Summary
Coronavirus disease 2019 (Covid‐19), caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection, is now pandemic. While most Covid‐19 patients will experience mild symptoms, a small proportion will develop severe disease, which could be fatal. Clinically, Covid‐19 patients manifest fever with dry cough, fatigue and dyspnoea, and in severe cases develop into acute respiratory distress syndrome (ARDS), sepsis and multi‐organ failure. These severe patients are characterized by hyperinflammation with highly increased pro‐inflammatory cytokines including IL‐6, IL‐17 and TNF‐alpha as well as C‐reactive protein, which are accompanied by decreased lymphocyte counts. Clinical evidence supports that gut microbiota dysregulation is common in Covid‐19 and plays a key role in the pathogenesis of Covid‐19. In this narrative review, we summarize the roles of intestinal dysbiosis in Covid‐19 pathogenesis and posit that the associated mechanisms are being mediated by gut bacterial metabolites. Based on this premise, we propose possible clinical implications. Various risk factors could be causal for severe Covid‐19, and these include advanced age, concomitant chronic disease, SARS‐CoV‐2 infection of enterocytes, use of antibiotics and psychological distress. Gut dysbiosis is associated with risk factors and severe Covid‐19 due to decreased commensal microbial metabolites, which cause reduced anti‐inflammatory mechanisms and chronic low‐grade inflammation. The preconditioned immune dysregulation enables SARS‐CoV‐2 infection to progress to an uncontrolled hyperinflammatory response. Thus, a pre‐existing gut microbiota that is diverse and abundant could be beneficial for the prevention of severe Covid‐19, and supplementation with commensal microbial metabolites may facilitate and augment the treatment of severe Covid‐19.
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- ARDS
acute respiratory distress syndrome
- CHR
corticotropin‐releasing hormone
- Covid‐19
coronavirus disease 2019
- CVD
cardiovascular disease
- DIC
disseminated intravascular coagulation
- GPR109a
G protein coupled receptor 109a
- HDACs
histone deacetylases
- isoDCA
isodeoxycholic acid
- isoalloLCA
isoallolithocholic acid; NK cells, natural killer cells; 3‐oxoLCA, 3‐oxolithocholic acid; PPAR‐gamma, peroxisome proliferator‐activated receptor gamma (PPARAQ7‐gamma); RSV, respiratory syncytial virus
- S
spike glycoprotein S
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
- Treg cells
regulatory T cells
- VDR
vitamin D receptor
1. INTRODUCTION
Covid‐19 (coronavirus disease 2019) is caused by SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus 2) that has spread worldwide, resulting in a huge impact on public health and the economy. SARS‐CoV‐2 is much more transmissible than SARS‐CoV, with tens of millions of people infected. 1 Although most SARS‐CoV‐2‐infected patients present with mild symptoms or are asymptomatic, severe Covid‐19 can be fatal. 2 The common respiratory symptoms are fever, dry cough, fatigue and dyspnoea, with additional manifestations such as increased production of phlegm/sputum, headache and haemoptysis. 3 , 4 , 5 , 6 In severe cases, patients present with acute respiratory distress syndrome (ARDS), sepsis and multi‐organ failure. These patients are characterized by hyperinflammation represented by a cytokine storm accompanied by lymphopenia. 7 Clinical studies have repeatedly reported that the risk factors associated with the severity of Covid‐19 include age‐associated dynamics such as the presence of pre‐existing chronic diseases, intestinal SARS‐CoV‐2 infection, extensive use of antibiotics and stress. 2 , 8 Understanding how SARS‐CoV‐2 causes severe Covid‐19 is critical to reduce morbidity.
SARS‐CoV‐2 is a positive‐sense single‐stranded RNA virus with an envelope. The whole genome sequence of SARS‐CoV‐2 virus has been mapped. 9 , 10 In 30‐kb nucleotides, at least 10 open reading frames have been identified, which encode multiple proteins for the virus to infect human cells which then progress to replication. 11 These include membrane proteins (spike glycoprotein S, envelope protein E and membrane protein M), nucleocapsid (N), and transcription complex nsps1–10 and nsps12–16. 12 Among them, S and N proteins have been structurally studied. 13 The structure of spike glycoprotein, which can bind to angiotensin‐converting enzyme 2 (ACE2) to facilitate the virus to enter host cells, explains why S protein has much higher affinity than that of SARS‐CoV for the same receptor. 13 , 14 Y‐shaped spike pairs are present (i.e., two heads with one stem) that render unusual freedom to the spike. N protein has a helical structure wound with RNA with five subunits per turn in a head‐to‐tail structure allowing the virus to resist environmental and physical challenges.
Recent studies have reported that the gut microbiota is dysregulated in Covid‐19. 15 , 16 , 17 Dysregulation of gut microbiota (dysbiosis) could be involved in the pathogenesis of Covid‐19 by accelerating hyperinflammation, a clinical characteristic of severe Covid‐19. The studies of gut dysbiosis in Covid‐19 could provide preventive and therapeutic opportunities. In this review, we summarise the evidence of gut dysbiosis in Covid‐19 pathogenesis, discuss the possible mechanisms, explore its links with other risk factors and speculate on clinical implications.
2. IMMUNE RESPONSES AND CLINICAL CHARACTERISTICS OF COVID‐19
Infection with SARS‐CoV‐2 initiates immune responses. The outcomes of the infection depend on the interaction of the extent of infection of the virus and the containment effect provided by the immune response to the virus. This interaction determines clinical manifestations of Covid‐19. 2 , 3 , 4 , 6 , 8 The virus enters the body from the respiratory system through binding to ACE2 on the surface of airway cells, particularly alveolar type II pneumocytes. The virus is also able to increase the expression of ACE2 to facilitate the infection. 18 The infection of the virus triggers an initial immune response, in which innate immune cells including monocytes, natural killer cells, macrophages and dendritic cells, as well as secreted cytokines are increased. Dendritic cells and macrophages present antigens to adaptive immune cells to elicit cellular and humoral immune responses. The virus could spread to many organs through the systemic circulation if the virus cannot be restricted to the lungs (Figure 1). As ACE2 is found in many organs, SARS‐CoV‐2 could potentially infect many sites including the heart, blood vessels, gut, lung, kidney, testis and brain. 19 Damage to major organs could cause severe morbidity and mortality. It has been proposed that SARS‐CoV‐2 may also initially infect the intestines and spread to other organs but this route has not been verified yet.
FIGURE 1.
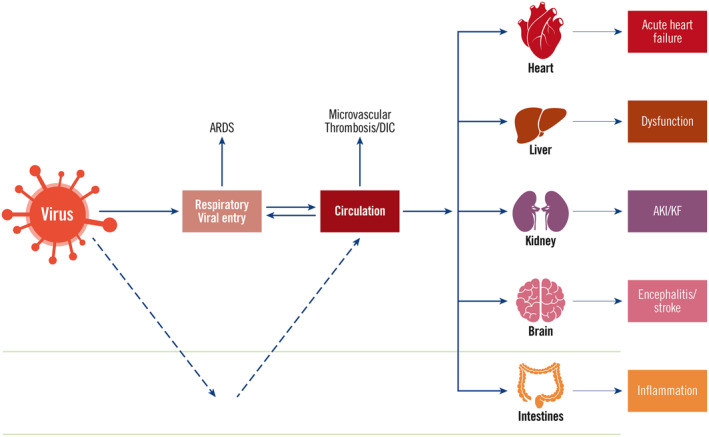
Viral entry, dissemination and damage to various organs. Severe acute respiratory syndrome coronavirus 2 virus initially infects respiratory system. If not controlled, it could enter the circulation system and subsequently infect various organs such as heart, liver, kidney, brain and intestines. The virus may also initially gain entry into the digestive system, and could disseminate to other organs. The damage to major organs can cause severe consequences such as acute respiratory distress syndrome, microvascular thrombosis, disseminated intravascular coagulation, acute heart failure, liver dysfunction, acute kidney injury/kidney failure, encephalitis/stroke and intestinal inflammation
In non‐severe Covid‐19 patients, adequate immune responses are elicited to eliminate viruses, allowing patients to achieve complete recovery. A case report examined the changes of clinical manifestations and immune responses in a recovered Covid‐19 patient. 20 At the early stage of infection, clonally expanded CD8+ T‐cells were enriched to clear the cells infected by SARS‐CoV‐2 viruses. Elicited B‐cells by viral antigens secreted antibodies IgG and IgM to neutralize viruses. Other studies showed that these antibodies were mainly against N and S proteins. IgM and IgG can be detected 5–17 and 6–14 days after symptoms appear. 21 , 22 Liao et al. 23 reported immune adaptations from bronchoalveolar lavage fluid from Covid‐19 cases. In moderate cases, inflammatory M1‐like macrophages were increased adequately, indicating the inflammatory responses were still under control.
