Nabais et al. use an egg explant system overexpressing Plk4 to study the spatiotemporal and biochemical regulation of de novo centriole assembly. They show that the onset and kinetics of biogenesis depend on Plk4 concentration, requiring the matrix that surrounds centrioles.
Abstract
Centrioles form centrosomes and cilia. In most proliferating cells, centrioles assemble through canonical duplication, which is spatially, temporally, and numerically regulated by the cell cycle and the presence of mature centrioles. However, in certain cell types, centrioles assemble de novo, yet by poorly understood mechanisms. Herein, we established a controlled system to investigate de novo centriole biogenesis, using Drosophila melanogaster egg explants overexpressing Polo-like kinase 4 (Plk4), a trigger for centriole biogenesis. We show that at a high Plk4 concentration, centrioles form de novo, mature, and duplicate, independently of cell cycle progression and of the presence of other centrioles. Plk4 concentration determines the temporal onset of centriole assembly. Moreover, our results suggest that distinct biochemical kinetics regulate de novo and canonical biogenesis. Finally, we investigated which other factors modulate de novo centriole assembly and found that proteins of the pericentriolar material (PCM), and in particular γ-tubulin, promote biogenesis, likely by locally concentrating critical components.
Introduction
“[...] the problem which has interested cytologists and embryologists for many years, namely, whether an ordinarily self-duplicating body may, under certain conditions, seem to be created de novo” (Dirksen, 1961).
It was not long after Boveri and Van Beneden independently discovered centrosomes in the late 1890s that scientists began proposing that centrioles were not always assembled through duplication (Harvey, 1936; Yatsu, 1905). The fascinating discovery that such an elaborate yet fully functional structure can form without a template raised a variety of questions regarding the regulation of organelle biogenesis, many of which stay relevant to this date. And while decades of research have contributed to our current understanding of the regulation of pro-centriole assembly next to an already mature, mother structure, much less is known regarding the “unguided” de novo centriole formation.
Centrioles are cylindrical microtubule (MT)–based structures that assemble centrosomes and cilia in eukaryotic cells. The animal centrosome is typically composed of two centrioles surrounded by pericentriolar material (PCM), a membraneless compartment that contains hundreds of proteins organized within distinct domains, which are responsible for anchoring and nucleating MTs (reviewed in Joukov and De Nicolo, 2019). Centriole biogenesis is usually tightly regulated to ensure a correct copy number and to prevent a variety of human diseases (Bettencourt-Dias et al., 2011; Godinho and Pellman, 2014; Godinho et al., 2014; Levine et al., 2017; Marteil et al., 2018; Lopes et al., 2018). In proliferating cells, centriole biogenesis occurs through a canonical pathway synchronous with cell cycle progression, called centriole duplication. Centrioles begin assembling at the G1–S transition, whereby a single procentriole forms at the proximal side of each of the two mother centrioles. During mitosis, centrioles undergo a centriole-to-centrosome conversion through the recruitment of Cep135/Bld10, Cep295/Ana1, and Cep152/Asterless (Asl), becoming competent for duplication in the next cell cycle (Fu et al., 2016; Izquierdo et al., 2014; Wang et al., 2011; Tsuchiya et al., 2016). After mitosis, one centrosome is segregated to each daughter cell. This process entails that the location, timing, and number of procentrioles assembled in cycling cells is determined by older/mature centrioles (Banterle and Gönczy, 2017; Breslow and Holland, 2019).
Polo-like kinase 4 (Plk4), also called Sak in fruit flies, is a major player in centriole biogenesis in most animal cells (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Kleylein-Sohn et al., 2007). Depletion or inhibition of its kinase activity prevents centriole formation, while overexpression leads to the formation of multiple centrioles (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Wong et al., 2015). Plk4 activity and function are regulated by its concentration, which is known to be very low in human cultured cells (Bauer et al., 2016). Full Plk4 activity is accomplished by trans-autophosphorylation of a conserved T-loop residue within its catalytic domain, which triggers kinase activation through a positive feedback mechanism (Lopes et al., 2015). Other centrosomal proteins also regulate Plk4 activity, such as its substrates STIL/Ana2 and Cep152/Asl (Klebba et al., 2015a, b; Moyer et al., 2015; Zitouni et al., 2016; McLamarrah et al., 2018; Boese et al., 2018; Aydogan et al., 2019). Moreover, at a high concentration, Plk4 self-assembles into nanoscale condensates in Xenopus extracts and in human cultured cells, which may also regulate centriole assembly (Montenegro Gouveia et al., 2018; Yamamoto and Kitagawa, 2019; Park et al., 2019).
Centrioles can also form de novo in a variety of cell types (reviewed in Nabais et al., 2017), but the regulation of this process remains largely unknown. De novo centriole assembly occurs naturally in organisms that lack centrosomes and generate centrioles to nucleate motile cilia, such as land plants that produce ciliated sperm (Renzaglia and Garbary, 2001), several unicellular organisms that alternate between nonflagellated and flagellated life cycle states, and animal multiciliated cells, where many centrioles are produced at once (reviewed in Nabais et al., 2017). In most animals, centrioles are lost during female oogenesis and are provided by the sperm upon fertilization, as they are needed for embryo development (Rodrigues-Martins et al., 2008; Varmark et al., 2007). However, centrioles form de novo in rodents during early embryogenesis (Gueth-Hallonet et al., 1993; Courtois et al., 2012) and in parthenogenetic insects that can reproduce without fertilization (Riparbelli et al., 1998; Tram and Sullivan, 2000; Riparbelli and Callaini, 2003; Riparbelli et al., 2005; Ferree et al., 2006). In the latter, multiple centrosomes form spontaneously in the egg at late stages of meiosis, two of which are captured for spindle formation and embryo development, thus replacing the centrioles that are otherwise inherited from the sperm (Tram and Sullivan, 2000).
Centrioles can also form de novo in cells that undergo physical, chemical, or genetic perturbations. Proliferating cells are capable of assembling centrioles de novo, but only after their centrosomes have been physically or chemically removed (Khodjakov et al., 2002; La Terra et al., 2005; Uetake et al., 2007; Wong et al., 2015). Although in these cases there is no strict control over the number of centrioles formed, it has been proposed that resident centrioles negatively regulate de novo centriole biogenesis (Marshall et al., 2001), and that such an inhibitory effect can be accomplished by having a single centriole in the cell (La Terra et al., 2005; Lambrus et al., 2015).
In Drosophila melanogaster tissue culture cells, evolutionary conserved centriolar components, such as Sas6, Sas4, and Bld10, are critical for both canonical and de novo assembly (Rodrigues-Martins et al., 2007), suggesting that centrioles assembled by both pathways share their core composition but perhaps differ in their triggering. Despite the widespread circumstances in which centrioles form de novo, the regulation and role of older centrioles on this process have not been addressed. This is in part due to the lack of a controlled model system suitable for high-resolution time-lapse imaging and amenable to experimental perturbations.
In this study, we investigated the spatio-temporal kinetics of de novo centriole assembly, including the effect of preassembled centrioles on the biogenesis of new ones, by visually tracking this process in cytoplasmic extracts where Plk4 is up-regulated. The fly egg is ideal to study centriole assembly since all the proteins necessary for the first centrosome and nuclear cycles are maternally inherited and, in the absence of fertilization, centrioles are not present. Therefore, centrosomes detected in unfertilized eggs result from de novo assembly and not from duplication from paternally inherited centrioles (Peel et al., 2007; Rodrigues-Martins et al., 2007). Here, we accomplished, for the first time, live imaging of de novo centriole assembly with high spatial resolution in single-egg cytosolic explants (Telley et al., 2013; de-Carvalho et al., 2018). We demonstrate that, at an elevated Plk4 concentration, centrioles form de novo and duplicate. Both pathways are concurrent as we show that de novo centriole formation occurs independently of preexisting centrioles. These results reveal that in D. melanogaster eggs upon Plk4 overexpression, resident centrioles do not inhibit de novo biogenesis, unlike in human cells and mouse developing embryos. We show that Plk4 modulates the kinetics of centriole assembly in a concentration-dependent manner that is suggestive of a switch-like molecular mechanism occurring in the cytosol. Finally, we find that the PCM, in particular γ-tubulin and Cep152/Asl, regulates de novo biogenesis, suggesting that a local environment of concentrated centriolar and PCM components is required for de novo centriole assembly.
Results
An assay to investigate centriole biogenesis live with high spatio-temporal resolution
De novo centriole assembly has been poorly studied in live samples due to the lack of a suitable system where the process can be triggered and documented in a timely manner. Overexpressing Plk4 drives de novo bona fide centriole biogenesis, validated by EM, in unfertilized D. melanogaster eggs (Rodrigues-Martins et al., 2007), but the onset of the process and its spatio-temporal kinetics were unknown. Reasons behind this knowledge gap are mostly imaging-related, for the axial depth is optically limited and greatly impaired by the light-scattering properties of the egg yolk. Therefore, it is currently impossible to visualize processes that take place deep inside the fruit fly egg, which would otherwise be the ideal system to address critical questions concerning centriole biogenesis.
We adapted for this purpose a cell-free assay that overcomes these limitations by generating cell cortex–free microscale explants that can be fully imaged, while retaining the native characteristics of the cytoplasm in vivo (Fig. 1 A; Telley et al., 2013; de-Carvalho et al., 2018). Using this assay, we observed de novo centriole biogenesis, at high spatio-temporal resolution (Fig. 1, B and C; and Video 1). Germline-specific mild overexpression of Plk4, assessed by Western blotting (Table S1, Fig. S1, A and B), triggers the formation of multiple centrioles in cytoplasmic explants initially devoid of centrioles, demonstrating that post-meiotic D. melanogaster egg extracts are competent for centriole biogenesis (Fig. 1 C) and recapitulate observations in eggs that were not extracted from, as reported previously (Rodrigues-Martins et al., 2007). Importantly, explants enable visualization of the first steps of de novo biogenesis, which normally occurs deep inside the egg. Moreover, we never observed centrosome disassembly during our time-lapse recordings, showing these structures are stable. While it is not possible to visualize the microtubule-organizing centers (MTOCs) in cytoplasmic explants by EM since the explants are imbedded in halocarbon oil, which is not compatible with sample processing, validation by EM had previously been performed in intact eggs overexpressing Plk4 using the same genetic constructs (Rodrigues-Martins et al., 2007). Given that Drosophila egg explants retain and recapitulate fundamental developmental and cytoplasmic properties (Telley et al., 2012, 2013), we can expect that the structures observed here are bona fide centrioles. Therefore, these extracts offer a powerful assay to investigate the regulation of centriole assembly.
Figure 1.
Visualization of centrosome biogenesis in Drosophila egg extract. (A) Drosophila egg extract is prepared by rupturing the membrane and aspirating the cytoplasm with a micropipette. The content is deposited as a droplet on functionalized glass surface. (B) Each explant is followed by 3D time-lapse imaging, documenting centriole formation over time. (C) Maximum intensity z projections from a fluorescence time-lapse of a droplet of cytosolic extract isolated from a Drosophila egg overexpressing Plk4. Centrioles are absent in the first time point and form de novo throughout the experiment detected as spots (Spd2, in green) associated with a MT array (magenta; arrowheads, numbers indicate the order of birth), reported by the MT associated protein Jupiter. Signals in the two channels are detected almost simultaneously, without observing any clear trend of one signal appearing before the other one. The larger green circles are yolk, and the high background is caused by other lipid granules that are highly autofluorescent in the green spectrum, and that cannot be avoided. The insets depict the first centrosomes formed de novo in this time-lapse. The numbers represent their order of appearance. Example of n = 68 explants. Time is reported as minutes:seconds.
Video 1.
In support of Fig. 1: Centriole biogenesis in a D. melanogaster egg explant. Maximum intensity z projection from a time-lapse video of a cytosolic explant isolated from an unfertilized Drosophila egg overexpressing Plk4, acquired on a spinning-disk confocal microscope. Centrioles are absent in the first time point and form de novo throughout the experiment detected as spots (Spd2, in green) associated with MT asters (magenta), reported by the MT associated protein Jupiter. Time (minutes:seconds) is shown at the top left. Playback rate is 5 frames per second (fps).
Figure S1.
In support of Figs. 1 and 2: Measurement of concentration fold-change of Plk4 in overexpression lines. (A) Insertion of a fluorescent tag into Drosophila Plk4 endogenous locus. Schematic representation of the WT dmPlk4 locus (WT) and of the dmPlk4 locus after successful tag integration by homologous recombination (HR). A donor plasmid carrying the mNeonGreen reporter and a small linker (dark green) flanked by 1 kbp homology arms was used for homologous recombination. The UTRs are shown in gray, and the coding sequences are depicted in orange. The arrows indicate the position of the screening primers dmPLK4 5′UTR 3 FW and dmPLK4 1exon Rev, which are located outside the homology arms. The same strategy was used for mNeonGreen and GFP tags, generating two lines that were used at different parts of this paper. The GFP knock-in was used as WT control in measurement of Plk4 expression level. The inset shows the integration of a fluorescent tag into Plk4 endogenous locus (HR Plk4) by Western blot, causing a migration shift of the PCR product in the agarose gel compared with the untagged Plk4 locus (WT Plk4). (B) Western blot analysis of Plk4 concentration for endogenous expression and for overexpression constructs. Two (out of four) representative Western blots are shown. We emphasize that the detection of endogenous Plk4 with a Western blot approach is extremely challenging. In fact, most studies so far have only detected Plk4 by means of affinity-\ tag or fluorescent reporter and/or under an overexpression scenario. We were able to visualize the endogenous Plk4 tagged with mEGFP using αGFP antibody. Plk4 overexpression was visualized with a GFP–Plk4 overexpression (o.e.) construct, whose extract shows similar centriole biogenesis results as the nontagged Plk4 overexpression construct used in most parts of this work. We also made extract from flies overexpressing nondegradable (ND) Plk4, which accumulates in embryos and serves as a positive control (pUASp–ND-Plk4–EGFP; Cunha-Ferreira et al., 2013). WT embryos (w1118) were loaded as negative control as they do not have GFP-tagged protein. The black arrowhead points at GFP-tagged Plk4 constructs, while the white arrowhead points at an unspecific signal also present in WT embryos. We register 3.2 ± 1.9 times higher Plk4 concentration in the extract of embryos overexpressing Plk4 as compared with the WT (n = 4). Inter-experiment variability is largely due to systematic errors of Western blot quantification, but also due to the endogenous concentration of Plk4 being near the detection limit. Despite the variability, this quantitation is in line with our dilution results (Fig. 4 B); at 1/5 dilution of the Plk4-overexpressing extract, we detect very few de novo events, suggesting that with further dilution, the kinetics converges toward WT conditions where de novo events are not observed. Note that while the V32 driver for protein expression used in our experiments normally leads to high levels of protein expression, pUAS–GFP–Plk4 is likely being down-regulated through targeted degradation, in contrast to pUAS–ND-Plk4–GFP, which accumulates to higher levels. Ab, antibody; Chr., chromosome.
We tested explants of several fluorescent protein fly lines, namely Ana1–dTomato, GFP–Plk4, Asl–mCherry, and Spd2–GFP (Table S1). We chose Spd2–GFP as our routine centrosome reporter because its fluorescence signal was brighter and more photostable across explants than all the others tested, and in our experience, this line does not perturb centriole biogenesis (unpublished data; de-Carvalho et al., 2020).
De novo–formed centrioles mature and acquire the ability to duplicate in the absence of cell cycle progression
It was previously proposed that in both human cells (La Terra et al., 2005; Lambrus et al., 2015) and D. melanogaster eggs (Rodrigues-Martins et al., 2007), centrioles that form de novo can then duplicate canonically. However, this was never confirmed directly. Centriole duplication is thought to depend on centriole maturation, a process called centriole-to-centrosome conversion, which requires the recruitment of Ana1 and Asl to the mother centriole and is coupled to cell cycle progression (Wang et al., 2011; Izquierdo et al., 2014; Fu et al., 2016; Chang et al., 2016). Centriole duplication is also known to be coupled to cell cycle progression, which does not occur in eggs (Horner et al., 2006; Vardy and Orr-Weaver, 2007; Deneke et al., 2019). Thus, we asked whether centrioles form de novo mature, recruiting Plk4, Ana1, and Asl, and undergo bona fide centriole duplication (Fig. 2, Video 2, Video 3, Video 4, and Video 5). Surprisingly, we observed that recently assembled centrioles can recruit Plk4, the trigger of biogenesis (Fig. 2, A and B, top panel), Ana1 (Fig. 2, C and D, top panel), and Asl (Fig. 2, E and F, top panel) in the absence of cell cycle progression.
Figure 2.
Centrioles assemble de novo, mature, and duplicate within the same explants, in the absence of cell-cycle progression. Images show maximum-intensity z projections from time-lapse videos of cytoplasmic explants extracted from noncycling unfertilized eggs overexpressing Plk4. Newly assembled centrosomes load Plk4 (A and B), Ana1 (C and D), Asl (E and F), and Spd2 (G, H, and J) shown in green, and nucleate MTs as reported by the MT-associated protein Jupiter (magenta). The larger green blobs result from yolk autofluorescence, highly noticeable in the Plk4 and Spd2 panels. (B, D, and F) Centrioles formed de novo also duplicate, which was inferred from changes in the intensity profile along the axis across the centrosomal signal (bottom plots); from a symmetrical Gaussian curve to a Gaussian mixture, suggesting the presence of more than one diffraction-limited structure (centriole). A uni- or bimodal Gaussian distribution was fitted to each de novo and canonical intensity profile, respectively (dashed lines represent modes from fit). The coefficient of determination (R2) is presented for each fit. Scale bars, 0.5 µm. (G) Centrioles form de novo and canonically over time; therefore, both biogenesis pathways co-occur. Centriole duplication was inferred from the change in the intensity profile across the Spd2 signal (H, bottom plots). Uni- or bimodal Gaussian fitting as in B–F. Colors represent one centrosome that first assembled de novo and later duplicated. (I) The duplication time depicted in the graph is the time elapsed between the documentation of the first centriole formed de novo (unimodal density) and the detection of a centriole pair (bimodal density). The horizontal line and error bars represent the median and interquartile range (n = 66 explants from different eggs). (K) Insets of the first three centrosomes formed de novo in time-lapse (J) and their corresponding normalized and bleach-corrected intensity of Spd2 (L) and Jupiter reporting MTs (M), plotted over time. Time is reported in minutes:seconds. Scale bar, 0.5 µm. Norm., normal.
