Abstract
PTEN and p53 are highly mutated in many cancers. These two tumor suppressors have critical functions in the nucleus, such as DNA repair, cell cycle progression, and genome maintenance. However, the in vivo functional relationship of nuclear PTEN and p53 is unknown. Here, we analyzed the liver of mice in which nuclear PTEN and p53 are individually or simultaneously depleted. We found that nuclear PTEN loss greatly upregulates p53 expression upon oxidative stress, while the loss of p53 potentiates stress-induced accumulation of PTEN in the nucleus. Next, we examined oxidative stress-induced DNA damage in hepatocytes, and found that nuclear PTEN loss aggravated the damage while p53 loss did not. Notably, mice lacking nuclear PTEN had increased hepatocellular carcinoma under oxidative stress, while mice lacking p53 in hepatocytes had accelerated hepatocellular carcinoma and intrahepatic cholangiocarcinoma. The formation of cholangiocarcinoma appears to involve the transformation of hepatocytes into cholangiocarcinoma. Simultaneous loss of nuclear PTEN and p53 exacerbated both types of liver cancers. These data suggest that nuclear PTEN and p53 suppress liver cancers through distinct mechanisms.
Keywords: nuclear PTEN, p53, DNA damage, liver cancer, hepatocellular carcinoma, cholangiocarcinoma
1. Introduction
Phosphatase and tensin homolog deleted on chromosome ten (PTEN) has various functions as a tumor suppressor at multiple intracellular locations, including the plasma membrane and nucleus [1–3]. At the plasma membrane, PTEN dephosphorylates phosphatidylinositol (3,4,5)-trisphosphate to phosphatidylinositol (4,5)-bisphosphate and regulates the PI3K-AKT-mTOR signaling pathway in cell proliferation and survival [4–7]. In contrast, in the nucleus, previous studies using in vitro cell culture systems have shown that PTEN controls DNA repair, cell cycle progression, and genome stability, independently of its lipid phosphatase activity [1,2,8–11]. p53, another critical tumor suppressor that is highly mutated in many cancers, also functions in the nucleus as a “guardian of the genome” [12,13]. p53 is activated in response to DNA damage and protects the integrity of the nuclear genome [12–14]. In response to a variety of cellular stresses, p53 regulates the expression of many genes for DNA damage response, cell cycle arrest, cell senescence, and apoptosis [15,16].
Previous studies have revealed the inter-dependency of PTEN and p53. For example, PTEN regulates p53 protein levels by inhibiting its Mdm2-mediated degradation [17,18]. Also, PTEN binds p53 and modifies its transcriptional activities under hypoxia [19]. Conversely, p53 upregulates PTEN transcription, and p53 deficiency leads to the loss of PTEN [20–22]. PTEN deficiency can overcome p53-dependent cellular senescence and enhances tumor formation [23]. Highlighting their additive effects, the formation of undifferentiated soft tissue sarcoma has been reported in smooth muscle lineage-specific PTEN and p53 double heterozygous knockout mice [24]. Furthermore, prostate-specific conditional double knockout of PTEN and p53 in mice generates lethal, invasive prostate cancers at young ages, while the single knockouts do not [23]. Although these studies show important physical and functional interplays between total PTEN and p53, the specific relationships between nuclear PTEN and p53 remain unknown.
Recently, to investigate the physiological role of nuclear PTEN in vivo, we have created genetically-engineered mice using the CRISPR/Cas9 genome editing, in which the amounts of PTEN in the nucleus are greatly decreased [25,26]. Using this nuclear PTEN deficient mouse line, we have shown that the loss of nuclear PTEN accelerates the formation of hepatocellular carcinoma in response to carcinogen and oxidative stress [25,26]. We also show that oxidative stress increases the localization of PTEN in the nucleus in hepatocytes through inhibition of nuclear export of PTEN [26]. In the absence of nuclear PTEN, oxidative stress-induced DNA damage was exacerbated in the liver [26]. Here, to study the functional relationship between nuclear PTEN and p53, we analyzed liver cancer induced by carcinogen and oxidative stress in mice that lack nuclear PTEN and p53.
