Summary
The novel coronavirus disease‐2019 (Covid‐19) public health emergency has caused enormous loss around the world. This pandemic is a concrete example of the existing gap between availability of advanced diagnostics and current need for cost‐effective methodology. The advent of the loop‐mediated isothermal amplification (LAMP) assay provided an innovative tool for establishing a rapid diagnostic technique based on the molecular amplification of pathogen RNA or DNA. In this review, we explore the applications, diagnostic effectiveness of LAMP test for molecular diagnosis and surveillance of severe acute respiratory syndrome coronavirus 2. Our results show that LAMP can be considered as an effective point‐of‐care test for the diagnosis of Covid‐19 in endemic areas, especially for low‐ and middle‐income countries.
Keywords: coronavirus, Covid‐19, diagnostic, loop‐mediated isothermal amplification (LAMP), POC, SARS‐CoV‐2
Abbreviations
- CoV
coronavirus
- DETECTR
DNA Endonuclease‐Targeted CRISPR Trans‐Reporter
- Flongle
flow cell dongle
- LAMP
loop‐mediated isothermal amplification
- PCR
polymerase chain reaction
- POCT
point‐of‐care test
- RT
reverse transcription
- SARS
severe acute respiratory syndrome
1. INTRODUCTION
The recent outbreak of the new coronavirus (severe acute respiratory syndrome coronavirus‐2 [SARS‐CoV‐2]), firstly reported from Wuhan, China, in December 2019, has spread across the globe in the last few months, causing more than 40 million infections and over one million deaths. SARS‐CoV‐2 belongs to the subgenus sarbecovirus, orthocoronavirinae subfamily. Like other betacoronaviruses, the genome of SARS‐CoV‐2 has five typical open reading frame (ORFs) on the same coding strand. 1
On 30 January 2020, the World Health Organisation (WHO) declared the Covid‐19 outbreak as a global public health emergency. The virus spreads so rapidly that it has attracted global attention, and rapid and simple diagnosis is one of the most effective ways to control and prevent the disease.
Diagnosis of Covid‐19 based on clinical symptoms, but diagnosis at early stages of infection, is difficult as patients can remain asymptomatic and there are no specific initial manifestations. Symptoms may appear in few days or up to 2 weeks after exposure. 2 Chest computed tomography and reverse transcription polymerase chain reaction (RT‐PCR) have been used for the coronavirus pneumonia clinical diagnosis. 3 RT‐PCR protocols have been rapidly developed for the quantitative and qualitative detection of SARS‐CoV‐2 in respiratory fluid, sputum, nasopharyngeal swabs and blood samples. 4 However, the application of RT‐PCR requires well‐equipped laboratories, experienced personnel and optimal conditions, the minimum total run time of an RT‐PCR test is 3–5 h from collecting the sample to result reporting. 5
To address these challenges, a rapid, specific, sensitive, robust and quantitative diagnostic tool would be highly desirable. Loop‐mediated isothermal amplification (LAMP) could be used for high‐throughput screening applications in both referral and local laboratories. Recently, different studies have presented the applicability of the LAMP as an effective tool for simple and rapid detection of pathogens. 5 , 6 , 7 , 8
Since the advent of the Covid‐19 pandemic, various studies have developed several LAMP assay prototypes for SARS‐CoV‐2 diagnosis. This work presents a review of the actual status, diagnosis robustness and perspectives of the LAMP test for this purpose.
1.1. Developments and features of the LAMP technique
During the year 2000, Notomi et al. 9 developed a new molecular technique, LAMP, as a simple field and cost‐effective diagnostic tool. 9 The LAMP technique uses a DNA polymerase from Bacillus stearothermophilus (Bst) that has polymerase and reverse transcriptase activity. Technically, LAMP uses two inner primers (FIP and BIP) and two outer primers (F3 and B3) that can recognize a total of six distinct regions in the target DNA. Two extra loop primers are also employed (LF and LB) to accelerate amplification and improve detection performance (Figure 1). 10 The DNA synthesis initiation by multiple primers makes the technique highly specific. 11 The Bst polymerase properties enable amplification using a normal water bath or heating block maintained at a fixed temperature avoiding the use of thermal cycler machines. Additionally, LAMP amplification product can be visualized using agarose gel electrophoresis and/or colorimetric naked‐eye detection systems 12 and real‐time fluorimetry. 13 , 14 Therefore, one of the major advantages of LAMP is its possible deployment in the field and in resource‐limited settings.
FIGURE 1.
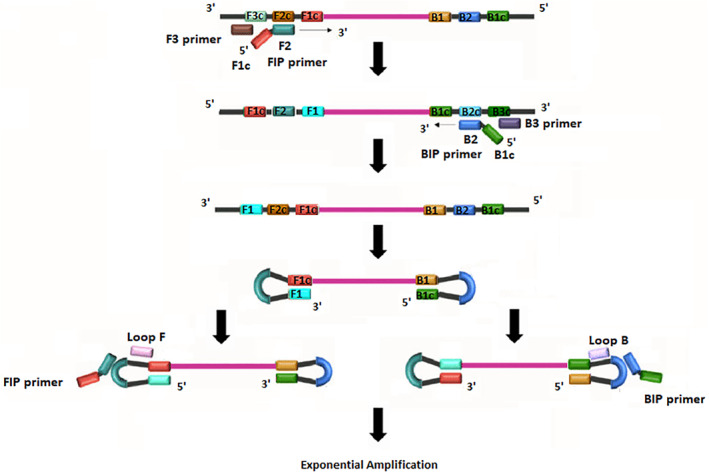
Schematic principle of loop‐mediated isothermal amplification
1.2. Search strategies and data sources
We went through articles in bioRxiv, Web of Science, PubMed, Embase, Google Scholar, Scopus and medRxiv, with different keywords: Covid‐19 LAMP assay, SARS‐CoV‐2, molecular diagnosis, Rapid diagnosis, RT‐PCR and LAMP, Covid‐19 diagnosis, Coronavirus RNA, Coronavirus Pneumonia and related words. The selection criteria covered the studies interested Covid‐19 LAMP development with or without clinical test studies.
