Graphical abstract
Keywords: Macroalgae, Biorefinery, Alginate, Vibrio sp. SP1, Carotenoids, Genome sequencing
Highlights
-
•
A novel bacterium Vibrio sp. SP1 rapidly consuming alginate was isolated.
-
•
Whole genome sequence of this strain was generated.
-
•
It was engineered to express heterologous enzymes for carotenoids production.
-
•
Direct conversion of brown macroalgae into carotenoids was demonstrated.
Abstract
Macroalgae is regarded as a promising third-generation marine biomass that can be utilized as a sustainable feedstock for bio-industry due to the high sugar level and absence of lignin. Alginate, composed of 1,4-linked D-mannuronate (M) and L-guluronate (G), is one of the major carbohydrates in brown macroalgae. It is difficult to be assimilated by most industrial microorganisms. Therefore, developing engineered microorganisms that can utilize alginate as a feedstock in order to produce natural products from marine biomass is critical. In this study, we isolated, characterized, and sequenced Vibrio sp. SP1 which rapidly grows using alginate as a sole carbon source. We further engineered this strain by introducing genes encoding enzymes under the control of synthetic expression cassettes to produce lycopene and β-carotene which are attractive phytochemicals owing to the antioxidant property. We confirmed that the engineered Vibrio sp. SP1 could successfully produce 2.13 ± 0.37 mg L−1 of lycopene, 2.98 ± 0.43 mg L−1 of β -carotene, respectively, from 10 g L−1 of alginate as a sole carbon source. Furthermore, our engineered strain could directly convert 60 g L−1 of brown macroalgae Sargassum fusiforme into 1.23 ± 0.21 mg L−1 of lycopene without any pretreatment which had been vitally required for the previous productions. As the first demonstrated strain to produce high-value product from Sargassum, Vibrio sp. SP1 is evaluated to be a desirable platform for the brown macroalgae-based biorefinery.
1. Introduction
Selection of the biomass to be utilized is one of the major issues in microbial bioproduction industry in terms of economic efficiency and environmental concerns. Currently, the resources of biomass are expanded and categorized by its properties. First-generation biomass is based on food resources such as sugarcane and corn. Although they contain high level of sugars that can be efficiently utilized by industrial microbes, ethical controversy such that meals for developing countries are used for other purposes still remains [1], [2]. Utilizing lignocellulosic biomass as second-generation biomass (e.g. wood and grass) does not cause that kind of problem, but high thermal energy is required for the saccharification process due to the presence of lignin [3], [4], and by-products tend to be toxic to the cell [5]. From these aspects, algae, known as third-generation biomass, are suitable resources for solving problems related to land biomass [6], [7]. Moreover, a recent study showed that the brown macroalgae Sargassum is widespread and increasing in quantity in the Atlantic [8], meaning it could be another potential feedstock that could be used for the biorefinery.
Rapid cellular growth and flexibility in fermentable carbon sources are the desirable properties in choosing chassis for bioproduction. Among diverse species, the marine bacteria Vibrio species [9], [10], [11], [12] are known to have the shortest doubling time. Especially when it comes to industrial application, Vibrio bacterium is getting gradual attention since its characteristics such as high biomass yield, fast growth, and resistance to salinity have advantages over other bacteria. Among various Vibrio species, the most commonly studied and engineered strain is Vibrio natriegens [13]. Metabolic engineering for the bioproduction enabled the strain to produce high value-added chemicals such as melanin [14], selenium nanoparticle [15], β -carotene and violacein [16]. However, despite the merit of the strain, it lacks ability to assimilate third generation biomass, alginate, a prominent sugar in brown macroalgae.
As a fast-growing marine bacterium, alginate utilizing Vibrio strains have been steadily reported to be isolated from thalassic environment [17], [18], [19]. Due to the lack of genetic tools for such non-conventional strains, recent studies have shown the possibility to convert alginate into biofuels using conventionally well-known microorganisms such as Escherichia coli and Saccharomyces cerevisiae by introducing heterologous enzymes originated from Vibrio species [20], [21]. Direct genetic modification of non-conventional host was also endeavored. Newly isolated Vibrio sp. dhg [22] was characterized to utilize alginate and produce diverse high-value added products, while exhibiting fast growing profile and engineering potential. Further investigation revealed that enzymes mediating the methylerythritol-4-phosphate (MEP) pathway found in the strain exhibits better activity than those of E. coli, implying Vibrio species could be an promising workhorse when producing isoprenoid or carotenoid metabolites [23]. Likewise, the accumulation of microbial and genetic resources is expected to enrich the realm of genetic compartments regarding metabolic engineering of Vibrio species for the production of value-added products. Considering the limitless immensity of the marine micro-ecology, broader investigation to find industrially applicable marine microorganism and its genetic resources are booming and has a long way to be carried on.
Here, we isolated fast-growing marine bacterium which is capable of not only decomposing but also metabolizing alginate as well as brown macroalgae as a sole carbon source. Computational analysis of data from whole genome sequencing disclosed the genetic resources regarding alginate decomposition, utilization and endogenous pathways. For further verification to engineer this bacteria, genetic compartments such as promoters and replication of origins were examined by expression of a fluorescent protein. At last, we verified the potential of Vibrio sp. SP1 as a platform for brown macroalgae-based biorefinery by demonstrating that this engineered strain can directly convert the brown macroalgae Sargassum fusiforme into lycopene and β -carotene, which are well-known chemicals with anti-oxidative activities for food, nutraceuticals, cosmetic industries [24], [25].
2. Material and methods
2.1. Bacterial strain and reagents
All strains and plasmids used in this study are listed in Table S1. Escherichia coli Mach-T1R was used for routine cloning process. Cloning materials for restriction endonucleases, Phusion DNA polymerase, NEBuilder HiFi DNA Assembly Cloning kit, and Quick ligation kit were purchased from New England Biolabs (NEB, Ipswich, MA, USA). EmeraldAmp polymerase chain reaction (PCR) Master Mix was supplied by Takara Bio Inc. (Takara Bio Inc., Shiga, Japan). Plasmids were prepared by GeneAll DNA Plasmid SV kit (Geneall, Seoul, Republic of Korea). The PCR purification kit and Gel extraction kit were purchased from Zymo Research (Zymo Research, Irvine, CA, USA). All primers used in this study were synthesized by Bioneer (Bioneer, Daejon, Republic of Korea).
2.2. Culture condition and medium composition
For bacterial isolation, a consortium, a symbiotic culture extracted from the intestine of a starfish, which was discovered from the seashore in Incheon, Republic of Korea, was aerobically cultured at 30 °C with shaking at 250 rpm (rotation per minute) in 20 mL of alginate minimal medium (pH 6.8) containing 10 g L−1 of alginate, 30 g L−1 of NaCl, 5 g L−1 of (NH4)2SO4, 1 mM of MgSO4, 2 g L−1 of K2HPO4 and 10 mL L−1 of trace metal solution (Table S2). After a series of subcultures from overnight cultured broth, the enriched consortium was spread on the agar plate containing alginate for the isolation of single colonies. We then picked the earliest visible colony for isolation and identification. The culture condition and medium composition for the comparative growth profiling of Vibrio sp. SP1 on other sugar sources was identical to alginate minimal medium, except for the type of sugar source, where 10 g L−1 of alginate were replaced by 10 g L−1 of glucose, mannitol, sucrose, fructose, and glycerol, respectively.