In severe Covid‐19 patients, IFN‐I and IFN‐III are reduced, while neutrophils and macrophages are increased with greatly increased pro‐inflammatory cytokines such as TNF‐alpha, IL‐6, IL‐17 and C‐reactive protein. 24 This cytokine storm is accompanied by CD4+ T‐cell, CD8+ T‐cell exhaustion and decreased macrophages. Bronchoalveolar lavage fluid investigations showed increased M1‐like macrophages and decreased CD8+ T‐cells that were proliferating rather than differentiating. 23 These hyperinflammatory responses are the key features of severe Covid‐19, leading to increased mortality. Moreover, the cytokines IL‐6 and IL‐17 have been correlated with infection severity and mortality. 25 These greatly increased cytokines could cause ARDS and multi‐organ failure (Figure 1). 26
Inflammation of vascular endothelial cells plays a key role in the pathogenesis of severe Covid‐19, 27 because it can progress to microvascular thrombosis and disseminated intravascular coagulation. Microvascular thrombosis could present in any organ, which is an important cause for multiple‐organ failure. 28 Major organ failure also results from hyperinflammation, leading to fatal damage such as acute heart failure, acute kidney injury/kidney failure and encephalitis/stroke. As the cytokine storm‐associated hyperinflammation is a key factor for severe Covid‐19, understanding the associated mechanisms and inhibition of the key elements could reduce the severity of Covid‐19.
In a recent publication, Leisman et al. 29 compared the cytokine levels in Covid‐19 and other cytokine storms. The authors found that IL‐6 levels in Covid‐19 were much lower than that caused by sepsis, cytokine release syndrome and Covid‐19‐unrelated ARDS. This arose a controversial opinion about the importance of the cytokine storm in multi‐organ failure in Covid‐19. Indeed, the multi‐organ failure in Covid‐19 is caused by various factors such as direct viral infection, microvascular thrombosis and pro‐inflammatory cytokines. However, the cytokine storm may play a central role as the formation of vascular thrombosis—a major pathological characteristic found in post‐mortem of Covid‐19–has inflammation involvement through activation of the coagulation pathway. 7 , 30 In addition, there are interactions among cytokines as well as between cytokines and complements, which may have a synergistic effect on an organ. 7 The cytokine storm together with the microvascular thrombosis in Covid‐19 could greatly accelerate SARS‐CoV‐2‐caused organ damage. Therefore, even the hyperinflammation has not reached a very high level as sepsis, cytokine release syndrome and Covid‐19‐unrelated ARDS, it could cause severe disease already.
How immune responses become uncontrolled is not clearly elucidated although some associated mechanisms have been proposed. Infection of SARS‐CoV‐2 can attract macrophages, dendritic cells and neutrophils, which secrete cytokines for controlling the viral insult. The binding of virus particles results in endo‐phagocytosis and downregulation of ACE2, causing inflammation and thrombosis. 31 Whether increased ACE2 expression by the virus could reduce the effect of ACE2 endo‐phagocytosis is not clear. 18 Activation of the complements C5a and C5b‐9 is also involved in SARS‐CoV‐2‐caused inflammation. 25 Guo et al. 32 identified a specific type of monocyte, which was responsible for the cytokine storm. The cells were not presented in healthy subjects but accounted for 98.3% of the total monocytes in severe Covid‐19 and only 12.1% in the remission of Covid‐19 patients. These cells highly expressed pro‐inflammatory cytokines, chemokine and inflammasome genes. Other risk factors such as underlying chronic diseases may accelerate already present inflammatory responses, leading to uncontrolled cytokine secretion, but the associated mechanisms are not well explained.
Clinical observations have identified many risk factors which are associated with the severity of Covid‐19 such as advanced age, chronic diseases including diabetes, hypertension and cardiovascular disease (CVD). 33 A study with 1482 hospitalized patients showed that among all comorbidities, 49.7% were hypertensive, 48.3% were obese, 34.6% presented with chronic liver disease, 28.3% with diabetes and 27.8% with CVD (https://www.cdc.gov/mmwr). Another study also revealed that hypertension (30%), diabetes (19%) and CVD (8%) adversely affected the clinical outcome of the infection. 34 How these factors are involved in the pathogenesis to cause severe Covid‐19 morbidity and mortality is of significant importance in the prevention and treatment of severe Covid‐19. We posit that an important plausible mechanism may implicate intestinal dysbiosis in chronic conditions that could accelerate SARS‐CoV‐2‐induced inflammation, leading to hyperinflammation‐related morbidity and mortality.
3. DYSREGULATION OF THE INTESTINAL MICROBIOTA IN COVID‐19
Initial evidence of intestinal dysbiosis in Covid‐19 emerged from frequently reported gut‐associated symptoms. 35 , 36 , 37 Several studies showed that gastrointestinal manifestations including diarrhoea, anorexia and nausea occurred in about 50%–60% of patients. 35 , 38 , 39 Subsequently, intestinal dysbiosis was demonstrated in Covid‐19. 15 By comparing gut microbiota in 30 hospitalized Covid‐19 patients and 30 healthy participants, it was reported that intestinal dysbiosis was associated with the Covid‐19 patients. 15 The bacterial diversity was decreased with higher abundance of opportunistic pathobiont bacterial groups such as Streptococcus, Rothia, Veillonella and Actinomyces, and lower abundance of commensal bacteria. Zuo et al. 16 also studied 15 hospitalized Covid‐19 patients and found persistent alterations of faecal microbiome. Compared to healthy patients, Covid‐19 positive patients had increased opportunistic pathogens and decreased beneficial commensal bacteria.
Gut dysbiosis has been associated with the severity of Covid‐19. Using a proteomic risk score based on 20 identified blood protein biomarkers, a recent study correlated and hence predicted the progression of severe Covid‐19 in older populations. 5 Most of the risk proteins were associated with inflammatory factors including IL‐1beta, IL‐6, TNF‐alpha and high‐sensitivity C‐reactive protein. Furthermore, gut microbiota was associated with inflammatory factors with Bacteroides, Streptococcus and Clostridiales genera being negatively related to pro‐inflammatory factors, while Ruminococcus, Blautia and Lactobacillus genera positively related to pro‐inflammatory factors, indicating the involvement of gut dysbiosis in the severity of Covid‐19. Zuo et al. 16 showed that the abundance of Coprobacillus, Clostridium ramosum and Clostridium hathewayi were positively correlated with dysbiosis and the severity of Covid‐19,while Faecalibacterium prausnitzii inversely correlated with the disease severity in 15 hospitalized patients. Indeed, patients with intestinal symptoms were associated with significantly increased risk of admittance to intensive care unit and the rate of mortality than those without gut symptoms. 40
Various reasons could explain intestinal dysbiosis in Covid‐19 (Figure 2). SARS‐CoV‐2 can also infect enterocytes in the intestines, causing gut dysbiosis and inflammation. Faecal viruses have been detected even in patients without gut symptoms. A recent meta‐analysis also showed a detection rate of 43.7% of SARS‐CoV‐2 RNAs in Covid‐19 patients' faecal specimens and much higher detection rates in severely infected patients. 41 Using intestinal organoids, studies demonstrated that SARS‐CoV‐2 was able to infect enterocytes. 42 , 43 , 44
FIGURE 2.
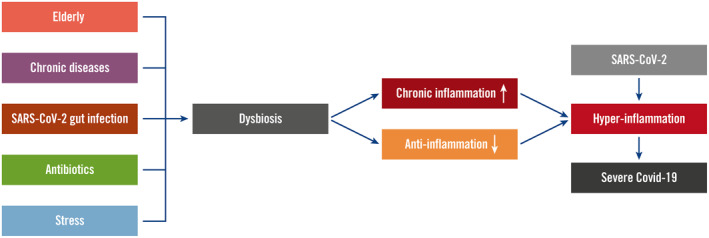
Gut dysbiosis in coronavirus disease 2019 (Covid‐19). Various risk factors of Covid‐19 including elderly, chronic diseases, severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) gut infection, use of antibiotics and stress can cause gut dysbiosis. Dysbiosis contributes to hyperinflammation by increased intestinal and systemic chronic inflammation and decreased anti‐inflammation mechanisms. These facilitate the form of hyperinflammation after SARS‐CoV‐2 infection, resulting in severe Covid‐19
Chronic diseases with major organ involvement such as the heart, liver, kidney and brain, linked with diabetes, obesity and hypertension have been identified as risk factors responsible for the severity of Covid‐19. Gut dysbiosis has been well demonstrated to play key roles in these metabolic diseases through chronic inflammation. 45 , 46 , 47 Thus, disturbed gut microbiota compositions in these diseases could perpetuate the reported increased severity of Covid‐19.
