Abstract
The ongoing SARS‐CoV‐2 pandemic with over 80 million infections and more than a million deaths worldwide represents the worst global health crisis of the 21th century. Beyond the health crisis, the disruptions caused by the COVID‐19 pandemic have serious global socio‐economic consequences. It has also placed a significant pressure on the scientific community to understand the virus and its pathophysiology and rapidly provide anti‐viral treatments and procedures in order to help the society and stop the virus spread. Here, we outline how advanced microscopy technologies such as high‐throughput microscopy and electron microscopy played a major role in rapid response against SARS‐CoV‐2. General applicability of developed microscopy technologies makes them uniquely positioned to act as the first line of defence against any emerging infection in the future.
1. INTRODUCTION
In the last 20 years, three members of the betacoronaviruses have emerged from zoonotic reservoirs able to infect humans—severe acute respiratory syndrome coronavirus (SARS‐CoV) in 2002 (Zhong et al., 2003), Middle East respiratory syndrome coronavirus (MERS‐CoV) in 2012 (De Wit, Van Doremalen, Falzarano, & Munster, 2016; Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, 2012) and severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) in December 2019 (Wu et al., 2020; Zhou et al., 2020). SARS‐CoV‐2 was first detected in Wuhan, Hubei province, China, from a cluster of atypical pneumonia diseases (later named coronavirus disease 2019, COVID‐19). While the spread of SARS‐CoV and MERS‐CoV has been successfully contained (De Wit et al., 2016), SARS‐CoV‐2 caused a global pandemic with over a million deaths and devastating socio‐economic consequences. SARS‐CoV‐2 also induced mobilisation of the global scientific community on an unprecedented scale in efforts to understand the virus, the underlying host–pathogen interactions, its patho‐physiology and the immunological responses. Many of these efforts were aimed at developing anti‐viral drugs, vaccines or therapeutic modalities in order to stop the virus spread and relieve the socio‐economic pressure. A PubMed query ‘SARS‐CoV‐2’ returned 52,926 scientific articles in the period from January 1, 2020 to December 31, 2020, which amounts to an incredible 145 articles per day.
Microscopy is a fundamental technology in modern bio‐medical research because it is the only technology able to quantitatively address complex spatio‐temporal dynamics of living systems at a sufficient resolution to provide the most realistic representations of the biological systems. Indeed, one can find light and electron microscopy imaging data in the majority of scientific articles published in biomedical fields (Jambor et al., 2020). Additionally, microscopy has played a key role in infectious disease research since the discovery of the first microorganisms (Laketa, 2018). Microscopy was essential for the discovery of infectious agents by direct observation as well as for testing the compliance with Koch postulates in order to identify new pathogens. It has also had a major role in infectious diseases diagnostics (Laketa, 2018). The importance of microscopy in infectious disease research has continued in modern times, which is illustrated in the ongoing SARS‐CoV‐2 pandemic. The very first articles describing the isolation, identification and characterisation of SARS‐CoV‐2 strongly relied on light and electron microscopy data (in addition to next generation sequencing) to provide evidence of SARS‐CoV‐2 emergence (Zhou, Yang, et al., 2020). This demonstrates how microscopy‐derived evidence is still an essential component in identification of new pathogens.
Electron microscopy (EM) has long been the method of choice for direct visualisation of viruses. While recent advances in super‐resolution microscopy allowed to resolve the molecular distribution of viral proteins on the virion surface (Chojnacki et al., 2012; Muranyi, Malkusch, Müller, Heilemann, & Kräusslich, 2013), EM still remains the only technique able to provide structural and morphological information. Due to its central role in virus research, EM has been extensively exploited during the 2020 COVID‐19 pandemic to study different aspects of SARS‐CoV‐2 infection and pathogenesis. In addition to their scientific value, digitally enhanced EM images of SARS‐CoV‐2 isolated virions or virions bound to the host cell surface are used by the news media to accompany the daily news describing the evolution of the COVID‐19 pandemic. Some of those images together with accurate illustrations generated from EM structural studies achieved widespread recognition among the general public and became an important resource to raise awareness on coronavirus diseases, fight disinformation and promote health recommendations to fight COVID‐19 (Goodsell, Voigt, Zardecki, & Burley, 2020).
This review will focus on a discussion of the advanced microscopy technology modalities that have had the biggest impact and have the highest potential to limit the SARS‐CoV‐2 pandemic. A special emphasis is given to technologies whose employment directly enables transition from the basic research towards more translational aspects. With this in mind, we centre our discussion around two advanced microscopy technologies—(a) high‐throughput microscopy and (b) electron microscopy with their roles in diagnostics, drug discovery and virus characterisation.
2. HIGH‐THROUGHPUT MICROSCOPY IN SARS‐COV‐2 RESEARCH
2.1. High‐throughput microscopy in anti‐viral drug development
High‐throughput microscopy is a method that typically involves automated microscopy, robotics and quantification by image analysis in order to test a large number of reagents for their activity in biological assays. It can incorporate various imaging modalities and generally employs cell‐based assays combined with fluorescently labelled structures or molecular components. The method has been extensively used in academia to perform genome‐wide genetic screens using RNA interference or overexpression (Boutros, Heigwer, & Laufer, 2015), as well as in industry at all stages of the drug discovery and development processes (Bickle, 2010). In the context of infectious disease research, it plays an important role in studying host–pathogen interactions and in the discovery of anti‐microbial compounds (Brodin & Christophe, 2011). The main advantage of high‐throughput microscopy over traditional biochemical methods in drug discovery is the fact that employment of cellular assays is more predictive of the in vivo situation compared to biochemical assays, especially concerning cell penetration and susceptibility to intracellular metabolism. In addition, the ability to multiplex many readouts in one assay allows assessment of toxicity, modes of action and multiple drug profiling, essentially replacing the need for secondary and tertiary assays (Simm et al., 2018).
With the emergence of SARS‐CoV‐2, it is more obvious than ever why rapid drug development is necessary. The key to utilising high‐throughput microscopy approaches in SARS‐CoV‐2 research and anti‐viral drug discovery is the development of appropriate fluorescent reporters (Figure 1). These fluorescent reporters need to be specific and should not interfere with the biological process under investigation. The first reported modification of the recombinant SARS‐CoV‐2 genome was the insertion of a fluorescent protein to enable SARS‐CoV‐2 basic research and microscopy‐based screening for anti‐viral compounds (Thi Nhu Thao et al., 2020; Xie et al., 2020). The insertion of a sequence encoding for green fluorescent protein (GFP) into a SARS‐CoV‐2 genome by Thi Nhu Thao et al. resulted in partial deletion of ORF7a protein and led to reduced SARS‐CoV‐2 replication kinetics compared to the wild‐type virus. Nevertheless, it was demonstrated that, despite reduced kinetics, the reporter could be used in anti‐viral screening applications as well as virus neutralisation assays (Thi Nhu Thao et al., 2020). On the other hand, insertion of the gene encoding for the mNeonGreen fluorescent protein replacing the entire ORF7a produced a virus with similar replication kinetics to the wild‐type virus (Xie et al., 2020).
FIGURE 1.
