Abstract
Recent phylogenomic analyses based on the maternally inherited plastid organelle have enlightened evolutionary relationships between the subfamilies of Orchidaceae and most of the tribes. However, uncertainty remains within several subtribes and genera for which phylogenetic relationships have not ever been tested in a phylogenomic context. To address these knowledge-gaps, we here provide the most extensively sampled analysis of the orchid family to date, based on 78 plastid coding genes representing 264 species, 117 genera, 18 tribes and 28 subtribes. Divergence times are also provided as inferred from strict and relaxed molecular clocks and birth–death tree models. Our taxon sampling includes 51 newly sequenced plastid genomes produced by a genome skimming approach. We focus our sampling efforts on previously unplaced clades within tribes Cymbidieae and Epidendreae. Our results confirmed phylogenetic relationships in Orchidaceae as recovered in previous studies, most of which were recovered with maximum support (209 of the 262 tree branches). We provide for the first time a clear phylogenetic placement for Codonorchideae within subfamily Orchidoideae, and Podochilieae and Collabieae within subfamily Epidendroideae. We also identify relationships that have been persistently problematic across multiple studies, regardless of the different details of sampling and genomic datasets used for phylogenetic reconstructions. Our study provides an expanded, robust temporal phylogenomic framework of the Orchidaceae that paves the way for biogeographical and macroevolutionary studies.
Subject terms: Evolution, Plant sciences
Introduction
Orchidaceae, with ca. 25,000 species and ~ 800 genera1,2 are one of two of the most diverse and widely distributed flowering plant families on Earth and have captivated the attention of scientists for centuries3. The family has a striking morphological and ecological diversity and evolved complicated interactions with fungi, animals and other plants4,5 and a diverse array of sexual systems6–8. Numerous efforts have been made to understand the natural history, evolution and phylogenetic relationships within the family2,7,9–13. To date, there are seven nuclear genome sequences available, i.e., Apostasia shenzhenica14, Dendrobium catenatum15, D. officinale16, Gastrodia elata17, Phalaenopsis equestris18, a Phalaenopsis hybrid cultivar19, P. aphrodite20, Vanilla planifolia21, 221 complete plastid genomes and 2,678 sequence read archives for Orchidaceae in NCBI (accessed 22 August 2020) .
Phylogenomic approaches have been implemented to infer relationships between major orchids clades in deep and recent time2,10,12,13,22,23, but extensive uncertainties remain regarding the phylogenetic placement of several subtribes24. This knowledge-gap stems from a dearth of both taxonomic and genomic sampling efforts that would be required to comprehensively cover all major orchid clades (subtribes/groups of genera). Givnish et al.2 published the first well-supported analysis of Orchidaceae based on plastid phylogenomics. They performed a maximum likelihood (ML) analysis of 75 genes from the plastid genome of 39 orchid species, covering 22 subtribes, 18 tribes and five subfamilies. This robust but taxonomically under-sampled study corroborated relationships of the subfamilies and tribes, observed in previous studies10–13.
Previous orchid studies have failed to resolve relationships in rapidly diversifying clades24–27 because of reduced taxon and data sampling28. This is particularly true for Cymbidieae and Pleurothallidinae, the two most species-rich groups in which generic relationships are largely the product of rapid diversification29 that is difficult to resolve using only a few loci26,30. Cymbidieae comprise 10 subtribes, ~ 145 genera and nearly 3800 species1, 90% of which occur in the Neotropics29. Four of these subtribes are among the most species-rich in the Andean and Chocoan region (Maxillariinae, Oncidiinae, Stanhopeinae and Zygopetaliinae31). Pleurothallidinae include ~ 5500 exclusively Neotropical species in 47 genera. Pleurothallid orchids are one of the most prominent components of the cloud forest flora in the northern and central Andes and Central America32, also being very well represented in the humid forests of eastern Brazil.
Another group in which phylogenetic relationships are unresolved is Orchidoideae1,33. This group comprises four mostly terrestrial tribes, 25 subtribes and over 3,600 species. The subfamily occurs on all continents except the Antarctic. Previous efforts to disentangle the phylogenetic relationships in the subfamily have mostly relied on a small set of nuclear and plastid markers34, and more recently on extensive plastid coding sequence data2.
The wide geographical range of these groups in the tropics and temperate regions and their striking vegetative and reproductive morphological variability make them ideal model clades for disentangling the contribution of abiotic and biotic drivers of orchid diversification across biomes. Occurring from alpine ecosystems to grasslands, they have conquered virtually all ecosystems available in any elevational gradient35–37, showing independent transitions to terrestrial, rupicolous and epiphytic habit. Moreover, they have evolved a diverse array of pollination systems38–40, rewarding species offering scent, oil and nectar, and even food- and sexual deceptive species41,42. However, the absence of a solid phylogenetic framework has precluded the study of how such systems evolved and the diversification dynamics of Cymbidieae, Pleurothallidinae and Orchidoideae more broadly.
Phylogenetic analyses are crucial to understanding the drivers of diversification in orchids, including the mode and tempo of morphological evolution43. High-throughput sequencing and modern comparative methods have enabled the production of massive molecular datasets to reconstruct evolutionary histories and thus provide unrivalled knowledge on plant phylogenetics44. Here, we present the most densely sampled plastid analysis of Orchidaceae, including data from 51 newly sequenced plastid genomes,. We apply two general approaches: (a) maximum likelihood phylogenetic analysis conducted on 78 plastid coding regions to inform relationships; (b) Bayesian inference in combination with strict and relaxed molecular clocks and a birth–death model applied to a subset of the plastid coding regions to produce a temporal framework of the orchid family. Our study expands the current generic representation for the Orchidaceae and clarifies previously unresolved phylogenetic relations within the Cymbidieae, Pleurothallidinae and Orchidoideae. The results reported here provide a robust framework for the orchid family and new insights into relationships at both deep and shallow phylogenetic levels.