In addition, the extensive use of antibiotics is a causal factor for gut dysbiosis when prescribed for the prevention of secondary bacterial infections in Covid‐19. It is well known that the use of antibiotics can cause gut microbiota alterations and subsequent transluminal mucosa transfer of endotoxins (e.g., lipopolysaccharides [LPS]) from opportunistic pathogens. 48 For example, antibiotics have been associated with Clostridium difficile infections, and faecal microbiome transplantation has been used successfully for the treatment of the disease. 49 , 50
Psychological distress and stress that can present in the form of depression and anxiety can also be risk factors for the severity of Covid‐19. Batty et al. 50 identified mental and cognition to be risk factors for hospitalized Covid‐19; lower cognition doubled Covid‐19 hospitalization. 50 Adam et al. 51 also reported that mental disorders increased the rate of viral infections such as the common cold by 44%, where the causal agent is another type of coronavirus. An early study by Cohen et al. 52 demonstrated that psychological distress increased coronavirus infections in healthy subjects who were inoculated with either one of five respiratory viruses including coronavirus‐type 229E through nasal drops. Gut dysbiosis is well known to exist in chronically stressed individuals, 53 and probiotics have been used to relieve stress in clinical trials. 54
The risk factors that are causal for intestinal dysbiosis could be associated with increased chronic low‐grade gut and systemic inflammation with decreased anti‐inflammatory effects (Figure 2). 55 Therefore, in infections with SARS‐CoV‐2, gut dysbiosis could present with additive effects in respect of inflammation that leads to hyperinflammation.
4. POSSIBLE MECHANISMS: THE INVOLVEMENT OF BACTERIAL METABOLITES
The effects that could be mediated by the intestinal microbiota may involve various commensal bacterial metabolites. Many bacterial metabolites such as short‐chain fatty acids, bile acids and amino acids have been identified and their effects studied on hosts. Among them, butyrate, a four‐carbon short‐chain fatty acid, is most extensively investigated. Butyrate exerts an important anti‐inflammatory effect through multiple mechanisms (Figure 3). It can activate regulatory T‐cells (Treg cells) through its receptor G protein coupled receptor 109a (GPR109a) to downregulate cytotoxic T‐cells and facilitate M2 macrophage activation to reduce pro‐inflammatory cytokines to anti‐inflammatory cytokine ratios. 56 , 57 , 58 Thus, given that butyrate participates in immunoregulation biochemical pathways in both the gut and peripheral tissues, 54 , 58 decreased production of butyrate could facilitate the progression of hyperinflammation. Butyrate can also reduce the activation of multiple pro‐inflammatory signalling pathways through inhibition of histone deacetylases (HDACs) and HDAC‐independent mechanisms. 59 In addition, butyrate has antibacterial and antiviral effects through promoting the secretion of mucins and antimicrobial peptide defensins. 58 Butyrate also maintains the intestinal barrier and thus prevents the translocation of bacteria/endotoxins, which cause systemic inflammation. 60 Therefore, reduced levels of butyrate could lead to a low‐grade inflammation systemically. Other short‐chain fatty acids, acetate and propionate, have also been shown to have protective effect on respiratory viral infections. 61 , 62 Antunes et al. 62 showed that high‐fibre diet was able to decrease respiratory syncytial virus (RSV) load in mice and intaking of acetate in drinking water for mice produced a similar effect. Acetate bound to its receptor GPR43, leading to increased production of type I interferon. Lynch et al. 61 found that propionate stimulated Treg cells to reduce RSV infectivity in a mouse model. Their roles in SARS‐CoV‐2 infection warrant research as well.
FIGURE 3.
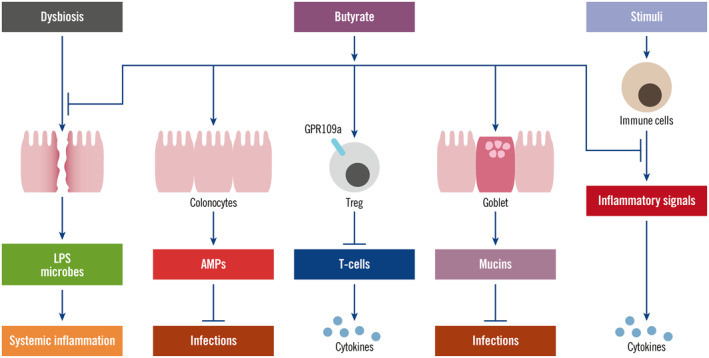
Anti‐inflammatory effect of butyrate. Butyrate exerts anti‐inflammatory effect through multiple mechanisms. Butyrate can reduce dysbiosis‐caused gut leakage, thus block the translocation of lipopolysaccharides and microbes, inhibiting systemic inflammation. Butyrate also increases colonocytes to secrete antimicrobial peptides to reduce microbial infections. Butyrate can activate regulatory T‐cells, which subsequently inhibit T‐cell activation, reducing cytokine production. It can also increase goblet cells to secrete mucins to protect from microbial infections. Through inhibition of multiple pro‐inflammatory pathways, butyrate can reduce the cytokine production by immune cells under effects of stimuli
In addition to butyrate, bile acids are well‐recognized signalling molecules. They are produced in the liver and further metabolized by the gut microbiota. Recent studies showed that epimerization derivatives of bile acids, isodeoxycholic acid (isoDCA), isoallolithocholic acid (isoalloLCA) and 3‐oxolithocholic acid (3‐oxoLCA), can modulate inflammatory responses through activation of Treg cells and inhibition of Th17 cells. 63 , 64 In an in vitro system with co‐culture of naïve T‐cells and dendritic cells, isoDCA can stimulate differentiation of Treg cells and exhibit an anti‐inflammatory effect. 63 The isoDCA reduced the secretion of IL‐6 and TNF‐alpha by dendritic cells which was stimulated by Toll‐like receptor agonists. Treatment of dendritic cells by isoDCA also decreased the expression of genes related to pro‐inflammatory factors including Tlr7, Tlr12, Nlrc5, Stat2, Stat6, Irf1 and Irf7, and increased the expression of genes for suppression of NF‐κB, MAPK, cytokine‐receptor signalling including Nfkbia, Dusp1, Dusp5 and Socs1. Hang et al. 65 also found that a derivative of LCA, isoalloLCA, promoted Treg cell differentiation, while 3‐oxoLCA inhibited Th17 cell differentiation. 64 Gut microbiota symbionts are necessary to maintain bile‐acid‐induced Treg differentiation. 65
Other bacterial metabolites such as tryptophan may also be involved in inflammation. In gut dysbiosis, the metabolism of tryptophan into kynurenine is increased due to activated indoleamine 2,3 dioxygenase, a rate‐limiting enzyme in the kynurenine pathway. Kynurenine is a pro‐inflammatory metabolite of the amino acid L‐tryptophan, and kynurenine/tryptophan is used to indicate the inflammatory states. 66 , 67 Indeed, it has been shown that altered tryptophan metabolism is involved in the pathogenesis of Covid‐19 and correlated with IL‐6 levels. 68
5. GUT DYSBIOSIS LINKED TO RISK FACTORS OF SEVERE COVID‐19
The intestinal microbiota and commensal bacterial metabolites play important roles in regulating pro‐ and anti‐inflammatory actions. Dietary factors, antibiotics and other medications (e.g., proton‐pump inhibitors), ageing, chronic diseases and lifestyle stressors are risk factors associated with gut dysbiosis. 69 Therefore, gut dysbiosis and altered bacterial metabolites can explain, at least in part, how intestinal microbiota dysbiotic shifts could mediate risk‐factor‐increased severe Covid‐19 (Figure 4).
FIGURE 4.

Interactions between gut dysbiosis and organs in coronavirus disease 2019. Gut dysbiosis affects multiple organs to form bidirectional gut–organs axes. Bile acids produced in the liver are secreted into gut, which are metabolized by gut bacteria and reabsorbed into the liver to form enterohepatic circulation. Other bacterial metabolites such as butyrate can also be absorbed into the liver. In gut dysbiosis, lipopolysaccharides and microbes are able to cross gut barrier to enter the liver. After processing in the liver, they enter to circulation system. In case of liver disease, there are also activated immune cells and cytokines. All these can be secret into the intestines and other organs. Chronic diseases in cardiovascular system, lung, brain and other organs could also produce and secrete immune cells and cytokine into circulation system
5.1. Ageing
Advanced age is almost a universally reported risk factor from Covid‐19 reports. Individuals in the older age bracket have been identified as a key factor associated with mortality rates. 34 With increasing age, Covid‐19‐caused mortality has an increased prevalence. 70 In a recent publication, Verity et al. 71 established a model to estimate the mortality rates in different age groups. 71 It was estimated that the Covid‐19 mortality rate for those aged less than 60 years was 0.32%, for those aged between 60 and 80 years, sharply increased to 6.4%, while for those aged more than 80 years reached a prevalence of mortality of 13.4%. Du et al. 72 reported the clinical features of 85 Covid‐19 death cases. The report revealed that the average age was 65.8 years; most had comorbidities including hypertension, diabetes and coronary heart disease.
