. Preventive vaccines are often designed to induce specific adaptive immunity, chiefly humoral immunity, before the occurrence of disease, which is normally caused by infection with a virus or bacteria. Thus, it can be used to reduce the incidence of viral or bacterial infection-induced carcinoma. Typical examples of preventive vaccines are those for HBV or HPV. Although a proper adaptive immune response is believed to be the primary reason for the effectiveness of these vaccines, it has been reported that immediate innate immunity other than time-consuming adaptive immunity is principally responsible for the spontaneous regression of cancer.148,149
Therapeutic vaccines are usually designed to elicit protective T cells. However, Maxime Thoreau et al. demonstrated that cooperation between T cells and macrophages is required to achieve the effects of a therapeutic vaccine. A denser presence of macrophages along with tumor regression has shown to precede the infiltration of CD8+ T cells.150 Numerous approaches choose synthetic peptides, recombinant proteins, whole tumor cells, viral vectors, bacteria or nucleic acids as vaccination candidates to activate T cells via antigen-presenting cells, which are mostly dendritic cells.151 Among these, some regimens that used GM-CSF as an adjuvant generated obvious immune responses.151,152 Sipuleucel-T was the first therapeutic vaccine approved by the FDA to be used in a particular group of prostate cancer patients. A fusion protein combining a targeting tumor antigen prostate acid phosphatase with GM-CSF was used to induce antigen-specific T cells. It prolonged the survival of patients in a few clinical trials.153 A STING agonist formulated with GM-CSF showed remarkable antitumor efficacy in multiple established tumors.154 Some tumor cells used as whole-cell vaccines can secrete GM-CSF.155,156 In addition, oncolytic virotherapy, which increases the targeting of cancer cells through virus infection, could induce antitumor immune responses, especially in cells that had been engineered to express GM-CSF.157,158 GM-CSF is combined for the purpose of enhancing DC functions and limiting Treg regulation. However, GM-CSF could also induce M1 macrophage polarization and activate macrophages to exert an antitumor function.40,159,160 In another virus-related tumor immunotherapy study, Danyang Wang et al. used an NF-κB-activating gene expression adeno-associated virus system to express an artificial neoantigen on the tumor cell surface, which could be targeted by specific immune cells. When they chose calreticulin, a signal to promote phagocytic uptake, the cancer cells could be engulfed by macrophages.161 In addition, exosomes derived from M1- but not M2-polarized macrophages boosted the antitumor vaccine by eliciting a release of Th1 cytokines and a stronger antigen-specific cytotoxic T-cell response.162 Xu et al. reported that a listeria-based tumor vaccine benefited anti-PD-1 therapy against hepatocellular carcinoma by skewing macrophage polarization.163
Antibodies
Checkpoint inhibitors, such as nivolumab (Opdivo) and pembrolizumab (Keytruda), are monoclonal antibodies. In addition, many other monoclonal antibodies have been approved for clinical cancer immunotherapy by the FDA. Rituximab and trastuzumab are examples of these monoclonal antibodies. Rituximab is used in B-cell lymphoma by targeting CD20. B lymphoma cells are more sensitive to macrophages in the presence of rituximab.164 Its combination with cyclophosphamide induced nearly complete tumor elimination in resistant bone marrow by activating macrophages.165 After blocking the CD47-SIRPα axis, rituximab-induced macrophage phagocytosis was augmented in nongerminal center B diffuse large B-cell lymphoma patients.166 Trastuzumab is an HER2-targeting antibody that has shown promising efficacy in breast cancer therapy. It has been reported that antibody-dependent cell phagocytosis mediated by macrophages is the main cause of the effectiveness of trastuzumab plus CD47 blockade.167 By binding with Fcγ receptors on macrophages, trastuzumab triggered macrophage phagocytic killing, and this function was augmented after increasing the expression of Fcγ receptors on macrophages.168 In addition, trastuzumab resistance was overcome by shifting macrophages from the M2 to M1 phenotype.169
Adoptive cell therapy
Adoptive cell therapy is also a very promising therapy that induces tumor regression by transferring specific immune cells to the tumor-bearing host. These cells may come from the host itself or some other donors. They are commonly manipulated to possess better effector functions and proliferate to a sufficient number in vitro before administration.170 Typical examples include T cells with engineered chimeric antigen receptors (CAR-Ts) or gene-modified T-cell receptors (TCR-Ts). In 2006, the adoptive transfer of TCR-engineered lymphocytes, which recognize an antigen named MART-1, caused tumor regression in metastatic melanoma patients.171 In 2010, administration of CAR-T cells against CD19 efficiently eliminated B cells in a patient with follicular lymphoma.172 However, insufficient infiltration into solid tumors is a major limitation for these T-cell-based immunotherapies. Local low-dose irradiation increased T-cell recruitment by inducing M1-phenotype macrophage differentiation.173 Cytokine release syndrome is considered to be closely related to the efficacy of adoptive cell therapy, but serious cytokine release syndrome may lead to death. It has been reported that cytokine release syndrome induced by CAR-T-cell transfer is mediated by macrophages.174 Inhibiting or neutralizing GM-CSF abolished macrophage-derived cytokines, which released syndrome-related cytokines and enhanced CAR-T cell functions.175,176 Therefore, taking the response of macrophages into account may benefit adoptive modified T-cell therapy. Modified macrophages with the chimeric antigen receptor (CARMA) have also been tested by Klichinsky et al. The first generation of chimeric antigen receptors, which combine the scFv of anti-CD19, anti-mesothelin, or anti-HER2 antibodies with a CD3 intracellular domain, has been constructed. This CARMA displayed a strong tumoricidal function in preclinical models.177
Small-molecule inhibitors
Because of several advantages, such as oral bioavailability, the relatively low cost, ease of crossing physiological barriers or access to intracellular targets, small-molecule drugs are complementary and synergistic with other immune-oncology therapies.178 Numerous small-molecule inhibitors have been proven to suppress tumors by targeting macrophage-associated molecules. For example, IDO is a poor prognosis indicator that is often highly expressed in macrophages, dendritic cells, and tumor cells. Small-molecule inhibitors targeting IDO have been tested in clinical trials to reestablish positive immune responses.179,180 ARG1 is a cytosolic enzyme that plays a key role in the immunosuppressive function of TAMs. Compounds inhibiting arginase have shown potential in tumor suppression.181 RRX-001, a small-molecule inhibitor, downregulated not only CD47 on cancer cells but also SIRPα on macrophages and showed hypotoxicity but strong antitumor activity in clinical trials.182 In addition, small-molecule inhibitors have great potential in combination with other oncotherapy strategies. Inhibition of Bcl-2 family members improved the efficacy of CAR-T therapy in B-cell malignancy.183 PI3K-γ inhibitors, such as IPI-549, overcome immune checkpoint resistance by reshaping the tumor microenvironment, including switching macrophage polarization from the M2 to M1 phenotype.184 Small-molecule inhibitors targeting CXCR2 on neutrophils and CCR2 on macrophages improve the chemotherapeutic effects in pancreatic ductal adenocarcinoma models.185 PLX-3397, a small-molecule inhibitor of CSF1R, cKIT, and FLT3 has been demonstrated to decrease tumor burden by reducing M2 macrophages in combination with adoptive cell transfer immunotherapy or other small-molecule inhibitors.186,187 FAK is indispensable for the migration and stable protrusion formation of macrophages. Small-molecule inhibitors against FAK have shown promising antitumor activity, especially when combined with chemotherapy and immunotherapy strategies.188
Prospect: macrophages are a promising target in future cancer immunotherapy
To date, great endeavors to boost T cell-directed anticancer immune responses have been made. As reported, the incidence of cancerogenesis is low in invertebrates with no T or B cells, indicating that innate immune cells are of great importance for preventing the initiation and development of cancer.189–191 In addition to their supporting role in all kinds of immunotherapies, macrophages may become a promising target in future cancer immunotherapy.33,192 Many targets and pharmacological agents related to macrophages in oncotherapy have been summarized in recent reviews.128,193 We updated the typical macrophages-targeting agents that have been registered for cancer-related clinical trials (excluding projects those are in the status of terminated, withdrawn, unknown, not yet recruiting) in Table 2. The potential and promising strategies targeting macrophages have been categorized into six types based on their objectives in Fig. 4. There are several advantages to target macrophages in cancer immunotherapy. Low infiltration is a major barrier for T-cell-based anticancer therapy, and macrophages account for ~30–50% of infiltrating immune cells in the tumor microenvironment. As mentioned above, circulating monocytes are a major source of infiltrating macrophages in tumors, and the accessibility of peripheral blood mononuclear cells makes it easy to operate if a macrophage-based therapy strategy is adopted in the clinic.
Table 2.