2. Materials and Methods
2.1. Animals
All animal work was performed according to the guideline established by the Johns Hopkins University Committee on Animal Care. Nuclear deficient PTEN (ndPTEN) mice that carry the K13R and D384V mutations have been previously described [25,26]. p53flox/flox mice (stock#: 008462) and albumin-Cre recombinase mice (stock#: 003574) were obtained from the Jackson laboratory, and bred to produce liver-specific p53 knockout mice (Alb-Cre::p53flox/flox, p53-hKO). To induce liver damage, we intraperitoneally injected 20% CCl4/mineral oil (10 ml/kg body weight) (289116, Sigma-Aldrich) into mice at 2–3 months of age. Male mice were used in all of the experiments in this study. To induce liver cancer, we intraperitoneally injected DEN (25 mg/kg body weight; N0258, Sigma-Aldrich) into mice at 2 weeks of age, and then intraperitoneally injected 20% CCl4/mineral oil (2.5 ml/kg bodyweight) twice per week from 6 to 14 week of age [26–28].
2.2. Antibodies
The primary antibodies used were: PTEN (9559, Cell Signaling Technology), GAPDH (MA5–15738, Invitrogen), Ki67 (M7249, DAKO), CK19 (MABT913, Sigma-Aldrich), phospho-H2AX (S139) (ab11174, Abcam), and active caspase-3 (AF835, R&D System). The fluorescently-labeled secondary antibodies were obtained from Invitrogen: Alexa488 anti-rabbit IgG (A21206) and Alexa568 anti-rat IgG (A11077). HRP-conjugated anti-rat IgG (474–1612, KPL) and anti-rabbit (NA934V, GE Healthcare) were used.
2.3. Quantitative RT-PCR
mRNAs were prepared from livers using a RNeasy mini kit (74104, Qiagen). cDNAs were synthesized using a ReadyScript cDNA synthesis kit (RDRT, Sigma-Aldrich). Quantitative real-time PCR (RT-PCR) was performed using a real-time PCR detection system (CFX98, BioRad), and PowerUp SYBR Green Master Mix kit (A25742, Applied Biosystems). The following primers were used. Albumin: forward 5′-TGCTTTTTCCAGGGGTGTGTT-3′, reverse 5′-TTACTTCCTGCACTAATTTGGCA-3′. p53: 5′-AGTCCTTTGCCCTGAACTGC-3′, 5′-TTTACGCCCGCGGATCTTGA-3′. Jagged-1: 5′-AACTGTTGTGGTGGAGTCCG-3′, 5′-GCAAAGTGTAGGACCTCGGC-3′. c-myc: 5′-TGTACCTCGTCCGATTCCAC-3′, 5′-CATCTTCTTGCTCTTCTTCAGAGTC-3′. CyclinD1, 5′-CAAAATGCCAGAGGCGGATG-3′, 5′-CCAGGGCCTTGACCGGG-3′. CTGF: 5′-CCAATGCACTTGCCTGGATG-3′, 5′-TCCAGTCGGTAGGCAGCTA-3′. Cyr61: 5′-CTGAAGAGGCTTCCTGTCTTTG-3′, 5′-GATCCGGGTCTCTTTCACCA-3′.
2.4. Western blotting
Livers were homogenized in RIPA buffer (9806, Cell Signaling Technology) containing complete protease inhibitor cocktail (1697498001, Roche) and phosphatase inhibitor cocktails (P5726 and P0044, Sigma-Aldrich). Lysates were centrifuged at 16,000 × g for 10 min at 4°C, and the supernatants were collected. Proteins were separated using SDS-PAGE and then transferred onto Immobilon-FL (IPFL00010, EMD Millipore). After blocking in 3% BSA/PBS containing 0.1% Tween 20 for 1 h at room temperature, the membranes were incubated with the primary antibodies overnight at 4°C. After washing, immunocomplexes were visualized using appropriate fluorescent-labeled secondary antibodies and detected using a PharosFX Plus molecular imager (Bio-Rad).