Ageing has been closely associated with chronic, low‐grade, sterile inflammation–inflammaging. 73 In inflammaging, innate immune cells such as macrophages are continuously activated with increased secretion of pro‐inflammatory cytokines including IL‐6, IL‐8 and IL‐1β, as well as a balance towards immunosenescence. 73 The compositions of Treg cells are changed in an aged population with increased natural Treg cells but decreased inducible Treg cells. 74 , 75 The dysfunctions of older Treg cells are indicated by increased incidences of excessive immune diseases such as autoimmune diseases and cancer. Therefore, SARS‐CoV‐2 infections in aged individuals may accelerate inflammaging, leading to hyperinflammation.
A dysregulated gut microbiota has been recognized to play a central role in inflammaging. 73 A review that investigated the available evidence with regard to ageing, inflammaging and the intestinal microbiota adverse shifts, reported that an aged‐type gut microbiota could be correlated with inflammaging. 76 With increasing age, gut microbiota present with deficits of beneficial inputs. Aged individuals most probably display increased proteobacteria and decreased butyrate‐producing bacteria such as Faecalibacterium prauznitzii, and thus present with a chronic deficit of intestinal microbiome‐elaborated butyrate. 73 Increased proteobacteria is positively correlated with pro‐inflammatory cytokines IL‐6 and IL‐8. 76 The low levels of butyrate in aged individuals may contribute to chronic low‐grade inflammation.
5.2. Diabetes and obesity
Both diabetes and obesity are factors that can predispose individuals for severe Covid‐19. 77 , 78 Therefore, diabesity (i.e., diabetes with concomitant obesity) presents a higher risk factor compared to either diabetes or obesity alone. Diabetes is the third most prevalent comorbidity of Covid‐19, increasing the adverse outcomes by two to three folds, and mortality by more than three folds. 78 Diabetes increases susceptibility to infections due to reduced immune responses by poor glycaemic control. 79 A recent study showed that glucose increased SARS‐CoV‐2 viral replication in monocytes and stimulated production of pro‐inflammatory cytokines IFN‐alpha, IFN‐beta, IFN‐lamda, IL‐6 and IL‐1beta. 18 This has demonstrated to be caused by increased glycosis through increased expression of HIF‐1alpha target genes GLUT‐1, PFKFB3, PKM2 and LDH‐A, which are responsible for glucose transport and glycolytic pathway. 18 Co‐culture of SARS‐CoV‐2‐infected monocytes with T‐cells or pulmonary epithelial cells caused dysfunction of T‐cells and apoptosis of epithelial cells, which was reversed by inhibition of HIF‐alpha, revealing impairment of T‐cells and epithelial by high levels of glucose. 18 Impaired immunity in diabesity also reduces systemic viral clearance. In addition, increased ACE2 expression by elevated glucose levels and anti‐diabetic agents such as thiazolidinedione 18 , 79 could facilitate infection with SARS‐CoV‐2. Importantly, in diabesity, there is chronic inflammation with increased IL‐6 and TNF‐alpha, 80 which increases the susceptibility to SARS‐CoV‐2‐induced hyperinflammation. In obesity, low‐grade chronic inflammation in visceral adipose tissue has been associated with increased inflammatory immune cells and cytokines including CD8+ T‐cells, macrophages, neutrophils and natural killer cells. 81 , 82 In addition, Treg cells and M2 macrophages have been reported as decreased. 83 Manipulation of peroxisome proliferator‐activated receptor gamma to increase Treg cell number could have therapeutic effects on obesity and insulin resistance. 84
Inflammatory responses in diabetes and obesity have been linked to gut dysbiosis. 85 , 86 Colonization of the gut with an obese microbiota in germ‐free mice markedly increased the body weight. 85 The inflammation that accompanies intestinal dysbiosis in diabetes can be reduced through the administration of probiotic lactic acid bacteria through increased butyrate production. 86 The administration of probiotics can also reduce the blood levels of glucose and increase glucose tolerance. 86 , 87 Butyrate not only exerts anti‐inflammatory effect but also regulates glucose metabolism. Butyrate is necessary for the secretion of glucagon‐like peptide‐1 (GLP‐1), which promotes insulin secretion but has a short half‐life of 2 min 88 Thus, butyrate could be effective for the treatment of obesity and diabetes. 89 Administration of butyric acid normalized hyperglycaemia in a diabetes mouse model. 90 The effect of metformin has been associated with increased butyrate‐producing bacteria. 87 , 91 In diabetes, gut microbiota is dysregulated that causes reduced butyrate production. 92 , 93 Administration of metformin restores the bacteria with increased Akkermansia muciniphila, Subdoligranulum variabile, Escherichia spp and decreased Intestininibactor bartlettii, as well as increased butyrate production. 94 , 95 , 96 Therefore, metformin not only decreases the glucose levels but also reduces inflammation through increasing butyrate production. However, the outcomes of clinical studies are controversial. Several studies reported the beneficial effects of metformin on diabetic Covid‐19 patients, 97 , 98 , 99 while other studies showed no effects or even worse outcomes. 100 , 101 The discrepancy could be caused by the selection of patients such as difference in blood glucose levels. 101 It could be important to have the same blood glucose levels for selected patients for comparison as glucose greatly increases ACE2 expression to enhance viral infectivity. A prevention strategy of diabetes by Faecalibacterium prausnitzii transplantation has also been proposed. 102 However, whether probiotics and butyrate could reduce the adverse effect of diabetes or diabesity in severe Covid‐19 is not well studied.
5.3. Cardiovascular disease
The heart is a major organ that is susceptible to assault by the SARS‐CoV‐2 and CVD increases the severity of Covid‐19. In a meta‐analysis, it was shown that CVD increased the rate to develop severe disease by five times. 103 SARS‐CoV‐2 infection can cause myocardial injury indicated by increased troponin. ACE2 is highly expressed in the heart. Although it is lower than that in the kidney and small intestine, it is higher than that in the lung. 104 Chen et al. 3 examined eight types of cells in the heart that included cardiomyocytes, endothelial cells, macrophages, fibroblasts, pericytes, smooth muscle cells, T‐cells and neuron‐like cells, and found that ACE2 was highly expressed in the pericytes, which accounted for about 10% of the total cells in the heart. 104 The pericytes are located outside the endothelial cells of capillaries and thus infections by SARS‐CoV‐2 could promote insufficient blood supply to cardiomyocytes, which in turn may increase the expression of ACE2. Studies have revealed that Ischaemic cardiomyopathy increases ACE2 expression by a factor of 1.8‐fold compared to non‐diseased hearts. 105 , 106 Increased expression of ACE2 by therapeutic agents for CVD has also been reported. 107
ACE2 is also expressed in endothelial cells, and SARS‐CoV‐2 infection causes endotheliitis. 27 , 108 In an in vitro experiment, SARS‐CoV‐2 was demonstrated to infect engineered human blood vessel organoids. 109 The dysfunction of endothelial cells could cause coagulation abnormality, which affects multiple organs. Endothelial cell dysfunction usually leads to vasoconstriction and thus, organ ischaemia, inflammation and tissue oedema. 110 Inflammatory cytokines produced by other infected organs may accelerate the endothelial cell dysfunction (Figure 4).
Gut microbiota has been well demonstrated to play a critical role in heart disease through the gut–heart axis (Figure 4). 111 A study has shown that CVD patients with SARS‐CoV‐2 infections have a higher rate of gut leakage and inflammasome activation. 112 Gut dysbiosis has also been considered as a pathogenic factor in hypertension through bacterial metabolites, sympathetic nervous system stimulation and endotoxaemia. 113 Plasma butyrate levels are inversely correlated with hypertension, 114 , 115 while oral supplementation of butyrate decreases endothelial dysfunction and macrophage activation in a mouse atherosclerotic model. 116 Butyrate can bind GPR41 and GPR43 receptors on endothelial cells to increase functionalities. 117 Decreased production of butyrate is a characteristic of CVD. 118 Other bacterial metabolites have also been reported to be involved in the pathogenesis of CVD. 118 Therefore, dysbiosis in CVD may cause inflammation and decreased anti‐inflammatory capability, which facilitates the formation of hyperinflammation in Covid‐19.
5.4. Chronic pulmonary disease
Gut dysbiosis could explain chronic pulmonary diseases as a risk factor of severe Covid‐19. The link of lung diseases with gut microbiota is also well recognized, giving rise to the gut–lung axis (Figure 4). 119 , 120 Gut dysbiosis is a common occurrence in many pulmonary conditions and diseases and is involved in the pathogenesis of allergies, asthma, cystic fibrosis, lung cancer and chronic obstructive pulmonary disease. 121 A dysbiotic gut promotes inflammatory profiles in lung conditions and reduces the regulation of pro‐ and anti‐inflammatory activities. 121 Alternatively, lung inflammation such as viral infections can disturb the gut microbiota that progresses to dysbiosis by releasing pro‐inflammatory cytokines into systemic circulation and subsequently the intestines.
