Graphical abstract
Keywords: Glycosyl hydrolase, Secondary binding site, Molecular dynamics, Crystallographic structure, GH11 xylanase, Paenibacillus sp
Highlights
-
•
PxXyn11B is the first crystal structure of a GH11 from Paenibacillus.
-
•
X-ray structure of PxXyn11B reveals its molecular architecture and a second binding site (SBS).
-
•
MD simulations shed light on structural determinants of uncommon PxXyn11B cooperative behavior.
-
•
Our results reveal molecular mechanism of SBS communication with the active site in GH11 scaffold settings.
Abstract
Glycoside hydrolases (GHs) are essential for plant biomass deconstruction. GH11 family consist of endo-β-1,4-xylanases which hydrolyze xylan, the second most abundant cell wall biopolymer after cellulose, into small bioavailable oligomers. Structural requirements for enzymatic mechanism of xylan hydrolysis is well described for GH11 members. However, over the last years, it has been discovered that some enzymes from GH11 family have a secondary binding sites (SBS), which modulate the enzymes activities, but mechanistic details of the molecular communication between the active site and SBS of the enzymes remain a conundrum. In the present work we structurally characterized GH11 xylanase from Paenibacillus xylanivorans A57 (PxXyn11B), a microorganism of agricultural importance, using protein crystallography and molecular dynamics simulations. The PxXyn11B structure was solved to 2.5 Å resolution and different substrates (xylo-oligosaccharides from X3 to X6), were modelled in its active and SBS sites. Molecular Dynamics (MD) simulations revealed an important role of SBS in the activity and conformational mobility of PxXyn11B, demonstrating that binding of the reaction products to the SBS of the enzyme stabilizes the N-terminal region and, consequently, the active site. Furthermore, MD simulations showed that the longer the ligand, the better is the stabilization within active site, and the positive subsites contribute less to the stabilization of the substrates than the negative ones. These findings provide rationale for the observed enzyme kinetics, shedding light on the conformational modulation of the GH11 enzymes via their SBS mediated by the positive molecular feedback loop which involve the products of the enzymatic reaction.
1. Introduction
Plant biomass is an important resource for production of energy, agro-industrial manufacture, wood-based industry, and biofuels [1]. Plant cell walls from different sources (monocot grasses, woody dicots, herbaceous dicots, and softwoods) have different compositions, but all contain three main fractions: cellulose, hemicellulose and lignin [2], [3]. Despite of a huge abundance of plant biomass, only 2% of this resource is currently used by humans [2]. The most abundant polysaccharide in cell walls is cellulose, making up 40% to 50% of the wall material, and the second largest fraction, representing 25% to 35% of the walls, comprises the hemicelluloses [2]. A large portion, of hemicellulose is constituted by xylan [4], which can be decorated with glucuronic acid and/or arabinose [5].
According to Carbohydrate-Active Enzymes database (CAZy), endo-β-1,4-xylanases (EC.3.2.1.8) are found in GH families 3, 5, 8, 9, 10, 11, 12, 16, 26, 30, 43, 44, 51, 62, 98, and 141 [6], [7], but the vast majority of characterized xylanases are from GH families 10 and 11 [7]. GH11 xylanases are endo-acting retaining enzymes which employ a classical Koshland double-displacement mechanism [8].
Although the canonical catalytic mechanism for GH11 enzymes is well described in structural and biochemical terms [9], [10], discovery of a secondary binding site (SBS) on the surface of the GH11 xylanases raised new and interesting questions regarding the molecular mechanism by which this binding site modulates catalysis promoted by GH11 members [11]. Previous studies attributed some putative functions to the SBS which include (i) targeting the enzyme to the substrate, (ii) guiding the substrate into the active site, (iii) altering enzyme specificity and (iv) acting as an allosteric site [12]. To date, only a handful of GH11 crystal structures identified glycans bound to the SBS, including the endo-xylanases from Bacillus subtilis (PDB id 2QZ3), Aspergillus niger (PDB id 2QZ2) [12], [13] and Bacillus circulans (BcX) (PDB id 1XNB) [14], but molecular mechanism by which binding to the SBS influences the active site of the enzyme remains unknown.
Formerly described as GH11XynB, PxXyn11B, a single domain xylanase from the recently described genus Paenibacillus xylanivorans [15], is active on beechwood xylan (85 IU/mg) and wheat arabinoxylan (131.5 IU/mg), displaying a very peculiar kinetic characteristic [16]. Instead of a typical hyperbolic curve expected for a Michaelis-Menten enzyme kinetics, a sigmoidal non-Michaelian curve was observed with a Hill coefficient (ηH) of 4, suggesting a high degree of cooperativity [16]. This is uncommon for GH11 members. Similar findings have been also described for a xylanase–xylose binding protein chimera, with a Hill coefficient (ηH > 2.0) [17] and a GH11 xylanase from Pseudozyma brasiliensis, PbXynA [18]. As discussed by Ghio et al. (2018) [16], this result could suggest the possible presence of secondary binding sites (SBSs) in PxXyn11B. Despite appealing biochemical evidence, detailed molecular mechanisms by which the interactions of xylo-oligosaccharides (XOS) with the SBS could result in an increase in enzyme activity in a cooperative fashion are not yet understood.
Ludwiczek and coworkers showed for the Bacillus circulans GH11 (BcX) that the enzyme binds XOS in the active site and SBS with similar dissociation constants (KD), but marked differences in the koff rates [14]. The authors also found that mutations in the SBS increased the affinity for XOS in the active site, suggesting some degree of cross-talk between the sites and indicating some cooperativity [14]. These findings indicate that the increase in the enzyme activity due to the interactions between XOS and SBS could be mediated by a decrease in the Michaelis constant KM.
An activation of the enzymes by their products has a considerable biotechnological relevance. Since many GH enzymes are strongly inhibited by their hydrolytic products [19], their use in industrial applications is restricted to a relatively small number of enzymes known or engineered to be more tolerant to product inhibition and/or to the conditions when the products are (partially) removed from the reaction [17]. On the other hand, for the GH11 enzymes with the SBS, XOS produced during the enzymatic hydrolysis of xylan causes the enzyme activation via interaction with the SBS. This could potentially result in an increased efficiency when used in bioprocesses under industrially relevant conditions. Given the importance of GH11 xylanases for a number of biotechnological applications, this effect can be of a general relevance for industrial application of GH11 enzymes.