Video 2.
In support of Fig. 2: Centrioles assemble de novo, recruit different centrosomal molecules, and duplicate. Maximum intensity z projection from time-lapse videos of explants from noncycling unfertilized Drosophila eggs overexpressing Plk4, acquired on a spinning-disk confocal microscope. Shown is centriole biogenesis reported by centrosomal protein Plk4 (in green) and the MT-associated protein Jupiter (magenta). The larger green blobs result from yolk autofluorescence, highly noticeable in this Plk4 video. Time (minutes:seconds) is shown at the top left. Playback rate is 5 fps.
Video 3.
In support of Fig. 2: Centrioles assemble de novo, recruit different centrosomal molecules, and duplicate. Maximum intensity z projection from time-lapse videos of explants from noncycling unfertilized Drosophila eggs overexpressing Plk4, acquired on a spinning-disk confocal microscope. Shown is centriole biogenesis reported by centrosomal protein Ana1 (in green) and the MT-associated protein Jupiter (magenta). Time (minutes:seconds) is shown at the top left. Playback rate is 5 fps.
Video 4.
In support of Fig. 2: Centrioles assemble de novo, recruit different centrosomal molecules, and duplicate. Maximum intensity z projection from time-lapse videos of explants from noncycling unfertilized Drosophila eggs overexpressing Plk4, acquired on a spinning-disk confocal microscope. Shown is centriole biogenesis reported by centrosomal protein Asl (in green) and the MT-associated protein Jupiter (magenta). Time (minutes:seconds) is shown at the top left. Playback rate is 5 fps.
Video 5.
In support of Fig. 2: Centrioles assemble de novo, recruit different centrosomal molecules, and duplicate. Maximum intensity z projection from time-lapse videos of explants from noncycling unfertilized Drosophila eggs overexpressing Plk4, acquired on a spinning-disk confocal microscope. Shown is centriole biogenesis reported by centrosomal protein Spd2 (in green) and the MT-associated protein Jupiter (magenta). Time (minutes:seconds) is shown at the top left. Playback rate is 5 fps.
Next, we investigated if centrioles that formed de novo also duplicate, as predicted by their ability to mature and recruit Plk4. In our assay, single centrioles are first detected as radially symmetrical intensity spots with Gaussian intensity profile (Fig. 2, B, D, F, and G). Over time, a single Plk4, Ana1, Asl, and Spd2 Gaussian intensity profile can evolve into a mixture of at least two Gaussian distributions (Fig. 2, B, D, F, and H), consistent with the presence of more than one centriole and canonical duplication. We next took advantage of 3D-Structured Illumination Microscopy imaging, which provides approximately twice the spatial resolution of confocal microscopy, to further characterize the MTOC structures in the explants. We observed that Spd2–GFP forms a ring at the center of the MT aster, with an inner diameter of 230–320 nm when viewed in cross-section (Fig. S2 A, insets). Previous studies have demonstrated that Spd2 forms toroids at the centrosome in D. melanogaster syncytial embryos, whereby Spd2 projections extend from a central hollow structure, which presumably contains a single centriole (Conduit et al., 2015). In our experiments, smaller structures form adjacent to older centrioles that previously formed de novo, providing further evidence for canonical duplication in this system (Fig. S2 A, insets). The fact that duplication is a property that has only been observed for centrioles, and not other MTOCs such as condensates (Montenegro Gouveia et al., 2018), strongly supports our findings that the structures observed here are bona fide centrioles structurally and functionally. Importantly, in 97% (66/68) of our time-lapse recordings captured by confocal microscopy, we observed the duplication of the first centriole within 2–3 min after its de novo assembly (Fig. 2 I, scatter plot).
Figure S2.
In support of Fig. 2: Temporal analysis of de novo centriole biogenesis at extended spatial resolution. (A) Visualization of centrosome biogenesis in a Drosophila egg extract by 3D-Structured Illumination Microscopy. Maximum-intensity z projections from a time-lapse acquisition of an unfertilized egg explant overexpressing Plk4. Centrioles (insets) are detected as barrel-shaped structures surrounded by the PCM component Spd2 (green) associated with a MT array (magenta), reported by the MT-associated protein Jupiter. Insets are single-plane images of three different centrosomes. Scale bar, 0.5 µm. Centrioles formed de novo can duplicate. Time is reported as minutes:seconds. (B) Time-lapse (top row) of an egg explant overexpressing GFP–Plk4, in which centriole form de novo over time (arrows) as shown in magnified views (bottom). Numbers indicate sequence of formation. Inter-event times are shown in Fig. S4 A. (C) Time-lapse (top row) of an egg explant overexpressing Asl–mCherry, in which centrioles form de novo over time (arrows) as shown in magnified views (bottom). Numbers indicate sequence of formation; after de novo event #1, two centrioles formed concomitantly within the temporal resolution (#2 and #2′′) followed by another event (#3). Inter-event times are shown in Fig. S4 B. (D) Histogram of frame-to-frame (instantaneous) displacements of first event centrosomes. Most of the centrosomes performed random movement and only in rare cases they moved away in a directed fashion from the explant boundaries. (E) Comparison of centrosome movement versus distance between registered biogenesis. Cumulative distribution functions of frame-to-frame displacement (black) in comparison with all subsequent inter-event distances as presented in Fig. 3. This comparison shows that the probability for a biogenesis event to quickly displace a distance typically seen between biogenesis events is extremely low. Any subsequent event after the first biogenesis is unlikely a duplication-and-run event.
We next asked whether centrioles are fully converting to centrosomes, maturing also in their ability to nucleate MTs. Indeed, we observed that as they age, centrioles continue incorporating centrosomal proteins and increase their MTOC capacity, which is reported by the intensity of the MT-associated protein Jupiter (Fig. 2, J–M).
Centriole-mediated regulation of centriole biogenesis
Interpretation of earlier experiments led to the model that existing centrioles play a dominant role in centriole assembly and negatively regulate de novo centriole biogenesis in other model systems (Marshall et al., 2001; La Terra et al., 2005; Uetake et al., 2007; Lambrus et al., 2015). Whether centrioles can release an inhibitory signal is unknown. On the other hand, it has been suggested that centrioles can act as catalyzers of centriole biogenesis by concentrating centriole components and therefore preventing biogenesis elsewhere (Marshall et al., 2001; Lopes et al., 2015). In particular, given that Plk4 activity is regulated by Plk4 trans-autoactivation, it was suggested that its sequestering at the centrosome would keep Plk4 activity low in the cytoplasm and prevent de novo biogenesis (Lopes et al., 2015).
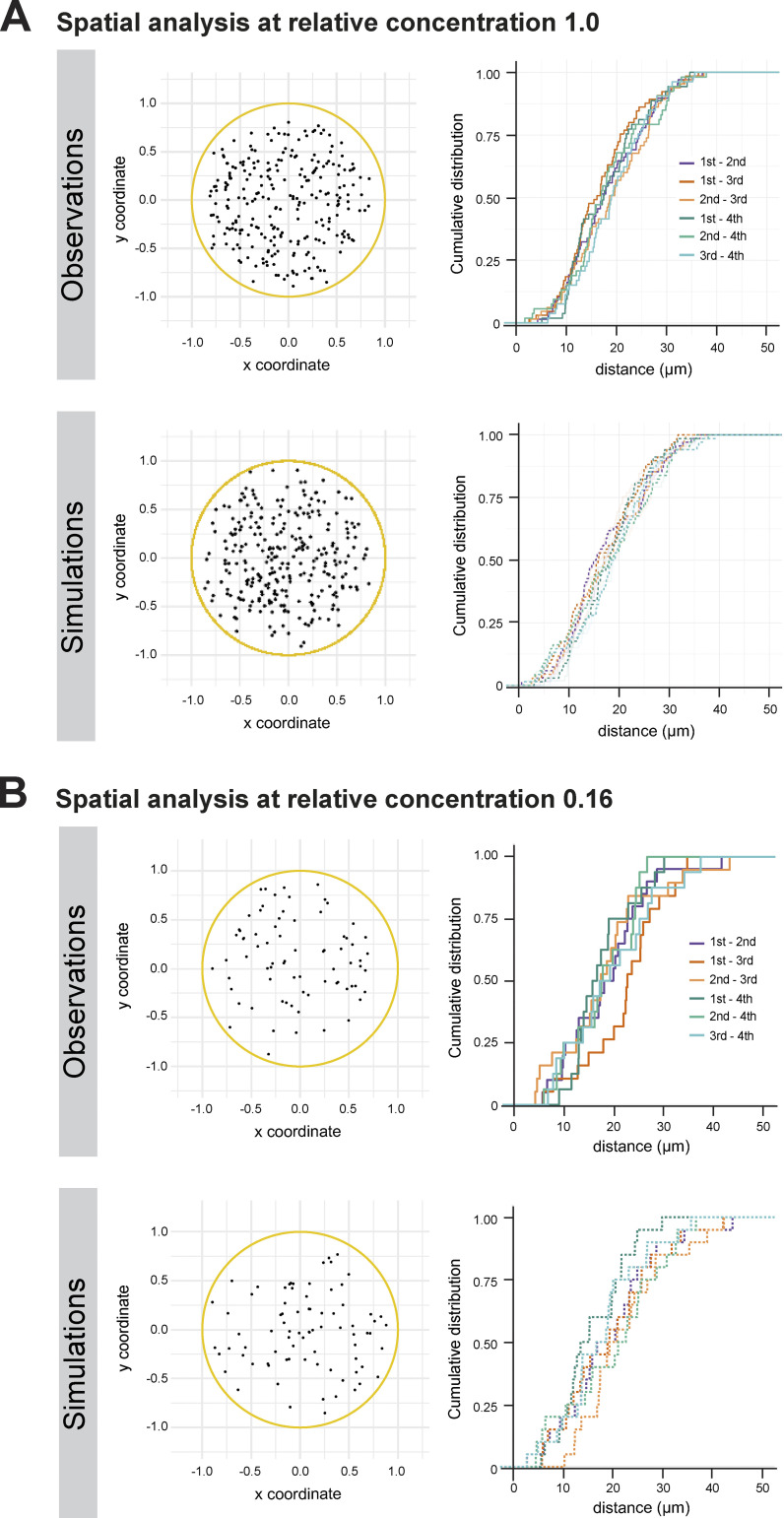
We investigated whether the appearance of the first centriole can prevent further de novo formation. Surprisingly, despite the assembly of centrioles and their duplication, we continue to see de novo formation (Fig. 2, G and H), challenging the view that existing centrioles have a context-independent inhibitory effect in centriole biogenesis. It is highly unlikely that these newly appearing centrioles formed by duplication and then subsequently moved away, because the displacement that centrioles generally undergo within one frame (Fig. S2 D) is 10 times smaller than the average distance between existing centrioles that were first detected as single spots (Fig. S2 E). We then analyzed the spatio-temporal kinetics of de novo biogenesis in our assay by investigating if centrioles impact the place and timing of other de novo events. Once the first centrosome had formed, we assessed if older centrioles affect the biogenesis of others, e.g., by promoting (triggering effect) or repressing (inhibitory effect) the assembly of new ones either globally or in their vicinity (Fig. 3, A and E). We did not observe a statistical difference in the pairwise inter-event distance between the first four centrioles formed de novo (Kruskal–Wallis mean rank test; Fig. 3 B; and Fig. S3 A, top). However, we noticed that new centrioles form, on average, >10 µm away from previous ones, regardless of centriole rank and explant size (Fig. 3, B and C), raising the question whether this process is spatially random or if there is any spatial regulation (e.g., an inhibitory effect) imposed by older centrioles on the birth of neighbors. To test these hypotheses, we generated stochastic models with similar geometric constraints as the cytosolic explants, allowing us to compare observed and simulated data. By measuring the simulated inter-event distances between four random events, independent and uniformly distributed within 3D spaces of similar geometry, we could derive that the experimental observations in explants do not significantly deviate from a random spatial process (Fig. 3 D; and Fig. S3 A, bottom).
Figure 3.
Spatio-temporal kinetics of de novo centriole biogenesis. (A) Schematic representation of the experimental data analysis for distances. The first four centrosomes formed de novo in the explants were tracked in 3D using the intensity signal from the Jupiter (MT reporter) channel (first tracking round) and Spd2 (centrosomal reporter) channel (second tracking round) combined. For each of the de novo birth events, an XYZT coordinate matrix was retrieved, from which the inter-event distances were calculated. Experimental n = 68 explants/eggs. (B) Scatter plot of observed inter-event distances for all pairwise combinations of the first four de novo biogenesis events. Horizontal lines and error bars represent median and interquartile distance, respectively. (C) Cumulative distribution functions (CDF) of inter-event distance. Distributions were not significantly (ns) different (Kruskal–Wallis mean rank test, P = 0.467). (D) In silico simulations were performed to test if the observed experimental data deviates from a theoretical scenario in which all four birth events occurred at independent and identically distributed random positions with a uniform probability density distribution, within explants with similar geometry as in the experiments. Four random events were obtained in 100 simulations of 68 explants. The graph depicts the median CDF of all experimentally observed (obs, solid line) and all simulated (sim, dashed line) inter-events distances, while the gray envelope indicates the 95% confidence interval (from quantile 0.025 to 0.975) for the simulated data. The experimental observations do not deviate from random simulations. (E) Schematic representation of the experimental data analysis for time. For each of the four de novo birth events, an XYZT coordinate matrix was retrieved, from which the inter-event time were calculated. Experimental n = 68 explants/eggs. (F) Scatter plot of observed inter-event time between the first four de novo biogenesis events. Horizontal lines and error bars represent median and interquartile range, respectively. The first event time is significantly different (**) from subsequent inter-event times (Kruskal–Wallis mean rank test, P = 0.0047). Note that this first event time exhibits high (systematic) variability due to an ill-defined time reference (see Materials and methods). (G) Cumulative distribution functions of observed (continuous) and in silico obtained inter-event time (dashed). Simulations were performed to test if the observed experimental data deviates from a theoretical scenario where all four birth events occurred independently at a constant rate within an explant with similar geometry as in the experiments. Four random events were obtained in 100 simulations of 68 explants. In the simulation, the first event rate of birth was approximated to the inter-event time between the first and second events. The graph depicts the median CDF of the experimentally observed (obs, continuous line) and simulated (sim, dashed line) waiting times between the first and second, second and third, and third and fourth events, while the gray envelope indicates the 95% confidence interval (from quantile 0.025 to 0.975) for the simulations. The observed and simulated waiting time distributions do not overlap, and differ more as centriole number increases, suggesting that the rate of biogenesis is increasing over time. (H) Estimation of the experimental birth rates using maximum likelihood estimation fitting. An exponential distribution with rate λ > 0 was fitted by maximum likelihood estimation to the CDF of each observed waiting time. The estimated rate of de novo centriole assembly is represented in the graph as a function of the number of centrioles previously/already present in the volume.
Figure S3.
In support of Fig. 3: Spatial analysis of de novo centriole biogenesis in fly explants at different Plk4 concentrations. (A) De novo centriole biogenesis in fly explants at high Plk4 concentration. Left: 2D z projections of the positions of centrioles at the moment they were first detected in the explants, 254 centrioles measured in 68 explants (Observations) and 272 centrioles from 68 in silico explants (Simulations). All coordinates were normalized to the measured explant diameter. Right: Distributions of observed and simulated inter-event distances measured in 3D for the first four centrosomes formed de novo in the explants. (B) De novo centriole biogenesis at lower Plk4 concentration. Left: z projections of the positions of centrioles at the moment they were first detected in the explants, 75 centrioles measured in 20 explants (Observations) and 80 centrioles from 20 in silico explants (Simulations). All coordinates were normalized to the measured explant diameter. Right: Distributions of observed and simulated inter-event distances measured in 3D for the first four centrosomes formed de novo in the explants, at the lowest Plk4 overexpression (0.16 relative concentration of Plk4). The gray envelope indicates the 95% confidence interval (from quantile 0.025 to 0.975) for the simulated data.
According to our measurements, older centrioles have only a short-range effect on the biogenesis of new centrioles, in which they promote canonical duplication, which occurs by definition in very close proximity (Fig. 2, B, D, F, and H). Importantly, older centrioles do not determine the place of de novo assembly elsewhere in the cytosol. Centrioles behave as autonomous entities at the initial stages of de novo assembly on the scale of tens of micrometers. While we cannot exclude that Plk4 overexpression may be sufficient to override inhibitory signals potentially generated from centrioles, our results rather suggest that de novo centriole biogenesis is not influenced by the mere presence of other recently assembled centrioles. Considering the absence of a local effect, we propose that biochemical changes at the level of the entire cytosol are responsible for stochastic de novo centriole formation. To obtain further insight, we next investigated the temporal kinetics of de novo biogenesis.