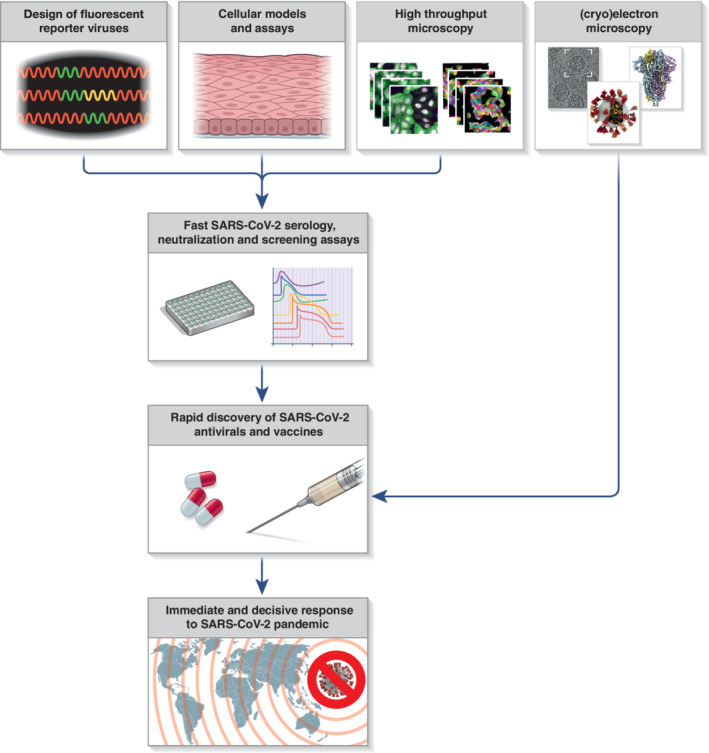
A scheme depicting the contribution of advanced microscopy and related technologies to rapid response to SARS‐CoV‐2 pandemic
Another milestone in application of high‐throughput microscopy to fight the SARS‐CoV‐2 pandemic is the development of an appropriate cellular assay to monitor virus infectivity (Figure 1). As it is common for all coronaviruses, viral protease 3CLpro is cleaving the viral polyproteins into individual non‐structural proteins, and it is essential for the viral replication (Harcourt et al., 2004; Thiel et al., 2003). Froggatt et al. developed a cellular reporter system based on flipGFP protein that changes fluorescent properties upon cleavage by the viral protease 3CLpro (Froggatt, Heaton, & Heaton, 2020). Changes in fluorescence reflect 3CLpro activity and, in turn, can be used to evaluate SARS‐CoV‐2 infection. Also relying on the 3CLpro activity, a parallel assay was developed that employed an endoplasmic reticulum (ER)‐anchored GFP molecule tagged with a 3CLpro cleavage site and nuclear localization signal sequence, which promotes GFP translocation from the ER to the nucleus upon SARS‐CoV‐2 infection (Pahmeier et al., 2020). Importantly, both assays allow for anti‐viral drug screening in fixed or living cells (Froggatt et al., 2020; Pahmeier et al., 2020). In addition, these assays enable discovery of direct inhibitors of 3CLpro, one of the main targets for pharmacological intervention against SARS‐CoV‐2, to be conducted under biosafety level 1 or 2 (BSL1 or BSL2) containment, which is critical for rapid drug discovery. Repurposing of known and clinically evaluated drugs is another avenue for accelerating discovery and deployment of anti‐SARS‐CoV‐2 therapies. For this reason, small‐ and large‐scale microscopy‐based screens of clinical‐stage or Food and Drug Administration–approved small molecules for their potential to inhibit SARS‐CoV‐2 replication have been conducted, identifying >100 anti‐SARS‐CoV‐2 candidate therapeutic drugs (Jeon et al., 2020; Riva et al., 2020).
2.2. High‐throughput microscopy in SARS‐CoV‐2 antibody diagnostics
Testing for SARS‐CoV‐2 infection and tracking of its transmission are essential to control viral spread (Figure 1). Standardised quantitative RT‐PCR tests have proven to be a good method to detect acutely infected individuals (Corman et al., 2020). However, there is a large number of undocumented SARS‐CoV‐2 cases arising from limitations in testing capacity and asymptomatic individuals (Mizumoto, Kagaya, Zarebski, & Chowell, 2020; Qiu et al., 2020). One way to track the true extent of the virus spread in the population is to monitor the presence of SARS‐CoV‐2‐specific antibodies using serological tests. In addition, these tests can provide information on the levels of anti‐viral antibodies, the serological response against different viral proteins, as well as the durability of anti‐viral immune responses, which are all key elements in developing successful vaccines and vaccination strategies.
Immuofluorescence microscopy using virus‐infected cells as a specimen is a classical approach in virology to assess the presence of specific anti‐viral antibodies and has been applied to previous coronavirus infections such as SARS‐CoV and MERS‐CoV (Chan et al., 2004; Manopo et al., 2005; Meyer, Drosten, & Müller, 2014). The first publications on antibody assessment of COVID‐19 patients relied on immunofluorescence microscopy (Wölfel et al., 2020). This is not surprising since immunofluorescence microscopy‐based assays do not depend on particular commercial solutions and can be rapidly deployed from the moment the first isolate of the pathogen has been obtained and a susceptible cell model discovered. The major disadvantage of using immunofluorescence microscopy in serology is its limited throughput capacity due to manual microscopy handling steps during image acquisition and sample evaluation based on visual inspection of micrographs which is subjective and not quantitative. These limitations have been addressed by Pape et al., where a high‐throughput microscopy‐compatible procedure was developed together with a robust machine learning‐based image analysis pipeline yielding an assay with slightly better specificity/sensitivity than a commercially available ELISA assay at the time (Pape et al., 2020). Microscopy‐based serological tests have been used in large population SARS‐CoV‐2 serology studies mainly as a confirmatory assay to provide increased specificity of detection in the setting of low antibody prevalence (Stringhini et al., 2020; Tönshoff et al., 2021). Increased specificity potential of the microscopy‐based assays compared to ELISA and others is probably due to expression of viral antigens in the cellular context ensuring correct protein folding and appropriate post‐translational modifications. A study using 293 T‐cells expressing the spike protein of SARS‐CoV‐2 and using FACS for a readout has shown increased specificity of that assay compared to other approaches (Grzelak et al., 2020).
The presence of specific anti‐viral antibodies in patient sera does not always correlate with neutralising capacity. Therefore, additional assays are used to assess the antibody neutralisation capacity, often by using time‐consuming plaque reduction neutralisation tests (Perera et al., 2020). Muruato et al. have developed a microscopy‐based virus neutralisation assay (Muruato et al., 2020) using SARS‐CoV‐2 construct harbouring mNeonGreen fluorescent protein (Xie et al., 2020). This approach has shortened the assay turnaround time by several days and increased sensitivity compared to a standard plaque assay. This is an important step towards the successful vaccine development, and the assay has been used to support clinical trials for COVID‐19 vaccine candidates (Mulligan et al., 2020). Furthermore, neutralising antibodies as well as COVID‐19 convalescent patient plasma have shown clinical benefits (Shen et al., 2020) underscoring the importance of fast neutralisation capacity assessment via microscopy‐based assays.