Cancer-associated clinical trials (excluding projects those are in the status of terminated, withdrawn, unknown, not yet recruiting) using typical macrophage-targeting agents
| Target | Agent | Organization | ClinicalTrials.gov Identifier | Tumors | Other interventions | Phase |
|---|---|---|---|---|---|---|
| CSF1 | Lacnotuzumab (MCS110) | Novartis Oncology | NCT02435680 | Advanced triple-negative breast cancer | Carboplatin, gemcitabine | II |
| NCT01643850 | Pigmented villonodular synovitis | None | II | |||
| NCT03694977 | Gastric cancer | PDR001 | II | |||
| CCL2 | Carlumab (CNTO 888) | Centocor Research & Development | NCT01204996 | Solid tumors | Standard of care | I |
| NCT00992186 | Prostate cancer | None | II | |||
| SIRPα | TTI-622 | Trillium Therapeutics | NCT03530683 | Advanced relapsed or refractory lymphoma or myeloma | Rituximab, PD-1 inhibitor, proteasome-inhibitor regimen | I |
| CC-95251 | Celgene | NCT03783403 | Advanced solid and hematologic cancer | None | I | |
| BI 765063 (OSE-172) | OSE Immunotherapeutics | NCT03990233 | Advanced solid tumors | BI 754091 | I | |
| FSI-189 | Gilead Sciences | NCT04502706 | Relapsed/refractory non-Hodgkin lymphoma | None | I | |
| TIE2 | CEP-11981 (ESK981) | Karmanos Cancer Institute | NCT04159896 | Prostate cancer | Nivolumab | II |
| NCT00875264 | Advanced cancer | None | I | |||
| NCT03456804 | Prostate cancer | None | II | |||
| Regorafenib (BAY 73-4506) | Bayer | NCT04170556 | Hepatocellular carcinoma | Nivolumab | I/II | |
| NCT04476329 | Hepatocellular carcinoma | None | II | |||
| Arry-614 | Array BioPharma | NCT01496495 | Myelodysplastic syndromes | None | I | |
| NCT00916227 | Myelodysplastic syndromes | None | I | |||
| Arginase | INCB001158 (CB1158) | Incyte | NCT03910530 | Advanced solid tumors | None | I |
| NCT02903914 | Advanced/metastatic solid tumors | Pembrolizumab | I/II | |||
| NCT03314935 | Solid tumors | Oxaliplatin, leucovorin, 5-fluorouracil, gemcitabine, cisplatin, paclitaxel | I/II | |||
| NCT03837509 | Multiple myeloma | Daratumumab | I/II | |||
| HER2 | CAR-macrophage | Carisma Therapeutics lnc. | NCT04660929 | HER2 overexpressing solid tumors | None | I |
| GC vitamin D-binding protein | EF-022 | Efranat | NCT02052492 | Solid tumors | None | I |
| CD40 | SEA-CD40 | Seattle Genetics | NCT02376699 | Solid tumors | Pembrolizumab | I |
| APX005M | Apexigen | NCT03389802 | Pediatric CNS | None | I | |
| NCT04130854 | Locally advanced rectal adenocarcinoma | None | II | |||
| NCT02482168 | Non-small-cell lung cancer, melanoma, urothelial carcinoma, head and neck cancer | None | I | |||
| NCT03165994 | Esophageal cancer, gastroesophageal cancer | Radiation therapy, paclitaxel, carboplatin | II | |||
| NCT03719430 | Soft tissue sarcoma | Doxorubicin | II | |||
| NCT03214250 | Metastatic pancreatic Adenocarcinoma | Nivolumab, nab-paclitaxel, gemcitabine | I/II | |||
| NCT04337931 | Melanoma | None | II | |||
| NCT02706353 | Melanoma | Pembrolizumab | I/II | |||
| CP-870,893 | VLST Corporation | NCT01103635 | Metastatic melanoma | Tremelimumab (anti- CTLA-4) | I | |
| Selicrelumab (R07009879) | Roche | NCT02760797 | Advanced solid tumors | Anti-PD-L1 | I | |
| NCT02665416 | Advanced solid tumors | Bevacizumab or vanucizumab | I | |||
| NCT02304393 | Solid tumors | Atezolizumab | I | |||
| NCT02588443 | Pancreatic ductal adenocarcinoma | Gemcitabine, nab-paclitaxel | I | |||
| CDX-1140 | Celldex Therapeutics | NCT04491084 | Non-small-cell lung cancer, lung cancer | CDX-301 | I/II | |
| NCT04520711 | Malignant epithelial neoplasms | TCR-T, pembrolizumab | I | |||
| NCT04616248 | Unresectable or metastatic breast cancer | Poly ICLC, radiation therapy, recombinant Flt3 ligand | I | |||
| NCT04364230 | Melanoma | 6MHP, NeoAg-mBRAF, Poly ICLC | I/II | |||
| NCT03329950 | Advanced malignancies | CDX-301, pembrolizumab, chemotherapy | I | |||
| Dacetuzumab (SGN-40) |
Genentech, Inc. Seagen Inc. |
NCT00525447 | Multiple myeloma | Lenalidomide, dexamethasone | I | |
| NCT00079716 | Multiple myeloma | None | I | |||
| NCT00435916 | Large B-cell diffuse lymphoma, non-Hodgkin lymphoma | None | II | |||
| NCT00103779 | Non-Hodgkin lymphoma | None | I | |||
| NCT00655837 | Large B-cell diffuse lymphoma, non-Hodgkin lymphoma | Rituximab, gemcitabine | I | |||
| NCT00556699 | Non-Hodgkin’s lymphoma | Rituximab | I | |||
| NCT00664898 | Multiple myeloma | Bortezomib | I | |||
| NCT00283101 | Lymphocytic, chronic leukemia | None | I/II | |||
| Lucatumumab (HCD122) | Novartis Pharmaceuticals | NCT00670592 | Non-Hodgkin’s lymphoma, Hodgkin’s lymphoma | None | I/II | |
| NCT01275209 | Follicular lymphoma | None | I | |||
| NCT00231166 | Multiple myeloma | None | I | |||
| 2141 V-11 | Rockefeller University | NCT04547777 | Glioma | None | I | |
| NCT04059588 | Solid tumor, skin cancer | D2C7-IT | I | |||
| ADC-1013 (JNJ-64457107) | Janssen Research & Development, LLC | NCT02829099 | Advanced solid neoplasms | None | I | |
| LVGN7409 | Lyvgen Biopharma Holdings Limited | NCT04635995 | Cancer | None | I | |
| Chi Lob 7/4 | Cancer Research UK | NCT01561911 | Neoplasms, lymphoma, non-Hodgkin, B cell | None | I | |
| NG-350A | PsiOxus Therapeutics | NCT03852511 | Metastatic cancer, epithelial tumor | None | I | |
| BTK | Ibrutinib (PCI-32765) | Pharmacyclics LLC | NCT02599324 | Renal cell, urothelial, gastric, colon, pancreatic adenocarcinoma | None | Ib/II |
| NCT01478581 | Multiple myeloma | Dexamethasone | I | |||
| NCT01752426 | Leukemia | heavy water (2H2O) | I, II | |||
| NCT01236391 | Mantle cell lymphoma | None | II | |||
| NCT01105247 | B-cell chronic lymphocytic leukemia, small lymphocytic lymphoma | None | I, II | |||
| NCT01614821 | Waldenstrom’s macroglobulinemia | None | II | |||
| NCT01292135 | B-cell chronic lymphocytic leukemia, small lymphocytic lymphoma | None | I | |||
| NCT01520519 | Leukemia | Rituximab | II | |||
| NCT01109069 | B-cell lymphoma, chronic lymphocytic leukemia | None | II | |||
| NCT01217749 | Chronic lymphocytic leukemia | Ofatumumab | I, II | |||
| NCT02403271 | Non-small-cell lung cancer, breast cancer, pancreatic cancer | Durvalumab | I, II | |||
| NCT01646021 | Mantle cell lymphoma | Temsirolimus | III | |||
| NCT01855750 | Lymphoma | Rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone | II | |||
| NCT01980628 | Marginal zone lymphoma, B-cell lymphoma | None | II | |||
| NCT01589302 | Prolymphocytic leukemia, small lymphocytic lymphoma, chronic lymphocytic leukemia | None | II | |||
| NCT01325701 | Diffuse large cell B lymphoma | None | II | |||
| NCT01578707 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | Ofatumumab | III | |||
| NCT01722487 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | Chlorambucil | III | |||
| NCT02436668 | Metastatic pancreatic adenocarcinoma | Gemcitabine, nab-paclitaxel | III | |||
| NCT01980654 | Follicular lymphoma, B-cell lymphoma, non-Hodgkin’s lymphoma | Rituximab | II | |||