1.3. Application of LAMP technique to SARS‐CoV‐2
Many scientific groups are interested in the development of the LAMP assay as a molecular tool for the development of a point‐of‐care (POC) test amplifying coronavirus RNA. Until July 2020, a total of 42 LAMP assays were developed and evaluated using clinical or simulated respiratory samples (Table 1).
TABLE 1.
Overview of LAMP assays for diagnosis of Covid‐19 reported in previous studies
| Author | Location | Publishing date | Gene target | Type of samples | Sampling | Visual detection | Sensitivity/specificity |
|---|---|---|---|---|---|---|---|
| Lamb et al. 15 | USA | 14/02/2020 | ORF1ab | Synthetized | N/A | SYBR Green I | *** |
| El‐Tholoth et al. 16 | USA | 19/02/2020 | ORF1ab | Synthetized | N/A | LCV dye | 100%–100% |
| Yu et al. 17 | China | 24/02/2020 | ORF1ab | Respiratory samples | 43 | SYBR Green/Finder | 97.6%–100% |
| Zhang et al. 18 | UK | 29/02/2020 | ORF1a, N, A | Nasopharyngeal swab | 7 | pH indicator dyes | 100%–100% |
| Yang et al. 19 | China | 02/03/2020 | ORF1ab, E and N | Nasopharyngeal swab | 208 | Fluorescent calcein | 99%–99% |
| Broughton et al. 20 | USA | 06/03/2020 | N, E | Nasopharyngeal swab | 78 | Lateral flow strip | 95%–100% |
| Nguyen et al. 21 | Denmark | 14/03/2020 | N/A | N/A | N/A | N/A | *** |
| Jiang et al. 22 | China | 15/03/2020 | N | Nasopharyngeal swab | 260 | RT‐monitoring | 91.4%–99.5% |
| Zhu et al. 23 | China | 17/03/2020 | ORF1ab, N | Nasopharyngeal swab | 129 | Dye streptavidin coated polymer nanoparticles | 100%–100% |
| Lu et al. 54 | China | 22/03/2020 | RdRp | Synthetized | N/A | WarmStart® Colorimetric LAMP | 100%–100% |
| Park et al. 24 | Korea | 27/03/2020 | Nsp3, S, ORF8 | Synthetized | N/A | Hydroxy‐naphtol‐blue | *** |
| Österdahl et al. 50 | UK | 01/04/2020 | ORF1a | Nasopharyngeal swab | 24 | RT‐monitoring | 80%–73% |
| Yan et al. 25 | China | 02/04/2020 | ORF1ab, S | Respiratory samples | 130 | Fluorescent calcein | 100%–100% |
| Butt et al. 26 | Pakistan | 08/04/2020 | ORF1a, N | Nasopharyngeal swab | 70 | WarmStart colorimetric LAMP | 95%–100% |
| Schmid‐Burgk et al. 27 | USA | 08/04/2020 | ORF1a, N, A | N/A | N/A | Agarose gel | *** |
| González et al. 51 | Mexico | 09/04/2020 | N | Synthetized | N/A | WarmStart colorimetric LAMP | *** |
| Bhadra et al. 28 | USA | 13/04/2020 | ORF1a, N, A | N/A | N/A | Fluorogenic oligonucleotide strand exchange | *** |
| Huang et al. 29 | UK | 14/04/2020 | ORF1ab, N, S | Nasopharyngeal swab | 16 | WarmStart colorimetric LAMP | 100%–100% |
| Baek et al. 30 | Korea | 20/04/2020 | N | Nasopharyngeal swab | 154 | WarmStart colorimetric LAMP | 100%–98.7% |
| Wang D 31 | China | 21/04/2020 | N | Synthetized | N/A | EvaGreen | *** |
| Sun et al. 32 | USA | 21/04/2020 | Equine model | N/A | EvaGreen | *** | |
| Kashir and Yaqinuddin 10 | Saudi Arabia | 23/04/2020 | N/A | N/A | N/A | N/A | *** |
| Rabe et al. 33 | USA | 23/04/2020 | ORF1a, N, A | Nasopharyngeal swab, saliva | N/A | WarmStart colorimetric LAMP | *** |
| Lee et al. 34 | Australia | 28/04/2020 | N | Nasopharyngeal swab | 157 | RT‐monitoring | 87%–100% |
| Mohon et al. 35 | Canada | 07/05/2020 | RdRp, S | Nasopharyngeal swab | 120 | Colorimetric fluorescence indicator | 97.6%–98.7 |
| Ben‐Assa et al. 36 | Israel | 07/05/2020 | ORF1a, N, A | Nasopharyngeal swab/saliva | 180 | WarmStart colorimetric LAMP | 25.9%–100% |
| Dao Thi et al. 37 | Germany | 09/05/2020 | ORF1a, N, A | Nasopharyngeal swab | 95 | WarmStart colorimetric LAMP | 92%–99.7% |
| Lalli et al. 38 | USA | 11/05/2020 | ORF1a, N, A | Saliva | 6 | WarmStart colorimetric LAMP | *** |