3. ELECTRON MICROSCOPY IN COVID‐19 RESEARCH
3.1. Electron microscopy as diagnostic tool for SARS‐CoV‐2
Negative staining, in which the sample is directly applied on carbon‐coated EM grids, stained with a thin layer of heavy metal salts and then observed by transmission electron microscopy (TEM) is a robust technique to screen, detect and identify infectious agents. Combined with immunolabelling, it has been used as a diagnostic technique to identify viruses in body fluids and faeces (Roingeard, Raynal, Eymieux, & Blanchard, 2019). Direct visualisation by EM is especially useful in the absence of specific molecular probes and assays such as when a novel pathogen emerges. Indeed, EM contributed to the identification of the viral pathogen responsible for the 2003 SARS epidemic, the first epidemic of the 21th century, confirming the presence of viruses‐like particles showing the ‘crown’ of glycoproteins on their surface, characteristic of the Coronaviridae, both in supernatants of cells inoculated with patients saliva and in lung secretion fluids from bronchoalveolar lavage samples (Drosten et al., 2003; Ksiazek et al., 2003).
SARS‐CoV‐2 viral RNA and proteins have been identified in bodily fluids, such as blood and urine, as well as in secretions and stool from COVID‐19‐positive patients, suggesting systemic spread of SARS‐CoV‐2 infection in different organs (Bradley et al., 2020). Analysis of COVID‐19 patient tissues collected either from biopsies or from autopsy specimens has indicated the presence of viral RNA and proteins in several compartments other than the respiratory tracts, such as kidneys, liver, heart, gastrointestinal tract, olfactory mucosa and brain (reviewed in [Trypsteen, Van Cleemput, van Snippenberg, Gerlo, & Vandekerckhove, 2020]). To substantiate these findings, several reports showed TEM micrographs of the infected tissues with morphological alterations and viral particles resembling those reported in cell culture models (Bradley et al., 2020; Dittmayer et al., 2020; Martines et al., 2020; Meinhardt et al., 2020) (Figure 2). However, common cellular structures such as clathrin‐coated pits, multivesicular bodies containing several intraluminal vesicles and ribosome‐decorated ER membranes can be misinterpreted as SARS‐CoV‐2 virions (Hopfer et al., 2021). Additionally, poor ultrastructure preservation due to autolysis of autoptic samples can lead to ambiguous identification of viral structures (Hopfer et al., 2021; Neil et al., 2020). Thus, controversies arose concerning the correct interpretation of cellular architecture and viral‐associated structures in several articles reporting EM images obtained from COVID‐19 patient samples (Dittmayer et al., 2020; Hopfer et al., 2021; Miller & Goldsmith, 2020). While infectious SARS‐CoV‐2 viruses can be isolated from patients' nasopharyngeal or oropharyngeal swabs, no solid evidence of direct visualisation by EM of SARS‐CoV‐2 virions in these specimens has been produced. In addition, even if virus‐like particles can be visualised, specific immunolabelling using SARS‐CoV‐2 antisera should be performed in order to demonstrate the presence of bona fide SARS‐CoV‐2 virions. Thus, direct visualisation of SARS‐CoV‐2 from patient swabs remains an open challenge.
FIGURE 2.
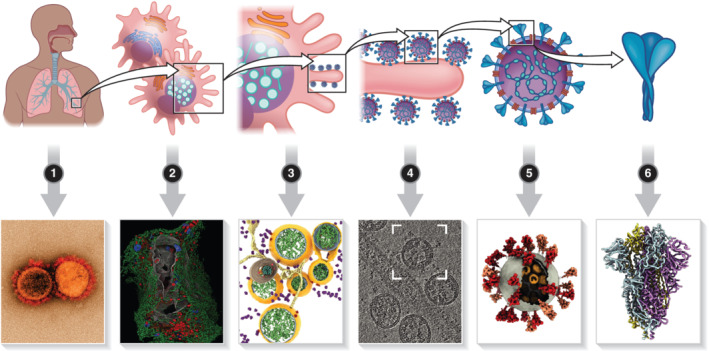
Electron microscopy in COVID‐19 research. 1) Electron microscopy analysis of COVID‐19 patient biological fluids or tissue has been used to identify SARS‐CoV‐2 virions and infected cells; 2) the large field of view provided by FIB‐SEM allowed to define the morphological alterations induced by SARS‐CoV‐2 infection on the whole infected cells, revealing the network of connection between the cellular organelles and the viral ROs; 3) high‐resolution electron tomography and in situ cryo‐FIB‐SEM have been used to investigate the biogenesis and structural organisation of SARS‐CoV‐2 replication organelles; 4–6) cryo‐EM was used to characterise the molecular architecture of SARS‐CoV‐2 virions and the structure, distribution and assembly of macro‐molecular complexes such as the spike protein trimers and the ribonucleoproteins. Image in panel (1) Courtesy: National Institute of Allergy and Infectious Diseases. Images in panels (2–5) adapted from (Cortese et al., 2020; Wolff et al., 2020; Klein et al., 2020; Yao et al., 2020; Walls et al., 2020) respectively
3.2. Electron microscopy analysis of SARS‐CoV‐2 mechanisms of replication and pathogenesis
EM of embedded specimens such as infected cell culture or tissues can provide information on the viral replication cycle as well as the cytopathogenic effects induced by virus infection. A substantial body of literature has been produced in the past years describing the host‐cell morphological alterations induced by members of the Coronaviridae family using TEM of thin sections and electron tomography (ET) techniques (Knoops et al., 2008; Maier et al., 2013; Snijder et al., 2020). ET in particular, which involves the acquisition of a series of tilted 2D projections that are later reconstructed to retrieve the 3D volumetric information, allowed for the definition of the ultrastructural architecture in infected cells, providing useful insight into the different steps of the viral replication cycle (Romero‐Brey & Bartenschlager, 2014, 2015). Upon infection, the coronavirus positive‐sense single‐strand RNA genome is directly translated in association with the host rough ER membranes. Expression of the viral proteins, such as the replicase complex responsible for viral RNA replication, induces an extensive remodelling of the cellular endomembrane systems, generating the specialised membrane compartments often referred to as viral replication organelles (ROs). EM imaging of coronaviruses‐infected cells showed the presence of double‐membrane vesicles (DMVs) together with a network of smooth ER tubules designated ‘convoluted membranes’ or ‘zippered ER’ due to the narrow luminal space (Figure 2). In addition, smaller double‐membrane spherules, open vesicles originating from wrapping of the zippered ER were also observed. Due to the spatial proximity of these elements composing the coronaviruses' ROs, it was difficult to precisely define the structures harbouring viral RNA synthesis. Metabolic labelling combined with quantitative electron microscopy ultimately showed that the synthesis of novel RNA was associated with the DMVs, indicating a central role for these structures in the viral replication cycle (Snijder et al., 2020).
Time‐resolved comparative EM analysis of SARS‐CoV‐1‐ and SARS‐CoV‐2‐infected cells showed striking parallels between the two viruses. Similar virus‐induced structures developed at comparable time‐points post infection in VeroE6 African green monkey cells (Ogando et al., 2020). Semi‐automated ET acquisition and 3D reconstruction performed on infected lung epithelial cells provided additional information on the structural organisation of SARS‐CoV‐2 ROs and allowed for the development of a model describing the biogenesis of DMVs (Cortese et al., 2020). First, DMVs are formed within rough ER membranes. Subsequently, DMV growth stretches and pulls the ER membranes generating the narrow‐spaced zippered ER. Due to their structural role in connecting the DMVs with the host ER membranes, the zippered ER has been recently redefined as ER ‘connectors’ (Cortese et al., 2020).