| NCT01973387 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | Rituximab | III | |||
| NCT01611090 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | Bendamustine, hydrochloride, rituximab | III | |||
| NCT02401048 | Diffuse large B-cell lymphoma, follicular lymphoma | MEDI4736 | I, II | |||
| NCT02639910 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | Tafasitamab, idelalisi, venetoclax | II | |||
| NCT02902965 | Multiple myeloma | Bortezomib dexamethasone | II | |||
| NCT01744691 | Chronic lymphocytic leukemia with 17p deletion, small lymphocytic lymphoma with 17p deletion | None | II | |||
| NCT02264574 | Chronic lymphocytic leukemia, small-cell lymphoma | Obinutuzumab, chlorambucil | III | |||
| NCT02514083 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | Fludarabine | II | |||
| Acalabrutinib (ACP-196) | Acerta Pharma BV | NCT02112526 | Activated B-cell diffuse large B-cell lymphoma | None | I | |
| NCT02180724 | Waldenström macroglobulinemia | None | II | |||
| NCT02213926 | Mantle cell lymphoma | None | II | |||
| NCT02211014 | Multiple myeloma | None | I | |||
| Zanubrutinib (BGB-3111) | BeiGene | NCT03206970 | Mantle cell lymphoma | None | II | |
| NCT03206918 | Chronic lymphocytic leukemia, small lymphocytic lymphoma | None | II | |||
| CSF1R | Pexidartinib (PLX-3397) | Plexxicon | NCT02371369 | Tenosynovial giant cell tumor | None | III |
| NCT02472275 | Intermediate- or high-risk prostate cancer | None | I | |||
| NCT02584647 | Sarcoma, malignant peripheral nerve shealth tumors | Sirolimus | I | |||
| NCT01596751 | Metastatic breast cancer | Eribulin | Ib/II | |||
| NCT02777710 | Pancreatic or colorectal cancers | Durvalumab | I | |||
| NCT02734433 | Advanced solid tumors | None | I | |||
| NCT03158103 | Gastrointestinal stromal tumor | MEK162 | I | |||
| BLZ945 | Novartis | NCT02829723 | Advanced solid tumors | PDR001 | I | |
| ARRY-382 | Array Biopharma | NCT01316822 | Metastatic cancer | None | I | |
| NCT02880371 | Advanced solid tumors | Pembrolizumab | II | |||
| Edicotinib (JNJ-40346527) | Johnson & Johnson | NCT03177460 | Prostate cancer | None | I | |
| IMC-CS4(LY3022855) | Eli Lilly | NCT01346358 | Advanced solid tumors | None | I | |
| NCT02265536 | Advanced breast, prostate cancer | None | I | |||
| NCT02718911 | Solid tumor | Durvalumab, tremelimumab | I | |||
| NCT03101254 | Melanoma | Vemurafenib cobimetinib | I & II | |||
| NCT03153410 | Pancreatic ductal adenocarcinoma | Cyclophosphamide, pembrolizumab, GVAX | I | |||
| Cabiralizumab (FPA008) | Five Prime Therapeutics | NCT02471716 | Tenosynovial giant cell tumor | None | II | |
| NCT03927105 | Peripheral T-cell lymphoma | Nivolumab | II | |||
| NCT03502330 | Melanoma, non-small-cell lung cancer, renal cell carcinoma | APX005M nivolumab | I | |||
| NCT04331067 | Triple-negative breast cancer | Nivolumab | Ib/II | |||
| NCT03158272 | Advanced malignancy | Nivolumab | I | |||
| NCT02526017 | Advanced solid tumors | Nivolumab | I | |||
| Emactuzumab (RO5509554) | Hoffman La Roche | NCT02323191 | Advanced solid tumors | Atezolizumab | I | |
| NCT02760797 | Advanced solid tumors | RO7009789 | I | |||
| NCT01494688 | Advanced solid tumors | Paclitaxel | I | |||
| NCT03708224 | Advanced head and neck squamous cell carcinoma | Atezolizumab | II | |||
| NCT03193190 | Pancreatic ductal adenocarcinoma | Additional therapies | I/II | |||
| TPX-0022 | Turning Point Therapeutics, Inc. | NCT03993873 | Advanced solid tumor | None | I | |
| DCC-3014 | Deciphera Pharmaceuticals LLC | NCT04242238 | Sarcoma | Avelumab | I | |
| NCT03069469 | Advanced malignant neoplasm | None | I & II | |||
| Q702 | Qurient Co., Ltd. | NCT04648254 | Solid tumor | None | I | |
| SNDX-6532 | Syndax | NCT03238027 | Solid tumor | Durvalumab | I | |
| NCT04301778 | Unresectable intrahepatic cholangiocarcinoma | Durvalumab | II | |||
| CD47 | Magrolimab (Hu5F9-G4) | Gilead Sciences | NCT02216409 | Solid tumor | None | I |
| NCT03248479 | Hematological Malignancies | Azacitidine | I | |||
| NCT02678338 | Acute myeloid leukemia, myelodysplastic syndrome | None | I | |||
| NCT03527147 | Non-Hodgkin’s lymphoma | AZD9150 acalabrutinib AZD6738 rituximab AZD5153 | I | |||
| NCT04599634 | B-cell malignancies | Obinutuzumab venetoclax | I | |||
| NCT02953782 | Advanced solid malignancies and colorectal carcinoma | Cetuximab | I | |||
| NCT03558139 | Ovarian cancer | Avelumab | I | |||
| NCT03248479 | Hematological malignancies | Azacitidine | I | |||
| NCT04541017 | T-cell lymphoma | Mogamulizumab | I/II | |||
| NCT03922477 | Acute myeloid leukemia | Atezolizumab | I | |||
| NCT04435691 | Acute myeloid leukemia | Azacitidine, venetoclax | I/II | |||
| NCT03869190 | Urothelial carcinoma | Atezolizumab, enfortumab, vedotin, niraparib | I/II | |||
| NCT02953509 | Non-Hodgkin lymphoma | Rituximab, gemcitabine, oxaliplatin | I/II | |||
| NCT04313881 | Myelodysplastic syndromes | Azacitidine | III | |||
| TTI-621 | Trillium Therapeutics | NCT02890368 | Solid tumors and mycosis fungoides | PD-1/PD-L1 inhibitor, pegylated interferon-α2a, radiation, talimogene laherparepvec | I | |
| NCT02663518 | Small-cell lung cancer | None | I | |||
| AO-176 | Arch Oncology | NCT03834948 | Solid tumor | Paclitaxel | I/II | |
| NCT04445701 | Multiple myeloma | Bortezomib, dexamethasone | I/II | |||
| IBI322 | Innovent Biologics (Suzhou) Co., Ltd | NCT04328831 | Advanced malignancies | None | I | |
| NCT04338659 | Advanced malignancies | None | I | |||
| ZL1201 | Zai Lab (Shanghai) Co., Ltd. | NCT04257617 | Locally advanced solid tumor | None | I | |
| CC-90002 | Celgene | NCT02367196 | Hematologic neoplasms | Rituximab | I | |
| HX009 | Waterstone Hanxbio Pty Ltd | NCT04097769 | Advanced solid tumor | None | I | |
| IBI188 | Innovent Biologics (Suzhou) Co. Ltd. | NCT03717103 | Advanced malignancies | Rituximab | I | |
| NCT03763149 | Advanced malignancies | None | I | |||
| SRF231 | Surface Oncology | NCT03512340 | Advanced solid cancers, hematologic cancers | None | I | |
| AK117 | Akesobio Australia Pty Ltd | NCT04349969 | Neoplasms malignant | None | I | |
| IMC-002 | ImmuneOncia Therapeutics Inc. | NCT04306224 | Solid tumor, lymphoma | None | I | |
| CCR2 | BMS-813160 | Bristol-Myers Squibb | NCT03184870 | Colorectal/pancreatic cancer | Chemotherapy or nivolumab | Ib/II |
| NCT03496662 | Pancreatic cancer | Nivolumab abraxane, gemcitabine | I/II | |||
| NCT03767582 | Pancreatic cancer | Radiation therapy, nivolumab, GVAX | I/II | |||
| NCT04123379 | Non-small-cell lung cancer, hepatocellular carcinoma | Nivolumab, BMS-986253 | II | |||
| NCT02996110 | Advanced cancer | Nivolumab, ipilimumab, relatlimab, BMS-986205 | II | |||
| CCX872-B | ChemoCentryx | NCT03778879 | Pancreatic cancer | Radiation therapy | II | |
| MLN1202 | Millenium | NCT01015560 | Bone metastases | None | II | |
| PF-04136309 | Pfizer | NCT02732938 | Metastatic pancreatic ductal adenocarcinoma | Nab-paclitaxel, Gemcitabine | II |
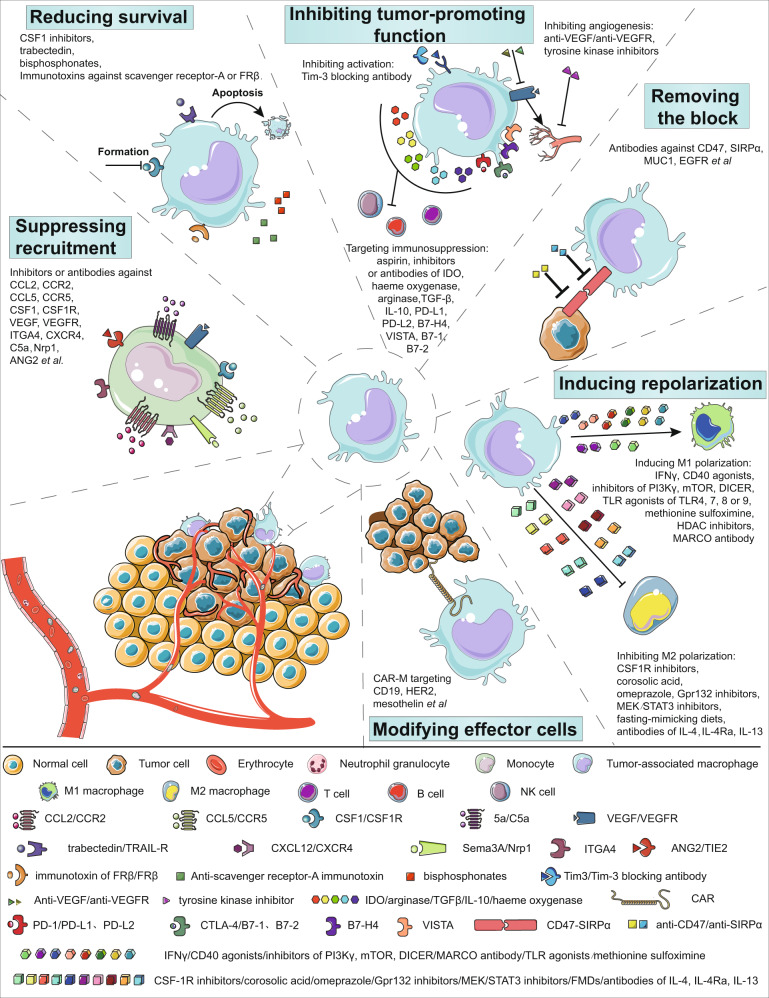
Fig. 4.