The kinetics of de novo biogenesis
We measured how long it takes for the first four de novo centrioles to appear in the explants (Fig. 3 E). We detected, on average, a long lag phase until the birth of the first event, independently of the centrosome reporter used (Fig. S2, B and C; and Fig. S4, A and B, for quantifications), after which the process seemingly accelerated. In fact, we observe that the rate of de novo centriole biogenesis then becomes in the range of one every 2 min (Fig. 3 F). We proceeded to test in silico if the rate at which these first events occur is also indicative of them being independent, as suggested by our previous spatial analysis.
Figure S4.
In support of Fig. 4: Temporal analysis of de novo biogenesis. (A) Scatter plot of observed inter-event time between the first four de novo biogenesis events in the GFP–Plk4 reporter fly line. Note that the fly line overexpresses two copies of Plk4, one of which is tagged with GFP. This higher expression level is a possible explanation for the slightly shorter time until the first de novo event. Horizontal lines and error bars represent median and interquartile range, respectively (n = 9). (B) Scatter plot of observed inter-event time between the first four de novo biogenesis events in the Asl–mCherry reporter fly line overexpressing Plk4. Horizontal lines and error bars represent median and interquartile range, respectively (n = 10). (C) Estimation of the mean centriole biogenesis times at the highest Plk4 concentration (1, in blue) and at the lowest Plk4 overexpression (0.16, in orange) by maximum likelihood estimation fitting of a simple exponential model to data shown in Fig. 4 C. (D) Estimation of the waiting time until the first de novo event and inter-event time between the first and subsequent de novo events, at high (1, in blue) and the lowest (0.16, in orange) concentration of Plk4, after fitting data shown in Fig. 4 C. (E) Model of Plk4 autoactivation and dephosphorylation based on data from (Lopes et al., 2015). Plk4 trans-autophosphorylates to become fully active, transitioning from an enzyme with basal activity, B form, to an activated form phosphorylated on its T-loop residue, A* form. Highly phosphorylated Plk4, A** form, is also active but is targeted for degradation (Cunha-Ferreira et al., 2013; Guderian et al., 2010; Holland et al., 2012; Klebba et al., 2013). Dark arrows indicate the forward phosphorylation reaction flux, while red arrows indicate the reverse dephosphorylation flux catalyzed by a putative counteracting phosphatase. The leftmost dark arrow marks the synthesized Plk4 that enters the system, while the dashed lines refer to Plk4 degradation. Green arrows depict the Plk4 forms that catalyze the forward flux. (F) A nonlinear balance between phosphorylation and dephosphorylation activities generates a Plk4 critical threshold, as a function of its concentration. Therefore, total concentration (active and inactive) of Plk4 in cells likely affects the timing at which a critical concentration threshold is overcome and triggers centriole assembly (Lopes et al., 2015). (G) Fitting of Plk4 autoactivation and dephosphorylation model to data measured in explants at different Plk4 concentrations. The gray gradient represents different concentrations of Plk4. The different concentrations were prepared experimentally by mixing the cytoplasm from high overexpression eggs (taken as the unit 1, black) with cytoplasm from WT eggs, in different proportions such that the dilutions are 0.5, 0.33, and 0.16 relative concentrations. The dots are the relative frequency of explants containing at least one de novo formed centriole for the different concentrations of Plk4: 1 (n = 56), 0.5 (n = 62), 0.33 (n = 39), and 0.16 (n = 25). The lines are the solution of the model of Plk4 trans-autophosphorylation. The continuous lines are the solution of the ordinary differential equation model, and the staircase lines are the results of stochastic simulations under the same parameter settings. The Plk4 activity in the higher concentration (denoted K) was adjusted, whereas the activities in the dilutions were set in relative terms (0.16 K, 0.33 K, and 0.5K). The modeling and simulations, as well as the remaining parameters and values, are described in the Materials and methods (Statistics and mathematical modeling). Notice that as Plk4 concentration decreases, so does the number of explants where centriole biogenesis occurs within 40 min of time-lapse recording.
Assuming independent de novo biogenesis events with a constant rate, computer simulations predict that all inter-event times should follow a similar distribution (Fig. 3 G). However, not all of the observed inter-event distributions were within the confidence interval of the simulation. Moreover, the difference was more noticeable at higher number of centrioles (see second to third and third to fourth, Fig. 3 G). Finally, maximum likelihood estimation of birth rate indicated a linear increase with centriole number (Fig. 3 H).
Altogether, our results demonstrate that the rate of de novo centriole formation progressively increases in time. Assuming that our detection of new centrioles by microscopy is seemingly immediate, we may describe their kinetics as biphasic: an initial lag phase preceding the formation of the first centriole(s), when the probability of centriole assembly is very low, and a subsequent phase where events occur in a burst. Not detecting any effect of the first centriole on the spatial distribution of following centrioles (Fig. 3, A–D) suggests that concentration and further auto-activation of Plk4 locally at the first centriole is unlikely to induce or prevent subsequent de novo events. Different factors may explain this kinetics. For instance, the delay could depend on how long it takes for Plk4 to accumulate and become activated at multiple sites. Other factors independent of Plk4, such as the activity of a yet unidentified inhibitor, or the time elapsed since the end of meiosis, may also play a role.
Plk4 concentration modulates the kinetics of assembly of the first but not subsequent centrioles
We asked whether the timing of centriole assembly depends on Plk4 concentration and established a titration assay for Plk4 using egg cytoplasm. With the exception of Plk4, WT eggs have all the same components at presumably similar concentrations as Plk4-overexpressing eggs. Thus, mixing egg cytoplasm from these two genetic backgrounds dilutes only Plk4 within a range of full overexpression and endogenous levels. We measured the spatial organization and temporal kinetics of de novo centriole biogenesis for a series of dilutions, in which the larger dilution (0.16) should be close to endogenous levels (Fig. S1 B).
Reassuringly, even at low concentrations of Plk4 close to endogenous levels, de novo biogenesis was indistinguishable from a random spatial process (Fig. 4 A and Fig. S3 B), corroborating our previous observations that de novo centriole biogenesis is not affected by the mere presence of other recently born centrioles, either through inhibition or activation. We then proceeded to investigate the kinetics of de novo biogenesis at different dilutions. We found that all tested dilutions, 0.5, 0.33, and 0.16 relative concentration, delay the onset of de novo centriole assembly (Fig. 4, B and C). The delay is dilution-dependent; centrosome formation occurs in all explants at the highest Plk4 concentration, saturating within 25 min. Importantly, saturation is not reached within the observation time at lower Plk4 concentrations (Fig. 4 B), and the onset of de novo centriole assembly occurs progressively later at larger dilutions, i.e., lower concentrations (Fig. 4 C), demonstrating that Plk4 concentration is a determinant for the onset of biogenesis. Strikingly, our results also show that the time from first to second de novo biogenesis event (Fig. 4 C; and Fig. S4, C and D) and the kinetics of centriole duplication (Fig. 4 D) are concentration independent.
Figure 4.
Plk4 concentration modulates the onset of centrosome biogenesis. (A) Inter-event distance at low Plk4 concentration. Cumulative distribution of observed (obs) and simulated (sim) inter-event distances measured in 3D for the first four centrosomes formed de novo in the explants, at the lowest Plk4 overexpression (0.16 relative concentration of Plk4). The gray envelope indicates the 95% confidence interval (from quantile 0.025 to 0.975) for the simulated data. (B) Plk4 titrations were performed by mixing WT and Plk4 overexpressing eggs at different ratios. Time of onset of de novo centriole biogenesis is shown as cumulative distribution function for four relative concentrations of Plk4. Lower concentrations delay the initiation of de novo centriole biogenesis, with a large majority of the individual explants not forming centrioles, at lower concentrations, during the observation time. (C) Time to the first de novo event, and inter-event time between the first and second de novo events in mixed explants with different concentrations of Plk4. In all dilutions tested, the time for the first event to occur is longer, while the first to second inter-event time is unaffected. Median with interquartile range is presented for n = 56, n = 62, n = 39, and n = 25 explants at 1, 0.5, 0.33, and 0.16 relative concentration of Plk4, respectively. (D) The duplication time of the first centriole formed de novo is similar at high (1) and low Plk4 concentration (0.16). Centrioles formed de novo duplicate, on average, 3 min after their biogenesis, at both high (1, n = 44 centrioles) and the lowest (0.16 Plk4 dilution, n = 20 centrioles) concentrations of Plk4 investigated. The horizontal lines and error bars represent the respective median and interquartile distance. The duplication time is not statistically different between the two conditions (Mann–Whitney test, P = 0.59).
The observed kinetics is reminiscent of a bistable process (Tyson and Novak, 2001; Charvin et al., 2010), switching from nonpermissive to permissive states for centriole biogenesis in a Plk4 concentration-dependent manner. Bistability is typically shown by cell cycle transitions; they rely on accumulation of a signal or activating enzyme, and the moment a critical threshold is crossed, the kinetics becomes essentially irreversible (Tyson and Novak, 2001; Charvin et al., 2010). We wondered whether our experimental observations, in particular the results from the dilution experiment, could be explained by the biochemistry of Plk4, as Plk4 undergoes auto-activation. Thus, we formulated a mathematical model of this process and linked it to centriole biogenesis. Full Plk4 activity is accomplished by trans-autophosphorylation of a conserved T-loop residue within its catalytic domain, which triggers kinase activation through a positive feedback mechanism (Fig. S4 E: from B to A*, and A to A** forms; Lopes et al., 2015). The expected kinetics of Plk4 activation may greatly depend on its concentration and, thus, on overcoming a critical concentration threshold (Fig. S4 F). Numerical simulations were largely in agreement with our experimental observations, supporting that biogenesis is controlled by a Plk4 activity threshold (Fig. S4 G).
In summary, our experiments are in agreement with a model entailing nonlinear accumulation of active Plk4, which is needed to drive centriole biogenesis. However, other, more indirect molecular mechanisms such as inhibition by Plk4 of a negative regulator of centriole biogenesis cannot be ruled out at this stage. Our results also lead to the hypothesis that in WT eggs, where de novo biogenesis does not occur, Plk4 concentration must be very low and undergoes limited oligomerization in the cytosol, which can prevent auto-activation until the sperm centriole enters the egg and locally concentrates Plk4, leading to centriole duplication.
Plk4 robust regulation under endogenous conditions
The concentration and the oligomerization state of Plk4 in the cytoplasm have never been studied in Drosophila. Therefore, we decided to investigate these biochemical parameters in the early fly embryo using fluorescence correlation spectroscopy (FCS). FCS is a technique with single-molecule sensitivity and, therefore, ideal for quantification of low-abundance proteins present at picomolar to nanomolar concentrations inside the cell. FCS measurements of Plk1, also a member of the Polo-like kinase family, revealed distinct diffusion coefficients for Plk1 in the cytoplasm that correlated with its kinase activity during different cell cycle stages (Mahen et al., 2011). Therefore, we conducted in vivo FCS to determine Plk4 concentration, diffusion, and oligomerization in syncytial embryos.
We tagged both Plk4 alleles with fluorescent reporters by CRISPR (Table S2, Table S3, Fig. S1 A, Fig. S5 A, and Video 6). First, we characterized the biophysical properties of the tag mNeonGreen in buffer and injected it into control (RFP–β-tubulin expressing) embryos for reference (Fig. S5, B and C). Next, we performed FCS in mNeonGreenPlk4 expressing embryos. We could detect bursts of green fluorescence above background signal (Fig. S5 D, i). More importantly, the mNeonGreen-Plk4 traces generated clear autocorrelation function (ACF) curves, whereas the background fluorescence measured in RFP–β-tubulin expressing embryos did not autocorrelate (Fig. S5 D, ii). For mNeonGreen-Plk4, the normalized ACFs were best fitted, with minimal residuals, to a two-component diffusion model, and this fit was corroborated by the distribution obtained from the maximum entropy method fit (MEMfit; Fig. 5 A and Table S4). Two fractions of diffusing mNeonGreen-Plk4 were detected in the cytoplasm: one diffusing at 17.17 µm2/s, which is similar to the fluorophore mNeonGreen alone (Fig. S5 C), and another, slower fraction diffusing at 1.49 µm2/s (Fig. 5 A and Table S4). While the first fraction probably refers to Plk4 monomers, the second cannot be explained by homo-oligomerization alone, suggesting that a fraction of Plk4 may associate with quasi-immobile substrates in the cytosol.
Figure S5.
In support of Fig. 5: FCS analysis of Drosophila embryos. (A) Maximum intensity z projections from a time-lapse video of a syncytial D. melanogaster embryo expressing endogenous mNeonGreen-Plk4 (green) and MT reporter RFP–β-tubulin (magenta). Plk4 localizes at the centrosomes (high-intensity tubulin spots) in interphase. Larger green dots result from yolk auto-fluorescence. At time point t = 00:00, the embryo is in metaphase of nuclear cycle 11. The insets show the progression of a single nucleus and its daughters throughout one cell cycle. The cell cycle stage is indicated above each image. Time is reported as minutes:seconds. The asterisk indicates an abnormal mitotic spindle. (B) FCS measurements of purified mNeonGreen fluorophore in a buffer supporting viability of the cytoplasm (Telley et al., 2013). (C) FCS measurements of mNeonGreen after injection into the cytosol of syncytial embryos expressing RFP–β-tubulin. The graphs show the normalized, fitted ACFs (blue dots and light blue curve), with SD (shaded area) and MEMfit (red line). The time lags (diffusion times) determined using the two fitting methods shown next to the MEMfit curves are in agreement. The peak at the fast time scale corresponds to the triplet state of the fluorophore (9.48 × 10−6 s in solution; 22 × 10−6 s in the cytoplasm), whereas the second peak in the slower time scale corresponds to the 3D diffusion of mNeonGreen, from which a diffusion coefficient D was calculated (1.59 × 10−4 s, D = 85.21 µm2/s in solution; 6.54 × 10−4 s, D = 20.72 µm2/s in the cytoplasm). The residuals obtained from the best fit are shown below the ACF graphs. (D) Single-molecule mNeonGreen–Plk4 quantifications in the cytosol of the syncytial fly embryo. i: Intensity traces of mNeonGreen–Plk4 (black) and background noise (gray). Of note, intensity bursts of mNeonGreen–Plk4 are well distinguishable from background noise (inset). ii: Raw ACFs from multiple independent FCS measurements. While the intensity of background acquisitions as measured in RFP–tubulin expressing embryos does not auto-correlate, traces from mNeonGreen–Plk4 expressing embryos exhibit significant autocorrelation.
Video 6.
In support of Fig. 5: mNeonGreen–Plk4 localization in a syncytial Drosophila embryo. Time-lapse video of an embryo expressing homozygous mNeonGreen–Plk4 (endogenously labeled by CRISPR, in green) and RFP–tubulin (magenta), acquired on a spinning-disk confocal microscope, through nuclear cycles 10–13. The video is a bleach-corrected maximum-intensity z projection. Time (minutes:seconds) is shown at the top left. Playback rate is 5 fps.
Figure 5.
Single-molecule mNeonGreen-Plk4 quantifications in the cytosol of the syncytial fly embryo by FCS. (A) Normalized fitted ACF (Fit, light blue dashed line), with SD (shaded area) and MEMfit distributions (Distribution, red line) for mNeonGreen-Plk4 in the cytoplasm. Based on the two fitting methods, three times scales were determined: the fastest time scale peak corresponds to the triplet state of the fluorophore (7.85 × 10−6 s); whereas the second and third slower time scales correspond to distinct 3D diffusional mobility of mNeonGreen-Plk4 in the cytoplasm, from which the diffusion coefficients (D) were calculated (fastest fraction: 7.89 × 10−4 s, D = 17.2 µm2/s; slower fraction: 9.11 × 10−3 s, D = 1.49 µm2/s). The residuals from the fitted data (Fit) are shown below the graphs. (B) Plk4 undergoes limited oligomerization in the cytosol of the Drosophila blastoderm embryo. The mNeonGreen distribution was fitted to a Weibull distribution, which has a peak value of 4,100 Hz. Next, the mNeonGreen–Plk4 data were fitted with an additional Weibull distribution (one for monomer-like and another for oligomer-like). The second mNeonGreen-Plk4 distribution peaks at 18,450 Hz. From this analysis, it follows that the overall normalized brightness (intensity per particle, mean ± SD) for mNeonGreen–Plk4 in the cytoplasm is higher than for the single mNeonGreen monomer injected into the cytoplasm at a similar concentration, indicating that Plk4 is present both as a monomer (around 30.1% of its diffusing pool) and as low-order oligomers (69.9% of diffusing mNeonGreen–Plk4 pool).
Next, we calculated the total concentration of mNeonGreen-Plk4 in the cytosol and determined its oligomeric state using the brightness of injected mNeonGreen monomer as calibration (Fig. S5 C). We confirmed that Plk4 concentration in the cytosol is very low, around 7.55 nM, and an estimate for diffusion in the cytosol suggests coexistence of monomeric and oligomeric forms (Fig. 5 B). More precisely, 30.1% of diffusing Plk4 is detected as a monomer, while around 69.9% forms low-order oligomers, likely dimers and at most tetramers (Fig. 5 B).
Altogether, the FCS results indicate that Plk4 is indeed a very low abundance protein that undergoes limited oligomerization within the cytosol, in early developing Drosophila embryos. Thus, it is likely that in WT eggs, the nanomolar concentration and the low oligomeric presence of Plk4 prevent de novo centriole assembly. Only when centrioles are provided by the sperm can Plk4 be concentrated, initiating centriole duplication. The change in the kinetics of de novo centriole assembly in response to Plk4 concentration agrees with the current body of knowledge in the centrosome field. Collectively, the data suggest that centriole formation is heavily regulated by the time-dependent concentration of several centrosomal molecules in a single place (Rale et al., 2018; Takao et al., 2019). But what helps to concentrate these centrosomal molecules? Recent studies suggest that the PCM may play an important role (Loncarek et al., 2008; Ito et al., 2019; Mercey et al., 2019a, 2019b).