In the present study, we determined the first crystal structure of a GH11 xylanase from Paenibacillus genus and conducted MD simulations to provide structural insights for the role of SBS in PxXyn11B enzymatic activity.
2. Material and methods
2.1. Cloning, expression and purification
Gene cloning and protein expression was previously described in Ghio et al. [16]. Briefly, Escherichia coli strain DH5α was used as a recipient for transformations during plasmid construction and plasmid propagation and storage, while E. coli Rosetta (DE3) (Novagen, Darmstadt, Germany) was used for protein expression.
A pre-inoculum of E. coli BL21 containing the pJExpress (NdeI/XbaI) PxXyn11B plasmid was started in LB-ampicilin (100 μg/mL) and incubated at 37 °C and 200 rpm in a shaker, overnight. Then, 5 mL of the pre-culture was used to inoculate 500 mL of the expression media. After reaching an OD600 of 0.6 at 37 °C and 200 rpm, the protein expression was induced with 0.5 mM isopropyl β-D-thiogalactopyranoside (IPTG) at 37 °C under continuous agitation at 200 rpm for 16 h. The culture was centrifuged at 4000×g at 4 °C for 20 min and the cell pellet was resuspended in a lysis buffer (20 mM Tris-HCl, 20 mM KCl, 1 mM EDTA, 0,1% (v/v) Triton-X and 10 mM Imidazole, pH 8.0). The resuspended cells were disrupted by two cycles of freezing in liquid nitrogen followed by thawing under room temperature. Next, lysozyme was added, on ice, to a final concentration of 1 mg/mL. After 30 min, six 10-second cycles of sonication on ice bath using a 550 Sonic Dismembrator Sonifier (Fisher Scientific, Hampton, USA) at 28% of the maximal amplitude were performed. The lysate was clarified by centrifugation at 10,000×g for 30 min at 4 °C to remove cell debris and the supernatant was used for PxXyn11B purification.
The supernatant was applied to a 5 mL Ni-NTA Superflow column (Qiagen, Hilden, Germany), previously equilibrated with the lysis buffer. Three volumes of wash buffer (20 mM Tris-HCl, 20 mM KCl, 1 mM EDTA, 0,1% (v/v) Triton-X, 20 mM Imidazole, pH 7.0) were used to wash the column. The protein was eluted with two elution buffers, both with 20 mM Tris-HCl, pH 8.0, 20 mM KCl and 1 mM EDTA, but with different imidazole concentration: the first buffer had 250 mM while the second had 500 mM of imidazole. The resulting protein fractions were pooled and concentrated in a 50 kDa molecular cut-off concentrator to reduce imidazole concentration to 20 mM. Purification was evaluated by 15% sodium dodecyl sulfate gel electrophoresis (SDS-PAGE) analysis under denaturing conditions [20] and subsequently stained with Coomassie Brilliant Blue R-250. The purified enzyme was further concentrated using a 10 kDa Vivaspin Concentrator (GE-Healthcare, Chicago, IL, USA) at 1500×g and its concentration was determined by measuring absorption at 280 nm using Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, EUA) and applying the calculated extinction coefficient (Ɛ = 75860 M−1 cm−1).
2.2. Enzymatic assays
In order to confirm the integrity and consequently activity of PxXyn11B, the enzymatic activity was carried out using 1% xylan from beechwood (Sigma, San Luis, MO, USA) as a substrate, in a final reaction volume of 0.2 mL at 400 rpm, for 15 min in a Thermomixer (Eppendorf, Hamburg, Germany). Reducing sugars released from xylan hydrolysis were measured using the 3,5-dinitrosalicylic acid assay. The amount (µmol) of xylose equivalents released were determined by absorbance at 540 nm with a xylose curve as standard reference. The enzymatic assay was conducted in triplicates accompanied by a control measured in the absence of PxXyn11B.
2.3. Crystallization, data collection, and processing
For crystallization assays the purified PxXyn11B was concentrated to 10 mg/mL. The crystallization trials were set using a crystallization robot Honeybee (DIGILAB, Massachusetts, USA) and the crystals were grown by sitting-drop vapor-diffusion technique with different protein-to-mother liquor ratios, using Crystal Screen kit (Hampton Research, California. USA). The crystals grew in crystallization conditions consisting of 1.6 M Sodium citrate tribasic dihydrate pH 6.5 at 20 °C, with 2 µL of protein mixed with 1 µL of buffer in the crystallization drop.
The resulting PxXyn11B crystals were flash-frozen in liquid nitrogen after bathing in ethylene glycol (30% v/v). X-ray diffraction data were collected at the W01B-MX2 beamline of the Brazilian National Synchrotron Light Laboratory (Campinas, Brazil) [21], using a Pilatus2M detector (Dectris, Philadelphia, USA), with an exposure time of 2 s per image and an oscillation angle of 0.2°. The diffraction image data were indexed, integrated using XDS package [22], and scaled with AIMLESS [23].
2.4. X-ray structure solution
Molecular replacement was performed with BALBES [24], using GH11 xylanase from Bacillus subtillis (PDB id 2Z79, [13]), which has 85.2% of amino acid sequence similarity. The resulting structure was then submitted to Phenix AutoBuild [25] and the refinement cycles were performed using Coot [26] and phenix.refine [27] programs. The model quality was evaluated using Molprobity [28], and the atomic coordinates have been deposited at the Protein Data Bank (PDB) under the accession code 7KV0.
2.5. Structural analysis
2.5.1. Structural comparisons
For structural comparison purposes, PxXyn11B crystal structure was superimposed with GH11 xylanases from T. reesei complexed with xylohexaose (X6) in the active site (PDB id 4HK8, [29]), and a crystallographic GH11 structure of Bacillus subtilis mutant E172A with SBS in complex with xylotetraose (XBS1, PDB id 2QZ3, [13]). The UCSF Chimera visualization software [30] was used for superimpositions and comparisons.