2.3. Genome sequencing, assembly, annotation and taxonomic analysis
To generate for long reads data, genomic DNA (gDNA) from Vibrio sp. SP1 was extracted with genomic DNA extraction kit (GeneAll, Seoul, Republic of Korea). The concentration and purity of the genomic DNA were measured by Nanodrop™ (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis. The sequencing library was constructed by using SMRTbell Template Prep Kit (Pacific Biosciences, Menlo Park, CA, USA), qualified by Bioanalyzer DNA 12,000 chip (Agilent Technologies, Santa Clara, CA, USA), and sequenced by the Pacific Biosciences (PacBio) RSII sequencer (Pacific Biosciences, Menlo Park, CA, USA). We obtained a total of 141,551 reads with mean length of 9,924 bases for 1,404,837,926 bases and de novo assembled the reads using RS HGAP Assembly (v3.0). Then, Illumina short-read sequencer (MiniSeq) was used to polish the putative errors in the draft genome. A gDNA library (300 bp average) for short reads was prepared using the KAPA HyperPlus Kit according to the manufacturer’s instructions (KAPA Biosystems, Wilmington, MA, USA) and sequenced by the MiniSeq 300-cycle mid-output kit (Illumina, San Diego, CA, USA). Paired-end reads obtained from MiniSeq were mapped to the genome generated from PacBio data using the breseq pipeline with default options [26]. The quality of obtained output genome was validated by CheckM to have 100% of completeness and 0.41% of contamination level [27]. After validation, the complete genome sequence was uploaded to Rapid Annotation using Subsystem Technology (RAST) server [28] for the annotation and the circular form of complete genome was created using DNAplotter [29]. The sequencing data were deposited to the open database (GenBank accession no. CP060589 and CP060590). To further specify the taxonomic characteristics, UBCG [30] and MEGA-X [31] were used to generate phylogenetic tree. Specific naming of the newly isolated strain as ‘Vibrio sp. SP1′ was followed by the taxonomic analysis which conforms to the ‘International Code of Nomenclature of Prokaryotes (2008 revision)’ [32].
2.4. Plasmid construction and identification
In order to test the applicability of various types of plasmid origins, five different replication origins (p15A, pMB1, CloDF13, RSF, and pUC) were constructed to have either chloramphenicol or tetracycline resistance gene. All detailed sequences of primers used in this study is presented in Table S3. To test origin of replication and construct modified plasmids with appropriate selectable marker for Vibrio sp. SP1, chloramphenicol resistance gene (CmR) from pACYCDuet-1 was amplified with AvrII_C-F and BamHI_C-R and inserted in pCDFDuet-1 after digestion with AvrII and BamHI, yielding pCDF-C. To prepare pRSF-C, chloramphenicol resistance gene from pdCas9 and pRSFDuet-1 backbone were amplified using BsaI_CAT_F, BsaI_CAT_R and BsaI_pRSF_F, BsaI_pRSF_R followed by BsaI digestion and ligation, resulting in pRSF-C. For pUC-C preparation lacI-ECK120029600 terminator fragment was prepared with overlap extension PCR using Bsu36I_term_F, overlap_term_R, overlap_lacI_F and lacI_BamHI_R then ligated to amplified pMD19 backbone, which was prepared by Bsu36I_pMD19_F and BamHI_pMD19_R, after digested with BamHI and Bsu36I. Multiple cloning site was amplified from pACYCDuet-1 using Bsu36I_MCS_F and duet_check_R and ligated to pMD19-lacI-ECK120029600 vector after digestion with Bsu36I. Resulted vector was Amplified using BsaI_pMD19_F and BsaI_pMD19_R which was ligated with CmR from pdCas9 after digested by BsaI. Site directed mutagenesis was conducted using Site_deletion_F and Site_deletion_R to remove redundant enzyme site, yielding pUC-C at last. For the identification of positive colonies, check primers which specifically binds to each origin of replication were used as follows: p15A using check1-F and check1-R, CloDF13 using check2-F and check2-R, pMB1 using check1-F and check3-R, RSF using check3-F and check4-R, and pUC using check1-F and check1-R.
To construct pTac-sGFP for testing compatibility of isopropyl β -D-1-thiogalactopyranoside (IPTG)-inducible promoter, we amplified fragment containing tac promoter from coding sequence of synthesized superfolder-green fluorescent protein (sGFP) using primers Ptac_BamHI-F and sGFP_AvrII-R. The amplified DNA fragment was inserted into pACYCDuet-1 by cleaving with BamHI and AvrII, creating pTac-sGFP. For constitutive promoters, DNA fragments of sGFP with J23100, J23108, and J23117 were firstly amplified by an overhang primer sGFP-F1, and then secondly amplified with J23100_BamHI-F, J23108_BamHI-F and J23117_BamHI-F respectively, both using sGFP_AvrII_R as reverse primer. The cloning strategy was identical with constructing pTac-sGFP, yielding pJ23100-sGFP, pJ23108-sGFP and pJ23117-sGFP.
To complete the carotenoid biosynthetic pathway in Vibrio sp. SP1, three heterologous genes (crtE encoding geranylgeranyl diphosphate (GGPP) synthase, crtB encoding phytoene synthase and crtI encoding phytoene desaturase) from Lamprocystis purpurea were introduced for lycopene production, and additional crtY gene encoding lycopene cyclase from Pantoea ananatis was introduced for β -carotene production. For the plasmid of lycopene production, p1EBI plasmid which encodes crtE-crtB-crtI gene under same Ptac promoter in a polycistronic manner was used [23]. To prepare β -carotene plasmid, p1EBIY, vector fragment was amplified using the primers of crtY_v-F and crtY_v-R with p1EBI as a template. The gene of crtY from Pantoea anantis amplified using crtY-F and crtY-R primers was used to generate p1EBIY. Finally, all amplified fragments were assembled by using NEBuilder HiFi DNA Assembly Cloning kit. pRSF-EBI and pRSF-EBIY were additionally constructed by inserting fragments cleaved from p1EBI and p1EBIY respectively into pRSF-C with HindIII and Bsu36I digestion. Same strategy was applied to construct pUC-EBI and pUC-EBIY except that pUC-C was used for the backbone vector.
2.5. Transformation of Vibrio sp. SP1
All plasmids were introduced into Vibrio sp. SP1 based on previously demonstrated electroporation method for Vibrio natriegens [33]. Briefly, a single colony of Vibrio sp. SP1 was inoculated into 5 mL of fresh Brain Heart Infusion (BHI) supplemented with v2 salt (11.92 g L−1 NaCl, 0.3 g L−1 KCl, and 2.2 g L−1 MgCl2) liquid medium (BHIv2). After overnight culture grown in medium, the seed was diluted to OD600 0.05 in 50 mL of fresh BHIv2 medium. When the cell density reached OD600 0.7 corresponding to exponential phase, cell was harvested and incubated for 10 min on ice. Then, cell was centrifuged at 4 °C with centrifugation 14,000 × g and washed three times using electroporation buffer (680 mM of sucrose and 7 mM of K2HPO4; pH 7.0). Finally, 1 μg of plasmid DNA was electroporated under the following conditions: 1.1 kV, 25 μF and 200 Ω in 1 mm cuvette using Gene Pulser Xcell (Bio-Rad, Hercules, CA, USA). After recovery culture for 1 h in 700 mL of BHIv2 medium, whole cells were spread on alginate agar plate or LB agar plate supplemented with additional 5 g L−1 NaCl (total of 15 g L−1 NaCl) containing antibiotics.