| Anahtar et al. 39 | USA | 18/05/2020 | ORF1a, N | Nasopharyngeal swab | 62 | WarmStart colorimetric LAMP | 87.5%–100% |
| Ganguli et al. 40 | USA | 21/05/2020 | ORF1ab, N, S | Synthetized | RT‐monitoring | *** | |
| Hu et al. 41 | China | 23/05/2020 | S | Nasopharyngeal swab | 205 | Hydroxy‐naphtol‐blue | 88.89%–99.00% |
| Tran et al. 42 | Vietnam | 25/05/2020 | ORF1ab, N | Synthetized | N/A | WarmStart colorimetric LAMP | 100%–100% |
| Haq et al. 43 | Pakistan | 29/05/2020 | ORF1ab, N, S | Nasopharyngeal swab | 72 | WarmStart colorimetric LAMP | 100%–100% |
| Li et al. 56 | China | 03/06/2020 | ORF1ab, N | Synthetized | N/A | RT‐monitoring | *** |
| Lau et al. 44 | Malaysia | 03/06/2020 | N | Nasopharyngeal swab | 49 | Hydroxy‐naphtol‐blue | 100%–100% |
| Zhang et al. 18 | UK | 03/06/2020 | N, E | Synthetized | N/A | WarmStart colorimetric LAMP | 87.5%–100 |
| Kellner et al. 45 | Austria | 23/06/2020 | ORF1ab, N, E | Nasopharyngeal swab | N/A | Hydroxy‐naphtol‐blue | 100%–100% |
| Matsumura et al. 46 | Japan | 24/06/2020 | N/A | Nasopharyngeal swab, oropharyngeal, sputum | 155 | RT‐monitoring | 80.9%–100% |
| Eckel et al. 47 | Germany | 03/07/2020 | N/A | Nasopharyngeal swab | 109 | RT‐monitoring | 17%–88.7% |
| Ooi et al. 48 | Singapore | 03/07/2020 | N, E | Synthetized | N/A | Lateral flow strip | *** |
| Nagura‐Ikeda et al. 49 | Japan | 07/07/2020 | N/A | Saliva | 103 | RT‐monitoring | 70.9%–100% |
Note: *** = Not Applicable.
Abbreviations: LAMP, loop‐mediated isothermal amplification; ORF, open reading frame; RT, reverse transcription.
The first studies were reported by Lamb et al. 15 and El‐Tholoth et al. 16 The objective of the first study 15 was to develop a fast screening diagnostic test. Simulated samples were generated by spiking biological samples with a fraction of the SARS‐CoV‐2 nucleic sequence. Primers were designed based on the publicly available SARS‐CoV‐2 data and also compared to other coronavirus sequences. To select the optimal conditions for reverse transcription‐LAMP (RT‐LAMP), different set‐up modifications were evaluated, diverse primer sets, several ranges of temperatures (55–65°C) and incubation times (20–45 min) were also assessed. The best amplification conditions were reached at 63°C for 30 min. To determine the LAMP detection limit, titred virus was serially diluted. The specificity of the LAMP assays was tested by testing samples extracted from different pathogens, including viruses, fungi and bacteria.
Similarly, El‐Tholoth et al. 16 reported the design of a two‐step LAMP (Covid‐19 Penn‐RAMP) achieved in closed tubes with colorimetric or fluorescence detection. When testing purified targets, the LAMP assay performance was different to conventional RT‐PCR assays, showing 10‐fold higher sensitivity. 16
Since these first two publications, the number of studies has grown, and so many countries are now focusing on this technique (Figure 2).
FIGURE 2.
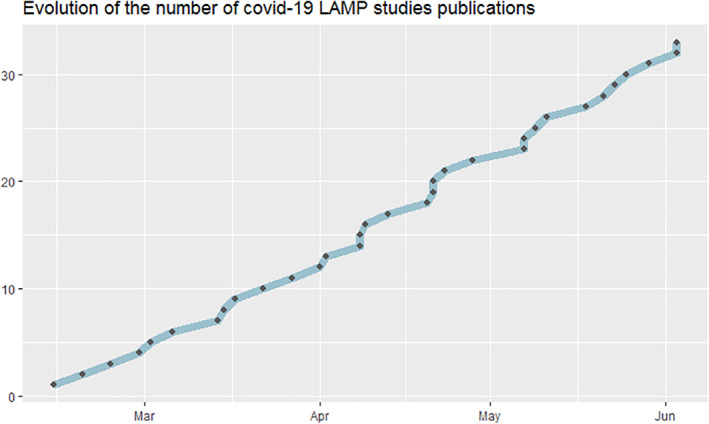
Evolution of the number of Covid‐19 loop‐mediated isothermal amplification studies
2. THE SARS‐CoV‐2 LAMP ASSAYS STUDIES
2.1. Geographic distribution of developed LAMP
Like the distribution of the pandemic, the highest number of studies related to LAMP was registered in China and the United States (10) (Figure 3). Then, the United Kingdom with four publications. Germany, Japan, Pakistan and Korea published two studies each; one of the Korean studies developed and evaluated RT‐LAMP assay using samples collected from Covid‐19 patients. The results revealed high agreement with the RT‐PCR tested on 154 clinical samples. 30
FIGURE 3.
