Stuani et al. demonstrate that IDH mutant AML cells display an enhanced mitochondrial phenotype, which is not reversed by IDH mutant inhibitors. This study provides a scientific rationale for combinatory mitochondrial-targeted therapies to treat IDH mutant AML patients, especially those unresponsive to or relapsing from IDH mutant inhibitors.
Abstract
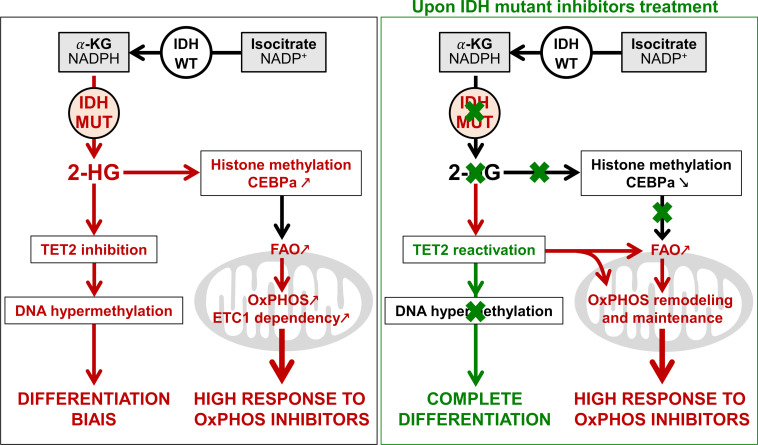
Mutations in IDH induce epigenetic and transcriptional reprogramming, differentiation bias, and susceptibility to mitochondrial inhibitors in cancer cells. Here, we first show that cell lines, PDXs, and patients with acute myeloid leukemia (AML) harboring an IDH mutation displayed an enhanced mitochondrial oxidative metabolism. Along with an increase in TCA cycle intermediates, this AML-specific metabolic behavior mechanistically occurred through the increase in electron transport chain complex I activity, mitochondrial respiration, and methylation-driven CEBPα-induced fatty acid β-oxidation of IDH1 mutant cells. While IDH1 mutant inhibitor reduced 2-HG oncometabolite and CEBPα methylation, it failed to reverse FAO and OxPHOS. These mitochondrial activities were maintained through the inhibition of Akt and enhanced activation of peroxisome proliferator-activated receptor-γ coactivator-1 PGC1α upon IDH1 mutant inhibitor. Accordingly, OxPHOS inhibitors improved anti-AML efficacy of IDH mutant inhibitors in vivo. This work provides a scientific rationale for combinatory mitochondrial-targeted therapies to treat IDH mutant AML patients, especially those unresponsive to or relapsing from IDH mutant inhibitors.
Graphical Abstract
Introduction
Changes in intermediary and energy metabolism provide the flexibility for cancer cells to adapt their metabolism to meet energetic and biosynthetic requirements for proliferation (Boroughs and DeBerardinis, 2015; Vander Heiden and DeBerardinis, 2017). Manipulating glycolysis, glutaminolysis, fatty acid β-oxidation (FAO), or oxidative phosphorylation (OxPHOS) markedly reduces cell growth in vitro and in vivo and sensitizes acute myeloid leukemia (AML) cells to drugs (Samudio et al., 2010; Škrtić et al., 2011; Scotland et al., 2013; Matre et al., 2016; Farge et al., 2017; Sharon et al., 2019). The importance of the metabolic reprogramming in this disease is further illustrated by recurrent mutations in the genes of two crucial metabolic enzymes, isocitrate dehydrogenases (IDH) 1 and 2, present in >15% of AML patients. Most of these mutations are at arginine (R) residues at codon 132 for IDH1 (IDH1 R132) and at 140 and 172 for IDH2 (IDH2 R140 and IDH2 R172; Mardis et al., 2009; Marcucci et al., 2010; Papaemmanuil et al., 2016).
The impact of IDH mutation and the related accumulation of the oncometabolite (R)-2-hydroxyglutarate (2-HG) have been well documented in leukemic transformation and AML biology (Figueroa et al., 2010; Sasaki et al., 2012; Losman et al., 2013; Kats et al., 2014; Inoue et al., 2016; Elkashef et al., 2017; Jiang et al., 2017; Turcan et al., 2018). As IDH mutations are early events in oncogenesis and are systematically conserved at relapse (Corces-Zimmerman and Majeti, 2014; Shlush et al., 2014; Ok et al., 2019), IDH1/2 mutated enzymes represent attractive therapeutic targets, and small molecules specifically inhibiting the mutated forms of these enzymes have been developed (Rohle et al., 2013; Wang et al., 2013; Okoye-Okafor et al., 2015; Yen et al., 2017; Stein et al., 2017; DiNardo et al., 2018; Pollyea et al., 2019; Stein et al., 2019; Chaturvedi et al., 2020). Both IDH2m and IDH1m inhibitors promote differentiation and reduce DNA and histone methylation levels as well as significantly decrease 2-HG concentration (Rohle et al., 2013; Yen et al., 2017; Stein et al., 2017). Overall response rates for ivosidenib (IDH1 mutant inhibitor; IDH1mi) and enasidenib (IDH2mi) are highly encouraging with up to 30% or 40% in monotherapy in phase 1/2 clinical trials for newly diagnosed or relapsed/refractory AML patients, respectively (Stein et al., 2017; DiNardo et al., 2018; Pollyea et al., 2019; Roboz et al., 2020). These results led to the US Food and Drug Administration approvals of enasidenib in August 2017 and ivosidenib in July 2018 for relapsed or refractory adult AML patients with IDH mutation. However, several mechanisms of resistance to these targeted therapies have already been identified (Amatangelo et al., 2017; DiNardo et al., 2018, 2015; Stein et al., 2017; Pollyea et al., 2019; Choe et al., 2020). Many nonresponders also displayed a significant decrease in the amount of 2-HG, suggesting that alternative mechanisms may compensate for 2-HG to drive tumor progression (DiNardo et al., 2015; Stein et al., 2017; Amatangelo et al., 2017; Stein et al., 2019; Harding et al., 2018; Wang et al., 2020 Preprint). Notably, clonal heterogeneity, second-site mutation, or multiple pathways involved in signaling, especially RAS signaling, have recently been shown to be responsible for relapse in patients treated with IDHmi (DiNardo et al., 2018; Intlekofer et al., 2018; Quek et al., 2018; Harding et al., 2018; Choe et al., 2020; Wang et al., 2020 Preprint). These observations suggest that targeting IDH mutant (IDHm) activity is not sufficient to achieve a durable clinical response in most patients and that new combinatory approaches need to be designed.
Given the crucial roles of WT IDH1/2 in cell metabolism (e.g., Krebs cycle, OxPHOS, cytosolic and mitochondrial redox, and anabolism including lipid biosynthesis) and in human disease, a better understanding of the contribution of oncogenic IDH mutations to metabolism and metabolic homeostasis is expected to lead to new therapeutic strategies. Several studies have demonstrated that IDHm cancer cells exhibit some metabolic specificities, in particular related to redox homeostasis through nicotinamide adenine dinucleotide (phosphate)NAD(P)+ availability (Tateishi et al., 2015; Hollinshead et al., 2018; Gelman et al., 2018; Mugoni et al., 2019). However, none of these studies have definitively shown how metabolic changes elicited by IDH mutations modulate cell proliferation and drug resistance or impact therapeutic response in AML. In particular, the role of metabolism in resistance to IDHmi has not yet been comprehensively studied in AML. Although existing literature in the field described several vulnerabilities to mitochondrial inhibitors in IDH1/2 mutant cells from solid tumors and AML (Grassian et al., 2014; Izquierdo-Garcia et al., 2014; Cuyàs et al., 2015; Matre et al., 2016; Chan et al., 2015; Baccelli et al., 2019), no studies have demonstrated why IDHm cells are more sensitive to mitochondrial inhibitors in AML. We therefore hypothesized that mitochondrial OxPHOS plays a crucial role in IDHm biology and in the response of AML patients with IDH mutation to IDHmi.
In the present study, we performed multi-omics and functional approaches using two engineered AML cell lines (HL60 and MOLM14), 10 patient-derived xenografts (PDXs) models from two clinical sites (Toulouse University Hospital [TUH] and University of Pennsylvania [UPENN]), and 111 patient samples from three clinical sites (TUH, Institut Paoli-Calmettes [IPC], and MD Anderson Cancer Center [MDACC]) to test this hypothesis and to expressly understand the mitochondrial reprogramming induced by IDH1 mutation and its role in the response to IDHmi. We uncovered that mitochondrial oxidative metabolism is enhanced in IDHm cells in vitro and in vivo. This AML-specific metabolic behavior mechanistically occurred through the increase in methylation-driven CEBPα-induced FAO of IDH1m cells. Furthermore, IDH1mi that reduced 2-HG oncometabolite and CEBPα methylation failed to reverse FAO and OxPHOS. This was due to the enhanced activation of peroxisome proliferator-activated receptor-γ coactivator-1 (PGC1α) upon IDH1mi. Accordingly, OxPHOS inhibitors improved anti-AML efficacy of IDHmi in vivo.
Results
A higher susceptibility of IDH1m AML cells to mitochondrial inhibitors is due to their enhanced OxPHOS activity
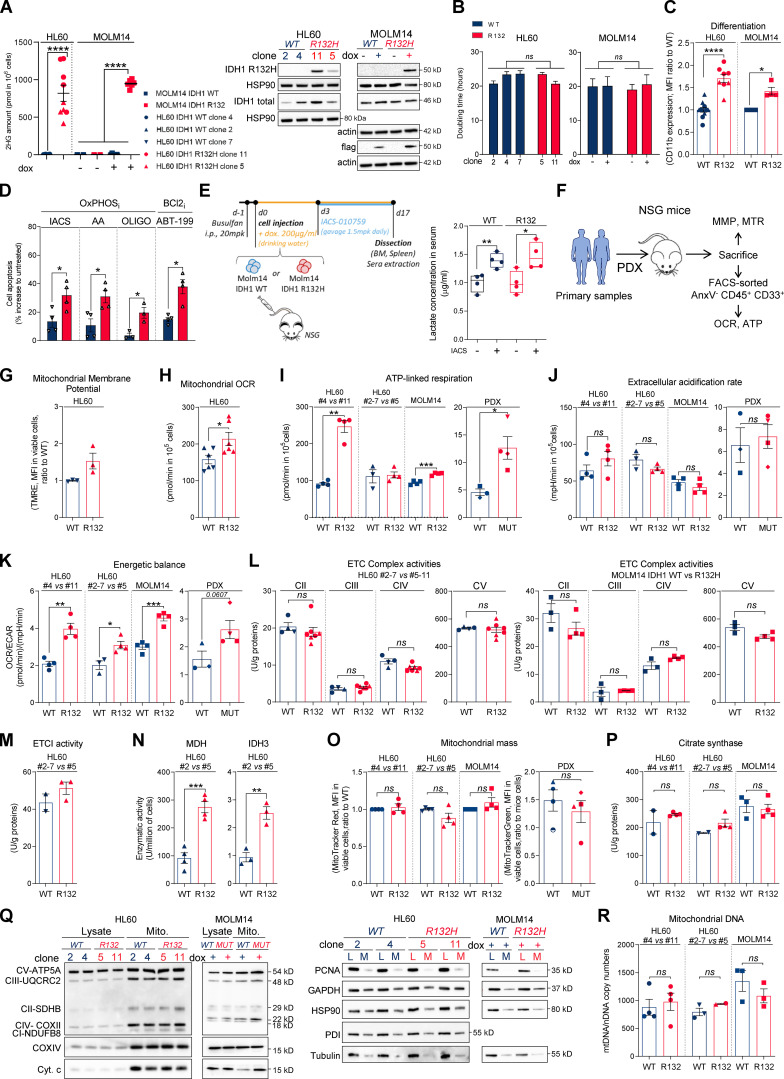
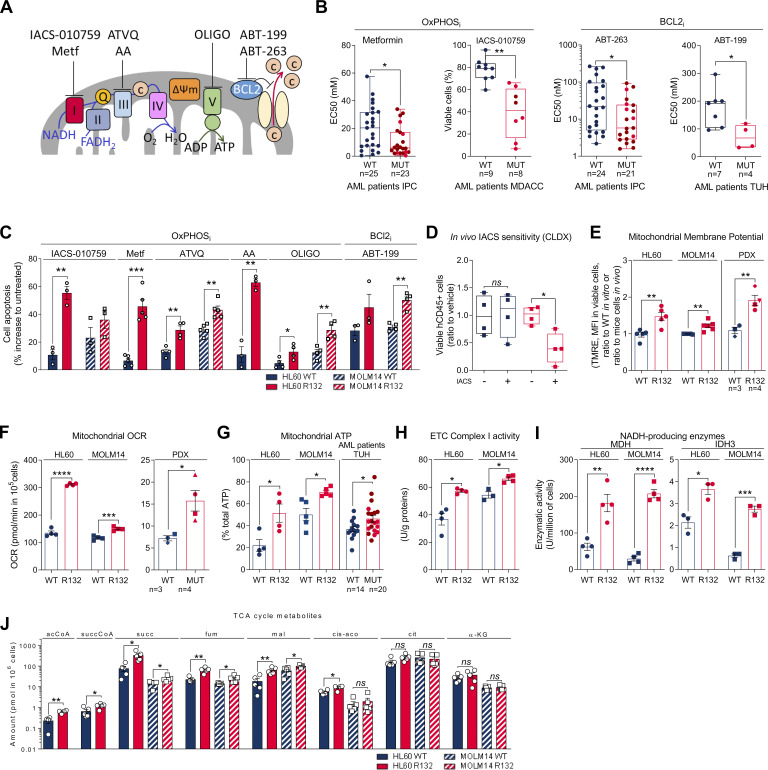
First, we confirmed a higher sensitivity of IDH1/2 mutant cells from primary AML patient samples (WT, n = 41; MUT, n = 36; TUH, IPC, MDACC; Table S1) and two genetically diverse cell lines (Fig. S1, A–C) to mitochondrial inhibitors, including inhibitors of electron transport chain (ETC) complex I (IACS-010759; Molina et al., 2018; metformin), ETC complex III (antimycin A [AA]; atovaquone [ATVQ]), ETC complex V (oligomycin), and BCL2 (ABT-199, ABT-263; Fig. 1, A–C; and Fig. S1 D). We also showed this result in vivo in NOD/LtSz-SCID/IL-2Rγchainnull (NSG)–immunodeficient mice xenografted with IDH1 WT and R132 mutant MOLM14 cells and then treated with IACS-010759 for 2 wk at 1.5 mg/kg/d (Fig. S1 E). Of note, this ETC complex I inhibitor induced an increased level of lactate in the serum of both IDH1 WT and R132 xenografted mice, confirming the systemic biological activity of this type of inhibitor (e.g., metabolic shift toward glycolysis with lactic acidosis or Pasteur effect; Fig. S1 E). However, only mice engrafted with IDH1 R132H cells showed a significant reduction in the total cell tumor burden following IACS treatment in vivo (Fig. 1 D).
Figure S1.
IDH1m cells exhibit a higher sensitivity to OxPHOSi and BCL2i due to their enhanced mitochondrial capabilities and OxPHOS activity in AML. (A) 2-HG concentration was determined from fresh HL60 (different clones) and MOLM14 IDH1 WT and R132H cells (left panel). Total lysates of HL60 (different clones) and MOLM14 IDH1 WT and R132H were immunoblotted with the corresponding antibodies (right panel; representative of three independent experiments). Error bars indicate mean ± SEM of at least five independent experiments. Each point is the mean of three technical replicates. (B) Doubling time of HL60 (different clones) and MOLM14 IDH1 WT and R132H treated or not with doxycycline (dox). Error bars indicate mean ± SEM of at least three independent experiments. (C) Intensity of CD11b staining (median) HL60 (different clones) and MOLM14 IDH1 WT and R132H treated with doxycycline. Error bars indicate mean ± SEM of at least three independent experiments. (D) Apoptosis induction following 48 h IACS-010759 (100 nM), 48 h metformin (10 mM), AA (10 µM), ATVQ (20 µM), oligomycin (OLIGO; 2 µM), and ABT-199 (200 nM) in other clones of HL60 IDH1 WT (clone 2) or R132H (clone 5). Error bars indicate mean ± SEM of at least three independent experiments. (E) Experimental scheme detailing administration time of IACS-010759 by gavage in MOLM14 CLDX. In this model, IDH1 R132 mutation or IDH1 WT overexpression is induced by doxycycline (left panel). Lactate concentration in the serum of mice engrafted with MOLM14 IDH1 WT and R132H AML cells and treated or not with IACS corresponding to Fig. 1D (right panel). (F) Schematic diagram of the in vivo strategy applied for the experiments described in Fig. 1, Fig. S1, Fig. 4, and Fig. S4. (G) TMRE assay in other clones of HL60 IDH1 WT (clones 2 and 7) or R132H (clone 5) to estimate MMP. Error bars indicate mean ± SEM (n = 3 independent experiments). (H) Mitochondrial OCR in other clones of HL60 IDH1 WT (clones 2 and 7) or R132H (clone 5). Error bars indicate mean ± SEM of at least three independent experiments. Each point is the mean of three technical replicates. (I) ATP-linked respiration of different clones of HL60 and MOLM14 IDH1 WT or R132H measured in vitro (n ≥ 3, independent experiments) and ex vivo in PDXs after cell sorting (three patients IDH1 WT and four patients IDH1 MUT). See also Table S1 for patient information. Error bars indicate mean ± SEM. Each point is the mean of three technical replicates. (J) ECAR of different clones of HL60 and MOLM14 IDH1 WT or R132H measured in vitro (n ≥ 3, independent experiments) and ex vivo in PDXs (three patients IDH1 WT and four patients IDH1 MUT; see also Table S1). Error bars indicate mean ± SEM. Each point is the mean of three technical replicates. (K) Energetic balance corresponding to the ratio between OCR and ECAR of different clones of HL60 and MOLM14 IDH1 WT or R132H measured in vitro (n ≥ 3, independent experiments) and ex vivo in PDXs (three patients IDH1 WT and four patients IDH1 MUT; see also Table S1). Error bars indicate mean ± SEM. Each point is the mean of three technical replicates. (L) Mitochondrial ETC complex II to ETC complex V complex activities in different clones of HL60 (left panel) and MOLM14 (right panel) IDH1 WT and R132H. Error bars indicate mean ± SEM of at least three independent experiments. (M) Mitochondrial ETC complex I activity in other clones of HL60 IDH1 WT (clones 2 and 7) or R132H (clone 5). Error bars indicate mean ± SEM of at least two independent experiments. (N) NADH-producing enzyme activities of MDH and IDH3 in other clones of HL60 IDH1 WT (clone 2) and R132H (clone 5). Error bars indicate mean ± SEM of at least three independent experiments. (O) Mitochondrial mass assay (MTR in cell lines or MTG in PDXs) in different clones of HL60 and MOLM14 IDH1 WT or R132H measured in vitro (n ≥ 4) and ex vivo from PDXs (four patients IDH1 WT and four patients IDH1 MUT). Error bars indicate mean ± SEM. (P) Citrate synthase enzymatic activities measured after 24 h in different clones of HL60 and MOLM14 IDH1 WT or R132H. Error bars indicate mean ± SEM of at least two independent experiments. (Q) Total lysates and lysates of purified mitochondria of different clones of HL60 and MOLM14 IDH1 WT or R132H were immunoblotted with the indicated antibodies related to OxPHOS (left panel) and to assess quality of the mitochondrial (Mito.) extraction (right panel). (R) mtDNA copy numbers in different clones of HL60 and MOLM14 IDH1 WT or R132H. nDNA, nuclear DNA. Error bars indicate mean ± SEM of at least two independent experiments. For each panel, groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. BM, bone marrow.
Figure 1.
