Abstract
Initial COVID-19 vaccine candidates were based on the original sequence of SARS-CoV-2. However, the virus has since accumulated mutations, among which the spike D614G is dominant in circulating virus, raising questions about potential virus escape from vaccine-elicited immunity. Here, we report that the D614G mutation modestly reduced (1.7–2.4-fold) SARS-CoV-2 neutralization by BNT162b2 vaccine-elicited mouse, rhesus, and human sera, concurring with the 95% vaccine efficacy observed in clinical trial.
Subject terms: SARS-CoV-2, RNA vaccines
Main text
Despite the proofreading function in coronavirus replication1, the pandemic spread of SARS-CoV-2 in naive human populations selects for mutations that may alter viral transmission and/or pathogenesis. One prominent mutation in circulating SARS-CoV-2 is the spike D614G substitution2. The D614G mutation was rare before March 2020, but became dominant as the pandemic continued, reaching >74% prevalence globally by June 20203. Recent studies demonstrate that the D614G mutation does not enhance viral pathogenesis in animal models, but increases viral replication in the upper respiratory tract, and causes more efficient transmission4,5. These animal results are consistent with the clinical findings that patients infected with the G614 virus do not develop more severe disease, but produce more virus in their nasopharyngeal swabs than the patients infected with the D614 virus2. Mechanically, the D614G mutation promotes the spike receptor-binding domain (RBD) adopting an open conformation for binding the angiotensin-converting enzyme 2 (ACE2) receptor, resulting in a higher virion infectivity and thermal stability3. Such structural change may affect antigenicity and/or virus entry.
The initial COVID-19 vaccine candidates targeted the original D614 SARS-CoV-2 strains. Thus, there is a sequence mismatch between the vaccine D614 spike antigen and the predominantly circulating G614 virus, raising the potential for virus escape from vaccine-elicited immunity. BNT162b2 is a lipid nanoparticle-formulated mRNA vaccine candidate that encodes the full-length D614 spike of SARS-CoV-26. In a recently completed phase III clinical trial, BNT162b2 achieved 95% efficacy against COVID-19 and had a good safety profile7. To determine whether BNT162b2-elicited sera neutralize G614 SARS-CoV-2, we tested serum specimens collected during preclinical and clinical studies of BNT162b27.
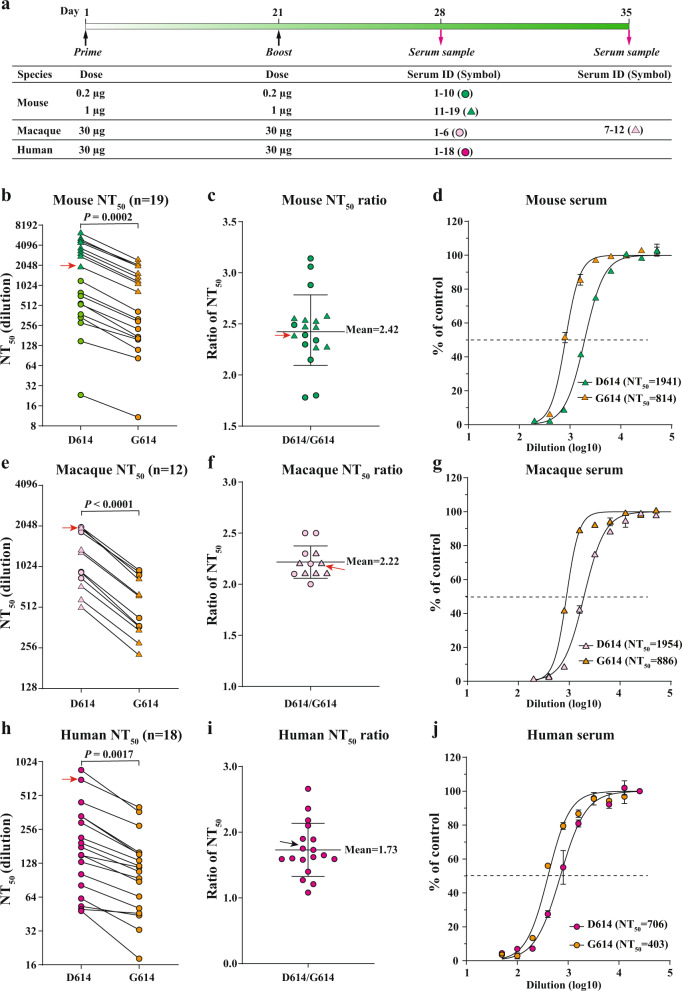
We selected three panels of serum specimens with a wide range of neutralizing titers (Fig. 1a), including BNT162b2-vaccinated mouse sera (19 samples; Fig. 1b–d), rhesus macaque sera (12 samples; Fig. 1e–g), and human sera (18 samples; Fig. 1h–j). The neutralizing titers of each serum were determined against a pair of isogenic D614 and G614 mNeonGreen reporter SARS-CoV-2s8. The mNeonGreen reporter virus-based neutralization assay was previously shown to deliver results comparable to the gold standard plaque reduction neutralization test9. To ensure the consistency of neutralization testing, we infected Vero cells with D614 or G614 reporter virus at equal infection multiplicities (~37%). Each serum was tested head-to-head against both viruses in duplicates on the same 96-well plate. The neutralizing titers against the D614 virus were, on average, 2.4, 2.2, and 1.7 times those against the G614 virus for mouse (Fig. 1c), macaque (Fig. 1f), and human sera (Fig. 1i), respectively. The largest difference in neutralizing titer for an individual serum between the two viruses was <3.2-fold, which was observed in a mouse (Fig. 1c). The observed differences are statistically significant (Fig. 1b, e, h). Collectively, the results indicate that the D614G substitution modestly reduces the inhibition of SARS-CoV-2 by antibodies elicited by the D614 spike antigen.