2.6. Molecular dynamics simulations
The molecular dynamics simulations of PxXyn11B were carried out using the AMBER18 [31], [32], [33]. The crystal structure of PxXyn11B without water molecules was used as starting model for MD simulations. Complexes of PxXyn11B bound to X4 in active site with and without X3 in SBS were created using the same starting structure. The protonation states at pH 6.5 and in the presence of 150 mM NaCl system were estimated using H++ server [34]. The apo, xylotetraose-bound and xylotetraose-bound with xylotriose in the SBS structures were solvated in octahedral boxes with a solvent padding of 12 Å. Chloride ions were added to reach neutrality in the simulation box. The simulations were set up using the AMBERFF 14SB force field [35], TIP3P water model [36] and GLYCAM06 carbohydrate force field [37] for the xylosides. Each system was minimized in 20,000 steps and then heated to 300 K in 50 ps in the NVT ensemble. Afterwards, the system density was equilibrated in 50 ps in the NPT ensemble. Harmonic constraints were kept for minimization, heating and density equilibration with harmonic weights of 10.0, 10.0 and 5.0 kcal/(mol. Å2), respectively. Finally, the system was equilibrated for 2 ns without restraints. Productive simulations were run for 500 ns in the NPT ensemble. A timestep of 2 fs was used and the covalent bonds involving hydrogen atoms were constrained using the SHAKE algorithm. The MD trajectories were analyzed using VMD [38], CPPTRAJ [39] and scripts/small codes developed in-house.
3. Results and discussion
3.1. Cloning, expression, and purification
PxXyn11B was expressed and purified with Ni+2 affinity chromatography. Next, a gel filtration chromatography was performed to ensure the sample purity. The theoretical mass of PxXyn11B predicted based on its amino acid sequence is 20.8 kDa and the purified protein migrated as a single band on SDS PAGE, with the expected molecular weight and a purity required for structural and biochemical studies (Supplementary Material).
3.2. PxXyn11B crystallographic structure determination and analysis
3.2.1. Synchrotron data collection, refinement, model building and comparison with homologous structures
The crystallographic diffraction data were collected at the Brazilian National Synchrotron Laboratory (LNLS) [21], using an angular step of 0.2 degrees per image, resulting in 1800 images for 360° collection. Processing the data with XDS revealed that PxXyn11B crystal belongs to C2221 space group.
Molecular Replacement was performed with BALBES using B. subtillis GH11 xylanase structure (PDB id 2Z79) as a template, resulting in a single solution with MR score of 0.66 and a Q-factor of 0.845. The structure was refined up to 2.5 Å, leading to a decrease in the Rfree and Rwork. Both collection and refinement parameters are given in Table 1.
Table 1.
PxXyn11B Crystallographic data collection and model refinement statistics.
| Dataset | PxXyn11B |
| Data collection | LNLS MX2 |
| Wavelength (Å) | 1.458 |
| Space group | C 2 2 21 |
| Cell Parameters | |
| a, b, c (Å) | 62.426 62.995 183.200 |
| Resolution range (Å) | 45.80–2.50 (2.59–2.50) |
| Rpim | 0.0593 (0.5444) |
| CC1/2 | 0.996 (0.564) |
| Total reflections | 164,962 (15,900) |
| Unique reflections | 12,880 (1266) |
| Multiplicity | 12.8 (12.6) |
| Completeness (%) | 99.86 (99.53) |
| Mosaicity | 0.3 |
| <I/σI> | 10.30 (1.39) |
| Refinement | |
| No. mol. asymmetric cell unit | 2 |
| Residues per molecule | 186 |
| Rwork | 0.2149 (0.3094) |
| Rfree | 0.2480 (0.3254) |
| Overall CC | 0.999 (0.849) |
| Ligands | EDO |
| B-factors (Å2) | |
| Mean | 45.65 |
| Macromolecules | 45.67 |
| Ligands | 47.95 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.56 |
| Ramachandran | |
| Ramachandran favored (%) | 96.47 |
| Ramachandran allowed (%) | 3.53 |
| Ramachandran outliers (%) | 0.00 |
| Rotamer outliers (%) | 1.01 |
| MolProbity Overall Score | 1.52 |
*Values in parentheses are for highest-resolution shell. Structure is based on single crystal.
Rpim = ∑h [1/(nh-1)]1/2 ∑i|<Ih> - Ih,i|/∑h ∑I Ih,i.
Rwork = Σhkl(||Fobs|-|Fcalc||)/Σhkl|Fobs|, where |Fobs| and |Fcalc| are the observed and calculated structure factor amplitudes, respectively.
PxXyn11B has two molecules in the asymmetric unit, but since their comparison indicated no significant differences and the superimposition of molecules A and B showed a RMSD of 0.212 Å, further analyses and representations were performed with molecule A. Each monomer (Fig. 1a) has a beta jelly roll fold, typical for GH-C clan [40], formed by 11 β strands and one alpha helix, and a cleft of approximately 2097 Å3, where the catalytic site is located, according to PDBsum [41].
Fig. 1.
PxXyn11B crystal structure. (A) 3D structure of PxXyn11B colored from blue (N-terminal region) to red (C-terminal region). α helix and β strands, as defined by PDBSum are indicated, and also the catalytic residues. (B) Side-view of (A), with N-terminal, C-terminal, Palm, Fingers, Thumb and Cord parts of the protein indicated. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
PxXyn11B has the canonical ‘right‐hand’ β‐sandwich fold of family GH11 xylanases, in which the β‐strands thread back and forth between the two packed β‐sheets to form the finger and palm domains [42] (Fig. 1b). An extended loop between the β8 and β9 strands assembles itself in so‐called thumb domain, and a loop between the β6 and β7 strands forms a cord between the two β‐sheet domains.
In order to compare PxXyn11B structure with other enzymes from GH11 family, the protein sequence was submitted to BLASTp [43] and eight homologous sequences were selected (Table 2). The sequences of the selected enzymes were aligned to PxXyn11B using Clustal Omega [44] and a structure-based sequence alignment was performed with ESPript [45] (Fig. 2).
Table 2.