IDH1m cells exhibit a higher sensitivity to OxPHOSi and BCL2i due to their enhanced mitochondrial capabilities and OxPHOS activity in AML. (A) Schematic representation of the ETC and BCL2 with OXPHOSi and BCL2i used in this study. Metf, metformin. (B) Plots of effective half-maximal concentration (EC50) values from ATP viability assays of metformin and ABT-263 after 48 h, from Annexin V–positive cell assays of ABT-199 after 24 h and percentage of viable cells after 72 h of IACS-010759 in primary samples with WT or mutant (MUT) IDH1 (red circles) or IDH2 (burgundy circles). See also Table S1 for patient information. (C) Apoptosis induction following IACS-010759 (100 nM during 48 h for HL60 and during 6 d for MOLM14; n = 3), 48 h metformin (10 mM; n = 5), AA (10 µM; n = 3), ATVQ (20 µM for HL60; n = 4 and 40 µM for MOLM14; n = 5), oligomycin (OLIGO; 2 µM; n = 4), and ABT-199 (200 nM; n = 3 for HL60 and n = 4 for MOLM14) in HL60 and MOLM14 IDH1 WT or R132H. (D) Total number of human viable AML cells expressing CD45 in vehicle compared with IACS-treated MOLM14 IDH1 WT and R132 mice in bone marrow and spleen (n = 5/group). (E) Assessment of MMP using TMRE assay in HL60 and MOLM14 IDH1 WT or R132H measured in vitro (n = 5, independent experiments) and in vivo in PDXs (three patients IDH1 WT and four patients IDH1 MUT). See also Table S1 for patient information. (F) Mitochondrial OCR of HL60 and MOLM14 IDH1 WT or R132H measured in vitro (n = 4, independent experiments) and ex vivo in PDXs after cell sorting (three patients IDH1 WT and four patients IDH1 MUT). See also Table S1 for patient information. Each point is the mean of three technical replicates. (G) Mitochondrial ATP in HL60 (n = 4) and MOLM14 (n = 5) IDH1 WT and R132H and in patients with IDH WT (n = 14) or MUT IDH1 (red circles) or IDH2 (burgundy circles; n = 21). See also Table S1 for patient information. Each point is the mean of three technical replicates. (H) Mitochondrial ETC complex I activity in HL60 and MOLM14 IDH1 WT and R132H (n ≥ 4). (I) NADH-producing enzyme activities of MDH (n = 4) and IDH3 (n = 3) in HL60 and MOLM14 IDH1 WT and R132H. (J) Acetyl-CoA (acCoA), succinyl-CoA (succCoA), succinate (succ), fumarate (fum), malate (mal), cis-aconitate (cis-aco), citrate (cit), and α-KG amounts measured over 24-h culture in HL60 and MOLM14 IDH1 WT and R132H (n ≥ 4). Each point is the mean of two technical replicates. For each panel, HL60 IDH1 WT is represented in blue by circles (clone 4), whereas R132H is represented in red by circles (clone 11). Results with other clones are described in Fig. S1. MOLM14 is represented by squares, blue for IDH1 WT and red for IDH1 R132H (both induced by doxycycline). Error bars indicate mean ± SEM of at least three independent experiments. Groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To better understand why IDH1m cells have a higher sensitivity to mitochondrial inhibition, we extensively analyzed several biochemical, enzymatic, and functional features relative to mitochondrial metabolism in IDHm versus WT AML cells from two genetically diverse AML cell lines in vitro, 10 PDXs in vivo (Fig. S1 F), and 33 primary AML samples ex vivo. Mitochondrial membrane potential (MMP), basal- and ATP-linked oxygen consumption rate (OCR), and ATP content were all significantly enhanced in IDHm AML cells in vitro and in vivo (Fig. 1, E–G; and Fig. S1, G–I). Hence, since extracellular acidification rate (ECAR), mainly associated with lactate production rate through glycolysis, remained unaffected (Fig. S1 J), the cellular energetic balance was also significantly increased in IDH1m AML cells (Fig. S1 K). Importantly, ETC complex I (and not other ETC complexes) activity, NADH-producing enzyme activity of tricarboxylic acid cycle (TCA) enzymes such as malate dehydrogenase (MDH2) and IDH3, and concentrations of Krebs cycle intermediates (except citrate and α-KG) were also enhanced in IDH1m (Fig. 1, H–J; and Fig. S1, L–N), indicating an increase in mitochondrial NADH availability, mitochondrial activities, and OxPHOS dependency, specifically in IDH1m AML cells. Interestingly, this was not due to an increase in mitochondrial biogenesis as shown by unaffected mitochondrial mass, citrate synthase activity, protein expression of ETC complexes, or ratio between mitochondrial and nucleic DNA (Fig. S1, O–R). Of note, another HL60 clone expressing less IDH1 R132 mutation and producing less 2-HG (Boutzen et al., 2016; Fig. S1 A; clone 5) recapitulated this higher mitochondrial inhibitor sensitivity and enhanced OxPHOS phenotype to a lower extent (Fig. S1). This reinforces the link between IDH1 mutation and increased mitochondrial capabilities.
Methylation- and CEBPα-dependent mitochondrial FAO is increased in IDH1m cells
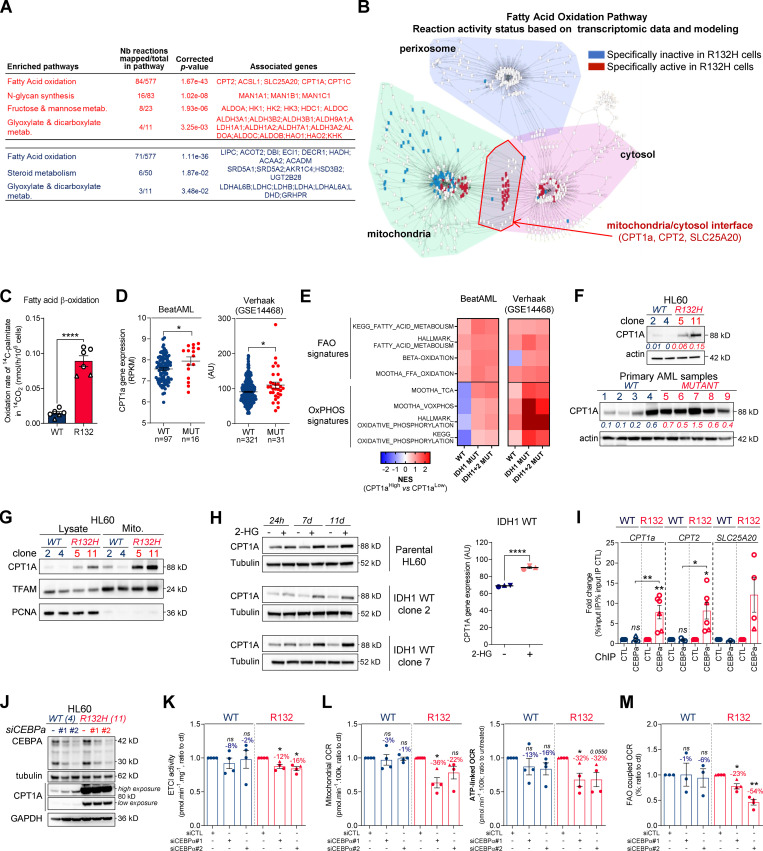
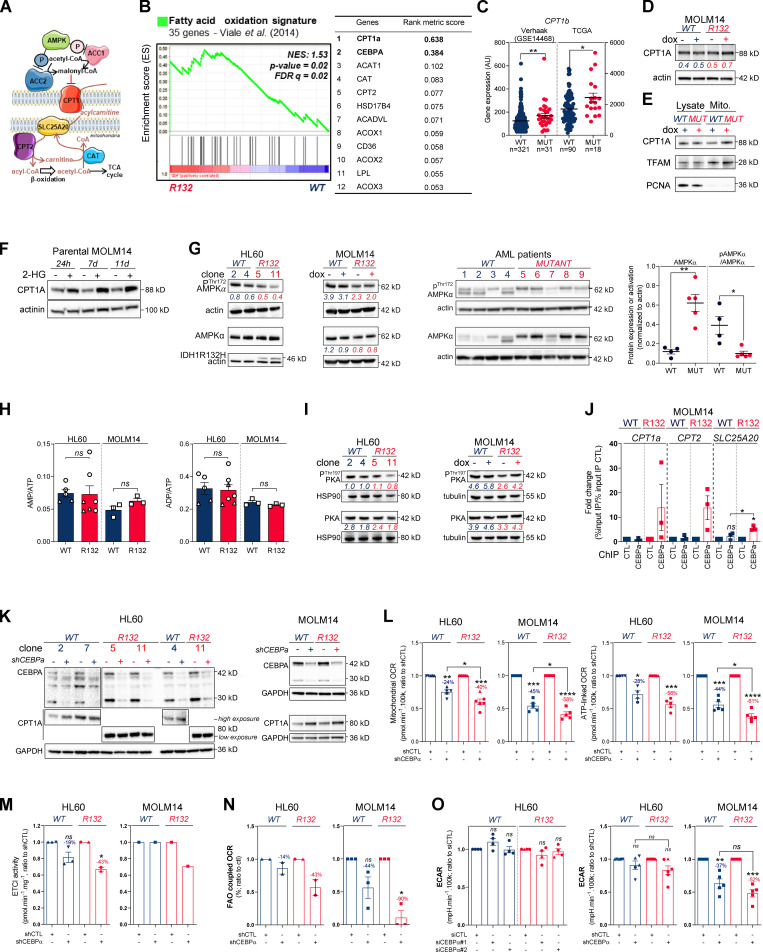
To further understand how mitochondrial reprogramming is induced by IDH1 mutation, we next performed a computational analysis of the metabolic network of IDH1m cells based on human genome scale metabolic network reconstruction Recon2 (7,440 metabolic reactions; Thiele et al., 2013). To reconstruct active leukemic metabolic networks of IDH1 WT and mutant AML cells at a global level, we integrated transcriptomic data and proliferation rates and applied metabolic constraints (Jerby et al., 2010; Poupin et al., 2018; Fig. 2 A). This analysis identified a significant enrichment of active reactions in various carbon metabolic pathways (N-glycan synthesis, fructose and mannose metabolism, and dicarboxylate metabolism; Fig. 2 A) in IDH1m cells. This further uncovered a major change in FAO in these cells (Fig. 2, A and B), especially CPT1a, CPT2, and SLC25A20, which mediate the transport of acyl-carnitines of different length from the cytosol to the mitochondrial matrix to initiate the process of FAO (Fig. S2 A). Accordingly, FAO rate was significantly higher in IDH1m AML cells using 14C-palmitate assay (Fig. 2 C).
Figure 2.
Methylation- and CEBPα-dependent mitochondrial FAO is increased in IDH1m cells. (A) Comparison of the predicted activity of reactions in the metabolic network of HL60 IDH1 WT versus R132H cells. Predictions of reaction activity or inactivity were made using the Recon2 metabolic network reconstruction and transcriptomic data from HL60 IDH1 WT and R132H. Pathway enrichment was performed on the set of reactions identified as specifically active (red) or specifically inactive (blue) in R132H cells. Corrected P values were obtained by performing a hypergeometric test followed by a Bonferroni correction. (B) Visualization of modulated reactions within the FAO pathway of the Recon2 metabolic network. Reactions predicted to be specifically active (red) or inactive (blue) in R132H using the computational modeling approach were mapped using the MetExplore web server (Chazalviel et al., 2018). (C) 14C palmitate oxidation by HL60 IDH1 WT clone 4 (circle; n = 4) and 2 (triangle; n = 2) and R132H clone 11 (circle; n = 4) and 5 (triangle; n = 2) to assessFAO rate. Each point is the mean of two technical replicates. (D) CPT1a gene expression across AML patient samples from BeatAML (Tyner et al., 2018) and GSE14468 (Verhaak cohort) datasets in function of their IDH1 status. Groups were compared using unpaired nonparametric Mann-Whitney test (*, P < 0.05). (E) Normalized ESs following GSEA analysis of patients with high or low expression of CPT1a (median as the reference) in IDH WT, IDH1m, or IDH1+2 mutant across AML transcriptomes from two independent cohorts, BeatAML and Verhaak (GSE14468). (F) Total lysates of HL60 IDH1 WT and R132H (representative of three independent experiments) and total lysates of primary samples IDH1 WT or MUT were immunoblotted with the indicated antibodies. See also Table S1 for patient information. (G) Total lysates (Lysate) and lysates of purified mitochondria (Mito.) of HL60 IDH1 WT and R132H were immunoblotted with the indicated antibodies (representative of two independent experiments). (H) Total lysates of parental HL60 and HL60 IDH1 WT clones 2 and 7 treated with exogenous 2-HG (100 µM) during 24 h, 7 d, and 11 d were immunoblotted with the indicated antibodies (left panel; representative of two independent experiments). CPT1a gene expression in HL60 IDH1 WT clones 2, 4, and 7 treated with exogenous 2-HG (100 µM) during 2 d (Boutzen et al., 2016). (I) qChIP experiments showing the relative recruitment of CEBPα on CPT1a, CPT2, and SLC25A20 locus in mutant IDH1 R132H versus IDH1 WT HL60, as indicated. Results are represented as the relative ratio between the mean value of IP chromatin (calculated as a percentage of the input) with the indicated antibodies and the one obtained with a control irrelevant antibody. HL60 IDH1 WT is represented in blue by circles (clone 4; n = 4) and triangles (clone 2; n = 2), whereas R132H is represented in red by circles (clone 11; n = 4) and triangles (clone 5; n = 2). (J) Total lysates of HL60 IDH1 WT and R132H transfected with siRNA control or targeting CEBPα were immunoblotted to confirm the knockdown and to measure CPT1a protein expression. This confirmation was performed for each siRNA experiment (n = 4). (K) Mitochondrial ETC complex I activity in HL60 IDH1 WT and R132H transfected with siRNA control or targeting CEBPα (n = 4, independent experiments). (L) Mitochondrial OCR and ATP-linked OCR of HL60 IDH1 WT or R132H transfected with siRNA control or targeting CEBPα (n = 4, independent experiments). Each point is the mean of three technical replicates. (M) FAO-coupled OCR of HL60 IDH1 WT or R132H transfected with siRNA control or targeting CEBPα (n ≥ 3, independent experiments). Each point is the mean of three technical replicates. For each panel except D as indicated, groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Error bars indicate mean ± SEM. AU, arbitrary units; NES, normalized ES.
Figure S2.
Methylation- and CEBPα-dependent mitochondrial FAO is increased in IDH1m cells. (A) Schematic representation of FAO and its regulation. (B) GSEA of FAO signature identified by Viale et al. (2014) performed using transcriptomes of HL60 IDH1 R132H clones compared with WT already published (Boutzen et al., 2016) and associated genes with top rank metric scores. (C) CPT1b gene expression across AML patient samples from GSE14468 (Verhaak cohort) and TCGA datasets in function of their IDH1 status. Groups were compared using unpaired nonparametric Mann-Whitney test. (D) Total lysates of MOLM14 IDH1 WT and R132H were immunoblotted with the indicated antibodies (representative of two independent experiments). (E) Total lysates (lysate) and lysates of purified mitochondria (Mito.) of MOLM14 IDH1 WT and R132H were immunoblotted with the indicated antibodies (representative of two independent experiments). (F) Total lysates of parental MOLM14 treated with exogenous 2-HG (50 µM) during 24 h, 7 d, and 11 d were immunoblotted with the indicated antibodies (representative of two independent experiments). (G) Lysates of different clones of HL60 and MOLM14 (left panel) and of AML primary samples (right panel) IDH1 WT or mutant (MUT) were immunoblotted with the indicated antibodies and quantified. (H) AMP/ATP (left panel) and ADP/ATP (right panel) ratios determined by metabolomics in HL60 IDH1 WT (clones 4 and 2) and R132H (clones 11 and 5) and in MOLM14 IDH1 WT and R132H cells. Error bars indicate mean ± SEM of at least three independent experiments. (I) Lysates of different clones of HL60 and MOLM14 IDH1 WT or R132H were immunoblotted with the indicated antibodies and quantified. (J) qChIP experiments showing the relative recruitment of CEBPα on CPT1a, CPT2, and SLC25A20 locus in mutant IDH1 R132H versus IDH1 WT MOLM14, as indicated. Results were represented as the relative ratio between the mean value of IP chromatin (calculated as a percentage of the input) with the indicated antibodies and the one obtained with a control irrelevant antibody. Error bars indicate mean ± SEM of at least three independent experiments. (K) Total lysates of HL60 and MOLM14 IDH1 WT and R132H transfected with shRNA control or targeting CEBPα were immunoblotted to confirm the knockdown and measure CPT1a protein expression. This confirmation was performed for each siRNA experiment (n ≥ 3). (L) Mitochondrial OCR and ATP-linked OCR of HL60 and MOLM14 IDH1 WT or R132H transfected with shRNA control or targeting CEBPα (n ≥ 4, independent experiments). Error bars indicate mean ± SEM. Each point is the mean of three technical replicates. (M) Mitochondrial ETC complex I activity in HL60 and MOLM14 IDH1 WT and R132H transfected with shRNA control or targeting CEBPα. Error bars indicate mean ± SEM of at least two independent experiments for HL60. For MOLM14, only one independent measure was performed. (N) FAO-coupled OCR of HL60 and MOLM14 IDH1 WT or R132H transfected with shRNA control or targeting CEBPα (n ≥ 2, independent experiments). Error bars indicate mean ± SEM. Each point is the mean of three technical replicates. (O) ECAR of HL60 and MOLM14 IDH1 WT or R132H transfected with siRNA or shRNA control or targeting CEBPα (n ≥ 4, independent experiments). For each panel, HL60 IDH1 WT is represented in blue by circles (clone 4), up triangles (clone 2), and down triangles (clone 7), whereas R132H is represented in red by circles (clone 11) or up triangles (clone 5). MOLM14 is represented by squares, blue for IDH1 WT and red for IDH1 R132H (both induced by doxycycline). Error bars indicate mean ± SEM. Each point is the mean of three technical replicates. For all panels except C as indicated, groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. AU, arbitrary units; CTL, control; dox, doxycycline; FDR, false discovery rate.
Furthermore, we performed a gene set enrichment analysis (GSEA) with a curated FAO gene signature (Viale et al., 2014) and found that this signature was enriched in IDH1m AML cells (Fig. S2 B). The most highly expressed gene in IDH1m cells was CPT1a, the key component of initiating FA shuttling into mitochondria. Consistent with this, CPT1a and its isoform CPT1b mRNA levels were also significantly up-regulated in two independent AML cohorts (Fig. 2 D and Fig. S2 C). Moreover, GSEA analysis of the transcriptomes of AML patients with IDH WT, IDH1, or IDH1/2 mutation revealed higher enrichment of FA metabolism and OxPHOS gene signatures in CPT1aHIGH patients with IDH mutations compared to IDH WT in two independent cohorts (Fig. 2 E). Furthermore, CPT1A protein was significantly increased in total cell lysates and in mitochondria isolated from IDH1m cells and in IDH1m primary samples compared with IDH1 WT cells (Fig. 2, F and G; and Fig. S2, D and E). Finally, parental HL60 and MOLM14 cell lines as well as HL60 engineered IDH1 WT clones treated with exogenous 2-HG displayed increased levels of CPT1A protein and CPT1a gene expression (Fig. 2 H and Fig. S2 F). This strengthens the observation that CPT1a plays a crucial role in FA metabolism and OxPHOS, specifically in 2-HG–producing IDHm AML cells.
The regulation of FAO is complex and involves many different signaling pathways and allosteric regulation (Fig. S2 A). We next examined AMP kinase (AMPK), a master regulator of energy and metabolic homeostasis. Surprisingly, whereas the AMPK protein level was increased in IDH1m cell lines and patients, its activation by phosphorylation was decreased (Fig. S2 G). Furthermore, we did not observe an increase in the AMP/ATP ratio in mutants compared with WT (Fig. S2 H), suggesting that AMP, the primary allosteric activator of AMPK, does not directly activate AMPK in these cells. No increase in ADP/ATP ratio was detected, also suggesting that neither AMP nor ADP enhanced the canonical phosphorylation of AMPK Thr172 via LKB1 (Shackelford and Shaw, 2009) in IDH1m AML cells (Fig. S2 G). These data showed that AMPK was not activated directly by AMP or via phosphorylation in mutant cells, indicating that the changes seen in FAO reflect an AMPK-independent mechanism. Similarly, the protein kinase A (PKA) pathway, another known energy-sensing pathway able to regulate FAO (Lim et al., 2013; Gerhart-Hines et al., 2011), did not appear to be differentially activated and thus is not involved in the observed FAO dysregulation (Fig. S2 I).
We previously demonstrated that IDH1 mutation and its 2-HG product dysregulate CEBPα (Boutzen et al., 2016), a well-known transcriptional regulator of several genes involved not only in myeloid differentiation but also in glucose and lipid metabolism in adipocytes. Moreover, CEBPα was the second-most highly expressed gene of the FAO gene signature in IDH1m cells (Fig. S2 B). Thus, we performed quantitative chromatin immunoprecipitation (qChIP) assays to assess CEBPα binding to promoters of genes encoding FA transporters. We observed that the recruitment of endogenous CEBPα to promoters of CPT1a, CPT2, and SLC25A20 increased specifically in IDH1m cells (Fig. 2 I and Fig. S2 J). CEBPα silencing with two different siRNAs or a constitutive shRNA led to a reduction of ETC complex I enzyme activity, mitochondrial basal OCR, mitochondrial ATP turnover, and FAO-coupled OCR slightly in IDH1 WT and to a significantly greater extent in IDH1m AML cells (Fig. 2, J–M; and Fig. S2, K–N), while ECAR remained similar between IDH1 WT and R132H cells (Fig. S2 O). Together, these results indicate that IDH1m AML cells display a gene signature specific for FA shuttling and a high FAO activity in a CPT1a- and CEBPα-dependent manner driven by 2-HG production that supports mitochondrial activity.