Results
Phylogenetic relationships and divergence times in the orchid family
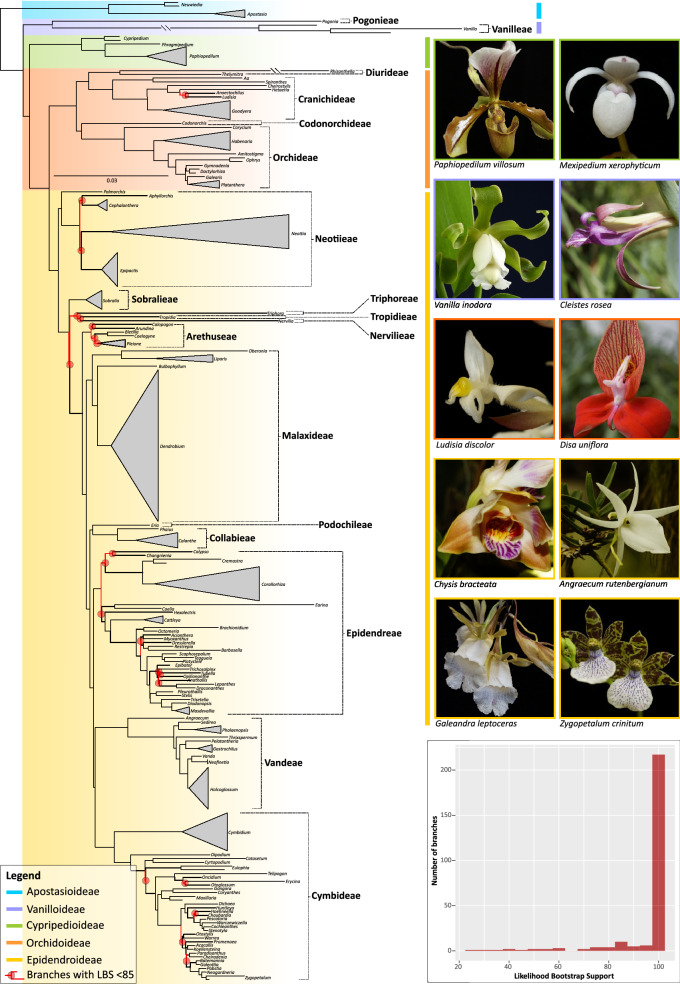
The maximum likelihood (ML) tree derived from the 78 plastid genes is provided in Fig. 1. Two hundred-and-thirty-one branches were recovered as strongly supported (i.e. likelihood bootstrap percentage [LBP] = 85–100), of which 209 attained maximum support. Only 26 branches recovered LBPs between 25 and 84 (Fig. 1, inset). Unsupported relationships were restricted to Epidendroideae and Orchidoideae but were more frequent in Epidendroideae and often linked to low levels of sequence variation. Here, poorly supported relationships occurred mostly towards the backbone of the tribes Arethuseae, Cymbidieae, Epidendreae and Neottieae and Tropidieae + Nervilieae and the most recent common ancestor (MRCA) of Arethuseae, Malaxideae, Podochilieae, Collabieae, Epidendreae, Vandeae and Cymbidieae. Intrageneric relationships were robustly supported, with only two instances for which few branches were recovered as poorly supported (Dendrobium: 3; Cymbidium: 1; Fig. S1).
Figure 1.
Maximum Likelihood phylogeny of the orchid family inferred from 78 coding plastid genes. Likelihood bootstrap support values (LBS) < 85% at nodes are highlighted in red together with their corresponding subtending branches. Orchid genera, tribes and subfamilies are indicated in the phylogeny together with photographs of selected representative species per subfamily. (Inset): Bar plot showing the frequency of LBS values at branches as computed by bin intervals of 5 units. Photos: O. Pérez-Escobar & D. Bogarín.
Absolute times of divergence under strict and relaxed clocks for Orchidaceae, subfamilies and most tribes are provided in Table 1 (phylogenetic trees with mean ages and intervals of confidence produced under both clock models are provided on Figs. S2, S3). Strict and relaxed molecular clocks revealed similar ages of divergence for the majority of the MRCAs of main orchid clades. Yet, we found stark differences in the length of the 95% highest posterior density intervals (HPD) derived from both models, with the relaxed clock producing larger HPDs (Tables 1, S1, S2; Figs. S2, S3). Under the strict and relaxed clocks, Orchidaceae diversified first during the late Cretaceous (88.1 my ± 3; 89.1 my ± 9, respectively). The largest differences on the MRCA ages occurred in Epidendroideae (44 my ± 2 vs 60 my ± 10 under a strict and relaxed clock models, respectively) and Vanilloideae (80 my ± 4 vs 67 my ± 9). A complete account of mean and median ages, HPDs, branch lengths and rate values estimated for chronograms derived from strict and relaxed molecular clock models are provided on Tables S1, S2.
Table 1.
Absolute age estimations of main orchid lineages under strict and relaxed clock models.
| MRCA | Strict clock | Relaxed clock | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stem branch length | Mean age | Age (95% HPD lower bound) | Age (95% HPD upper bound) | 95% HDP length | Age (median) | Stem branch length | Mean age | Age (95% HPD lower bound) | Age (95% HPD upper bound) | 95% HDP length | Age (median) | |
| 270—Orchidaceae | 35.35 | 88.15 | 84.78 | 91.71 | 6.93 | 88.15 | 23.90 | 89.81 | 80.99 | 95.91 | 14.92 | 88.15 |
| 271—Apostasioideae | 48.38 | 39.77 | 37.71 | 41.88 | 4.17 | 39.75 | 53.39 | 36.42 | 8.59 | 62.25 | 53.65 | 36.15 |
| 276—Vanilloideae | 3.95 | 80.96 | 77.63 | 84.29 | 6.66 | 80.95 | 12.65 | 67.69 | 52.73 | 80.94 | 28.20 | 68.29 |
| 280—Cypripedioideae | 38.52 | 31.54 | 29.70 | 33.23 | 3.52 | 31.53 | 38.39 | 38.48 | 21.66 | 55.84 | 34.18 | 37.59 |
| 289—Orchidoideae | 5.43 | 57.76 | 55.38 | 60.33 | 4.96 | 57.75 | 8.42 | 60.02 | 48.91 | 70.72 | 21.80 | 60.05 |
| 291—Diurideae | 1.42 | 54.36 | 51.85 | 56.86 | 5.01 | 54.35 | 12.37 | 40.85 | 29.52 | 52.02 | 22.51 | 40.65 |
| 292—Cranichideae | 9.76 | 46.02 | 43.86 | 48.08 | 4.22 | 46.02 | 11.63 | 41.59 | 29.62 | 53.64 | 24.02 | 41.42 |
| 304—Orchideae | 15.30 | 38.48 | 36.35 | 40.61 | 4.25 | 38.46 | 13.19 | 41.11 | 29.85 | 52.68 | 22.84 | 41.00 |
| 320—Epidendroideae | 18.67 | 44.52 | 42.54 | 46.58 | 4.05 | 44.50 | 8.22 | 60.23 | 48.38 | 71.60 | 23.22 | 60.27 |
| 321—Neottieae | 8.89 | 35.63 | 33.79 | 37.55 | 3.76 | 35.62 | 3.97 | 56.26 | 43.08 | 69.24 | 26.17 | 56.47 |
| 347—Sobralieae | 33.06 | 7.67 | 6.99 | 8.38 | 1.39 | 7.66 | 41.83 | 11.51 | 5.72 | 19.00 | 13.28 | 10.93 |
| 356—Arethuseae | 16.85 | 19.77 | 18.46 | 21.03 | 2.56 | 19.77 | 23.58 | 22.53 | 12.55 | 34.78 | 22.23 | 21.66 |
| 363—Malaxideae | 2.22 | 33.74 | 32.24 | 35.24 | 3.00 | 33.74 | 7.85 | 36.99 | 27.49 | 46.87 | 19.39 | 36.74 |
| 410—Collabieae | 14.60 | 17.32 | 16.00 | 18.57 | 2.57 | 17.32 | 13.47 | 22.47 | 10.72 | 35.05 | 24.33 | 22.08 |
| 416—Epidendroideae | 2.56 | 32.18 | 30.81 | 33.63 | 2.82 | 32.17 | 5.21 | 35.48 | 28.12 | 42.53 | 14.41 | 35.41 |
| 460—Vandeae | 8.91 | 25.60 | 24.13 | 27.05 | 2.92 | 25.59 | 9.31 | 30.77 | 22.20 | 39.32 | 17.12 | 30.76 |
| 484—Cymbidieae | 2.94 | 31.57 | 30.18 | 32.97 | 2.79 | 31.57 | 4.67 | 35.42 | 28.23 | 42.89 | 14.65 | 35.32 |
Phylogenetic informativeness of plastid genes
Phylogenetic informativeness plots are provided on Fig. S4 (see Tables S3, S4 for a detailed account of PI per-site and net values for each assessed locus). Per-site and net phylogenetic informativeness (PI) analyses recovered both ycf1 as the most informative locus, which attained the highest values at a reference time (phylogenetic depth) of 0.51. On average, plastid loci attained their highest PI value at a reference time of 0.85 (SD = 0.16). In contrast, the highest PI values of the 10 most informative loci occurred at an average reference time of 0.63 (SD = 0.11) and 0.80 (SD = 0.17) for per-site and net PI calculations.