Enzymes used in structural comparison with PxXyn11B.
| Microorganism | Enzyme | Identity (%) | Query (%) | RMSD (Å) | PDB id | Ref |
|---|---|---|---|---|---|---|
| Panibacillus xylanivorans A59 | PxXyn11B | – | – | – | 7KV0 | This study |
| B. subtillis | XBS1 | 85.33 | 96 | 0.343 | 2QZ3 | Vandermarliere 2008 |
| Thermopolyspora flexuosa | NFX | 66.47 | 89 | 0.643 | 1M4W | Hakulinen 2003 |
| Thermobifida fusca | TfGH11 | 65.22 | 96 | 0.667 | 3ZSE | Van Bueren 2012 |
| Streptomyces lividans | XlnB2 | 63.69 | 94 | 0.468 | 5EJ3 | Gagné 2016 |
| Escherichia coli | EnXyn11A | 60.44 | 94 | 0.594 | 2VGD | Vardakou 2008 |
| Chaetomium thermophilum | CTX | 59.12 | 94 | 0.554 | 1H1A | Hakulinen 2003 |
| Trichoderma reesei | XynII | 53.04 | 94 | 0.542 | 4HK8 | Wan 2015 |
| Aspergillus niger | AnXyn11A | 43.45 | 86 | 0.833 | 6QE8 | Schroder 2019 |
Fig. 2.
Structure-based sequence alignment. (A) The sequences of AnXyn11A, CTX, XynII, XBS1, EnXyn11A, TfGH11, NFX and XlnB2 were aligned with PxXyn11B sequence, and the NTR (N-terminal region), catalytic residues and SBS residues are highlighted. (B) Structural superposition of the aforementioned enzymes. Navy blue: AnXyn11A; Purple: CTX; Dark green: XynII; Dark cyan: PxXyn11B; Dark red: XBS1; Goldenrod: EnXyn11A; Orange: TfGH11; Red: NFX; Cyan: XlnB2. (C) Side view of (B). (D) Back view of (B). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As shown in Fig. 2, there are some differences among the enzymes, specially concerning their N-terminal region (NTR). This region is longer in some enzymes, such as XlnB2, EnXyn11A and AnXyn11A. Other GH11 xylanases also have longer NTR, although not as long as in the previous ones. It has been widely accepted that N-terminal region (NTR) of GH11 is of importance for xylanase thermostability [46]. For example, GH11 xylanases from Nonomuraea flexuosa and Neocallimastix patriciarum have high thermostability and long NTRs, with the last one having the longest NTR reported for GH11 [46], [47], [48].
Comparing the SBS within GH11 xylanase sequences, it is possible to observe that several residues play key roles in this binding site (Fig. 2a). The amino acid sequences alignment analysis reveals that N54 is only present in PxXyn11B and XBS1, while the same position is identified as S82 in AnXyn11A. Other important residues, N139 and W183, are only present in PxXyn11B and XBS1 as well. This indicates that N54, N139 and W183 residues may be responsible for the formation of SBS. The comparison of the enzyme structures, regarding percentage of identity, query coverage and root mean square deviations (RMSD) is shown in Table 2.
3.2.2. Catalytic site analysis
In order to analyze the active site of PxXyn11B, the enzyme was superimposed to Trichoderma reesei GH11 (PDB id 4HK8, [29]) complexed with xylohexaose (X6), bound to its active site (Fig. 3).
Fig. 3.
Structural analysis of PxXyn11B active site. (A) PxXyn11B molecular surface showing a X6 molecule modelled based on the structural superimposition with Trichoderma reesei Xyn II complexed with X6 (PDB id 4HK8) (B) Superimposition of PxXyn11B (dark cyan) and T. reesei GH11 (PDB id 4HK8, dark green). (C) Subsites in PxXyn11B, with respective residues. Red triangles indicate the cleavage site. Xylanopyroside residues are depicted as yellow stars, in accordance with the Symbolic nomenclature of Glycans [53]. (D) Zoom into the active sites, with the residues displayed with the respective colors. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The enzymatic hydrolysis of the glycosidic bond requires a proton donor and a nucleophile/base (Glu 170 and Glu 77 in PxXyn11B, respectively). The hydrolysis by glycoside hydrolases can occur through two different mechanisms: retention or inversion of anomeric configuration of the substrate [8]. In both the retaining and the inverting mechanisms, the position of the proton donor is identical, within hydrogen-bonding distance of the glycosidic oxygen, but in retaining enzymes, the nucleophilic catalytic base is in a close vicinity of the sugar anomeric carbon [10], [49]. GH11 xylanases, such as PxXyn11B, follow the retaining type hydrolytic mechanism [50].
The active site of PxXyn11B comprises 6 subsites, similar to that of GH11 from Trichoderma reesei (PDB id 4HK8). According to Madan and Lee [51], three amino acid residues are not conserved within the active site: Y96, Y171 and S16. In PxXyn11B, they were identified as Y87, Y164 and Q6, i.e., only the last residue is not conserved.
The analysis of the active site also indicates the presence of two important residues previously described in literature: Y87 and Y172 at the +2 and +3 subsites. The crystal structure indicates that these Tyr residues contribute to the enzyme reaction by stabilizing the positive subsites. In line with this finding, alanine substitutions of Y180 at the +2 subsite and Y96 at the +3 subsite was shown to reduce the enzymatic activities by as much as 95% in GH11 from Thermomyces lanuginosus, TlXynA [52].
3.2.3. SBS analysis
The secondary (or substrate) binding site was previously reported for bacterial GH11 structures for only few enzymes, including GH11 xylanase from Bacillus subtilis (PDB id: 2QZ3) [12], [13], Aspergillus niger (PDB id 2QZ2) [12], [13] and Bacillus circulans (BcX) (PDB id 1XNB) [14]. For comparison purposes, PxXyn11B was superimposed to the Bacillus subtilis xylanase structure, and the residues within SBS were analyzed.
As can be observed in the alignment shown in the Fig. 2 and also in Fig. 4, PxXyn11B has a strictly conserved SBS, also present in Bacillus subtilis xylanase structure (PDB id: 2QZ3). In addition to the sequence and structure similarity between PxXyn11B and the Bacillus subtillis enzymes, their previous kinetic characterization revealed a sigmoidal curve, typical for cooperative enzymes and similar to the one previously observed for PbXynA [18]. Furthermore, Hill coefficients of 4 and 6 were determined for PxXyn11B [16] and PbXynA [18], respectively. Therefore, we set out to evaluate the putative roles of the interaction of XOS in the SBS and their role in for the enzyme activity using molecular dynamics (MD) simulations.