IDHmi reverse 2-HG production but maintain high OxPHOS phenotype and mitochondrial metabolism
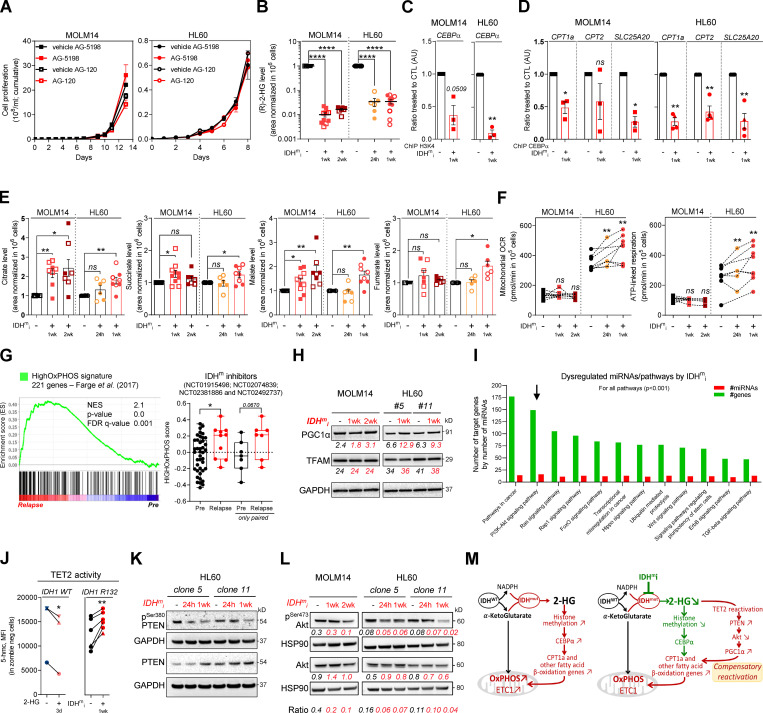
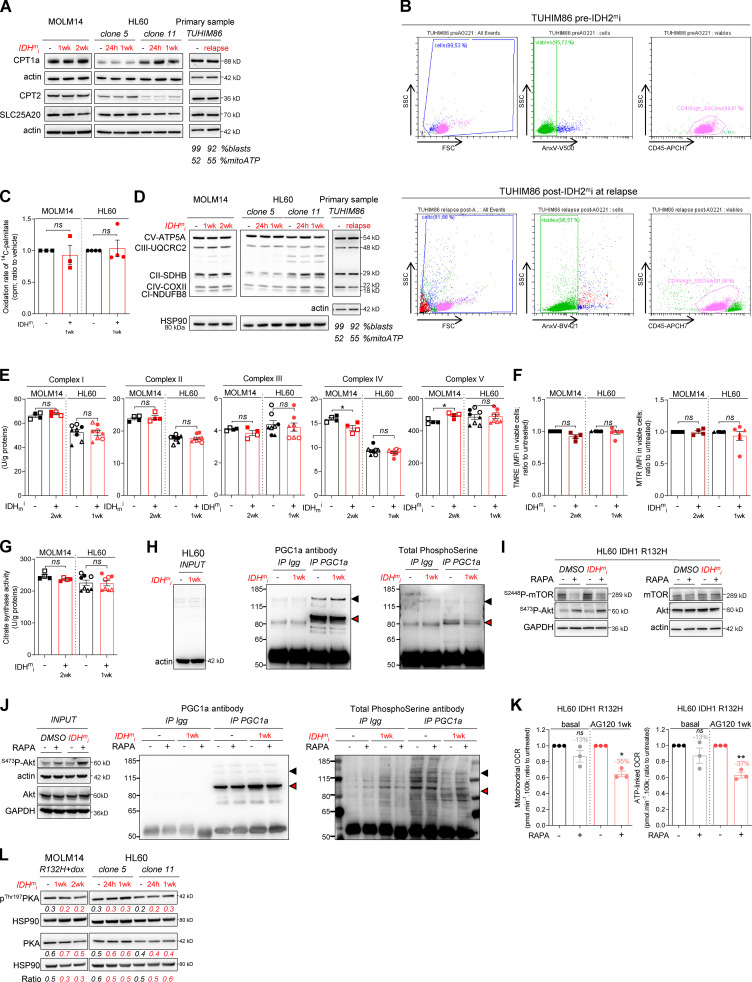
As IDHm cells exhibited higher mitochondrial activity than WT cells, we investigated the impact of IDHmi (US Food and Drug Administration–approved ivosidenib AG-120 or its preclinical version AGI-5198 for IDH1 and enasidenib AG-221 for IDH2) on FAO and OxPHOS. First, if we did not observe any change in cell proliferation rates following IDHmi (Fig. 3 A), the level of 2-HG was significantly reduced as early as 24 h after the beginning of the treatment (Fig. 3 B). Moreover, IDHmi reversed occupancy of the H3K4me3-activating locus in the CEBPα promoter (Fig. 3 C) that we previously showed was significantly increased in IDH1-R132H mutant HL60 and MOLM14 cells (Boutzen et al., 2016). IDHmi also prevented the recruitment of endogenous CEBPα to promoter of CPT1a, CPT2, and SLC25A20 (Fig. 3 D). However, neither the corresponding protein amounts (including in one IDHm AML patient before enasidenib treatment and at relapse) nor the FAO rate were impacted following treatment with IDH1mi (Fig. S3, A–C).
Figure 3.
IDHmi reverse 2-HG production but do not necessarily decrease high OxPHOS phenotype and mitochondrial metabolism. (A) Cumulative cell proliferation through time in MOLM14 and HL60 IDH1 R132H treated with vehicle (DMF) or AG-5198 (2 µM) and vehicle (DMSO) or AG-120 (2 µM; n = 4, independent experiments). (B) 2-HG levels normalized to IS measured over 24-h culture in MOLM14 and HL60 IDH1 R132H following 24-h, 1-wk, or 2-wk treatment with AG-5198 (2 µM, plain symbols) or AG-120 (2 µM, empty symbols; n ≥ 3, independent experiments). (C) qChIP experiments showing the relative recruitment of histone H3 trimethylation at lysine 4 (H3K4me3) on CEBPα promoter in MOLM14 and HL60 IDH1 R132H following 1-wk treatment with AG-5198 (2 µM). Results are represented as the relative ratio between the mean value of IP chromatin (calculated as a percentage of the input) with CEBPα antibody and the one obtained with a control irrelevant antibody, normalized to the untreated condition (n = 3, independent experiments). (D) qChIP experiments showing the relative recruitment of CEBPα on CPT1a, CPT2, and SLC25A20 locus in MOLM14 and HL60 IDH1 R132H following 1-wk treatment with AG-5198 (2 µM). Results are represented as the relative ratio between the mean value of IP chromatin (calculated as a percentage of the input) with the indicated antibodies and the one obtained with a control irrelevant antibody, normalized to the untreated condition. HL60 IDH1 R132H is represented by circles (clone 11; n = 3) and triangles (clone 5; n = 1). (E) Citrate, succinate, malate, and fumarate levels normalized to IS measured over 24-h culture in MOLM14 and HL60 IDH1 R132H following 24-h, 1-wk, or 2-wk treatment with AG-5198 (2 µM, plain symbols) or AG-120 (2 µM, empty symbols; n ≥ 3, independent experiments). (F) Mitochondrial OCR and ATP-linked OCR of MOLM14 and HL60 IDH1 R132H in vehicle (DMF) and after 24-h, 1-wk, or 2-wk treatment with AG-5198 (2 µM; n ≥ 3, independent experiments). (G) GSEA of HighOxPHOS signature (identified by Farge et al., 2017) performed using transcriptomes of patients harboring an IDH mutation included in clinical trials for IDHmi and published (GSE153348) with associated score. (H) Total lysates of MOLM14 and HL60 IDH1 R132H following 1-wk or 2-wk treatment with AG-5198 (2 µM) were immunoblotted with the indicated antibodies relative to mitochondrial regulation. Immunoblot representative of three independent experiments. (I) Bioinformatics analysis of differentially expressed miRNAs following AG-5198 (2 µM) treatment of HL60 and MOLM14 IDH1m cells for 1 wk. The graph is showing the top KEGG pathways of biological function of the targets of all differentially expressed miRNAs between untreated and treated cells. This enrichment pathways analysis utilizes the union of targeted genes by the selected miRNAs before the statistical calculation. For all these analyses, a P value threshold <0.001 was used. The arrow highlights the pathway of interest in this study (PI3K/Akt). (J) Intensities of 5-OH-methylcytosine staining (median) in HL60 IDH1 WT (circle for clone 4, n = 1; up triangle for clone 2, n = 1; and down triangle for clone 7, n = 1) treated with exogenous 2-HG (100 µM) during 3 d and in HL60 IDH1 R132H (circle for clone 11, n = 5; and triangle for clone 5, n = 1) treated 1 wk with AG-120 (2 µM). (K) Total lysates of HL60 IDH1 R132H following 24-h or 1-wk treatment with AG-120 (2 µM) were immunoblotted with the indicated antibodies. (L) Total lysates of MOLM14 and HL60 IDH1 R132H following 24-h, 1-wk, or 2-wk treatment with AG-5198 (2 µM) were immunoblotted with the indicated antibodies relative to signaling proteins and were quantified. Ratio of phosphorylated to total form of Akt was measured to assess the activation of the pathway. (M) Schematic diagram of metabolic reprogramming induced by IDH1 mutation in AML cells and its impact on OxPHOS status through FAO regulation at the steady state and upon treatment with IDHmi. For each panel, groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars indicate mean ± SEM. AU, arbitrary units; CTL, control; FDR, false discovery rate; NES, normalized ES.
Figure S3.
IDHmi reverse 2-HG production but do not necessarily decrease high OxPHOS phenotype and mitochondrial metabolism. (A) Total lysates of MOLM14 and HL60 IDH1 R132H following 24-h, 1-wk, or 2-wk treatment with AG-5198 (2 µM) were immunoblotted with the indicated antibodies relative to FAO proteins. Total lysates of one primary sample IDH2 MUT before IDH1mi (AG221) and at relapse was immunoblotted with the indicated antibodies. Percentage of blasts determined as CD45dim/SSClow–positive cells and percentage of mitochondrial ATP (mitoATP) are given for both time points. See gating strategy in B and Table S1 for patient information. (B) Gating strategy used to assess the percentage of AML blasts in primary AML specimen TUHIM86 before treatment with IDHmi and at relapse. Human peripheral blood mononuclear cells are gated based on the forward (FSC) and side scatter (SSC). Dead cells are excluded with Annexin V staining. AML blast gate is CD45dim and SSClow. (C) 14C palmitate oxidation by MOLM14 and HL60 IDH1 R132H following 1-wk treatment with AG-5198 (2 µM) to assess FAO rate. Error bars indicate mean ± SEM of three independent experiments. Each point is the mean of three technical replicates. The results are given in mean counts per minute (cpm) and normalized to the untreated condition. (D) Total lysates of MOLM14 and HL60 IDH1 R132H following 24-h, 1-wk, or 2-wk treatment with AG-5198 (2 µM) were immunoblotted with the indicated antibodies relative to ETC proteins (representative of at least three independent experiments). Total lysates of one primary sample IDH2 MUT before IDH1mi (AG221) and at relapse were immunoblotted with the indicated antibodies. (E) Mitochondrial ETC complex activities in different clones of HL60 and MOLM14 IDH1 R132H following 1- or 2-wk treatment with AG-5198 (2 µM, plain circles) or AG-120 (2 µM, empty circles). Error bars indicate mean ± SEM of at least four independent experiments. (F) MMP (TMRE assay) and mitochondrial mass (MTR stain) in viable cells measured in different clones of HL60 and MOLM14 IDH1 R132H following 1 or 2 wk of treatment with AG-5198 (2 µM), respectively. Error bars indicate mean ± SEM of at least three independent experiments. (G) Citrate synthase enzymatic activity measured after 24 h in HL60 and MOLM14 IDH1 R132H following 1- or 2-wk treatment with AG-5198 (2 µM, plain circles) or AG-120 (2 µM, empty circles), respectively. Error bars indicate mean ± SEM of at least four independent experiments. (H) IP of PGC1α was followed by immunoblotting using total phosphoserine antibody (right panel) in HL60 IDH1 R132H following 1-wk treatment with control (DMSO) or AG-120 (2 µM). Immunoblots of the inputs confirmed same amount of proteins loaded in the two conditions (left panel). The arrowheads highlight the bands corresponding to PGC1a. (I) Akt was activated through short mTOR inhibition with rapamycin (4 h, 100 nM) in HL60 IDH1 R132H following 1-wk treatment with AG-120 (2 µM). Corresponding total lysates were immunoblotted with the indicated antibodies. (J) IP of PGC1α was followed by immunoblotting using total phosphoserine antibody (right panel) in HL60 IDH1 R132H following 1-wk treatment with control (DMSO) or AG-120 (2 µM) and treated with rapamycin (4 h, 100 nM). Immunoblots of the inputs confirmed same amount of proteins loaded in the two conditions and activation of Akt with rapamycin (left panel). The arrowheads highlight the bands corresponding to PGC1a. (K) Mitochondrial OCR and ATP-linked OCR of HL60 IDH1 R132H following 1-wk treatment with control (DMSO) or AG-120 (2 µM) and treated with rapamycin (4 h, 100 nM). For each panel, HL60 IDH1 WT is represented in blue by circles (clone 4), up triangles (clone 2), and down triangles (clone 7), whereas R132H is represented in red by circles (clone 11) or up triangle (clone 5). MOLM14 is represented by squares, blue for IDH1 WT and red for IDH1 R132H (both induced by doxycycline). Error bars indicate mean ± SEM of three independent experiments. Each point is the mean of three technical replicates. (L) Total lysates of two different clones of HL60 and MOLM14 IDH1 R132H following 24-h, 1-wk, or 2-wk treatment with AGI-5198 (2 µM) were immunoblotted with the indicated antibodies. For each panel, groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01. CI, complex I; CII, complex II; CIII, complex III, CIV, complex IV; CV, complex V.
As FAO is one of the major biochemical pathways that support mitochondrial function and OxPHOS, especially in AML (Samudio et al., 2010; Farge et al., 2017), we then assessed several mitochondrial activities upon IDHmi treatment. IDHmi maintained ETC complex activities or protein amounts (Fig. S3, D and E), MMP, and mitochondrial mass (Fig. S3, F and G). Interestingly, the amounts of mitochondrial TCA cycle intermediates, OCR, and ATP-linked OCR were maintained in MOLM14 IDHm and even increased in HL60 IDHm upon IDHmi (Fig. 3, E and F). Of particular interest, GSEA of transcriptomic data (Gene Expression Omnibus [GEO] accession no. GSE153348) from four clinical trials conducted in MDACC (NCT01915498, enasidenib for IDH2m patients; NCT02074839, ivosidenib for IDH1m patients; NCT02381886, IDH305 for IDH1m patients; and NCT02492737, AG-881 for IDH1 or IDH2m patients; Wang et al., 2020 Preprint) showed an enrichment in gene signature associated with high OxPHOS function (Farge et al., 2017) in 10 patients at relapse compared with 41 patients before IDHmi treatment (including seven paired specimens; Fig. 3 G). Collectively, these results underpin the role of OxPHOS phenotype in the nongenetic mechanism of adaptive resistance to IDHmi.
Since IDHmi reversed 2-HG production and CEBPα-related FAO features but maintained OxPHOS phenotype, we next sought to identify the compensation that might explain this regulatory shift from CEBPα-dependent CPT1a-driven FAO to allosteric and/or posttranscriptional mechanisms less dependent on transcriptional effects of CEBPα. We analyzed PGC1α, which is a master regulator of mitochondrial biogenesis, OxPHOS, and FA biosynthesis and degradation (Wu et al., 1999; Bhalla et al., 2011). PGC1α protein amount was increased following IDHmi treatment while mitochondrial transcription factor A (TFAM) remained unaffected (Fig. 3 H). TFAM plays a crucial role in maintaining structure and copy number of mitochondrial DNA (mtDNA) and is therefore critical for efficient transcription of mtDNA genes, including 13 OxPHOS proteins (Virbasius and Scarpulla, 1994; Parisi and Clayton, 1991). PGC1α is dynamically regulated by various signaling pathways, transcriptional activators, and posttranslational modifications such as phosphorylation, deacetylation, acetylation, and methylation.
To gain better insight into PGC1α regulation by IDH1mi, we first performed transcriptomic analysis of IDH1 R132H HL60 and MOLM14 cells following treatment with IDH1mi. Interestingly, we found that several microRNAs (miRNAs) and their target signaling pathways were dysregulated by the treatment (Fig. 3 I). Using the DIANA miRPath v.3.0 tool (Vlachos et al., 2012), PI3K-Akt, FoxO, and RAS signaling pathways were enriched and targeted by more than half of miRNAs (11–16 miRNAs) in all of the 16 modulated miRNAs (Table S2). Furthermore, several studies have shown that IDHm cells inhibit TET2 activity in a 2-HG–dependent manner and that loss of TET2 results in reduced PTEN expression and a subsequent hyperactivation of the Akt pathway (Figueroa et al., 2010; Xu et al., 2011; Palam et al., 2018). Accordingly, we found that exogenous 2-HG inhibited TET2 activity in IDH1 WT cells (Fig. 3 J). By contrast, IDHmi reversed TET2 inhibition, increased total PTEN expression, and decreased inhibitory phosphorylation at Ser380 of PTEN in IDH1m cells (Fig. 3, J and K). Of note, a prominent repressive phosphorylation at the serine 570 of PGC1α is mediated by PI3K-Akt (Puigserver et al., 2003; Li et al., 2007). PGC1α immunoprecipitation (IP) confirmed that PGC1α level was increased after treatment with IDHmi. We further observed that serine phosphorylation profiling of PGC1α IP was decreased after IDHmi treatment, reinforcing the idea that IDHmi decreased the inhibitory phosphorylation of PGC1α and maintained OxPHOS metabolism (Fig. S3 H). Moreover, we found that Akt phosphorylation was decreased upon IDHmi treatment (Fig. 3 L), resulting in reduced activation of this pathway. Thus, to strengthen the mechanistic link between Akt and PGC1α phosphorylation, we forced the activation of Akt through short mTOR inhibition with rapamycin after IDHmi (Fig. S3 I) and performed PGC1α IP (Fig. S3 J). Consistently, IDHmi led to a decrease in serine-phosphorylation of PGC1α, and rapamycin reversed this effect only after IDH1mi. Importantly, rapamycin decreased mitochondrial OCR and ATP turnover more significantly after IDH1mi, supporting that Akt pathway is a key regulator of PGC1α and OxPHOS activities after IDH1mi (Fig. S3 K). By contrast, IDHmi treatment did not lead to the activation of PGC1α via enhanced phosphorylation of PKA, another putative energetic sensor (Fig. S3 L). Altogether, these results showed that, while reducing 2-HG and CEBPα activity, IDHmi restored TET2 activity and PTEN expression leading to Akt inhibition and reduced PGC1α phosphorylation that favors a transcriptional dependency shift of FAO from CEBPa toward PGC1a to maintain FAO and OxPHOS phenotypes (Fig. 3 M).
Treatment with OxPHOS inhibitors enhances antileukemic effects of IDHmi in vivo
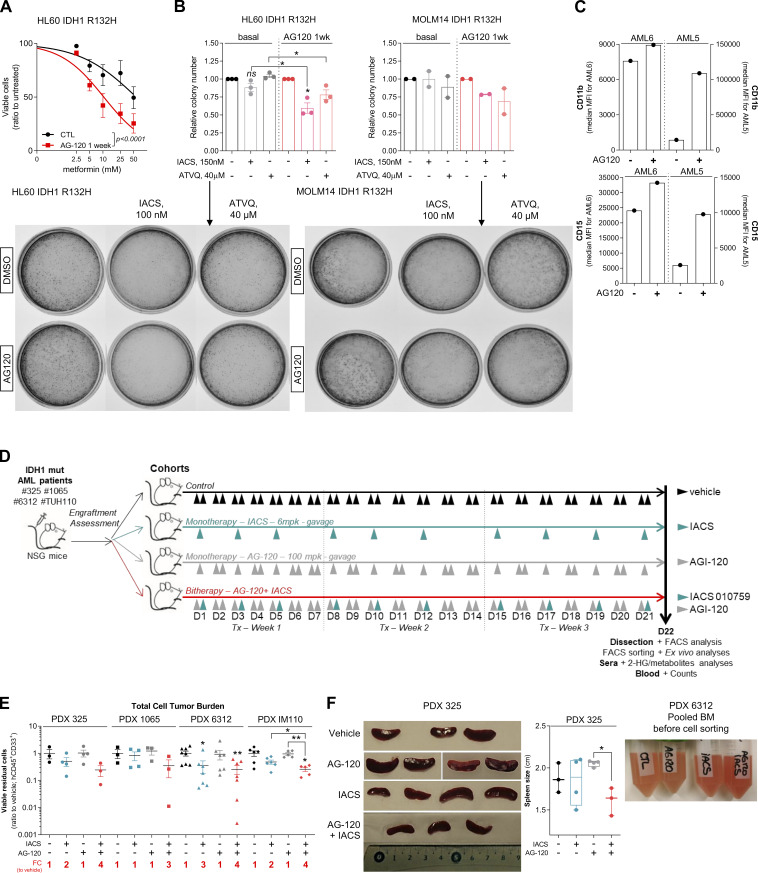
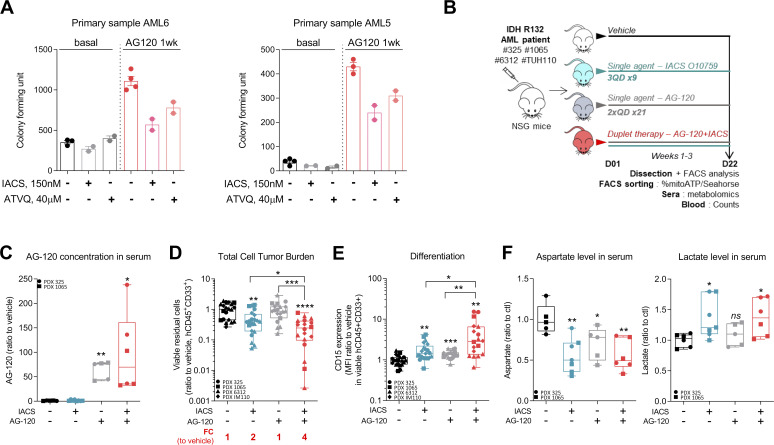
OxPHOS activation upon IDHmi treatment reinforces the validity of specifically treating these patients with a combination of IDHm and OxPHOS inhibitors. We first assessed this combination in vitro and in methylcellulose assays using our engineered cell lines and primary samples. We found that IDH1 R132H cell lines pretreated with IDH1mi were significantly more sensitive to ETC complex I inhibitor metformin in vitro (Fig. S4 A). Furthermore, ETC complex I inhibitor IACS-010759 and ETC complex III inhibitor ATVQ decreased colony-forming ability at a greater and significant extent after IDH1mi compared with nontreated mutant cells (Fig. S4 B). Interestingly, while IDH1mi led to a marked increase in differentiation in both primary samples harboring the IDH1 mutation as expected (Fig. S4 C), and therefore to more colony-forming units, IACS-010759 and ATVQ treatments significantly decreased the number of colonies only after IDH1mi (Fig. 4 A).