Discussion
A robust temporal phylogenomic framework for the orchid family
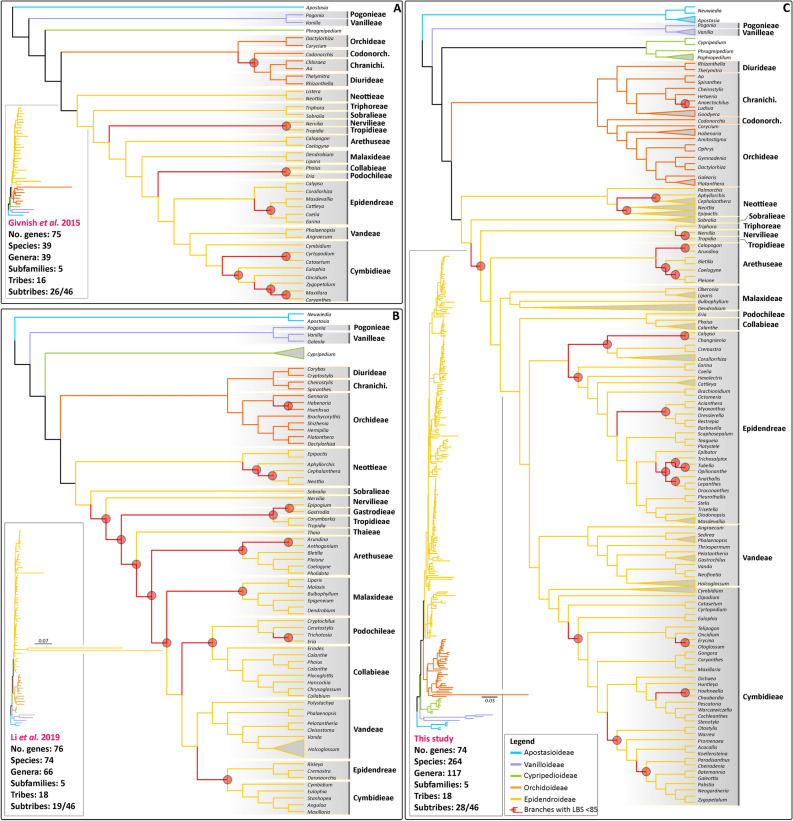
Previous phylogenomic studies of the orchid family included up to 74 species representing 18 tribes, 19 subtribes and 66 genera28. Our study sampled 264 species from all subfamilies, representing 18 tribes (out of 22), 28 subtribes (out of 46) and 74 genera (~ 10% of the currently recognised genera; Fig. 2). In general, our phylogenomic frameworks are in agreement with previously published family-wide orchid analyses either inferred from dozens of markers2,13 or from a handful of loci30. Here, representativeness of Cymbidieae and Epidendreae, two of the most prominent tropical Epidendroideae45 clades, have increased from eight to 32 genera and six to 30, respectively2,28. In particular, relationships inferred from extensive plastid data within Zygopetaliinae (Cymbidieae) and Pleurothallidinae (Epidendreae) are presented for the first time. Our 78-coding sequence plastid ML analysis led to similar results as reported by Givnish et al.2, Niu et al.13 and Li et al.28 but with an overall clear increase in support: 22% of branches with LBS < 85 in Givnish et al.2 and 21% in Li et al.28 vs 11.5% in this study. This is particularly evident in relationships inferred within Orchidoideae, Cymbidieae, Epidendreae and Collabieae. For the latter, high support for the previously unresolved relationship of Podochilieae + Collabieae2,28 was attained for the first time.
Figure 2.
A comparison of the main plastid topologies of the orchid family published to date. (A) Givnish et al.2 inference based on 75 plastid genes and 39 orchid species; (B) Li et al.28 inference based on 76 plastid genes and 76 orchid species; (C) This study: 78 plastid and 264 orchid species. LBP at nodes are highlighted in red together with their corresponding subtending branches. (Inset): trees with branch lengths proportional to substitutions/site.
The absolute age estimates derived from our strict and relaxed molecular clocks and five of the most informative plastid loci are in line with previous nuclear-plastid multi-locus and phylogenomic plastid-only chronograms2,46,47. Nonetheless, our ML tree also identifies intricate relationships that have been consistently recovered as unsupported in several studies. These include poorly supported basal branches in Epidendroideae representing Sobralieae, Nervilieae and Tiphoreae28,48, Arundina + remainder of Arethuseae28, and the position of Eulophiinae in the Cymbidieae26,29,49 (Fig. 2). Uncertainty around the phylogenetic position of these clades might be due to limited taxon sampling in this and previous studies. Alternatively, intragenomic conflict50–52 and lack of phylogenetic informativeness required to sort out relationships derived from rapid diversifications22,53,54 in plastid DNA sequences (regardless of whether whole plastid genome datasets are employed55) might hamper the phylogenetic placement of clades with robust support.
Improved support of phylogenetic relationships within Cymbidieae
Multiple studies have inferred evolutionary relationships in Cymbidieae from morphological and molecular characters29,30. Relationships among subtribes have recently been estimated using the plastid genes psaB, rbcL, matK and ycf1 combined with the low-copy nuclear gene Xdh26. Here, Cymbidiinae was sister to the remainder of Cymbidieae. Poorly supported and incongruent relationships were found among Catasetinae, Eulophiinae and Eriopsidinae, however, when compared with the topologies obtained by Whitten et al.30, Freudenstein & Chase48 and Pérez-Escobar et al.7.
The most complete taxonomic sampling conducted to date under a plastid phylogenomic framework2 included 8 of 11 subtribes of Cymbidieae, but some inter-subtribal relationships were unresolved: Stanhopeinae (20 genera), Maxillariinae (12 genera), Zygopetalinae (36 genera), Oncidiinae (65 genera) and Eulophiinae (13 genera). A clade formed by Stanhopeinae and Maxillariinae had poor support (LBP = 62) and their relationship to Zygopetaliinae also had low support (LBP = 72). The relationship between Eulophiinae and a clade of Stanhopeinae, Maxillariinae, Zygopetalinae and Oncidiinae also had poor support (LBP = 42). One of the outcomes of our expanded sampling (nine subtribes) is the improvement of support in Cymbidieae, more specifically for branches of some groups involved in rapid diversifications that historically have been problematic to resolve2,30. In particular, Maxillariinae + Stanhopeinae and Catasetinae + Cyrtopodiinae are now both strongly supported (LBP = 100). In addition, our results also support the placement of Dipodium (Dipodiinae) as sister to the rest of Cymbidieae, a relationship which was previously recovered from a few loci26. However, our plastid phylogenomic framework is still incomplete due to absence of representatives of Eriopsidiinae and Coleopsidinae.