Fig. 1. The effect of spike D614G substitution on SARS-CoV-2 neutralization.
a Immunization and sample collection. Mice, macaques, and humans were immunized with BNT162b2 at indicated doses on days 1 and 21. Serum samples were collected on days 28 or 35 for neutralization testing. Samples from different groups are indicated by different symbols. b, e, h Neutralizing titers of mouse (b), macaque (e), and human sera (h) against D614 and G614 mNeonGreen SARS-CoV-2. Sera from BNT162b2-vaccinated mice (n = 19), macaques (n = 12), and humans (n = 18) were tested for neutralizing titers. The NT50 values are plotted. Symbols represent sera from individual specimens. The P values were calculated using the Student’s t test. c, f, i Ratios of NT50 values between D614 and G614 viruses. Symbols represent individual sera; the midline represents the geometric mean; and error bars represent standard deviation. d, g, j Representative neutralizing curves. The representative specimens are indicated by red arrows for each species group. Symbols represent individual replicates. Solid lines represent the fitted curve. The dotted line indicates 50% viral inhibition.
The current findings using the authentic isogenic D614 and G614 SARS-CoV-2s confirm the previous results using a pseudovirus neutralization test10. Sera from clinical trial participants immunized with BNT162b2 twice, 21 days apart at doses of 1, 10, and 30 μg, had neutralization titers against pseudovirus bearing D614 SARS-CoV-2 spikes that were 1.7 times those against pseudovirus bearing G614 spikes10, matching the authentic virus neutralization results reported here.
Previous studies that examined the effect of spike D614G substitution on neutralization activities reached different conclusions. However, in all cases, the differences in neutralization titers between the D614 and G614 viruses were modest. One study compared the neutralizing titers of 25 human convalescent sera against D614 or G614 nanoluciferase SARS-CoV-24. The neutralization titers against G614 were 0.8–5.1-fold lower than those against D614 virus, in agreement with our results. However, because the study examined antisera elicited by natural infection with viruses of unknown genotype, the response could have been elicited by either D614 or G614 virus4. Another study that examined human convalescent sera found that, regardless of whether the infecting virus had a D614 or G614 spike, neutralization titers against G614 pseudoviruses were 1.9–2.0 times those against D614 pseudoviruses11. The same study also analyzed sera from mice, rhesus macaques, and humans who were immunized with RNAs encoding different versions of spike antigen. The results showed that pseudovirus neutralization titers against G614 pseudovirus were 3.9–6.5 times those against D614 pseudovirus11. The discrepancy from different studies could be due to different assay systems and conditions (e.g., contradictory pseudovirus results from two studies10,11), underlining the importance of harmonizing the assays in the COVID-19 research.
Thus, the consensus appears to be that the effect of spike D614G mutation on neutralization is modest, small enough that different neutralization assays and laboratories can reach different conclusions about whether vaccine candidate-elicited or infection-elicited neutralization is greater against D614 or G614 virus. Depending on which studies are compared, one could conclude that authentic virus neutralization assays and pseudovirus neutralization assays give comparable or different results. Since the D164G substitution promotes an open conformation of spike RDB for binding to receptor ACE23, the modest difference in neutralization titer may be caused by the improved spike function (i.e., cellular attachment and virus entry) rather than antigenicity. Thus, cautions should be taken when comparing the neutralization titers among different variants.