2.5. Immunofluorescence microscopy
Mice were anesthetized by intraperitoneal injection of Avertin and fixed by cardiac perfusion of ice-cold 4% paraformaldehyde in PBS [25]. The livers were dissected and further fixed in 4% paraformaldehyde in PBS for 3 h at 4°C. The samples were incubated in PBS containing 30% sucrose overnight and frozen in optimal cutting temperature (OCT) compound (23–730-571, Fisher Scientific) in a Tissue-Tek Cryomold (4566, Sakura Finetek USA). Frozen tissue blocks were sectioned and mounted on Superfrost Plus Microscope Slides (12–550-15, Fisher Scientific). Sections were subjected to antigen retrieval with 1 mM EDTA using a microwave oven and incubated with the primary antibodies at 4°C overnight. After washes, the samples were incubated with appropriate fluorescently-labeled secondary antibodies at room temperature for 1 h. DAPI (1 μg/ml) was used to stain nuclear DNA.
2.6. Histology
4% paraformaldehyde-fixed livers were embedded in paraffin at the Johns Hopkins School of Medicine Pathology Core. Paraffin sections were cut, and hematoxylin and eosin stained at the Pathology Core. To analyze fibrosis, paraffin sections were stained using a Picro Sirius Red Stain Kit (ab150681, Abcam) according to the manufacturer’s instructions. To immunostain CK19, Ki67, and active caspase-3, paraffin sections were deparaffinized, subjected to antigen retrieval with 1 mM EDTA using a pressure cooker for 15 min, and incubated with the primary antibody at 4°C overnight. After washes with PBS, the samples were incubated with HRP-conjugated secondary antibody for 1 h at room temperature and stained with DAB (ab64238, Abcam). Hematoxylin (MHS16, Sigma-Aldrich) was used to counterstain nuclei. The samples were viewed using an Olympus BX51 microscope equipped with a DP-70 color camera.
3. Results
3.1. The loss of p53 enhances oxidative stress-induced nuclear localization of PTEN in hepatocytes
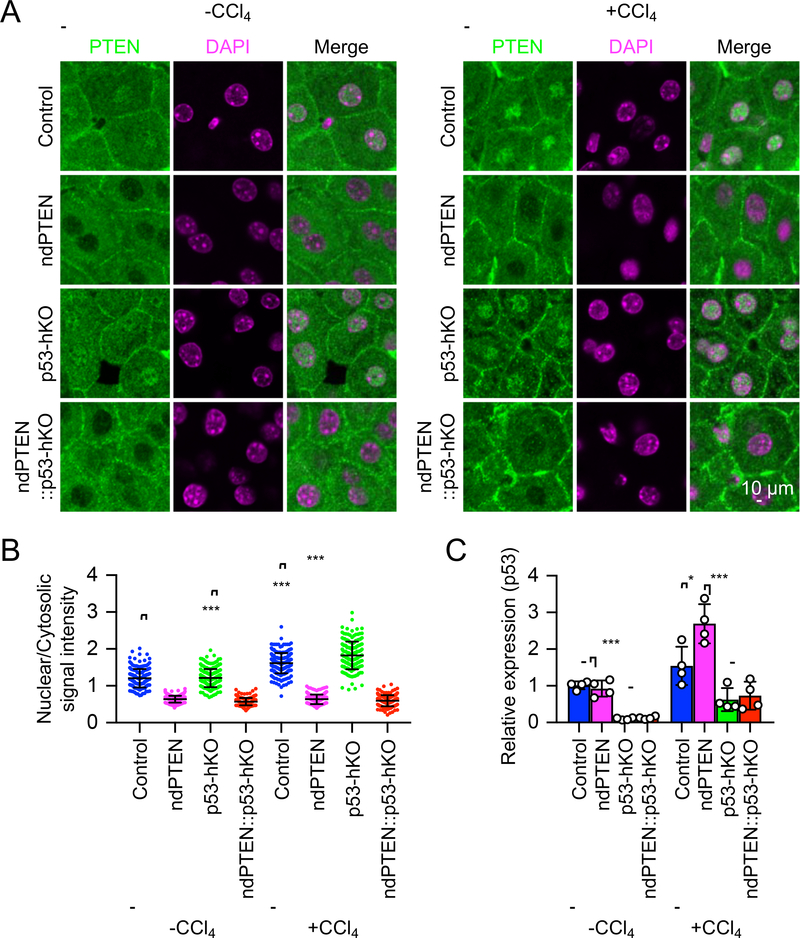
To investigate the role of p53 for the nuclear localization of PTEN, we examined the localization of PTEN in the liver of control mice (p53flox/flox), nuclear PTEN deficient mice (ndPTEN), hepatocyte-specific p53 knockout mice (Alb-Cre::p53flox/flox, p53-hKO), and ndPTEN and p53-hKO double mutant mice (ndPTEN::Alb-Cre::p53flox/flox, ndPTEN::p53-hKO) after intraperitoneal injection of a hepatotoxin, carbon tetrachloride (CCl4) that produces oxidative stress