2.6. Plasmid copy number determination
Method was adapted from the previous work and modified accordingly [34]. Absolute copy number of DNAs were measured via quantitative PCR (qPCR) using StepOnePlus Real-time PCR system (Thermo Fisher Scientific, Waltham, MA, USA) and primers amplifying each target. Reaction mixtures were prepared in triplicates using Accupower 2X greenstar qPCR Master Mix (Bioneer, Daejon, Republic of Korea), using purified DNAs or cell lysates as templates. All culture was incubated in 30 °C, 250 rpm shaking chamber.
To develop standard curves, concentrations of freshly purified plasmids and PCR-amplified partial rpoA containing flanking regions were measured using Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and serially diluted. All diluted standard was subjected to qPCR with qPCR_target_F and qPCR_target_R primers where target corresponds to p15A, CloDF13, RSF, pUC and rpoA. Standard curves were plotted with log10 of template molecules and cycle of threshold (Ct) values.
To calculate plasmid copy number, cultures of either plasmid bearing Vibrio sp. SP1 or E. coli K-12 W3110 were grown overnight in LBv2 or LB, respectively, with 34 mg L−1 chloramphenicol. The seed cultures were inoculated to initial OD600 of 0.05 and grown for 6–8 h, which were regarded to be in stationary phase. 1 mL of the cultures were centrifuged and washed with chilled electroporation buffer or deionized water, respectively. Then cells were diluted with deionized water to 20 – 100 fold and boiled for 10 min at 95 °C to make cell lysates. Cell lysates were subjected to qPCR and absolute copy number of plasmid or gDNA were obtained substituting Ct values for standard curve equations. Subsequently, ratio of plasmid to gDNA copies was considered as plasmid copy number.
2.7. Fluorescence measurement
Expression of sGFP was analyzed by Hidex Sense Microplate Reader (Hidex, Turku, Finland) using transformed Vibrio sp. SP1 with pTac-sGFP, pJ23100-sGFP, pJ23108-sGFP and pJ23117-sGFP. Each strains were cultivated for overnight in 5 mL of glucose minimal medium (10 g L−1 glucose, 10 g L−1 NaCl, 5 g L−1 (NH4)2SO4, 1 mM of MgSO4, 100 mM K2HPO4, 100 mM KH2PO4 and 10 mL L−1 trace metal solution) supplemented with 12 μg mL−1 of chloramphenicol. The overnight cultures were 100-fold diluted for refreshment, and reinoculated as OD600 0.1 after it reached 1.0. For the induction of tac promoter, 0.1 mM of IPTG was added to the culture broth when OD600 reached 0.4. The fluorescence intensity of 150 μL broth was detected using excitation and emission wavelength of 485 nm and 535 nm, respectively, after 6 h and 12 h of cultivation. The optical density of the cell was measured using 600 nm wavelength at Jenway 7300 Visible Spectrophotometer (Jenway, UK). Subsequently, the specific fluorescence was obtained by normalizing the fluorescence value by cell density and subtracting it from the autofluorescence of the cells bearing empty plasmid backbone.
2.8. Quantification of alginate from purified sugar and raw material
To measure the amount of alginate, modified carbazole sulfuric acid method was used [35]. Briefly, 0.025 M of sodium tetraborate·10H2O in absolute sulfuric acid and 0.125% (w/v) of carbazole in absolute ethanol were used. In glass test tube, samples were diluted to a concentration of 0 to 1 g L−1. Subsequently, 1 mL of sodium tetraborate solution was added and mixed vigorously. After waiting for 5 min for the reaction to be stabilized on ice, 40 μL of carbazole solution was added and each 100 μL of samples was then quantified at the wavelength of 530 nm.
2.9. Culture condition and quantification of carotenoids production
To measure carotenoids amounts, each recombinant strain harboring plasmids for the production of lycopene (p1EBI), β -carotene (p1EBIY) and backbone (pACYCDuet-1) as negative control were cultivated in the alginate minimal media with addition of 5 μg mL−1 of chloramphenicol. In brief, the overnight cultures in 5 mL of alginate minimal media were refreshed by inoculation into the same medium. When optical density of the cells at 600 nm reached exponential phase, the refreshed seed was inoculated at OD600 0.05 in 250 mL baffled flask containing 20 mL medium. The cells were cultured aerobically at 30 °C with continuous shaking (250 rpm). When optical density of the cells reached 0.4 at 600 nm wavelength, 0.1 mM of IPTG was added for inducing gene expression. At each time points, the 1 mL of culture was harvested to quantify lycopene. The culture experiments were conducted with biological triplicates. For lycopene extraction, the harvested cell pellet was lysed by adding 1 mL of absolute acetone. The tubes were incubated at 55 °C for 15 min and intermittently vortexed to enhance the extraction. The cell debris was removed by centrifugation at 19,000 × g for 15 min at room temperature and only supernatant containing products was obtained. The analysis of products was carried out by high-performance liquid chromatography (HPLC) equipped with an Agilent Poroshell 120 EC-C18 column (4 μm, 4.6 × 150 mm, Agilent Technologies, Santa Clara, CA, USA). The analysis was conducted with a mobile phase of acetonitrile-methanol (the ratio 65:35 v/v) at a flow rate of 1 mL/min. The column temperature was set at room temperature during analysis. The concentration of products was measured by UV/Vis detector with absorbance at a wavelength of 450 nm. The analytical standards for authentic lycopene and β -carotene were purchased from Sigma-Aldrich (Sigma-Aldrich, St. Louis, MO, USA). To produce lycopene in 3 L minimal medium culture, aerobic fed-batch culture was carried out at 5 L of bioreactor from Fermentec (Fermentec, Cheongju, Republic of Korea). Powder of Sargassum fusiforme was bought from Wando, Republic of Korea. 4 g L−1 of purified alginate was supplied to initiate the culture for the first 6 h with 4 L min−1 of filtered air (O2 20% and N2 80%) and 500 rpm for efficient agitation. After depletion of initial sugar, 20 g L−1 of brown seaweed powder was fed into bioreactor directly every 4 h during entire cultivation.