Despite the dominance of circulating G614 virus, mRNA-based vaccine candidates that encode D614 antigens have achieved 94–95% efficacies, leaving little room for improvement, at least in the initial months after immunization7. However, future mutations could be more impactful on vaccine development. The introduction of SARS-CoV-2 to mink farms has led to mink/human transmission. Mutations recovered from the mink/human transmission might attenuate vaccine efficacy12. Vaccine efficacy against the mink strains as well as the newly emerged U.K. and South African strains should be thoroughly investigated13–15. Once human immunity to SARS-CoV-2 becomes widespread through infection or immunization, immune selection could accelerate the rate of antigenic change. To ensure vaccine research and development stays ahead of viral evolution, we must closely monitor the emergence of new variants with altered transmission, pathogenicity, or immune susceptibility. For influenza virus, a well-accepted system of serological criteria for strain changes has been established16. As correlates of vaccine-mediated protection against COVID-19 emerge, laboratory results using reliable assays and interpretations, rather than disease re-emergence, can guide decisions on future strain changes for vaccine improvement.
Methods
Cells
Vero cells (ATCC®CCL-81) were purchased from the American Type Culture Collection (ATCC, Bethesda, MD), and maintained in a high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT) and 1% penicillin/streptomycin at 37 °C with 5% CO2. All culture medium and antibiotics were purchased from ThermoFisher Scientific (Waltham, MA). The cells were tested negative for mycoplasma.
mNG SARS-CoV-2
The virus stocks of D614 and G614 mNG SARS-CoV-2 were produced using an infectious cDNA clone of SARS-CoV-2 in which the ORF7 of the viral genome was replaced with reporter mNG gene5,8. After rescue from the genome-length viral RNA-electroporated cells, the viral stocks were prepared by amplifying the mNG SARS-CoV-2 on Vero E6 cells for one round. Both the D614 and G614 mNG viruses were prepared under an identical condition17. The titers of the virus stocks were determined by a standard plaque assay. All procedures involving handling infectious SARS-CoV-2 were performed at the BSL-3 facility at UTMB.
Serum
The serum samples of vaccine BNT162b2-immunized mouse, monkey and human subjects were obtained from Pfizer. All sera were heat-inactivated at 56 °C for 30 min prior to testing.
mNG SARS-CoV-2 reporter neutralization assay
The neutralizing activities of each serum against D614 or G614 mNG SARS-CoV-2 were measured using a similar protocol to that described previously9. Vero CCL-81 cells (1.2 × 104) in 50 µl of phenol red-free DMEM (Gibco) containing 2% FBS (Hyclone) and 100 U/ml penicillin-streptomycin (P/S; Gibco) were seeded in each well of black µCLEAR flat-bottom 96-well plate (Greiner Bio-one™). The cells were incubated overnight at 37 °C with 5% CO2. On the following day, each serum was twofold serially diluted in 2% FBS and 100 U/ml P/S DMEM and aliquoted at 30 μl per well in a 96-well plate. Afterwards, 30 μl of D614 or G614 mNG SARS-CoV-2 was added to each well of the serum plate. The serum-virus plate was incubated at 37 °C for 1 h. The virus-serum mixtures were transferred to the Vero cell plate with the final multiplicity of infection (MOI) of 2. For each serum, the starting dilution was 1/20 with nine two-fold dilutions to the final dilution of 1/5120. After incubating the infected cells at 37 °C for 18 h, 25 μl of Hoechst 33342 Solution (400-fold diluted in Hank’s Balanced Salt Solution; Gibco) were added to each well to stain cell nuclei. The plate was sealed with Breath-Easy sealing membrane (Diversified Biotech), incubated at 37 °C for 20 min, and quantified for mNG fluorescence on CytationTM 7 (BioTek). The raw images were acquired using ×4 objective, and processed using the default setting. The total cells (indicated by nucleus staining) and mNG-positive cells were quantified for each well. Infection rates were determined by dividing the mNG-positive cell number by the total cell number. Relative infection rates were obtained by normalizing the infection rates of serum-treated groups to those of non-serum-treated controls. The curves of the relative infection rates versus the serum dilutions (log10 values) were plotted using Prism 8 (GraphPad). A nonlinear regression method was used to determine the fold dilution that neutralized 50% of mNG fluorescence (NT50). Each serum was tested in duplicates.