in the livers [26–28]. Immunofluorescence microscopy showed that PTEN is located both in the cytosol and nucleus of hepatocytes in control mice without the injection, and this nuclear localization was increased by the oxidative stress (Fig. 1A and B). In p53-hKO mice, the basal level of PTEN nuclear localization was unaffected. In contrast, its oxidative-stress induced nuclear accumulation was further enhanced compared to control mice (Fig. 1A and B). As negative controls, in ndPTEN mice and ndPTEN::p53-hKO double mutant mice, the nuclear localization of PTEN was greatly decreased at both the basal and stress conditions (Fig. 1A and B). Therefore, stress-induced PTEN’s nuclear accumulation is amplified in the absence of p53.
Figure 1. Oxidative stress induces nuclear PTEN accumulation and p53 upregulation in livers.
(A) Control, ndPTEN, p53-hKO, and ndPTEN::p53-hKO mice were subjected to intraperitoneal injection of 20% CCl4. At 2 days after CCl4 injection, cryosections of livers were analyzed by immunofluorescence microscopy using anti-PTEN antibodies. DAPI was used to stain nuclear DNA. (B) The intensity of PTEN signals in the nucleus was quantified relative to that in the cytosol. Bars are average ± SD (n = 161–221 hepatocytes from three mice). (C) Levels of p53 mRNA in the livers were quantified relative to those of albumin mRNA after CCl4 injection. Values were normalized to control mice without CCl4 treatment. Bars are average ± SD (n = 4 livers). One-way ANOVA with post-hoc Tukey was performed in (B and C): *p < 0.05, ***p < 0.001.
3.2. Nuclear PTEN deficiency potentiates stress-induced upregulation of p53 in hepatocytes
To determine the effects of nuclear PTEN deficiency on oxidative stress-induced expression of p53, we examined the levels of p53 mRNAs after CCl4 injection. In control mice, p53 mRNA expression was increased after oxidative stress and further enhanced in ndPTEN mice (Fig. 1C). As negative controls, in p53-hKO mice and ndPTEN::p53-hKO mice, the levels of p53 mRNAs were substantially decreased in livers (Fig. 1C). The remaining p53 mRNAs were likely derived from cells other than hepatocytes in the liver, such as Kupffer, immune, and endothelial cells. p53 mRNA levels appeared to be slightly increased in these cells after CCl4 injection independently of nuclear PTEN. Thus, nuclear PTEN deficiency accelerates oxidative stress-induced p53 upregulation in hepatocytes.