Development of specialised instrumentation, including novel sample preparation techniques and software for automated image acquisition, has streamlined the process for imaging large biological specimens. In particular, this enabled focus‐ion beam scanning electron microscopy (FIB‐SEM), a technique that combines cycles of automatic etching of the specimen surface with sequential imaging by SEM, to achieve isotropic high‐resolution imaging of large volumes, from whole cells to entire organisms (Xu et al., 2017). Integrative microscopy studies that coupled FIB‐SEM and 3D reconstruction of the whole infected cells together with ET, live cell imaging and super‐resolution light microscopy have defined additional SARS‐CoV‐2‐induced cellular modifications that alter host‐cell homeostasis and function. These studies demonstrate that SARS‐CoV‐2 replication affects not only the ER but also numerous other cellular organelles including causing fragmentation of the Golgi apparatus, alterations in the morphology and molecular composition of the mitochondria, increase in peroxisome number and extensive remodelling of the cytoskeleton to form a cage‐like structure surrounding the viral ROs (Cortese et al., 2020) (Figure 2). Similar alterations of the cytoskeletal network have been previously described for Zika virus, another positive‐strand RNA virus (Cortese et al., 2017), suggesting that studying the ultrastructure of the infected cells can allow for the identification of unexpected parallels between distantly related viruses with the potential to provide novel targets for broad‐spectrum anti‐viral therapies.
3.3. Cryo‐electron microscopy of SARS‐CoV‐2 in vaccine and anti‐viral drug development
Analysis of vitrified specimens, preserved in a frozen‐hydrated status by means of rapid cooling with cryogenic liquid under high pressure, can provide near‐atomic resolved structural details of the fully hydrated biological material in its native conformation. The high resolution achieved through cryo‐EM approaches can provide information on the structural–functional relationship of the different viral components, expanding our understanding of viral infection and pathogenesis. The first cryo‐EM structure related to SARS‐CoV‐2, the 3.5 Å‐resolution trimeric spike ectodomain, was deposited in the Protein Data Bank (PDB) at the end of February 2020, less than 2 months after the first genome sequence of SARS‐CoV‐2 was reported (Wrapp et al., 2020) (Figure 2). Following this first entry, the number of deposited structures increased rapidly reaching up to 185 entries by the end of December 2020. Most of the structures, up to 143, describe different structural conformations of the spike glycoprotein (S protein) that decorates the SARS‐CoV‐2 virus envelope producing the characteristic crown‐like appearance on the virion surface. It is not surprising that most of the efforts are focused on the structural characterisation of SARS‐CoV‐2 spike glycoprotein, since it is responsible for receptor binding and entry into the host cell and, therefore, the main component of most COVID‐19 vaccines (Krammer, 2020). SARS‐CoV‐2 S protein is also the primary antigenic component responsible for inducing host immune response, including neutralising antibody production and protective immunity against viral infection (Krammer, 2020). Cryo‐EM studies have been instrumental in defining the structure of the S protein under different conformational states (pre and post fusion), under different pH conditions, in complex with its cellular receptor (angiotensin convertase enzyme 2 [ACE2]), and with neutralising antibodies or antibody fragments (Benton et al., 2020; Cai et al., 2020; Custódio et al., 2020; Lv et al., 2020; Zhou et al., 2020). Moreover, several studies used cryo‐electron tomography to define the structure of the S protein and other SARS‐CoV‐2 structural proteins in situ, using purified virus preparations or cells infected directly on EM grids (Ke et al., 2020; Turoňová et al., 2020; Yao et al., 2020) (Figure 2). These in situ studies revealed the presence of flexible hinges in the stalk region of the S protein homotrimer that allowed the protein to be tilted up to 90° towards the viral membrane. The large conformational space might influence the interaction between the S protein and the ACE2 receptor or hinder the access of antibodies to epitopes located in the stalk region. In addition, they allowed for a precise definition of the glycan shell that coats the S protein surface and might therefore shield important epitopes from the binding of neutralising antibodies (Ke et al., 2020; Turoňová et al., 2020). Together, this structural information has provided one of the major focal points for the rapid development of vaccines.
The viral genome encodes for several enzymes that are essential for viral genome replication. Such proteins represent the ideal target for developing anti‐viral compounds. Indeed, several drugs currently available to suppress otherwise deadly viruses such as hepatitis C virus and HIV target essential enzymatic activities including virus‐encoded polymerases and proteases (Arts & Hazuda, 2012; Götte & Feld, 2016). Accurate high‐resolution structures are instrumental in guiding and supporting rational drug design and in explaining the biological functions of bioactive compounds. Cryo‐EM was used to determine the structure of SARS‐CoV‐2 replication and transcription complex (RTC), a large macromolecular machinery formed by the viral RNA–dependent RNA polymerase (RdRp) nsp12, together with several other non‐structure proteins. This structure was resolved in complex with an RNA template and the nucleoside analogue remdesivir, the only currently approved direct anti‐viral that inhibits viral replication (Yin et al., 2020). The 2.5 Å resolution of the RTC complex allowed for identification of key residues within the RdRp active site that are involved in the interaction with the nucleoside analogue and revealed how remdesivir incorporation into the primer strand is responsible for the delayed termination of RNA chain elongation. Moreover, the authors highlighted structural similarities between the SARS‐CoV‐2 RdRp and polymerase complexes from other positive‐strand RNA viruses, suggesting a conserved mechanism of genome replication and thus providing structural models for future development of broad‐spectrum anti‐virals.
Finally, a major breakthrough in in situ structural microscopy was provided by combining cryo‐ET with FIB milling. Conventional cryo‐EM is limited to specimens thinner than approximately 300 nm thin but cannot be used to visualise entire cells whose thickness far exceeds this limit. In cryo‐FIB approaches, thin lamellae are milled at cryogenic temperatures within specific regions of the vitrified specimen, allowing for subsequent analysis through transmission electron microscopy (TEM) and tomography. This allows to image the cell interior and obtain structural information of macromolecular complexes in their cellular context (Villa, Schaffer, Plitzko, & Baumeister, 2013). In situ cryo‐ET performed on cryo‐FIB‐milled lamellae, allowed for the visualisation of ROs, from chemically inactivated SARS‐CoV‐2, in close‐to‐native conditions (Klein et al., 2020). Bundles of branched double‐stranded RNA filaments were observed within the DMV interior. Since the viral genome forms a double‐stranded RNA intermediate form during its replication, this finding supports the hypothesis that the DMV interior is the site of viral genome replication. Additionally, in several regions, the inner and outer DMVs membranes clamped together to form pore‐like openings. This observation is in line with an elegant in‐situ cryo‐ET study of murine hepatitis virus, a murine coronavirus, that showed the presence of several proteinaceous pores on the surface of the DMVs (Wolff et al., 2020). Subtomogram averaging indicated that this double‐membrane‐spanning molecular complex was mainly composed of the non‐structural protein nsp3, one of the viral proteases, arranged in a six‐fold symmetry around the channel opening. With this combined data, we can begin to build a model where newly synthesized viral genomes, exported through the pore, would associate with ribosomes to be translated or with the nucleocapsid (N) protein, to form ribonucleocapsids (RNP). The RNPs are then assembled in progeny virions through budding either into the Golgi cisternae or in membranes derived from the ER–Golgi intermediate compartment (ERGIC). Moreover, due to its multifunctional role as viral protease and ROs ‘gatekeeper’, compounds that target nsp3 could inhibit multiple essential functions at once, making this protein an ideal candidate for development of novel anti‐viral strategies.