3. Results and discussion
3.1. Isolation and characterization of alginate-assimilating bacterium
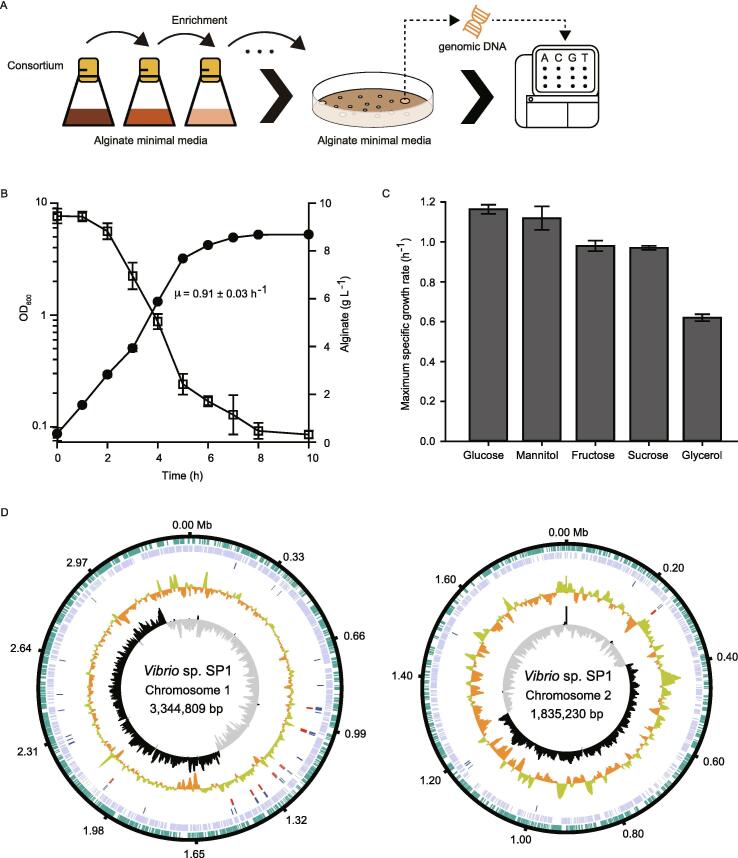
Alginate-assimilating bacterium was successfully isolated by the serial culture of the consortium in alginate minimal media (Fig. 1A). The isolated bacterium could efficiently utilize alginate as a sole carbon source with high specific growth rate (Fig. 1B, μ = 0.91 ± 0.03 h−1). Its comparable growth rate against Vibrio sp. dhg suggested that both bacteria have similar alginate decomposing capability [22]. Moreover, it could also metabolize other biomass-derived carbons sources such as glucose, mannitol, sucrose, fructose, and glycerol with high specific growth rate (Fig. 1C). Notably, when grown in glucose minimal medium, the maximum specific growth rate of this bacterium (μ = 1.16 ± 0.02 h−1) was more than twice as high as that of E. coli W3110 (μ = 0.5644 h−1). In rich media, Vibrio sp. SP1 showed short doubling time of 20.84 ± 0.29 min, similar to that of Vibrio natriegens ATCC 14,048 or Vibrio sp. dhg (Fig. S1).
Fig. 1.
(A) The experimental flow from isolation to sequencing of novel bacterium (B) A time-course data of growth and carbohydrate consumption of Vibrio sp. SP1 using 10 g L−1 of alginate minimal media. Closed circle and open square represent OD600 and amount of alginate, respectively. (C) Maximum specific growth rates were measured in various carbon sources. (D) Circular genome map of Vibrio sp. SP1. From the outer to inner circle: the direction of protein coding sequences (plus strand for green and minus strand for light purple), tRNAs (blue), rRNAs (red), GC content (orange/yellow) and GC skew (black/gray). The map of complete genome sequence was visualized by DNAplotter. The error bars represent the standard deviations of biological triplicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In order to further characterize this bacterium, we conducted whole genome sequencing by combining PacBio platform for generating draft genome and Illumina platform for compensating errors such as repetitive or misreading sequences [36]. As shown in Table S4, a total of 41 errors identified by mapping short reads to PacBio data were corrected to generate the final genome. The draft genome of this strain contained two circular chromosomes of 3,344,809 bp and 1,835,230 bp with GC content of 44.8% and 44.6%, respectively (Fig. 1D and Table 1). Based on the RAST annotation, there was a total of 4,654 protein coding sequences (CDS) and 166 RNAs including 129 tRNAs and 37 rRNAs in two chromosomes (Table 1). Among 4,654 annotated genes, 3,202 genes in the genome were categorized with specific subsystem features (Table 2). Since it was hard to discriminate among Vibrio species due to the high similarity in 16 s rDNA sequences, we referred to the list of the closest neighbors (Fig. S2A) based on the RAST database server and phylogenetic tree was drawn by UBCG [30] analysis. MEGA-X [31] visualization tool presented that the isolated bacteria was relatively close to Vibrio alginolyticus strains but with split branch (Fig. S2B). Therefore, we named this novel strain Vibrio sp. SP1 following the international criteria [32]. Further comparative genomic analysis presented the identity of Vibrio sp. SP1 with Vibrio sp. dhg, fast-growing Vibrio strain recently isolated to also utilize alginate, as 79.80% (Fig. S3).
Table 1.
The summary of genomic contents of Vibrio sp. SP1.
| Attributes | Chromosome |
|
|---|---|---|
| 1 | 2 | |
| Length (bp) | 3,344,809 | 1,835,230 |
| GC content (%) | 44.8 | 44.6 |
| CDS | 3,000 | 1,654 |
| tRNAs | 116 | 13 |
| rRNAs | 34 | 3 |
Table 2.
Distribution of subsystem categories and features of Vibrio sp. SP1 genome based on the RAST annotation server.