3.3. Deficiency of nuclear PTEN, but not p53, increases stress-induced DNA damage in hepatocytes
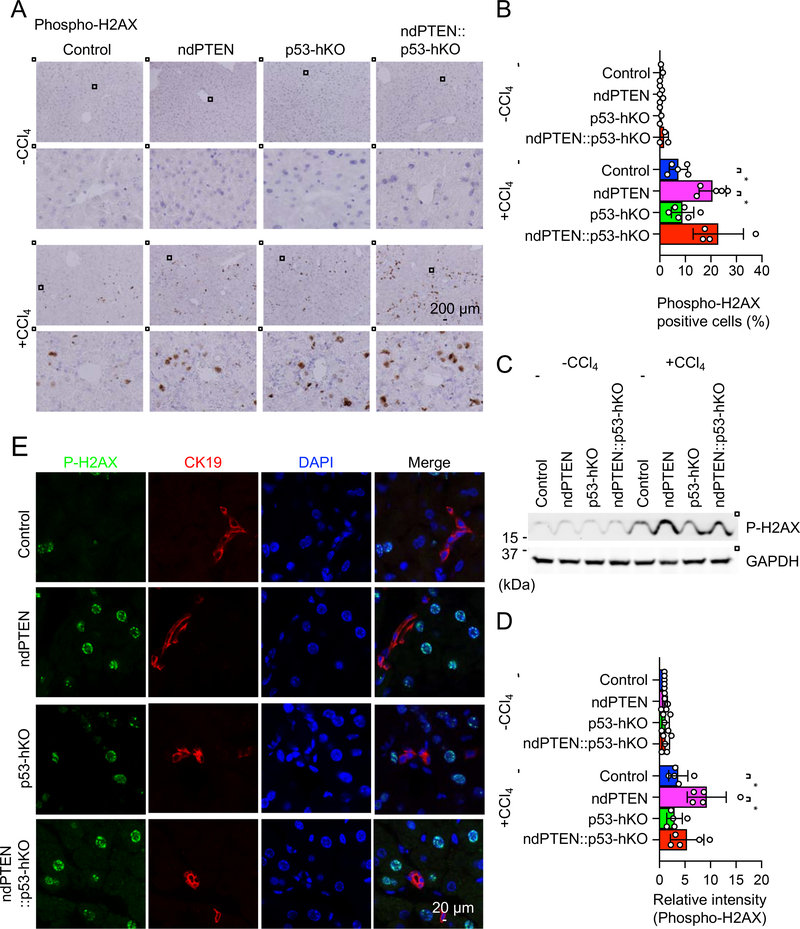
To compare the role of nuclear PTEN and p53 for DNA repair after oxidative stress in the liver, we examined DNA damage using immunofluorescence microscopy with an antibody to serine139-phosphorylated histone H2AX (phospho-H2AX), which is a well-established DNA damage marker [8]. Without CCl4 injection, we found only negligible levels of phospho-H2AX-positive cells in control, ndPTEN, p53-hKO, and ndPTEN::p53-hKO mice (Fig. 2A and B). After the CCl4 injection, more phospho-H2AX-positive cells were found in the control mice (Fig. 2A and B). As reported previously [26], ndPTEN mice showed a significant increase in the percentage of phospho-H2AX positive cells compared to control mice (Fig. 2A and B). In contrast, p53-hKO mice showed levels of phospho-H2AX positive cells similar to control mice after CCl4 injection (Fig. 2A and B). Therefore, p53 is not required for DNA repair after CCl4 injection in the liver. Further supporting this notion, ndPTEN::p53-hKO mice displayed an increase in the percentage of phospho-H2AX-positive cells to an extent similar to that of ndPTEN mice after CCl4 injection (Fig. 2A and B). These immunofluorescence data were biochemically confirmed by Western blotting of isolated livers using an antibody to phospho-H2AX (Fig. 2C and D).
Figure 2. Oxidative stress induces DNA damage in ndPTEN mice, but not p53-hKO mice.
(A) Paraffin sections of livers from the indicated mice were analyzed by immunohistochemistry using anti-phospho-H2AX (serine139) antibodies at 2 days after CCl4 injection. Nuclei were counterstained with hematoxylin. (B) Percentages of phospho-H2AX-positive cells were quantified. Bars are average ± SD (n = 3–6 livers). (C and D) Livers were isolated from the indicated mice after CCl4 injection and analyzed by Western blotting with antibodies to phospho-H2AX and GAPDH. (D) The intensity of phospho-H2AX signals was quantified relative to that of GAPDH. Bars are average ± SD (n = 5 livers). (E) Cryosections of livers were analyzed by immunofluorescence microscopy using antibodies of CK19 and phospho-H2AX. One-way ANOVA with post-hoc Tukey was performed in (B and D): *p < 0.05.