4. CONCLUSION
The ongoing SARS‐CoV‐2 pandemic has illustrated the importance of a rapid response to emerging infectious diseases. SARS‐CoV‐2 has spread globally within a few months of its first observation, leading the WHO to declare a pandemic in March 2020. Important public health decisions have had to be made on a weekly, if not daily basis, and the biomedical field as well as the other scientific fields have had to tailor their technologies and procedures to meet these new demands. With an unprecedented combined effort, the scientific community has risen to the challenge by rapidly accumulating knowledge on the new virus and developing tests, procedures and medical treatments in order to restrain the SARS‐Cov‐2 pandemic. This effort is exemplified by the 180 vaccines currently in various stages of development (Krammer, 2020) and the first vaccines being administered to the general population within 1 year of the first infection report. Microscopy has played a major role in these efforts as a cornerstone technology in biomedical and infectious diseases research. In this review, we outlined the advanced microscopy technologies, such as high‐throughput screening and (cryo) electron microscopy, as vital technologies in the fight against SARS‐CoV‐2 (Figure 1). Techniques for rapid viral genome cloning and fluorescent protein tagging (Thi Nhu Thao et al., 2020; Xie et al., 2020) together with cellular model system (Froggatt et al., 2020; Pahmeier et al., 2020) and high‐throughput microscopy have enabled the testing of reagents for their inhibitory effect against SARS‐CoV‐2. Cryo‐electron tomography has been used to determine virion and viral protein structures within months of the first observed SARS‐CoV‐2 infections, enabling structure‐guided drug design and giving rise to therapeutic antibodies with affinities in femtomolar range and neutralisation capacity in picomolar range (Schoof et al., 2020). Studies on SARS‐CoV‐2‐induced cellular ultrastructural changes have been vital in understanding viral impact on infected host cells and have provided clues for potential therapeutic procedures (Cortese et al., 2020; Ogando et al., 2020; Wolff et al., 2020). Microscopy‐based serological assays enabled specific, sensitive and quantitative assessment of SARS‐CoV‐2‐specific antibodies and proved to be especially useful in situations where commercial assays are either not yet developed or in short supply due to a high demand (Pape et al., 2020). Moreover, the employment of microscopy‐based virus neutralisation assays paved the way to fast vaccine development (Mulligan et al., 2020; Muruato et al., 2020). Currently, the major bottleneck for wider employment of microscopy technologies in the response to SARS‐CoV‐2 is the deficiency of microscopy infrastructure under enhanced biosafety containment (biosafety level 2, 3 or 4), especially in hospital settings, as well as the requirement for high technical expertise.
While in recent years, molecular techniques, such as PCR, ELISA and next generation sequencing, have replaced EM as the primary approach to identify pathogens, the ‘open view’ granted by EM observation still offers several advantages. For instance, it allows to investigate a sample without prior assumptions about the nature of the observed pathogen. In addition, in scenarios where a novel pathogen arises, the absence of specific probes can hamper the ability of molecular and serological assays to detect the novel agent, while direct visualisation through EM can allow for its rapid identification. However, high expertise is needed for proper identification of the viral ultrastructural morphologies, and great care has to be taken to avoid misidentifying common cellular structures as virions. The COVID‐19 pandemic has clearly demonstrated the importance of rapid communication of scientific data to provide early visibility and broad dissemination among the scientific community and the general public. Nevertheless, one has to be careful to prevent ambiguous information finding their way into the scientific literature. Given the technical requirement for both proper image acquisition and the complexity of image analysis with advanced microscopy techniques, it is vital to take special care when interpreting these studies. The publication of potentially ambiguous EM micrographs derived from COVID‐19 patient samples has prompted researchers to request that authors and editors conduct more rigorous assessment of EM data in order to prevent the spread of misleading or false information (Dittmayer et al., 2020; Miller & Goldsmith, 2020). In this direction, dedicated web resources have been developed to sort, organise and curate COVID‐19‐related information deposited in the public databases (Brzezinski et al., 2021), such as the structural data of SARS‐CoV‐2 proteins deposited in the PDB server. Overall, the pressure that the current pandemic has exerted on the scientific community favoured the long‐sought evolution towards a more open, transparent and direct way of communicating the research results. For the microscopy community, this should translate to the practise of making raw datasets available for published work. This will allow not only for a more transparent evaluation during the peer‐review process, but also permits other groups to re‐analyse those datasets, thus fostering novel and potentially unanticipated discoveries. Cortese et al. have taken a step in this direction by sharing all the raw data associated with our EM analysis of SARS‐CoV‐2‐infected cells. Data have been uploaded on EMPIAR, a public repository for EM datasets where direct access and visualisation through the MoBIE plugin of the FIJI software, a framework for sharing and browsing of multimodal big image data (Vergara et al., 2020) was provided. Big‐data viewers such as MoBIE do not require the download of the data to visualise the dataset content, a great advantage considering that EM data can easily reach terabytes size, and thus combine date share with a user‐friendly experience.
In summary, during SARS‐CoV‐2 pandemic, advanced microscopy technologies have emerged to be especially relevant for rapid response by facilitating drug and vaccine discovery, serological testing and the accumulation of knowledge surrounding virus pathophysiology. Furthermore, many of the developed microscopy‐based procedures are not necessarily SARS‐CoV‐2 specific but can be made generally applicable. This makes microscopy technologies uniquely positioned to act as the first line of defence against any emerging infection in the future.
ACKNOWLEDGEMENTS
We are grateful to Dr. Christopher J. Neufeldt for the useful suggestions and the critical reading of our manuscript. We would like to thank Deutsches Zentrum fuer Infektionsforschung (DZIF) (VL: project TTU 04.705) for funding. Individual images used in the Figure 1 courtesy of medical illustrations database—https://smart.servier.com/. The authors declare they have no conflicts of interest.
Cortese M, Laketa V. Advanced microscopy technologies enable rapid response to SARS‐CoV‐2 pandemic. Cellular Microbiology. 2021;23:e13319. 10.1111/cmi.13319
Funding information Deutsches Zentrum für Infektionsforschung, Grant/Award Number: TTU 04.705
Contributor Information
Mirko Cortese, Email: mirko.cortese@med.uni-heidelberg.de.