Strategies for targeting macrophages for tumor immunotherapy. These strategies are categorized into six types based on their objectives. Agents or drugs are listed as examples in the subcategory for one of their main effects. This may not be the only effective because of their complex mechanisms. (1) Suppression of macrophage recruitment;81,82,205 molecules on monocytes/macrophages, such as CCR2, CCR5, VEGFR, CSF1R, ITGA4, and C5a, contribute to the infiltration of macrophages into tumors. Inhibitors or antibodies against them or some of their ligands (such as CCL2, CCL5, VEGF, and CSF1) could suppress the recruitment of macrophages. Reduced angiogenesis caused by targeting Nrp1 and ANG2 could also result in a decrease in macrophage recruitment. (2) Reduction of macrophage survival.205–208 As CSF1 is a crucial signal for the differentiation of macrophages, CSF1 inhibitors restrain the formation of macrophages. Trabectedin could also be used to reduce the survival of macrophages by inducing apoptosis. Immunotoxins targeting scavenger receptor-A or folate receptor β (FRβ) can deplete TAMs, and bisphosphonates are metabolic analogs that reduce macrophages. (3) Inhibition of tumor-promoting functions;205,209–211 Tim-3 blocking antibody is reported to regulate the activation of TAMs. By inhibiting angiogenesis, anti-VEGF, anti-VEGFR, and tyrosine kinase inhibitors could weaken the protumoral function of TAMs. TAMs contribute to an immunosuppressive microenvironment by expressing indoleamine-pyrrole 2,3-dioxygenase (IDO), heme oxygenase, arginase, TGFβ, IL-10, prostaglandins, and so on. Aspirin reduces the generation of prostaglandins. Blocking immune checkpoints (PD-L1, PD-L2, B7-H4, VISTA, B7-1, and B7-2) on macrophages could relieve the function of other immune cells. (4) Removal of the macrophage blockade;207,212,213 interactions between CD47 on tumors and SIRPα on macrophages help tumor cells evade macrophage phagocytosis. Antibodies against CD47 or SIRPα could remove the blockage. In addition, antibodies against MUC1 and EGFR inhibit SIRPα. (5) Induction of repolarization;43,113,193,207,210,214–222 M1 polarization of TAMs is associated with antitumor responses, while M2 polarization is associated with protumor activities. Several factors can induce M1 polarization, including IFNγ, CD40 agonists, inhibitors of PI3Kγ/mTOR/DICER, agonists of TLR4/7/8/9, methionine sulfoximine, histone deacetylase (HDAC) inhibitors, and antibodies against macrophage receptors with collagenous structures (MARCOs). In contrast, factors inhibiting M2 polarization, such as CSF1R inhibitors, corosolic acid, omeprazole, Gpr132 inhibitors, MEK/STAT3 inhibitors, fast-mimicking diets, and antibodies against IL-4, IL-4Rα, and IL-13, can also reduce the tumor burden. (6) Modification of effector cells.177 Chimeric antigen receptor macrophages (CAR-Ms) similar to CAR-T cells have been used to enhance tumoricidal functions. Targets, such as CD19, HER2, and mesothelin, have been explored
Currently, it is generally believed that cancer cells originate from endogenous cells in humans. Even if numerous tumor-specific antigens have been identified, most specific antigens still exist in a few normal cells. In contrast, not all cancer cells express just one specific antigen because of tumor heterogeneity. Clearance of specific antigen-expressing cancer cells may only result in temporary and limited antitumor efficacy. Nevertheless, as a type of innate immune cell, macrophages can exert a tumor-suppressive function without targeting one specific antigen.194,195
Macrophages are a double-edged sword in the tumor microenvironment. As a prominent component of tumor stromal cells, macrophages can gather around blood vessels, induce angiogenesis, and promote tumor invasion. On the other hand, they could also phagocytose cancer cells and remodel the tumor microenvironment. Fortunately, the polarization of macrophages can be repolarized. The transformation from M2- to M1-phenotype macrophages is sufficient to cause a tumor-suppressive effect.194–196 Of note, the polarization of macrophages is independent of T cells, while M1 macrophages are able to induce Th1 immune responses, and M2 macrophages can trigger Th2 immune responses.197 This provides an opportunity to target macrophages in cancer immunotherapy. More importantly, the direction of macrophages to T or B cells does not rely on the existence of tumor-specific antigens. While IFN-γ from M1 macrophages is an incentive for Th1 responses, TGF-β and IL-10-derived M2 macrophages cause the generation of Treg cells.32,113,197
Trogocytosis is a process in which a tumor-derived antigen is transferred to Fcγ receptor-expressing lymphocytes with the help of certain antibodies. It has been demonstrated that tumor cells decrease the expression of specific antigens by delivering them to CAR-T cells or NK cells, leading to fratricide T cells or NK cells.198,199 Trogocytosis has also been discovered between tumor cells and macrophages and is partially responsible for tumor immune escape.200,201 However, Velmurugan et al. reported that persistent trogocytosis of macrophages eventually leads to the killing of antibody-opsonized tumor cells. They explained that these discrepancies might be caused by limited contact time between two types of cells and the lack of competing endogenous antibodies under physiological conditions.202 Moreover, macrophages are capable of presenting antigens. Proteins that have been passed to the plasma membrane by trogocytosis might be more likely to be processed and presented than cytosolic proteins.
In addition, as mentioned above, macrophages from different sources may exert different functions. This offers an opportunity for more accurately targeted immunotherapy. For example, CCR2+Ly6C+ inflammatory monocytes can be recruited to pulmonary metastasis sites by CCL2 secreted by tumor cells and then differentiate into Ly6C− macrophages that promote metastasis.101 Selectively targeting this group of monocytes may reduce metastasis without damaging the homeostasis maintaining functions of residual macrophages.
Macrophages also have advantages in certain types of cancer. Approximately 20% of nonparenchymal cells in the liver are macrophages. Macrophages in different locations function differently. By stimulating adaptive immune responses, they exert tumoricidal or protumoral and, in general, protumoral functions.203 It has been summarized in a previous review that targeting pathogenic macrophages is a promising option for patients with liver disease.204 Moreover, ascites is a common pathological phenomenon in liver cancer that is often accompanied by a poor prognosis. Integrated single-cell RNA sequencing revealed that lymphocytes in ascites are similar to those in peripheral blood, while myeloid cells in ascites are more likely to originate from tumor-infiltrating myeloid cells. This notion was further confirmed by RNA velocity and phylogenetic trees of macrophages from various tissues. According to this study, intratumoral macrophage-based immunotherapy for hepatocellular carcinoma can not only resolve tumor burden in situ but also relieve ascites.
Thus, macrophages provide a force to be considered in tumor immunotherapy. Research on macrophages might open a new door for oncotherapy. To address various malignancies, more strategies based on or combined with macrophages need to be explored in the future.
Acknowledgements
Several elements used for figures in this review were downloaded from https://smart.servier.com. The Servier Medical Art by Servier is licensed under a Creative Commons Attribution 3.0 Unported License. This work was supported by the National Natural Science Foundation of China (NSFC) (No. 81672914), (No. 81472654) (Y. Luo), (No. 81601374) (Z. Duan), and the Fundamental Research Funds for the Central Universities (3332020033, Z. Duan).
Author contributions
Z.D. wrote the paper and Y.L. revised it.
Competing interests
The authors declare no competing interests.
References
- 1.van Furth R, et al. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull. World Health Organ. 1972;46:845–852. [PMC free article] [PubMed] [Google Scholar]
- 2.Xuetao Cao, W. H. Medical Immunology, third edn. (People’s Medical Publishing House, 2015).
- 3.Hashimoto D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019;19:369–382. doi: 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat. Immunol. 2012;13:1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortez-Retamozo V, et al. Origins of tumor-associated macrophages and neutrophils. Proc. Natl Acad. Sci. USA. 2012;109:2491–2496. doi: 10.1073/pnas.1113744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shand FH, et al. Tracking of intertissue migration reveals the origins of tumor-infiltrating monocytes. Proc. Natl Acad. Sci. USA. 2014;111:7771–7776. doi: 10.1073/pnas.1402914111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 10.Robben PM, LaRegina M, Kuziel WA, Sibley LD. Recruitment of Gr-1+ monocytes is essential for control of acute toxoplasmosis. J. Exp. Med. 2005;201:1761–1769. doi: 10.1084/jem.20050054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shechter R, et al. Infiltrating blood-derived macrophages are vital cells playing an anti-inflammatory role in recovery from spinal cord injury in mice. PLoS Med. 2009;6:e1000113. doi: 10.1371/journal.pmed.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunay IR, et al. Gr1(+) inflammatory monocytes are required for mucosal resistance to the pathogen Toxoplasma gondii. Immunity. 2008;29:306–317. doi: 10.1016/j.immuni.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez FO, Helming L, Gordon S. Alternative activation of macrophages: an immunologic functional perspective. Annu. Rev. Immunol. 2009;27:451–483. doi: 10.1146/annurev.immunol.021908.132532. [DOI] [PubMed] [Google Scholar]
- 14.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nandi S, et al. The CSF-1 receptor ligands IL-34 and CSF-1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev. Biol. 2012;367:100–113. doi: 10.1016/j.ydbio.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 18.Menezes S, et al. The heterogeneity of Ly6C(hi) monocytes controls their differentiation into iNOS(+) macrophages or monocyte-derived dendritic cells. Immunity. 2016;45:1205–1218. doi: 10.1016/j.immuni.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mass E, et al. Specification of tissue-resident macrophages during organogenesis. Science. 2016;353:6304. doi: 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier EL, et al. Systemic analysis of PPARgamma in mouse macrophage populations reveals marked diversity in expression with critical roles in resolution of inflammation and airway immunity. J. Immunol. 2012;189:2614–2624. doi: 10.4049/jimmunol.1200495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosas M, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.N AG, et al. The nuclear receptor LXRalpha controls the functional specialization of splenic macrophages. Nat. Immunol. 2013;14:831–839. doi: 10.1038/ni.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saeed S, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmad I, Valverde A, Ahmad F, Naqvi AR. Long noncoding RNA in myeloid and lymphoid cell differentiation, polarization and function. Cells. 2020;9:269. doi: 10.3390/cells9020269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolter J, Kierdorf K, Henneke P. Origin and differentiation of nerve-associated macrophages. J. Immunol. 2020;204:271–279. doi: 10.4049/jimmunol.1901077. [DOI] [PubMed] [Google Scholar]
- 26.Varol C, Mildner A, Jung S. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 2015;33:643–675. doi: 10.1146/annurev-immunol-032414-112220. [DOI] [PubMed] [Google Scholar]
- 27.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat. Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bian Z, et al. Deciphering human macrophage development at single-cell resolution. Nature. 2020;582:571–576. doi: 10.1038/s41586-020-2316-7. [DOI] [PubMed] [Google Scholar]
- 29.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills CD, Lenz LL, Harris RA. A breakthrough: macrophage-directed cancer immunotherapy. Cancer Res. 2016;76:513–516. doi: 10.1158/0008-5472.CAN-15-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klink, M. Interaction of Immune and Cancer Cells (Chemical Industry Press Co, Ltd, 2016).