CCl4 is converted to CCl3O2 radical by CYP2E1 in hepatocytes, which is toxic to hepatocytes [29]. However, it is unclear whether CCl3O2 radical also induces DNA damage in biliary epithelial cells (i.e., cholangiocytes). To address this question, we performed immunofluorescence microscopy of livers using antibodies to phospho-H2AX and CK19, a marker for cholangiocytes. In contrast to hepatocytes, CK19-positive cholangiocytes are not stained by anti-phospho-H2AX antibodies (Fig. 2E). These data suggest that CCl4 leads to DNA damage in hepatocytes, but not cholangiocytes.
3.4. Deficiencies of nuclear PTEN and p53 induce different types of liver cancers in response to oxidative stress
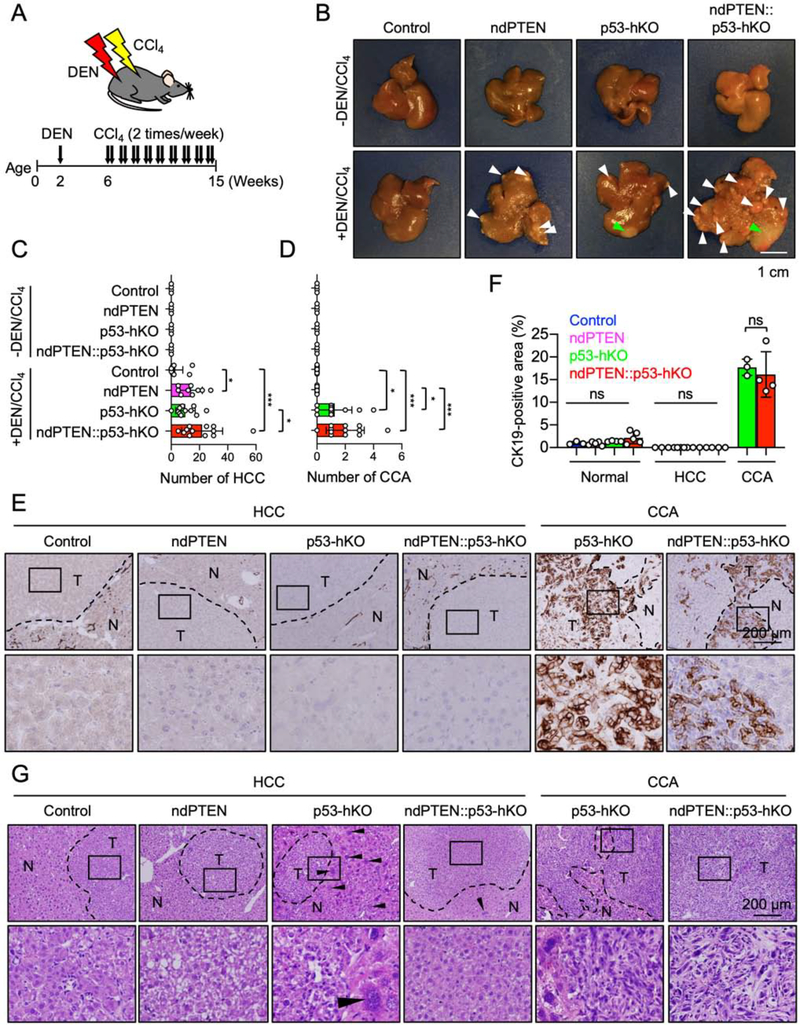
To examine the role of nuclear PTEN and p53 for tumorigenesis, we used a preclinical platform of liver cancer induced by a single injection of a carcinogen, N-nitrosodiethylamine (DEN) and followed by repeated injections of CCl4 (Fig. 3A) [26]. In control mice, only a small number of hepatocellular carcinomas (HCCs) were found at 15 weeks (Fig. 3B and C). Conversely, the formation of HCC was enhanced in both ndPTEN mice and p53-hKO mice (Fig. 3B and C). Importantly, the occurrence of HCC was highest in ndPTEN::p53-hKO mice (Fig. 3B and C). Furthermore, to our surprise, p53-hKO mice, but not control or ndPTEN mice, developed a bile duct cancer, intrahepatic cholangiocarcinoma (CCA) (Fig. 3B and D), which was stained by anti-CK19 antibodies (Fig. 3E and F) and histologically distinguishable from HCC (Fig. 3G). The frequency of CCA was further increased in ndPTEN::p53-hKO mice (Fig. 3B and D). These data suggest that nuclear PTEN and p53 play distinct roles in the suppression of HCC and CCA.