Fig. 4.
Superimposition of PxXyn11B and B. subtilis xylanase A (PDB id: 2QZ3) structures. (A) Front view, showing catalytic site and SBS in the back. (B) Side view of (A), with residues in active site indicated. (C) Zoom in the SBS region, with residues involved highlighted and distances between PxXyn11B residues and xylopyranoside residues indicated.
3.3. Molecular dynamics simulations
The experimentally determined crystal structure of PxXyn11B was used for MD simulations. For a more thorough investigation, three systems were prepared and simulated: an apo enzyme, as determined by the crystallographic structure; the enzyme bound to xylotetraose (X4) in its active site, and the enzyme bound to X4 in the active site and a xylotriose (X3) molecule in the SBS. In all cases, a 0.5 μs productive simulation was analyzed using CPPTRAJ [39], AmberEnergy++ [54] and also AMBERTOOLS.
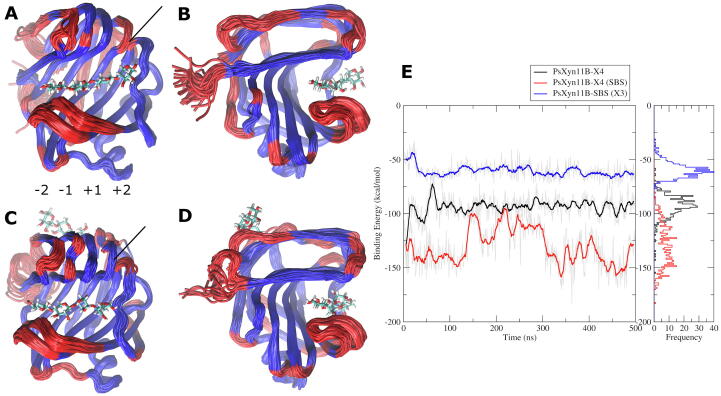
The analyses of the simulations revealed that PxXyn11B is a remarkably stable enzyme, with only small fluctuations in the structure being observed during the simulations. Using the package MDLovoFit [55], we mapped the most stable core of the enzyme (the 30% less mobile fraction of the enzyme structure), that includes most of the β-strands. This rigid portion of the protein is shown in blue cartoon in the Fig. 5, while the mobile fraction of the enzyme is given in red in the figure.
Fig. 5.
(A and B) Two views of PxXyn11B bound to xylotetraose (X4). The figures show the superposition of 500 frames taken at 1-ns intervals during the equilibrium MD simulation. The sugar subsites (−2 to +2) are indicated below the panel (A). (C and D) Two views of PxXyn11B bound to X4 in the active site and to a xylotriose (X3) in the SBS. Both X3 and X4 are shown in sticks. The lines indicate the change in mobility close to strand β5. (E) Interaction (potential) energy computed between the enzyme and the substrate (X4) for the binary complex (black line) and for the ternary complex (red line), i.e., the enzyme bound to X3 in the SBS. For the sake of comparison, the interaction energies between the enzyme and the X3 in the SBS are also shown in blue lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The mapping of the atomic fluctuation across the enzyme structure also indicates a slightly reduced mobility for the enzyme bound to X3 in the SBS as compared to the enzyme bound to X4 in the active site but with an empty SBS (Supplementary Material). The analysis shows that the C-terminal portion of the enzyme is dramatically stabilized by the interactions with X3 bound to the SBS. This stabilization propagates to the C-terminal β-strands, in particular β11, the last β-strand in the structure and that encompasses the active site and contains the catalytical residue Glu170 and Tyr172, known to be part of the positive subsites +1 and +2 (Fig. 5). Furthermore, the palm and the thumb region show higher stabilization when X3 is bound to SBS. It is important to highlight that the initial enzyme coordinates were exactly the same for the simulations of the apo enzyme and X4- and X3-bound enzyme systems. This finding suggests that the stabilization of the terminal portion of the enzyme through interactions with XOS can directly reduce mobility of the substrate via stabilization of the residues in the active site. This result is also supported by the slight decrease in the interaction energy between the enzyme and the substrate (X4) when the simulation of the complex PxXyn11B-X4 is compared to the ternary complex PxXyn11B-X4-X3SBS (Fig. 5). We found that the average interaction (potential) energy between the enzyme and the xylotriose bound in SBS was approximately −57 kcal/mol, while the substrate (xylotetraose) binds to the enzyme with an interaction (potential) energy of −95 and −125 kcal/mol for the binary and ternary complex, respectively. This data suggests a better fit of the substrate in the active site when the site is stabilized by the interaction of XOS in the SBS in the protein surface. In line with these findings, binding energy estimate based on the MM-GBSA as implemented in MM-PBSA.py [56] showed an interaction energy of −49.7 ± 0.4 kcal/mol for the interaction of X4 with GH11-X3, while for the binary complex (X4-GH11), an interaction energy of −35.4 ± 0.2 kcal/mol was computed from MD data, reinforcing a notion of better interaction of the enzyme with the substrate in the presence of XOS in the SBS.
Another significant point is that the thumb-loop, known for its importance in GH11 catalysis, since its movement has a key role in glycosylation/deglycosylation and product release [57], is also stabilized by the X3 in SBS (Supplementary Material). Is it also worth of note that the reported findings, based on classical MD simulations, are consistent with the NMR measurements of Bacillus circulans xylanase performed by Ludwiczek and coworkers [14], who reported an increase in the affinity for the substrate after binding of XOS to the SBS. Moreover, Paes and coworkers [57] established higher binding-energy values for the positive subsites, with −17.7 and −13.2 kcal/mol for subsites +1 and +2, respectively, than for negative subsites, with −14.0 and −23.8 kcal/mol for subsites −1 and −2, respectively. This is consistent with our simulation, where the xylopyranoside residues in positive subsites demonstrated higher mobility than the ones located in negative subsites. Furthermore, the total interaction energy of the binary complex, with Thermobacillus xylanilyticus GH11 and the ligand, was calculated as −86.3 kcal/mol, which is consistent with the energy computed for PxXyn11B binary complex, but higher than the energy found for PxXyn11B ternary complex.