- 32.Mills CD. Anatomy of a discovery: M1 and M2 macrophages. Front. Immunol. 2015;6:212. doi: 10.3389/fimmu.2015.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mills CD. M1 and M2 macrophages: oracles of health and disease. Crit. Rev. Immunol. 2012;32:463–488. doi: 10.1615/CritRevImmunol.v32.i6.10. [DOI] [PubMed] [Google Scholar]
- 34.Mills CD, Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. J. Innate Immun. 2014;6:716–726. doi: 10.1159/000364945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol. 2011;11:750–761. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 36.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41:49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petty AJ, Yang Y. Tumor-associated macrophages: implications in cancer immunotherapy. Immunotherapy. 2017;9:289–302. doi: 10.2217/imt-2016-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J. Pathol. 2013;229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 39.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Investig. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapouri-Moghaddam A, et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 41.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Movahedi K, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70:5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 45.Franklin RA, Li MO. Ontogeny of tumor-associated macrophages and its implication in cancer regulation. Trends Cancer. 2016;2:20–34. doi: 10.1016/j.trecan.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Y, Cao X. The origin and function of tumor-associated macrophages. Cell Mol. Immunol. 2015;12:1–4. doi: 10.1038/cmi.2014.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. npj Breast Cancer. 2016;2:1–12. doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 49.Biswas SK, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 50.Salmaninejad A, et al. Tumor-associated macrophages: role in cancer development and therapeutic implications. Cell. Oncol. 2019;42:591–608. doi: 10.1007/s13402-019-00453-z. [DOI] [PubMed] [Google Scholar]
- 51.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers. 2014;6:1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J. Hematol. Oncol. 2017;10:58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gordy C, Pua H, Sempowski GD, He YW. Regulation of steady-state neutrophil homeostasis by macrophages. Blood. 2011;117:618–629. doi: 10.1182/blood-2010-01-265959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008;8:958–969. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shevach EM, Rosenthal AS. Function of macrophages in antigen recognition by guinea pig T lymphocytes. II. Role of the macrophage in the regulation of genetic control of the immune response. J. Exp. Med. 1973;138:1213–1229. doi: 10.1084/jem.138.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 57.Li Q, Barres BA. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018;18:225–242. doi: 10.1038/nri.2017.125. [DOI] [PubMed] [Google Scholar]
- 58.Hulsmans M, et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169:510–522 e520. doi: 10.1016/j.cell.2017.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lavine KJ, et al. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc. Natl Acad. Sci. USA. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leid J, et al. Primitive embryonic macrophages are required for coronary development and maturation. Circ. Res. 2016;118:1498–1511. doi: 10.1161/CIRCRESAHA.115.308270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kang K, et al. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7:485–495. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Odegaard JI, et al. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang W, et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59:347–357. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yamamoto S, et al. A subset of cerebrovascular pericytes originates from mature macrophages in the very early phase of vascular development in CNS. Sci. Rep. 2017;7:3855. doi: 10.1038/s41598-017-03994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hibbs JB, Jr., Vavrin Z, Taintor RR. L-arginine is required for expression of the activated macrophage effector mechanism causing selective metabolic inhibition in target cells. J. Immunol. 1987;138:550–565. [PubMed] [Google Scholar]
- 66.Mills CD. Macrophage arginine metabolism to ornithine/urea or nitric oxide/citrulline: a life or death issue. Crit. Rev. Immunol. 2001;21:399–425. doi: 10.1615/CritRevImmunol.v21.i5.10. [DOI] [PubMed] [Google Scholar]
- 67.Ran S, Montgomery KE. Macrophage-mediated lymphangiogenesis: the emerging role of macrophages as lymphatic endothelial progenitors. Cancers. 2012;4:618–657. doi: 10.3390/cancers4030618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rao S, et al. Obligatory participation of macrophages in an angiopoietin 2-mediated cell death switch. Development. 2007;134:4449–4458. doi: 10.1242/dev.012187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stefater JA, 3rd, et al. Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature. 2011;474:511–515. doi: 10.1038/nature10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spiric Z, Eri Z, Eric M. Significance of vascular endothelial growth factor (VEGF)-C and VEGF-D in the progression of cutaneous melanoma. Int. J. Surg. Pathol. 2015;23:629–637. doi: 10.1177/1066896915583694. [DOI] [PubMed] [Google Scholar]
- 71.Jeong H, et al. Tumor-associated macrophages enhance tumor hypoxia and aerobic glycolysis. Cancer Res. 2019;79:795–806. doi: 10.1158/0008-5472.CAN-18-2545. [DOI] [PubMed] [Google Scholar]
- 72.Feng J, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2020;39:126. doi: 10.1186/s13046-020-01629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Srinivasan S, et al. Hypoxia-induced expression of phosducin-like 3 regulates expression of VEGFR-2 and promotes angiogenesis. Angiogenesis. 2015;18:449–462. doi: 10.1007/s10456-015-9468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Squadrito ML, De Palma M. Macrophage regulation of tumor angiogenesis: implications for cancer therapy. Mol. Asp. Med. 2011;32:123–145. doi: 10.1016/j.mam.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 75.Jetten N, et al. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis. 2014;17:109–118. doi: 10.1007/s10456-013-9381-6. [DOI] [PubMed] [Google Scholar]
- 76.Yeo EJ, et al. Myeloid WNT7b mediates the angiogenic switch and metastasis in breast cancer. Cancer Res. 2014;74:2962–2973. doi: 10.1158/0008-5472.CAN-13-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 78.Zhou H, Binmadi NO, Yang YH, Proia P, Basile JR. Semaphorin 4D cooperates with VEGF to promote angiogenesis and tumor progression. Angiogenesis. 2012;15:391–407. doi: 10.1007/s10456-012-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Ramirez-Pedraza M, Fernandez M. Interplay between macrophages and angiogenesis: a double-edged sword in liver disease. Front. Immunol. 2019;10:2882. doi: 10.3389/fimmu.2019.02882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gurevich DB, et al. Live imaging of wound angiogenesis reveals macrophage orchestrated vessel sprouting and regression. EMBO J. 2018;37:e97786. doi: 10.15252/embj.201797786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Casazza A, et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell. 2013;24:695–709. doi: 10.1016/j.ccr.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 82.Mazzieri R, et al. Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells. Cancer Cell. 2011;19:512–526. doi: 10.1016/j.ccr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 83.Belgiovine C, D’Incalci M, Allavena P, Frapolli R. Tumor-associated macrophages and anti-tumor therapies: complex links. Cell Mol. Life Sci. 2016;73:2411–2424. doi: 10.1007/s00018-016-2166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Du Cheyne C, Tay H, De Spiegelaere W. The complex TIE between macrophages and angiogenesis. Anat. Histol. Embryol. 2019;49:585–596. doi: 10.1111/ahe.12518. [DOI] [PubMed] [Google Scholar]
- 85.Albini A, Bruno A, Noonan DM, Mortara L. Contribution to tumor angiogenesis from innate immune cells within the tumor microenvironment: implications for immunotherapy. Front. Immunol. 2018;9:527. doi: 10.3389/fimmu.2018.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang W, et al. miR-100 maintains phenotype of tumor-associated macrophages by targeting mTOR to promote tumor metastasis via Stat5a/IL-1ra pathway in mouse breast cancer. Oncogenesis. 2018;7:97. doi: 10.1038/s41389-018-0106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 88.Bonde AK, Tischler V, Kumar S, Soltermann A, Schwendener RA. Intratumoral macrophages contribute to epithelial-mesenchymal transition in solid tumors. BMC Cancer. 2012;12:35. doi: 10.1186/1471-2407-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mason SD, Joyce JA. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lan J, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79:146–158. doi: 10.1158/0008-5472.CAN-18-0014. [DOI] [PubMed] [Google Scholar]
- 92.Yin Z, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:310. doi: 10.1186/s13046-019-1313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Linde N, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018;9:21. doi: 10.1038/s41467-017-02481-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kitamura T, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J. Exp. Med. 2015;212:1043–1059. doi: 10.1084/jem.20141836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Deventer HW, Palmieri DA, Wu QP, McCook EC, Serody JS. Circulating fibrocytes prepare the lung for cancer metastasis by recruiting Ly-6C+ monocytes via CCL2. J. Immunol. 2013;190:4861–4867. doi: 10.4049/jimmunol.1202857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nosaka T, et al. Alveolar macrophages drive hepatocellular carcinoma lung metastasis by generating leukotriene B4. J. Immunol. 2018;200:1839–1852. doi: 10.4049/jimmunol.1700544. [DOI] [PubMed] [Google Scholar]
- 103.Sharma SK, et al. Pulmonary alveolar macrophages contribute to the premetastatic niche by suppressing antitumor T cell responses in the lungs. J. Immunol. 2015;194:5529–5538. doi: 10.4049/jimmunol.1403215. [DOI] [PubMed] [Google Scholar]
- 104.Kimura Y, et al. The innate immune receptor Dectin-2 mediates the phagocytosis of cancer cells by Kupffer cells for the suppression of liver metastasis. Proc. Natl Acad. Sci. USA. 2016;113:14097–14102. doi: 10.1073/pnas.1617903113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Du R, et al. HIF1alpha induces the recruitment of bone marrow-derived vascular modulatory cells to regulate tumor angiogenesis and invasion. Cancer Cell. 2008;13:206–220. doi: 10.1016/j.ccr.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lin EY, et al. Vascular endothelial growth factor restores delayed tumor progression in tumors depleted of macrophages. Mol. Oncol. 2007;1:288–302. doi: 10.1016/j.molonc.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Linde N, et al. Vascular endothelial growth factor-induced skin carcinogenesis depends on recruitment and alternative activation of macrophages. J. Pathol. 2012;227:17–28. doi: 10.1002/path.3989. [DOI] [PubMed] [Google Scholar]