Figure S4.
Treatment with an OxPHOS inhibitor enhances antileukemic effects of IDHmi in IDH1m primary samples ex vivo and in vivo. (A) Percentage of viable cells measured by flow cytometry (% AnxV neg) of HL60 IDH1 R132H following 1-wk pretreatment with control (DMSO) or AG-120 (2 µM) and treated with increasing concentrations of metformin. Error bars indicate mean ± SEM of at least three independent experiments. (B) Relative colony number (ratio to ctl) assessed in methylcellulose assays after a 6-d incubation of HL60 and MOLM14 IDH1 R132H pretreated 1 wk with control (DMSO) or AG-120 (2 µM) and then treated with IACS or ATVQ. Corresponding representative photographs of colony-formation units stained using MTT assay (10 mg/ml; 2 h, 37°C). The pictures of each well were taken at the same time. The numbers of colonies with ≥20 cells were counted manually under the microscope. (C) CD11b and CD15 intensities staining measured by flow cytometry of the two primary samples AML5 and AML6 after a 17-d and a 27-d incubation with AG-120, respectively. (D) Experimental scheme detailing exhaustive administration times of IACS-010759 and AG-120 by gavages in PDX models of AML. (E) Total number of human viable AML cells expressing CD45 and CD33 in mono or duplet therapies compared with vehicle for each IDH1 R132 PDXs 325, 1065, 6312, and TUH110 in bone marrow and spleen. Fold change (FC) between the mean of each group and the mean of vehicle was calculated. (F) Photos and corresponding spleen size from mice of PDX 325. Photos of the different groups were taken separately to allow direct and quick crushing but with the same ruler and dimensions to allow comparison (left panel). Photo of the pooled BM of the mice of each treatment group of PDX 6312 before cell sorting (right panel). For each panel, groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01. BM, bone marrow; ctl, control; Tx, treatment.
Figure 4.
Treatment with an OxPHOS inhibitor enhances antileukemic effects of IDHmi in vivo. (A) Number of colony-forming units following methylcellulose assays in two primary samples after a 17-d (AML6) and a 27-d incubation (AML5) with the indicated treatments. The numbers of colonies with ≥20 cells were counted manually under the microscope. See also Table S1 for patient information. (B) Experimental scheme detailing administration time of IACS-010759 and AG-120 by gavages in PDX models of AML. See also Table S1 for patient information. (C) AG-120 concentration in mice sera of PDXs 325 and 1065. (D) Total number of human viable AML cells expressing CD45 and CD33 in mono or duplet therapies compared with vehicle IDH1 R132 PDXs 325, 1065, 6312, and TUH110 in bone marrow and spleen. Fold change (FC) between the mean of each group and the mean of vehicle was calculated. (E) Intensity of CD15 staining (median) in bone marrow in mono and duplet therapies compared with vehicle-treated IDH1 R132 PDXs 325, 1065, 6312, and TUH110. (F) Aspartate and lactate levels normalized to control group in mice sera of IDH1 R132 PDXs 325 and 1065. For C–F, groups were compared with vehicle (no bracket) or other groups (corresponding brackets) with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Error bars indicate mean ± SEM. ctl, control.
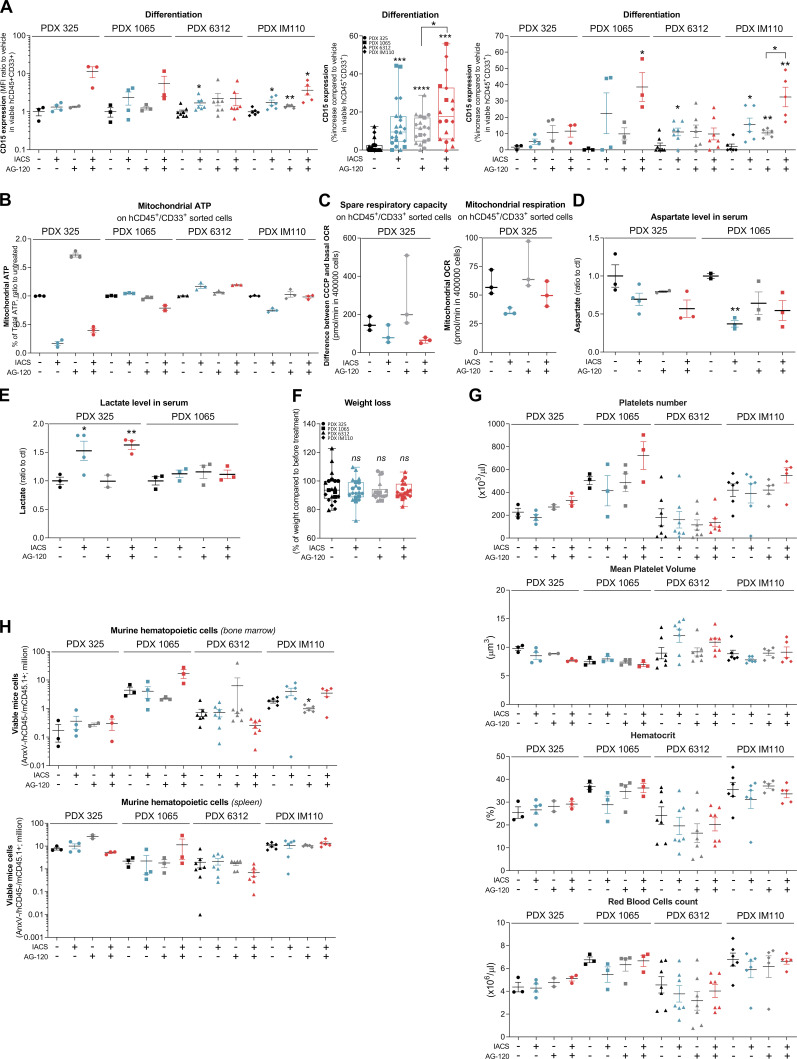
We thus evaluated different single- or duplet-therapeutic approaches in vivo with IDH1mi AG-120 (150 mg/kg, twice every day, 3 wk) and ETC complex I inhibitor IACS-010759 (6 mg/kg, every other day, 3 wk) in four IDH1 R132 PDX models (PDXs #325, #1065, #6312, and #TUH110; Fig. 4 B and Fig. S4 D). We measured AG-120 concentration in the serum of the mice of the corresponding groups (Fig. 4 C). Total cell tumor burden was significantly reduced in the duplet therapy compared with vehicle and single therapies (Fig. 4 D and Fig. S4 E). Additionally, spleen size was lower, and we observed an indicative color change in AML cell suspension from bone marrow in mice treated with the duplet therapy (Fig. S4 F). As observed in AML primary cells after IDHmi (Okoye-Okafor et al., 2015; Yen et al., 2017; Chaturvedi et al., 2017), expression of the myeloid differentiation marker CD15 was significantly increased by AG-120 monotherapy (Fig. 4 E and Fig. S5 A). Addition of IACS-010759 further enhanced CD15 expression (Fig. 4 E and Fig. S5 A). Interestingly, mitochondrial ATP content and respiratory capacities were maintained or even increased after AG-120 alone in vivo, confirming the results obtained in vitro (Fig. S5, B and C). Adding IACS reversed these enhanced OxPHOS capabilities in the duplet therapy in vivo in two PDXs (Fig. S5, B and C). Analysis of mice serum metabolomes showed that aspartate level was significantly reduced in all treated groups, in particular in the ones with IACS, while lactate level was enhanced with IACS treatment, including in the duplet therapy (Fig. 4 F; and Fig. S5, D and E). The ETC has been shown to be important in aspartate production required for pyrimidine synthesis and cell proliferation (Birsoy et al., 2015; Sullivan et al., 2015).
Figure S5.
Effect of the combination of OxPHOS inhibitor and IDHmi on differentiation, mitochondrial readouts, mice weight, complete blood count, and murine hematopoietic cells in IDH1m PDXs in vivo. (A) Intensity of CD15 staining (median) in bone marrow in mono and duplet therapies compared with vehicle treated for each IDH1 R132 PDXs 325, 1065, 6312, and TUH110 (left panel). Percentage of CD15-positive cells in bone marrow in mono and duplet therapies compared with vehicle treated for IDH1 R132 PDXs 325, 1065, 6312, and TUH110 (pooled in middle panel and separated in right panel). (B) Percentage of mitochondrial ATP contribution to total ATP after FACS sorting of human viable cells expressing CD45 and CD33 in bone marrow of IDH1 R132H PDXs 325, 1065, 6312, and TUH110 mice treated with mono and duplet therapies compared with vehicle. Error bars indicate mean ± SEM of at least two technical replicates. (C) Spare respiratory capacity and mitochondrial OCR after FACS sorting of human viable cells expressing CD45 and CD33 in bone marrow of IDH1 R132H PDXs 325 treated with mono and duplet therapies compared with vehicle. Error bars indicate mean ± SEM of three technical replicates. (D and E) Aspartate (D) and lactate (E) levels normalized to control group in mice sera of IDH1 R132 PDXs 325 and 1065. (F) Change in body weight of mice during the different treatments of PDXs 325, 1065, 6312, and TUH110 mice compared with vehicle. Error bars indicate mean ± SEM. (G) Change in platelets and red blood cell features during the different treatments of PDXs 325, 1065, 6312, and TUH110 mice compared with vehicle. Errors bar indicate mean ± SEM. (H) Total number of murine viable AML cells expressing CD45.1 in negative CD45 population in mono or duplet therapies compared with vehicle for each IDH1 R132 PDXs 325, 1065, 6312, and TUH110 in bone marrow and spleen. For each panel, PDX 325 is represented by circles, PDX 1065 by squares, PDX 6312 by triangles, and PDX TUH110 by lozenges. Groups were compared with unpaired two-tailed t test with Welch’s correction. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. ctl, control.
Increase in lactate production is a prominent feature of the Pasteur effect (e.g., increased glycolysis in response to the inhibition of mitochondrial ATP production). Hence, measuring aspartate and lactate levels in patient serum could help monitor the efficacy of their response to IDHm plus OxPHOS inhibitors. Of note, the duplet therapy did not display higher global (Fig. S5, F and G) and normal murine hematological (Fig. S5 H) toxicities compared with single agents. Collectively, these results not only confirmed that IDHmi does not reverse metabolic and mitochondrial features of IDHm AML cells in vivo but also showed that combining this drug with an ETC complex I inhibitor increased drug efficacy in PDX models of AML. Overall, the animal studies demonstrate that the combination of IDHmi and OxPHOS inhibitor is more efficient than single-agent therapies and enhances killing of AML cells.
Discussion
Through the production of (R)-2-HG, IDH mutations directly impact histone/DNA methylation and gene expression (Figueroa et al., 2010; Turcan et al., 2018; Guilhamon et al., 2013; Flavahan et al., 2016), cell proliferation, and differentiation bias (Losman et al., 2013; Kats et al., 2014; Boutzen et al., 2016; Lu et al., 2012). Furthermore, several studies have investigated the mitochondrial phenotype associated with IDHm cells in diverse solid cancers, and various results were reported, suggesting that oxidative metabolism driven by IDH mutation largely depends on the model studied. Notably, it was observed that IDH1 mutation conferred a higher sensitivity to metformin in a transformed mammary epithelial cell line (Cuyàs et al., 2015), resulted in increased oxidative TCA metabolism under hypoxia in HCT116 colon cancer cells (Grassian et al., 2014), and led to enhanced expression of genes involved in mitochondrial structure and function in cholangiocarcinoma (Farshidfar et al., 2017). However, IDH1m glioma cells had reduced ATP content and respiration due to inhibition of ATP synthase or hypersuccinylation by 2-HG (Fu et al., 2015; Li et al., 2015) and increased NADH-coupled proline biosynthesis, resulting in a ‘‘metabolic bypass’’ of ETC complex I (Hollinshead et al., 2018). Although these studies have advanced our understanding of the effect of IDH mutation on mitochondria, mitochondrial metabolism, and phenotype of IDHm cells have remained incomplete, especially in AML. In the current study, we first showed that IDH1m AML cells displayed a higher OxPHOS phenotype than WT cells without changes in mitochondrial biogenesis. Interestingly, growing evidence highlights that OxPHOS also is a crucial process in other AML subgroups, such as FLT3-ITD or EVI1 mutant, through different mechanisms and pathways. The metabolic dependency of FLT3-ITD AML cells for glutamine metabolism supports mitochondrial and redox functions (Gallipoli et al., 2018; Alvarez-Calderon et al., 2015; Gregory et al., 2018). In EVI1 mutant AML cells, mitochondrial function was specifically driven by the creatine kinase pathway (Fenouille et al., 2017). Altogether, our findings confirm a marked ETC complex I dependency of AML cells (Scotland et al., 2013; Farge et al., 2017; Molina et al., 2018; Baccelli et al., 2019) and uncover the crucial role of OxPHOS in the AML subgroup with IDH mutations. Accordingly, IDHm AML cells exhibited enhanced vulnerabilities to various small molecules targeting mitochondrial OxPHOS, such as inhibitors of ETC complexes I, III, and V. As described in the literature (Chan et al., 2015; Konopleva et al., 2016; Bisaillon et al., 2020), we also observed that IDHm AML cells were more sensitive to other types of mitochondrial inhibitor, such as ABT-199.
Consistent with the OxPHOS hyperactivation, we further demonstrated that FAO and CPT1a expression were increased in IDH1m AML cells in a CEBPα-dependent manner. Interestingly, treatment of IDH WT AML cells with 2-HG also increased CPT1a expression. However, while IDHmi systematically abrogated 2-HG production, it maintained or even increased mitochondrial activities (including TCA cycle, OxPHOS capacities) in IDHm AML cell lines and primary patients in vitro and in vivo. Transcriptomic data of AML patient cells harboring IDH mutations obtained from clinical trials for the assessment of IDHmi (NCT01915498, NCT02074839, NCT02381886, and NCT02492737) showed a significant increase in OxPHOS gene signature at relapse. This implies that the maintenance of mitochondrial activities could be a major nongenetic mechanism of adaptive resistance to this targeted therapy. Mechanistically, we showed that IDHmi restored TET2 activity and PTEN expression, induced the inactivation of Akt, and preserved PGC1α, resulting in the maintenance of FAO and OxPHOS phenotypes. This suggests that PGC1α expression and Akt phosphorylation might be useful readouts to monitor the response of IDHmi by flow cytometry in clinics (Scharping et al., 2016). Interestingly, the RAS/MAPK signaling pathway might be activated in nonresponding AML patients or at relapse (Amatangelo et al., 2017; Wang et al., 2020 Preprint; Choe et al., 2020). On the other hand, several studies have shown that mitochondrial metabolism is enhanced in RAS-driven cancers and myeloproliferative neoplasm (Viale et al., 2014; Kong et al., 2019). Of note, N-RAS mutant HL60 cells with IDH1 mutation were more OxPHOS inducible upon treatment with IDHmi than RAS WT MOLM14 cells with IDH1 mutation in our study. The RAS signaling pathway was also targeted by the majority of the miRNAs dysregulated by IDHmi. These observations may link genetic alterations that drive primary therapeutic resistance, RAS signaling, and OxPHOS metabolism, especially in the context of Akt inactivation.
While the small-molecule IDHmi enasidenib (IDH2) and ivosidenib (IDH1) elicit partial responses or complete remission in AML patients, ∼60% of patients are nonresponders (Stein et al., 2017; DiNardo et al., 2018; Pollyea et al., 2019; Roboz et al., 2020). In these clinical settings, several resistance mechanisms have been reported, such as the emergence of second-site IDH2 mutations preventing the binding of the inhibitor (Intlekofer et al., 2018; Choe et al., 2020) or IDH isoform switch from cytosolic IDH1 mutation to mitochondrial IDH2 mutation (Harding et al., 2018; Choe et al., 2020; Wang et al., 2020 Preprint). Therefore, the development of new combinatory therapies with IDHmi is required. Considering the increased OxPHOS features of IDHm AML cells following treatment with IDHmi, we evaluated the antileukemic activity of combining OxPHOS inhibitor (such as IACS-010759) with IDH1mi. Although IACS as single agent had limited antileukemic activity, its association with AG-120 impaired mitochondrial activities, enhanced differentiation, and increased the antileukemic efficacy in PDX models of AML, which may represent a beneficial alternative for AML patients unresponsive to IDHmi.
There are several limitations to our studies that should be taken into consideration. First, the potential toxicity in humans of the duplet combination of a mitochondrial inhibitor with IDHmi should be carefully considered throughout future clinical development. Second, the experimental design protocol for the animal experiments could be challenged. Indeed, we decided to directly treat the mice with both AG-120 and OxPHOS inhibitors for 3 wk, while different therapeutic designs could be considered, such as induction therapy with AG-120 and IACS, then maintenance with AG-120 monotherapy or treatment with AG-120, and then consolidation with IACS. Third, differences in the localization of the two enzymes (cytosolic and mitochondrial), in their capabilities to form dimers with the WT counterpart, or in the amount of 2-HG produced could have pronounced effects on the metabolic reprogramming or the mechanisms of response induced by IDHmi. Lastly, Wang et al. very recently found that AML stemness might be a major driver of primary resistance to IDHmi (Wang et al., 2020 Preprint). Thus, a better understanding of the effects of the combination of OxPHOS inhibitor and IDHmi on leukemic stem cells would help move this duplet therapy into the clinic.
Finally, our study supports the merit of future clinical trials testing the combination of IDHmi and mitochondrial inhibitors with cytarabine, azacitidine, venetoclax, or RAS/MAPK inhibitors. Because this proposed therapeutic strategy could overcome different newly identified mechanisms of resistance to IDHmi, this would be especially relevant as alternative therapeutic approaches for the treatment of those patients who are unresponsive to or are relapsing from IDHmi.
Materials and methods
Primary AML samples
Primary AML patient specimens are from four clinical sites (UPENN, Philadelphia, PA; MDACC at University of Texas, Houston; TUH, Toulouse, France; and IPC, Marseille, France).
For TUH, frozen samples of bone marrow or peripheral blood were obtained from patients diagnosed with AML after signed informed consent in accordance with the Declaration of Helsinki and stored at the HIMIP (Hémopathies malignes de l’Inserm en Midi-Pyrénées) collection (BB-0033-00060). According to French law, the HIMIP biobank collection has been declared to the Ministry of Higher Education and Research (DC 2008–307, collection 1 for HIMIP) and a transfer agreement (AC 2008–129) obtained after approbation by the Comité de Protection des Personnes Sud-Ouest et Outremer II (ethical committee). Clinical and biological annotations of the samples have been declared to the CNIL (Comité National Informatique et Libertés; i.e., Data processing and Liberties National Committee). For the IPC clinical site, ex vivo drug sensitivity was performed on previously frozen (HEMATO-BIO-IPC 2013–015 clinical trial, NCT02320656) or fresh (CEGAL-IPC-2014-012, NCT02619071 clinical trial) mononuclear cell samples from 49 AML patients after informed consent (Table S1). Both trials have been approved by a properly constituted institutional review board (Comité de Protection des Personnes) and by the French National Security Agency of Medicine and Health Products. The samples are subjected to next-generation sequencing to screen for mutations within a selected panel of ∼150 actionable genes in AML (i.e., known to be of prognostic value and/or druggable). For UPENN, AML samples were obtained from patients diagnosed with AML in accordance with US Common Rules at the Stem Cell and Xenograft Core Facility at the UPENN School of Medicine and with informed consent in accordance with institutional guidelines. Peripheral blood or bone marrow samples were frozen in FCS with 10% DMSO and stored in liquid nitrogen. The percentage of blasts was determined by flow cytometry and morphological characteristics before purification.