One other novelty of our study is the inference of relationships in Zygopetalinae, a subtribe in which relationships have previously been poorly understood56. The most extensively sampled analysis of Zygopetalinae inferred from plastid markers (matK-ycf1)30 included 60 species and 27 genera, but relationships between most genera attained only low support. Our expanded molecular, but taxonomically reduced, matrix (i.e. 20 genera and 21 species) produced greater support for the backbone relationships in the subtribe, including the radiation of the Huntleya clade (Dichaea, Huntleya, Chaubardia and the Chondrorhyncha complex56,57). Nonetheless, relationships between the Huntleya grade (i.e. Huntleya clade + Cryptarrhena) and the remainder of Zygopetalinae still remains unresolved.
Our phylogenetic analyses further place for the first time in the orchid tree of life Cheiradenia and Hoehneella with moderate to strong support (Figs. 1, S1). Cheiradenia is a monospecific genus restricted to the lowland wet forests of Venezuela and Guyana and northern Brazil, whereas Hoehneella includes two species exclusively distributed in the Brazilian evergreen wet forests of the states of Espírito Santo and São Paulo58. Referring to the similarity of both vegetative and floral reproductive characters, Pupulin58 hypothesised that Cheiradenia should be closely related to members of the Zygopetalum clade (e.g. Koellensteina, Paradisanthus), with Hoehneella being related to the Huntleya clade (i.e. Huntleya and Chaubardia). Our ML tree supports both assumptions, placing Cheiradenia as sister to Paradisanthus with maximum support and Hoehneella as sister to Chaubardia in a moderately supported clade (83 LBP: Figs. 1, S1). Koellensteina kellneriana (the taxonomic type of the genus) clustered with Acacallis and not with Otostylis and Paradisanthus, and therefore we confirm that Koellensteina in the strict sense is related to Acacallis. In addition, Otostylis is recovered as sister to Warrea and not to Paradisanthus as previously suggested by Williams et al.56 based on a weakly supported placement. Our results also highlight the extensive and independent terrestrial and epiphytic habit transitions occurring in this clade, as most sister genera show different habit types.
Novel and robust relationships in the most rapidly diversifying subtribe Pleurothallidinae
One of the most spectacular Neotropical plant diversifications is perhaps that of the Pleurothallidinae, for it involves the evolution of ~ 5000 species that have conquered virtually all biogeographical regions in the American tropics32,45. The rapid radiation of Pleurothallidinae occurring in the last ~ 20 Myrs29 is associated with the evolution of a diverse suite of pollination systems ranging from food deception59 to pseudocopulation60 linked to dipterans61,62 and a complex array of reproductive and vegetative morphologies22,32. Understanding of relationships in the subtribe has relied mostly on relatively small number of markers63–65, which have informed with some confidence the phylogenetic placement and monophyly of genera in Pleurothallidinae, yet basal branches in these trees have often lacked good support.
Several attempts have been conducted to estimate generic relationships in the subtribe, most of which have relied on nuclear rITS and plastid matK markers64. A synthesis of the phylogenetic relationships in the subtribe based on such studies was conducted by Karremans66. Here, a cladogram depicting the commonest topologies of relationships between genera was provided and nine clades were defined (termed “affinities” by the author) but without considering the magnitude of the support for these (see Fig. 2 in Karremans66). Our plastid phylogenomic analysis recovered well-supported relationships in Pleurothallidinae that are mostly in line with previously published studies29,63. However, these previous trees based on a handful of DNA nuclear and plastid markers yielded poor resolution and low support for backbone branches as well as infrageneric relationships. In contrast, our plastid phylogenomic inferences recovered high support along the backbone, thus recovering novel placements. Some of these noteworthy well-supported relationships are the position of Acianthera as sister to Myoxanthus and Dresslerella as sister to Barbosella + Restrepia (Figs. 1, S1).
Acianthera includes over 300 species distributed throughout the American tropics and subtropics64,67,68, is often retrieved as sister to the remainder of Pleurothallidinae with moderate support63. Karremans66 used a series of “affinities” to describe groups of genera affiliated with a core genus and thus described the “Acianthera affinity” as the frequent clustering of several genera allied with Acianthera64. Our study contradicts Karreman’s66 concept of the Acianthera affinity by placing with high support Acianthera in the Restrepia affinity as sister to Myoxanthus. Dresslerella was previously recovered with low support as sister to the remaining genera in the Restrepia affinity (Barbosella, Echinosepala, Myoxanthus, Restrepia, Restrepiella and Restrepiopsis). In contrast, our analysis robustly places Dreslerella as sister to Restrepia and Barbosella, a result that does not support the monophyly of the Restrepia affinity.
Although estimates of the ancestral distribution of the Pleurothallidinae are still uncertain, most of the early divergent Pleurothallidinae and their sister groups are found in the Antilles or Brazil29. The remarkable relationship recovered here for Acianthera + Myoxanthus could yield more clues about the biogeographic history and evolution of the subtribe because Brazil harbours a high species diversity of Acianthera and some of the early divergent clades in Myoxanthus (particularly the species close to M. lonchophyllus), whereas Myoxanthus is notably absent in the Antilles. In addition, other early divergent clades such as Octomeria and Barbosella are more diverse in Brazil. These clades share the lack of stem annulus as a morphological symplesiomorphy, a character that later appears in more diverse groups such as Masdevallia + Dracula, Lepanthes, and Pleurothallis + Stelis69. Members of these clades probably diversified after a migration to the mountainous areas of the northern Andes ca 16 ± 5 Ma and together account for almost 80% of the species in the subtribe29. The modern range extends mostly along the Andean and Central American mountain ranges. Here, another noteworthy relationship is that the less diverse Specklinia clade (Scaphosepalum + Platystele) was recovered as sister to the most species-rich clades of the subtribe (Masdevallia, Lepanthes, and Pleurothallis). In previous phylogenetic analyses Specklinia clade was recovered as sister to Pleurothallis29.
Likewise, relationships between early divergent members in the Lepanthes affinity (Anathallis, Draconanthes, Epibator, Lepanthes, Opilionanthe, Trichosalpinx and Tubella) were largely weakly supported, demonstrating the need for increased taxon sampling, principally in Lepanthopsis and Tubella32. In particular, the early diversification of the Lepanthes affinity (> 1500 spp.), inferred to have occurred around 8 Ma, has been linked to colonisation of newly formed environments in the Andean Cordillera, accelerated mountain uplift and the evoluton of specific pollination systems (pseudocopulation and food mimicry60).