This bidirectional interaction may also exist between SARS‐CoV‐2 lung infections and gut dysbiosis. Pre‐existing gut dysbiosis can cause low‐grade systemic inflammation, which could then accelerate the inflammation in the lung caused by SARS‐CoV‐2 lung infection. The pro‐inflammatory cytokines released from lung inflammation may transfer to the gut through the systemic circulation, accelerating gut dysbiosis. Therefore, it forms a feed‐forward regulation. It has been proposed that targeting the gut–lung axis could be used for anti‐inflammation therapy in Covid‐19. 122 , 123 , 124
5.5. Chronic liver disease
The investigation about the overall effect of chronic liver disease on the severity of Covid‐19 has resulted in controversial outcomes. By comparing Covid‐19 with liver disease and non‐liver disease in 2780 patients, Singh et al. 125 found that pre‐existing chronic liver disease, particularly cirrhosis, increased the severity and mortality from Covid‐19. 126 Several other studies have also shown that liver disease is associated with severe Covid‐19. 127 Even simple hepatic steatosis can increase the severity of Covid‐19. 128 However, in a pooled study, Lippi et al. 129 showed no association between chronic liver disease and the severity and mortality from Covid‐19 infections. This contrast could be explained by the selection of patients studied. Different liver diseases may have various impacts. As shown in Singh's study, 125 the effects of cirrhosis and non‐cirrhosis on Covid‐19 is significantly different. 126 Simple hepatic steatosis and non‐alcoholic steatohepatitis may also need to be distinguished as pre‐existing inflammatory conditions could be important in the severity of Covid‐19. A recent meta‐analysis confirmed that liver injury was common in Covid‐19, reaching 25%, with a worsening clinical outcome. 130
The effect of chronic liver disease on the severity of Covid‐19 could be explained by the gut–liver axis (Figure 4). Intestinal dysbiosis is reported in chronic liver disease, which leads to altered levels of bacterial metabolites such as butyrate and primary and secondary bile acids. Therefore, there is increased risk of local and systemic low‐grade inflammation and decreased anti‐inflammatory capacity in the gut, which increases the severity of Covid‐19. In non‐alcoholic fatty liver disease, butyrate is decreased, leading to increased inflammation both in the liver and intestines. 131
There are bidirectional interactions between the gut microbiota and the liver (Figure 4). 132 The bile acids produced in the liver are important for the maintenance of a balanced microbial ecosystem in the gut. The detergent effect of bile acids can inhibit bacterial overgrowth and as such bile acids control pathobiont proliferation. Furthermore, the metabolism of bile acids by gut bacteria is an important factor that maintains a normal bile acid pool. Bile acids are signalling molecules, which regulate bile acid biosynthesis in the liver as well as the physiological processes of other organs. The intestinal dysbiosis in chronic liver disease affects the composition of the bile acid pool, which in turn further disrupts the gut microbiota. How the anti‐inflammatory bile acids, namely isoDCA and isoalloLCA, change in chronic liver disease and how they contribute to the effect of chronic liver disease on the severity of Covid‐19 at present remain unknown.
An important function of the liver is to detoxify environmental and other ingested chemicals that have been absorbed from the intestines. Liver detoxification processes could be impaired in various chronic liver diseases that could progress the accumulation of pro‐inflammatory chemicals such as LPS in the systemic circulation, which subsequently enter various end organs (e.g., heart and brain).
5.6. Stress
The gut–brain axis exhibits a bidirectional flow of interactions in neurological diseases between the brain and gut microbiota (Figure 4). Lifestyle stressors could increase the severity of Covid‐19 133 as the adverse effect of stress on the common cold has shown. 52 Moreover, these effects could be mediated by gut dysbiosis.
Stress can increase mast cell secretion of pro‐inflammatory cytokines such as IL‐6 and TNF‐alpha through increasing production of hypothalamic and amgydala corticotropin‐releasing hormone. 134 This causes gut dysbiosis with a concomitant reduced level of butyrate production. The effect of gut dysbiosis causes adverse effects on Covid‐19 similar to other unfavourable conditions. In addition, Covid‐19 could cause stress, which accelerates the severity of Covid‐19, giving rise to a feed‐forward loop. Therefore, lifestyle stressors could be a critical risk factor for severe Covid‐19 that warrants further investigation.
6. PREVENTIVE AND THERAPEUTIC IMPLICATIONS
Understanding the important roles of the gut microbiota in the pathogenesis of Covid‐19 could have important implications in the prevention and treatment of the disease. A healthy gut microbiota can maintain an immune system that is in equilibrium ready to neutralize Covid‐19 viral assaults. Hence a pre‐existing balanced pro‐ and anti‐inflammatory gut of microbial metabolites could potentially avoid hyperinflammation after Covid‐19 and thus prevent severe Covid‐19. Various approaches which could improve the gut microbiota could be used beneficially, particularly in the vulnerable populations. The aged or those with underling chronic diseases may greatly benefit from a gut microbiota that may be improved with the administration of probiotics, prebiotics and synbiotics.
Improvement in gut microbiota profiles could be useful in reducing the possibility to trigger hyperinflammation in those Covid‐19 patients presenting with advanced age. As noted previously, gut microbiota profiles that produce higher levels of butyrate and bile acid derivatives that stimulate Treg cells and M2 macrophages will provide an efficient immune brake in the prevention of cytokine storms. The roles of the gut microbiota and butyrate in the prevention and treatment of hyperinflammation in Covid‐19 warrant dedicated focused studies.
The approaches to improve gut microbiota profiles for the treatment of Covid‐19 could be difficult in those patients who have dysbiotic gut caused by SARS‐CoV‐2 infections of intestinal cells and who have extensively administered antibiotics. Alternatively, microbial metabolites could be used directly such as the administrations of butyrate and ursodeoxycholic acid. 135 , 136 , 137 Given that butyrate has an anti‐inflammatory effect through various mechanisms, in Covid‐19, butyrate may reduce hyperinflammation. Studies have shown that high levels of gut butyrate‐producing bacteria are associated with reduced respiratory viral infections in kidney transplant recipients. 138 However, butyrate is metabolized in the human body rapidly, leading to low bioavailability. Therefore, specific dosages to be used could be of importance for achieving an anti‐inflammatory effect while avoiding weakening the antiviral effect of immune responses. Although as yet to be studied, IsoDCA could also be used in the treatment of Covid‐19.
Gut microbiota and bacterial metabolites could also be involved in the prevention and treatment efficacy of other agents used in the treatment of Covid‐19, such as vitamin D. Low levels of vitamin D are now known to be associated with the severity of Covid‐19; low concentrations of vitamin D and its metabolite 25‐hydroxy vitamin D are inversely correlated with the severity of Covid‐19. 139 , 140 Gut microbiota can affect the blood levels of 25‐hydroxy vitamin D and vitamin D receptor (VDR) expression. 139 After binding of vitamin D, VDR regulates genes to increase innate immunity and secretion of antivirus defensin, which cleave virus membrane. 141 Activation of VDR also increases ratios of Th2/Th1 and Treg/Th17 to facilitate anti‐inflammatory effects. 142 In animal experiments, vitamin D metabolite 1,25‐dihydroxyvitamin D binds to VDR to reduce renin–angiotensin system, and thus decrease the inflammatory status. 143 On the other hand, activated VDR affects gut microbiota composition. 144 It also increases antimicrobial peptide expression and gut barrier integrity. 139 Increased expression of antimicrobial peptides by vitamin D in the respiratory tract protects the lungs from viral infections. 130 Vitamin D in combination with magnesium and vitamin B12 are able to reduce oxygen support and intensive care in Covid‐19. 131 Combination use of vitamin D and bacterial metabolites may warrant controlled clinical studies.
7. CONCLUSIONS
Intestinal dysbiosis is common in patients presenting with high risk factors of severe Covid‐19. Intestinal dysbiosis could mediate, at least partially, the pathogenesis of severe Covid‐19. The gut commensal bacterial cohort elaborated metabolites are important in maintaining a regulated pro‐ and anti‐inflammatory states in the host. Loss of equilibrium in immunity with partial dependence on a eubiotic gut microbiota could facilitate the occurrence of hyperinflammation, leading to severe Covid‐19 and increased mortality. Improvement of intestinal microbiota profiles and administrations of commensal bacterial metabolites could prevent severe Covid‐19 and lead to novel therapeutic strategies.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
AUTHOR CONTRIBUTIONS
Jiezhong Chen drafted the original manuscript with assistance from Luis Vitetta. All authors contributed to further improvement of the manuscript and approved the final version of the manuscript.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Paul Griffiths, Professor of Virology, University College London, for reviewing, suggestions and editing.