Vibor Laketa, Email: vibor.laketa@med.uni-heidelberg.de.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study
REFERENCES
- Arts, E. J. , & Hazuda, D. J. (2012). HIV‐1 antiretroviral drug therapy. Cold Spring Harbor Perspectives in Medicine, 2(4), a007161–a007161. 10.1101/cshperspect.a007161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton, D. J. , Wrobel, A. G. , Xu, P. , Roustan, C. , Martin, S. R. , Rosenthal, P. B. , … Gamblin, S. J. (2020). Receptor binding and priming of the spike protein of SARS‐CoV‐2 for membrane fusion. Nature, 588(7837), 327–330. 10.1038/s41586-020-2772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickle, M. (2010). The beautiful cell: High‐content screening in drug discovery. Analytical and Bioanalytical Chemistry, 398(1), 219–226. 10.1007/s00216-010-3788-3 [DOI] [PubMed] [Google Scholar]
- Boutros, M. , Heigwer, F. , & Laufer, C. (2015). Microscopy‐based high‐content screening. Cell, 163(6), 1314–1325. 10.1016/j.cell.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Bradley, B. T. , Maioli, H. , Johnston, R. , Chaudhry, I. , Fink, S. L. , Xu, H. , … Marshall, D. A. (2020). Histopathology and ultrastructural findings of fatal COVID‐19 infections in Washington state: A case series. The Lancet, 396(10247), 320–332. 10.1016/S0140-6736(20)31305-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin, P. , & Christophe, T. (2011). High‐content screening in infectious diseases. Current Opinion in Chemical Biology, 15(4), 534–539). Elsevier Current Trends. 10.1016/j.cbpa.2011.05.023 [DOI] [PubMed] [Google Scholar]
- Brzezinski, D. , Kowiel, M. , Cooper, D. R. , Cymborowski, M. , Grabowski, M. , Wlodawer, A. , … Minor, W. (2021). Covid‐19.bioreproducibility.org: A web resource for SARS‐CoV‐2‐related structural models. Protein Science, 30(1), 115–124. 10.1002/pro.3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, Y. , Zhang, J. , Xiao, T. , Peng, H. , Sterling, S. M. , Walsh, R. M. , … Chen, B. (2020). Distinct conformational states of SARS‐CoV‐2 spike protein. Science, 369(6511), eabd4251. 10.1126/science.abd4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, P. K. S. , Ng, K. C. , Chan, R. C. W. , Lam, R. K. Y. , Chow, V. C. Y. , Hui, M. , … Tam, J. S. (2004). Immunofluorescence assay for serologic diagnosis of SARS. Emerging Infectious Diseases, 10(3), 530–532. 10.3201/eid1003.030493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chojnacki, J. , Staudt, T. , Glass, B. , Bingen, P. , Engelhardt, J. , Anders, M. , … Kräusslich, H.‐G. (2012). Maturation‐dependent HIV‐1 surface protein redistribution revealed by fluorescence nanoscopy. Science, 338(September), 524–529. [DOI] [PubMed] [Google Scholar]
- Corman, V. M. , Landt, O. , Kaiser, M. , Molenkamp, R. , Meijer, A. , Chu, D. K. , … Drosten, C. (2020). Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Eurosurveillance, 25(3), 1–8. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese, M. , Goellner, S. , Acosta, E. G. , Neufeldt, C. J. , Oleksiuk, O. , Lampe, M. , … Bartenschlager, R. (2017). Ultrastructural characterization of Zika virus replication factories. Cell Reports, 18(9), 2113–2123. 10.1016/j.celrep.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese, M. , Lee, J.‐Y. , Cerikan, B. , Neufeldt, C. J. , Oorschot, V. M. J. , Köhrer, S. , … Bartenschlager, R. (2020). Integrative imaging reveals SARS‐CoV‐2‐induced reshaping of subcellular morphologies. Cell Host & Microbe, 28(6), 853–866.e5. 10.1016/j.chom.2020.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custódio, T. F. , Das, H. , Sheward, D. J. , Hanke, L. , Pazicky, S. , Pieprzyk, J. , … Löw, C. (2020). Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS‐CoV‐2. Nature Communications, 11(1), 5588. 10.1038/s41467-020-19204-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wit, E. , Van Doremalen, N. , Falzarano, D. , & Munster, V. J. (2016). SARS and MERS: Recent insights into emerging coronaviruses. Nature Reviews Microbiology, 14(8), 523–534. 10.1038/nrmicro.2016.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmayer, C. , Meinhardt, J. , Radbruch, H. , Radke, J. , Heppner, B. I. , Heppner, F. L. , … Laue, M. (2020). Why misinterpretation of electron micrographs in SARS‐CoV‐2‐infected tissue goes viral. The Lancet, 396(10260), e64–e65. 10.1016/S0140-6736(20)32079-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten, C. , Günther, S. , Preiser, W. , van der Werf, S. , Brodt, H.‐R. , Becker, S. , … Doerr, H. W. (2003). Identification of a novel coronavirus in patients with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1967–1976. 10.1056/NEJMoa030747 [DOI] [PubMed] [Google Scholar]
- Froggatt, H. M. , Heaton, B. E. , & Heaton, N. S. (2020). Development of a fluorescence‐based, high‐throughput SARS‐CoV‐2 3CL pro reporter assay. Journal of Virology, 94(22), e01265–20. 10.1128/JVI.01265-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodsell, D. S. , Voigt, M. , Zardecki, C. , & Burley, S. K. (2020). Integrative illustration for coronavirus outreach. PLoS Biology, 18(8), e3000815. 10.1371/journal.pbio.3000815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götte, M. , & Feld, J. J. (2016). Direct‐acting antiviral agents for hepatitis C: Structural and mechanistic insights. Nature Reviews Gastroenterology & Hepatology, 13(6), 338–351. 10.1038/nrgastro.2016.60 [DOI] [PubMed] [Google Scholar]
- Grzelak, L. , Temmam, S. , Planchais, C. , Demeret, C. , Tondeur, L. , Huon, C. , … van der Werf, S. (2020). A comparison of four serological assays for detecting anti–SARS‐CoV‐2 antibodies in human serum samples from different populations. Science Translational Medicine, 12(559), eabc3103. 10.1126/scitranslmed.abc3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt, B. H. , Jukneliene, D. , Kanjanahaluethai, A. , Bechill, J. , Severson, K. M. , Smith, C. M. , … Baker, S. C. (2004). Identification of severe acute respiratory syndrome coronavirus Replicase products and characterization of papain‐like protease activity. Journal of Virology, 78(24), 13600–13612. 10.1128/JVI.78.24.13600-13612.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfer, H. , Herzig, M. C. , Gosert, R. , Menter, T. , Hench, J. , Tzankov, A. , … Miller, S. E. (2021). Hunting coronavirus by transmission electron microscopy—A guide to SARS‐CoV‐2‐associated ultrastructural pathology in COVID‐19 tissues. Histopathology, 78(3), 358–370. 10.1111/his.14264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambor, H. , Antonietti, A. , Alicea, B. , Audisio, T. L. , Auer, S. , Bhardwaj, V. , … Weissgerber, T. L. (2020). Creating clear and informative image‐based figures for scientific publications. BioRxiv. 10.1101/2020.10.08.327718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon, S. , Ko, M. , Lee, J. , Choi, I. , Byun, S. Y. , Park, S. , … Kim, S. (2020). Identification of antiviral drug candidates against SARS‐CoV‐2 from FDA‐approved drugs. Antimicrobial Agents and Chemotherapy, 64(7), e00819–20. 10.1128/AAC.00819-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke, Z. , Oton, J. , Qu, K. , Cortese, M. , Zila, V. , McKeane, L. , … Briggs, J. A. G. (2020). Structures and distributions of SARS‐CoV‐2 spike proteins on intact virions. Nature, 588(7838), 498–502. 10.1038/s41586-020-2665-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, S. , Cortese, M. , Winter, S. L. , Wachsmuth‐Melm, M. , Neufeldt, C. J. , Cerikan, B. , … Chlanda, P. (2020). SARS‐CoV‐2 structure and replication characterized by in situ cryo‐electron tomography. Nature Communications, 11(1), 5885. 10.1038/s41467-020-19619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops, K. , Kikkert, M. , van den Worm, S. H. E. , Zevenhoven‐Dobbe, J. C. , van der Meer, Y. , Koster, A. J. , … Snijder, E. J. (2008). SARS‐coronavirus replication is supported by a Reticulovesicular network of modified endoplasmic reticulum. PLoS Biology, 6(9), e226. 10.1371/journal.pbio.0060226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer, F. (2020). SARS‐CoV‐2 vaccines in development. Nature, 586(7830), 516–527. 10.1038/s41586-020-2798-3 [DOI] [PubMed] [Google Scholar]