Patient TUHIM86 (Fig. 3) relapsed following bone marrow allograft and “3+7” in June 2018, when we collected a blood sample corresponding to the sample before IDHmi. Then, following a temporary authorization for use, the patient was treated with two cycles of Vidaza and five cycles of enasidenib until remission. After two more cycles of enasidenib, the patient relapsed in February 2019, when we collected a blood sample corresponding to the sample after IDHmi.
Mice and mouse xenograft model
Animals were used in accordance with a protocol reviewed and approved by the Institutional Animal Care and Use Committee of Région Midi-Pyrénées (France). NSG mice were produced at the Genotoul Anexplo platform at Toulouse, France, using breeders obtained from Charles River Laboratories. Mice were housed in sterile conditions using high-efficiency particulate arrestance filtered microisolators and fed with irradiated food and sterile water.
Mice (6–9 wk old) were sublethally treated with busulfan (20 mg/kg for MOLM14 and 30 mg/kg for PDXs) 24 h before injection of leukemic cells. Leukemia samples were thawed at room temperature, washed twice in PBS, and suspended in Hank’s Balanced Salt Solution at a final concentration of 0.2–10 × 106 cells for PDXs and 2 × 106 for cell line–derived xenograft (CLDX) per 200 µl of Hank’s Balanced Salt Solution per mouse for tail vein injection. Transplanted mice were treated with antibiotic (Baytril) for the duration of the experiment for PDXs. Daily monitoring of mice for symptoms of disease (ruffled coat, hunched back, weakness, and reduced mobility) determined the time of killing for injected animals with signs of distress. If no signs of distress were seen, mice were initially analyzed for engraftment 8 wk after injection except where otherwise noted. Mice were randomly assigned to the different treatment arms based on the blast counts in blood determined 3–8 d before the beginning of the treatment. The randomization was based on the number of mice with validated engraftment (higher than 20 blasts/µl of blood), sex, and body weight. The duration of treatment was determined before starting each experiment unless the animals died before the end of treatment or were euthanized for ethical reasons. During the treatments, total body weight was monitored every 2 d.
In vivo mice treatment CLDX
All mice were treated with doxycycline (200 µg/ml) and sucrose (10 µg/ml) supplemented to the drinking water on the third day after cell injection until the sacrifice of the mice to induce IDH1 WT or IDH1 R132 expression.
For IACS-10759 treatment, IACS-10759 was solubilized in water containing 0.5% methylcellulose before administration to mice. NSG mice were treated daily by gavage of 1.5 mg/kg IACS-10759 (according to weight loss of mice) from day 3 after cell injection for 14 d. For control, NSG mice were treated by daily gavage of vehicle. IACS-10759 was kindly provided by J.R. Marszalek. Mice were monitored for toxicity and were provided nutritional supplements as needed.
In vivo mice treatment PDXs
8–18 wk (PDXs) after AML cell transplantation and when mice were engrafted (tested by flow cytometry on peripheral blood or bone marrow aspirates), NSG mice were treated as described below:
For IACS-10759 treatment, IACS-10759 was solubilized in water containing 0.5% methylcellulose before administration to mice. NSG mice were treated three times a week by gavage of 6 mg/kg IACS-10759 (according to weight loss of mice) for 21 d. For control, NSG mice were treated by daily gavage of vehicle. IACS-10759 was kindly provided by J.R. Marszalek.
For AG-120 treatment, AG-120 (ivosidenib; AGIOS Pharmaceuticals) was solubilized in water containing 0.5% methylcellulose and 0.2% Tween 80 before administration to mice. NSG mice were treated twice a day by gavage of 150 mg/kg AG-120 for 21 d. For control, NSG mice were treated twice a day by gavage of vehicle.
Mice were monitored for toxicity and provided nutritional supplements as needed.
Assessment of leukemic engraftment
NSG mice were humanely killed in accordance with European ethics protocols. Bone marrow (mixed from tibias and femurs) and spleen were dissected and flushed in Hank’s Balanced Salt Solution with 1% FBS. Mononuclear cells from bone marrow and spleen were labeled with mCD45.1-PerCP-Cy5.5 (560580), hCD45-APC (555485) and hCD44-PECy7 (560533) or hCD33-PE (555450; all antibodies from BD Biosciences), and Annexin V-V500 (561501) to determine the fraction of viable human blasts (AnnxV-hCD45+mCD45.1−hCD44+ or AnnxV-hCD45+mCD45.1−hCD33+ cells) using flow cytometry. All antibodies used for cytometry were used at concentrations between 1/50 and 1/200 depending on specificity and cell density. Analyses were performed on a CytoFLEX flow cytometer with CytExpert software (Beckman Coulter) and FlowJo 10.2 (Tree Star). The number of AML cells/µl peripheral blood and number of AML cells in total cell tumor burden (in bone marrow and spleen) were determined by using CountBright beads (Invitrogen) using described manufacturer protocol.
Statistical analyses
Statistical analyses were conducted using Prism software v8.0 (GraphPad Software). For in vitro and in vivo studies, statistical significance was determined by the two-tailed unpaired Student’s t test with Welch’s correction. For transcriptomic analysis of cohorts, statistical significance was determined by the nonparametric Mann-Whitney test. A P value <0.05 was considered statistically significant. For all figures, ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Unless otherwise indicated, all data represent the mean ± SEM from at least three independent experiments. Box-and-whisker plots display all the individual data points as well as the corresponding median. For in vitro metabolomic analysis, Seahorse, and ATP assays, each biological replicate represents the mean of at least two technical replicates.
Cell culture
Unless otherwise specified, all AML cell lines were cultured in minimum essential medium-α (MEMα, 22561–021; Gibco) supplemented with 10% FBS (Invitrogen) in the presence of 100 U/ml of penicillin and 100 µg/ml of streptomycin (1% penicillin/streptomycin [P/S]) and were incubated at 37°C with 5% CO2. The cultured cells were diluted every 2 to 3 d and maintained in an exponential growth phase. Clones from the HL60 cell line expressing either IDH1 WT (#2, #4, and #7) or IDH1 R132H (#3, #5, and #11) were previously generated by our team and are GFP positive (Boutzen et al., 2016). MOLM14 was obtained from M. Carroll in 2011 and engineered to express IDH1 WT or R132H as described below. These cell lines have been routinely tested for mycoplasma contamination in the laboratory.
1-Octyl ester of (R)-2-HG was synthesized at the Organic Synthesis Core (Memorial Sloan Kettering Cancer Center, New York, NY) as previously described (Boutzen et al., 2016) and dissolved in DMSO (maximum [max], 1/1,000 final dilution). Octylalcohol (Sigma-Aldrich) solution in DMSO at the same concentration of (R)-2-HG was used as a control solution to ensure that only the effect of (R)-2-HG–free acid on the cells was observed. AGI-5198 (EMD Millipore) was dissolved in dimethylfuran (max, 1/1,000 final dilution). AG-120 (MedChem) was dissolved in DMSO (max, 1/1,000 final dilution).
Generation of MOLM14 cells with IDH1 mutation by retroviral transduction
At day 0, 4 × 106 293T cells were plated in DMEM+10% FBS and 1% P/S. At day 1, medium was removed from 293T cells, the cells were washed once with PBS, and 7.5 ml OPTIMEM+10% FBS (without P/S) was added. DNA mix for transfection was prepared as follows: for each 10-cm plate, (i) Solution A: 230 µl OPTIMEM (without FBS and P/S) + 20 µl lipofectamine 2000, incubated 5 min at room temperature; (ii) Solution B: 250 µl OPTIMEM + 6 µg DNA of interest + 4 µg p8.91 + 2 µg pVSVG (envelope vector), mixed gently. Solution A was added to solution B (lipofectamine on DNA) without mixing (no bubbles), and the mix was incubated 15 min at room temperature, then added on 293T cells dropwise. After 3–6 h of incubation at 37°C, the supernatant was removed, and 10 ml DMEM+10% FBS+P/S was added. At day 3, after 48-h culture, the lentiviral supernatants were filtered (0.45 µm) and then used straightaway or stored at −80°C. For lentiviral infection by spinoculation, 2 × 106 AML cells were incubated with 2 ml lentiviral supernatant in a 6-well plate. Polybrene at 8 µg/ml was added in each well, then the plate was centrifuged 60 min, 800 g, 37°C. After 4–6 h, the supernatant was removed; cells were washed once with PBS, then diluted in classical culture media. 3 d after lentiviral infection, the cells were selected with hygromycin B (300 µg/ml) for generation of MOLM14 IDH WT or R132H.
Plasmid references
For IDH1 WT (doxycycline inducible), pSLIK-IDH1-FLAG (Addgene Plasmid #66802) was used. For IDH1 R132H (doxycycline inducible), pSLIK-IDH1-R132H-FLAG (Addgene Plasmid #66803) was used. To induce IDH1 WT or R132H expression, cells were treated every 2 d with 0.5 µg/ml doxycycline.
Generation of HL60 and MOLM14 IDH1 WT and R132H knockdown for CEBPα with shRNA and siRNA
HL60 and MOLM14 IDH1 WT and R132H constitutive shCTL and shCEBPα were obtained by retroviral transduction as described for the generation of MOLM14 cells with IDH1 mutation (5′>3′): shRNA control, purchased from Sigma-Aldrich (SHC002 MISSION pLKO.1-puro Non-mammalian shRNA control; 5′-CCGGCAACAAGATGAAGAGCACCAACTC-3′); shRNA CEBPA, purchased from Sigma-Aldrich (SHCLNG-NM_004364, TRCN0000007306; 5′-CCGGGCACGAGACGTCCATCGACATCTCGAGATGTCGATGGACGTCTCGTGCTTTTT-3′).
The transfection of siRNA into HL60 cells was performed using the Neon transfection system (Life Technologies) according to the manufacturer’s recommendations (1400V, 30 s, 1 pulse). After transfection, cells were seeded at 1 × 106 cells/ml for 24 h in culture media. 1.4 µM of siRNA was used per condition for 15 million cells. RNA interference-mediated gene knockdown was achieved using prevalidated Sigma-Aldrich siRNAs (5′>3′) for CEBPα#1 (5′-CGGACUUGGUGCGUCUAAG-3′), and CEBPα#2 (5′-GCAACUCUAGUAUUUAGGA-3′), and Control (5′-UAGCAAUGACGAAUGCGUA-3′).
Chemograms
The drug screening library included 78 substances consisting of conventional chemotherapeutics and a broad range of targeted oncology compounds. Each drug was plated on 96-well plates at four concentrations covering a 1,000-fold concentration range. 10–20,000 patient-derived mononuclear cells were seeded per well in 96-well plates and incubated in the presence of compounds in a humidified environment at 37°C and 5% CO2. After 48 h of treatment, cell viability was measured using the CellTiter-Glo luminescent assay (Promega), and the data were normalized to negative control wells (dimethyl sulfoxide only). Effective half-maximal concentration values were deduced from dose–response curves obtained using GraphPad Prism 6 software (GraphPad Software, Inc.). In this study, only data obtained for metformin and ABT-263 are displayed.
FACS-based cell sorting
Human AML cells from AML-xenografted mice were stained with AnnexinV-V500 and the following fluorescent conjugated antibodies: hCD45-APCH7 (641399) and hCD33-PE (555450; all antibodies from BD Biosciences) by MoFlo Astrios (Beckman Coulter) and FACSMelody (BD Biosciences).
Measurement of oxygen consumption in AML cultured cells in vitro or PDXs ex vivo
Measurement of oxygen consumption was performed using Seahorse Assay All XF assays with the XFe24 Analyzer (Seahorse Bioscience). The day before the assay, the sensor cartridge was placed into the calibration buffer medium (1 ml) supplied by Seahorse Biosciences to hydrate overnight. Seahorse XFe24 plate wells were coated with 50 µl of Cell-Tak (Cat #354240; Corning) solution at a concentration of 22.4 µg/ml and kept at 4°C overnight. Then, Cell-Tak–coated Seahorse plate wells were rinsed with distillated water (700 µl), and AML cells were plated at a density of 2 × 105 cells per well (in vitro experiments) or 4 × 105 cells per well (in vivo experiments after cell sorting) in 50 µl XF base minimal DMEM media containing 5.6 mM glucose, 1 mM pyruvate, and 2 mM glutamine (Seahorse media). The plate was spanned at 80 g for 5 min, and then 450 µl of Seahorse media was added into each well carefully. After 1-h incubation at 37°C in CO2-free atmosphere, basal OCR (as a respiration indicator) and ECAR (as a glycolysis indicator) were measured using the XFe24 analyzer with Seahorse Cell Mito Stress Assay–recommended parameters.
The ATP-linked respiration rate was determined by the difference between the starting OCR and the OCR after adding oligomycin (first injection; 1-µM final concentration). The Spare Respiratory Capacity was calculated as the difference between maximum respiration obtained after adding carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP; second injection; 2-µM final concentration) and basal OCR. The mitochondrial respiration rate was determined by the difference between the starting OCR and the OCR after adding AA/rotenone mixture (third injection; 1 µM for each drug).
ATP analysis
ATP was measured using the Promego Cell Titer Glo kit (G9242) and corresponding manufacturer’s protocol. In summary, following treatment, 24,000 cells (cell lines) or 40,000 cells (primary samples) were resuspended in 80 µl and distributed in a 96-well plate. Cells were then treated in replicates of three with 20 µl control (PBS) or with sodium iodoacetate (IA; final concentration 100 µM) both alone and in combination with oligomycin (OA; final concentration 1.6 µM) + AA (final concentration 100 µM). Following 1-h incubation at 37°C with 5% CO2, 100 µl of Cell Titer Glo reaction mix was added to each well for a final volume of 200 µl. Plates were then analyzed for luminescence with the microplate reader CLARIOstar (BMG Labtech). Total ATP was calculated as the difference between PBS and IA + (OA+AA) conditions. Mitochondrial ATP was calculated as the percentage of total ATP corresponding to the difference between IA and IA + (OA+AA).
Enzymatic activity measurements
MDH and IDH enzymatic activities were measured as previously described (Gibon et al., 2004). Protocols were optimized for AML cell lines in order to determine the right amount of cells for each assay.
NAD-MDH
The activity of MDH was measured using a continuous assay, in which 2,000 extracted cells were mixed with 100 µl of 100 mM Tricine/KOH, pH 8, 0.1 mM EDTA, 0.05% Triton 100%, NADH 1.2 mM, and oxaloacetate 0 (blanks) or 1.125 mM (substrate saturation). Absorbance was measured at 340 nm and at 37°C until rates were stabilized.
NAD-IDH
The activity of IDH was measured using a continuous assay, in which 8,000 extracted cells were mixed in 100 µl of 100 mM Tricine/KOH, pH 8, 8 mM MgCl2, 0.05% Triton 100%, NAD+ 3 mM, and isocitrate 0 (blanks) or 3 mM (substrate saturation). Absorbance was measured at 340 nm and at 37°C until rates were stabilized.
Respiratory chain enzyme activities were measured on a Roche Diagnostics/Hitachi Modular P analyzer based on methods described previously (Kramer and Nowak, 1988; Krähenbühl et al., 1996) and adapted to match apparatus requirements. The activities of all the complexes in each sample were normalized by the amount of protein to allow sample comparison.
Complex I activity
Briefly, reduced nicotinamide adenine dinucleotide phosphate–ubiquinone reductase activity (complex I) was measured by following the disappearance of nicotinamide adenine dinucleotide phosphate using rotenone as a specific inhibitor to ensure the specificity of the assay.
Complex II activity
Complex II activity, succinate-ubiquinone reductase, was assayed through the reduction of 2,6-dichlorophenolindophenol, a final electron acceptor, after the addition of succinate.
Complex III activity
The activity of complex III, ubiquinone–cytochrome c reductase, was determined by assaying the rate of reduction of cytochrome c.
Complex IV activity
The cytochrome c oxidase (complex IV) activity was based on the same assay as for complex III using potassium cyanide to inhibit the activity of this enzyme.
Complex V activity
Complex V activity was measured according to a method coupling ADP production to NADH disappearance through the conversion of phosphoenolpyruvate into pyruvate, then into lactate (Rustin et al., 1993).
Citrate synthase activity
The activity of citrate synthase was assayed as described previously (Itoh and Srere, 1970) with the reduction of 5′-5′-dithiobis 2-nitrobenzoic acid caused by the deacetylation of acetyl-CoA.
Mitochondrial and nuclear genome copy number analysis
Total DNA (nuclear and mitochondrial) was extracted and purified from 2 × 106 cells using QIAamp DNA Mini Kit (51304; Qiagen) according to the manufacturer’s instructions. Next, the concentration and purity of DNA were determined. mtDNA is quantified by taking the ratio between a target mitochondrial gene and a reference nuclear gene (mtDNA/nDNA) using quantitative real time PCR, as described previously (Gonzalez-Hunt et al., 2016). Briefly, a RT-PCR using primers targeting a mitochondrial gene (mito fragment of tRNA-Leu [UUR] gene) and a nuclear gene (nuc fragment of β2-microglobulin gene) designed by Venegas and Halberg (2012) was performed. Then relative mtDNA content was calculated using the comparative Ct method with the following formula:
Cell death assay
After treatment, 5 × 105 cells were washed with PBS and resuspended in 200 µl of Annexin-V binding buffer (BD Biosciences). 2 μl of Annexin-V-BV421 or V500 (BD Biosciences) and 7-amino-actinomycin D (Sigma-Aldrich) were added for 15 min at room temperature in the dark. All samples were analyzed using the CytoFLEX flow cytometer.
Measurement of MMP and mitochondrial mass
MMP in viable cells (in vitro cell lines) or in viable human CD45+CD33+ blasts (in vivo PDXs) was measured by flow cytometry using a tetramethylrhodamine ethyl ester (TMRE) probe. Rapidly, cells were incubated with 200 nM TMRE and for in vivo experiments with CD45 and CD45.1 antibodies. After 20-min incubation at 37°C, cells were washed twice and labeled with anxV before flow cytometry analysis. Negative control was obtained by incubating cells with 50 µM FCCP for 15 min before TMRE and antibody labeling. MMP corresponds to the mean fluorescence intensity (MFI) of TMRE in viable cells in the TMRE+ population (gate designed with FCCP samples). To represent different results from PDXs on the same figure, the MMPs of human blasts of PDX models (MFI in hCD45+ TMRE+) were normalized to MMPs of mice cells (MFI in mCD45.1+ TMRE+) for each sample in vivo.
Mitochondrial mass in viable cells (in vitro cell lines) or in viable human hCD45+ blasts (in vivo PDXs) was measured by flow cytometry using MitoTracker Red (MTR; for in vitro experiment, as HL60 IDH clones are GFP positive) or MitoTracker Green (MTG; for PDXs) probes. Briefly, cells were incubated with 20 nM MTR for in vitro experiments and with 100 nM MTG and CD45, CD45.1 antibodies for in vivo experiments for 20 min at 37°C. Then, cells were washed twice and labeled with anxV and analyzed by flow cytometry. Mitochondrial mass corresponds to the MFI of MTR or MTG in viable cells in vitro. To represent different results from PDXs on the same figure, MMPs of human blasts of PDX models (MFI in hCD45+) were normalized to MMPs of mice cells (MFI in mCD45.1+) for each sample in vivo.
Measurement of differentiation in PDXs
Differentiation in viable human CD45+CD33+ blasts (in vivo PDXs) was measured by flow cytometry using the following fluorescently conjugated antibodies: APC conjugated–CD15 (BD Biosciences) and PE conjugated–CD11b (Beckman Coulter). Rapidly, cells were incubated with CD15, CD11b, CD45, and CD33 antibodies. After 20-min incubation at 37°C, cells were washed twice and labeled with anxV before flow cytometry analysis.
Metabolomic analysis for in vitro analyses
For sampling of central metabolites by fast filtration (Fig. 1), 3 million cells were harvested by vacuum filtration (nylon, 0.45-µm pore size, 47 mm; Sartorius) and washed with the same volume of NaCl (0.9%, 4°C) solution (the whole filtration procedure including the washing was completed in <20 s). Metabolism was quenched by placing the filter containing the cells in a centrifugation tube filled with 3 ml of a cold (−80°C) mixture of 80% methanol and 20% water and 200 µl isotope-dilution mass spectrometry (MS) for absolute quantification. After 15 min at −80°C, the samples were centrifuged (2,000 g, −10°C, 5 min), and the supernatant was collected. The cell pellet was extracted twice more with 1 ml of the quenching solution and centrifuged 5 min, and the 2 ml supernatant was added to the former one. The samples were lyophilized, dissolved in 200 µl water, centrifuged (2,000 g, 4°C, 5 min) to remove the debris and then stored at −80°C until liquid chromatography (LC)/MS analysis.