Another novel placement concerns Teagueia (diverse in Colombia, Ecuador and Peru70–72), which resembles Platystele73. Karremans74 had suggested a close relationship between Teagueia and Scaphosepalum, but our results place Teagueia as sister to Platystele with high support, thus corroborating the long-standing hypotheses of their sister relationship based on the similarity of their reproductive structures72,73.
Evolutionary relationships in Orchidoideae
Our study provides a well-supported tree for Orchidoideae. Our ML inference supports the findings of Pridgeon et al.35 in which Diurideae is sister to Cranichideae and Codonorchideae to Orchideae. Our findings differ from Givnish et al.2 and Salazar et al.34, in which Diurideae/Cranichideae are sister to Codonorchideae, with Orchideae sister to all these (Fig. 2). Givnish et al.2 included all four tribes but only six of 21 subtribes of Orchidoideae, and the relationship of Diurideae to Cranichideae was poorly supported.
Conclusions
This study presents a well-resolved, more densely sampled and strongly supported analysis of Orchidaceae and their absolute times of divergence than all previous similar studies. For deep branches and recent diversifications in Cymbidieae and Epidendreae, support is improved, yet several recalcitrant branches that historically have been challenging to resolve were also recovered as poorly supported (e.g. early divergent taxa in the Epidendroideae, initial radiation of the Lepanthes affinity in Pleurothallidinae). Similarly, our analyses provide a well-supported result for Orchidoideae. Although taxon sampling was sufficient to resolve the relationships between the major clades in the family, sampling of unrepresented genera and representatives of Eriopsidiinae, Goodyerinae, and Coleopsidinae would further enhance our understanding of phylogenetic relationships.
Material and methods
Sampling, DNA extraction and sequencing
Two-hundred and sixty-four species representing 117 genera, 28 subtribes and 18 tribes were sampled in this study. For 51 species plastid genomes were sequenced. Table S5 provides voucher information and accession numbers of plastid genomes sourced from NCBI. Fresh leaves were stored in silica gel for subsequent DNA extraction using a CTAB method75. Total DNA was purified with silica columns and then eluted in Tris-EDTA76. DNA samples were adjusted to 50 ng/uL and sheared to fragments of approximately 500 bp.
High-throughput sequencing
The library preparation, barcoding and sequencing (Illumina HiSeqX) were conducted at Rapid Genomics LLC (Gainesville, FL, USA), BGI Genomics (Shenzhen, China) and Genewiz GmbH (Leipzig, Germany). Pair-end reads of 150 bp were obtained for fragments with insert size of 300–600 bp. Overhangs were blunt ended using T4 DNA polymerase, Klenow fragment and T4 polynucleotide kinase. Subsequently, a base 'A' was added to the 3′ end of the phosphorylated blunt DNA fragments. DNA fragments were ligated to adapters, which have a thymine (T) overhang. Ligation products were gel-purified by electrophoresis to remove all unbound adapters or split adapters that were ligated together. Ligation products were then selectively enriched and amplified by PCR. For each sample, between one and 10 million paired-end reads were generated.
Plastid genome assembly and annotation
Raw sequences were quality filtered using Trimmomatic77 in order to eliminate sequencing artefacts, improve uniformity in the read length (> 40 bp) and ensure quality (Phred score> 20) for further analysis. Filtered sequences were processed with BBNorm78 to normalize coverage by down-sampling reads over high-depth areas of the genomes (maximum depth coverage 900 × and minimum depth 6x). This step creates a flat coverage distribution in order to improve read assembly. Subsequently, overlapping reads were merged into single reads using BBmerge79 in order to accelerate the assembly process. Overlapping of paired reads was evaluated with Flash80 to reduce redundancy. Merged reads were used to carry out the whole genome de novo assembly with SPAdes (hash length 3,355,77)81.
To produce contiguous, linear plastid genome sequences we relied on a refence-based and de-novo approaches. The reference based approach was conducted on MIRA v. 482, a software that maps read data against a consensus sequence of a reference assembly (simple mapping). MIRA has been useful for assembling complicated genomes with many repetitive sequences83–85. MIRA produces BAM files as output, which were subsequently used to generate consensus sequences in SAMTOOLS86. We sourced 11 reference plastomes from the NCBI repository that represent related species, namely: Cattleya crispata, Goodyera fumata, Masdevallia picturata, M. coccinea, Oncidium sphacelatum and Sobralia callosa. The de-novo assembly approach relied on GetOrganelle87, using the recommended default settings for assemblies of green-plant plastid genomes.
Newly sequenced and datamined plastid genomes were annotated through the Chlorobox portal of the Max Planck Institute88. Sequences were uploaded as fasta files, and running parameters were established as follow: BLAST protein search identity = 65%, BLAST rRNA, tRNA, DNA search identity = 85%, genetic code = bacterial/plant plastid, max intron length = 3,000, options = allow overlaps. Apostasia wallichii, Masdevallia picturata, Oncidium sphacelatum, Sobralia callosa and Goodyera fumata were set as the ‘Server Reference’ and Cattleya liliputana was set as the ‘Custom Reference’ for CDS and tRNA, rRNA, primer, other DNA or RNA specifications.
Phylogenetic analysis
A set of 78 plastid genes was used to reconstruct phylogenetic relationships in Orchidaceae. These were aligned89 using MAFFT 790 and subsequently concatenated (proportions of missing data per species is provided on Table S5). This step was performed at the supercomputing centre APOLO, EAFIT University, Medellín, Colombia. Phylogenetic reconstruction based on maximum likelihood (ML) was implemented in RAxML v. 8.091, using 1,000 bootstrap replicates and the GTR + GAMMA model. Absolute age estimation analyses relied on fossil and secondary calibration points, strict and molecular clocks and a birth/death model implemented in BEAST v. 1.892. The fossil constraint was added to the MRCA of Dendrobium following Xiang et al.93 using a normal distribution with mean value of 21.07 and a standard deviation (SD) of 3.0. Following Givnish et al.2, the two secondary calibration points were added to the root of the tree and MRCA of the Orchidaceae, using a normal distribution and mean values of 123.48 (SD = 2.0) and 90 (SD = 2.0). Because dating analyses conducted on dozens of gene alignments and hundreds of terminals are extremely computationally greedy, we estimated absolute ages on the five most phylogenetically informative genes (see below) and by constraining the tree topology to the ML tree derived from RAxML. For each clock model, we conducted two MCMC analyses with 250 million generations each with a sampling frequency of 10,000 generations. The convergence of the strict and relaxed molecular clocks parameters was confirmed on the software TRACER v1.6. (http://tree.bio.ed.ac.uk/software/tracer/). Maximum clade credibility trees were summarised from the MCMC trees in the program TreeAnnotator v.1.8. of the software BEAST. The individual gene alignments employed for ML and Bayesian phylogenetic infereces are freely available at 10.6084/m9.figshare.14068892
Phylogenetic informativeness profiles
To estimate the phylogenetic informativeness (PI) of plastid genes we calculated the per-site and net values for each assessed locus with the HyPhy substitution rates algorithm for DNA sequences94 using in the web application PhyDesign http://phydesign.townsend.yale.edu/). The input files were the consensus ML ultrametric tree converted with the function chronos of the R-package APE (http://ape-package.ird.fr/) using an smoothing rate of 1 and a relaxed clock model, and the partitioned concatenated gene alignments.