- Ksiazek, T. G. , Erdman, D. , Goldsmith, C. S. , Zaki, S. R. , Peret, T. , Emery, S. , … Anderson, L. J. (2003). A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1953–1966. 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- Laketa, V. (2018). Microscopy in infectious disease research—Imaging across scales. In Journal of Molecular Biology (430, 17, pp. 2612–2625). Academic Press. 10.1016/j.jmb.2018.06.018 [DOI] [PubMed] [Google Scholar]
- Lv, Z. , Deng, Y. Q. , Ye, Q. , Cao, L. , Sun, C. Y. , Fan, C. , … Wang, X. (2020). Structural basis for neutralization of SARS‐CoV‐2 and SARS‐CoV by a potent therapeutic antibody. Science, 369(6509), 1505–1509. 10.1126/SCIENCE.ABC5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier, H. J. , Hawes, P. C. , Cottam, E. M. , Mantell, J. , Verkade, P. , Monaghan, P. , … Britton, P. (2013). Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. MBio, 4(5), e00801–e00813. 10.1128/mBio.00801-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manopo, I. , Lu, L. , He, Q. , Chee, L. L. , Chan, S. W. , & Kwang, J. (2005). Evaluation of a safe and sensitive spike protein‐based immunofluorescence assay for the detection of antibody responses to SARS‐CoV. Journal of Immunological Methods, 296(1–2), 37–44. 10.1016/j.jim.2004.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martines, R. B. , Ritter, J. M. , Matkovic, E. , Gary, J. , Bollweg, B. C. , Bullock, H. , … Zaki, S. R. (2020). Pathology and pathogenesis of SARS‐CoV‐2 associated with fatal coronavirus disease, United States. Emerging Infectious Diseases, 26(9), 2005–2015. 10.3201/eid2609.202095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhardt, J. , Radke, J. , Dittmayer, C. , Franz, J. , Thomas, C. , Mothes, R. , … Heppner, F. L. (2020). Olfactory transmucosal SARS‐CoV‐2 invasion as a port of central nervous system entry in individuals with COVID‐19. Nature Neuroscience, 24, 168–175. 10.1038/s41593-020-00758-5 [DOI] [PubMed] [Google Scholar]
- Meyer, B. , Drosten, C. , & Müller, M. A. (2014). Serological assays for emerging coronaviruses: Challenges and pitfalls. Virus Research, 194, 175–183. 10.1016/j.virusres.2014.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S. E. , & Goldsmith, C. S. (2020). Caution in identifying coronaviruses by electron microscopy. Journal of the American Society of Nephrology, 31(9), 2223–2224. 10.1681/ASN.2020050755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto, K. , Kagaya, K. , Zarebski, A. , & Chowell, G. (2020). Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID‐19) cases on board the diamond princess cruise ship, Yokohama, Japan, 2020. Eurosurveillance, 25(10), 2000180. 10.2807/1560-7917.ES.2020.25.10.2000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan, M. J. , Lyke, K. E. , Kitchin, N. , Absalon, J. , Gurtman, A. , Lockhart, S. , … Jansen, K. U. (2020). Phase I/II study of COVID‐19 RNA vaccine BNT162b1 in adults. Nature, 586(7830), 589–593. 10.1038/s41586-020-2639-4 [DOI] [PubMed] [Google Scholar]
- Muranyi, W. , Malkusch, S. , Müller, B. , Heilemann, M. , & Kräusslich, H. G. (2013). Super‐resolution microscopy reveals specific recruitment of HIV‐1 envelope proteins to viral assembly sites dependent on the envelope C‐terminal tail. PLoS Pathogens, 9(2), e1003198. 10.1371/journal.ppat.1003198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruato, A. E. , Fontes‐Garfias, C. R. , Ren, P. , Garcia‐Blanco, M. A. , Menachery, V. D. , Xie, X. , & Shi, P.‐Y. (2020). A high‐throughput neutralizing antibody assay for COVID‐19 diagnosis and vaccine evaluation. Nature Communications, 11(1), 4059. 10.1038/s41467-020-17892-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil, D. , Moran, L. , Horsfield, C. , Curtis, E. , Swann, O. , Barclay, W. , … Roufosse, C. (2020). Ultrastructure of cell trafficking pathways and coronavirus: How to recognise the wolf amongst the sheep. The Journal of Pathology, 252(4), 346–357. 10.1002/path.5547 [DOI] [PubMed] [Google Scholar]
- Ogando, N. S. , Dalebout, T. J. , Zevenhoven‐Dobbe, J. C. , Limpens, R. W. A. L. , van der Meer, Y. , Caly, L. , … Snijder, E. J. (2020). SARS‐coronavirus‐2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. Journal of General Virology, 101, 925–940. 10.1099/jgv.0.001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahmeier, F. , Neufeldt, C. J. , Cerikan, B. , Prasad, V. , Pape, C. , Laketa, V. , … Cortese, M. (2020). A versatile reporter system to monitor virus infected cells and its application to dengue virus and SARS‐CoV‐2. Journal of Virology, 17, 2020. 10.1128/JVI.01715-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, C. , Remme, R. , Wolny, A. , Olberg, S. , Wolf, S. , Cerrone, L. , … Laketa, V. (2020). Microscopy‐based assay for semi‐quantitative detection of SARS‐CoV‐2 specific antibodies in human sera. BioEssays, 43(3), 2000257. 10.1002/bies.202000257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera, R. A. , Mok, C. K. , Tsang, O. T. , Lv, H. , Ko, R. L. , Wu, N. C. , … Peiris, M. (2020). Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), march 2020. Eurosurveillance, 25(16), 2000421. 10.2807/1560-7917.ES.2020.25.16.2000421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu, H. , Wu, J. , Hong, L. , Luo, Y. , Song, Q. , & Chen, D. (2020). Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID‐19) in Zhejiang, China: An observational cohort study. The Lancet Infectious Diseases, 20(6), 689–696. 10.1016/S1473-3099(20)30198-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva, L. , Yuan, S. , Yin, X. , Martin‐Sancho, L. , Matsunaga, N. , Pache, L. , … Chanda, S. K. (2020). Discovery of SARS‐CoV‐2 antiviral drugs through large‐scale compound repurposing. Nature, 586(7827), 113–119. 10.1038/s41586-020-2577-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roingeard, P. , Raynal, P.‐I. , Eymieux, S. , & Blanchard, E. (2019). Virus detection by transmission electron microscopy: Still useful for diagnosis and a plus for biosafety. Reviews in Medical Virology, 29(1), e2019. 10.1002/rmv.