Chen J, Hall S, Vitetta L. Altered gut microbial metabolites could mediate the effects of risk factors in Covid‐19. Rev Med Virol. 2021;31(5):e2211. doi: 10.1002/rmv.2211
[Correction added on 18 February 2021, after first online publication. One of the corresponding author's ORCID ID and email address were updated in this version.]
Contributor Information
Jiezhong Chen, Email: Jiezhong_chen@medlab.co.
Luis Vitetta, Email: luis.vitetta@sydney.edu.au.
DATA AVAILABILITY STATEMENT
Data sharing is not applicable for this narrative review.
REFERENCES
- 1. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019‐nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689‐697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goyal P, Choi JJ, Pinheiro LC, et al. Clinical characteristics of Covid‐19 in New York City. N Engl J Med. 2020;382(24):2372‐2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Young BE, Ong SWX, Kalimuddin S, et al. Epidemiologic features and clinical course of patients infected with SARS‐CoV‐2 in Singapore. JAMA. 2020;323(15):1488‐1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buonaguro FM, Ascierto PA, Morse GD, et al. Covid‐19: time for a paradigm change. Rev Med Virol. 2020;30(5):e2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS‐CoV‐2. Gene Rep. 2020;19:100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McHugh J. Arginine restriction attenuates bone loss in arthritis. Nat Rev Rheumatol. 2020;16(3):128. [DOI] [PubMed] [Google Scholar]
- 13. Yao H, Song Y, Chen Y, et al. Molecular architecture of the SARS‐CoV‐2 virus. Cell. 2020;183(3):730‐738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai W, He L, Zhang X, et al. Characterization of the receptor‐binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020;17(6):613‐620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gu S, Chen Y, Wu Z, et al. Alterations of the gut microbiota in patients with COVID‐19 or H1N1 influenza. [published online ahead of print Jan 4, 2020]. Clin Infect Dis. 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID‐19 during time of hospitalization. Gastroenterology. 2020;159(3):944‐955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang T, Chakraborty S, Saha P, et al. Gnotobiotic rats reveal that gut microbiota regulates colonic mRNA of Ace2, the receptor for SARS‐CoV‐2 infectivity. Hypertension. 2020;76(1):e1‐e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Codo AC, Davanzo GG, Monteiro LB, et al. Elevated glucose levels favor SARS‐CoV‐2 infection and monocyte response through a HIF‐1α/glycolysis‐dependent axis. Cell Metabol. 2020;32(3):437‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS‐CoV‐2 infection. Eur J Intern Med. 2020;76:14‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thevarajan I, Nguyen THO, Koutsakos M, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non‐severe COVID‐19. Nat Med. 2020;26(4):453‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. EOM, Byrne P, Walsh KA, et al. Immune response following infection with SARS‐CoV‐2 and other coronaviruses: a rapid review. Rev Med Virol. 2020: e2162. 10.1002/rmv.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID‐19). Clin Infect Dis. 2020;71(15):778‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liao M, Liu Y, Yuan J, et al. Single‐cell landscape of bronchoalveolar immune cells in patients with COVID‐19. Nat Med. 2020;26(6):842‐844. [DOI] [PubMed] [Google Scholar]
- 24. Blanco‐Melo D, Nilsson‐Payant BE, Liu WC, et al. Imbalanced host response to SARS‐CoV‐2 drives development of COVID‐19. Cell. 2020;181(5):1036‐1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mahmudpour M, Roozbeh J, Keshavarz M, Farrokhi S, Nabipour I. COVID‐19 cytokine storm: the anger of inflammation. Cytokine. 2020;133:155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID‐19. Nat Med. 2020;26(7):1017‐1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID‐19. Lancet. 2020;395(10234):1417‐1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ackermann M, Verleden SE, Kuehnel M, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid‐19. N Engl J Med. 2020;383(2):120‐128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leisman DE, Ronner L, Pinotti R, et al. Cytokine elevation in severe and critical COVID‐19: a rapid systematic review, meta‐analysis, and comparison with other inflammatory syndromes. Lancet Respir Med. 2020;8(12):1233‐1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gu SX, Tyagi T, Jain K, et al. Thrombocytopathy and endotheliopathy: crucial contributors to COVID‐19 thromboinflammation. Nat Rev Cardiol. 2020. 10.1038/s41569-020-00469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franco R, Rivas‐Santisteban R, Serrano‐Marín J, Rodríguez‐Pérez AI, Labandeira‐García JL, Navarro G. SARS‐CoV‐2 as a factor to disbalance the renin‐angiotensin system: a suspect in the case of exacerbated IL‐6 production. J Immunol. 2020;205(5):1198‐1206. [DOI] [PubMed] [Google Scholar]
- 32. Guo C, Li B, Ma H, et al. Single‐cell analysis of two severe COVID‐19 patients reveals a monocyte‐associated and tocilizumab‐responding cytokine storm. Nat Commun. 2020;11(1):3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Costa FF, Rosário WR, Ribeiro Farias AC, de Souza RG, Duarte Gondim RS, Barroso WA. Metabolic syndrome and COVID‐19: an update on the associated comorbidities and proposed therapies. Diabetes Metab Syndr. 2020;14(5):809‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin L, Jiang X, Zhang Z, et al. Gastrointestinal symptoms of 95 cases with SARS‐CoV‐2 infection. Gut. 2020;69(6):997‐1001. [DOI] [PubMed] [Google Scholar]
- 36. Park SK, Lee CW, Park DI, et al. Detection of SARS‐CoV‐2 in fecal samples from patients with asymptomatic and mild COVID‐19 in Korea. Clin Gastroenterol Hepatol. 2020. 10.1016/j.cgh.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ng SC, Tilg H. COVID‐19 and the gastrointestinal tract: more than meets the eye. Gut. 2020;69(6):973‐974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sulaiman T, Algharawi AA, Idrees M, et al. The prevalence of gastrointestinal symptoms among patients with COVID‐19 and the effect on the severity of the disease. JGH Open. 2020. 10.1002/jgh3.12415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elmunzer BJ, Spitzer RL, Foster LD, et al. Digestive manifestations in patients hospitalized with COVID‐19. Clin Gastroenterol Hepatol. 2020;S1542‐3565(20):31371–31379. 10.1016/j.cgh.2020.09.041. [DOI] [Google Scholar]
- 40. Woodruff M, Ramonell R, Cashman K, et al. Critically ill SARS‐CoV‐2 patients display lupus‐like hallmarks of extrafollicular B cell activation. medRxiv; 2020. 10.1101/2020.04.29.20083717. [DOI] [Google Scholar]
- 41. Wong MC, Huang J, Lai C, Ng R, Chan FKL, Chan PKS. Detection of SARS‐CoV‐2 RNA in fecal specimens of patients with confirmed COVID‐19: a meta‐analysis. J Infect. 2020;81(2):e31‐e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lamers MM, Beumer J, van der Vaart J, et al. SARS‐CoV‐2 productively infects human gut enterocytes. Science. 2020;369(6499):50‐54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou J, Li C, Liu X, et al. Infection of bat and human intestinal organoids by SARS‐CoV‐2. Nat Med. 2020;26(7):1077‐1083. [DOI] [PubMed] [Google Scholar]
- 44. Dickson I. Organoids demonstrate gut infection by SARS‐CoV‐2. Nat Rev Gastroenterol Hepatol. 2020;17(7):383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dosoky NS, May‐Zhang LS, Davies SS. Engineering the gut microbiota to treat chronic diseases. Appl Microbiol Biotechnol. 2020;104(18):7657‐7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruff WE, Greiling TM, Kriegel MA. Host‐microbiota interactions in immune‐mediated diseases. Nat Rev Microbiol. 2020;18(9):521‐538. [DOI] [PubMed] [Google Scholar]
- 47. Illiano P, Brambilla R, Parolini C. The mutual interplay of gut microbiota, diet and human disease. FEBS J. 2020;287(5):833‐855. [DOI] [PubMed] [Google Scholar]
- 48. Roshan N, Clancy AK, Borody TJ. Faecal microbiota transplantation is effective for the initial treatment of Clostridium difficile infection: a retrospective clinical review. Infect Dis Ther. 2020;9(4):935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol. 2016;13(9):508‐516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rubin TA, Gessert CE, Aas J, Bakken JS. Fecal microbiome transplantation for recurrent Clostridium difficile infection: report on a case series. Anaerobe. 2013;19:22‐26. [DOI] [PubMed] [Google Scholar]
- 51. Adam Y, Meinlschmidt G, Lieb R. Associations between mental disorders and the common cold in adults: a population‐based cross‐sectional study. J Psychosom Res. 2013;74(1):69‐73. [DOI] [PubMed] [Google Scholar]
- 52. Cohen S, Tyrrell DA, Smith AP. Psychological stress and susceptibility to the common cold. N Engl J Med. 1991;325(9):606‐612. [DOI] [PubMed] [Google Scholar]