Supplementary Information
Acknowledgements
We would like to thank Esteban Urrea for helping with the bioinformatics pipelines. We thank Norris Williams and the late Mark Whitten (University of Florida) for collecting and preparing the specimens. Kurt Neubig from Southern Illinois University provided the sequences of 11 new samples. We also thank Janice Valencia for critical feedback on the paper, Juan David Pineda Cardenas for advising about computational resources used through EAFIT and Juan Carlos Correa for computational advices at BIOS. Martha Charitonidou provided constructive feedback on figure design. The University of Costa Rica provided access to the genetic material for the projects B8257 and B6140. Finally, we would like to thank IDEA WILD for supporting with photographic equipment and Sociedad Colombiana de Orquideología for supporting M. A. Serna-Sánchez with a grant to conduct her undergraduate studies. C.F.H received a PhD fellowship from FAPESP (11/08308-9, 13/19124-1). O.A.P.E. is supported by the Swiss Orchid Foundation and the Sainsbury Orchid Fellowship at the Royal Botanic Gardens, Kew. A.A. acknowledges financial support from the Swedish Research Council (2019-05191), the Swedish Foundation for Strategic Research (FFL15-0196) and the Royal Botanic Gardens, Kew.
Author contributions
M.A.S.S., O.A.P.E., and T.A. designed research; O.A.P.E., M.A.S.S., T.A., C.F.H. and A.A. generated new data; M.A.S.S., O.A.P.E., M.F.T.J., A.C.A.Y., J.E.A.G. and S.D. performed all analyses; O.A.P.E., D.B., M.W.C., M.A.S.S., S.D. and T.A. wrote the manuscript, with contributions from all authors.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: The original version of this Article contained an error in Figure 1 where the image showing "Paphiopedilum villosum" was incorrectly labelled as "Apostasia shenzhenica".
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maria Alejandra Serna-Sánchez, Oscar A. Pérez-Escobar and Diego Bogarín.
Change history
7/6/2021
A Correction to this paper has been published: 10.1038/s41598-021-93674-y
Contributor Information
Oscar A. Pérez-Escobar, Email: o.perez-escobar@kew.org
Tatiana Arias, Email: tatiana.arias48@tdea.edu.co.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-83664-5.
References
- 1.Chase MW, et al. An updated classification of orchidaceae: updated classification of orchidaceae. Bot. J. Linn. Soc. 2015;177:151–174. [Google Scholar]
- 2.Givnish TJ, et al. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc. R. Soc. B Biol. Sci. 2015;282:1553. doi: 10.1098/rspb.2015.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darwin Ch. On the Various Contrivances by which British and Foreign Orchids are Fertilised by Insects, and on the Good Effects of Intercrossing, 1st Issue. 1. London: John Murray; 1862. [PMC free article] [PubMed] [Google Scholar]
- 4.Fay MF, Chase MW. Orchid biology: from Linnaeus via Darwin to the 21st century. Ann. Bot. 2009;104:359–364. doi: 10.1093/aob/mcp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramírez SR, et al. Asynchronous diversification in a specialized plant-pollinator mutualism. Science. 2011;333:1742–1746. doi: 10.1126/science.1209175. [DOI] [PubMed] [Google Scholar]
- 6.Borba EL, Barbosa AR, de Melo MC, Gontijo SL, de Oliveira HO. Mating systems in the Pleurothallidinae (Orchidaceae): evolutionary and systematic implications. Lankesteriana. 2011;11:207–211. [Google Scholar]
- 7.Pérez-Escobar OA, et al. Multiple geographical origins of environmental sex determination enhanced the diversification of Darwin’s favourite orchids. Sci. Rep. 2017;7:12878. doi: 10.1038/s41598-017-12300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez-Escobar OA, Gottschling M, Whitten WM, Salazar G, Gerlach G. Sex and the Catasetinae (Darwin’s favourite orchids) Mol. Phylogenet. Evol. 2016;97:1–10. doi: 10.1016/j.ympev.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Bateman R, Rudall P. Evolutionary and morphometric implications of morphological variation among flowers within an inflorescence: a case-study using European orchids. Ann. Bot. 2006;98:975–993. doi: 10.1093/aob/mcl191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong W-L, et al. Molecular evolution of chloroplast genomes of orchid species: insights into phylogenetic relationship and adaptive evolution. Int. J. Mol. Sci. 2018;19:716. doi: 10.3390/ijms19030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freudenstein JV, Chase MW. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot. 2015;115:665–681. doi: 10.1093/aob/mcu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J, et al. Comparative chloroplast genomes of photosynthetic orchids: insights into evolution of the Orchidaceae and development of molecular markers for phylogenetic applications. PLoS ONE. 2014;9:e99016. doi: 10.1371/journal.pone.0099016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu Z, et al. The complete plastome sequences of four orchid species: insights into the evolution of the orchidaceae and the utility of plastomic mutational hotspots. Front. Plant Sci. 2017;8:715. doi: 10.3389/fpls.2017.00715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang G-Q, et al. The Apostasia genome and the evolution of orchids. Nature. 2017;549:379–383. doi: 10.1038/nature23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang G-Q, et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016;6:19029. doi: 10.1038/srep19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan L, et al. The Genome of Dendrobium officinale illuminates the biology of the important traditional chinese orchid herb. Mol. Plant. 2015;8:922–934. doi: 10.1016/j.molp.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 17.Yuan Y, et al. The Gastrodia elata genome provides insights into plant adaptation to heterotrophy. Nat. Commun. 2018;9:1615. doi: 10.1038/s41467-018-03423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai J, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2014;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 19.Huang J-Z, et al. The genome and transcriptome of Phalaenopsis yield insights into floral organ development and flowering regulation. PeerJ. 2016;4:e2017. doi: 10.7717/peerj.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao Y-T, et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018;16:2027–2041. doi: 10.1111/pbi.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, et al. Genomics-based diversity analysis of Vanilla species using a Vanilla planifolia draft genome and Genotyping-By-Sequencing. Sci. Rep. 2019;9:3416. doi: 10.1038/s41598-019-40144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogarín D, et al. Anchored Hybrid Enrichment generated nuclear, plastid and mitochondrial markers resolve the Lepanthes horrida (Orchidaceae: Pleurothallidinae) species complex. Mol. Phylogenet. Evol. 2018;129:27–47. doi: 10.1016/j.ympev.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Escobar OA, et al. Resolving relationships in an exceedingly young Neotropical orchid lineage using Genotyping-by-sequencing data. Mol. Phylogenet. Evol. 2020;144:106672. doi: 10.1016/j.ympev.2019.106672. [DOI] [PubMed] [Google Scholar]