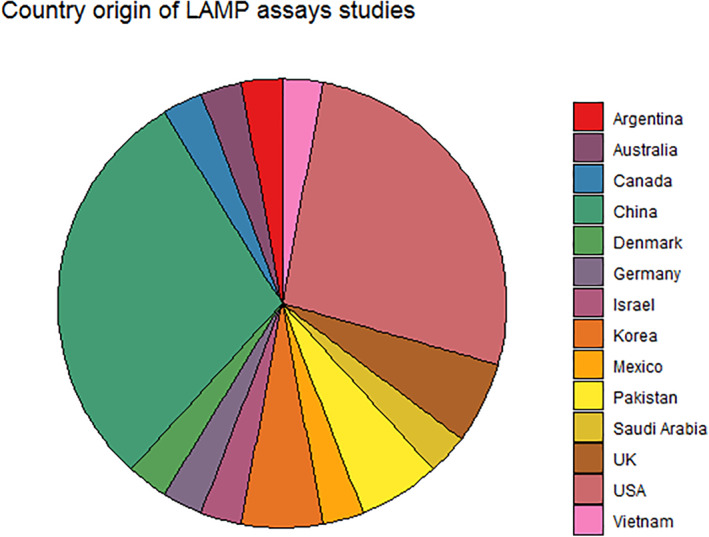
Country origin of loop‐mediated isothermal amplification assays studies
Only seven studies were reported from Europe (UK, Germany and Denmark) despite the large number of infections and deaths (WHO). Two assays were tested on biological samples, 37 , 50 the UK researchers piloted an RT‐LAMP assay on nasopharyngeal samples from 21 residents in a dependency care home, with two index Covid‐19 cases, and compared it to RT‐PCR. The German researchers used purified RNA and crude pharyngeal samples and revealed that RT‐LAMP assays have excellent specificity, despite a lower sensitivity compared to RT‐PCR.
Haq et al. 43 and Butt et al. 26 implemented the RT‐LAMP protocol for the qualitative detection of viral RNA. Extracted RNA from 70 nasopharyngeal swabs was analysed using RT‐PCR and RT‐LAMP. The second study from Pakistan developed and validated an RT‐LAMP to propose a potential RT‐PCR alternative for rapid testing of suspected Covid‐19 individuals. Comparative RT‐LAMP assay assessment showed good specificity and sensitivity.
A single study was reported from two other countries. Lee et al. 34 from Australia evaluated their LAMP assay on 157 clinical specimens previously screened by E‐gene RT‐PCR and revealed a specificity and sensitivity of 100% and 87%. A Canadian study reported that the validation of LAMP targeting the S gene compared to RT‐PCR reference, exhibited a negative percent agreement (NPA) and positive percent agreement (PPA) of 98.72% and 97.62%, respectively. RT‐LAMP targeting S and RdRp gene showed an NPA 100% and PPA of 91.97% when discrepant samples were included. 35
The first Latin American study 51 demonstrated the mixed use of a colorimetric embodiment of LAMP. This strategy was used to amplify and detect SARS‐CoV‐2 RNA using a set of in house designed initiators that target the N protein.
2.2. Gene targets
The LAMP specificity and sensitivity depend generally on the primer sets used; hence, when designing the primers, care must be taken. Despite the fact it is difficult to choose a valid and specific target for amplification (species‐specific target and/or highly conserved region), it is essential to confirm that the primers are specific and amplifies the selected target; for that, preliminary optimisation is required before final primer validation. Although the Eiken Primer Explorer software was the most used tool for LAMP primer design, proper primers can be manually designed. For the detection of SARS‐CoV‐2 RNA different target regions were selected. Most of the studies targeted the conserved sequence of nucleocapsid gene (N gene) and ORF1ab (Figure.4) because of their high homology and their divergence from the other coronaviruses. 29 The N gene is at the 3'‐end of the SARS‐CoV‐2 RNA, 52 ORF1ab encodes the replicase polyprotein and it is about 21‐kb long. 53 Dao Thi et al. 37 compared several primers and selected one primer set to detect N gene as the best. The same target was selected by Badhra et al. 28 and Zhang et al. 18 This corresponds to the results of Viehweger et al. 27 who reported that the N gene has the highest read coverage of all coronavirus genes after they sequenced RNA from cell cultures infected with coronavirus HCoV‐229E. Using RNA isolated in vitro, they confirmed that the RT‐LAMP detection limit using N gene primers is 100 copies. 54
FIGURE 4.
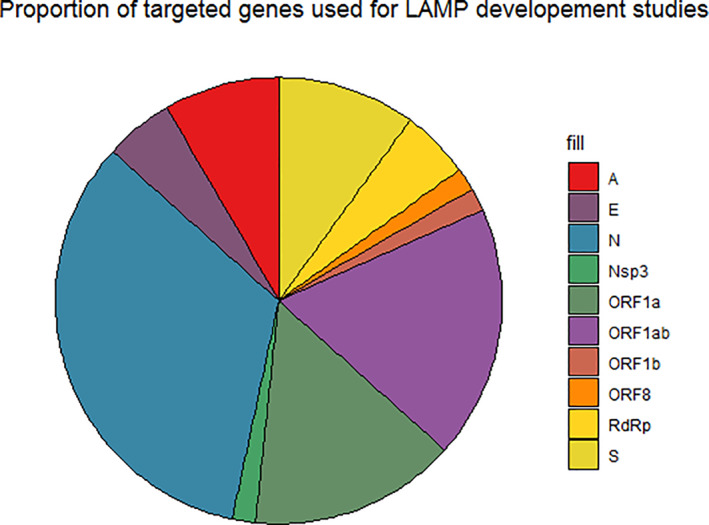
Proportion of targeted genes used for loop‐mediated isothermal amplification development studies
2.3. LAMP sensitivity, specificity and application
One of the major advantages of the LAMP technique is the high degree of sensitivity and specificity. Since the first tests, articles reported the successful application of RT‐LAMP assays to detect SARS‐CoV‐2 RNA in patient samples, demonstrating that 1–10 copies of viral RNA in a sample can lead to a successful detection, 10–100 fold more sensitive than classical RT‐PCR. The LAMP assays tested on clinical samples showed a high specificity (80%–100%) and sensitivity (73%–100%), applying repeated RT‐PCR as reference.