- 53. Dinan TG, Cryan JF. Mood by microbe: towards clinical translation. Genome Med. 2016;8(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang N, Zhang Y, Li M, et al. Efficacy of probiotics on stress in healthy volunteers: a systematic review and meta‐analysis based on randomized controlled trials. Brain Behav. 2020;10(9):e01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang J, Chen WD, Wang YD. The relationship between gut microbiota and inflammatory diseases: the role of macrophages. Front Microbiol. 2020;11:1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U. S. A. 2014;111(6):2247‐2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Arpaia N, Campbell C, Fan X, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature. 2013;504(7480):451‐455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chen J, Vitetta L. The role of butyrate in attenuating pathobiont‐induced hyperinflammation. Immune Netw. 2020;20(2):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen J, Zhao KN, Vitetta L. Effects of intestinal microbial‐elaborated butyrate on oncogenic signaling pathways. Nutrients. 2019;11(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fachi JL, Felipe JS, Pral LP, et al. Butyrate protects mice from Clostridium difficile‐induced colitis through an HIF‐1‐dependent mechanism. Cell Rep. 2019;27(3):750‐761. [DOI] [PubMed] [Google Scholar]
- 61. Lynch JP, Werder RB, Loh Z, et al. Plasmacytoid dendritic cells protect from viral bronchiolitis and asthma through semaphorin 4a‐mediated Treg expansion. J Exp Med. 2018;215(2):537‐557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Antunes KH, Fachi JL, de Paula R, et al. Microbiota‐derived acetate protects against respiratory syncytial virus infection through a GPR43‐type 1 interferon response. Nat Commun. 2019;10(1):3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Campbell C, McKenney PT, Konstantinovsky D, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581(7809):475‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hang S, Paik D, Yao L, et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature. 2019;576(7785):143‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Song X, Sun X, Oh SF, et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature. 2020;577(7790):410‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haroon E, Welle JR, Woolwine BJ, et al. Associations among peripheral and central kynurenine pathway metabolites and inflammation in depression. Neuropsychopharmacology. 2020;45(6):998‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Badawy AA, Guillemin G. The plasma [Kynurenine]/[Tryptophan] ratio and indoleamine 2,3‐Dioxygenase: time for appraisal. Int J Tryptophan Res. 2019;12:1178646919868978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Thomas T, Stefanoni D, Reisz JA, et al. COVID‐19 infection alters kynurenine and fatty acid metabolism, correlating with IL‐6 levels and renal status. JCI Insight. 2020;5(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2014;7(1):17‐44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Onder G, Rezza G, Brusaferro S. Case‐fatality rate and characteristics of patients dying in relation to COVID‐19 in Italy. JAMA. 2020;323(18):1775‐1776. [DOI] [PubMed] [Google Scholar]
- 71. Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model‐based analysis. Lancet Infect Dis. 2020;20(6):669‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Du Y, Tu L, Zhu P, et al. Clinical features of 85 fatal cases of COVID‐19 from Wuhan. A retrospective observational study. Am J Respir Crit Care Med. 2020;201(11):1372‐1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Franceschi C, Garagnani P, Parini P, Giuliani C, Santoro A. Inflammaging: a new immune‐metabolic viewpoint for age‐related diseases. Nat Rev Endocrinol. 2018;14(10):576‐590. [DOI] [PubMed] [Google Scholar]
- 74. Churov AV, Mamashov KY, Novitskaia AV. Homeostasis and the functional roles of CD4(+) Treg cells in aging. Immunol Lett. 2020;226:83‐89. [DOI] [PubMed] [Google Scholar]
- 75. Schmitt V, Rink L, Uciechowski P. The Th17/Treg balance is disturbed during aging. Exp Gerontol. 2013;48(12):1379‐1386. [DOI] [PubMed] [Google Scholar]
- 76. Shintouo CM, Mets T, Beckwee D, et al. Is inflammageing influenced by the microbiota in the aged gut? A systematic review. Exp Gerontol. 2020;141:111079. [DOI] [PubMed] [Google Scholar]
- 77. Hill MA, Mantzoros C, Sowers JR. Commentary: COVID‐19 in patients with diabetes. Metabolism. 2020;107:154217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kwok S, Adam S, Ho JH, et al. Obesity: a critical risk factor in the COVID‐19 pandemic. Clin Obes. 2020;10(6):e12403. 10.1111/cob.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guarisco G, Leonetti F. Covid‐19 and diabesity: when a pandemia cross another pandemia. Eat Weight Disord. 2020. 10.1007/s40519-020-00958-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chee YJ, Tan SK, Yeoh E. Dissecting the interaction between COVID‐19 and diabetes mellitus. J Diabetes Investig. 2020;11(5):1104‐1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McNelis JC, Olefsky JM. Macrophages, immunity, and metabolic disease. Immunity. 2014;41(1):36‐48. [DOI] [PubMed] [Google Scholar]
- 82. Nishimura S, Manabe I, Nagasaki M, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15(8):914‐920. [DOI] [PubMed] [Google Scholar]
- 83. Feuerer M, Herrero L, Cipolletta D, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930‐939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cipolletta D, Feuerer M, Li A, et al. PPAR‐γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486(7404):549‐553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027‐1031. [DOI] [PubMed] [Google Scholar]
- 86. Wang G, Si Q, Yang S, et al. Lactic acid bacteria reduce diabetes symptoms in mice by alleviating gut microbiota dysbiosis and inflammation in different manners. Food Funct. 2020;11(7):5898‐5914. [DOI] [PubMed] [Google Scholar]
- 87. Palacios T, Vitetta L, Coulson S, et al. Targeting the intestinal microbiota to prevent type 2 diabetes and enhance the effect of metformin on glycaemia: a randomised controlled pilot study. Nutrients. 2020;12(7):2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mandaliya DK, Seshadri S. Short chain fatty acids, pancreatic dysfunction and type 2 diabetes. Pancreatology. 2019;19(2):280‐284. [DOI] [PubMed] [Google Scholar]
- 89. Vallianou N, Stratigou T, Christodoulatos GS, Dalamaga M. Understanding the role of the gut microbiome and microbial metabolites in obesity and obesity‐associated metabolic disorders: current evidence and perspectives. Curr Obes Rep. 2019;8(3):317‐332. [DOI] [PubMed] [Google Scholar]
- 90. Jiao W, Zhang Z, Xu Y, et al. Butyric acid normalizes hyperglycemia caused by the tacrolimus‐induced gut microbiota. Am J Transpl. 2020;20(9):2413‐2424. [DOI] [PubMed] [Google Scholar]
- 91. Maniar K, Singh V, Moideen A, Bhattacharyya R, Chakrabarti A, Banerjee D. Inhalational supplementation of metformin butyrate: a strategy for prevention and cure of various pulmonary disorders. Biomed Pharmacother. 2018;107:495‐506. [DOI] [PubMed] [Google Scholar]
- 92. Qin J, Li Y, Cai Z, et al. A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55‐60. [DOI] [PubMed] [Google Scholar]
- 93. Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99‐103. [DOI] [PubMed] [Google Scholar]
- 94. Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262‐266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment‐naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850‐858. [DOI] [PubMed] [Google Scholar]
- 96. Mardinoglu A, Boren J, Smith U. Confounding effects of metformin on the human gut microbiome in type 2 diabetes. Cell Metabol. 2016;23(1):10‐12. [DOI] [PubMed] [Google Scholar]
- 97. Scheen AJ. Metformin and COVID‐19: From cellular mechanisms to reduced mortality. Diabetes Metab. 2020;46(6):423‐426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Crouse A, Grimes T, Li P, Might M, Ovalle F, Shalev A. Metformin use is associated with reduced mortality in a diverse population with COVID‐19 and diabetes. medRxiv. 2020. 10.1101/2020.07.29.20164020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Luo P, Qiu L, Liu Y, et al. Metformin treatment was associated with decreased mortality in COVID‐19 patients with diabetes in a retrospective analysis. Am J Trop Med Hyg. 2020;103(1):69‐72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Do JY, Kim SW, Park JW, Cho KH, Kang SH. Is there an association between metformin use and clinical outcomes in diabetes patients with COVID‐19? Diabetes Metab. 2020. 10.1016/j.diabet.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Gao Y, Liu T, Zhong W, et al. Risk of metformin in patients with type 2 diabetes with COVID‐19: a preliminary retrospective report. Clin Transl Sci. 2020;13(6):1055–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ganesan K, Chung SK, Vanamala J, Xu B. Causal relationship between diet‐induced gut microbiota changes and diabetes: a novel strategy to transplant Faecalibacterium prausnitzii in preventing diabetes. Int J Mol Sci. 2018;19(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zheng Z, Peng F, Xu B, et al. Risk factors of critical & mortal COVID‐19 cases: a systematic literature review and meta‐analysis. J Infect. 2020;81(2):e16‐e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chen L, Li X, Chen M, Feng Y, Xiong C. The ACE2 expression in human heart indicates new potential mechanism of heart injury among patients infected with SARS‐CoV‐2. Cardiovasc Res. 2020;116(6):1097‐1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Goulter AB, Goddard MJ, Allen JC, Clark KL. ACE2 gene expression is up‐regulated in the human failing heart. BMC Med. 2004;2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Burrell LM, Risvanis J, Kubota E, et al. Myocardial infarction increases ACE2 expression in rat and humans. Eur Heart J. 2005;26(4):369‐375. [DOI] [PubMed] [Google Scholar]