Ion chromatography–MS analysis of intracellular central metabolites was performed as described previously (Scotland et al., 2013). Briefly, analysis was performed by high-performance anion exchange chromatography (Dionex ICS 2000 system) coupled to a triple quadrupole QTrap 4000 (Applied Biosystems) mass spectrometer. All samples were analyzed in the negative mode giving [M-H]− ions, which were monitored in the multiple reaction monitoring mode with a multiple reaction monitoring detection window of 600 s, a target Scan Time of 2 s, a pause of 5.007 ms, and a duration of 74.8 min. The following electrospray ionization source parameters were applied: ion spray voltage, −3.3 kV; curtain gas, 20 psi; nebulizer gas pressure, 40 psi; desolvation gas pressure, 50 psi; and temperature nebulization, 650°C. The injection volume was 15 µl. Samples were injected on a Dionex IonPac AS11 column (250 × 2 mm) equipped with a Dionex AG11 guard column (50 × 2 mm). Mobile phase was composed of a KOH gradient that varied as follows: 0 min, 0.5; 1 min, 0.5; 9.5 min, 4.1; 14.6 min, 4.1; 24 min, 9.65; 36 min, 60; 36.1 min, 90; 43 min, 90; 43.1 min, 0.5; and 45 min, 0.5. Data treatment was done with Analyst Software 3.5 (Thermo Fisher Scientific).
Sampling of acyl-CoAs by fast filtration (Fig. 1) was performed as follows: 10 million cells were harvested by vacuum filtration (nylon, 0.45-µm pore size, 47 mm; Sartorius) and washed with the same volume of NaCl (0.9%, 4°C) solution (the whole filtration procedure including the washing was completed in <20 s). Metabolism was quenched by placing the filter containing the cells in a centrifugation tube filled with 10 ml of a cold (−20°C) mixture of 80% methanol and 20% water with 125 mM formic acid (pH 3). The samples were vortexed and sonicated 10 s successively three times and kept 10 min on ice, then centrifuged (2,000 g, −10°C, 5 min), and the supernatant was transferred to glass tubes. The samples were evaporated with a RotaVap overnight and resolubilized with 50 µl of 25 mM ammonium formate with 2% methanol, then vortexed and sonicated, transferred to Eppendorf and centrifuged (20,000 g, 10 min, 4°C), and finally transferred into vials and stored at −80°C until analysis.
Analysis of intracellular acyl-CoAs was performed by LC (HPLC U3000; Dionex) coupled with an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization probe. MS analyses were performed in the positive FTMS mode at a resolution of 60,000 (at m/z 400) with the following source parameters: capillary temperature was 250°C, source heater temperature was 390°C, sheath gas flow rate was 50, auxiliary gas flow rate was 10, S-Lens RF level was 70%, and source voltage was 4 kV. Samples were injected on a Phenomenex Kinetex C-18 column (100 mm × 3.0 mm; 1.7-µm particle size). Solvent A was 50 mM ammonium formate, and solvent B was methanol at a flow rate of 400 µl min−1. The solvent B was varied as follows: 0 min, 2%; 3 min, 2%; 23 min, 25%; 23.1 min, 80%; 25 min, 80%; 25.1 min, 2%; and 32 min, 2%. The volume of the injection was 5 µl. Identification was determined by extracting the accurate mass of acyl-CoAs with a mass accuracy of 5 ppm. The internal standard (IS) used corresponds to U-13C–acyl-CoAs (isotope-dilution MS method). Data treatment was done with TraceFinder Software (Thermo Fisher Scientific).
LC/MS analysis of intracellular 2-HG (Fig. S1) and TCA cycle intermediates (Fig. 3) was performed as follows: Frozen cell pellets (containing 3 × 106 cells) were suspended in 750 µl methanol/water/IS solution (500 µl methanol [Sigma-Aldrich] and 250 µl water [Milli-Q]), spiked with 2-morpholinoenthanesulfonic acid (MES; Cat #341–01622; Dojindo) at 10-µM final concentration). The samples were transferred to lysing tubes (Cat #VK05-2ml; Bertin Instruments), prechilled at −80°C for 1 h, and subjected to 40-s lysing using the Precellys 24 instrument (Bertin Instruments). Next, 400 µl of chloroform (Sigma-Aldrich) was added to the samples, followed by vortexing for 10 s and centrifugation (15,000 g, 15 min, 4°C). The resulting upper phase was collected and lyophilized, while the middle protein-containing phase was collected, lysed in 200 µl 1% SDS and 50 mM-Tris buffer, and quantified (BCA Protein Assay Kit, Cat #23225; Thermo Fisher Scientific). The lyophilized upper phase was dissolved in methanol/water/formic acid solution (375 µl methanol [Sigma-Aldrich]; 125 µl water [Milli-Q]; and 0.5 µl formic acid [Sigma-Aldrich]) and then loaded on a Captiva ND Lipid plate (Cat #A59640002; Agilent Technologies). The filtrate was collected, divided in two separate vials, and lyophilized. One vial was further used for 2-HG measurements, while the other for quantification of TCA metabolites. For 2-HG analysis, lyophilized samples were suspended in 50 µl diacetyl-L–tartaric anhydride (50 mg/ml; Sigma-Aldrich) solution composed of 4 volumes acetonitrile and 1 volume acetic acid for derivatization. The samples were then placed at 75°C for 2 h, followed by lyophilization and final solubilization in 50 µl MS-grade water (Sigma-Aldrich). For TCA analysis, lyophilized samples were dissolved in 50 µl MS-grade water. 10 μl of each sample was injected on a UPLC column Acquity HSS T3, 2.1 × 100 mm (Cat #186003539; Waters) coupled to triple-quadrupole MS (LCMS-8050; Shimadzu). Samples were run using a gradient of the following solvents: solvent A (2 mM ammonium formate solution in water, pH 3.5) and solvent B (methanol/0.1% formic acid). The gradient was 1 min/100%A; 2.75 min/20%A; 4.25 min/20%A; 5.00 min/10%A; 6.00 min/97%A; and 10.00 min/97%A. The flow rate was 0.6 ml/min, column oven set at 40°C. For 2-HG analysis, the following transitions were monitored: 2-HG (RT: L-2-HG 2.1 min/D-2-HG 2.3 min): 363.0 m/z > 147.1 m/z (collision energy [CE]: 10V)/129.1 m/z (CE: 24V); lactate (RT: L-lactate 2.3 min/D-lactate 2.4 min): 305.0 m/z > 89.1 m/z (CE: 7V); and MES (IS, RT: 0.6 min): 194.0 m/z > 80.2 m/z (CE: 24V). For TCA analysis, the following transitions were monitored: malate (RT: 0.67 min) 132.8 m/z > 115.20 m/z (CE: 16V)/132.8 m/z > 71.2 m/z (CE: 17V), 132.8 m/z > 73.2 m/z (CE: 17V); succinate (RT: 0.64 min) 116.9 m/z > 73.1 m/z (CE: 14V)/116.9 m/z > 99.9 m/z (CE 26V)/116.9 m/z > 99.0 m/z (CE 14V); fumarate (RT: 0.64 min) 114.8 m/z > 27.1 m/z (CE: 25V)/114.8 m/z > 71.1 m/z (CE: 13V); citrate (RT: 0.63 min) 190.9 m/z > 111.2 m/z (CE: 14V)/190.9 m/z > 87.2 m/z (CE: 17V), 190.9 m/z > 85.2 m/z (CE: 17V); α-ketoglutarate (RT: 0.67 min) 144.9 m/z > 101.0 m/z (CE: 11V)/144.9 m/z > 57.2 m/z (CE: 12V)/144.9 m/z > 100.0 m/z (CE: 28V); and MES (same as above). The identities of the enantiomers were confirmed using pure standards (all purchased from Sigma-Aldrich). For data analysis, areas under the respective peaks were integrated using Lab Solutions software (version 5.9; Shimadzu). Each target compound area was then normalized (divided) by the area of the IS to produce area ratio or normalized area.
Metabolomic analysis in mice serums following mono/duplet therapies
Sampling of serum
Mice blood was collected into EDTA-coated tubes and kept at 4°C after dissection. The tubes were centrifuged at 3,000 rpm for 5 min at 4°C, and the serum was collected and stored at −80°C.
Extraction of metabolites
Metabolite extraction was performed on frozen serum (−80°C) of mice. 50 µl was mixed with 200 µl frozen methanol, then sonicated for 5 min and mixed at 4°C for 20 min. Next, samples were vortexed 2 min, centrifuged at 15,000 g for 30 min at 4°C, and dried under nitrogen with Turbovap (Biotage) at 30°C. Finally, samples were resuspended in 45 µl of mobile phase A (see below), sonicated, vortexed, and centrifuged at 4°C for 5 min at 20,000 g. The resulting supernatants were then transferred into vials and further mixed with 105 μl of acetonitrile and 5 µl of an external standard mixture containing 20 µg/ml of 13C5-2-HG, 13C5-α-KG, 13C4-succinic acid, and 13C4-aspartic acid.
Intracellular metabolite quantification
LC–high resolution mass spectrometry quantification of 2-HG, aspartic acid, and lactic acid was performed using an Ultimate 3000 chromatographic system coupled with a Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific) fitted with an electrospray source. Chromatographic separation was performed on a Sequant ZIC-pHILIC column (5 µm, 2.1 × 150 mm; Merck) maintained at 45°C. Mobile phase A consisted of an aqueous buffer of 10 mM of ammonium acetate, while mobile phase B was made of 100% acetonitrile. Chromatographic elution was achieved at a flow rate of 200 µl/min. After injection of 10 µl of sample, elution started with an isocratic step of 2 min at 30% A, followed by a linear gradient from 30% to 60% of phase A in 5 min. The chromatographic system was then rinsed for 5 min at 0% B, and the run ended with an equilibration step of 8 min. The column effluent was directly introduced into the electrospray source of the mass spectrometer, and analyses were performed in the negative ion mode. Source parameters were as follows: capillary voltage, −2.5 kV; capillary temperature, 350°C; and sheath gas and auxiliary gas pressures set at 35 and 10 arbitrary units, respectively. The detection was performed from m/z 50 to 600 at a resolution of 70,000 (full width at half-maximum at m/z 200) using an AGC target of 1e6 and a maximum injection time of 250 ms. 2-HG, aspartic acid, and lactic acid were detected as deprotonated [M-H]− species at m/z 147.0299, 132.0302, and 89.0244, respectively. 2-HG and aspartic acid were quantified by isotope dilution using the corresponding 13C-labeled homologues as ISs, while lactic acid was quantified using 13C4-succinic acid.
AG-120 quantification in mice sera
The concentration of AG-120 in plasma was determined using nonvalidated LC with tandem MS methods. A 10-µl aliquot of plasma was mixed with 60 µl of acetonitrile containing the IS for protein precipitation. The mixture was vortexed at 800 rpm for 4 min and centrifuged at 4,000 rpm for 10 min at 4°C. A 50-µl aliquot of supernatant was mixed with 50 µl of Milli-Q water. A 10-µl aliquot was injected onto ACE C18 (3 µm, 50 × 2.1 mm) set at 40°C. AG-120 was eluted using a standard gradient of 0.1% formic acid in water and 0.1% formic acid in acetonitrile at a flow rate of 0.6 ml/min. The following mass transition, 583.2→214.0, was used to measure AG-120. Calibration standards and quality control samples were prepared in blank mouse plasma for analyzing AG-120 concentrations in plasma.
Data were acquired using Analyst 1.6.3 (AB Sciex). The standard curve had a coefficient of determination (R2) value >0.98 in a linear regression with 1/X2 weighting. The quality control samples had a precision and accuracy within 20% of theoretical values. The peak area ratios of analyte relative to IS were used for AG-120 quantitation. Linearity was achieved in the AG-120 concentration range from 1 ng/ml to 2,500 ng/ml.
Mitochondria purification
Mitochondria were purified by differential centrifugation. Briefly, 50 × 106 cells were washed three times and resuspended in mitochondria isolation buffer (MIB; 200 mM mannitol, 70 mM sucrose, 0.5 mM EGTA, 10 mM Hepes, pH 7.2, and 0.2% FA-free BSA [A6003, Sigma-Aldrich]) supplemented with 1× protease/phosphatase inhibitor cocktail (PPC1010; Sigma-Aldrich). Next, cells were ground using a 1-ml Tissue Grinder Dounce (357538; Wheaton) and centrifuged at 600 g for 10 min twice. Then, supernatant was centrifuged at 10,400 g for 10 min. The pellet was resuspended in MIB and centrifuged at 10,400 g for 10 min; this step was repeated three times. Finally, the pellet was resuspended in MIB and centrifuged at 16,000 g for 2 min to obtain the mitochondria fraction. During all the purification, samples were always maintained at 4°C. To assess the purity of the mitochondria fraction, an immunoblot analysis of mitochondria (TFAM), nuclear (PCNA), endoplasmic reticulum (PDI), cytoskeleton (tubulin), and cytosol (GAPDH, HSP90) protein expression in total lysate and mitochondria fraction was performed.
Western blot analysis of total proteins and mitochondria purified proteins
For immunoblot assay, cells, purified mitochondria, or total lysate from mitochondria purification was first subjected to lysis in NuPAGE LDS Sample Buffer. Total protein content from every sample was measured using Pierce BCA Protein Assay Kit according to the manufacturer’s recommendations. Sample lysates were then loaded onto NuPAGE 4–12% Bis-Tris Protein Gels (10, 12, or 15 wells; between 10 and 40 µg proteins). After electrophoresis (1 h, 160V), proteins were transferred to nitrocellulose membranes (70 min, 110V). The transblotted membranes were blocked for 1 h with Tris-buffered saline 0.1%, Tween 20%, 5% BSA, or 10% milk and then probed with appropriate primary antibodies (dilution as recommended by manufacturers in 5% BSA or 10% milk) overnight at 4°C. Next, the membranes were washed three times for a total of 30 min and then incubated with secondary antibodies (dilution factor = 5,000 in 5% BSA or 10% milk) at room temperature for 45 min. After another three washes of 10 min, immunoreactive bands were visualized by enhanced chemiluminescence (ECL Supersignal West Pico; Thermo Fisher Scientific) with a Syngene camera. Quantification of chemiluminescent signals was done with the GeneTools software from Syngene and normalized to their corresponding loading control. Antibodies for immunoblotting were purchased from the following sources: Akt (Cat #9272S; RRID:AB_329827), PSer473Akt (Cat #4060; RRID:AB_2315049), AMPKα (Cat #5831; RRID: AB_ 10622186), PThr172AMPKα (Cat #2535; RRID: AB_ 3312502535), CEBPα (Cat #8178; RRID:AB_11178517), COXIV (Cat #4850; RRID:AB_2085424), CPT1A (Cat #12252; RRID: AB_2797857), FLAG (Cat #14793; RRID:AB_ 2572291), GAPDH (Cat #5174; RRID:AB_ 10622025), HSP90 (Cat #4874; RRID:AB_ 21212144), IDH1 (Cat #8137; RRID:AB_ 10950504), mTOR (Cat #2983; RRID:AB_2105622), PSer2448mTOR (Cat #5536; RRID:AB_10691552), PCNA (Cat #2586; RRID:AB_ 2160343), PGC1a (Cat #2178; RRID:AB_823600), PKA (Cat #5842; RRID:AB_ 10706172), PThr197PKA (Cat #5661; RRID:AB_10707163), PTEN (Cat #9188; RRID:AB_2253290), and tubulin (Cat #2148; RRID:AB_2288042) from Cell Signaling Technology; PSer380PTEN (Cat #2143-S) from Epitomics; Total OxPHOS cocktail CI-II-III-V (Cat #ab110411; RRID:AB_2756818), and TFAM (Cat #ab119684; RRID: AB_10900340) from Abcam; Actin (Cat #MAB1501; RRID:AB_ 2223041) from Millipore; IDH1 R132H (Cat #DIA-H09; RRID:AB_ 2335716) from Dianova; CPT2 (Cat #HPA028201; RRID:AB_10599177), and SLC25A20 (Cat #HPA029863; RRID:AB_10602893) from Sigma-Aldrich. HRP conjugate anti-rabbit (W4011) and anti-mouse (W4021) secondary antibodies were purchased from Promega.
Determination of FAO
For Fig. 2 C, cells were incubated in duplicate for 3 h at 37°C with [1-14C] palmitate (0.1 mCi/ml; PerkinElmer) and cold palmitate (80 µM). Incubation buffer also contained 125 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1.25 mM KH2PO4, 1.25 mM MgSO4, 25 mM NaHCO3, 10 mM L-Carnitine, and 3% FA-free BSA (A6003; Sigma-Aldrich). After incubation, FAO was measured by 14CO2 trapped in 300 µl of 1 M benzethonium hydroxide. Radioactivity of 14CO2 was determined by liquid scintillation counting and normalized for cell number.
For Fig. S3 C, FAO was measured in biological tri- or quadriplicates by quantifying the production of 3H2O from [9,10-3H]palmitate (NET043001MC; Perkin Elmer). Briefly, the day before, cells were diluted at 0.4 million cells/ml and treated with AG-5198 or vehicle (dimethylformamide; DMF). Then, the day of the experiment, 200 µl of cells was collected after homogenization of the flasks, and an aliquot was kept to determine the number of cells in each flask by Trypan blue exclusion method. Then, the cells contained in the 200 µl collected were washed two times with Dulbecco’s PBS and incubated with 200 µl of [9,10(n)-3H]palmitic acid (60 Ci/mmol; NEN) bound to FA-free albumin (final concentration 125 µM) containing 1 mM carnitine. After 3-h incubation at 37°C, the mixture was removed and added to a tube containing 200 µl of cold 10% (wt/vol) TCA. The tubes were centrifuged for 10 min at 2,200 g at 4°C, and aliquots of supernatants (350 µl) were removed, mixed with 55 µl of 6 M NaOH, and applied to ion-exchange resin (DOWEX). The columns were washed twice with 750 µl of water, and the eluates were counted (five technical countings). Palmitate oxidation rates were expressed as counts per minute per cell and normalized to the number of cells contained in the 200 µl collected from each flask.
FAO-coupled OCR (for Fig. 2 M and Fig. S2 N) was performed as follows: To determine the part of the OCR dependent on FAO, the same protocol as for the measure of mitochondrial respiration already described was performed, except that wells were duplicated and 3 µM of etomoxir was added to half of them after 45 min of resting and 15 min before the OCR measurement.
5-OH-methylcytosine measures using flow cytometry
We used the protocol previously described by Figueroa et al. (2010). Briefly, 106 HL60 cells (IDH1 WT or R132H) was used for each condition. After wash with PBS, cells were stained with Zombie Aqua (#423101; Biolegend; 1/1,000e) for 15 min at room temperature, then washed with PBS. Cells were fixed and permeabilized with IntraPrep Permabilization Reagent (#A07803; Beckman Coulter) as described by the manufacturer. Cells were then treated with 2N HCL for 20 min at room temperature in the dark and subsequently neutralized with 100 mM Tris-HCl, pH 8.0, for 20 min. After wash with PBS, cells were incubated with anti–5-OH-methylcytosine (#39770, Active Motif; 1:400) or IgG Rabbit antibody (#2729S; Cell Signaling; 1:400) for 30 min at room temperature in the dark. After wash with PBS, cells were incubated with secondary antibodies coupled with ALEXA FLUOR 555 (#A21428; Invitrogen) for 30 min in the dark. Cells were then washed three times in PBS and analyzed with CytoFlex. Data analysis was done using CytExpert.
ChIP assays
ChIP assays were mainly performed as previously described (Riscal et al., 2016). Briefly, 107 cells were cross-linked in 1% formaldehyde/1% paraformaldehyde for 5 min, followed by addition of 125 mM glycine to stop the reaction. Cells were then washed in PBS, resuspended in lysis buffer (10 mM Tris, pH 8, 140 mM NaCl, 0.1% SDS, 0.5% Triton X-100, 0.05% NaDoc, 1 mM EDTA, 0.5 mM EGTA, and protease inhibitors), and chromatin was sheared by sonication. qChIPs were performed by incubating cell lysates (Input) with 20 µl of protein G–Dynabeads and 5 µg of antibody. The same amount of rabbit IgGs (Santa Cruz) was used for control ChIP experiments. After overnight incubation, washing, reverse cross-linking, and treatment with both RNase A and Proteinase K, proteins were removed with phenol/chloroform extraction, and DNA was recovered using the NucleoSpin Extract II kit. Input and IP DNA was then analyzed by quantitative PCR using the SYBR Green Master mix on a LightCycler 480 SW 1.5 apparatus (Roche). Results are represented as the mean value of at least three independent experiments of IP chromatin (calculated as a percentage of the input) with the indicated antibodies after normalization by a control ChIP performed with rabbit IgGs.
For CEBPα Chip (CEBP binding site predicted by Genomatix): CPT1a: (forward) 5′-CATGAAAGAGTCAGGGGTGAA-3′, (reverse) 5′-GTGCGCATAAAATCGCTTAG-3′; CPT2: (forward) 5′-TGAGGGTTCTGAGGTTCCTG-3′, (reverse) 5′-ACAGCCTGCCCAGGTGAG-3′; SLC25A20: (forward) 5′-ACCAGCCAAAACCCATCAG-3′, (reverse) 5′-CTCACACATGCCCCTCTTCT-3′; and for H3K27/H3K4 Chip (CEBP binding site predicted by Genomatix): available upon request.