One principal limitation of LAMP assays for the diagnosis of SARS‐CoV‐2 arises from their dependence on time intensive and laboratory‐based procedures for viral isolation, lysis and removal of possible inhibiting materials. Recently, respiratory samples were directly tested by the variplex SARS‐CoV‐2 ready to use test based on LAMP method. However, the variplex test system failed to accurately detect SARS‐CoV‐2 without RNA extraction. 47
To avoid the time‐consuming and expensive sample preparation step and further increase the sensitivity, several studies focused on innovative approaches in sample preparation. Anahtar et al. 39 reported that RT‐LAMP can be applied directly from a nasopharyngeal sample; furthermore, LAMP sensitivity has risen by 30% with chemical RNase inactivation using TCEP/EDTA and heat‐mediated lysis. Moreover, this inactivation step reduces the sample infectivity as well decreasing the risk for laboratory personnel. Rabe et al. 33 developed a rapid process capable of inactivating virions and endogenous nucleases; this inactivation was coupled with a purification protocol. The purification and inactivation protocols, associated with RT‐LAMP, increased the sensitivity to at least one viral SARS‐CoV‐2 RNA copy per microliter.
An Austrian group presented several improvements that overcome the limitations of the SARS‐CoV‐2 RT‐LAMP. They adopted a method to reduce the risk of carry‐over contamination, improve the validity of the assay and result detection by using hydroxynaphtholblue as colorimetric visualisation. Additionally, they benchmarked a pipette‐free method that enables sensitive and specific identification of SARS‐Co‐V‐2. 45 , 44 Similarly, Zhang et al. 18 reported the use of guanidine hydrochloride and combined primer sets; Bhadra et al. 28 used sequence‐specific fluorogenic oligonucleotide strand exchange probes, to increase rapidity and sensitivity in colorimetric LAMP.
2.4. Combining LAMP with innovative techniques
Various studies chose to combine LAMP with other techniques to improve its efficiency. Broughton et al. 20 report the use of a CRISPR–Cas12 based assay named SARS‐CoV‐2 DNA endonuclease‐targeted CRISPR trans reporter (DETECTR) to detect SARS‐CoV‐2 RNA from patient samples. This assay performs in parallel reverse transcription and isothermal amplification using RT–LAMP. Studies reported that CRISPR‐based assays are not considering viral genome mutations and RNA editing in human cells, Ooi et al. 48 presented the VaNGuard (variant nucleotide guard) to avoid this inconvenience. This test can detect the virus when its genome or transcriptome has evolved or has been edited by deaminases in infected human cells. 48
Another innovative approach followed by Zhu et al. 23 was the development of a one‐step RT‐LAMP coupled with nanoparticle‐based biosensor assay. The SARS‐CoV‐2 RT‐LAMP‐NBS nearly equipment‐free platform makes it a useful diagnosis tool for in field, clinic, public health and primary care laboratories, especially for resource‐poor regions. 23
Schmid‐Burgk et al. 55 proposed LAMP‐Seq, a barcoded RT‐LAMP protocol that enables sample pooling and significantly reduce cost and organisational efforts. A Chinese team proposed a LAMP‐based method combined with nanopore Flongle (flow cell dongle) rapid real‐time sequencing workflow to detect SARS‐CoV‐2 in both laboratory and wild‐caught environment. Nanopore Flongle was developed to be a quick, accessible and cost‐efficient real‐time sequencing. The nanopore Flongle workflow was used for genome analysis and identification of SARS‐CoV‐2 after 30‐min sequencing. 56
To improve the ease of use, portability and reproducibility, a Mexican team 51 demonstrated the use of a three‐dimensional printed incubation chamber for commercial PCR tubes and a colorimetric LAMP reaction for the detection of coronavirus RNA.