- 109.Quaranta V, et al. Macrophage-derived granulin drives resistance to immune checkpoint inhibition in metastatic pancreatic cancer. Cancer Res. 2018;78:4253–4269. doi: 10.1158/0008-5472.CAN-17-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nielsen SR, et al. Macrophage-secreted granulin supports pancreatic cancer metastasis by inducing liver fibrosis. Nat. Cell Biol. 2016;18:549–560. doi: 10.1038/ncb3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou J, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol. Res. 2018;6:1578–1592. doi: 10.1158/2326-6066.CIR-17-0479. [DOI] [PubMed] [Google Scholar]
- 112.Li X, et al. Harnessing tumor-associated macrophages as aids for cancer immunotherapy. Mol. Cancer. 2019;18:177. doi: 10.1186/s12943-019-1102-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sun D, et al. M2-polarized tumor-associated macrophages promote epithelial-mesenchymal transition via activation of the AKT3/PRAS40 signaling pathway in intrahepatic cholangiocarcinoma. J. Cell Biochem. 2020;121:2828–2838. doi: 10.1002/jcb.29514. [DOI] [PubMed] [Google Scholar]
- 115.Kuwada K, et al. The epithelial-to-mesenchymal transition induced by tumor-associated macrophages confers chemoresistance in peritoneally disseminated pancreatic cancer. J. Exp. Clin. Cancer Res. 2018;37:307. doi: 10.1186/s13046-018-0981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dudas J, Ladanyi A, Ingruber J, Steinbichler TB, Riechelmann H. Epithelial to mesenchymal transition: a mechanism that fuels cancer radio/chemoresistance. Cells. 2020;9:428. doi: 10.3390/cells9020428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhu X, et al. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J. Exp. Clin. Cancer Res. 2019;38:81. doi: 10.1186/s13046-019-1095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zheng P, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017;36:53. doi: 10.1186/s13046-017-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu S, et al. Activated HIF1alpha of tumor cells promotes chemoresistance development via recruiting GDF15-producing tumor-associated macrophages in gastric cancer. Cancer Immunol. Immunother. 2020;69:1973–1987. doi: 10.1007/s00262-020-02598-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Halbrook CJ, et al. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 2019;29:1390–1399 e1396. doi: 10.1016/j.cmet.2019.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Buchholz SM, et al. Depletion of macrophages improves therapeutic response to gemcitabine in murine pancreas cancer. Cancers. 2020;12:1978. doi: 10.3390/cancers12071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nowak M, Klink M. The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells. 2020;9:1299. doi: 10.3390/cells9051299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chiang CS, et al. Irradiation promotes an m2 macrophage phenotype in tumor hypoxia. Front. Oncol. 2012;2:89. doi: 10.3389/fonc.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fu E, et al. M2 macrophages reduce the radiosensitivity of head and neck cancer by releasing HBEGF. Oncol. Rep. 2020;44:698–710. doi: 10.3892/or.2020.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Meng Y, et al. Blockade of tumor necrosis factor alpha signaling in tumor-associated macrophages as a radiosensitizing strategy. Cancer Res. 2010;70:1534–1543. doi: 10.1158/0008-5472.CAN-09-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang EY, et al. Carcinoembryonic antigen as a marker of radioresistance in colorectal cancer: a potential role of macrophages. BMC Cancer. 2018;18:321. doi: 10.1186/s12885-018-4254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rahal OM, et al. Blocking interleukin (IL)4- and IL13-mediated phosphorylation of STAT6 (Tyr641) decreases M2 polarization of macrophages and protects against macrophage-mediated radioresistance of inflammatory breast cancer. Int. J. Radiat. Oncol. Biol. Phys. 2018;100:1034–1043. doi: 10.1016/j.ijrobp.2017.11.043. [DOI] [PubMed] [Google Scholar]
- 128.Beltraminelli T, De Palma M. Biology and therapeutic targeting of tumour-associated macrophages. J. Pathol. 2020;250:573–592. doi: 10.1002/path.5403. [DOI] [PubMed] [Google Scholar]
- 129.Wu Q, et al. Macrophage biology plays a central role during ionizing radiation-elicited tumor response. Biomed. J. 2017;40:200–211. doi: 10.1016/j.bj.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J. Immunother. Cancer. 2018;6:8. doi: 10.1186/s40425-018-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gordon SR, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ansell SM, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin’s lymphoma. N. Engl. J. Med. 2015;372:311–319. doi: 10.1056/NEJMoa1411087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Prima V, Kaliberova LN, Kaliberov S, Curiel DT, Kusmartsev S. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc. Natl Acad. Sci. USA. 2017;114:1117–1122. doi: 10.1073/pnas.1612920114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Noguchi T, et al. Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol. Res. 2017;5:106–117. doi: 10.1158/2326-6066.CIR-16-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sun NY, et al. Blockade of PD-L1 enhances cancer immunotherapy by regulating dendritic cell maturation and macrophage polarization. Cancers. 2019;11:1400. doi: 10.3390/cancers11091400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gubin MM, et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell. 2018;175:1014–1030 e1019. doi: 10.1016/j.cell.2018.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen D, et al. Chloroquine modulates antitumor immune response by resetting tumor-associated macrophages toward M1 phenotype. Nat. Commun. 2018;9:873. doi: 10.1038/s41467-018-03225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Meng Y, et al. Galactan isolated from Cantharellus cibarius modulates antitumor immune response by converting tumor-associated macrophages toward M1-like phenotype. Carbohydr. Polym. 2019;226:115295. doi: 10.1016/j.carbpol.2019.115295. [DOI] [PubMed] [Google Scholar]
- 139.Ma Q, et al. PlGF signaling and macrophage repolarization contribute to the anti-neoplastic effect of metformin. Eur. J. Pharmacol. 2019;863:172696. doi: 10.1016/j.ejphar.2019.172696. [DOI] [PubMed] [Google Scholar]
- 140.Rodell CB, et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng. 2018;2:578–588. doi: 10.1038/s41551-018-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peranzoni E, et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc. Natl Acad. Sci. USA. 2018;115:E4041–e4050. doi: 10.1073/pnas.1720948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Arlauckas SP, et al. In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci. Transl. Med. 2017;9:389. doi: 10.1126/scitranslmed.aal3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Majeti R, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Willingham SB, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl Acad. Sci. USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Weiskopf K, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J. Clin. Investig. 2016;126:2610–2620. doi: 10.1172/JCI81603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Barkal AA, et al. Engagement of MHC class I by the inhibitory receptor LILRB1 suppresses macrophages and is a target of cancer immunotherapy. Nat. Immunol. 2018;19:76–84. doi: 10.1038/s41590-017-0004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Barkal AA, et al. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019;572:392–396. doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wiemann B, Starnes CO. Coley’s toxins, tumor necrosis factor and cancer research: a historical perspective. Pharmacol. Ther. 1994;64:529–564. doi: 10.1016/0163-7258(94)90023-X. [DOI] [PubMed] [Google Scholar]
- 149.Thomas JA, Badini M. The role of innate immunity in spontaneous regression of cancer. Indian J. Cancer. 2011;48:246–251. doi: 10.4103/0019-509X.82887. [DOI] [PubMed] [Google Scholar]
- 150.Thoreau M, et al. Vaccine-induced tumor regression requires a dynamic cooperation between T cells and myeloid cells at the tumor site. Oncotarget. 2015;6:27832–27846. doi: 10.18632/oncotarget.4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Santos PM, Butterfield LH. Dendritic cell-based cancer vaccines. J. Immunol. 2018;200:443–449. doi: 10.4049/jimmunol.1701024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Walter S, et al. Multipeptide immune response to cancer vaccine IMA901 after single-dose cyclophosphamide associates with longer patient survival. Nat. Med. 2012;18:1254–1261. doi: 10.1038/nm.2883. [DOI] [PubMed] [Google Scholar]
- 153.Cheever MA, Higano CS. PROVENGE (Sipuleucel-T) in prostate cancer: the first FDA-approved therapeutic cancer vaccine. Clin. Cancer Res. 2011;17:3520–3526. doi: 10.1158/1078-0432.CCR-10-3126. [DOI] [PubMed] [Google Scholar]
- 154.Fu J, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci. Transl. Med. 2015;7:283ra252. doi: 10.1126/scitranslmed.aaa4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Agarwalla P, Barnard Z, Fecci P, Dranoff G, Curry WT., Jr Sequential immunotherapy by vaccination with GM-CSF-expressing glioma cells and CTLA-4 blockade effectively treats established murine intracranial tumors. J. Immunother. 2012;35:385–389. doi: 10.1097/CJI.0b013e3182562d59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dranoff G, et al. Vaccination with irradiated tumor cells engineered to secrete murine granulocyte-macrophage colony-stimulating factor stimulates potent, specific, and long-lasting anti-tumor immunity. Proc. Natl Acad. Sci. USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Chiocca EA, Rabkin SD. Oncolytic viruses and their application to cancer immunotherapy. Cancer Immunol. Res. 2014;2:295–300. doi: 10.1158/2326-6066.CIR-14-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Senzer NN, et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009;27:5763–5771. doi: 10.1200/JCO.2009.24.3675. [DOI] [PubMed] [Google Scholar]
- 159.Halstead ES, et al. GM-CSF overexpression after influenza a virus infection prevents mortality and moderates M1-like airway monocyte/macrophage polarization. Respir. Res. 2018;19:3. doi: 10.1186/s12931-017-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang D, Dai W, Wang J. A cell-specific nuclear factor-kappa b-activating gene expression strategy for delivering cancer immunotherapy. Hum. gene Ther. 2019;30:471–484. doi: 10.1089/hum.2018.093. [DOI] [PubMed] [Google Scholar]
- 162.Cheng L, Wang Y, Huang L. Exosomes from M1-polarized macrophages potentiate the cancer vaccine by creating a pro-inflammatory microenvironment in the lymph node. Mol. Ther. 2017;25:1665–1675. doi: 10.1016/j.ymthe.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xu G, et al. Listeria-based hepatocellular carcinoma vaccine facilitates anti-PD-1 therapy by regulating macrophage polarization. Oncogene. 2019;39:1429–1444. doi: 10.1038/s41388-019-1072-3. [DOI] [PubMed] [Google Scholar]
- 164.Lefebvre ML, Krause SW, Salcedo M, Nardin A. Ex vivo-activated human macrophages kill chronic lymphocytic leukemia cells in the presence of rituximab: mechanism of antibody-dependent cellular cytotoxicity and impact of human serum. J. Immunother. 2006;29:388–397. doi: 10.1097/01.cji.0000203081.43235.d7. [DOI] [PubMed] [Google Scholar]
- 165.Roghanian A, et al. Cyclophosphamide enhances cancer antibody immunotherapy in the resistant bone marrow niche by modulating macrophage FcgammaR expression. Cancer Immunol. Res. 2019;7:1876–1890. doi: 10.1158/2326-6066.CIR-18-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bouwstra R, et al. CD47 expression defines efficacy of rituximab with CHOP in non-germinal center B-cell (non-GCB) diffuse large B-cell lymphoma patients (DLBCL), but not in GCB DLBCL. Cancer Immunol. Res. 2019;7:1663–1671. doi: 10.1158/2326-6066.CIR-18-0781. [DOI] [PubMed] [Google Scholar]