IP of PGC1α
For IP, 25 million cells were washed with PBS and lysed using 340 µl Lysis Buffer (20 mmol/L TRIS, pH 7.5, 5 mmol/liter EGTA, 150 mmol/liter NaCl, 0.5% NP-40, phosphatase, and protease inhibitors 1×) on ice for 20 min, followed by centrifugation at 16,000 g at 4°C for 15 min. 40 µl of the supernatant was kept for the input fractions, and the rest was incubated overnight with 5 µg IgG mouse antibody (#sc-2025; Santa Cruz) or PGC1α antibody (#ST1202; Sigma-Aldrich) at 4°C overnight, followed by precipitation using magnetic DynaBead G (Invitrogen; prewashed twice with lysis buffer) for 1 h at 4°C. 40 µl of the supernatant was kept for the supernatant fraction, and the rest was discarded. Then, the beads were washed three times with lysis buffer and 50 µl of LDS 2× buffer, and 10 µl reducing agent was added. The samples were heated 4 min at 95°C, and 12 µl was used right away for immunoblots of total phosphoserine (#P5747; Sigma-Aldrich).
Clonogenic assays
AML cells (1,000 for cell lines and 100,000 for primary samples) were plated in tri- or quadriplicate in 35-mm Petri dishes in H4230 Stem Cell Technologies methylcellulose medium (#H4230) supplemented with 10% of 5637 cell line–conditioned medium, 10% IMDM, and the different reagents. After an incubation of 7 d for cell lines and between 14 d and 30 d for primary samples in a humidified CO2 incubator, the numbers of colonies with ≥20 cells were counted manually under the microscope. One dish served for photographs, one other was collected for flow cytometry, and the rest were potentially replated. For photographs, 400 µl of 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT) at 10 mg/ml was gently added to the methylcellulose and incubated for an additional 2 h at 37°C in a humidified CO2 incubator.
Transcriptomic analysis
Genome-scale metabolic network analysis for the prediction of the reaction activity
We used the computational method iMat proposed by the Shlomi group (Zur et al., 2010) in order to identify the metabolic reactions that are specifically active in HL60 IDH1 WT and IDH1 R132H cells, respectively. Under the hypothesis that reactions associated with significantly highly expressed genes are more likely to be active (i.e., to carry a nonzero flux), the principle of the iMat method is to find, within the global metabolic human network, a consistent network of reactions (i.e., a network in which all reactions can be active) that includes a maximal number of reactions associated with highly expressed genes and a minimal number of reactions associated with not significantly expressed genes. Using this method, the activity of a reaction is not only predicted from the expression level of its associated gene but is also inferred from the activity of downstream and upstream reactions in the network.
We used the gene expression data obtained by microarray analyses for HL60 IDH1 WT and IDH1 R132H cells (E-MTAB-4327; Boutzen et al., 2016) and the generic human metabolic network reconstruction Recon2 (Thiele et al., 2013; version 2.04, downloaded from http://vmh.uni.lu/#downloadview), in which we added the reaction for the production of the oncometabolite 2-HG. The transcripts were classified in two categories (highly expressed or not expressed) based on their expression level using the DABG (Detection Above BackGround) method from the BioConductor package “oligo” in R. This information was mapped to the Recon2 network reactions by using the Gene-Protein-Reaction association rules defined in the network reconstruction (Thiele et al., 2013) to identify the reactions associated with highly or not expressed genes.
We then applied the iMat algorithm, which consists of solving a mixed integer linear programming optimization problem, to find steady-state flux distributions that (1) maximize the number of reactions whose predicted flux is consistent with the measured expression level of their associated gene(s) (i.e., a nonzero flux for a reaction associated with a highly expressed gene and a zero flux for a reaction associated with a nonsignificantly expressed gene) and (2) comply with stoichiometric and thermodynamic constraints. One predicted flux distribution corresponds to a set of active reactions (i.e., reactions carrying a nonzero flux) representing a consistent and fully connected subnetwork. We modified the original version of the algorithm by adding some constraints to account for the measured growth rates of the cells and the uptake and secretion rates of metabolites calculated from exometabolomic data.
Several distinct alternative subnetworks having the same adequacy with experimental data were identified for each cell type (IDH1 WT and IDH1 R132H). Reactions were considered as specifically active (respectively inactive) in IDH1 R132H cells if they were predicted to be active (respectively inactive) in all possible subnetworks obtained for IDH1 R132H cells but either active or inactive in IDH1 WT subnetworks. Pathway enrichment statistics were performed on these two sets of reactions using a one-tailed exact Fisher test, with a Bonferroni correction for multiple tests, using the metabolic pathways defined in Recon2. Visualization was achieved using the MetExplore webserver (Chazalviel et al., 2018).
Publicly accessible transcriptomic databases of AML patients used in this study
GEO accession no. GSE14468 (Verhaak cohort; Verhaak et al., 2009)
Data were from Affymetrix Human Gene GeneChip U133 Plus 2 arrays. Patients who did not receive intensive chemotherapy from FAB M3 or with TET2 or CEBPα mutations were removed (as it had been shown that TET2 and CEBPα are regulated epigenetically by 2-HG). Therefore, 321 IDH1 WT and 31 IDH1 or 68 IDH1+2 mutated patients were considered.
BeatAML cohort (Tyner et al., 2018)
Data were from RNA sequencing experiments. Patients with TET2 or CEBPα mutations were removed as well as patients with no information on IDH status. Therefore, 97 IDH1 WT and 16 IDH1 or 26 IDH1+2 mutated patients were considered.
TCGA cohort (Ley et al., 2013)
Data were from Affymetrix Human Gene GeneChip U133 Plus 2 arrays. Patients who did not receive intensive chemotherapy or were from FAB M3 were removed. Therefore, 90 IDH1 WT and 18 IDH1m patients were considered.
GEO accession no. GSE153348 (Wang et al., 2020 Preprint)
Data were from RNA sequencing experiments using the HiSeq 2000 platform on longitudinal samples during the IDHmi therapy. Transcriptomes from 41 patients harboring an IDH mutation before treatment and 10 patients at relapse (seven paired pretreatment/relapse) from four clinical trials (NCT01915498 [enasidenib for IDH2m patients]; NCT02074839 [ivosidenib for IDH1m patients]; NCT02381886 [IDH305 for IDH1m patients]; and NCT02492737 [AG-881 for IDH1m or IDH2m patients]) were analyzed. To score a gene signature S within each sample, we applied the “gene sets overall expression” proposed by Jerby-Arnon et al. (2018). This method first z-scores the genes across all samples and estimates an enrichment score (ES) by averaging out z-scores of all genes that belong to S. To account for noise, the ES is normalized by using random sets of genes compatible with S (i.e., same number of genes and same distribution of gene average expression).
GSEA was performed using the GSEA v2.0 tool developed by the Broad Institute (Subramanian et al., 2005). Signatures are available on the Broad Institute website and correspond to the ones used in our previous study (Farge et al., 2017). The normalized ESs were computed for the ranked genes from the CPT1aHIGH (above median) compared with CPT1aLOW (below median) in IDH WT, IDH1m, and IDH1+2 mutant primary samples (Fig. 2 E) and from IDH1m HL60 compared with WT cells (Fig. S2 H).
miRNA enrichment pathways analysis following AG-120 treatment
For prediction of miRNA targets, we used the freely available bioinformatics tool DIANA or DNA intelligent analysis (http://diana.imis.athena-innovation.gr/DianaTools). More precisely, we used DIANA miRPath v.3.0 for investigating the effect of AG-5198 treatment in IDH1m cells on the modulation of miRNAs associated to various biologically relevant pathways. The DIANA-miRPath web server was able to predict miRNA targets (in coding or 3′–untranslated regions) using the DIANA-microT-CDS algorithm. The default settings of P value 0.001 and threshold score 0.8 were used (Vlachos et al., 2012).
Complete blood count measurements
Platelet number, mean platelet volume, hematocrit, and red blood cell count were measured with 20 µl of mouse blood using a Yumizen H500 analyzer (HORIBA Ltd).
Online supplemental material
Fig. S1 shows that IDH1m cells exhibit a higher sensitivity to OxPHOSi and BCL2i due to their enhanced mitochondrial capabilities and OxPHOS activity in AML. Fig. S2 shows that methylation- and CEBPα-dependent mitochondrial FAO is increased in IDH1m cells. Fig. S3 shows that IDHmi reverse 2-HG production but do not necessarily decrease high OxPHOS phenotype and mitochondrial metabolism. Fig. S4 shows that treatment with an OxPHOS inhibitor enhances antileukemic effects of IDHmi in IDH1m primary samples ex vivo and in vivo. Fig. S5 shows the effect of the combination of OxPHOS inhibitor and IDHmi on differentiation, mitochondrial readouts, mice weight, complete blood count, and murine hematopoietic cells in IDH1m PDXs in vivo. Table S1 lists clinical and mutational features of AML patient samples used in this study. Table S2 lists the miRNAs dysregulated in IDH1m AML cells following IDHmi.
Supplementary Material
presents clinical and mutational features of AML patient samples used in this study.
lists miRNAs dysregulated in IDH1 mutant AML cells following IDHmi.
Acknowledgments
We thank The Cancéropoles PACA and GSO, the Network MetaboCancerGSO, Karine Marendziak, all members of mice core facilities (UMS006, ANEXPLO, Institut National de la Santé et de la Recherché Médicale [INSERM], Toulouse) for their support and technical assistance, and Audrey Sarry, Prof. Véronique De Mas, and Eric Delabesse for the management of the Biobank BRC-HIMIP (Biological Resources Centres–Inserm Midi-Pyrénées “Cytothèque des hémopathies malignes”), which is supported by Cancer Pharmacology of Toulouse-Oncopole and Région (CAPTOR). We thank Anne-Marie Benot, Muriel Serthelon, and Stéphanie Nevouet for their daily help with the administrative and financial management of the Sarry laboratory. We thank the Institut Paoli Calmettes Direction de la Recherche Clinique et de l'Innovation, the Hematology Department, and the IPC/Centre de Recherches en Cancérologie de Marseille Biological Resource Center for sample collection and processing. We also thank the patients and their families.
This work was also supported by grants from the Région Midi-Pyrénées (FLEXAML, J-E. Sarry), the Plan Cancer Biologie des Systèmes 2014 (FLEXAML; J-E. Sarry), the Laboratoire d'Excellence Toulouse Cancer (TOUCAN; contract ANR11-LABEX), the (MetaboCancerGSO; J-E. Sarry, J-C. Portais, and L. Le Cam) the Program Hospitalo-Universitaire en Cancérologie (CAPTOR; contract ANR11-PHUC0001), La Ligue Nationale de Lutte Contre le Cancer, the Fondation ARC, the Fondation Toulouse Cancer Santé, and the Association G.A.E.L. Metatoul (Metabolomics & Fluxomics Facilities, Toulouse, France; http://www.metatoul.fr) and LEMM are part of the national infrastructure MetaboHUB-ANR-11-INBS-0010 (The French National Infrastructure for Metabolomics and Fluxomics; http://www.metabohub.fr). Metatoul is supported by grants from the Région Midi-Pyrénées, the European Regional Development Fund, the SICOVAL, the Infrastructures en Biologie Santé et Agronomie (France), the Centre National de la Recherche Scientifique and the Institut National de la Recherche Agronomique. The project has been partly supported by Canceropole PACA, SIRIC (grant INCa-Inserm-DGOS 6038 2012-2017), and Canceropôle-SIRIC EmA (INCA 2017-024-COLLETTE and INCa 2017-024-MAL). C. Montersino was supported by SIRIC (grant INCa-Inserm-DGOS 6038 2012-2017) and Canceropôle-SIRIC EmA. E. Turtoi was supported by SIRIC-Montpellier Cancer Program (grant INCa-Inserm-DGOS_12553). N. Baran and M. Konopleva were supported by funding from Leukemia Spore grant P50 (CA100632-16). V. Pancaldi is supported by a chair of Bioinformatics in Oncology of the CRCT, and A. Hucteau has a fellowship from the PhD program of the Laboratoire d'Excellence Toulouse Cancer. Work in A. Turtoi’s laboratory is supported by a SIRIC Montpellier Cancer Grant (INCa_Inserm_DGOS_12553), the Foundation de France (grant no. 00078461), and a LabEx MabImprove Starting Grant.
Author contributions: L. Stuani, M. Sabatier, and J-E. Sarry designed the experiments. L. Stuani, M. Sabatier, E. Saland, G. Cognet, C. Bosc, E. Boet, N. Broin, C. Larrue, N. Baran, M.Y. Cissé, M. Conti, S. Loric, A. Zavoriti, C. Cassan, M. Hosseini, and L.K. Linares performed in vitro and ex vivo experiments. M. Sabatier, E. Saland, C. Bosc, G. Cognet, A. Sahal, E. Boet, T. Farge, P.-L. Mouchel, and M. Gotanègre performed in vivo experiments. C. Montersino and R. Castellano performed chemogrammes’ analysis. L. Stuani, F. Castelli, L. Gales, E. Turtoi, E. Chu-Van, A. Turtoi, and G. Cazals performed metabolomic analyses. N. Poupin, L. Fernando, and F. Jourdan performed genome-scale metabolic network analysis. L. Stuani, T. Farge, T. Kaoma, A. Hucteau, F. Wang, K. Takahashi, and V. Pancaldi performed transcriptomic and methylome analysis on publicly available cohorts. AML bone marrow and blood samples were provided by M. Carroll, N. Vey, and Y. Collette; P. Bories and C. Récher; and C.D. DiNardo, and M. Konopleva. J.R. Marszalek and M. Konopleva provided IACS-010759. B. Nicolay and S. Ronseaux provided and measured AG-120 in mice sera. L. Le Cam, F. Fenaille, A. Turtoi, Y. Gibon, C.D. DiNardo, M. Konopleva, L.K. Linares, F. Bellvert, and J-C. Portais managed the resources and shared their expertise. L. Stuani and J-E. Sarry wrote the manuscript, designed the figures, and managed the revision. N. Poupin, L. Le Cam, M.A. Selak, F. Fenaille, F. Jourdan, L.K. Linares, C. Récher, and J-C. Portais reviewed the manuscript. L. Stuani and J-E. Sarry directed the research.
References
- Alvarez-Calderon, F., Gregory M.A., Pham-Danis C., DeRyckere D., Stevens B.M., Zaberezhnyy V., Hill A.A., Gemta L., Kumar A., Kumar V., et al. 2015. Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations. Clin. Cancer Res. 21:1360–1372. 10.1158/1078-0432.CCR-14-2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatangelo, M.D., Quek L., Shih A., Stein E.M., Roshal M., David M.D., Marteyn B., Farnoud N.R., de Botton S., Bernard O.A., et al. 2017. Enasidenib induces acute myeloid leukemia cell differentiation to promote clinical response. Blood. 130:732–741. 10.1182/blood-2017-04-779447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccelli, I., Gareau Y., Lehnertz B., Gingras S., Spinella J.-F., Corneau S., Mayotte N., Girard S., Frechette M., Blouin-Chagnon V., et al. 2019. Mubritinib Targets the Electron Transport Chain Complex I and Reveals the Landscape of OXPHOS Dependency in Acute Myeloid Leukemia. Cancer Cell. 36:84–99.e8. 10.1016/j.ccell.2019.06.003 [DOI] [PubMed] [Google Scholar]
- Bhalla, K., Hwang B.J., Dewi R.E., Ou L., Twaddel W., Fang H.B., Vafai S.B., Vazquez F., Puigserver P., Boros L., and Girnun G.D.. 2011. PGC1α promotes tumor growth by inducing gene expression programs supporting lipogenesis. Cancer Res. 71:6888–6898. 10.1158/0008-5472.CAN-11-1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy, K., Wang T., Chen W.W., Freinkman E., Abu-Remaileh M., and Sabatini D.M.. 2015. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 162:540–551. 10.1016/j.cell.2015.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaillon, R., Moison C., Thiollier C., Krosl J., Bordeleau M.-E., Lehnertz B., Lavallée V.-P., MacRae T., Mayotte N., Labelle C., et al. 2020. Genetic characterization of ABT-199 sensitivity in human AML. Leukemia. 34:63–74. 10.1038/s41375-019-0485-x [DOI] [PubMed] [Google Scholar]
- Boroughs, L.K., and DeBerardinis R.J.. 2015. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 17:351–359. 10.1038/ncb3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutzen, H., Saland E., Larrue C., de Toni F., Gales L., Castelli F.A., Cathebas M., Zaghdoudi S., Stuani L., Kaoma T., et al. 2016. Isocitrate dehydrogenase 1 mutations prime the all-trans retinoic acid myeloid differentiation pathway in acute myeloid leukemia. J. Exp. Med. 213:483–497. 10.1084/jem.20150736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.M., Thomas D., Corces-Zimmerman M.R., Xavy S., Rastogi S., Hong W.-J., Zhao F., Medeiros B.C., Tyvoll D.A., and Majeti R.. 2015. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med. 21:178–184. 10.1038/nm.3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, A., Herbst L., Pusch S., Klett L., Goparaju R., Stichel D., Kaulfuss S., Panknin O., Zimmermann K., Toschi L., et al. 2017. Pan-mutant-IDH1 inhibitor BAY1436032 is highly effective against human IDH1 mutant acute myeloid leukemia in vivo. Leukemia. 31:2020–2028. 10.1038/leu.2017.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi, A., Goparaju R., Gupta C., Weder J., Klünemann T., Araujo Cruz M.M., Kloos A., Goerlich K., Schottmann R., Othman B., et al. 2020. In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site. Leukemia. 34:416–426. 10.1038/s41375-019-0582-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazalviel, M., Frainay C., Poupin N., Vinson F., Merlet B., Gloaguen Y., Cottret L., and Jourdan F.. 2018. MetExploreViz: web component for interactive metabolic network visualization. Bioinformatics. 34:312–313. 10.1093/bioinformatics/btx588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe, S., Wang H., DiNardo C.D., Stein E.M., de Botton S., Roboz G.J., Altman J.K., Mims A.S., Watts J.M., Pollyea D.A., et al. 2020. Molecular mechanisms mediating relapse following ivosidenib monotherapy in IDH1-mutant relapsed or refractory AML. Blood Adv. 4:1894–1905. 10.1182/bloodadvances.2020001503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman, M.R., and Majeti R.. 2014. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia. 28:2276–2282. 10.1038/leu.2014.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuyàs, E., Fernández-Arroyo S., Corominas-Faja B., Rodríguez-Gallego E., Bosch-Barrera J., Martin-Castillo B., De Llorens R., Joven J., and Menendez J.A.. 2015. Oncometabolic mutation IDH1 R132H confers a metformin-hypersensitive phenotype. Oncotarget. 6:12279–12296. 10.18632/oncotarget.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo, C., de Botton S., Pollyea D.A., Stein E.M., Fathi A.T., Roboz G.J., Collins R., Swords R.T., Flinn I.W., Altman J.K., et al. 2015. Molecular Profiling and Relationship with Clinical Response in Patients with IDH1 Mutation-Positive Hematologic Malignancies Receiving AG-120, a First-in-Class Potent Inhibitor of Mutant IDH1, in Addition to Data from the Completed Dose Escalation Portion. Blood. 126:1306. 10.1182/blood.V126.23.1306.1306 [DOI] [Google Scholar]
- DiNardo, C.D., Stein E.M., de Botton S., Roboz G.J., Altman J.K., Mims A.S., Swords R., Collins R.H., Mannis G.N., Pollyea D.A., et al. 2018. Durable Remissions with Ivosidenib in IDH1-Mutated Relapsed or Refractory AML. N. Engl. J. Med. 378:2386–2398. 10.1056/NEJMoa1716984 [DOI] [PubMed] [Google Scholar]