Recently, a study compared 12 molecular diagnostic assays for SARS‐CoV‐2 including eight commercial kits. All the assays presented a specificity of 100% after testing 155 respiratory samples. The RT‐PCR N2 assay kits of the Japanese National Institute of Infectious Disease (NIID) and the US Centers for Disease Control and Prevention (CDC) were the most efficient assays with 100% sensitivity. Despite the fact that the LAMP assays presented a sensitivity of 80.9%, it is by far the fastest technique (35 min). 46
2.5. LAMP challenges and potential improvement
The main objective behind the advent of LAMP as a POC test is making this tool a patient‐centric diagnostic technique. However, various challenges remain, specifically in terms of limitations in sensitivity, reliability of the assay, effectiveness with untreated patient samples and making RT‐LAMP valid on field. Based on the reviewed articles, the latter points should be taking into account. LAMP quantitative methods that were developed are hardly applicable as POC tests due to the need of relative costly equipment and well trained personnel. Considering validness, performing RT‐LAMP outside laboratories can probably lead to carryover cross‐contamination, resulting in potentially large numbers of false positives, this point was rarely discussed. In term of sensitivity, current RT‐LAMP assays are reported to be one to two times less sensitive than RT‐PCR, leading also to substantial numbers of false negatives. 39 , 20 Another advantage of LAMP is the insensitivity to possible inhibitors in crude templates. Hence, it is important to focus on sample preparation and develop solutions and buffers to stabilize DNA in the sample in order to ensure subsequent amplification. 11 Usually, amplicons are stable, which can lead to unintended carry‐over contamination. Thereby, it is recommended to select a closed tube detection system to avoid post‐amplification contamination. 11 To deploy more effectively LAMP in field settings and adapt it for processing a large numbers of samples, new protocols and kits may need to be developed as well as frontline personnel trainings.
The development and application of SARS‐CoV‐2 RT‐LAMP assays is matching the global objective of seeking simple, rapid and affordable tests. Focusing on the above‐cited points will certainly improve the applicability of the LAMP assay, which is a good diagnostic tool suitable for this globally spread disease. Furthermore, it is in line with the WHO guidelines for diagnostics in developing countries, being ASSURED: Affordable, Sensitive, Specific, User‐friendly, Rapid and robust, Equipment‐free and Deliverable to end users. 57
3. CONCLUSION
In this review, we discussed the diagnostic performances and advances of the LAMP assay to detect SARS‐CoV‐2. The study outputs revealed that the RT‐LAMP method has reliable application for SARS‐CoV‐2 diagnosis due to its simple application and low technical requirements, thus presenting a potentially effective test to help us to fight the ongoing Covid‐19 pandemic.
CONFLICT OF INTEREST
The author declares that there is no conflict of interest.
ACKNOWLEDGEMENT
The author would like to thank Lyndon Zass (Computational Biology Division, Department of Integrative Biomedical Sciences, University of Cape Town, South Africa) for critical reading of the manuscript.
Chaouch M. Loop‐mediated isothermal amplification (LAMP): an effective molecular point‐of‐care technique for the rapid diagnosis of coronavirus SARS‐CoV‐2. Rev Med Virol. 2021;31(6):e2215. 10.1002/rmv.2215
REFERENCES
- 1. Phan T. Novel coronavirus: From discovery to clinical diagnostics. Infect Genet Evol. 2020;79:104211. 10.1016/j.meegid.2020.104211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Singhal T. A review of coronavirus disease‐2019 (COVID‐19). Indian J Pediatr. 2020; 87: 281‐286. 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25:23‐30. 10.2807/1560-7917.Es.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J, Wang H, Mao L, et al. Rapid genomic characterization of SARS‐CoV‐2 viruses from clinical specimens using nanopore sequencing. Sci Rep. 2020;10:17492. 10.1038/s41598-020-74656-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Luo Z, Ang MJY, Chan SY, et al. Combating the coronavirus pandemic: early detection, medical treatment, and a concerted effort by the global community. Research (Wash D C). 2020;2020:6925296. 10.34133/2020/6925296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Notomi T, Mori Y, Tomita N, Kanda H. Loop‐mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol. 2015;53:1‐5. 10.1007/s12275-015-4656-9. [DOI] [PubMed] [Google Scholar]
- 7. Nzelu CO, Caceres AG, Guerrero‐Quincho S, et al. A rapid molecular diagnosis of cutaneous leishmaniasis by colorimetric malachite green‐loop‐mediated isothermal amplification (LAMP) combined with an FTA card as a direct sampling tool. Acta Trop. 2016;153:116‐119. 10.1016/j.actatropica.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 8. Bao H, Zhao Y, Wang Y, et al. Development of a reverse transcription loop‐mediated isothermal amplification method for the rapid detection of subtype H7N9 avian influenza virus. BioMed Res Int. 2014;2014:525064. 10.1155/2014/525064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Notomi T, Okayama H, Masubuchi H, et al. Loop‐mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:E63. 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kashir J, Yaqinuddin A. Loop mediated isothermal amplification (LAMP) assays as a rapid diagnostic for COVID‐19. Med Hypotheses. 