| Subsystem Features | Counts | Ratio (%) |
|---|---|---|
| Cofactors, Vitamins, Prosthetic Groups, Pigments | 287 | 8.96 |
| Cell Wall and Capsule | 120 | 3.75 |
| Virulence, Disease and Defense | 96 | 3.00 |
| Potassium metabolism | 38 | 1.19 |
| Photosynthesis | 0 | 0.00 |
| Miscellaneous | 39 | 1.22 |
| Phages, Prophages, Transposable elements, Plasmids | 4 | 0.12 |
| Membrane Transport | 245 | 7.65 |
| Iron acquisition and metabolism | 58 | 1.81 |
| RNA Metabolism | 197 | 6.15 |
| Nucleosides and Nucleotides | 94 | 2.94 |
| Protein Metabolism | 280 | 8.74 |
| Cell Division and Cell Cycle | 41 | 1.28 |
| Motility and Chemotaxis | 129 | 4.03 |
| Regulation and Cell signaling | 96 | 3.00 |
| Secondary Metabolism | 4 | 0.12 |
| DNA Metabolism | 106 | 3.31 |
| Fatty Acids, Lipids, and Isoprenoids | 116 | 3.62 |
| Nitrogen Metabolism | 44 | 1.37 |
| Dormancy and Sporulation | 4 | 0.12 |
| Respiration | 141 | 4.40 |
| Stress Response | 166 | 5.18 |
| Metabolism of Aromatic Compounds | 8 | 0.25 |
| Amino Acids and Derivatives | 428 | 13.37 |
| Sulfur Metabolism | 33 | 1.03 |
| Phosphorus Metabolism | 53 | 1.66 |
| Carbohydrates | 375 | 11.71 |
| Total | 3202 | 100 |
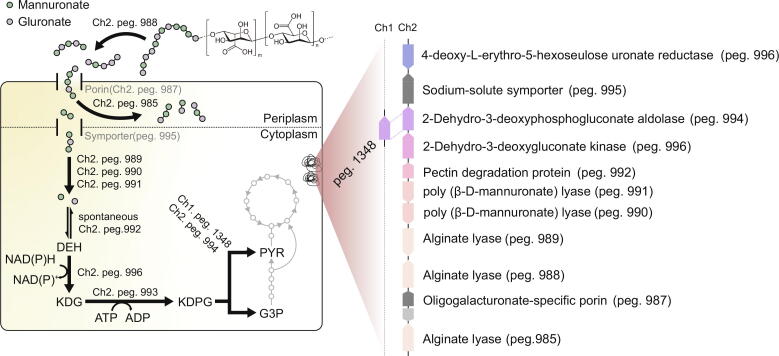
According to previous studies, it has been suggested that alginate can be utilized by the following metabolic pathway [37]. Briefly, alginate polymer which is composed of 1,4-linked D-mannuronate (M) and L-guluronate (G) is sequentially digested by extracellular, periplasmic, and cytosolic alginate lyases during uptake by porins and symporters. Conveyed monomers are then converted to 4-deoxy-L-erythro-5-hexoseulose uronic acid (DEH) spontaneously or with the help of pectin degradation protein [38]. Reductase and kinase further convert DEH to 2-keto-3-deoxy-6-phosphogluconate (KDPG). Finally, aldolase, as a glycolytic enzyme, splits KDPG into pyruvate (PYR) and D-glyceraldehyde 3-phosphate (G3P) leading to TCA cycle. We have confirmed that all enzymes participating in alginate assimilation are encoded in the genome of Vibrio sp. SP1 (Table S5, Fig. 2A). There were three alginate lyases (peg. 985, peg. 988, and peg. 989) and two poly (β -D-mannuronate) lyases (peg. 990 and peg. 991) which can only cleave M−M linked oligoalginate. Two out of five enzymes, peg. 985 and peg. 988, were predicted to have signal peptide sequences (MKLNLLVAAMAVTLPTLAIA and MKHIFFKSLLASSILLAVGCNS) [39], suggesting the presence of secreted alginate lyases (Fig. S4A). The others are assumed to cleave oligoalginates in cytoplasmic space. Despite the difference in cellular location, three alginate lyases identically had consensus sequences ((R/E) (S/T/N) EL, Q (I/V) H, YFKAG (V/I) YNQ)) which belong to Polysaccharide Lyase Family-7 (Fig. S4B). These genes in alginate decomposing pathway were highly conserved in amino acid sequences compared to other alginate metabolizing Vibrio species (Fig. S4C).
Fig. 2.
Genomic analysis of alginate assimilation pathway Graphic demonstration of alginate metabolizing pathway in Vibrio sp. SP1 based on sequenced genome data. Predicted secretion proteins, peg. 985 and peg. 988, were represented as extracellular and periplasmic alginate lyases for convenience. Most related genes were located in chromosome 2.
3.2. Development of genetic tools for Vibrio sp. SP1
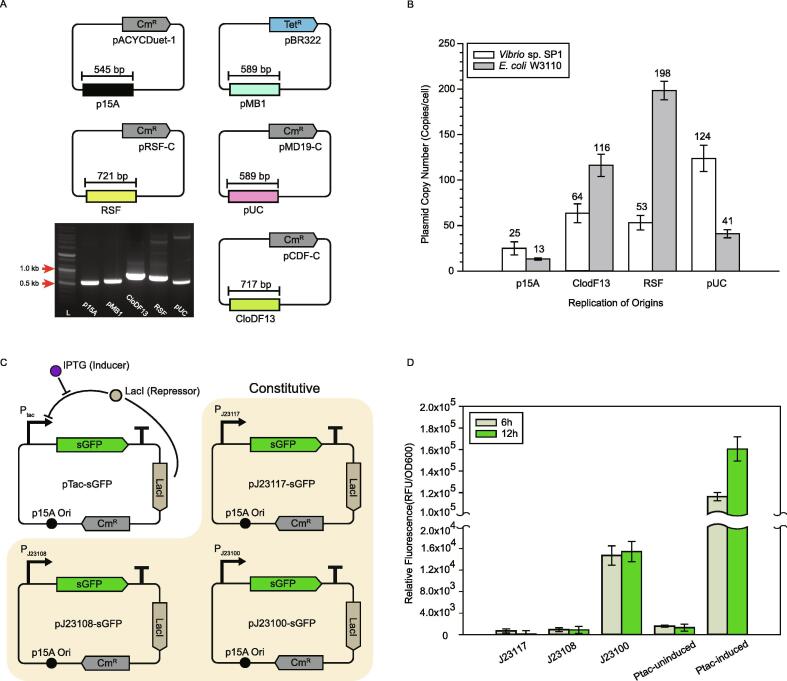
The availability of genetic systems is critical for introducing foreign DNA into a strain. Current plasmid transformation method for bacteria requires an appropriate selection marker. For the selection of proper antibiotics as a selection marker, minimal inhibitory concentration (MIC) was measured based on the growth of Vibrio sp. SP1. We decided to use chloramphenicol and tetracycline as selection antibiotics with concentrations of 5 μg mL−1 in LBv2 media (Table S6). Next, variable replication origins were also tested. Plasmids harboring origin p15A, pMB1, CloDF13, RSF, pUC and pSC101 were transformed into Vibrio sp. SP1. Transformants were obtained except for pSC101. PCR amplified origins verified by electrophoresis demonstrated that five tested origins were compatible in Vibrio sp. SP1 (Fig. 3A). To set criteria for choosing appropriate expression vector, we also measured plasmid copy number (PCN) for each replication origin using standard curves (Fig. S5). It was shown that p15A, CloDF13, RSF, and pUC have 25.0 ± 13.2, 63.7 ± 10.4, 53.2 ± 8.1 and 123.8 ± 14.5 copies per cell, respectively (Fig. 3B). Also, when the stability of the plasmid was measured, the most stable origin turned out to be p15A, followed by pUC and RSF, while CloDF13 was the least stable origin where 16.4% of the population lost its plasmid (Fig. S6). Among tested origins, p15A showed the highest homogeneity of expressed protein. We then tested the availability of the previously characterized constitutive (J23100, J23108 and J23117) and inducible (Ptac) promoters in Vibrio sp. SP1 by measuring the expression level of sGFP (Fig. 3C). Vibrio sp. SP1 harboring pJ23100-sGFP, pJ23108-sGFP and pJ23117-sGFP presented the expression level (RFU/OD600) to be 15,446.9 ± 1,874.1, 900.9 ± 654.8 and 85.9 ± 673.3) respectively after 12 h culture, exhibiting the identical order of strength (J23100 > J23108 > J23117) measured in E. coli (Fig. 3D) [40]. The induced recombinant Vibio sp. SP1 harboring pTac-sGFP showed 128-fold higher fluorescence value (160,507.0 ± 11,252.8) compared to the uninduced cell and it was the highest among the tested promoters as expected (Fig. 3D), indicating that the Ptac promoter and LacI-based repression system is working well in Vibrio sp. SP1.
Fig. 3.
Identification of applicable genetic tools for Vibrio sp. SP1. (A) Various plasmids harboring different replication origins were introduced into Vibrio sp. SP1. The image of agarose gel electrophoresis was obtained by running PCR products targeted to amplify each replication origin using transformed colonies. L denotes ladder marker. (B) Calculated PCN of Vibio sp. SP1 and E. coli W3110 was shown. (C) Schematic diagram for constitutive and inducible sGFP expression vector is represented. (D) Validation of heterologous sGFP expression using Ptac and J23 promoters in Vibrio sp. SP1. Ptac was either induced with 0.1 mM of IPTG or not. The error bars represent the standard deviations of biological triplicates.