PCM components promote the early steps of centriole de novo assembly
In D. melanogaster cultured cells, codepletion of the centriolar protein Ana2 and the PCM component D-pericentrin–like protein (D-Plp) additively impair centriole biogenesis, indicating that two alternative pathways, a centriolar and a PCM-mediated, may be involved (Ito et al., 2019). Moreover, in mouse ependymal cells without centrioles and specialized electron-dense deuterosomes that can feed centriole assembly, a correct number of centrioles can form de novo within pericentrin-rich areas (Mercey et al., 2019b). To test the role of the PCM in de novo centriole assembly, we performed perturbation experiments in D. melanogaster–cultured cells, since it is easier to knock down multiple genes in cultured cells than in the organism. To create an assay for de novo centriole assembly, we depleted centrioles through successive cell divisions in the presence of RNAi against Plk4 (Fig. 6 A). As cells proliferate in the absence of centriole duplication, centriole number is progressively reduced. This is followed by a recovery period, without RNAi against Plk4, where Plk4 translation is resumed and centrioles assemble de novo (Rodrigues-Martins et al., 2007).
Figure 6.
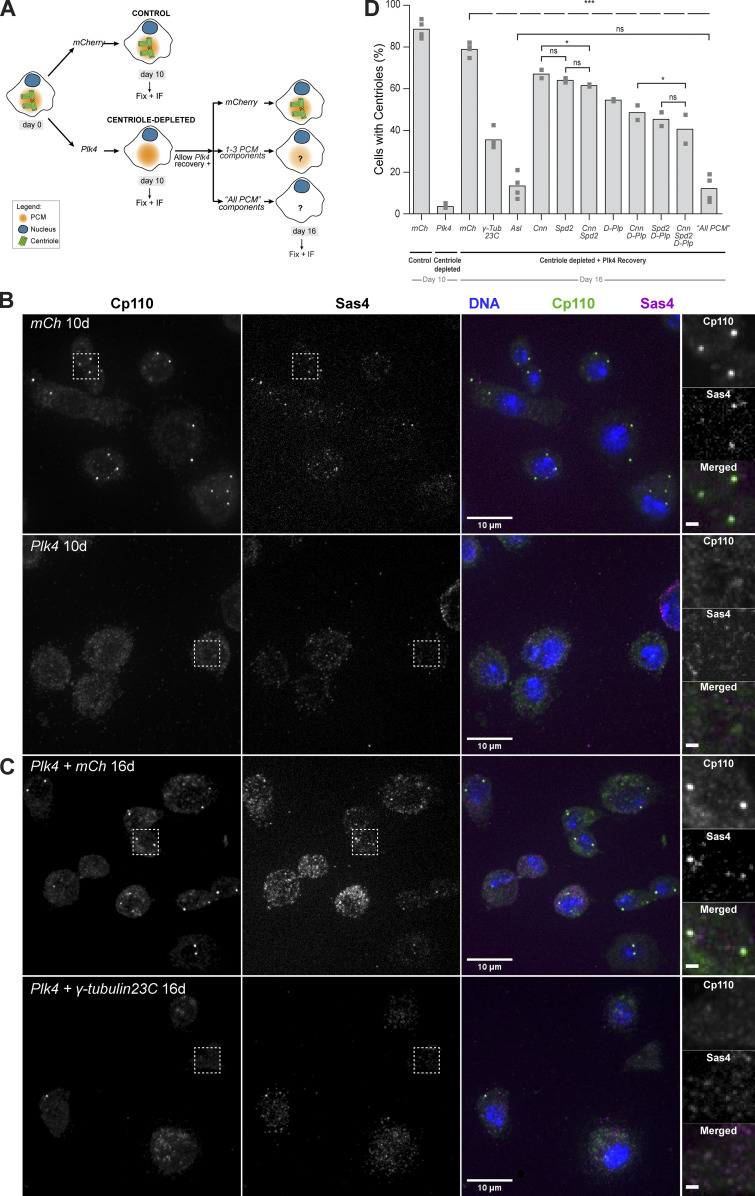
De novo centriole biogenesis is partially impaired in PCM-depleted Drosophila cells. (A) DMEL-cultured cells were treated with RNAi against Plk4 over the course of 12 d to deplete their centrioles. mCherry (mCh) RNAi was used as negative control. Cells treated with RNAi against Plk4 gradually lost centrioles during proliferation. On day 10, samples were obtained for fixation and staining, and centriole-depleted cells were treated with RNAi against individual PCM components. On day 12, Plk4 translation was allowed to recover. On day 16, cells were fixed and stained by immunofluorescence (IF). We targeted for individual PCM components—Cnn, Asl, D-Plp, Spd2, or γ-tubulin 23C—or combinations of these molecules previously shown to be essential for PCM assembly in cycling cells—Cnn + Spd2, Cnn + D-Plp, Spd2 + D-Plp, or Cnn + Spd2 + D-Plp (Gomez-Ferreria et al., 2007; Conduit et al., 2014; Lerit et al., 2015; Feng et al., 2017; Citron et al., 2018). Additionally, we depleted all four PCM components—Cnn + Asl + D-Plp + Spd2 (referred to as All PCM)—required for PCM maintenance (Pimenta-Marques et al., 2016). (B) Maximum-intensity z projections of fluorescence from DMEL cells after 10 d treatment with RNAi against Plk4 (top row) or mCherry (mCh, bottom row). Cells were stained for centriolar markers Sas4 (magenta) and Cp110 (green), in addition to DAPI against DNA (blue). Inset squares in each fluorescence channel are shown at higher magnification on the right (scale bar, 1 µm). Knock-down of Plk4 (bottom row) caused loss of centrioles, as reported by the absence of spot signals in the green and magenta channels. Note that it is common for a small fraction of untreated DMEL cultured cells to have either too many (more than four) or too few (less than two) centrioles (Bettencourt-Dias et al., 2005). This is found in most cell lines from D. melanogaster as they are permissive to those changes. In contrast to vertebrate cells, a p53-dependent cell cycle arrest does not occur in the presence of numerical centrosome abnormalities in these insect cells. (C) Maximum-intensity z projections of fluorescence from centriole-depleted DMEL cells after 6 d treatment with RNAi against mCherry (top row), allowing recovery of normal centriole number, or against γ-tubulin 23C (bottom row) leading to little or no recovery of centrioles. Staining, color code, and insets are as in B. (D) Quantification of cells with centrioles after 10 and 16 d of RNAi treatment. Centriole number was scored in >300 cells per treatment, per experiment. The bars represent the average of proportions obtained in two or four independent experiments (gray squares) for the conditions listed. Superscripts denote statistical significance in treatments. ns (not significant), P ≥ 0.05; *, P < 0.05; ***, P < 0.001 (Pearson’s χ2 test, and two-proportions z test on pooled data). The top dashed arch denotes statistical difference between the mCh (control) and every other condition. γ-tub, γ-tubulin.
After RNAi against Plk4 (Fig. 6 B), we further depleted PCM components, while allowing Plk4 translation to recover, which is sufficient to drive centriole de novo assembly in the mCherry-treated control cells (Fig. 6 C). After 10 d, only 3% of the cells treated with RNAi against Plk4 had centrioles, whereas in the mCherry-treated control, ∼80% of the cells had at least one centriole, as expected (Fig. 6 D; Rodrigues-Martins et al., 2007). Cells depleted of centrioles were then treated for 4 d with RNAi against individual—Cnn, Asl, D-Plp, or Spd2—or combinations of PCM molecules necessary for centriole assembly through the canonical duplication pathway: Cnn + Spd2, Cnn + D-Plp, Spd2 + D-Plp, or Cnn + Spd2 + D-Plp (Gomez-Ferreria et al., 2007; Conduit et al., 2014; Lerit et al., 2015; Feng et al., 2017; Citron et al., 2018; and Table S5). Additionally, we depleted all four PCM components, Cnn + Asl + D-Plp + Spd2 (referred to as All PCM), previously shown to be essential for centriole maintenance (Pimenta-Marques et al., 2016), and the downstream PCM protein, γ-tubulin, which is known to be at the core of MT nucleation across species, and to contribute to centriole duplication in Caenorhabditis elegans embryos and human cells (Dammermann et al., 2004; Kleylein-Sohn et al., 2007).
While cells treated with control mCherry double-stranded RNA (dsRNA) recovered centriole number within 4 d after ceasing Plk4 dsRNA treatment (indicating that centrioles formed de novo), only 10–15% of the cells treated with dsRNA against All PCM had centrioles (Fig. 6 D), suggesting an important role of the PCM in regulating de novo assembly. From all components of the PCM, Asl, which is known to be both substrate and recruiter of Plk4, generated the strongest phenotype, confirming observations from Dzhindzhev et al. (2010). De novo centriole formation was impaired by γ-tubulin 23C depletion, whereby only ∼35% of Plk4-depleted cells recovered a normal centriole number (Fig. 6 D). This result implies that γ-tubulin is important for de novo centriole biogenesis.
We proceeded to validate this observation in vivo and generated fly lines expressing shRNA against γ-tubulin 23C and γ-tubulin 37C—a maternally expressed gene, mostly abundant in early fly development (Tavosanis et al., 1997)—under control of the UASp/Gal4 system (Table S6). Fertilized eggs laid by females overexpressing the shRNA targeting γ-tubulin 37C do not develop (Table S7), and unfertilized eggs display spindle defects similar to those previously shown in oocytes from γ-tubulin 37C mutant females (asterisks in Fig. 7 A; Tavosanis et al., 1997), indicating that this RNAi construct is functional. We collected unfertilized eggs expressing RNAi targeting γ-tubulin 23C and/or 37C, while simultaneously overexpressing Plk4, under control of the V32-Gal4 driver. In the control, centrioles form de novo in 73% (22/30) of the eggs overexpressing Plk4 alone (Fig. 7 B). On the other hand, in the case of recombinant γ-tubulin 23C + 37C RNAi flies overexpressing Plk4, only 26% (14/54) of their eggs show centrioles, while individual γ-tubulin knock-downs display intermediate phenotypes (Fig. 7 B). Therefore, γ-tubulin depletion impairs de novo centriole assembly in vivo too.
Figure 7.
De novo centriole biogenesis is partially impaired in unfertilized eggs overexpressing Plk4 after depletion of γ-tubulin. (A) Maximum-intensity z projections of fluorescence from unfertilized eggs overexpressing Plk4 alone (control) or together with RNAi against γ-tubulin 23C, γ-tubulin 37C, or a combination of both. Eggs were stained against Bld10 (cyan), Ana1 (yellow) and tyrosinated α-tubulin (magenta). Centrioles were identified by colocalization of spot-like signals from at least two of the three reporters. Inset squares in each fluorescence channel are shown at higher magnification on the right (scale bar, 3 µm). Orange asterisks reveal putative meiotic defects, previously described to occur in oocytes from γ-tubulin 37C mutant females (Tavosanis et al., 1997). In these example images, the control shows signal spots in all channels, while in RNAi conditions, some reporter signals were present (white square) and others absent (orange square) in the fewer centrosome-like dots observed. Note that in the double knock-down condition (γ-tubulin 23C+37C) we did not detect any signal from tyrosinated α-tubulin despite the presence of some centrosome-like dots bearing centriolar reporters (two examples are shown, i and ii). (B) Quantification of eggs with centrioles depleted of γ-tubulin 37C alone, or in combination with depletion of γ-tubulin 23C. The presence of centrioles was scored by the concomitant detection of at least two centrosomal reporters. n = 30 eggs (control); n = 49 eggs (γ-tubulin 23C); n = 47 eggs (γ-tubulin 37C); n = 54 eggs (γ-tubulin 23C + 37C). ns (not significant), P ≥ 0.05; **, P < 0.01; *, P < 0.001 (Pearson’s χ2 test, and two-proportions z test on pooled data). α-Tub, α-tubulin.
Discussion
De novo centriole assembly is widely documented across the eukaryotic tree of life. Numerous studies reported its incidence and even its relationship with life-history traits in particular groups (Mizukami and Gall, 1966; Aldrich, 1967; Grimes, 1973a,b; Mir et al., 1984; Renzaglia and Garbary, 2001; Idei et al., 2013), but they have not addressed how de novo assembly is regulated in living cells and what the contribution of older centrioles to this process is. Herein, we demonstrate that cytosolic explants from post-meiotic D. melanogaster eggs overexpressing Plk4 are capable of de novo and canonical centriole biogenesis, offering the opportunity to investigate centriole formation at high spatio-temporal resolution by fluorescence microscopy and super-resolution techniques (Fig. 1). In these explants, Plk4 triggers stochastic formation of multiple centrioles. Our assay allowed us to study several important open questions regarding the regulation of de novo centriole biogenesis.
Canonical biogenesis is spatially and temporally robust
Current knowledge supports the need for timely extrinsic cues, provided by the cell cycle, to control biogenesis (Wang et al., 2011; Izquierdo et al., 2014; Fu et al., 2016; Tsuchiya et al., 2016). However, herein we observed that centrioles can be formed de novo and undergo time-dependent centriole-to-centrosome conversion, maturation, and duplication (Fig. 2), all in the absence of cell cycle transitions (Horner et al., 2006; Vardy and Orr-Weaver, 2007; Deneke et al., 2019). Surprisingly, we also observed that while de novo takes longer to start at low Plk4-overexpressing conditions, duplication time is similar at different concentrations of Plk4 (Fig. 4 D). This indicates that, despite the absence of a typical cell cycle reaction network, canonical biogenesis is both spatially and temporally robust. When it occurs, it always occurs close to an existing centriole within a conserved time interval. Hence, we propose that distinct biochemical reaction networks regulate de novo and canonical biogenesis, with the initiation of de novo biogenesis being more sensitive to Plk4 concentration.
Can nonlinear accumulation of active Plk4 in the cytosol explain de novo biogenesis?
In switch-like processes, systems typically undergo irreversible transitions upon crossing a critical threshold. It is possible that de novo assembly is a switch-like mechanism operating within the entire cytosol: the measured time of first de novo events is modulated by the concentration of Plk4 while the inter-event time is much less affected (Fig. 4, C and D; and Fig. S4, D and E), and the spatial distribution falls within random predictions at different Plk4 concentrations (Fig. S3). Theoretical modeling and simulations agree with the nonlinear kinetics of Plk4 trans-autoactivation in the cytosol (Lopes et al., 2015), suggesting that the burst in biogenesis occurs once a critical activity threshold is crossed (Fig. 4, and Fig. S4 C, also proposed by Lambrus et al., 2015, for the regulation of canonical duplication). Plk4 may also need to oligomerize and form condensates that become stable seeds, nucleation points for centriole assembly (Montenegro Gouveia et al., 2018; Yamamoto and Kitagawa, 2019; Park et al., 2019). Consistent with this, we observe a few oligomeric forms of Plk4 in the cytoplasm at extremely low concentrations of Plk4 (Fig. 5). We suspect that the concentration of active Plk4 increases over time at multiple sites in the cytosol, overcoming the activity of counteracting factors and driving centriole biogenesis almost simultaneously in independent locations in the explants. Once a critical threshold in molecular concentration is locally crossed, Plk4-driven centriole assembly is likely irreversibly catalyzed. In the future, it will be important to develop a molecular sensor to monitor Plk4 concentration and activity in space and time to understand the importance of each one of the factors in centriole biogenesis.
Which factors can help to locally promote centriole formation?
Besides local Plk4 concentration, other factors may play a role in regulating the location of de novo centriole assembly, breaking cytosolic homogeneity. Our experiments support an important role for the PCM in promoting de novo assembly, in particular the component Asl and its downstream effector γ-tubulin. In fact, depletion of γ-tubulin led to a strong reduction in de novo biogenesis, both in vitro and in vivo (Figs. 6 and 7). We also observed a mild role for D-Plp, which is enhanced by Cnn and Spd2, perhaps through their function in γ-tubulin recruitment (Fig. 6).
The PCM may generate protein scaffolds in the cytoplasm where centriolar proteins bind with higher affinity, therefore locally concentrating these molecules and forming stable seeds for centriole biogenesis. For example, CEP152/Asl has been shown to recruit and activate Plk4 (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Boese et al., 2018), while D-Plp promotes SAS6 recruitment (Ito et al., 2019). Moreover, γ-tubulin promotes MT nucleation, which may attract more components via motor-based transport or through entrapment of proteins with MT-binding capacity, such as Plk4 (Montenegro Gouveia et al., 2018). These diverse properties of the PCM may promote centriole biogenesis within biochemically confined environments in the cytoplasm. Our results likely provide an explanation why centrioles can form de novo within PCM aggregates, both in vertebrate multiciliated cells (Mercey et al., 2019b) and in cycling cells (Khodjakov et al., 2002).
Do preexisting centrioles influence the assembly of new ones?
Previous research had suggested that once centrioles form de novo in cells without centrioles, any other events of biogenesis would be “templated,” i.e., follow the canonical pathway (Marshall et al., 2001; La Terra et al., 2005; Uetake et al., 2007; Lambrus et al., 2015). Together, these studies suggest that centrioles negatively regulate the de novo pathway and/or play a dominant role in biogenesis by recruiting the centrosomal components that limit biogenesis. In fly egg explants, we observed that centrioles continue to form de novo long after the first centriole has assembled and duplicated (Fig. 2). Both pathways, de novo formation and canonical duplication, co-occur within the same cytoplasmic compartment, indicating that “older” centrioles and their duplication do not prevent biochemically de novo centriole assembly, even at low Plk4 overexpression (Fig. 4 and Fig. S3). Thus, it appears that these pathways are not inherently mutually inhibitory in the fly germline under these conditions. We compared our observations with in silico results obtained under similar spatial geometries. These indicate that recently formed centrioles can duplicate but do not influence de novo formation (Fig. 3 and Fig. S3).