2020;141:109786. 10.1016/j.mehy.2020.109786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nzelu CO, Kato H, Peters NC. Loop‐mediated isothermal amplification (LAMP): an advanced molecular point‐of‐care technique for the detection of leishmania infection. PLoS Negl Trop Dis. 2019;13:e0007698. 10.1371/journal.pntd.0007698.PNTD-D-19-00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nzelu CO, Gomez EA, Caceres AG, et al. Development of a loop‐mediated isothermal amplification method for rapid mass‐screening of sand flies for leishmania infection. Acta Trop. 2014;132:1‐6. 10.1016/j.actatropica.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 13. Ibarra‐Meneses AV, Cruz I, Chicharro C, et al. Evaluation of fluorimetry and direct visualization to interpret results of a loop‐mediated isothermal amplification kit to detect leishmania DNA. Parasit Vectors 2018;11:250. 10.1186/s13071-018-2836-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tomita N, Mori Y, Kanda H, Notomi T. Loop‐mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877‐882. 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 15. Lamb LE, Bartolone SN, Ward E, Chancellor MB. Rapid detection of novel coronavirus (COVID‐19) by reverse transcription‐loop‐mediated isothermal amplification. medRxiv. 2020. 10.1101/2020.02.19.20025155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. El‐Tholoth M, Bau HH, Song J. A single and two‐stage, closed‐tube, molecular test for the 2019 novel coronavirus (COVID‐19) at home, clinic, and points of entry. ChemRxiv. 2020. 10.26434/chemrxiv.11860137.v1. [DOI] [Google Scholar]
- 17. Yu L, Wu S, Hao X, et al. Rapid detection of COVID‐19 coronavirus using a reverse transcriptional loop‐mediated isothermal amplification (RT‐LAMP) diagnostic platform. Clin Chem. 2020;66(7):975–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Y, Odiwuor N, Xiong J, et al. Rapid molecular detection of SARS‐CoV‐2 (COVID‐19) virus RNA using colorimetric LAMP. medRxiv. 2020. 10.1101/2020.02.26.20028373. [DOI] [Google Scholar]
- 19. Yang W, Dang X, Wang Q, et al. Rapid detection of SARS‐CoV‐2 using reverse transcription RT‐LAMP method. MedRxiv. 2020; 10.1101/2020.03.02.20030130. [DOI] [Google Scholar]
- 20. Broughton JP, Deng X, Yu G, et al. Rapid detection of 2019 novel coronavirus SARS‐CoV‐2 using a CRISPR‐based DETECTR lateral Flow assay. medRxiv. 2020. 10.1101/2020.03.06.20032334. [DOI] [Google Scholar]
- 21. Nguyen T, Duong Bang D, Wolff A. 2019 novel coronavirus disease (COVID‐19): paving the road for rapid detection and point‐of‐care diagnostics. Micromachines (Basel). 2020;11(3):306. 10.3390/mi11030306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jiang M, Pan W, Arastehfar A, et al. Development and validation of a rapid single‐step reverse transcriptase loop‐mediated isothermal amplification (RT‐LAMP) system potentially to be used for reliable and high‐throughput screening of COVID‐19. MedRxiv. 2020; 10.1101/2020.03.15.20036376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhu X, Wang X, Han L, et al. Reverse transcription loop‐mediated isothermal amplification combined with nanoparticles‐based biosensor for diagnosis of COVID‐19. medRxiv. 2020. 10.1101/2020.03.17.20037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park GS, Ku K, Baek SH, et al. Development of reverse transcription loop‐mediated isothermal amplification (RT‐LAMP) assays targeting SARS‐CoV‐2. J Mol Diagn. 2020;22(6):729–735. 10.1101/2020.03.09.983064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yan C, Cui J, Huang L, et al. Rapid and visual detection of 2019 novel coronavirus (SARS‐CoV‐2) by a reverse transcription loop‐mediated isothermal amplification assay. Clin Microbiol Infect. 2020;26(6):773–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Butt AM, Siddique S, An X, Tong Y. Development of a dual‐gene loop‐mediated isothermal amplification (LAMP) detection assay for SARS‐CoV‐2: a preliminary study. medRxiv. 2020. 10.1101/2020.04.08.20056986. [DOI] [Google Scholar]
- 27. Viehweger A, Krautwurst S, Lamkiewicz K, et al. Direct RNA nanopore sequencing of full‐length coronavirus genomes provides novel insights into structural variants and enables modification analysis. Genome Res. 2019;29:1545‐1554. 10.1101/gr.247064.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhadra S, Riedel TE, Lakhotia S, Tran ND, Ellington AD. High‐surety isothermal amplification and detection of SARS‐CoV‐2, including with crude enzymes. bioRxiv. 2020. 10.1101/2020.04.13.039941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang WE, Lim B, Hsu CC, et al. RT‐LAMP for rapid diagnosis of coronavirus SARS‐CoV‐2. Microb Biotechnol. 2020;13:950‐961. 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baek YH, Um J, Antigua KJC, et al. Development of a reverse transcription‐loop‐mediated isothermal amplification as a rapid early‐detection method for novel SARS‐CoV‐2. Emerg microbes Infect. 2020;9:998‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang D. One‐pot Detection of COVID‐19 with Real‐time Reverse‐transcription Loop‐mediated Isothermal Amplification (RT‐LAMP) Assay and Visual RT‐LAMP Assay. BioRxiv. 2020; 10.1101/2020.04.21.052530. [DOI] [Google Scholar]
- 32. Sun F, Ganguli A, Nguyen J, et al. Smartphone‐based multiplex 30‐minute nucleic acid test of live virus from nasal swab extract. Lab on a Chip. 2020;20(9):1621–1627. [DOI] [PubMed] [Google Scholar]