3.3. Carotenoids production from Vibrio sp. SP1 using alginate and brown macroalgae
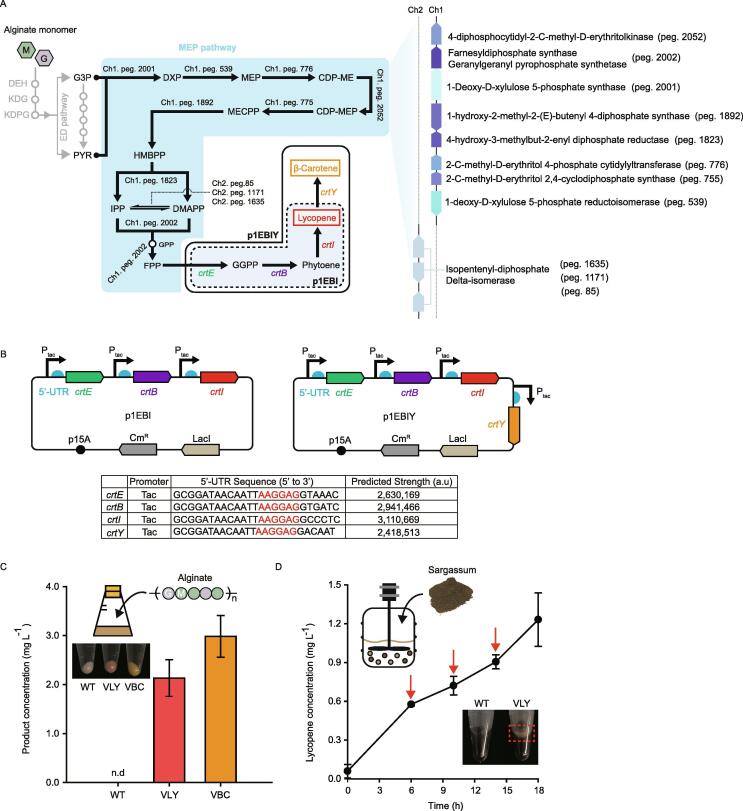
Assimilation of alginate in Vibrio sp. SP1 generates KDPG that is cleaved to G3P and PYR with equimolar ratio through Entner-Doudoroff (ED) pathway [41]. We hypothesized that it would be advantageous for Vibrio sp. SP1 to convert alginate into carotenoids because condensation of G3P and pyruvate is the first step of MEP pathway for the production of carotenoids (Fig. 4A). From genome sequence analysis using KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database [42], we confirmed that Vibrio sp. SP1 contains genes for MEP pathway to generate farnesyl pyrophosphate (FPP) (Fig. 4A and Table S7).
Fig. 4.
Carotenoids production using engineered Vibrio sp. SP1. (A) Illustration engineered carotenoid pathway using alginate. Blue area represents endogenous MEP pathway found in Vibrio sp. SP1. Dotted box represents p1EBI plasmid containing genes for lycopene production, and the outer white box with a solid line is for p1EBIY plasmid consisted of genes for β -carotene production. G and M denote L-guluronate and D-mannuronate, respectively. (B) Detailed plasmid maps of p1EBI and p1EBIY used for lycopene and β -carotene production, respectively. Synthetic 5′-UTR sequences of each gene were designed using UTR Designer and the predicted expression levels are shown. The red characters represent Shine-Dalgarno (SD) sequence. (C) The amounts of lycopene and β -carotene produced by engineered Vibrio sp. SP1 from 10 g L−1 of alginate are presented during the batch culture for 12 h. The schematic diagram and photograph in the upper left corner represent batch culture using alginate and cell pellet after 12 h, respectively. WT, wild-type bacteria with backbone plasmid; VLY, strain for lycopene production; VBC, strain for β -carotene production; n.d, not detected. (D) The amount of lycopene produced by VLY using Sargassum powder directly. Red arrows represent the time of the addition of brown seaweed powder. The photograph on lower right side shows cell pellets with seaweed powder of WT and VLY after 18 h. The error bars represent the standard deviations of biological triplicates. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
For the biosynthesis of lycopene from FPP, expression of three heterologous enzymes crtE, crtB and crtI was required and encoding genes were introduced into plasmid p1EBI harboring p15A origin [22]. Expression of single enzyme CrtY was additionally required for the conversion of lycopene to β -carotene, therefore crtY was introduced to p1EBI yielding p1EBIY (Fig. 4B) [42]. For optimal gene expression, each gene was expressed under the control of inducible Ptac promoter and synthetic 5′-UTRs designed by UTR Designer (http://sbi.postech.ac.kr/utr_designer) (Fig. 4B) [43]. From p1EBI and p1EBIY, we also cloned crtEBI or crtEBIY expression cassettes in RSF and pUC origin respectively and obtained four additional plasmids pRSF-EBI, pRSF-EBIY, pUC-EBI and pUC-EBIY. Vibrio sp. SP1 was then transformed with these six vectors. However, only p1EBI and p1EBIY transformed plates showed colored colonies, indicating the production of target products (Fig. 4C), while others seemed to be non-transformable. This could be explained by toxicity of crtEBI and crtEBIY genes when excessively overexpressed, destabilizing introduced expression system [44], [45], [46]. Therefore, we determined to use p15A among tested origins. The successfully transformed Vibrio sp. SP1 with p1EBI and p1EBIY were named as VLY and VLB, respectively, for the further experiments.
After 12 h cultivation with 0.1 mM IPTG, 2.13 ± 0.37 mg L−1 of lycopene from VLY and 2.98 ± 0.43 mg L−1 of β -carotene from VBC were produced using 10 g L−1 of alginate as a sole carbon source whereas the products were non-detectable in wild-type strain (Fig. 4C). Previous attempt to produce carotenoids from Vibrio natriegens yielded 0.6 mg L−1 of β -carotene from 4 g L−1 glucose [16]. Considering this strain had additional genetic engineering to integrate genes regarding MVA pathway from Lactobacillus acidophilus to replenish the amount of substrate and the substrate was glucose, the overall productivity of β -carotene from alginate in the engineered Vibrio sp. SP1 seems to be sufficiently competitive. Even comparing the production of lycopene from alginate with the formerly isolated and engineered Vibrio sp. dhg producing 0.47 mg L−1 in 9 h exhibits the promising potential prospective of Vibrio sp. SP1 as a cell factory [22].
Next, we tested if the brown macroalgae Sargassum fusiforme powder could be used as a feedstock directly for the production of lycopene in the engineered Vibrio sp. SP1 without any pretreatment. Given that alginate, despite its relatively low cost compared to monosaccharides, demands additional alkaline extraction from brown macroalgae, it was worthwhile to examine the ability to directly utilize brown macroalgae without pretreatment for possibility of industrial production. Unlike production from alginate, we scaled-up the reaction working volume to 3 L in 5 L bioreactor to obtain the detectable amount of production. Because of the dark pigments and lots of sediment of brown macroalgae, it was impossible to determine the exact amount of cell and carbon sources. From total of 60 g L−1 brown seaweed in bioreactor, engineered Vibrio sp. SP1 could produce 1.23 ± 0.21 mg L−1 of lycopene (Fig. 4D), demonstrating the first step of directly utilizing brown macroalgae Sargassum to produce value-added products.