- 167.Tsao LC, et al. CD47 blockade augmentation of trastuzumab antitumor efficacy dependent on antibody-dependent cellular phagocytosis. JCI Insight. 2019;4:24. doi: 10.1172/jci.insight.131882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shi Y, et al. Trastuzumab triggers phagocytic killing of high HER2 cancer cells in vitro and in vivo by interaction with Fcgamma receptors on macrophages. J. Immunol. 2015;194:4379–4386. doi: 10.4049/jimmunol.1402891. [DOI] [PubMed] [Google Scholar]
- 169.Xu M, et al. Intratumoral delivery of IL-21 overcomes anti-Her2/Neu resistance through shifting tumor-associated macrophages from M2 to M1 phenotype. J. Immunol. 2015;194:4997–5006. doi: 10.4049/jimmunol.1402603. [DOI] [PubMed] [Google Scholar]
- 170.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Morgan RA, et al. Cancer regression in patients after transfer of genetically engineered lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kochenderfer JN, et al. Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood. 2010;116:4099–4102. doi: 10.1182/blood-2010-04-281931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Klug F, et al. Low-dose irradiation programs macrophage differentiation to an iNOS(+)/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell. 2013;24:589–602. doi: 10.1016/j.ccr.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 174.Giavridis T, et al. CAR T cell-induced cytokine release syndrome is mediated by macrophages and abated by IL-1 blockade. Nat. Med. 2018;24:731–738. doi: 10.1038/s41591-018-0041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sterner RM, et al. GM-CSF inhibition reduces cytokine release syndrome and neuroinflammation but enhances CAR-T cell function in xenografts. Blood. 2019;133:697–709. doi: 10.1182/blood-2018-10-881722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Sachdeva M, Duchateau P, Depil S, Poirot L, Valton J. Granulocyte-macrophage colony-stimulating factor inactivation in CAR T-cells prevents monocyte-dependent release of key cytokine release syndrome mediators. J. Biol. Chem. 2019;294:5430–5437. doi: 10.1074/jbc.AC119.007558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Klichinsky M, et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020;38:947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Adams JL, Smothers J, Srinivasan R, Hoos A. Big opportunities for small molecules in immuno-oncology. Nat. Rev. Drug Discov. 2015;14:603–622. doi: 10.1038/nrd4596. [DOI] [PubMed] [Google Scholar]
- 179.Okamoto A, et al. Indoleamine 2,3-dioxygenase serves as a marker of poor prognosis in gene expression profiles of serous ovarian cancer cells. Clin. Cancer Res. 2005;11:6030–6039. doi: 10.1158/1078-0432.CCR-04-2671. [DOI] [PubMed] [Google Scholar]
- 180.Liu X, et al. Selective inhibition of IDO1 effectively regulates mediators of antitumor immunity. Blood. 2010;115:3520–3530. doi: 10.1182/blood-2009-09-246124. [DOI] [PubMed] [Google Scholar]
- 181.Rodriguez PC, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 182.Cabrales P. RRx-001 acts as a dual small molecule checkpoint inhibitor by downregulating CD47 on cancer cells and SIRP-alpha on monocytes/macrophages. Transl. Oncol. 2019;12:626–632. doi: 10.1016/j.tranon.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Karlsson H, et al. Combining CAR T cells and the Bcl-2 family apoptosis inhibitor ABT-737 for treating B-cell malignancy. Cancer Gene Ther. 2013;20:386–393. doi: 10.1038/cgt.2013.35. [DOI] [PubMed] [Google Scholar]
- 184.De Henau O, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature. 2016;539:443–447. doi: 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Nywening TM, et al. Targeting both tumour-associated CXCR2(+) neutrophils and CCR2(+) macrophages disrupts myeloid recruitment and improves chemotherapeutic responses in pancreatic ductal adenocarcinoma. Gut. 2018;67:1112–1123. doi: 10.1136/gutjnl-2017-313738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ngiow SF, et al. Co-inhibition of colony stimulating factor-1 receptor and BRAF oncogene in mouse models of BRAF(V600E) melanoma. Oncoimmunology. 2016;5:e1089381. doi: 10.1080/2162402X.2015.1089381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mok S, et al. Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer Res. 2014;74:153–161. doi: 10.1158/0008-5472.CAN-13-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Osipov A, Saung MT, Zheng L, Murphy AG. Small molecule immunomodulation: the tumor microenvironment and overcoming immune escape. J. Immunother. Cancer. 2019;7:224. doi: 10.1186/s40425-019-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Robert J. Comparative study of tumorigenesis and tumor immunity in invertebrates and nonmammalian vertebrates. Dev. Comp. Immunol. 2010;34:915–925. doi: 10.1016/j.dci.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang J, et al. Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res. 2015;75:306–315. doi: 10.1158/0008-5472.CAN-14-2819. [DOI] [PubMed] [Google Scholar]
- 191.Mills CD, Ley K, Buchmann K, Canton J. Sequential immune responses: the weapons of immunity. J. Innate Immun. 2015;7:443–449. doi: 10.1159/000380910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cavaillon JM. The historical milestones in the understanding of leukocyte biology initiated by Elie Metchnikoff. J. Leukoc. Biol. 2011;90:413–424. doi: 10.1189/jlb.0211094. [DOI] [PubMed] [Google Scholar]
- 193.Pathria P, Louis TL, Varner JA. Targeting tumor-associated macrophages in cancer. Trends Immunol. 2019;40:310–327. doi: 10.1016/j.it.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 194.O’Sullivan T, et al. Cancer immunoediting by the innate immune system in the absence of adaptive immunity. J. Exp. Med. 2012;209:1869–1882. doi: 10.1084/jem.20112738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mills CD, Shearer J, Evans R, Caldwell MD. Macrophage arginine metabolism and the inhibition or stimulation of cancer. J. Immunol. 1992;149:2709–2714. [PubMed] [Google Scholar]
- 196.Beatty GL, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000;164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 198.Hamieh M, et al. CAR T cell trogocytosis and cooperative killing regulate tumour antigen escape. Nature. 2019;568:112–116. doi: 10.1038/s41586-019-1054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nakamura K, et al. Fratricide of natural killer cells dressed with tumor-derived NKG2D ligand. Proc. Natl Acad. Sci. USA. 2013;110:9421–9426. doi: 10.1073/pnas.1300140110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Boross P, Jansen JH, Pastula A, van der Poel CE, Leusen JH. Both activating and inhibitory Fc gamma receptors mediate rituximab-induced trogocytosis of CD20 in mice. Immunol. Lett. 2012;143:44–52. doi: 10.1016/j.imlet.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 201.Beum PV, et al. Loss of CD20 and bound CD20 antibody from opsonized B cells occurs more rapidly because of trogocytosis mediated by Fc receptor-expressing effector cells than direct internalization by the B cells. J. Immunol. 2011;187:3438–3447. doi: 10.4049/jimmunol.1101189. [DOI] [PubMed] [Google Scholar]
- 202.Velmurugan R, Challa DK, Ram S, Ober RJ, Ward ES. Macrophage-mediated trogocytosis leads to death of antibody-opsonized tumor cells. Mol. Cancer Ther. 2016;15:1879–1889. doi: 10.1158/1535-7163.MCT-15-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu Y, Zheng L. Dynamic education of macrophages in different areas of human tumors. Cancer Microenviron. 2012;5:195–201. doi: 10.1007/s12307-012-0113-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Tacke F. Targeting hepatic macrophages to treat liver diseases. J. Hepatol. 2017;66:1300–1312. doi: 10.1016/j.jhep.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 205.Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017;14:399–416. doi: 10.1038/nrclinonc.2016.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Bak SP, Walters JJ, Takeya M, Conejo-Garcia JR, Berwin BL. Scavenger receptor-A-targeted leukocyte depletion inhibits peritoneal ovarian tumor progression. Cancer Res. 2007;67:4783–4789. doi: 10.1158/0008-5472.CAN-06-4410. [DOI] [PubMed] [Google Scholar]
- 207.Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 2018;17:887–904. doi: 10.1038/nrd.2018.169. [DOI] [PubMed] [Google Scholar]
- 208.Nagai T, et al. Targeting tumor-associated macrophages in an experimental glioma model with a recombinant immunotoxin to folate receptor beta. Cancer Immunol. Immunother. 2009;58:1577–1586. doi: 10.1007/s00262-009-0667-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Komohara Y, et al. The coordinated actions of TIM-3 on cancer and myeloid cells in the regulation of tumorigenicity and clinical prognosis in clear cell renal cell carcinomas. Cancer Immunol. Res. 2015;3:999–1007. doi: 10.1158/2326-6066.CIR-14-0156. [DOI] [PubMed] [Google Scholar]
- 210.Mehla K, Singh PK. Metabolic regulation of macrophage polarization in cancer. Trends Cancer. 2019;5:822–834. doi: 10.1016/j.trecan.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv. Drug Deliv. Rev. 2017;114:206–221. doi: 10.1016/j.addr.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tseng D, et al. Anti-CD47 antibody-mediated phagocytosis of cancer by macrophages primes an effective antitumor T-cell response. Proc. Natl Acad. Sci. USA. 2013;110:11103–11108. doi: 10.1073/pnas.1305569110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ring NG, et al. Anti-SIRPalpha antibody immunotherapy enhances neutrophil and macrophage antitumor activity. Proc. Natl Acad. Sci. USA. 2017;114:E10578–E10585. doi: 10.1073/pnas.1710877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Komohara Y, Fujiwara Y, Ohnishi K, Takeya M. Tumor-associated macrophages: potential therapeutic targets for anti-cancer therapy. Adv. Drug Deliv. Rev. 2016;99:180–185. doi: 10.1016/j.addr.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 215.Baer C, et al. Suppression of microRNA activity amplifies IFN-gamma-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 2016;18:790–802. doi: 10.1038/ncb3371. [DOI] [PubMed] [Google Scholar]
- 216.Georgoudaki AM, et al. Reprogramming tumor-associated macrophages by antibody targeting inhibits cancer progression and metastasis. Cell Rep. 2016;15:2000–2011. doi: 10.1016/j.celrep.2016.04.084. [DOI] [PubMed] [Google Scholar]