- 33. Rabe BA, Cepko C. SARS‐CoV‐2 detection using an isothermal amplification reaction and a rapid, inexpensive protocol for sample inactivation and purification. medRxiv. 2020. 10.1101/2020.04.23.20076877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee JYH, Best N, McAuley J, et al. Validation of a single‐step, single‐tube reverse transcription‐loop‐mediated isothermal amplification assay for rapid detection of SARS‐CoV‐2 RNA. bioRxiv. 2020. 10.1101/2020.04.28.067363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mohon AN, Oberding L, Hundt J, et al. Optimization and clinical validation of dual‐target RT‐LAMP for SARS‐CoV‐2. J Virol Methods. 2020;286:113972. 10.1016/j.jviromet.2020.113972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ben‐Assa N, Naddaf R, Gefen T, et al. SARS‐CoV‐2 On‐the‐Spot Virus Detection Directly From Patients. MedRxiv. 2020; 10.1101/2020.04.22.20072389. [DOI] [Google Scholar]
- 37. Dao Thi VL, Herbst K, Boerner K, et al. Screening for SARS‐CoV‐2 infections with colorimetric RT‐LAMP and LAMP sequencing. medRxiv. 2020. 10.1101/2020.05.05.20092288. [DOI] [Google Scholar]
- 38. Lalli MA, Chen X, Langmade SJ, et al. Rapid and extraction‐free detection of SARS‐CoV‐2 from saliva with colorimetric LAMP. MedRxiv. 2020; 10.1101/2020.05.07.20093542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Anahtar MN, McGrath GE, Rabe BA, et al. Clinical assessment and validation of a rapid and sensitive SARS‐CoV‐2 test using reverse‐transcription loop‐mediated isothermal amplification. medRxiv 2020. 10.1101/2020.05.12.20095638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ganguli A, Mostafa A, Berger J, et al. Rapid Isothermal Amplification and Portable Detection System for SARS‐CoV‐2. BioRxiv. 2020; 10.1101/2020.05.21.108381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu X, Deng Q, Li J, et al. Development and clinical application of a rapid and sensitive loop‐mediated isothermal amplification test for SARS‐CoV‐2 infection. Msphere. 2020;5(4). 10.1101/2020.05.20.20108530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tran DH, Hoang QC, Tran HT, et al. A comparative study of isothermal nucleic acid amplification methods for SARS‐CoV‐2 detection at point of care. BioRxiv. 2020; 10.1101/2020.05.24.113423. [DOI] [Google Scholar]
- 43. Haq F, Sharif S, Khurshid A, et al. Development optimization and validation of RT‐LAMP based COVID‐19 facility in Pakistan. bioRxiv. 2020. 10.1101/2020.05.29.124123. [DOI] [Google Scholar]
- 44. Lau YL, Ismail I, Mustapa NI, et al. Real‐time reverse transcription loop‐mediated isothermal amplification for rapid detection of SARS‐CoV‐2. PeerJ 2020;8:e9278. 10.7717/peerj.9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kellner MJ, Ross JJ, Schnabl J, et al. Scalable, rapid and highly sensitive isothermal detection of SARS‐CoV‐2 for laboratory and home testing. bioRxiv. 2020. 10.1101/2020.06.23.166397. [DOI] [Google Scholar]
- 46. Matsumura Y, Shimizu T, Noguchi T, Nakano S, Yamamoto M, Nagao M. Comparison of 12 molecular detection assays for SARS‐CoV‐2. bioRxiv. 2020. 10.1101/2020.06.24.170332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Eckel F, Kusters F, Drossel B, Konert M, Mattes H, Schopf S. Variplex test system fails to reliably detect SARS‐CoV‐2 directly from respiratory samples without RNA extraction. Eur J Clin Microbiol Infect Dis. 2020;39:2373‐2377. 10.1007/s10096-020-03983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ooi KH, Tay JWD, Teo SY, et al. A CRISPR‐based SARS‐CoV‐2 diagnostic assay that is robust against viral evolution and RNA editing. bioRxiv. 2020. 10.1101/2020.07.03.185850. [DOI] [Google Scholar]
- 49. Nagura‐Ikeda M, Imai K, Tabata S, et al. Clinical evaluation of self‐collected saliva by RT‐qPCR, direct RT‐qPCR, RT‐LAMP, and a rapid antigen test to diagnose COVID‐19. J Clin Microbiol. 2020;58(9):e01438. 10.1101/2020.06.06.20124123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Osterdahl MF, Lee KA, Ni Lochlainn M, et al. Detecting SARS‐CoV‐2 at point of care: preliminary data comparing Loop‐mediated isothermal amplification (LAMP) to PCR. medRxiv. 2020. 10.1101/2020.04.01.20047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gonzalez‐Gonzalez E, Lara‐Mayorga IM, Yee‐de Leon F, et al. Scaling diagnostics in times of COVID‐19: colorimetric loop‐mediated isothermal amplification (LAMP) assisted by a 3D‐printed incubator for cost‐effective and scalable detection of SARS‐CoV‐2. medRxiv. 2020. 10.1101/2020.04.09.20058651. [DOI] [Google Scholar]
- 52. Cui J, Li F, Shi Z‐L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Woo PC, Huang Y, Lau SK, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2:1804‐1820. 10.3390/v2081803viruses-02-01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu R, Wu X, Wan Z, Li Y, Jin X, Zhang C. A novel reverse transcription loop‐mediated isothermal amplification method for rapid detection of SARS‐CoV‐2. Int J Mol Sci. 2020;21:2826. 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schmid‐Burgk JL, Schmithausen RM, Li D, et al. LAMP‐seq: population‐scale COVID‐19 diagnostics using combinatorial barcoding. bioRxiv. 2020. 10.1101/2020.04.06.025635. [DOI] [Google Scholar]
- 56. Li J, Quan W, Yan S, et al. Rapid detection of SARS‐CoV‐2 and other respiratory viruses by using LAMP method with Nanopore Flongle workflow. bioRxiv. 2020. 10.1101/2020.06.03.131474. [DOI] [Google Scholar]
- 57. Mabey D, Peeling RW, Ustianowski A, Perkins MD. Diagnostics for the developing world. Nat Rev Microbiol. 2004;2:231‐240. 10.1038/nrmicro841.nrmicro841. [DOI] [PubMed] [Google Scholar]