- 107. Bansal M. Cardiovascular disease and COVID‐19. Diabetes Metab Syndr. 2020;14(3):247‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ferrario CM, Jessup J, Chappell MC, et al. Effect of angiotensin‐converting enzyme inhibition and angiotensin II receptor blockers on cardiac angiotensin‐converting enzyme 2. Circulation. 2005;111(20):2605‐2610. [DOI] [PubMed] [Google Scholar]
- 109. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181(4):905‐913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bonetti PO, Lerman LO, Lerman A. Endothelial dysfunction: a marker of atherosclerotic risk. Arterioscler Thromb Vasc Biol. 2003;23(2):168‐175. [DOI] [PubMed] [Google Scholar]
- 111. Trøseid M, Andersen G, Broch K, Hov JR. The gut microbiome in coronary artery disease and heart failure: current knowledge and future directions. EBioMedicine. 2020;52:102649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Hoel H, Heggelund L, Reikvam DH, et al. Elevated markers of gut leakage and inflammasome activation in COVID‐19 patients with cardiac involvement. J Intern Med. 2020. 10.1111/joim.13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Robles‐Vera I, Toral M, Duarte J. Microbiota and hypertension: role of the sympathetic nervous system and the immune system. Am J Hypertens. 2020;33(10):890–901. [DOI] [PubMed] [Google Scholar]
- 114. Kim S, Goel R, Kumar A, et al. Imbalance of gut microbiome and intestinal epithelial barrier dysfunction in patients with high blood pressure. Clin Sci. 2018;132(6):701‐718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. RRM, Marques FZ. Diet‐related gut microbial metabolites and sensing in hypertension. J Hum Hypertens. 2020. 10.1038/s41371-020-0388-3. [DOI] [PubMed] [Google Scholar]
- 116. Aguilar EC, Santos LC, Leonel AJ, et al. Oral butyrate reduces oxidative stress in atherosclerotic lesion sites by a mechanism involving NADPH oxidase down‐regulation in endothelial cells. J Nutr Biochem. 2016;34:99‐105. [DOI] [PubMed] [Google Scholar]
- 117. Robles‐Vera I, Toral M, de la Visitación N, Aguilera‐Sánchez N, Redondo JM, Duarte J. Protective effects of short‐chain fatty acids on endothelial dysfunction induced by angiotensin II. Front Physiol. 2020;11:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhu Y, Shui X, Liang Z, et al. Gut microbiota metabolites as integral mediators in cardiovascular diseases (Review). Int J Mol Med. 2020;46(3):936‐948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Wypych TP, Wickramasinghe LC, Marsland BJ. The influence of the microbiome on respiratory health. Nat Immunol. 2019;20(10):1279‐1290. [DOI] [PubMed] [Google Scholar]
- 120. Budden KF, Gellatly SL, Wood DL, et al. Emerging pathogenic links between microbiota and the gut‐lung axis. Nat Rev Microbiol. 2017;15(1):55‐63. [DOI] [PubMed] [Google Scholar]
- 121. Zhang D, Li S, Wang N, Tan HY, Zhang Z, Feng Y. The cross‐talk between gut microbiota and lungs in common lung diseases. Front Microbiol. 2020;11:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Conte L, Toraldo DM. Targeting the gut‐lung microbiota axis by means of a high‐fibre diet and probiotics may have anti‐inflammatory effects in COVID‐19 infection. Ther Adv Respir Dis. 2020;14:1753466620937170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Aktas B, Aslim B. Gut‐lung axis and dysbiosis in COVID‐19. Turk J Biol. 2020;44(3):265‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. He LH, Ren LF, Li JF, Wu YN, Li X, Zhang L. Intestinal flora as a potential strategy to fight SARS‐CoV‐2 infection. Front Microbiol. 2020;11:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Singh S, Khan A. Clinical characteristics and outcomes of coronavirus disease 2019 among patients with preexisting liver disease in the United States: a multicenter research network study. Gastroenterology. 2020;159(2):768‐771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Verma N, Duseja A, Singh V. Impact of pre‐existing chronic liver disease on the outcome of patients with COVID‐19 disease. Gastroenterology. 2020. 10.1053/j.gastro.2020.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zhou YJ, Zheng KI, Wang XB, et al. Metabolic‐associated fatty liver disease is associated with severity of COVID‐19. Liver Int. 2020;40(9):2160‐2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chen VL, Hawa F, Berinstein JA, et al. Hepatic steatosis is associated with increased disease severity and liver injury in coronavirus disease‐19. Dig Dis Sci. 2020. 10.1007/s10620-020-06618-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lippi G, de Oliveira MHS, Henry BM. Chronic liver disease is not associated with severity or mortality in coronavirus disease 2019 (COVID‐19): a pooled analysis. Eur J Gastroenterol Hepatol. 2020;33(1):114–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Kullar R, Patel AP, Saab S. Hepatic injury in patients with COVID‐19. J Clin Gastroenterol. 2020;54(10):841‐849. [DOI] [PubMed] [Google Scholar]
- 131. Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33(3):128‐133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chen J, Thomsen M, Vitetta L. Interaction of gut microbiota with dysregulation of bile acids in the pathogenesis of nonalcoholic fatty liver disease and potential therapeutic implications of probiotics. J Cell Biochem. 2019;120(3):2713‐2720. [DOI] [PubMed] [Google Scholar]
- 133. Batty GD, Deary IJ, Luciano M, Altschul DM, Kivimäki M, Gale CR. Psychosocial factors and hospitalisations for COVID‐19: prospective cohort study based on a community sample. Brain Behav Immun. 2020;89:569‐578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vanuytsel T, van Wanrooy S, Vanheel H, et al. Psychological stress and corticotropin‐releasing hormone increase intestinal permeability in humans by a mast cell‐dependent mechanism. Gut. 2014;63(8):1293‐1299. [DOI] [PubMed] [Google Scholar]
- 135. Anderson G, Reiter RJ. Melatonin: roles in influenza, Covid‐19, and other viral infections. Rev Med Virol. 2020;30(3):e2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Archer DL, Kramer DC. The use of microbial accessible and fermentable carbohydrates and/or butyrate as supportive treatment for patients with coronavirus SARS‐CoV‐2 infection. Front Med. 2020;7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Subramanian S, Iles T, Ikramuddin S, Steer CJ. Merit of an ursodeoxycholic acid clinical trial in COVID‐19 patients. Vaccines. 2020;8(2):320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lee JR, Huang J, Magruder M, et al. Butyrate‐producing gut bacteria and viral infections in kidney transplant recipients: a pilot study. Transpl Infect Dis. 2019;21(6):e13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ye K, Tang F, Liao X, et al. Does serum vitamin D level affect COVID‐19 infection and its severity? A case‐control study. J Am Coll Nutr. 2020:1‐8. [DOI] [PubMed] [Google Scholar]
- 140. Panagiotou G, Tee SA, Ihsan Y, et al. Low serum 25‐hydroxyvitamin D (25[OH]D) levels in patients hospitalized with COVID‐19 are associated with greater disease severity. Clin Endocrinol. 2020;93(5):629–630. [DOI] [PubMed] [Google Scholar]
- 141. Liu PT, Stenger S, Li H, et al. Toll‐like receptor triggering of a vitamin D‐mediated human antimicrobial response. Science. 2006;311(5768):1770‐1773. [DOI] [PubMed] [Google Scholar]
- 142. Daniel C, Sartory NA, Zahn N, Radeke HH, Stein JM. Immune modulatory treatment of trinitrobenzene sulfonic acid colitis with calcitriol is associated with a change of a T helper (Th) 1/Th17 to a Th2 and regulatory T cell profile. J Pharmacol Exp Ther. 2008;324(1):23‐33. [DOI] [PubMed] [Google Scholar]
- 143. Vaidya A, Williams JS. The relationship between vitamin D and the renin‐angiotensin system in the pathophysiology of hypertension, kidney disease, and diabetes. Metabolism. 2012;61(4):450‐458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wang J, Thingholm LB, Skiecevičienė J, et al. Genome‐wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat Genet. 2016;48(11):1396‐1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable for this narrative review.