- 217.Takenaka MC, et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 2019;22:729–740. doi: 10.1038/s41593-019-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lettieri-Barbato D, Aquilano K. Pushing the limits of cancer therapy: the nutrient game. Front. Oncol. 2018;8:148. doi: 10.3389/fonc.2018.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.O’Flanagan CH, Smith LA, McDonell SB, Hursting SD. When less may be more: calorie restriction and response to cancer therapy. BMC Med. 2017;15:106. doi: 10.1186/s12916-017-0873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Di Biase S, et al. Fasting-mimicking diet reduces HO-1 to promote T cell-mediated tumor cytotoxicity. Cancer Cell. 2016;30:136–146. doi: 10.1016/j.ccell.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Palmieri EM, et al. Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep. 2017;20:1654–1666. doi: 10.1016/j.celrep.2017.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Guerriero JL, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature. 2017;543:428–432. doi: 10.1038/nature21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hashimoto D, Miller J, Merad M. Dendritic cell and macrophage heterogeneity in vivo. Immunity. 2011;35:323–335. doi: 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Klein I, et al. Kupffer cell heterogeneity: functional properties of bone marrow derived and sessile hepatic macrophages. Blood. 2007;110:4077–4085. doi: 10.1182/blood-2007-02-073841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Elchaninov AV, Fatkhudinov TK, Vishnyakova PA, Lokhonina AV, Sukhikh GT. Phenotypical and functional polymorphism of liver resident macrophages. Cells. 2019;8:1032. doi: 10.3390/cells8091032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Dal-Secco D, et al. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. J. Exp. Med. 2015;212:447–456. doi: 10.1084/jem.20141539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Karlmark KR, et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology. 2009;50:261–274. doi: 10.1002/hep.22950. [DOI] [PubMed] [Google Scholar]
- 228.Holt MP, Cheng L, Ju C. Identification and characterization of infiltrating macrophages in acetaminophen-induced liver injury. J. Leukoc. Biol. 2008;84:1410–1421. doi: 10.1189/jlb.0308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Varol C, Yona S, Jung S. Origins and tissue-context-dependent fates of blood monocytes. Immunol. Cell Biol. 2009;87:30–38. doi: 10.1038/icb.2008.90. [DOI] [PubMed] [Google Scholar]
- 230.Sierro F, et al. A liver capsular network of monocyte-derived macrophages restricts hepatic dissemination of intraperitoneal bacteria by neutrophil recruitment. Immunity. 2017;47:374–388 e376. doi: 10.1016/j.immuni.2017.07.018. [DOI] [PubMed] [Google Scholar]
- 231.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 232.Taylor PR, et al. The beta-glucan receptor, dectin-1, is predominantly expressed on the surface of cells of the monocyte/macrophage and neutrophil lineages. J. Immunol. 2002;169:3876–3882. doi: 10.4049/jimmunol.169.7.3876. [DOI] [PubMed] [Google Scholar]
- 233.Sabatel C, et al. Exposure to bacterial CpG DNA protects from airway allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity. 2017;46:457–473. doi: 10.1016/j.immuni.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 234.Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, Perlman H. Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am. J. Respir. Cell Mol. Biol. 2013;49:503–510. doi: 10.1165/rcmb.2013-0086MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Bedoret D, et al. Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. J. Clin. Investig. 2009;119:3723–3738. doi: 10.1172/JCI39717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Gibbings SL, et al. Three unique interstitial macrophages in the murine lung at steady state. Am. J. Respir. Cell Mol. Biol. 2017;57:66–76. doi: 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat. Neurosci. 2011;14:1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 238.Nissen SK, et al. Alterations in blood monocyte functions in Parkinson’s disease. Mov. Disord. 2019;34:1711–1721. doi: 10.1002/mds.27815. [DOI] [PubMed] [Google Scholar]
- 239.Yang T, Guo R, Zhang F. Brain perivascular macrophages: recent advances and implications in health and diseases. CNS Neurosci. Ther. 2019;25:1318–1328. doi: 10.1111/cns.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 241.Kim WK, et al. CD163 identifies perivascular macrophages in normal and viral encephalitic brains and potential precursors to perivascular macrophages in blood. Am. J. Pathol. 2006;168:822–834. doi: 10.2353/ajpath.2006.050215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Borda JT, et al. CD163, a marker of perivascular macrophages, is up-regulated by microglia in simian immunodeficiency virus encephalitis after haptoglobin-hemoglobin complex stimulation and is suggestive of breakdown of the blood-brain barrier. Am. J. Pathol. 2008;172:725–737. doi: 10.2353/ajpath.2008.070848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fabriek BO, et al. CD163-positive perivascular macrophages in the human CNS express molecules for antigen recognition and presentation. Glia. 2005;51:297–305. doi: 10.1002/glia.20208. [DOI] [PubMed] [Google Scholar]
- 244.Aspelund A, et al. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J. Exp. Med. 2015;212:991–999. doi: 10.1084/jem.20142290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Galea I, et al. Mannose receptor expression specifically reveals perivascular macrophages in normal, injured, and diseased mouse brain. Glia. 2005;49:375–384. doi: 10.1002/glia.20124. [DOI] [PubMed] [Google Scholar]
- 246.Wang G, et al. Microglia/macrophage polarization dynamics in white matter after traumatic brain injury. J. Cereb. Blood Flow. Metab. 2013;33:1864–1874. doi: 10.1038/jcbfm.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hu X, et al. Microglia/macrophage polarization dynamics reveal novel mechanism of injury expansion after focal cerebral ischemia. Stroke. 2012;43:3063–3070. doi: 10.1161/STROKEAHA.112.659656. [DOI] [PubMed] [Google Scholar]
- 248.Goldmann T, et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat. Immunol. 2016;17:797–805. doi: 10.1038/ni.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Song E, Iwasaki A. Monocytes inadequately fill in for meningeal macrophages. Trends Immunol. 2019;40:463–465. doi: 10.1016/j.it.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Quinn JM, et al. Calcitonin receptor antibodies in the identification of osteoclasts. Bone. 1999;25:1–8. doi: 10.1016/S8756-3282(99)00094-0. [DOI] [PubMed] [Google Scholar]
- 251.Yoshida H, et al. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 252.Kong YY, et al. OPGL is a key regulator of osteoclastogenesis, lymphocyte development and lymph-node organogenesis. Nature. 1999;397:315–323. doi: 10.1038/16852. [DOI] [PubMed] [Google Scholar]
- 253.Sadahira Y, Mori M. Role of the macrophage in erythropoiesis. Pathol. Int. 1999;49:841–848. doi: 10.1046/j.1440-1827.1999.00954.x. [DOI] [PubMed] [Google Scholar]
- 254.Miyake Y, et al. Critical role of macrophages in the marginal zone in the suppression of immune responses to apoptotic cell-associated antigens. J. Clin. Investig. 2007;117:2268–2278. doi: 10.1172/JCI31990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Taylor PR, et al. Dectin-2 is predominantly myeloid restricted and exhibits unique activation-dependent expression on maturing inflammatory monocytes elicited in vivo. Eur. J. Immunol. 2005;35:2163–2174. doi: 10.1002/eji.200425785. [DOI] [PubMed] [Google Scholar]
- 256.You Y, et al. Marginal zone B cells regulate antigen capture by marginal zone macrophages. J. Immunol. 2011;186:2172–2181. doi: 10.4049/jimmunol.1002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.den Haan JM, Kraal G. Innate immune functions of macrophage subpopulations in the spleen. J. Innate Immun. 2012;4:437–445. doi: 10.1159/000335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Hanayama R, et al. Autoimmune disease and impaired uptake of apoptotic cells in MFG-E8-deficient mice. Science. 2004;304:1147–1150. doi: 10.1126/science.1094359. [DOI] [PubMed] [Google Scholar]
- 259.N AG, Castrillo A. Origin and specialization of splenic macrophages. Cell Immunol. 2018;330:151–158. doi: 10.1016/j.cellimm.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 260.Kohyama M, et al. Role for Spi-C in the development of red pulp macrophages and splenic iron homeostasis. Nature. 2009;457:318–321. doi: 10.1038/nature07472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Lloyd CM, Phillips AR, Cooper GJ, Dunbar PR. Three-colour fluorescence immunohistochemistry reveals the diversity of cells staining for macrophage markers in murine spleen and liver. J. Immunol. Methods. 2008;334:70–81. doi: 10.1016/j.jim.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 262.Mondor I, et al. Lymphatic endothelial cells are essential components of the subcapsular sinus macrophage niche. Immunity. 2019;50:1453–1466 e1454. doi: 10.1016/j.immuni.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Kuka M, Iannacone M. The role of lymph node sinus macrophages in host defense. Ann. N. Y Acad. Sci. 2014;1319:38–46. doi: 10.1111/nyas.12387. [DOI] [PubMed] [Google Scholar]
- 264.Gray EE, Cyster JG. Lymph node macrophages. J. Innate Immun. 2012;4:424–436. doi: 10.1159/000337007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Bain CC, et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat. Commun. 2016;7:1–14. doi: 10.1038/ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Cailhier JF, et al. Resident pleural macrophages are key orchestrators of neutrophil recruitment in pleural inflammation. Am. J. Respir. Crit. Care Med. 2006;173:540–547. doi: 10.1164/rccm.200504-538OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Miyanishi M, et al. Identification of Tim4 as a phosphatidylserine receptor. Nature. 2007;450:435–439. doi: 10.1038/nature06307. [DOI] [PubMed] [Google Scholar]
- 269.Cassado Ados A, D’Imperio Lima MR, Bortoluci KR. Revisiting mouse peritoneal macrophages: heterogeneity, development, and function. Front. Immunol. 2015;6:225. doi: 10.3389/fimmu.2015.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ghosn EE, et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc. Natl Acad. Sci. USA. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Fainaru O, et al. Runx3 regulates mouse TGF-beta-mediated dendritic cell function and its absence results in airway inflammation. EMBO J. 2004;23:969–979. doi: 10.1038/sj.emboj.7600085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hacker C, et al. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 2003;4:380–386. doi: 10.1038/ni903. [DOI] [PubMed] [Google Scholar]
- 273.Dupasquier M, et al. The dermal microenvironment induces the expression of the alternative activation marker CD301/mMGL in mononuclear phagocytes, independent of IL-4/IL-13 signaling. J. Leukoc. Biol. 2006;80:838–849. doi: 10.1189/jlb.1005564. [DOI] [PubMed] [Google Scholar]
- 274.Nguyen KD, et al. Alternatively activated macrophages produce catecholamines to sustain adaptive thermogenesis. Nature. 2011;480:104–108. doi: 10.1038/nature10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Odegaard JI, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zigmond E, Jung S. Intestinal macrophages: well educated exceptions from the rule. Trends Immunol. 2013;34:162–168. doi: 10.1016/j.it.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 277.Carlin LM, et al. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell. 2013;153:362–375. doi: 10.1016/j.cell.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hettinger J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat. Immunol. 2013;14:821–830. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]
- 279.Ruffell B, Affara NI, Coussens LM. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012;33:119–126. doi: 10.1016/j.it.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Doak GR, Schwertfeger KL, Wood DK. Distant relations: macrophage functions in the metastatic niche. Trends Cancer. 2018;4:445–459. doi: 10.1016/j.trecan.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Loyher PL, et al. Macrophages of distinct origins contribute to tumor development in the lung. J. Exp. Med. 2018;215:2536–2553. doi: 10.1084/jem.20180534. [DOI] [PMC free article] [PubMed] [Google Scholar]