To reconcile the previous published data and our own, we propose that, once Plk4 in the cytosol crosses a threshold of concentration and activity, centrioles may form at various locations independently. Some of the new centrioles may mature and recruit Plk4 and duplicate, but they cannot prevent the de novo assembly of centrioles in other places. In other cells that were artificially devoid of centrioles, upon restauration of the right conditions, many centrioles can form de novo, almost simultaneously, similar to what we observe here (Marshall et al., 2001; La Terra et al., 2005; Uetake et al., 2007; Lambrus et al., 2015; Wong et al., 2015). Given that in those systems centriole duplication takes longer, de novo formed centrioles may only acquire the competence to duplicate later, preventing simultaneous de novo and canonical duplication, as we described here. Our results further suggest that the spatio-temporal (local) concentration of Plk4 must be well regulated in cells to prevent supernumerary centriole formation.
How do our results fit with naturally occurring biogenesis?
A previous study had estimated 1,200–5,000 Plk4 molecules per cell in asynchronous human cells, from which around 70 molecules are loaded at the centrosome (Bauer et al., 2016). We generated flies labeled with mNeonGreen at Plk4 genomic loci (Fig. S1 A) and confirmed that the endogenous, diffusing pool of Plk4 is present at low concentration and undergoes limited self-association in the cytosol in early fly embryos (Fig. 5 B). These properties of Plk4 in the cytosol are unfavorable to de novo centriole assembly, ensuring that centrioles form in the right place by canonical biogenesis. Our measurements help building a quantitative framework for the transition of Plk4 molecules from the cytoplasm to the centriolar compartment, which ultimately controls centriole biogenesis.
Finally, our findings in D. melanogaster relate to the naturally occurring parthenogenetic development in other organisms, including some species of wasps, flies, and aphids (Riparbelli et al., 1998; Tram and Sullivan, 2000; Riparbelli and Callaini, 2003; Riparbelli et al., 2005; Ferree et al., 2006). In those cases of parthenogenesis, multiple functional centrosomes form spontaneously in the egg during meiosis, two of which assemble the first mitotic spindle and trigger normal development. In the case of Drosophila mercatorum, the centrosomes that assemble de novo can also duplicate, and they do so in a cell cycle–dependent manner (Riparbelli and Callaini, 2003). It would be relevant to determine if the burst in centrosome assembly coincides with an increase in global Plk4 concentration or activation in the egg of these species. Just like in our system, a highly variable number of MTOCs are assembled, suggesting the presence of weak control mechanisms against de novo centriole formation in the germline, once the eggs enter meiosis. Further studies aimed at documenting centrosome birth kinetics and their maturation in these natural systems may reveal more about the principles that govern de novo centriole formation and their conservation throughout species evolution.
In oocytes from some parthenogenetic Hymenoptera, maternal centrosomes form de novo close to cytoplasmic organelles highly enriched in γ-tubulin, called accessory nuclei (Ferree et al., 2006). Moreover, centrosome ablation in vertebrate CHO cells is followed by accumulation of γ-tubulin and pericentrin in nuclear envelope invaginations, hours before bona fide centrioles are detected (Khodjakov et al., 2002). Interestingly, if treated with nocodazole, acentriolar CHO cells are no longer capable of assembling centrioles de novo (Khodjakov et al., 2002). Therefore, besides substantiating previous studies, our work further suggests that the organization of PCM-rich foci likely represents the first, essential step for de novo centriole assembly. In the future, it will be important to understand in detail how PCM and MTs contribute to the early onset of centriole formation.
Materials and methods
Fly work and sample preparation
D. melanogaster stocks and husbandry
All D. melanogaster stocks used in this study are listed in Table S1. Transgenic mNeonGreen- and mEGFP-Plk4 flies were generated in-house by CRISPR/Cas9-mediated gene editing (Port et al., 2014). 20 bp gRNAs targeting the N-terminal region of Plk4, with 5′ BbsI-compatible overhangs, were ordered as single-stranded oligonucleotides (Sigma-Aldrich). The complementary oligonucleotides were annealed, phosphorylated, and cloned into BbsI-digested pCFD3-dU6:3gRNA expression plasmid (from Simon Bullock, MRC, Cambridge, UK). Plasmid DNA templates were designed for homologous recombination-mediated integration of the fluorescent proteins mNeonGreen or mEGFP between the 5′UTR and the first coding exon of Plk4. 1 kbp long 5′ and 3′ homology arms were PCR-amplified from genomic DNA isolated from y1,M{nanos-Cas9.P}ZH-2A,w* flies (Table S2; BDSC #54591). The mNeonGreen (Addgene plasmid #138329) and the mEGFP (Addgene plasmid #18696) coding sequences were PCR-amplified from plasmids using the primers listed in Table S2. All fragments were sub-cloned into the pUC19 plasmid (Stratagene) using restriction enzymes: 5′ homology arm, NdeI and EcoRI; fluorescent tag + linker, EcoRI and KpnI; and 3′ homology arm, KpnI and XbaI. Synonymous mutations were performed on the homology arms, removing the protospacer-adjacent motif sequence from the donor plasmid to prevent retargeting. The final donor template for homologous recombination-mediated integration was composed of a fluorescent reporter and a short flexible linker (see sequence in Table S2), flanked by 1 kbp homology arms. Two circular plasmids, pCFD3-Plk4-gRNA and mNeonGreen or mEGFP templates, were coinjected into nos-Cas9 embryos (BDSC #54591; Port et al., 2014). Injected flies were crossed to a balancer strain, and single-fly crosses were established from their offspring. The resulting generation was screened for positive integrations by PCR, using primers dmPLK4 5UTR 3 FW and dmPLK4 1exon Rev (Table S3). Homozygous mNeonGreen-Plk4 and mEGFP-Plk4 were crossed to pUb–RFP–β2-tubulin flies (gift from Yoshihiro Inoue; Kitazawa et al., 2014), establishing stable stocks.
We also generated flies expressing shRNAs against γ-tubulin 37C and 23C under the UASp promoter and crossed them with the V32-Gal4 (w*; P{maternal-αtubulin4-GAL::VP16}V2H, kindly provided by Daniel St. Johnston), at 25°C, to knock down both genes in the female germline. To generate γ-tubulin 37C and 23C constructs, sense and antisense oligos for each target gene were annealed and cloned into pWALIUM22 (Drosophila Genomics Resource Center), using NheI and EcoRI restriction enzyme sites (Table S6). Each construct was inserted into different landing sites on the third chromosome by PhiC31 integrase-mediated recombination (Table S6). Germline-specific Plk4 overexpression was accomplished by crossing flies carrying the pUASp-Plk4 construct (Rodrigues-Martins et al., 2007) and the V32-Gal4, at 25°C.
Centrosomes were visualized using the following centrosomal reporters: (1) pUb-Spd2–GFP (homemade construct, injected at BestGene); (2) Ana1-tdTomato (gift from Tomer Avidor-Reiss; Blachon et al., 2008); (3) pUASp-GFP-Plk4 (homemade construct, injected at BestGene); and (4) Asl-mCherry (gift from Jordan Raff; Conduit et al., 2015), in combination with either endogenous Jupiter-GFP (BDSC #6836) or endogenous Jupiter-mCherry (gift from Daniel St. Johnston; Lowe et al., 2014), as reporters for centrosomal MT nucleation.
Flies were maintained at 25°C in vials supplemented with 20 ml of culture medium (8% molasses, 2.2% beet syrup, 8% cornmeal, 1.8% yeast, 1% soy flour, 0.8% agar, 0.8% propionic acid, and 0.08% nipagin).
Testing UASp-RNAi lines for developmental lethality
To test for lethality effects of γ-tubulin 37C and γ-tubulin 23C shRNAs alone and recombined, each line was crossed to V32-Gal4 flies. Female progeny carrying the Gal4 and shRNA were crossed to w1118 males (10 females × 5 males per vial, four independent crosses), and the number of pupae in each vial was counted 9–10 d after each transfer (three technical repeats were performed). See results in Table S7.
Embryo/egg collections
For embryo collections, 3–4-d-old female and male flies were transferred to a cage coupled to a small apple juice agar plate (25% apple juice, 2% sucrose, 1.95% agar, and 0.1% nipagin), supplemented with fresh yeast paste. Embryos were collected for 1 h and aged for half an hour. For unfertilized egg collections, approximately one hundred 5–7-d-old virgin females were placed in the cage, and 20-min collections were performed. At this age, a large majority of eggs did not contain any centrosomes, nor did the explants from those eggs. All cages were maintained at 25°C, under 50–60% humidity. The embryos or eggs were dechorionated in 7% sodium hypochlorite solution (VWR), washed thoroughly in Milli-Q water, aligned, and immobilized on clean, poly-L-lysine (PLL)–functionalized coverslips, using a thin layer of heptane glue. Samples were covered with Voltalef grade H10S oil (Arkema).
Preparation of micropipettes and functionalized coverslips
High Precision 22 × 22 glass coverslips no. 1.5 (Marienfeld) were cleaned for 10 min in 3 M sodium hydroxide, followed by four dip-and-drain washes in Milli-Q water. Next, they were sonicated for 15 min in “Piranha” solution (H2SO4 and H2O2 [30% concentrated] mixed at a 3:2 ratio), followed by two washes in Milli-Q water, once in 96% ethanol, and twice again in Milli-Q water for 5 min each. Coverslips were spin-dried and subsequently treated for 20 min with PLL solution 0.01% (Sigma-Aldrich), followed by multiple dip-and-drain washes in Milli-Q water. The coverslips were spin-dried and stored in a clean and dry rack.
Glass capillaries (0.75 mm inner diameter, 1 mm outer diameter; Sutter Instrument) were forged into glass needles by pulling them on a vertical pipette puller (Narishige PC-10), using a one-step pulling protocol, at ∼55% heating power. Using a sharp scalpel, the tip of the capillary was cut, generating micropipettes with a 30–35 µm diameter pointed aperture (Telley et al., 2013).
Single-egg extract preparation
Cytoplasmic extraction from individual unfertilized eggs and explant deposition onto the surface of PLL-coated coverslips was performed on a custom-made micromanipulation setup coupled to an inverted confocal microscope, as previously described in Telley et al. (2013) and de-Carvalho et al. (2018). The size of the explants was manually controlled in order to produce explants measuring 40–80 µm in diameter and ∼10 µm in height, allowing time-lapse imaging of the entire explant volume.
Egg immunostaining and imaging
Unfertilized eggs overexpressing Plk4 and knocked down for γ-tubulin were collected from 5–7-d-old virgin females for 2 h at 25°C, and aged at 25°C for 4 h. The protocol was conducted according to Riparbelli and Callaini (2005). Briefly, aged eggs were rinsed in Milli-Q water + 0.1% Tween, dechorionated in 7% sodium hypochlorite solution (VWR), and washed extensively with Milli-Q water. Using a metal grid, dechorionated eggs were transferred into a scintillation flask containing 50% ice-cold methanol + 50% heptane. The vitelline membrane was removed by vigorously shaking the eggs for 3 min. Devitellinized eggs sank to the bottom of the lower methanol phase and were then collected into a 1.5-ml Eppendorf tube and fixed for 10 min in methanol at −20°C. Following fixation, the eggs were rehydrated in methanol:PBS series (70:30%, 50:50%, and 30:70%) for 5 min each, washed twice in PBS for 10 min, and incubated for 1 h in 1× Dulbecco’s PBS with 0.1% Triton X-100 and 1% BSA (D-PBSTB) at RT. Primary antibody incubations were performed overnight at 4°C with the following antibodies: rabbit anti-Bld10 (dilution 1:500; gift from Tim Megraw, Florida State University, Tallahassee, FL); rat anti-tubulin YL1/2 (dilution 1:50; Biorad) and guinea pig anti-Ana1 (dilution 1:500; kindly provided by Jordan Raff), diluted in D-PBSTB. Eggs were washed extensively in D-PBSTB and incubated with secondary antibodies for 2 h at RT: donkey anti-rabbit Alexa 555 (dilution 1:1,000; Molecular Probes), goat anti-rat Alexa 488 (dilution 1:1,000; Jackson Immunoresearch Laboratories), and donkey anti-guinea pig Alexa 647 (dilution 1:1,000; Jackson Immunoresearch Laboratories) in D-PBSTB. Eggs were washed twice in PSB with 0.1% Triton X-100 and twice in PBS, and mounted onto coverslips in Vectashield mounting media (Vector Laboratories).
Imaging was conducted at 18°C on a Nikon Eclipse Ti-E microscope equipped with a Yokogawa CSU-X1 Spinning Disk confocal scanner and a piezoelectric stage (Physik Instrumente) with a 220 µm travel range. 0.3 µm optical sections were recorded with an EMCCD Photometrics 512 camera using a Plan Fluor 40× 1.30 NA oil immersion objective, controlled with Metamorph 7.5 software. 491 nm, 561 nm, and 640 nm laser lines were used to excite the secondary antibodies. Egg counts were tested with a χ2 test against the null hypothesis that the outcome is random. Then, each test condition was compared with the control condition with a two-proportions z test with the null-hypothesis (H0) that the proportions of eggs with centrioles are equal versus the alternative hypothesis (HA) that the proportion in the test is smaller. The significance level for multiple testing was Bonferroni-corrected. The significance level was P = 0.01.
Image acquisition, processing, and analysis
Time-lapse explant imaging on the spinning disk confocal microscope
Centriole formation was followed by time-lapse imaging in explants initially devoid of centrosomes. Explants were imaged at 23°C using a Plan Apo VC 60× 1.2 NA water objective. We have acquired optical sections of 0.45 µm, carefully selecting the total number of stacks (30–35 planes) in order to cover the entire volume of each individual explant. The images were recorded with an Andor iXon3 888 EMCCD camera using a Yokogawa CSU-W1 Spinning Disk confocal scanner equipped with a piezoelectric stage (737.2SL; Physik Instrumente), installed on a Nikon Eclipse Ti-E microscope. Dual-color (488 nm and 561 nm excitation laser lines, at 100 ms exposure time), 15 s time-lapses were collected with Andor IQ3 software.
Image processing
Multi-stack, time-lapse calibrated images were deconvolved with Huygens (Scientific Volume Imaging, Netherlands) using a point spread function (PSF) automatically calculated from the dataset and run in batch mode, for each channel separately. 32-bit deconvolved images were converted to 16-bit and processed using Fiji (National Institutes of Health; Schindelin et al., 2012). Maximum-intensity z projections of both fluorescence emission channels were produced from the time-lapse acquisitions in Fiji, and selected time frames and insets were further processed with Photoshop CS6 (Adobe). Graphic representations were performed using GraphPad Prism software (version 5.0), and the final figures were assembled in Illustrator CS6 (Adobe).
Centrosome tracking
Centrosomes were tracked using the Fiji Plug-in TrackMate v3.5.1 (Jaqaman et al., 2008). Centrosomes were identified by the Spd2–GFP localization at the center of mass of the microtubule aster. Relying on this criteria, we performed the TrackMate analysis sequentially, starting with the Jupiter-mCherry channel. First, we applied a 3D Gaussian Blur filter to the images (sigma = 0.7 pixels), facilitating the particle detection on TrackMate using the Laplacian of Gaussian algorithm. The microtubule asters were automatically detected inside spheres of ∼0.7 µm in radius, adjusting the threshold value for each time-lapse video independently. Next, the first four de novo formed asters were manually tracked from the list of detected particles. A corrected XYZT coordinate matrix of the first de novo events was saved for each video and imported to MatLab R2016b (The MathWorks, Inc.). MatLab was used to build a 3D binary mask with spheres of radius r (where r ≥ microtubule aster size), centered at the detected coordinate points. This allowed bypassing incorrect particle detection caused by the large number of green auto-fluorescent yolk particles of intermediate signal intensity, therefore excluding them from the analysis early on. The resulting 3D masks were concatenated into 4D hyperstacks, using the Bio-Formats importer plugin in FIJI. The Spd2–GFP images were multiplied by the corresponding 4D binary masks, resulting in a 4D image retaining the pixel intensity values solely within the Jupiter-mCherry ROIs. Next, we used TrackMate to detect centrioles within spheres of 0.3 µm radius, combining sub-pixel localization and a Median filter. After detection, the particles were manually tracked. The final centrosome tracks were exported as an Excel MS spreadsheet.
Western blot analysis
16-bit TIFF images of the protein gels were imported to Fiji (National Institutes of Health; Schindelin et al., 2012). We then used the Gel Analysis tool to determine the density (intensity) of the bands on the Western blots. Briefly, a rectangular selection was first drawn around the first lane, and the same selection area was then propagated to the remaining lanes. Once all lanes were selected, the intensity profile per lane was plotted. Background intensity was discarded from each peak by drawing a line across the base of the distribution and closing off the peak. For each fly line, the area under the corresponding peak was measured and the results exported as an Excel MS spreadsheet. All measurements were conducted in parallel for α-GFP and α-actin, the loading control. The GFP-Plk4 intensity was divided by the intensity of the corresponding actin band. The fold-change in overexpression was determined by calculating the ratio between normalized Plk4 overexpression (pUASp-Plk4-GFP) and normalized endogenously labeled Plk4 (mEGFP-Plk4 knock-in). The reported fold-change is the mean ± SD of four independent experiments and gel band analyses.