- Elkashef, S.M., Lin A.-P., Myers J., Sill H., Jiang D., Dahia P.L.M., and Aguiar R.C.T.. 2017. IDH Mutation, Competitive Inhibition of FTO, and RNA Methylation. Cancer Cell. 31:619–620. 10.1016/j.ccell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farge, T., Saland E., de Toni F., Aroua N., Hosseini M., Perry R., Bosc C., Sugita M., Stuani L., Fraisse M., et al. 2017. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 7:716–735. 10.1158/2159-8290.CD-16-0441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farshidfar, F., Zheng S., Gingras M.-C., Newton Y., Shih J., Robertson A.G., Hinoue T., Hoadley K.A., Gibb E.A., Roszik J., et al. Cancer Genome Atlas Network . 2017. Integrative Genomic Analysis of Cholangiocarcinoma Identifies Distinct IDH-Mutant Molecular Profiles. Cell Rep. 19:2878–2880. 10.1016/j.celrep.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouille, N., Bassil C.F., Ben-Sahra I., Benajiba L., Alexe G., Ramos A., Pikman Y., Conway A.S., Burgess M.R., Li Q., et al. 2017. The creatine kinase pathway is a metabolic vulnerability in EVI1-positive acute myeloid leukemia. Nat. Med. 23:301–313. 10.1038/nm.4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., et al. 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 18:553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan, W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suvà M.L., and Bernstein B.E.. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 529:110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Chin R.M., Vergnes L., Hwang H., Deng G., Xing Y., Pai M.Y., Li S., Ta L., Fazlollahi F., et al. 2015. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell Metab. 22:508–515. 10.1016/j.cmet.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallipoli, P., Giotopoulos G., Tzelepis K., Costa A.S.H., Vohra S., Medina-Perez P., Basheer F., Marando L., Di Lisio L., Dias J.M.L., et al. 2018. Glutaminolysis is a metabolic dependency in FLT3ITD acute myeloid leukemia unmasked by FLT3 tyrosine kinase inhibition. Blood. 131:1639–1653. 10.1182/blood-2017-12-820035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman, S.J., Naser F., Mahieu N.G., McKenzie L.D., Dunn G.P., Chheda M.G., and Patti G.J.. 2018. Consumption of NADPH for 2-HG Synthesis Increases Pentose Phosphate Pathway Flux and Sensitizes Cells to Oxidative Stress. Cell Rep. 22:512–522. 10.1016/j.celrep.2017.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines, Z., Dominy J.E. Jr., Blättler S.M., Jedrychowski M.P., Banks A.S., Lim J.H., Chim H., Gygi S.P., and Puigserver P.. 2011. The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol. Cell. 44:851–863. 10.1016/j.molcel.2011.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon, Y., Blaesing O.E., Hannemann J., Carillo P., Höhne M., Hendriks J.H.M., Palacios N., Cross J., Selbig J., and Stitt M.. 2004. A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell. 16:3304–3325. 10.1105/tpc.104.025973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Hunt, C.P., Rooney J.P., Ryde I.T., Anbalagan C., Joglekar R., and Meyer J.N.. 2016. PCR-based analysis of mitochondrial DNA copy number, mitochondrial DNA damage, and nuclear DNA damage. Curr. Protoc. Toxicol. 67:11.1–: 25.. 10.1002/0471140856.tx2011s67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian, A.R., Parker S.J., Davidson S.M., Divakaruni A.S., Green C.R., Zhang X., Slocum K.L., Pu M., Lin F., Vickers C., et al. 2014. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 74:3317–3331. 10.1158/0008-5472.CAN-14-0772-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, M.A., Nemkov T., Reisz J.A., Zaberezhnyy V., Hansen K.C., D’Alessandro A., and DeGregori J.. 2018. Glutaminase inhibition improves FLT3 inhibitor therapy for acute myeloid leukemia. Exp. Hematol. 58:52–58. 10.1016/j.exphem.2017.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilhamon, P., Eskandarpour M., Halai D., Wilson G.A., Feber A., Teschendorff A.E., Gomez V., Hergovich A., Tirabosco R., Fernanda Amary M., et al. 2013. Meta-analysis of IDH-mutant cancers identifies EBF1 as an interaction partner for TET2. Nat. Commun. 4:2166. 10.1038/ncomms3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, J.J., Lowery M.A., Shih A.H., Schvartzman J.M., Hou S., Famulare C., Patel M., Roshal M., Do R.K., Zehir A., et al. 2018. Isoform Switching as a Mechanism of Acquired Resistance to Mutant Isocitrate Dehydrogenase Inhibition. Cancer Discov. 8:1540–1547. 10.1158/2159-8290.CD-18-0877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollinshead, K.E.R., Munford H., Eales K.L., Bardella C., Li C., Escribano-Gonzalez C., Thakker A., Nonnenmacher Y., Kluckova K., Jeeves M., et al. 2018. Oncogenic IDH1 Mutations Promote Enhanced Proline Synthesis through PYCR1 to Support the Maintenance of Mitochondrial Redox Homeostasis. Cell Rep. 22:3107–3114. 10.1016/j.celrep.2018.02.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, S., Li W.Y., Tseng A., Beerman I., Elia A.J., Bendall S.C., Lemonnier F., Kron K.J., Cescon D.W., Hao Z., et al. 2016. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell. 30:337–348. 10.1016/j.ccell.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer, A.M., Shih A.H., Wang B., Nazir A., Rustenburg A.S., Albanese S.K., Patel M., Famulare C., Correa F.M., Takemoto N., et al. 2018. Acquired resistance to IDH inhibition through trans or cis dimer-interface mutations. Nature. 559:125–129. 10.1038/s41586-018-0251-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, H., and Srere P.A.. 1970. A new assay for glutamate-oxaloacetate transaminase. Anal. Biochem. 35:405–410. 10.1016/0003-2697(70)90202-2 [DOI] [PubMed] [Google Scholar]
- Izquierdo-Garcia, J.L., Cai L.M., Chaumeil M.M., Eriksson P., Robinson A.E., Pieper R.O., Phillips J.J., and Ronen S.M.. 2014. Glioma cells with the IDH1 mutation modulate metabolic fractional flux through pyruvate carboxylase. PLoS One. 9:e108289. 10.1371/journal.pone.0108289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby, L., Shlomi T., and Ruppin E.. 2010. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 6:401. 10.1038/msb.2010.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon, L., Shah P., Cuoco M.S., Rodman C., Su M.J., Melms J.C., Leeson R., Kanodia A., Mei S., Lin J.R., et al. 2018. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell. 175:984–997.e24. 10.1016/j.cell.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, B., Zhang J., Xia J., Zhao W., Wu Y., Shi M., Luo L., Zhou H., Chen A., Ma H., et al. 2017. IDH1 Mutation Promotes Tumorigenesis by Inhibiting JNK Activation and Apoptosis Induced by Serum Starvation. Cell Rep. 19:389–400. 10.1016/j.celrep.2017.03.053 [DOI] [PubMed] [Google Scholar]
- Kats, L.M., Reschke M., Taulli R., Pozdnyakova O., Burgess K., Bhargava P., Straley K., Karnik R., Meissner A., Small D., et al. 2014. Proto-oncogenic role of mutant IDH2 in leukemia initiation and maintenance. Cell Stem Cell. 14:329–341. 10.1016/j.stem.2013.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, G., You X., Wen Z., Chang Y.I., Qian S., Ranheim E.A., Letson C., Zhang X., Zhou Y., Liu Y., et al. 2019. Downregulating Notch counteracts KrasG12D-induced ERK activation and oxidative phosphorylation in myeloproliferative neoplasm. Leukemia. 33:671–685. 10.1038/s41375-018-0248-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva, M., Pollyea D.A., Potluri J., Chyla B., Hogdal L., Busman T., McKeegan E., Salem A.H., Zhu M., Ricker J.L., et al. 2016. Efficacy and Biological Correlates of Response in a Phase II Study of Venetoclax Monotherapy in Patients with Acute Myelogenous Leukemia. Cancer Discov. 6:1106–1117. 10.1158/2159-8290.CD-16-0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krähenbühl, S., Schäfer T., and Wiesmann U.. 1996. Determination of the activities of the enzyme complexes of the electron transport chain in human fibroblasts. Clin. Chim. Acta. 253:79–90. 10.1016/0009-8981(96)06338-3 [DOI] [PubMed] [Google Scholar]
- Kramer, P., and Nowak T.. 1988. The preparation and characterization of Cr(III) and Co(III) complexes of GDP and GTP and their interactions with avian phosphoenolpyruvate carboxykinase. J. Inorg. Biochem. 32:135–151. 10.1016/0162-0134(88)80022-9 [DOI] [PubMed] [Google Scholar]
- Ley, T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J. Jr., Laird P.W., Baty J.D., et al. Cancer Genome Atlas Research Network . 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Monks B., Ge Q., and Birnbaum M.J.. 2007. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature. 447:1012–1016. 10.1038/nature05861 [DOI] [PubMed] [Google Scholar]
- Li, F., He X., Ye D., Lin Y., Yu H., Yao C., Huang L., Zhang J., Wang F., Xu S., et al. 2015. NADP(+)-IDH Mutations Promote Hypersuccinylation that Impairs Mitochondria Respiration and Induces Apoptosis Resistance. Mol. Cell. 60:661–675. 10.1016/j.molcel.2015.10.017 [DOI] [PubMed] [Google Scholar]
- Lim, J.H., Gerhart-Hines Z., Dominy J.E., Lee Y., Kim S., Tabata M., Xiang Y.K., and Puigserver P.. 2013. Oleic acid stimulates complete oxidation of fatty acids through protein kinase A-dependent activation of SIRT1-PGC1α complex. J. Biol. Chem. 288:7117–7126. 10.1074/jbc.M112.415729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman, J.-A., Looper R.E., Koivunen P., Lee S., Schneider R.K., McMahon C., Cowley G.S., Root D.E., Ebert B.L., and Kaelin W.G. Jr. 2013. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 339:1621–1625. 10.1126/science.1231677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C., Ward P.S., Kapoor G.S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C.R., Khanin R., Figueroa M.E., Melnick A., et al. 2012. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 483:474–478. 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci, G., Maharry K., Wu Y.-Z., Radmacher M.D., Mrózek K., Margeson D., Holland K.B., Whitman S.P., Becker H., Schwind S., et al. 2010. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 28:2348–2355. 10.1200/JCO.2009.27.3730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis, E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K., Koboldt D.C., Fulton R.S., Delehaunty K.D., McGrath S.D., et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361:1058–1066. 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matre, P., Velez J., Jacamo R., Qi Y., Su X., Cai T., Chan S.M., Lodi A., Sweeney S.R., Ma H., et al. 2016. Inhibiting glutaminase in acute myeloid leukemia: metabolic dependency of selected AML subtypes. Oncotarget. 7:79722–79735. 10.18632/oncotarget.12944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, J.R., Sun Y., Protopopova M., Gera S., Bandi M., Bristow C., McAfoos T., Morlacchi P., Ackroyd J., Agip A.A., et al. 2018. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 24:1036–1046. 10.1038/s41591-018-0052-4 [DOI] [PubMed] [Google Scholar]
- Mugoni, V., Panella R., Cheloni G., Chen M., Pozdnyakova O., Stroopinsky D., Guarnerio J., Monteleone E., Lee J.D., Mendez L., et al. 2019. Vulnerabilities in mIDH2 AML confer sensitivity to APL-like targeted combination therapy. Cell Res. 29:446–459. 10.1038/s41422-019-0162-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ok, C.Y., Loghavi S., Sui D., Wei P., Kanagal-Shamanna R., Yin C.C., Zuo Z., Routbort M.J., Tang G., Tang Z., et al. 2019. Persistent IDH1/2 mutations in remission can predict relapse in patients with acute myeloid leukemia. Haematologica. 104:305–311. 10.3324/haematol.2018.191148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okoye-Okafor, U.C., Bartholdy B., Cartier J., Gao E.N., Pietrak B., Rendina A.R., Rominger C., Quinn C., Smallwood A., Wiggall K.J., et al. 2015. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat. Chem. Biol. 11:878–886. 10.1038/nchembio.1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palam, L.R., Mali R.S., Ramdas B., Srivatsan S.N., Visconte V., Tiu R.V., Vanhaesebroeck B., Roers A., Gerbaulet A., Xu M., et al. 2018. Loss of epigenetic regulator TET2 and oncogenic KIT regulate myeloid cell transformation via PI3K pathway. JCI Insight. 3:e94679. 10.1172/jci.insight.94679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil, E., Gerstung M., Bullinger L., Gaidzik V.I., Paschka P., Roberts N.D., Potter N.E., Heuser M., Thol F., Bolli N., et al. 2016. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 374:2209–2221. 10.1056/NEJMoa1516192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi, M.A., and Clayton D.A.. 1991. Similarity of human mitochondrial transcription factor 1 to high mobility group proteins. Science. 252:965–969. 10.1126/science.2035027 [DOI] [PubMed] [Google Scholar]
- Pollyea, D.A., Tallman M.S., de Botton S., Kantarjian H.M., Collins R., Stein A.S., Frattini M.G., Xu Q., Tosolini A., See W.L., et al. 2019. Enasidenib, an inhibitor of mutant IDH2 proteins, induces durable remissions in older patients with newly diagnosed acute myeloid leukemia. Leukemia. 33:2575–2584. 10.1038/s41375-019-0472-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupin, N., Corlu A., Cabaton N.J., Dubois-Pot-Schneider H., Canlet C., Person E., Bruel S., Frainay C., Vinson F., Maurier F., et al. 2018. Large-Scale Modeling Approach Reveals Functional Metabolic Shifts during Hepatic Differentiation. J. Proteome Res. 18:204–216. 10.1021/acs.jproteome.8b00524 [DOI] [PubMed] [Google Scholar]
- Puigserver, P., Rhee J., Donovan J., Walkey C. J., Yoon J Cliff, Oriente F., Kitamura Y., Altomonte J., Dong Hengjiang, Accili D., et al. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 423:550–555. 10.1038/nature01667 [DOI] [PubMed] [Google Scholar]
- Quek, L., David M.D., Kennedy A., Metzner M., Amatangelo M., Shih A., Stoilova B., Quivoron C., Heiblig M., Willekens C., et al. 2018. Clonal heterogeneity of acute myeloid leukemia treated with the IDH2 inhibitor enasidenib. Nat. Med. 24:1167–1177. 10.1038/s41591-018-0115-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riscal, R., Schrepfer E., Arena G., Cissé M.Y., Bellvert F., Heuillet M., Rambow F., Bonneil E., Sabourdy F., Vincent C., et al. 2016. Chromatin-Bound MDM2 Regulates Serine Metabolism and Redox Homeostasis Independently of p53. Mol. Cell. 62:890–902. 10.1016/j.molcel.2016.04.033 [DOI] [PubMed] [Google Scholar]
- Roboz, G.J., DiNardo C.D., Stein E.M., de Botton S., Mims A.S., Prince G.T., Altman J.K., Arellano M.L., Donnellan W., Erba H.P., et al. 2020. Ivosidenib induces deep durable remissions in patients with newly diagnosed IDH1-mutant acute myeloid leukemia. Blood. 135:463–471. 10.1182/blood.2019002140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohle, D., Popovici-Muller J., Palaskas N., Turcan S., Grommes C., Campos C., Tsoi J., Clark O., Oldrini B., Komisopoulou E., et al. 2013. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 340:626–630. 10.1126/science.1236062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustin, P., Lebidois J., Chretien D., Bourgeron T., Piechaud J.-F., Rötig A., Sidi D., and Munnich A.. 1993. The investigation of respiratory chain disorders in heart using endomyocardial biopsies. J. Inherit. Metab. Dis. 16:541–544. 10.1007/BF00711676 [DOI] [PubMed] [Google Scholar]
- Samudio, I., Harmancey R., Fiegl M., Kantarjian H., Konopleva M., Korchin B., Kaluarachchi K., Bornmann W., Duvvuri S., Taegtmeyer H., and Andreeff M.. 2010. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Invest. 120:142–156. 10.1172/JCI38942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, M., Knobbe C.B., Itsumi M., Elia A.J., Harris I.S., Chio I.I.C., Cairns R.A., McCracken S., Wakeham A., Haight J., et al. 2012. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 26:2038–2049. 10.1101/gad.198200.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharping, N.E., Menk A.V., Moreci R.S., Whetstone R.D., Dadey R.E., Watkins S.C., Ferris R.L., and Delgoffe G.M.. 2016. The Tumor Microenvironment Represses T Cell Mitochondrial Biogenesis to Drive Intratumoral T Cell Metabolic Insufficiency and Dysfunction. Immunity. 45:374–388. 10.1016/j.immuni.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotland, S., Saland E., Skuli N., de Toni F., Boutzen H., Micklow E., Sénégas I., Peyraud R., Peyriga L., Théodoro F., et al. 2013. Mitochondrial energetic and AKT status mediate metabolic effects and apoptosis of metformin in human leukemic cells. Leukemia. 27:2129–2138. 10.1038/leu.2013.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford, D.B., and Shaw R.J.. 2009. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 9:563–575. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, D., Cathelin S., Mirali S., Di Trani J.M., Yanofsky D.J., Keon K.A., Rubinstein J.L., Schimmer A.D., Ketela T., and Chan S.M.. 2019. Inhibition of mitochondrial translation overcomes venetoclax resistance in AML through activation of the integrated stress response. Sci. Transl. Med. 11:eaax2863. 10.1126/scitranslmed.aax2863 [DOI] [PubMed] [Google Scholar]
- Shlush, L.I., Zandi S., Mitchell A., Chen W.C., Brandwein J.M., Gupta V., Kennedy J.A., Schimmer A.D., Schuh A.C., Yee K.W., et al. HALT Pan-Leukemia Gene Panel Consortium . 2014. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 506:328–333. 10.1038/nature13038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skrtić, M., Sriskanthadevan S., Jhas B., Gebbia M., Wang X., Wang Z., Hurren R., Jitkova Y., Gronda M., Maclean N., et al. 2011. Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell. 20:674–688. 10.1016/j.ccr.2011.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, E.M., DiNardo C.D., Pollyea D.A., Fathi A.T., Roboz G.J., Altman J.K., Stone R.M., DeAngelo D.J., Levine R.L., Flinn I.W., et al. 2017. Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood. 130:722–731. 10.1182/blood-2017-04-779405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, E.M., DiNardo C.D., Fathi A.T., Pollyea D.A., Stone R.M., Altman J.K., Roboz G.J., Patel M.R., Collins R., Flinn I.W., et al. 2019. Molecular remission and response patterns in patients with mutant-IDH2 acute myeloid leukemia treated with enasidenib. Blood. 133:676–687. 10.1182/blood-2018-08-869008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, L.B., Gui D.Y., Hosios A.M., Bush L.N., Freinkman E., and Vander Heiden M.G.. 2015. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 162:552–563. 10.1016/j.cell.2015.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi, K., Wakimoto H., Iafrate A.J., Tanaka S., Loebel F., Lelic N., Wiederschain D., Bedel O., Deng G., Zhang B., et al. 2015. Extreme Vulnerability of IDH1 Mutant Cancers to NAD+ Depletion. Cancer Cell. 28:773–784. 10.1016/j.ccell.2015.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele, I., Swainston N., Fleming R.M.T., Hoppe A., Sahoo S., Aurich M.K., Haraldsdottir H., Mo M.L., Rolfsson O., Stobbe M.D., et al. 2013. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31:419–425. 10.1038/nbt.2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan, S., Makarov V., Taranda J., Wang Y., Fabius A.W.M., Wu W., Zheng Y., El-Amine N., Haddock S., Nanjangud G., et al. 2018. Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nat. Genet. 50:62–72. 10.1038/s41588-017-0001-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyner, J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. 2018. Functional genomic landscape of acute myeloid leukaemia. Nature. 562:526–531. 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden, M.G., and DeBerardinis R.J.. 2017. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 168:657–669. 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venegas, V., and Halberg M.C.. 2012. Measurement of mitochondrial DNA copy number. Methods Mol. Biol. 837:327–335. 10.1007/978-1-61779-504-6_22 [DOI] [PubMed] [Google Scholar]
- Verhaak, R.G.W., Wouters B.J., Erpelinck C.A.J., Abbas S., Beverloo H.B., Lugthart S., Löwenberg B., Delwel R., and Valk P.J.M.. 2009. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 94:131–134. 10.3324/haematol.13299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale, A., Pettazzoni P., Lyssiotis C.A., Ying H., Sánchez N., Marchesini M., Carugo A., Green T., Seth S., Giuliani V., et al. 2014. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 514:628–632. 10.1038/nature13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius, J.V., and Scarpulla R.C.. 1994. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: a potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc. Natl. Acad. Sci. USA. 91:1309–1313. 10.1073/pnas.91.4.1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos, I.S., Kostoulas N., Vergoulis T., Georgakilas G., Reczko M., Maragkakis M., Paraskevopoulou M.D., Prionidis K., Dalamagas T., and Hatzigeorgiou A.G.. 2012. DIANA miRPath v.2.0: investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 40(W1):W498-504. 10.1093/nar/gks494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F., Travins J., DeLaBarre B., Penard-Lacronique V., Schalm S., Hansen E., Straley K., Kernytsky A., Liu W., Gliser C., et al. 2013. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 340:622–626. 10.1126/science.1234769 [DOI] [PubMed] [Google Scholar]
- Wang, F., Morita K., DiNardo C.D., Furudate K., Tanaka T., Yan Y., Patel K.P., MacBeth K.J., Wu B., Liu G., et al. 2020. Leukemia stemness and co-occurring mutations drive resistance to IDH inhibitors in acute myeloid leukemia Lead contact. bioRxiv. (Preprint posted October 27, 2020) 10.1101/2020.10.27.357111 [DOI] [PMC free article] [PubMed]
- Wu, Z., Puigserver P., Andersson U., Zhang C., Adelmant G., Mootha V., Troy A., Cinti S., Lowell B., Scarpulla R.C., and Spiegelman B.M.. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 98:115–124. 10.1016/S0092-8674(00)80611-X [DOI] [PubMed] [Google Scholar]
- Xu, W., Yang H., Liu Y., Yang Y., Wang P., Kim S.H., Ito S., Yang C., Wang P., Xiao M.T., et al. 2011. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 19:17–30. 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, K., Travins J., Wang F., David M.D., Artin E., Straley K., Padyana A., Gross S., DeLaBarre B., Tobin E., et al. 2017. AG-221, a First-in-Class Therapy Targeting Acute Myeloid Leukemia Harboring Oncogenic IDH2 Mutations. Cancer Discov. 7:478–493. 10.1158/2159-8290.CD-16-1034 [DOI] [PubMed] [Google Scholar]
- Zur, H., Ruppin E., and Shlomi T.. 2010. iMAT: an integrative metabolic analysis tool. Bioinformatics. 26:3140–3142. 10.1093/bioinformatics/btq602 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents clinical and mutational features of AML patient samples used in this study.
lists miRNAs dysregulated in IDH1 mutant AML cells following IDHmi.