In mechanistic terms, how does the binding of a X3 molecule to the SBS propagates as a stabilizing effect of the enzyme active site? One possible explanation for this stabilizing effect would be a change in the sugar conformation in the subsite −1, which is known to be relevant to enzyme activity. We measured the Cremer-Pople puckering angles for the −1 subsite in both simulations, i.e., with the X3 bound in the SBS and in the absence of a XOS in the SBS and in both scenarios we found the −1 xylose to be in the low energy 4C1 boat conformation, similar to the conformations observed in Michaelis complexes for other β-xylanases [58]. Thus, the distortion of the sugar conformation at the −1 subsite does not seem to be the determinant driving force in this context.
A deeper examination of the MD trajectories revealed a major repositioning in the side chains of Tyr172 and Asn62. These residues, that are located in the β-strands β11 and β5, which are the same β-strands that are directly stabilized by the interaction of the XOS in the SBS (see blue triangles in Fig. 2). In addition, these residues play an important role coordinating the interaction between the substrate and the enzyme positive subsites +1 to +3 (Fig. 3). In the absence of a ligand bound in the SBS, these interactions become looser, resulting in a decrease in the interaction energy (enzyme-substrate) and, most probably, less efficient catalysis (Fig. 6). Interestingly, these findings are in line with the previous report from Ludwiczek and coworkers [14], who revealed that the increase in the enzyme activity due to interaction in the SBS were due to a decrease in the Michaelis KM for soluble and insoluble substrates. The analysis of the simulation data then suggests that the mechanism involved in the activity enhancement includes direct stabilization of β-strands 11 and 5, with consequent stabilization of the residues in the positive subsites of the active site. The stabilization of these residues results in tighter interaction with the substrate, increasing the affinity (or the interaction energy), decreasing the KM and leading to augmented efficiency. In line with this hypothesis, site-directed mutagenesis of the amino acid residues from the SBSs of Bacillus subtilis and also Aspergillus niger xylanases enzymes reveal the KM values of the enzymes with impaired SBSs were strongly increased, indicating weaker substrate binding affinity [12].
Fig. 6.

Comparison between the binary complex PxXyn11B-X4 (blue) and the ternary complex PxXyn11B-X4/X3 (orange). Representative structures of the most populated cluster in the MD trajectory are compared. The hydrogen atoms were omitted for clarity. X4 and X3 are shown in sticks and the subsites are numbered in the bottom of the figure. The repositioning of N62, Q173 and Y172 in the binary complex create an additional space that allows xylopyranose residues in the positive site to fluctuate, loosening the interaction with the enzyme. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is also interesting to note that X2 and X3 are the major products released by GH11 xylanases and, frequently, competitive inhibitors of the enzymes. Although PxXynB inhibition by its end products, X2 and X3, was not studied explicitly, when PxXynB was combined with a GH43 β-xylosidase EcXyl43, a marked reduction in X3 production and an increase in the release of xylose both from xylan and wheat straw was observed [59]. This might indicate that the feedback regulation of GH11 xylanases by their end products are even more complex and involve participation of β-xylosidases.
Finally, we also simulated different substrates in the presence of a X3 molecule bound in SBS. Worth of note, in the simulation of PxXyn11B bound to X5 in the active site and X3 in the SBS, we observed that X3 escaped from the SBS and diffused to the solvent. After about 100 ns, the X3 molecule reached an additional site close to the thumb region and stayed bound at this new site until the end of the simulation (Supplementary Figs. S3 and S4). This new interaction also contributed to the stabilization of the substrate in the active site, as inferred from the interaction energies (Supplementary Fig. S3). These results suggest that more than one site may exist in the enzyme surface contributing to the enzyme stabilization and also to the substrate stabilization. In line with this finding, previous crystal structure of A. niger GH11 xylanase (PDB id: 2QZ2) revealed X3 molecule bound to the enzyme surface at the SBS which has a different location, quite distinct from the SBS found in Bacillus subtilis enzyme (PDB id: 2QZ3) [12], [13]. Site-directed mutagenesis of the SBSs of these two enzymes reveal that they strongly affect activity and binding affinity of both Bacillus subtilis and also Aspergillus niger xylanases [12]. All this collective evidence indicates that more than one surface XOS binding site might exist and contribute to the enzyme stabilization in GH11 xylanase structures.
Taken together, our structural studies and molecular dynamic simulations reveal an elegant biochemical feedback loop by which the main products of xylan hydrolytic reaction, X2 and X3, catalyzed by GH11 xylanase bind to its SBS and cause conformational changes which propagate to and stabilize the enzyme active site. This product-induced allosteric regulation could be a common for GH11 xylanases with the SBS evolved to regulate substrate affinity in single domain enzymes, somehow parallel to what is observed in CBM containing GHs [14].
4. Conclusions
X-ray structure of PxXyn11B and its amino acid sequence alignment reveal conservation of the enzyme SBS. The structural comparisons and MD simulations provide an explanation for the molecular communication between SBS and active site, shedding light on the molecular basis of the cooperative behavior of PxXyn11B. An elegant product-mediated positive feedback loop intermediates allosteric interaction between the active site and the SBS of the enzyme, involving structural stabilization of the N-terminal region of the enzyme. This molecular mechanism could be employed for molecular engineering of GH11 xylanases activation by their hydrolytic products.
CRediT authorship contribution statement
Lorenzo Briganti: Investigation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing. Caio Capetti: Investigation, Formal analysis, Data curation, Visualization, Writing - original draft. Vanessa O.A. Pellegrini: Investigation, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Project administration. Silvina Ghio: Investigation, Formal analysis, Data curation. Eleonora Campos: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Supervision. Alessandro S. Nascimento: Conceptualization, Methodology, Formal analysis, Data curation, Writing - original draft, Writing - review & editing, Supervision. Igor Polikarpov: Conceptualization, Methodology, Formal analysis, Writing - original draft, Writing - review & editing, Supervision, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This research used resources of the Brazilian Synchrotron Light Laboratory (LNLS), an open national facility operated by the Brazilian Centre for Research in Energy and Materials (CNPEM) for the Brazilian Ministry for Science, Technology, Innovations, and Communications (MCTIC). The beamline staffs are acknowledged for their assistance during the experiments. Research developed with the help from HPC resources provided by the Superintendence of Information Technology from University of Sao Paulo.