4. Conclusions
In this study, we isolated a novel bacterium Vibrio sp. SP1 which could efficiently degrade alginate and various carbohydrates. Using this strain, we could produce lycopene and β -carotene from alginate. Additionally, by fed-batch cultivation, 1.23 ± 0.21 mg L−1 of lycopene was successfully produced directly from 60 g L−1 of the brown seaweed Sargassum. To our knowledge, the strain is the first engineered bacterium to directly convert the brown seaweed Sargassum into lycopene without any pretreatment. Nevertheless, additional development of this strain using various genetic tools is mandatory to further increase titer, yield, and productivity of value-added products for brown macroalgae-based biorefinery in the future. Previously, elevation in bottleneck enzyme expression [47], heterologous expression of mevalonate (MVA) pathway which is absent in most bacteria [48], and deletion of competing pathways were endeavored and proved to increase their titer in several folds [49]. Introducing such extensive modifications would become a lot more easier owing to recently developed natural competent system in Vibrio species [50], [51].
5. Data availability
The complete genomic sequence of Vibrio sp. SP1 for this study was deposited at GenBank under the accession number CP060589 and CP060590.
CRediT authorship contribution statement
Sungwoo Park: Conceptualization, Formal analysis, Writing – Original draft. Sung Won Cho: Software, Validation, Writing - review & editing. Yungyu Lee: Investigation, Visualization, Writing - review & editing. Mincheol Choi: Data curation. Jina Yang: Resources. Hojun Lee: Methodology. Sang Woo Seo: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The plasmid p1EBI used in this research was kindly gifted by Prof. Gyoo Yeol Jung at Pohang University of Science and Technology (POSTECH). This research was supported by the Basic Science Research Program (NRF-2018R1C1B6001129), the Bio & Medical Technology Development Program (NRF-2018M3A9H3020459), and the Basic Research Laboratory Project (NRF-2018R1A4A1022513), through the National Research Foundation (NRF) of Korea, funded by the Ministry of Science and ICT (MSIT). SWS is partially supported by Creative-Pioneering Researchers Program through Seoul National University (SNU).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.007.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Tenenbaum DJ. Food vs. fuel: diversion of crops could cause more hunger 2008. [DOI] [PMC free article] [PubMed]
- 2.Renzaho A.M.N., Kamara J.K., Toole M. Biofuel production and its impact on food security in low and middle income countries: implications for the post-2015 sustainable development goals. Renew Sustain Energy Rev. 2017;78:503–516. [Google Scholar]
- 3.Sun Y.e., Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83(1):1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- 4.Lee H.R., Kazlauskas R.J., Park T.H. Mild pretreatment of yellow poplar biomass using sequential dilute acid and enzymatically-generated peracetic acid to enhance cellulase accessibility. Biotechnol Bioprocess Eng. 2017;22(4):405–412. [Google Scholar]
- 5.Jönsson L.J., Martín C. Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol. 2016;199:103–112. doi: 10.1016/j.biortech.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 6.John R.P., Anisha G.S., Nampoothiri K.M., Pandey A. Micro and macroalgal biomass: a renewable source for bioethanol. Bioresour Technol. 2011;102(1):186–193. doi: 10.1016/j.biortech.2010.06.139. [DOI] [PubMed] [Google Scholar]
- 7.Hong S.-J., Park Y.S., Han M.-A., Kim Z.-H., Cho B.-K., Lee H. Enhanced production of fatty acids in three strains of microalgae using a combination of nitrogen starvation and chemical inhibitors of carbohydrate synthesis. Biotechnol Bioprocess Eng. 2017;22(1):60–67. [Google Scholar]
- 8.Wang M., Hu C., Barnes B.B., Mitchum G., Lapointe B., Montoya J.P. The great Atlantic Sargassum belt. Science (80-) 2019;365(6448):83–87. doi: 10.1126/science.aaw7912. [DOI] [PubMed] [Google Scholar]
- 9.EAGON RG. Pseudomonas natriegens, a marine bacterium with a generation time of less than 10 minutes. J Bacteriol 1962;83. https://doi.org/10.1128/jb.83.4.736-737.1962. [DOI] [PMC free article] [PubMed]
- 10.Ulitzur S. Vibrio parahaemolyticus and Vibrio alginolyticus: short generation-time marine bacteria. Microb Ecol. 1974;1(1):127–135. doi: 10.1007/BF02512384. [DOI] [PubMed] [Google Scholar]
- 11.Hoffart E., Grenz S., Lange J., Nitschel R., Müller F., Schwentner A. High substrate uptake rates empower Vibrio natriegens as production host for industrial biotechnology. Appl Environ Microbiol. 2017;83(22) doi: 10.1128/AEM.01614-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calero P., Nikel P.I. Chasing bacterial chassis for metabolic engineering: a perspective review from classical to non-traditional microorganisms. Microb Biotechnol. 2019;12(1):98–124. doi: 10.1111/mbt2.2019.12.issue-110.1111/1751-7915.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoff J., Daniel B., Stukenberg D., Thuronyi B.W., Waldminghaus T., Fritz G. Vibrio natriegens: an ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ Microbiol. 2020;22(10):4394–4408. doi: 10.1111/emi.v22.1010.1111/1462-2920.15128. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z., Tschirhart T., Schultzhaus Z., Kelly E.E., Chen A., Oh E. Melanin produced by the fast-growing marine bacterium vibrio natriegens through heterologous biosynthesis: characterization and application. Appl Environ Microbiol. 2020;86(5) doi: 10.1128/AEM.02749-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández-Llamosas H., Castro L., Blázquez M.L., Díaz E., Carmona M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-16252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis G.A., Tschirhart T., Spangler J., Walper S.A., Medintz I.L., Vora G.J. Exploiting the feedstock flexibility of the emergent synthetic biology chassis vibrio natriegens for engineered natural product production. Mar Drugs. 2019;17.;17(12):679. doi: 10.3390/md17120679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Badur A.H., Jagtap S.S., Yalamanchili G., Lee J.-K., Zhao H., Rao C.V. Alginate lyases from alginate-degrading Vibrio splendidus 12B01 are endolytic. Appl Environ Microbiol. 2015;81(5):1865–1873. doi: 10.1128/AEM.03460-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doi H, Chinen A, Fukuda H, Usuda Y. Vibrio algivorus sp. nov., an alginate-and agarose-assimilating bacterium isolated from the gut flora of a turban shell marine snail. Int J Syst Evol Microbiol 2016;66. https://doi.org/10.1099/ijsem.0.001165. [DOI] [PubMed]