Statistics and mathematical modeling
Centrosome tracking data were imported in R version 3.4.1 for further analysis and modeling. The data were analyzed in two ways, one aiming at identifying possible spatial constraints in the positioning of the centrioles relative to each other within the explant at the time a centrosome is formed (neglecting time), and the other aimed at understanding temporal constraints (neglecting space). The data were analyzed statistically, and simulations were performed in an effort to understand the underlying principles. The details regarding sample size, statistical tests, and descriptive statistics are indicated in the respective figure legends and in the main text.
The experimental data were compared with simulated data by calculating the empirical cumulative distributions of each dataset (1 experimental and 100 simulated, each consisting of 68 explants) using the function ecdf from the stats package; and overlapping the median and 95% confidence interval (from the quantiles 0.025–0.975) of the simulated datasets’ cumulative distributions with the corresponding empirical distribution from the experimental dataset. Random numbers were generated using the function runif from the stats library.
For the spatial analysis, each time a new centriole appeared, the 3D pairwise distances between centrioles were calculated and labeled according to appearance relative to prior centrosomes in the explant. This allowed keeping track of event order and, if any spatial effect of existing centrosomes on the appearance of a new centrosome was present, we would be able to detect a difference in their pairwise distances. To test this, the function kruskal.test of the stats library was used to perform the Kruskal–Wallis rank-sum test on the pairwise distances and labels. To complement this analysis, we decided to compare the distributions of pairwise distances with those expected by a spatially null model whereby centrosomes appear randomly across the available space in the explant. To simulate this null model, sets of random points were simulated in sections of semi-spheres of similar geometry as each of the experimental explants, characterized by height and diameter . To this effect, a coordinate was generated which satisfied where was a random number between 0 and 1, by applying the optim function from the stats library with the “Brent” method, starting with This ensured that the coordinate was selected proportionally to the area of the circle it specifies. The two extremes, and correspond to the lowest and highest point of the explant, respectively. Subsequently, the coordinates and were generated, within the respective circle at coordinate by generating a random angle between 0 and and a random number between 0 and 1, resulting in and where and The pairwise distances between simulated points were calculated in the same way as for the experimental data, and the respective empirical cumulative distributions were computed and compared with the experimental empirical distribution, as described above.
For the temporal analysis, the waiting times between centrosome births were calculated from the data and labeled according to which centrosome had just formed. Accounting for a possible change of centrosome birth rate as a function of the number of existing centrosomes, centrosome birth rates were estimated from each of the observed distributions of waiting times by maximum likelihood using the fitdistr function from the MASS library. The experimental data were then compared with a temporal null model whereby centrosomes form at a constant rate in time, irrespective of the existence of other centrosomes and of the volume of the explant. To this effect, random samples of Poisson-distributed waiting times were generated using the rexp function of the stats library, using the rate estimated from the waiting times between the appearance of the first and second centrosomes. The empirical cumulative distributions of these waiting times were compared with those from experimental data, as described above.
The trans-autophosphorylation of Plk4 was modeled following Lopes et al. (2015). Briefly, it is assumed that Plk4 protein is produced with constant source rate s in basal activity form B. The phosphorylation of this B form in the T-loop results in a form A1 with higher catalytic activity. The phosphorylation of the A1 form the degron converts it to a A2 that is targeted for proteasome increasing its degradation rate but that keeps the same catalytic activity. The phosphorylation at the T-loop is catalyzed by either low activity B form or the high activities A1 and A2 forms, while only the latter are assumed to phosphorylate the degron of other Pkl4 forms. Both phosphorylation reactions can be reversed by the constant activity of a phosphatase. To keep the stochastic model as simple as possible, we neglected the small first order phosphorylation term in Lopes et al. (2015), corresponding to a putative phosphorylation of Plk4 by other (yet unidentified) kinases. The dynamics of the three Plk4 forms is described by the following set of differential equations:
with
The rate of de novo centriole formation in the explant is assumed to be proportional Plk4 activity () and therefore the probability that an explant has no centrioles F decreases in time according to
The system of four differential equations was solved numerically using the function ode of the package deSolve in R software.
The stochastic solutions for the same set of reactions were obtained by the Gillespie algorithm as implemented in the function ssa of the package GillespieSSA in R. Each simulation corresponded to an explant where the Plk4 trans-autophosphorylation was simulated independently. The biosynthesis of the first centriole was simulated as a single reaction event that removes a single “precursor” F with a propensity The simulated explant is assumed to form one centriole upon this event.
The model, formulated in differential equations and extended to a stochastic version, was used to reproduce the temporal evolution of the number of explants containing at least one centriole under different concentrations of Plk4. Experimentally, four activity levels of Plk4 were obtained by mixing the cytoplasm of eggs overexpressing Plk4 and WT, in different proportions with expected activities relative to the overexpressing egg of 1.0, 0.5, 0.33, and 0.12 (Fig. 5 B and Fig. S4 G). The corresponding levels of Plk4 activity were defined in the model through the source parameter The value of K and the remaining parameters were adjusted by solving the ordinary differential equations for variable F and visually comparing (1-F) with the experimental time course of the frequencies of explants with at least one centriole (Fig. S4 G). The adjusted parameters were then used to simulate the stochastic kinetics. The parameter values of the solutions illustrated in Fig. S4 G were and The value of N was set to 2,000 molecules for the Gillespie simulations and to the unit in the ordinary differential equations.
3D-structured illumination microscopy
Cytoplasmic explants were imaged at 20°C with a Plan Apo 60× NA 1.42 oil objective on a GE HealthCare Deltavision OMX system, equipped with two PCO Edge 5.5 sCMOS cameras and 488 nm and 568 nm laser lines. Spherical aberrations were minimized by matching the refractive index of the immersion oil (1.518, in this case) to that of the cytosol, providing the most symmetrical PSF. 15 s, multi-stack time-lapses were acquired, with 0.125 µm z steps and 15 frames (three angles and five phases per angle) per z section, using AcquireSR (GE Healthcare). Images were reconstructed in Applied Precision's softWorx software (GE Healthcare) with channel-specific Wiener filter settings (0.003–0.005 for the 488 nm excitation and 0.005–0.008 for the 568 nm excitation) and channel-specific optical transfer functions. Finally, the reconstructed images were aligned on softWorx and processed using Fiji (National Institutes of Health; Schindelin et al., 2012). Maximum-intensity z projections of both fluorescence emission channels were produced from the time-lapse acquisitions, and single-plane insets were cropped in Fiji. Selected time frames and insets were further processed with Photoshop CS6 (Adobe).
Biochemistry
Immunoblotting
The protocol used for total protein extraction from unfertilized D. melanogaster eggs was adapted from Prudêncio and Guilgur (2015). Eggs from different genetic backgrounds—w1118 (negative control), mEGFP-Plk4 (endogenously labeled Plk4), V32-Gal4/mEGFP-Plk4 (overexpression of labeled Plk4), and V32-Gal4/mGFP–Plk4-ND (positive control, overexpression of nondegradable Plk4, mutated on residues Ser293 and Thr297; Cunha-Ferreira et al., 2009)—were collected at 25°C, chemically dechorionated, and lysed in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, pH 8, 0.1% Nonidet P-40, 1× protease inhibitors, 200 mM NaF, 1 mM DTT, 150 mM β-glycerol phosphate, and 1 mM Na3VO3). Samples were snap-frozen in liquid nitrogen and sonicated on ice for 30 s at ∼22% amplitude, using the Misonix XL 2020 sonicator. Soluble fractions were cleared by two consecutive rounds of centrifugation at 16,000 relative centrifugal force for 10 min at 4°C, transferring the supernatant into a new Protein LoBind Eppendorf in between, while avoiding the upper lipid layer. The samples were denatured at 99°C for 10 min in 6× SDS loading buffer, supplemented with protease inhibitors. In all cases, 100–120 µg of total protein was run on Invitrogen NuPAGE 4–12% precast Bis-Tris polyacrylamide gels for SDS-PAGE (Thermo Fisher Scientific) and then transferred onto nitrocellulose membranes for Western blotting. Membranes were blocked using 5% milk powder in TBS with 0.1% Tween 20 (TBST) for 60 min at RT. Primary antibody incubation was performed overnight at 4°C using the following antibodies: anti-GFP rabbit (diluted 1:2,000 in 5% milk in TBST; Abcam) and anti-actin (diluted 1:2,000 in 5% milk in TBST; Sigma-Aldrich). The membranes were washed three times in TBST for 15 min. For detection of GFP-Plk4, secondary antibody anti-rabbit HRP (Bethyl Laboratories) was diluted 1:40,000 (in 2.5% milk in TBST), while for detection of actin, anti-rabbit HRP was diluted 1:20,000 (in 2.5% milk in TBST). The membranes were incubated with secondary antibodies for 90 min at RT and then washed twice in TBST and once in TBS. For detection of GFP-Plk4, membranes were incubated in WesternBright Sirius HRP substrate (Advansta), while detection of actin was performed by incubating membranes with Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific), following the manufacturer’s instructions. Finally, membranes were exposed for 10–30 s (in the case of GFP-Plk4) and for 3–5 min (in the case of actin) on an Amersham Imager 680 (GE Healthcare), and images were acquired with its charge-coupled device camera.
mNeonGreen purification
The mNeonGreen coding sequence was cloned with an N-terminal streptavidin-binding peptide tag and a flexible linker into the pETMz expression vector (gift from the Protein Expression and Purification Facility at the European Molecular Biology Laboratory (EMBL), Heidelberg, Germany), between NcoI and BamHI restriction sites. The 6xHis–Z-tag–TEV–streptavidin-binding peptide–linker–mNeonGreen protein was expressed in BL21 (Rosetta) Competent Escherichia coli at 25°C for 5 h. The grown liquid culture was harvested and centrifuged at 4,000 rpm for 25 min at 4°C. The pellet was resuspended in ice-cold lysis buffer containing 50 mM K-Hepes, pH 7.5, 250 mM KCl, 1 mM MgCl2, 1 mM DTT, 7 mM imidazole, 1× DNaseI, and 1× protease inhibitors. The sample was applied to a prechilled French press, equilibrated with lysis buffer, and run twice at a constant pressure (∼12 kPa). The cell lysate was collected in a flask on ice and ultracentrifuged at 4°C for 25 min at 50,000 rpm using a Ti-70 rotor (Beckman). The protein purification was done through affinity chromatography on a Ni-column (HiTrap chelating HP column, 1 ml; GE HealthCare). The column was loaded with a filtered solution of 100 mM nickel chloride, washed extensively with Milli-Q water, and equilibrated with wash buffer (50 mM K-Hepes, pH 7.5, 250 mM KCl, 1 mM MgCl2, 1 mM DTT, and 7 mM imidazole). The clarified lysate was applied to the column (at 1.5 ml/min), followed by 200 ml wash buffer. The protein was eluted at 1.5 ml/min with elution buffer: 50 mM K-Hepes, pH 7.5, 250 mM KCl, 1 mM MgCl2, 1 mM DTT, and 400 mM imidazole. 1-ml sample fractions were collected and kept at 4°C. The most concentrated samples were pooled together, and their N-terminal 6xHis–Z-tag was cleaved with TEV protease overnight at 4°C by treating with 150 U TEV/mg protein. The following day, the cleaved protein was passed through a column for size-exclusion chromatography to remove contaminants, the cleaved tag, and the TEV protease (with Tiago Bandeiras at Instituto de Biologia Experimental e Tecnológica, Oeiras, Portugal). Additionally, the elution buffer was exchanged to a storage buffer: 50 mM K-Hepes, pH 7.8, 100 mM KCl, 2 mM MgCl2, 1 mM DTT, and 1 mM EGTA. The HiLoad Superdex 75 16/60 (GE HealthCare) gel filtration column was equilibrated with storage buffer for 1 h. The sample was spun at 15,000 rpm for 15 min at 4°C, and the clear fraction was applied to the gel filtration column coupled to an ÄKTA avant chromatography system device at 1 ml/min. The cleaved mNeonGreen protein was concentrated approximately five times using Amicon 10K centrifugal filters. Pure glycerol was added at 5% vol/vol, and small aliquots were snap-frozen in liquid nitrogen and stored at −80°C.
Plk4 titration in cytoplasmic extract
Plk4 dilution was accomplished by mixing cytoplasm from flies with different genetic composition. Unfertilized eggs collected from females overexpressing Plk4 in the germline (genotype: V32-Gal4/pUb-Spd2-GFP; Jupiter-mCherry/pUASp-GFP-Plk4) were homogenized in unfertilized eggs from females without the transgenic pUASp element (genotype: V32-Gal4/pUb-Spd2-GFP; Jupiter-mCherry), where all components are at WT levels, specifically diluting overall Plk4 concentration in the cytoplasm. Different final Plk4 concentrations were achieved by mixing Plk4 overexpression:WT eggs at the following ratios: 6:0 (1 relative Plk4 concentration, control); 3:3 (0.5 relative Plk4 concentration); 2:4 (0.33 relative Plk4 concentration), and 1:5 (0.16 relative Plk4 concentration). Small explants were produced from the cytoplasmic mixtures, and images were acquired for 40 min. All time-lapse acquisitions within this section were performed at 1-min time intervals with 0.45-µm optical sections, using a Plan Apo VC 60× 1.2 NA water objective. The images were recorded at 23°C with an Andor iXon3 888 EMCCD camera using a Yokogawa CSU-W1 Spinning Disk confocal scanner equipped with a piezoelectric stage (737.2SL; Physik Instrumente), installed on a Nikon Eclipse Ti-E microscope. Dual-color images (488 nm and 561 nm excitation laser lines) were collected at 100 ms exposure time with Andor IQ3 software.
FCS data acquisition and analysis
Standard rhodamine 6G calibration
All FCS measurements were performed at 20°C on a point-scanning confocal microscope (Zeiss LSM780 Confocor3) equipped with a UV-VIS-IR C Achromat 40× 1.2 NA water-immersion objective and a gallium arsenide detector array wavelength selected between 491 and 561 nm. Before each experiment, the system was aligned using a high concentration and calibrated using a low concentration of rhodamin 6G solution in water. The known diffusion coefficient of rhodamine 6G (410 µm2/s; Majer and Zick, 2015) allowed us to determine the lateral beam waist (wxy = 232 nm) and the structure factor (S = 5.77) of the focused laser (PSF). The resultant volume of illumination is calculated through the following formula:
The values for wxy and S were used as constants in the subsequent model-based fittings of the ACFs, and the volume was used to calculate the concentration (see below).
Calibration with purified mNeonGreen
mNeonGreen fluorescent tag was first measured in a cytoplasm-compatible buffer. Fluorescence intensity in time [I(t)] was recorded as six iterations of 10 s. Each 10 s trace was autocorrelated into an ACF, G(τ), using the Zeiss onboard autocorrelator, which calculates the self-similarity through the following formula:
Here, <> denotes the time average, and τ is called the time lag. The resulting G(τ) curves of the fluorophores in buffer were readily fitted using a regular 3D diffusion model,
where N reflects the number of moving particles in the confocal volume and GT(τ) is the correlation function associated to blinking/triplet kinetics:
where T is the fraction of molecules in the dark state and τt the lifetime of the dark state. GD(τ) is the correlation function associated to diffusion, which in this case is simple Brownian diffusion in 3D:
These fittings allowed us to measure the number of molecules in the confocal volume and therefore their brightness (<I(t)>/N) together with the characteristic diffusion times (τD).
The above model fit is based on the assumption that there are only two characteristic time scales generating the ACF. To get a model free estimate of the number of time scales involved, we used a MEMfit of the combined and normalized ACFs of each experiment. MEMfit analyzes the FCS autocorrelation data in terms of a quasi-continuous distribution of diffusing components, making it an ideal model to examine the ACF of a highly heterogeneous system without prior knowledge of the amount of diffusing species.
To be able to quantify the brightness of individual fluorescent tags in an embryo, the purified mNeonGreen was injected into pUb–RFP–β2-tubulin dechorionated embryos. An anomalous coefficient had to be included to fit the resultant ACF:
For simple Brownian diffusion, a = 1, and the fit function is identical to the one used to fit the fluorophores in buffer. However, for fluorophores injected into the cytosol of embryos, the fitting algorithm gave an anomalous coefficient of a = 0.8. An anomalous coefficient <1 indicates constrained diffusion and could be caused by the more crowded environment in the yolk. In addition, the large amount of (uncorrelated) autofluorescence generated by the yolk leads to an underestimation of the brightness, therefore requiring a background correction factor. The background values were determined per excitation power from embryos lacking the Plk4 reporter. If the background itself does not autocorrelated, it has no influence on the obtained time scales in the data. Nevertheless, the background will impact the absolute number, N, and consequently also the calculated brightness. Therefore, all the measurements were background corrected via the formula
where BG is the measured background from embryos lacking the reporter fluorophore. Consequently, the corrected brightness was calculated as
Finally, any 1 ms–binned intensity trace that contained changes in average intensity (most likely arising from yolk spheres moving through the confocal spot during the measurement) was discarded from further analysis.
mNeonGreen-Plk4 measurements in embryos
For the measurements of mNeonGreen-Plk4, embryo staging was done based on the pUb–RFP–β2-tubulin reporter. We chose embryos at blastoderm stage, in division cycles 10 or 11. Before each FCS acquisition series, a large field-of-view image of the embryo was acquired. Six different 10-s-long intensity traces were measured at the internuclear cytoplasmic space of the syncytium. The 10-s measurement was long enough to obtain sufficient passage events and short enough to avoid each trace to be contaminated by events that do not arise from mNeonGreen-Plk4 diffusing in the cytosol.