Funding
This research was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) via grants 2015/13684-0, 2015/26722-8 and 2020/03983-9 and by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) via grants 423693/2016-6, 303988/2016-9 and 303165/2018-9. It was also funded by Brazil-Argentina bi-national Project PICT CABBIO 2016-4695 (Arg).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.03.002.
Contributor Information
Alessandro S. Nascimento, Email: asnascimento@ifsc.usp.br.
Igor Polikarpov, Email: ipolikarpov@ifsc.usp.br.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Scarlat N., Dallemand J.F., Monforti-Ferrario F., Nita V. The role of biomass and bioenergy in a future bioeconomy: policies and facts. Environ Dev. 2015;15:3–34. doi: 10.1016/j.envdev.2015.03.006. [DOI] [Google Scholar]
- 2.Pauly M., Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 3.Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Smith P.J., Wang H.T., York W.S., Peña M.J., Urbanowicz B.R. Designer biomass for next-generation biorefineries: Leveraging recent insights into xylan structure and biosynthesis. Biotechnol Biofuels. 2017;10:286. doi: 10.1186/s13068-017-0973-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silveira R.L., Stoyanov S.R., Gusarov S., Skaf M.S., Kovalenko A. Plant biomass recalcitrance: Effect of hemicellulose composition on nanoscale forces that control cell wall strength. J Am Chem Soc. 2013;135:19048–19051. doi: 10.1021/ja405634k. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen S.T.C., Freund H.L., Kasanjian J., Berlemont R. Function, distribution, and annotation of characterized cellulases, xylanases, and chitinases from CAZy. Appl Microbiol Biotechnol. 2018;102:1629–1637. doi: 10.1007/s00253-018-8778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombard V., Golaconda Ramulu H., Drula E., Coutinho P.M., Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koshland D.E. Stereochemistry and the mechanism of enzymatic reactions. Biol Rev. 1953;28:416–436. doi: 10.1111/j.1469-185X.1953.tb01386.x. [DOI] [Google Scholar]
- 9.MacLeod A.M., Lindhorst T., Withers S.G., Warren R.A.J. The Acid/Base catalyst in the exoglucanase/xylanase from Cellulomonas fimi is glutamic acid 127: evidence from detailed kinetic studies of mutants. Biochem. 1994;33:6371–6376. doi: 10.1021/bi00186a042. [DOI] [PubMed] [Google Scholar]
- 10.Davies G., Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure. 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 11.Cockburn D., Wilkens C., Ruzanski C., Andersen S., Nielsen J.W., Smith A.M. Analysis of surface binding sites (SBSs) in carbohydrate active enzymes with focus on glycoside hydrolase families 13 and 77-a mini-review. Sect Cell Mol Biol. 2014;69:705–712. doi: 10.2478/s11756-014-0373-9. [DOI] [Google Scholar]
- 12.Cuyvers S., Dornez E., Rezaei M.N., Pollet A., Delcour J.A., Courtin C.M. Secondary substrate binding strongly affects activity and binding affinity of Bacillus subtilis and Aspergillus niger GH11 xylanases. FEBS J. 2011;278:1098–1111. doi: 10.1111/j.1742-4658.2011.08023.x. [DOI] [PubMed] [Google Scholar]
- 13.Vandermarliere E., Bourgois T.M., Rombouts S., Van Campenhout S., Volckaert G., Strelkov S.V. Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-β-xylanases. Biochem J. 2008;410:71–79. doi: 10.1042/BJ20071128. [DOI] [PubMed] [Google Scholar]
- 14.Ludwiczek M.L., Heller M., Kantner T., McIntosh L.P. A secondary xylan-binding site enhances the catalytic activity of a single-domain family 11 glycoside hydrolase. J Mol Biol. 2007;373:337–354. doi: 10.1016/j.jmb.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 15.Ghio S., Sauka D.H., Ferrari A.E., Piccini F.E., Ontañon O.M., Campos E. Paenibacillus xylanivorans sp. nov., a xylan-degrading bacterium isolated from decaying forest soil. Int J Syst Evol Microbiol. 2019;69:3818–3823. doi: 10.1099/ijsem.0.003686. [DOI] [PubMed] [Google Scholar]
- 16.Ghio S., Ontañon O., Piccinni F.E., Marrero Díaz de Villegas R., Talia P., Grasso D.H. Paenibacillus sp. A59 GH10 and GH11 extracellular endoxylanases: application in biomass bioconversion. Bioenergy Res. 2018;11:174–190. doi: 10.1007/s12155-017-9887-7. [DOI] [Google Scholar]
- 17.Ribeiro L.F., Tullman J., Nicholes N., Silva S.R.B., Vieira D.S., Ostermeier M. A xylose-stimulated xylanase-xylose binding protein chimera created by random nonhomologous recombination. Biotechnol Biofuels. 2016;9:119. doi: 10.1186/s13068-016-0529-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borges T.A., Souza A.T.d., Squina F.M., Riaño-Pachón D.M., Santos R.A.C.D., Machado E. Biochemical characterization of an endoxylanase from Pseudozyma brasiliensis sp. nov. strain GHG001 isolated from the intestinal tract of Chrysomelidae larvae associated to sugarcane roots. Process Biochem. 2014;49:77–83. doi: 10.1016/j.procbio.2013.10.004. [DOI] [Google Scholar]
- 19.Polizeli M.L.T.M., Rizzatti A.C.S., Monti R., Terenzi H.F., Jorge J.A., Amorim D.S. Mini-review Xylanases from fungi: properties and industrial applications. Appl Microbiol Biotechnol. 2005;67:577–591. doi: 10.1007/s00253-005-1904-7. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Guimarães B.G., Sanfelici L., Neuenschwander R.T., Rodrigues F., Grizolli W.C., Raulik M.A. The MX2 macromolecular crystallography beamline: a wiggler X-ray source at the LNLS. J Synchrotron Radiat. 2009;16:69–75. doi: 10.1107/S0909049508034870. [DOI] [PubMed] [Google Scholar]
- 22.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr 2010;66:125–32. https://doi.org/10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed]
- 23.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr Sect D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr. Sect. D Biol. Crystallogr., vol. 64, International Union of Crystallography; 2007, p. 125–32. https://doi.org/10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed]