- 19.Doi H., Tokura Y., Mori Y., Mori K., Asakura Y., Usuda Y. Identification of enzymes responsible for extracellular alginate depolymerization and alginate metabolism in Vibrio algivorus. Appl Microbiol Biotechnol. 2017;101(4):1581–1592. doi: 10.1007/s00253-016-8021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wargacki A.J., Leonard E., Win M.N., Regitsky D.D., Santos C.N.S., Kim P.B. An engineered microbial platform for direct biofuel production from brown macroalgae. Science (80-) 2012;335(6066):308–313. doi: 10.1126/science.1214547. [DOI] [PubMed] [Google Scholar]
- 21.Enquist-Newman M., Faust A.M.E., Bravo D.D., Santos C.N.S., Raisner R.M., Hanel A. Efficient ethanol production from brown macroalgae sugars by a synthetic yeast platform. Nature. 2014;505(7482):239–243. doi: 10.1038/nature12771. [DOI] [PubMed] [Google Scholar]
- 22.Lim H.G., Kwak D.H., Park S., Woo S., Yang J.-S., Kang C.W. Vibrio sp. dhg as a platform for the biorefinery of brown macroalgae. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-10371-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M.J., Noh M.H., Woo S., Lim H.G., Jung G.Y. Enhanced lycopene production in escherichia coli by expression of two MEP pathway enzymes from vibrio sp. dhg. Catalysts. 2019;9(12):1003. doi: 10.3390/catal9121003. [DOI] [Google Scholar]
- 24.Sies H., Stahl W. Lycopene: antioxidant and biological effects and its bioavailability in the human. Proc. Soc. Exp. Biol. Med. 1998;218(2):121–124. doi: 10.3181/00379727-218-44285a. [DOI] [PubMed] [Google Scholar]
- 25.Giovannucci E., Ascherio A., Rimm E.B., Stampfer M.J., Colditz G.A., Willett W.C. Intake of carotenoids and retino in relation to risk of prostate cancer. J Natl Cancer Inst. 1995;87(23):1767–1776. doi: 10.1093/jnci/87.23.1767. [DOI] [PubMed] [Google Scholar]
- 26.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Eng. Anal. Multicell. Syst., Springer; 2014, p. 165–88. [DOI] [PMC free article] [PubMed]
- 27.Parks D.H., Imelfort M., Skennerton C.T., Hugenholtz P., Tyson G.W. Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A. The RAST Server: rapid annotations using subsystems technology. BMC Genom. 2008;9(1):75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carver T., Thomson N., Bleasby A., Berriman M., Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Na S.-I., Kim Y.O., Yoon S.-H., Ha S.-M., Baek I., Chun J. Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol. 2018;56(4):280–285. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms | Molecular Biology and Evolution | Oxford Academic. Mol Biol Evol 2018;35. [DOI] [PMC free article] [PubMed]
- 32.Parker C.T., Tindall B.J., Garrity G.M. International code of nomenclature of Prokaryotes. Int J Syst Evol Microbiol. 2019;69:S1. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 33.Weinstock M.T., Hesek E.D., Wilson C.M., Gibson D.G. Vibrio natriegens as a fast-growing host for molecular biology. Nat Methods. 2016;13(10):849–851. doi: 10.1038/nmeth.3970. [DOI] [PubMed] [Google Scholar]
- 34.Tschirhart T., Shukla V., Kelly E.E., Schultzhaus Z., NewRingeisen E., Erickson J.S. Synthetic biology tools for the fast-growing marine bacterium vibrio natriegens. ACS Synth Biol. 2019;8(9):2069–2079. doi: 10.1021/acssynbio.9b00176. [DOI] [PubMed] [Google Scholar]
- 35.Bitter T., Muir H.M. A modified uronic acid carbazole reaction. Anal Biochem. 1962;4(4):330–334. doi: 10.1016/0003-2697(62)90095-7. [DOI] [PubMed] [Google Scholar]
- 36.Koren S., Schatz M.C., Walenz B.P., Martin J., Howard J.T., Ganapathy G. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 2012;30(7):693–700. doi: 10.1038/nbt.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H.S., Lee C.-G., Lee E.Y. Alginate lyase: structure, property, and application. Biotechnol Bioprocess Eng. 2011;16(5):843–851. [Google Scholar]
- 38.Hobbs J.K., Lee S.M., Robb M., Hof F., Barr C., Abe K.T. KdgF, the missing link in the microbial metabolism of uronate sugars from pectin and alginate. Proc Natl Acad Sci U S A. 2016;113(22):6188–6193. doi: 10.1073/pnas.1524214113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Käll L., Krogh A., Sonnhammer E.L.L. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338(5):1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Registry of standard biological parts, Anderson promoter collection. http:// parts.igem.org/Promoters/Catalog/Anderson.
- 41.Conway T. The Entner-Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol Rev 1992;9:1–27. [DOI] [PubMed]
- 42.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo S.W., Yang J.-S., Kim I., Yang J., Min B.E., Kim S. Predictive design of mRNA translation initiation region to control prokaryotic translation efficiency. Metab Eng. 2013;15:67–74. doi: 10.1016/j.ymben.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Farmer W.R., Liao J.C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18(5):533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 45.Kim S.W., Keasling J.D. Metabolic engineering of the nonmevalonate isopentenyl diphosphate synthesis pathway in Escherichia coli enhances lycopene production. Biotechnol Bioeng. 2001 doi: 10.1002/1097-0290(20000220)72:4<408::AID-BIT1003>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 46.Yoon S.-H., Lee Y.-M., Kim J.-E., Lee S.-H., Lee J.-H., Kim J.-Y. Enhanced lycopene production in Escherichia coli engineered to synthesize isopentenyl diphosphate and dimethylallyl diphosphate from mevalonate. Biotechnol Bioeng. 2006;94(6):1025–1032. doi: 10.1002/(ISSN)1097-029010.1002/bit.v94:610.1002/bit.20912. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y., Nambou K., Wei L., Cao J., Imanaka T., Hua Q. Lycopene production in recombinant strains of Escherichia coli is improved by knockout of the central carbon metabolism gene coding for glucose-6-phosphate dehydrogenase. Biotechnol Lett. 2013;35(12):2137–2145. doi: 10.1007/s10529-013-1317-0. [DOI] [PubMed] [Google Scholar]
- 48.Yang C., Gao X., Jiang Y.u., Sun B., Gao F., Yang S. Synergy between methylerythritol phosphate pathway and mevalonate pathway for isoprene production in Escherichia coli. Metab Eng. 2016;37:79–91. doi: 10.1016/j.ymben.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z., Sun JingXin, Yang Q., Yang J. Metabolic engineering escherichia coli for the production of lycopene. Molecules. 2020;25(14):3136. doi: 10.3390/molecules25143136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dalia T.N., Hayes C.A., Stolyar S., Marx C.J., McKinlay J.B., Dalia A.B. Multiplex genome editing by natural transformation (MuGENT) for synthetic biology in Vibrio natriegens. BioRxiv. 2017 doi: 10.1101/122655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dalia T.N., Hayes C.A., Stolyar S., Marx C.J., McKinlay J.B., Dalia A.B. Multiplex Genome Editing by Natural Transformation (MuGENT) for Synthetic Biology in Vibrio natriegens. ACS Synth Biol. 2017;6(9):1650–1655. doi: 10.1021/acssynbio.7b0011610.1021/acssynbio.7b00116.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete genomic sequence of Vibrio sp. SP1 for this study was deposited at GenBank under the accession number CP060589 and CP060590.