- 24.Grace OM, Pérez-Escobar OA, Lucas EJ, Vorontsova MS, Lewis GP, Walker BE, Lohmann LG, Knapp S, Wilkie P, Sarkinen T, Darbyshire I, Lughadha EN, Monro A, Woudstra Y, Demissew S, Muasya AM, Díaz S, Baker WJ, Antonelli A. Botanical monography in the Anthropocene. Trends Plant Sci. 2021 doi: 10.1016/j.tplants.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 25.Jin W-T, et al. Phylogenetics of subtribe Orchidinae s.l. (Orchidaceae; Orchidoideae) based on seven markers (plastid matK, psaB, rbcL, trnL-F, trnH-psbA, and nuclear nrITS, Xdh): implications for generic delimitation. BMC Plant Biol. 2017;17:222. doi: 10.1186/s12870-017-1160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M-H, Zhang G-Q, Liu Z-J, Lan S-R. Subtribal relationships in Cymbidieae (Epidendroideae, Orchidaceae) reveal a new subtribe, Dipodiinae, based on plastid and nuclear coding DNA. Phytotaxa. 2016;246:37. [Google Scholar]
- 27.Nauheimer L, Schley RJ, Clements MA, Micheneau C, Nargar K. Australasian orchid biogeography at continental scale: molecular phylogenetic insights from the Sun Orchids (Thelymitra, Orchidaceae) Mol. Phylogenet. Evol. 2018;127:304–319. doi: 10.1016/j.ympev.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 28.Li Y-X, et al. Phylogenomics of Orchidaceae based on plastid and mitochondrial genomes. Mol. Phylogenet. Evol. 2019;139:106540. doi: 10.1016/j.ympev.2019.106540. [DOI] [PubMed] [Google Scholar]
- 29.Pérez-Escobar OA, et al. Recent origin and rapid speciation of Neotropical orchids in the world’s richest plant biodiversity hotspot. New Phytol. 2017;215:891–905. doi: 10.1111/nph.14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitten WM, Neubig KM, Williams NH. Generic and subtribal relationships in neotropical Cymbidieae (Orchidaceae) based on matK/ycf1 plastid data. Lankesteriana. 2014;13:375–392. [Google Scholar]
- 31.Pérez-Escobar OA, et al. The origin and diversification of the hyperdiverse flora in the Chocó biogeographic region. Front. Plant Sci. 2019;10:1328. doi: 10.3389/fpls.2019.01328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bogarín D, et al. Phylogenetic comparative methods improve the selection of characters for generic delimitations in a hyperdiverse Neotropical orchid clade. Sci. Rep. 2019;9:1–17. doi: 10.1038/s41598-019-51360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Górniak M, Paun O, Chase MW. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, Xdh: Congruence with organellar and nuclear ribosomal DNA results. Mol. Phylogenet. Evol. 2010;56:784–795. doi: 10.1016/j.ympev.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Salazar GA, Chase MW, Soto Arenas MA, Ingrouille M. Phylogenetics of Cranichideae with emphasis on Spiranthinae (Orchidaceae, Orchidoideae): evidence from plastid and nuclear DNA sequences. Am. J. Bot. 2003;90:777–795. doi: 10.3732/ajb.90.5.777. [DOI] [PubMed] [Google Scholar]
- 35.Bone RE, Cribb PJ, Buerki S. Phylogenetics of Eulophiinae (Orchidaceae: Epidendroideae): evolutionary patterns and implications for generic delimitation: evolutionary patterns in Eulophiinae. Bot. J. Linn. Soc. 2015;179:43–56. [Google Scholar]
- 36.Pérez-Escobar OA, et al. Andean mountain building did not preclude dispersal of lowland epiphytic orchids in the Neotropics. Sci. Rep. 2017;7:4919. doi: 10.1038/s41598-017-04261-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar GA, et al. Phylogenetic systematics of subtribe Spiranthinae (Orchidaceae: Orchidoideae: Cranichideae) based on nuclear and plastid DNA sequences of a nearly complete generic sample. Bot. J. Linn. Soc. 2018;186:273–303. [Google Scholar]
- 38.Martins A, et al. From tree tops to the ground: reversals to terrestrial habit in Galeandra orchids (Epidendroideae: Catasetinae) Mol. Phylogenet. Evol. 2018;127:952–960. doi: 10.1016/j.ympev.2018.06.041. [DOI] [PubMed] [Google Scholar]
- 39.Nunes C, et al. More than euglossines: the diverse pollinators and floral scents of Zygopetalinae orchids. Sci. Nat. 2017;104:92. doi: 10.1007/s00114-017-1511-3. [DOI] [PubMed] [Google Scholar]
- 40.Pansarin L, Pansarin E, Gerlach G, Sazima M. The natural history of Cirrhaea and the pollination system of Stanhopeinae (Orchidaceae) Int. J. Plant Sci. 2018 doi: 10.1086/697997. [DOI] [Google Scholar]
- 41.Cisternas MA, et al. Phylogenetic analysis of Chloraeinae (Orchidaceae) based on plastid and nuclear DNA sequences. Bot. J. Linn. Soc. 2012;168:258–277. [Google Scholar]
- 42.Ramirez SR, Roubik DW, Skov C, Pierce NE. Phylogeny, diversification patterns and historical biogeography of Euglossine orchid bees (Hymenoptera: Apidae) Biol. J. Linn. Soc. 2010;100:552–572. [Google Scholar]
- 43.Van Der Cingel NA. An Atlas of Orchid Pollination: European Orchids. Boca Raton: CRC Press; 2001. [Google Scholar]
- 44.Weitemier K, et al. Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Appl. Plant Sci. 2014;2:1400042. doi: 10.3732/apps.1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. Genera Orchidacearum: Vol. 5. Epidendroideae (Part Two) Oxford: Oxford University Press; 2009. [Google Scholar]
- 46.Chomicki G, et al. The velamen protects photosynthetic orchid roots against UV-B damage, and a large dated phylogeny implies multiple gains and losses of this function during the Cenozoic. New Phytol. 2015;205:1330–1341. doi: 10.1111/nph.13106. [DOI] [PubMed] [Google Scholar]
- 47.Ramírez SR, Gravendeel B, Singer RB, Marshall CR, Pierce NE. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature. 2007;448:1042–1045. doi: 10.1038/nature06039. [DOI] [PubMed] [Google Scholar]
- 48.Freudenstein JV, Chase MW. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot. 2015;115:665–681. doi: 10.1093/aob/mcu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pérez-Escobar OA. Molecular Phylogenetics, Evolution of Sexual Systems and Historical Biogeography of Darwin’s Favorite Orchids (Catasetinae) and Swan Orchids (Cycnoches Lindl) München: Ludwig-Maximilians Universität; 2016. [Google Scholar]
- 50.Soltis DE, Kuzoff RK. Discordance between nuclear and chloroplast phylogenies in the heuchera group author (saxifragaceae) Evolution. 1995;49:727–742. doi: 10.1111/j.1558-5646.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 51.van der Niet T, Peter Linder H. Dealing with incongruence in the quest for the species tree: a case study from the orchid genus Satyrium. Mol. Phylogenet. Evol. 2008;47:154–174. doi: 10.1016/j.ympev.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 52.Pérez-Escobar OA, Balbuena JA, Gottschling M. Rumbling orchids: how to assess divergent evolution between chloroplast endosymbionts and the nuclear host. Syst. Biol. 2016;65:51–65. doi: 10.1093/sysbio/syv070. [DOI] [PubMed] [Google Scholar]