- 25.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. Sect. D Biol. Crystallogr., vol. 64, International Union of Crystallography; 2007, p. 61–9. https://doi.org/10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed]
- 26.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan Q., Zhang Q., Hamilton-Brehm S., Weiss K., Mustyakimov M., Coates L. X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism. Acta Crystallogr Sect D Biol Crystallogr. 2014;70:11–23. doi: 10.1107/S1399004713023626. [DOI] [PubMed] [Google Scholar]
- 30.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C. UCSF chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 31.Case D.A., Cheatham T.E., Darden T., Gohlke H., Luo R., Merz K.M. The amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomon-Ferrer R., Götz A.W., Poole D., Le Grand S., Walker R.C. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 33.Le Grand S., Götz A.W., Walker R.C. SPFP: Speed without compromise - a mixed precision model for GPU accelerated molecular dynamics simulations. Comput Phys Commun. 2013;184:374–380. doi: 10.1016/j.cpc.2012.09.022. [DOI] [Google Scholar]
- 34.Gordon J.C., Myers J.B., Folta T., Shoja V., Heath L.S., Onufriev A. H++: A server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 37.Kirschner K.N., Youngye A.B., Tschampel S.M., González-Outeiriño J., Daniels C.R., Foley B.L. GLYCAM06: A generalizable biomolecular force field. Carbohydrates J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humphrey W., Dalke A., Schulten K. Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Roe D.R., Cheatham T.E. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9:3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 40.CAZypedia contributors. Glycoside Hydrolase Family 11 n.d. https://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_11 (accessed October 5, 2020).
- 41.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murakami M.T., Arni R.K., Vieira D.S., Degrève L., Ruller R., Ward R.J. Correlation of temperature induced conformation change with optimum catalytic activity in the recombinant G/11 xylanase a from Bacillus subtilis strain 168 (1A1) FEBS Lett. 2005;579:6505–6510. doi: 10.1016/j.febslet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 43.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb:2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han N., Miao H., Ding J., Li J., Mu Y., Zhou J. Improving the thermostability of a fungal GH11 xylanase via site-directed mutagenesis guided by sequence and structural analysis. Biotechnol Biofuels. 2017;10 doi: 10.1186/s13068-017-0824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hakulinen N., Turunen O., Jänis J., Leisola M., Rouvinen J. Three-dimensional structures of thermophilic β-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa: comparison of twelve xylanases in relation to their thermal stability. Eur J Biochem. 2003;270:1399–1412. doi: 10.1046/j.1432-1033.2003.03496.x. [DOI] [PubMed] [Google Scholar]
- 48.Cheng Y.S., Chen C.C., Huang C.H., Ko T.P., Luo W., Huang J.W. Structural analysis of a glycoside hydrolase family 11 xylanase from Neocallimastix patriciarum: Insights into the molecular basis of a thermophilic enzyme. J Biol Chem. 2014;289:11020–11028. doi: 10.1074/jbc.M114.550905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCarter J.D., Stephen Withers G. Mechanisms of enzymatic glycoside hydrolysis. Curr Opin Struct Biol. 1994;4:885–892. doi: 10.1016/0959-440X(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 50.Gebler J., Gilkes N.R., Claeyssens M., Wilson D.B., Beguin P., Wakarchuk W.W. Stereoselective hydrolysis catalyzed by related β-1,4-glucanases and β- 1,4-xylanases. J Biol Chem. 1992;267:12559–12561. doi: 10.1016/s0021-9258(18)42313-7. [DOI] [PubMed] [Google Scholar]
- 51.Madan B., Lee S.G. Sequence and structural features of subsite residues in GH10 and GH11 xylanases. Biotechnol Bioprocess Eng. 2018;23:311–318. doi: 10.1007/s12257-018-0105-z. [DOI] [Google Scholar]
- 52.Wu X., Zhang S., Zhang Q., Zhao Y., Chen G., Guo W. The contribution of specific subsites to catalytic activities in active site architecture of a GH11 xylanase. Appl Microbiol Biotechnol. 2020;104:8735–8745. doi: 10.1007/s00253-020-10865-9. [DOI] [PubMed] [Google Scholar]
- 53.Svensson B, Cockburn D, Williams S. Surface Binding Site - CAZypedia n.d. https://www.cazypedia.org/index.php/Surface_Binding_Site (accessed October 28, 2020).
- 54.Nascimento AS. AmberEnergy++. https://github.com/alessandronascimento/amberenergy (accessed November 10, 2020).
- 55.Martínez L., Kleinjung J. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS ONE. 2015;10:e0119264. doi: 10.1371/journal.pone.0119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller B.R., McGee T.D., Swails J.M., Homeyer N., Gohlke H., Roitberg A.E. MMPBSA.py: An efficient program for end-state free energy calculations. J Chem Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 57.Paës G., Cortés J., Siméon T., O'Donohue M.J., Tran V. Thumb-loops up for catalysis: a structure/function investigation of a functional loop movement in a GH11 xylanase. Comput Struct Biotechnol J. 2012;1:e201207001. doi: 10.5936/csbj.201207001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iglesias-Fernández J., Raich L., Ardèvol A., Rovira C. The complete conformational free energy landscape of β-xylose reveals a two-fold catalytic itinerary for β-xylanases. Chem Sci. 2015;6:1167–1177. doi: 10.1039/C4SC02240H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ontañon O.M., Ghio S., Marrero Díaz de Villegas R., Piccinni F.E., Talia P.M., Cerutti M.L. EcXyl43 β-xylosidase: molecular modeling, activity on natural and artificial substrates, and synergism with endoxylanases for lignocellulose deconstruction. Appl Microbiol Biotechnol. 2018;102:6959–6971. doi: 10.1007/s00253-018-9138-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.