From these measurements, the MEMfit method on the normalized ACF indicates three time scales for the tagged Plk4 molecules: a first time scale of 5–50 µs, corresponding to the triplet state dynamics that were similarly found in both the buffer as well as from fluorophores injected in the embryo; a second time scale of ∼0.8 ms, most likely coming from the diffusion of a Plk4 monomer (see similarity to mNeonGreen monomer in cytosol); and a third time scale of diffusion that is much slower, 9 ms. To fit the ACFs, the diffusional part of the fit function was associated with two components:
The fraction f corresponds to the fast-diffusing Plk4. The diffusion coefficient of each of the components can be calculated from the diffusion time scales τD via
In vitro experiments
D. melanogaster cell culture
Drosophila (DMEL; ATCC CRL-1963) cells were cultured in Express5 SFM (GIBCO) supplemented with 1× L-glutamine–penicillin–streptomycin. dsRNA synthesis was performed as previously described (Bettencourt-Dias et al., 2004). dsDNA sequences were first amplified by PCR from fly genomic DNA (D. melanogaster gene coding sequences) or plasmid DNA (mCherry construct; #632522; Clontech) using the primers listed in Table S5. The dsDNA was purified in nuclease-free water using the DNA Clean & Concentrator Kit (#D4014; Zymo Research) and transcribed using the T7 RiboMAX Express RNAi system (P1700; Promega). The dsRNA was quantified by running in 2% agarose gel with Invitrogen Low DNA Mass Ladder (#10068013; Thermo Fisher Scientific) and stored in single-usage aliquots at −20°C.
DMEL cells were plated and treated for 12 d with 40 µg dsRNA against Plk4 or mCherry (control), replacing the dsRNA every 4 d. Cells were fixed at day 10 to confirm centriole depletion, and treatment with dsRNA against PCM was initiated. Cells were then treated for 6 d with different amounts and combinations of dsRNA: 80 µg mCherry alone; 20 µg of individual PCM components (Cnn, Asl, D-Plp, Spd2, or γ-tubulin 23C); or combinations of two (Cnn + Spd2, Cnn + D-Plp, or Spd2 + D-Plp), three (Cnn + Spd2 + D-Plp), or four of these molecules (Cnn + Asl + D-Plp + Spd2, referred to as All PCM).
Immunostaning and imaging of D. melanogaster cultured cells
DMEL cells were plated onto clean glass coverslips and allowed to adhere for 1 h and 30 min. The media were removed, and cells were fixed at −20°C for 10 min in chilled methanol. Cells were permeabilized and washed in D-PBSTB for 1 h. Cells were incubated overnight at 4°C with primary antibodies—rat anti-Sas4 (dilution 1:500; kindly provided by David Glover, University of Cambridge, Cambridge, UK) and rabbit anti-CP110 (dilution 1:10,000; Metabion)—diluted in D-PBSTB. Cells were washed in D-PBSTB and incubated for 90 min at room temperature with secondary antibodies—donkey anti-rat Alexa 555 (dilution 1:1,000; Molecular Probes) and donkey anti-rabbit Alexa 647 (dilution 1:1,000; Jackson Immunoresearch Laboratories)—and DAPI (dilution 1:200) in D-PBSTB. Cells were washed and mounted with Dako Faramount Aqueous Mounting Medium (S3025; Agilent).
Cell imaging was conducted at 18°C on a Nikon Eclipse Ti-E microscope equipped with a Yokogawa CSU-X1 Spinning Disk confocal scanner. Images were recorded with a EMCCD Photometrics 512 camera. Optical sections of 0.3 µm thickness were acquired with a Plan Apo 100× 1.49 NA oil immersion objective using a piezoelectric stage (737.2SL; Physik Instrumente), controlled by Metamorph 7.5 software. The centriole number was scored in 300 cells per treatment, per independent experiment. Data are presented as mean ± SEM of two independent experiments. We tested all counts with a χ2 test against the null hypothesis that the outcome is random. Then, each 16-d test condition was compared with the 16-d mCherry control condition with a two-proportions z test and H0 that the proportions of cells with centrioles are equal versus HA that the proportion in the test is smaller. The significance level for multiple testing was Bonferroni-corrected. Significance level was P = 0.01. All images were processed with ImageJ (National Institutes of Health) and Adobe Photoshop CS6 (Adobe Systems), and the final figures were assembled in Adobe Illustrator CS6 (Adobe Systems).
Online supplemental material
Fig. S1 (in support of Figs. 1 and 2) reports measurement of concentration fold-change of Plk4 in overexpression lines. Fig. S2 (in support of Fig. 2) shows visualization of centrosome biogenesis in a Drosophila egg extract by 3D-Structured Illumination Microscopy, time-lapses of explants with different reporters, and comparison of centrosome movement versus distance between registered biogenesis. Fig. S3 (in support of Fig. 3) shows spatial analysis of de novo centriole biogenesis in fly explants. Fig. S4 (in support of Fig. 4) shows temporal analysis of de novo centriole biogenesis. Fig. S5 (in support of Fig. 5) shows FCS in Drosophila embryos. Table S1 lists D. melanogaster strains generated and/or used in this study. Table S2 lists oligonucleotides used for CRISPR-mediated knock-in of mNeonGreen and mEGFP into the endogenous D. melanogaster Plk4 locus. Table S3 shows sequencing and screening primers used to check the mNeonGreen-Plk4 and mGFP-Plk4 lines generated in this study. Table S4 shows FCS parameters determined from the model-based fittings. Table S5 lists primers used for dsRNA synthesis. Table S6 shows sequences of the oligonucleotides used to generate shRNA targeting different D. melanogaster gene products. Table S7 shows a lethality assay to determine the viability of the shRNA fly lines. Video 1 (in support of Fig. 1) shows centriole biogenesis in a D. melanogaster egg explant. Video 2, Video 3, Video 4, and Video 5 (in support of Fig. 2) show that centrioles assemble de novo, recruit different centrosomal molecules, and duplicate. Video 6 (in support of Fig. 5) shows mNeonGreen-Plk4 localization in a syncytial Drosophila embryo.
Supplementary Material
lists D. melanogaster strains generated and/or used in this study.
lists oligonucleotides used for CRISPR-mediated knock-in of mNeonGreen and mEGFP into the endogenous D. melanogaster Plk4 locus.
shows sequencing and screening primers used to check the mNeonGreen-Plk4 and mGFP-Plk4 lines generated in this study.
shows FCS parameters determined from the model-based fittings.
lists primers used for dsRNA synthesis.
shows sequences of the oligonucleotides used to generate shRNA targeting different D. melanogaster gene products.
shows a lethality assay to determine the viability of the shRNA fly lines.
Acknowledgments
We would like to thank the Central Imaging and Flow Cytometry Facility at the National Centre for Biological Sciences in Bangalore, where all FCS experiments were performed. We thank Tiago Bandeiras at Instituto de Biologia Experimental e Tecnológica, Oeiras, for the gel filtration chromatography conducted in his facility with Micael Freitas. We thank Tomer Avidor-Reiss (University of Toledo, Toledo, OH), Daniel St. Johnston (The Gurdon Institute, Cambridge, UK), Yoshihiro Inoue, and Jordan Raff (University of Oxford, Oxford, UK); for sharing transgenic fly lines. We thank David Glover (University of Cambridge, Cambridge, UK), Jordan Raff, and Tim Megraw (Florida State University, Tallahassee, FL) for providing antibodies. We acknowledge the technical support of Instituto Gulbenkian de Ciência (IGC)'s Advanced Imaging Facility, in particular Gabriel Martins and Nuno Pimpão Martins. We thank members of the Cell Cycle Regulation laboratory at IGC for giving feedback on earlier versions of the paper. Finally, we wish to thank the anonymous reviewers and the editor for helpful comments that greatly improved the paper, as well as Erin Tranfield (IGC) for proofreading the paper.
IGC’s Advanced Imaging Facility is supported by national Portuguese funding (PPBI-POCI-01-0145-FEDER-022122), co-financed by the Lisboa Regional Operational Program (Lisboa 2020), under the Portugal 2020 Agreement, through the European Regional Development Fund (FEDER) and Fundação para a Ciência e a Tecnologia (FCT, Portugal). We thank IGC’s Fly Facility and Fly Transgenesis Facility supported by Congento (LISBOA-01-0145-FEDER-022170), co-financed by Fundação para a Ciência e a Tecnologia and Lisboa 2020, under the Portugal 2020 Agreement (European Regional Development Fund). Transgenic fly stocks were obtained from the Bloomington Drosophila Stock Center (NIH742 P40OD018537). We acknowledge financial support from a Boehringer Ingelheim Fonds PhD Fellowship awarded to C. Nabais, a Human Frontiers Science Program Young Investigator Grant (RGY0083/2016) awarded to I.A. Telley supporting J. de–Carvalho, the Fundação para a Ciência e a Tecnologia supporting I.A. Telley (Investigador FCT IF/00082/2013), the European Commission FP7 grant (PEOPLE-2013-CIG Nº 818743) awarded to I.A. Telley, European Research Council grants (ERC-2010-StG-261344-CentriolStructure&Number and ERC-2015-CoG-683258-Birth&Death) awarded to M. Bettencourt-Dias, and the Calouste Gulbenkian Foundation. T. van Zanten acknowledges a European Molecular Biology Organization (EMBO) fellowship (ALTF 1519-2013) and a National Centre for Biological Sciences Campus fellowship. S. Mayor acknowledges a JC Bose Fellowship from Department Of Science & Technology (Government of India), support from the National Centre for Biological Sciences–Max Planck Lipid Centre, a grant from the Human Frontiers Science Program (RGP0027/2012), and support from The Wellcome Trust DBT India Alliance Margdarshi Fellowship (IA/M/15/1/502018).
The authors declare no competing financial interests.
Author contributions: Conceptualization: C. Nabais, I.A. Telley, and M. Bettencourt-Dias. Methodology: C. Nabais, J. de-Carvalho, and I.A. Telley (egg explant assay); and C. Nabais and T. van Zanten (FCS measurements). Software: D. Pessoa and J. Carneiro (design and implementation of model simulations). Validation: C. Nabais, J. de-Carvalho, D. Pessoa, T. van Zanten, S. Mayor, J. Carneiro, I.A. Telley, and M. Bettencourt-Dias. Investigation: C. Nabais and J. de-Carvalho (performing data collection egg explants); C. Nabais and T. van Zanten (performing data collection in FCS measurements); and D. Pessoa and J. Carneiro (collecting in silico data). Analysis: C. Nabais and I.A. Telley (experimental data from egg explants, eggs, and cell culture); T. van Zanten and S. Mayor (FCS data); and D. Pessoa and J. Carneiro (theoretical model). Resources: C. Nabais (CRISPR fly line, vectors and plasmid design, and recombinant protein purification); P. Duarte (genotyping); and I.A. Telley (design of micromanipulation microscope). Visualization of data: C. Nabais, D. Pessoa, T. van Zanten, and I.A. Telley. Writing – original draft: C. Nabais. Writing – review and editing: C. Nabais, S. Mayor, J. Carneiro, I.A. Telley, and M. Bettencourt-Dias. Supervision and coordination: I.A. Telley and M. Bettencourt-Dias.
References
- Aldrich, H.C. 1967. The ultrastructure of meiosis in three species of Physarum. Mycologia. 59:127–148. 10.1080/00275514.1967.12018400 [DOI] [PubMed] [Google Scholar]
- Aydogan, M.G., Steinacker T.L., Mofatteh M., Gartenmann L., Wainman A., Saurya S., Conduit P.T., Zhou F.Y., Boemo M.A., and Raff J.W.. 2019. A free-running oscillator times and executes centriole biogenesis. BioRxiv. doi: (Preprint posted January 3, 2019) 10.1101/510875v1 [DOI]
- Banterle, N., and Gönczy P.. 2017. Centriole Biogenesis: From Identifying the Characters to Understanding the Plot. Annu. Rev. Cell Dev. Biol. 33:23–49. 10.1146/annurev-cellbio-100616-060454 [DOI] [PubMed] [Google Scholar]
- Bauer, M., Cubizolles F., Schmidt A., and Nigg E.A.. 2016. Quantitative analysis of human centrosome architecture by targeted proteomics and fluorescence imaging. EMBO J. 35:2152–2166. 10.15252/embj.201694462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., Sinka R., Frenz L., and Glover D.M.. 2004. RNAi in Drosophila Cell Cultures. In Gene Silencing by RNA Interference: Technology and Application. Sohail M., editor. CRC Press, Inc.. pp. 147–166. [Google Scholar]
- Bettencourt-Dias, M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., and Glover D.M.. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207. 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias, M., Hildebrandt F., Pellman D., Woods G., and Godinho S.A.. 2011. Centrosomes and cilia in human disease. Trends Genet. 27:307–315. 10.1016/j.tig.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon, S., Gopalakrishnan J., Omori Y., Polyanovsky A., Church A., Nicastro D., Malicki J., and Avidor-Reiss T.. 2008. Drosophila asterless and vertebrate Cep152 Are orthologs essential for centriole duplication. Genetics. 180:2081–2094. 10.1534/genetics.108.095141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boese, C.J., Nye J., Buster D.W., McLamarrah T.A., Byrnes A.E., Slep K.C., Rusan N.M., and Rogers G.C.. 2018. Asterless is a Polo-like kinase 4 substrate that both activates and inhibits kinase activity depending on its phosphorylation state. Mol. Biol. Cell. 29:2874–2886. 10.1091/mbc.E18-07-0445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslow, D.K., and Holland A.J.. 2019. Mechanism and Regulation of Centriole and Cilium Biogenesis. Annu. Rev. Biochem. 88:691–724. 10.1146/annurev-biochem-013118-111153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.W., Hsu W.B., Tsai J.J., Tang C.J., and Tang T.K.. 2016. CEP295 interacts with microtubules and is required for centriole elongation. J. Cell Sci. 129:2501–2513. 10.1242/jcs.186338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvin, G., Oikonomou C., Siggia E.D., and Cross F.R.. 2010. Origin of irreversibility of cell cycle start in budding yeast. PLoS Biol. 8:e1000284. 10.1371/journal.pbio.1000284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citron, Y.R., Fagerstrom C.J., Keszthelyi B., Huang B., Rusan N.M., Kelly M.J.S., and Agard D.A.. 2018. The centrosomin CM2 domain is a multi-functional binding domain with distinct cell cycle roles. PLoS One. 13:e0190530. 10.1371/journal.pone.0190530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu, O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., and Hoffmann I.. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739. 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B., Dix C.I., Novak Z.A., Dobbie I.M., Schermelleh L., et al. 2014. A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife. 3:e03399. 10.7554/eLife.03399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit, P.T., Wainman A., Novak Z.A., Weil T.T., and Raff J.W.. 2015. Re-examining the role of Drosophila Sas-4 in centrosome assembly using two-colour-3D-SIM FRAP. eLife. 4:e08483. 10.7554/eLife.08483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois, A., Schuh M., Ellenberg J., and Hiiragi T.. 2012. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 198:357–370. 10.1083/jcb.201202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira, I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49. 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira, I., Bento I., Pimenta-Marques A., Jana S.C., Lince-Faria M., Duarte P., Borrego-Pinto J., Gilberto S., Amado T., Brito D., et al. 2013. Regulation of autophosphorylation controls PLK4 self-destruction and centriole number. Curr. Biol. 23:2245–2254. 10.1016/j.cub.2013.09.037 [DOI] [PubMed] [Google Scholar]
- Dammermann, A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., and Oegema K.. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. 10.1016/j.devcel.2004.10.015 [DOI] [PubMed] [Google Scholar]
- de-Carvalho, J., Deshpande O., Nabais C., and Telley I.A.. 2018. A cell-free system of Drosophila egg explants supporting native mitotic cycles. 144. 1st ed. Elsevier Inc. 233–257 pp. doi: . 10.1016/bs.mcb.2018.03.011 [DOI] [PubMed] [Google Scholar]
- de-Carvalho, J., Tlili S., Hufnagel L., Saunders T., and Telley I.. 2020. Aster repulsion drives local ordering in an active system. bioRxiv. doi: (Preprint posted December 8, 2020) 10.1101/2020.06.04.133579 [DOI]
- Deneke, V.E., Puliafito A., Krueger D., Narla A.V., De Simone A., Primo L., Vergassola M., De Renzis S., and Di Talia S.. 2019. Self-Organized Nuclear Positioning Synchronizes the Cell Cycle in Drosophila Embryos. Cell. 177:925–941.e17. 10.1016/j.cell.2019.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirksen, E.R. 1961. The presence of centrioles in artificially activated sea urchin eggs. J. Biophys. Biochem. Cytol. 11:244–247. 10.1083/jcb.11.1.244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev, N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., and Glover D.M.. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718. 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Feng, Z., Caballe A., Wainman A., Johnson S., Haensele A.F.M., Cottee M.A., Conduit P.T., Lea S.M., and Raff J.W.. 2017. Structural Basis for Mitotic Centrosome Assembly in Flies. Cell. 169:1078–1089.e13. 10.1016/j.cell.2017.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree, P.M., McDonald K., Fasulo B., and Sullivan W.. 2006. The origin of centrosomes in parthenogenetic hymenopteran insects. Curr. Biol. 16:801–807. 10.1016/j.cub.2006.03.066 [DOI] [PubMed] [Google Scholar]