Statistical analysis
The paired t test was used to compare the neutralizing activities of serum against D614 and G614 mNG SARS-CoV-2 in the software Prism 9 (GraphPad). Two-tailed P values were calculated and indicated in the corresponding figure panels.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank colleagues at Pfizer, BioNTech, and UTMB for helpful discussions and support during the study. We thank the Pfizer-BioNTech clinical trial C4591001 participants, from whom the post-immunization human sera were obtained, and the staff of the New Iberia Research Center, who provided the rhesus macaque post-immunization sera. We thank the many colleagues at Pfizer and BioNTech who developed and produced the BNT162b2 vaccine candidate. P.-Y.S. was supported by NIH grants AI134907 and UL1TR001439, and awards from the Sealy & Smith Foundation, Kleberg Foundation, the John S. Dunn Foundation, the Amon G. Carter Foundation, the Gilson Longenbaugh Foundation, and the Summerfield Robert Foundation.
Author contributions
Conceptualization: X.X. and P.-Y.S.; methodology: J.Z., X.X., C.R.F.G., and P.-Y.S; investigation: J.Z., X.X., C.R.F.G., K.A.S., I.K., K.T., M.C., D.C., P.R.D., and P.-Y.S; resources: M.C., D.C., P.R.D., and P.-Y.S; data curation: J.Z., X.X., C.R.F.G., and P.-Y.S; writing—original draft: J.Z., X.X., and P.-Y.S; writing—review & editing: J.Z., X.X., C.R.F.G., P.R.D., and P.-Y.S.; supervision: X.X., M.C., D.C., P.R.D., and P.-Y.S.; funding acquisition: P.-Y.S.
Data availability
The results supporting the findings in this study are available upon request from the corresponding authors.
Competing interests
The authors declare that there are no competing interests. X.X. and P.-Y.S. have filed a patent on the reverse genetic system and reporter SARS-CoV-2. K.A.S., I.K., K.T., M.C., D.C., and P.R.D. are employees of Pfizer and may hold stock options. J.Z., X.X., C.R.F.G., and P.-Y.S. received compensation from Pfizer to perform the neutralization assay.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Philip R. Dormitzer, Email: Philip.Dormitzer@pfizer.com
Pei-Yong Shi, Email: peshi@UTMB.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-021-00313-8.
References
- 1.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korber, B. et al. Tracking Changes in SARS-CoV-2 Spike: evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell10.1016/j.cell.2020.06.043 (2020). [DOI] [PMC free article] [PubMed]
- 3.Yurkovetskiy L, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183:739–751 e738. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science10.1126/science.abe8499 (2020). [DOI] [PMC free article] [PubMed]
- 5.Plante, J. A. et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature10.1038/s41586-020-2895-3 (2020). [DOI] [PMC free article] [PubMed]
- 6.Walsh, E. E. et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 10.1056/NEJMoa2027906 (2020). [DOI] [PMC free article] [PubMed]
- 7.Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med.10.1056/NEJMoa2034577 (2020). [DOI] [PMC free article] [PubMed]
- 8.Xie X, et al. An infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848 e843. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muruato AE, et al. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun. 2020;11:4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sahin, U. et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. Preprint at medRxiv10.1101/2020.12.09.20245175 (2020).
- 11.Weissman, D. et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe.10.1016/j.chom.2020.11.012 (2020). [DOI] [PMC free article] [PubMed]
- 12.Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science10.1126/science.abe5901 (2020). [DOI] [PMC free article] [PubMed]
- 13.Tegally, H. et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv10.1101/2020.1112.1121.20248640 (2020).
- 14.Xie, X. et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat. Med.10.1038/s41591-021-01270-4 (2021). [DOI] [PubMed]
- 15.Liu, Y. et al. Neutralizing activity of BNT162b2-elicited serum—preliminary report. N. Engl. J. Med.10.1056/NEJMc2102017 (2021). [DOI] [PMC free article] [PubMed]
- 16.Trombetta CM, Perini D, Mather S, Temperton N, Montomoli E. Overview of serological techniques for influenza vaccine evaluation: past, present and future. Vaccines. 2014;2:707–734. doi: 10.3390/vaccines2040707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie, X. et al. Engineering SARS-CoV-2 using a reverse genetic system. Nat. Protoc.10.1038/s41596-41021-00491-41598 (2021). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The results supporting the findings in this study are available upon request from the corresponding authors.