- 53.Fragoso-Martínez I, et al. A pilot study applying the plant Anchored Hybrid Enrichment method to New World sages (Salvia subgenus Calosphace; Lamiaceae) Mol. Phylogenet. Evol. 2017;117:124–134. doi: 10.1016/j.ympev.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Léveillé-Bourret É, Starr JR, Ford BA, Moriarty Lemmon E, Lemmon AR. Resolving rapid radiations within angiosperm families using anchored phylogenomics. Syst. Biol. 2018;67:94–112. doi: 10.1093/sysbio/syx050. [DOI] [PubMed] [Google Scholar]
- 55.Zhang R, et al. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst. Biol. 2020;69:613–622. doi: 10.1093/sysbio/syaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mark Whitten W, Williams NH, Dressler RL, Gerlach G, Pupulin F. Generic relationships of Zygopetalinae (Orchidaceae: Cymbidieae): combined molecular evidence. Lankesteriana. 2005;5:87–107. [Google Scholar]
- 57.Neubig KM, Williams NH, Whitten WM, Pupulin F. Molecular phylogenetics and the evolution of fruit and leaf morphology of Dichaea (Orchidaceae: Zygopetalinae) Ann. Bot. 2009;104:457–467. doi: 10.1093/aob/mcp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pupulin F. In: Genera Orchidacearum, Vol. 5, Epidendroideae (Part Two) Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN, editors. Oxford: Oxford University Press; 2009. pp. 456–546. [Google Scholar]
- 59.Bogarín D, et al. Pollination of Trichosalpinx (Orchidaceae: Pleurothallidinae) by biting midges (Diptera: Ceratopogonidae) Bot. J. Linn. Soc. 2018;186:510–543. [Google Scholar]
- 60.Blanco MA, Barboza G. Pseudocopulatory pollination in Lepanthes (Orchidaceae: Pleurothallidinae) by fungus gnats. Ann. Bot. 2005;95:763–772. doi: 10.1093/aob/mci090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Damon A, et al. A survey of pollination in remnant orchid populations in Soconusco, Chiapas, Mexico. Trop. Ecol. 2007;48:1–14. [Google Scholar]
- 62.Policha T, et al. Disentangling visual and olfactory signals in mushroom-mimicking Dracula orchids using realistic three-dimensional printed flowers. New Phytol. 2016;210:1058–1071. doi: 10.1111/nph.13855. [DOI] [PubMed] [Google Scholar]
- 63.Pridgeon AM, Solano R, Chase MW. Phylogenetic relationships in Pleurothallidinae (Orchidaceae): combined evidence from nuclear and plastid DNA sequences. Am. J. Bot. 2001;88:2286–2308. [PubMed] [Google Scholar]
- 64.Karremans AP, et al. Phylogenetic reassessment of Acianthera (Orchidaceae: Pleurothallidinae) Harvard Pap. Bot. 2016;21:171–187. [Google Scholar]
- 65.Bogarín D, Karremans AP, Fernández M. Genus-level taxonomical changes in the Lepanthes affinity (Orchidaceae, pleurothallidinae) Phytotaxa. 2018;340:128–136. [Google Scholar]
- 66.Karremans AP. Genera Pleurothallidinarum: an updated phylogenetic overview of Pleurothallidinae. Lankesteriana. 2016;16:219–241. [Google Scholar]
- 67.Damián A, Mitidieri N, Chiron G. A taxonomic synopsis of Acianthera (Orchidaceae: Pleurothallidinae) in Peru, including two new species. An. del Jard. Bot. Madrid. 2018;75:1–21. [Google Scholar]
- 68.Luer CA. A systematic method of classification of the pleurothallidinae versus a strictly phylogenetic method. Selbyana. 2002;23:57–110. [Google Scholar]
- 69.Stern WWL, Pridgeon A, Luer C. Stem structure and its bearing on the systematics of Pleurothallidinae (Orchidaceae) Bot. J. Linn. Soc. 1985;91:457–471. [Google Scholar]
- 70.Jost L, Shepard A. Two new species of Teagueia (Orchidaceae: Pleurothallidinae) from east-central Ecuador. Lankesteriana. Lankesteriana. 2011;11:9–14. [Google Scholar]
- 71.Luer CA. Icones Pleurothallidinarum VIII. Systematics of Lepanthopsis, Octomeria subgenus Pleurothallopsis, Restrepiella, Restrepiopsis, Salpistele, and Teagueia. Addenda to Platystele, Porroglossum, and Scaphosepalum. Monogr. Syst. Bot. Missouri Bot. Gard. 1991;39:1–161. [Google Scholar]
- 72.Luer CA. Icones Pleurothallidinarum XXIV. A First Century Of New Species of Stelis of Ecuador part one. Addenda to Lepanthes of Ecuador. Addenda to Barbosella, Dracula, Dresslerella, Lepanthopsis, Platystele, Pleurothallis, Restrepia, Scaphosepalum, Teagueia. Monogr. Syst. Bot. Missouri Bot. Gard. 2002;88:1–122. [Google Scholar]
- 73.Luer CA. Icones Pleurothallidinarum VII. Systematics of Platystele (Orchidaceae) Monogr. Syst. Bot. Missouri Bot. Gard. 1990;38:1–115. [Google Scholar]
- 74.Karremans AP, et al. Phylogenetic reassessment of Specklinia and its allied genera in the pleurothallidinae (Orchidaceae) Phytotaxa. 2016;272:1–36. [Google Scholar]
- 75.Doyle J, Doyle J. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- 76.Neubig, K. M. et al. in DNA Banking for the 21st Century: Proceedings of the US Workshop on DNA Banking (eds. Applequist, W. & Campbell, L.) 81–112 (William L. Brown Center, Missouri Botanical Garden, 2014).
- 77.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bushnell, B. BBMap. Available at https://sourceforge.net/projects/bbmap/ (2017).
- 79.Bushnell B, Rood J, Singer E. BBMerge—accurate paired shotgun read merging via overlap. PLoS ONE. 2017;12:e0185056. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chevreux, B., Wetter, T. & Suhai, S. Genome sequence assembly using trace signals and additional sequence information. In German Conference on Bioinformatics99, 45–56 (Citeseer, 1999).
- 83.Cock PJA, Grüning BA, Paszkiewicz K, Pritchard L. Galaxy tools and workflows for sequence analysis with applications in molecular plant pathology. PeerJ. 2013;1:e167. doi: 10.7717/peerj.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Parakhia MV, et al. Draft genome sequence of the endophytic bacterium Enterobacter spp. MR1, isolated from drought tolerant plant (Butea monosperma) Indian J. Microbiol. 2014;54:118–119. doi: 10.1007/s12088-013-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ward JA, Ponnala L, Weber CA. Strategies for transcriptome analysis in nonmodel plants. Am. J. Bot. 2012;99:267–276. doi: 10.3732/ajb.1100334. [DOI] [PubMed] [Google Scholar]
- 86.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jin JJ, et al. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tillich M, et al. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chan CX, Ragan MA. Next-generation phylogenomics. Biol. Direct. 2013;8:3. doi: 10.1186/1745-6150-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiang XG, et al. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J. Biogeogr. 2016;43:1310–1323. [Google Scholar]
- 94.Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: Hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.