- Fu, J., Lipinszki Z., Rangone H., Min M., Mykura C., Chao-Chu J., Schneider S., Dzhindzhev N.S., Gottardo M., Riparbelli M.G., et al. 2016. Conserved molecular interactions in centriole-to-centrosome conversion. Nat. Cell Biol. 18:87–99. 10.1038/ncb3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho, S.A., and Pellman D.. 2014. Causes and consequences of centrosome abnormalities in cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369:20130467. 10.1098/rstb.2013.0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho, S.A., Picone R., Burute M., Dagher R., Su Y., Leung C.T., Polyak K., Brugge J.S., Théry M., and Pellman D.. 2014. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 510:167–171. 10.1038/nature13277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria, M.A., Rath U., Buster D.W., Chanda S.K., Caldwell J.S., Rines D.R., and Sharp D.J.. 2007. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17:1960–1966. 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Grimes, G.W. 1973a. Origin and development of kinetosomes in Oxytricha fallax. J. Cell Sci. 13:43–53. [DOI] [PubMed] [Google Scholar]
- Grimes, G.W. 1973b. Morphological discontinuity of kinetosomes during the life cycle of Oxytricha fallax. J. Cell Biol. 57:229–232. 10.1083/jcb.57.1.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guderian, G., Westendorf J., Uldschmid A., and Nigg E.A.. 2010. Plk4 trans-autophosphorylation regulates centriole number by controlling betaTrCP-mediated degradation. J. Cell Sci. 123:2163–2169. 10.1242/jcs.068502 [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet, C., Antony C., Aghion J., Santa-Maria A., Lajoie-Mazenc I., Wright M., and Maro B.. 1993. γ-Tubulin is present in acentriolar MTOCs during early mouse development. J. Cell Sci. 105:157–166. [DOI] [PubMed] [Google Scholar]
- Habedanck, R., Stierhof Y.D., Wilkinson C.J., and Nigg E.A.. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146. 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Harvey, E.B. 1936. Parthenogenetic Merogony or Cleavage without Nuclei in Arbacia punctulata. Mar. Biol. Lab. 71:101–121. 10.2307/1537411 [DOI] [Google Scholar]
- Holland, A.J., Fachinetti D., Zhu Q., Bauer M., Verma I.M., Nigg E.A., and Cleveland D.W.. 2012. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 26:2684–2689. 10.1101/gad.207027.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner, V.L., Czank A., Jang J.K., Singh N., Williams B.C., Puro J., Kubli E., Hanes S.D., McKim K.S., Wolfner M.F., and Goldberg M.L.. 2006. The Drosophila calcipressin sarah is required for several aspects of egg activation. Curr. Biol. 16:1441–1446. 10.1016/j.cub.2006.06.024 [DOI] [PubMed] [Google Scholar]
- Idei, M., Osada K., Sato S., Nakayama T., Nagumo T., and Mann D.G.. 2013. Sperm ultrastructure in the diatoms Melosira and Thalassiosira and the significance of the 9 + 0 configuration. Protoplasma. 250:833–850. 10.1007/s00709-012-0465-8 [DOI] [PubMed] [Google Scholar]
- Ito, D., Zitouni S., Jana S.C., Duarte P., Surkont J., Carvalho-Santos Z., Pereira-Leal J.B., Ferreira M.G., and Bettencourt-Dias M.. 2019. Pericentrin-mediated SAS-6 recruitment promotes centriole assembly. eLife. 8:e41418. 10.7554/eLife.41418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo, D., Wang W.J., Uryu K., and Tsou M.F.B.. 2014. Stabilization of cartwheel-less centrioles for duplication requires CEP295-mediated centriole-to-centrosome conversion. Cell Rep. 8:957–965. 10.1016/j.celrep.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaqaman, K., Loerke D., Mettlen M., Kuwata H., Grinstein S., Schmid S.L., and Danuser G.. 2008. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 5:695–702. 10.1038/nmeth.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov, V., and De Nicolo A.. 2019. The Centrosome and the Primary Cilium: The Yin and Yang of a Hybrid Organelle. Cells. 8:701. 10.3390/cells8070701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., Rieder C.L., Sluder G., Cassels G., Sibon O., and Wang C.L.. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158:1171–1181. 10.1083/jcb.200205102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa, D., Matsuo T., Kaizuka K., Miyauchi C., Hayashi D., and Inoue Y.H.. 2014. Orbit/CLASP is required for myosin accumulation at the cleavage furrow in Drosophila male meiosis. PLoS One. 9:e93669. 10.1371/journal.pone.0093669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba, J.E., Buster D.W., Nguyen A.L., Swatkoski S., Gucek M., Rusan N.M., and Rogers G.C.. 2013. Polo-like kinase 4 autodestructs by generating its Slimb-binding phosphodegron. Curr. Biol. 23:2255–2261. 10.1016/j.cub.2013.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba, J.E., Buster D.W., McLamarrah T.A., Rusan N.M., and Rogers G.C.. 2015a. Autoinhibition and relief mechanism for Polo-like kinase 4. Proc. Natl. Acad. Sci. USA. 112:E657–E666. 10.1073/pnas.1417967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebba, J.E., Galletta B.J., Nye J., Plevock K.M., Buster D.W., Hollingsworth N.A., Slep K.C., Rusan N.M., and Rogers G.C.. 2015b. Two Polo-like kinase 4 binding domains in Asterless perform distinct roles in regulating kinase stability. J. Cell Biol. 208:401–414. 10.1083/jcb.201410105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn, J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., and Nigg E.A.. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202. 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- La Terra, S., English C.N., Hergert P., McEwen B.F., Sluder G., and Khodjakov A.. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722. 10.1083/jcb.200411126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus, B.G., Uetake Y., Clutario K.M., Daggubati V., Snyder M., Sluder G., and Holland A.J.. 2015. p53 protects against genome instability following centriole duplication failure. J. Cell Biol. 210:63–77. 10.1083/jcb.201502089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit, D.A., Jordan H.A., Poulton J.S., Fagerstrom C.J., Galletta B.J., Peifer M., and Rusan N.M.. 2015. Interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome function. J. Cell Biol. 210:79–97. 10.1083/jcb.201503117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine, M.S., Bakker B., Boeckx B., Moyett J., Lu J., Vitre B., Spierings D.C., Lansdorp P.M., Cleveland D.W., Lambrechts D., et al. 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev. Cell. 40:313–322.e5. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek, J., Hergert P., Magidson V., and Khodjakov A.. 2008. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10:322–328. 10.1038/ncb1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes, C.A.M., Jana S.C., Cunha-Ferreira I., Zitouni S., Bento I., Duarte P., Gilberto S., Freixo F., Guerrero A., Francia M., et al. 2015. PLK4 trans-Autoactivation Controls Centriole Biogenesis in Space. Dev. Cell. 35:222–235. 10.1016/j.devcel.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Lopes, C.A.M., Mesquita M., Cunha A.I., Cardoso J., Carapeta S., Laranjeira C., Pinto A.E., Pereira-Leal J.B., Dias-Pereira A., Bettencourt-Dias M., and Chaves P.. 2018. Centrosome amplification arises before neoplasia and increases upon p53 loss in tumorigenesis. J. Cell Biol. 217:2353–2363. 10.1083/jcb.201711191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, N., Rees J.S., Roote J., Ryder E., Armean I.M., Johnson G., Drummond E., Spriggs H., Drummond J., Magbanua J.P., et al. UK Drosophila Protein Trap Screening Consortium . 2014. Analysis of the expression patterns, subcellular localisations and interaction partners of Drosophila proteins using a pigP protein trap library. Development. 141:3994–4005. 10.1242/dev.111054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahen, R., Jeyasekharan A.D., Barry N.P., and Venkitaraman A.R.. 2011. Continuous polo-like kinase 1 activity regulates diffusion to maintain centrosome self-organization during mitosis. Proc. Natl. Acad. Sci. USA. 108:9310–9315. 10.1073/pnas.1101112108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majer, G., and Zick K.. 2015. Accurate and absolute diffusion measurements of Rhodamine 6G in low-concentration aqueous solutions by the PGSE-WATERGATE sequence. J. Chem. Phys. 142:164202. 10.1063/1.4919054 [DOI] [PubMed] [Google Scholar]
- Marshall, W.F., Vucica Y., and Rosenbaum J.L.. 2001. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr. Biol. 11:308–317. 10.1016/S0960-9822(01)00094-X [DOI] [PubMed] [Google Scholar]
- Marteil, G., Guerrero A., Vieira A.F., de Almeida B.P., Machado P., Mendonça S., Mesquita M., Villarreal B., Fonseca I., Francia M.E., et al. 2018. Over-elongation of centrioles in cancer promotes centriole amplification and chromosome missegregation. Nat. Commun. 9:1258. 10.1038/s41467-018-03641-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLamarrah, T.A., Buster D.W., Galletta B.J., Boese C.J., Ryniawec J.M., Hollingsworth N.A., Byrnes A.E., Brownlee C.W., Slep K.C., Rusan N.M., and Rogers G.C.. 2018. An ordered pattern of Ana2 phosphorylation by Plk4 is required for centriole assembly. J. Cell Biol. 217:1217–1231. 10.1083/jcb.201605106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercey, O., Al Jord A., Rostaing P., Mahuzier A., Fortoul A., Boudjema A.R., Faucourt M., Spassky N., and Meunier A.. 2019a. Dynamics of centriole amplification in centrosome-depleted brain multiciliated progenitors. Sci. Rep. 9:13060. 10.1038/s41598-019-49416-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercey, O., Levine M.S., LoMastro G.M., Rostaing P., Brotslaw E., Gomez V., Kumar A., Spassky N., Mitchell B.J., Meunier A., and Holland A.J.. 2019b. Massive centriole production can occur in the absence of deuterosomes in multiciliated cells. Nat. Cell Biol. 21:1544–1552. 10.1038/s41556-019-0427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir, L., Wright M., and Moisand A.. 1984. Variations in the number of centrioles, the number of microtubule organizing centers 1 and the percentage of mitotic abnormalities in Physarum polycephalum amoebae. Protoplasma. 120:20–35. 10.1007/BF01287614 [DOI] [Google Scholar]
- Mizukami, I., and Gall J.. 1966. Centriole replication. II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J. Cell Biol. 29:97–111. 10.1083/jcb.29.1.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montenegro Gouveia, S., Zitouni S., Kong D., Duarte P., Ferreira Gomes B., Sousa A.L., Tranfield E.M., Hyman A., Loncarek J., and Bettencourt-Dias M.. 2018. PLK4 is a microtubule-associated protein that self-assembles promoting de novo MTOC formation. J. Cell Sci. 132:jcs219501. 10.1242/jcs.219501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer, T.C., Clutario K.M., Lambrus B.G., Daggubati V., and Holland A.J.. 2015. Binding of STIL to Plk4 activates kinase activity to promote centriole assembly. J. Cell Biol. 209:863–878. 10.1083/jcb.201502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabais, C., Pereira S.G., and Bettencourt-Dias M.. 2017. Noncanonical Biogenesis of Centrioles and Basal Bodies. Cold Spring Harb. Symp. Quant. Biol. 82:123–135. 10.1101/sqb.2017.82.034694 [DOI] [PubMed] [Google Scholar]
- Park, J.E., Zhang L., Bang J.K., Andresson T., DiMaio F., and Lee K.S.. 2019. Phase separation of Polo-like kinase 4 by autoactivation and clustering drives centriole biogenesis. Nat. Commun. 10:4959. 10.1038/s41467-019-12619-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel, N., Stevens N.R., Basto R., and Raff J.W.. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17:834–843. 10.1016/j.cub.2007.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta-Marques, A., Bento I., Lopes C.A.M., Duarte P., Jana S.C., and Bettencourt-Dias M.. 2016. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science. 353:aaf4866. 10.1126/science.aaf4866 [DOI] [PubMed] [Google Scholar]
- Port, F., Chen H.M., Lee T., and Bullock S.L.. 2014. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA. 111:E2967–E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudêncio, P., and Guilgur L.G.. 2015. Protein Extraction from Drosophila Embryos and Ovaries. Bio Protoc. 5:e1459. 10.21769/BioProtoc.1459 [DOI] [Google Scholar]
- Rale, M.J., Kadzik R.S., and Petry S.. 2018. Phase Transitioning the Centrosome into a Microtubule Nucleator. Biochemistry. 57:30–37. 10.1021/acs.biochem.7b01064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzaglia, K.S., and Garbary D.J.. 2001. Motile Gametes of Land Plants: Diversity, Development, and Evolution. Crit. Rev. Plant Sci. 20:107–213. 10.1080/20013591099209 [DOI] [Google Scholar]
- Riparbelli, M.G., and Callaini G.. 2003. Drosophila parthenogenesis: a model for de novo centrosome assembly. Dev. Biol. 260:298–313. 10.1016/S0012-1606(03)00243-4 [DOI] [PubMed] [Google Scholar]
- Riparbelli, M.G., and Callaini G.. 2005. The meiotic spindle of the Drosophila oocyte: the role of centrosomin and the central aster. J. Cell Sci. 118:2827–2836. 10.1242/jcs.02413 [DOI] [PubMed] [Google Scholar]
- Riparbelli, M.G., Stouthamer R., Dallai R., and Callaini G.. 1998. Microtubule organization during the early development of the parthenogenetic egg of the hymenopteran Muscidifurax uniraptor. Dev. Biol. 195:89–99. 10.1006/dbio.1997.8841 [DOI] [PubMed] [Google Scholar]
- Riparbelli, M.G., Tagu D., Bonhomme J., and Callaini G.. 2005. Aster self-organization at meiosis: a conserved mechanism in insect parthenogenesis? Dev. Biol. 278:220–230. 10.1016/j.ydbio.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins, A., Riparbelli M., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science. 316:1046–1050. 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- Rodrigues-Martins, A., Riparbelli M., Callaini G., Glover D.M., and Bettencourt-Dias M.. 2008. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 7:11–16. 10.4161/cc.7.1.5226 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an Open-Source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao, D., Yamamoto S., and Kitagawa D.. 2019. A theory of centriole duplication based on self-organized spatial pattern formation. J. Cell Biol. 218:3537–3547. 10.1083/jcb.201904156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavosanis, G., Llamazares S., Goulielmos G., and Gonzalez C.. 1997. Essential role for γ-tubulin in the acentriolar female meiotic spindle of Drosophila. EMBO J. 16:1809–1819. 10.1093/emboj/16.8.1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley, I.A., Gáspár I., Ephrussi A., and Surrey T.. 2012. Aster migration determines the length scale of nuclear separation in the Drosophila syncytial embryo. J. Cell Biol. 197:887–895. 10.1083/jcb.201204019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telley, I.A., Gáspár I., Ephrussi A., and Surrey T.. 2013. A single Drosophila embryo extract for the study of mitosis ex vivo. Nat. Protoc. 8:310–324. 10.1038/nprot.2013.003 [DOI] [PubMed] [Google Scholar]
- Tram, U., and Sullivan W.. 2000. Reciprocal inheritance of centrosomes in the parthenogenetic hymenopteran Nasonia vitripennis. Curr. Biol. 10:1413–1419. 10.1016/S0960-9822(00)00795-8 [DOI] [PubMed] [Google Scholar]
- Tsuchiya, Y., Yoshiba S., Gupta A., Watanabe K., and Kitagawa D.. 2016. Cep295 is a conserved scaffold protein required for generation of a bona fide mother centriole. Nat. Commun. 7:12567. 10.1038/ncomms12567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson, J.J., and Novak B.. 2001. Regulation of the eukaryotic cell cycle: molecular antagonism, hysteresis, and irreversible transitions. J. Theor. Biol. 210:249–263. 10.1006/jtbi.2001.2293 [DOI] [PubMed] [Google Scholar]
- Uetake, Y., Loncarek J., Nordberg J.J., English C.N., La Terra S., Khodjakov A., and Sluder G.. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176:173–182. 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy, L., and Orr-Weaver T.L.. 2007. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev. Cell. 12:157–166. 10.1016/j.devcel.2006.10.017 [DOI] [PubMed] [Google Scholar]
- Varmark, H., Llamazares S., Rebollo E., Lange B., Reina J., Schwarz H., and Gonzalez C.. 2007. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 17:1735–1745. 10.1016/j.cub.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Wang, W.J., Soni R.K., Uryu K., and Tsou M.F.B.. 2011. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 193:727–739. 10.1083/jcb.201101109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.L., Anzola J.V., Davis R.L., Yoon M., Motamedi A., Kroll A., Seo C.P., Hsia J.E., Kim S.K., Mitchell J.W., et al. 2015. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 348:1155–1160. 10.1126/science.aaa5111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S., and Kitagawa D.. 2019. Self-organization of Plk4 regulates symmetry breaking in centriole duplication. Nat. Commun. 10:1810. 10.1038/s41467-019-09847-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsu, N. 1905. The formation of centrosomes in enucleated egg-fragments. J. Exp. Zool. 2:287–312. 10.1002/jez.1400020206 [DOI] [Google Scholar]
- Zitouni, S., Francia M.E., Leal F., Montenegro Gouveia S., Nabais C., Duarte P., Gilberto S., Brito D., Moyer T., Kandels-Lewis S., et al. 2016. CDK1 prevents unscheduled PLK4-STIL complex assembly in centriole biogenesis. Curr. Biol. 26:1127–1137. 10.1016/j.cub.2016.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists D. melanogaster strains generated and/or used in this study.
lists oligonucleotides used for CRISPR-mediated knock-in of mNeonGreen and mEGFP into the endogenous D. melanogaster Plk4 locus.
shows sequencing and screening primers used to check the mNeonGreen-Plk4 and mGFP-Plk4 lines generated in this study.
shows FCS parameters determined from the model-based fittings.
lists primers used for dsRNA synthesis.
shows sequences of the oligonucleotides used to generate shRNA targeting different D. melanogaster gene products.
shows a lethality assay to determine the viability of the shRNA fly lines.