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero‐Brey, I. , & Bartenschlager, R. (2014). Membranous replication factories induced by plus‐Strand RNA viruses. Viruses, 6(7), 2826–2857. 10.3390/v6072826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero‐Brey, I. , & Bartenschlager, R. (2015). Viral infection at high magnification: 3D electron microscopy methods to analyze the architecture of infected cells. Viruses, 7(12), 6316–6345. 10.3390/v7122940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof, M. , Faust, B. , Saunders, R. A. , Sangwan, S. , Rezelj, V. , Hoppe, N. , … Manglik, A. (2020). An ultrapotent synthetic nanobody neutralizes SARS‐CoV‐2 by stabilizing inactive spike. Science, 370(6523), 1473–1479. 10.1126/science.abe3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, C. , Wang, Z. , Zhao, F. , Yang, Y. , Li, J. , Yuan, J. , … Liu, L. (2020). Treatment of 5 critically ill patients with COVID‐19 with convalescent plasma. JAMA, 323(16), 1582–1589. 10.1001/jama.2020.4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm, J. , Klambauer, G. , Arany, A. , Steijaert, M. , Wegner, J. K. , Gustin, E. , … Ceulemans, H. (2018). Repurposing high‐throughput image assays enables biological activity prediction for drug discovery. Cell Chemical Biology, 25(5), 611–618.e3. 10.1016/j.chembiol.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder, E. J. , Limpens, R. W. A. L. , de Wilde, A. H. , de Jong, A. W. M. , Zevenhoven‐Dobbe, J. C. , Maier, H. J. , … Bárcena, M. (2020). A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biology, 18(6), e3000715. 10.1371/journal.pbio.3000715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringhini, S. , Wisniak, A. , Piumatti, G. , Azman, A. S. , Lauer, S. A. , Baysson, H. , … Guessous, I. (2020). Seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in Geneva, Switzerland (SEROCoV‐POP): A population‐based study. The Lancet, 396(10247), 313–319. 10.1016/S0140-6736(20)31304-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Nhu Thao, T. , Labroussaa, F. , Ebert, N. , V'kovski, P. , Stalder, H. , Portmann, J. , … Thiel, V. (2020). Rapid reconstruction of SARS‐CoV‐2 using a synthetic genomics platform. Nature, 582(7813), 561–565. 10.1038/s41586-020-2294-9 [DOI] [PubMed] [Google Scholar]
- Thiel, V. , Ivanov, K. A. , Putics, Á. , Hertzig, T. , Schelle, B. , Bayer, S. , … Ziebuhr, J. (2003). Mechanisms and enzymes involved in SARS coronavirus genome expression. Journal of General Virology, 84(9), 2305–2315. 10.1099/vir.0.19424-0 [DOI] [PubMed] [Google Scholar]
- Tönshoff, B. , Müller, B. , Elling, R. , Renk, H. , Meissner, P. , Hengel, H. , … Kräusslich, H.‐G. (2021). Prevalence of SARS‐CoV‐2 infection in children and their parents in Southwest Germany. JAMA Pediatrics. 10.1001/jamapediatrics.2021.0001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trypsteen, W. , Van Cleemput, J. , van Snippenberg, W. , Gerlo, S. , & Vandekerckhove, L. (2020). On the whereabouts of SARS‐CoV‐2 in the human body: A systematic review. PLoS Pathogens, 16(10), e1009037. 10.1371/journal.ppat.1009037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turoňová, B. , Sikora, M. , Schürmann, C. , Hagen, W. J. H. , Welsch, S. , Blanc, F. E. C. , … Beck, M. (2020). In situ structural analysis of SARS‐CoV‐2 spike reveals flexibility mediated by three hinges. Science, 370(6513), 203–208. 10.1126/science.abd5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergara, H. , Pape, C. , Meechan, K. , Zinchenko, V. , Genoud, C. , Wanner, A. , … Arendt, D. (2020). Whole‐body integration of gene expression and single‐cell morphology. BioRxiv. 10.1101/2020.02.26.961037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa, E. , Schaffer, M. , Plitzko, J. M. , & Baumeister, W. (2013). Opening windows into the cell: Focused‐ion‐beam milling for cryo‐electron tomography. Current Opinion in Structural Biology, 23(5), 771–777. 10.1016/j.sbi.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Walls, A. C , Park, Y. J , Tortorici, M. A , Wall, A , McGuire, A. T , & Veesler, D. (2020). Structure, Function, and Antigenicity of the SARS‐CoV‐2 Spike Glycoprotein. Cell, 181(2), 281–292. 10.1016/j.cell.2020.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Müller, M. A. , … Wendtner, C. (2020). Virological assessment of hospitalized patients with COVID‐2019. Nature, 581(7809), 465–469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- Wolff, G. , Limpens, R. W. A. L. , Zevenhoven‐Dobbe, J. C. , Laugks, U. , Zheng, S. , de Jong, A. W. M. , … Bárcena, M. (2020). A molecular pore spans the double membrane of the coronavirus replication organelle. Science, 369(6509), 1395–1398. 10.1126/science.abd3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp, D. , Wang, N. , Corbett, K. S. , Goldsmith, J. A. , Hsieh, C.‐L. , Abiona, O. , … McLellan, J. S. (2020). Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science, 367(6483), 1260–1263. 10.1126/science.abb2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y.‐M. , Wang, W. , Song, Z.‐G. , … Zhang, Y.‐Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. , Muruato, A. , Lokugamage, K. G. , Narayanan, K. , Zhang, X. , Zou, J. , … Shi, P.‐Y. (2020). An infectious cDNA clone of SARS‐CoV‐2. Cell Host & Microbe, 27(5), 841–848.e3. 10.1016/j.chom.2020.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, C. S. , Hayworth, K. J. , Lu, Z. , Grob, P. , Hassan, A. M. , García‐Cerdán, J. G. , … Hess, H. F. (2017). Enhanced FIB‐SEM systems for large‐volume 3D imaging. ELife, 6. e25916. 10.7554/eLife.25916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, H. , Song, Y. , Chen, Y. , Wu, N. , Xu, J. , Sun, C. , … Li, S. (2020). Molecular architecture of the SARS‐CoV‐2 virus. Cell, 183(3), 730–738.e13. 10.1016/j.cell.2020.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, W. , Mao, C. , Luan, X. , Shen, D.‐D. , Shen, Q. , Su, H. , … Xu, H. E. (2020). Structural basis for inhibition of the RNA‐dependent RNA polymerase from SARS‐CoV‐2 by remdesivir. Science, 368(6498), 1499–1504. 10.1126/science.abc1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. M. E. , & Fouchier, R. A. M. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367(19), 1814–1820. 10.1056/nejmoa1211721 [DOI] [PubMed] [Google Scholar]
- Zhong, N. S. , Zheng, B. J. , Li, Y. M. , Poon, L. L. M. , Xie, Z. H. , Chan, K. H. , … Guan, Y. (2003). Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People's Republic of China, in February, 2003. Lancet, 362(9393), 1353–1358. 10.1016/S0140-6736(03)14630-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, P. , Yang, X.‐L. , Wang, X.‐G. , Hu, B. , Zhang, L. , Zhang, W. , … Shi, Z.‐L. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, 579(7798), 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, T. , Tsybovsky, Y. , Gorman, J. , Rapp, M. , Cerutti, G. , Chuang, G.‐Y. , … Kwong, P. D. (2020). Cryo‐EM structures of SARS‐CoV‐2 spike without and with ACE2 reveal a pH‐dependent switch to mediate Endosomal positioning of receptor‐binding domains. Cell Host & Microbe, 28(6), 867–879.e5. 10.1016/j.chom.2020.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study