Figure 3. Deficiencies of nuclear PTEN and p53 differentially accelerate liver tumorigenesis in response to oxidative stress.
(A) Experimental design for DEN/CCl4-induced liver cancer in mice. Control, ndPTEN, p53-hKO, and ndPTEN::p53-hKO mice were subjected to intraperitoneal injection of DEN at 2 weeks of age. Mice were further treated with CCl4 twice per week from 6 to 14 weeks of age and were analyzed at 15 weeks. (B) Images of livers. White arrowheads indicate HCC, and green arrowheads indicate CCA. (C and D) The number of HCC in (C) and CCA in (D). Bars are average ± SD (n = 4 mice untreated and 7 mice DEN/CCl4-treated). (E) Immunohistochemistry using anti-CK19 antibodies. (F) CK19-positive regions were quantified. Bars are average ± SD (n = 3–5 livers). Nuclei were counterstained with hematoxylin. (G) Liver sections were subjected to hematoxylin and eosin staining. Arrowheads indicate giant hepatocytes. One-way ANOVA with post-hoc Tukey was performed in (C, D, and F): *p < 0.05, ***p < 0.001.
To examine the effects of DEN and CCl4 on tissue injuries, we analyzed fibrosis using Sirius Red staining (Fig. 4A and B). We found similar, low levels of fibrosis in the livers of control, ndPTEN, p53-hKO, and ndPTEN::p53-hKO mice in non-tumor and HCC regions (Fig. 4A and B). In contrast, fibrosis was dramatically and similarly increased in CCA regions of p53-hKO and ndPTEN::p53-hKO mice (Fig. 4A and B). Consistent with these tissue injuries, we found increased apoptosis in CCA regions in p53-hKO and ndPTEN::p53-hKO mice using immunohistochemistry with anti-active-caspase-3 antibodies (Fig. 4C and D). Therefore, liver injuries caused by DEN and CCl4 treatments were indistinguishable in each non-tumor or tumor region in the four groups of mice and were not likely the reason to induce different types and amounts of liver cancers.
Figure 4. Fibrosis is induced in CCA in p53-hKO and ndPTEN::p53-hKO mice.
(A) Paraffin liver sections were subjected to Sirius Red staining. T: Tumors, N: Non-tumor regions. (B) Fibrotic regions were quantified based on Sirius Red staining in no-tumor (normal), HCC, and CCA regions. Bars are average ± SD (n = 3–5 livers). (C) Immunohistochemistry of active caspase-3. Nuclei were counterstained with hematoxylin. (D) Active caspase-3-positive cells were quantified. Bars are average ± SD (n = 3–5 livers). (E) Ki67 immunohistochemistry. Nuclei were counterstained with hematoxylin. (F) Ki67-positive cells were quantified. Bars are average ± SD (n = 3–5 livers). One-way ANOVA with post-hoc Tukey was performed in (B, D, and F): ns, not significant.
In addition, immunohistochemistry of Ki67, a marker for cell proliferation, showed similar increases in percentages of Ki67-positive cells in HCC and CCA regions (Fig. 4E and F). These data suggest that once initiated, rates of cell proliferation in HCC and CCA are similar regardless of the presence or absence of nuclear PTEN and p53.
It has been reported that Wnt/ß-catenin signaling and Notch signaling are involved in HCC and CCA, respectively [30–32]. In addition, Hippo/YAP signaling plays an important role in liver cancer formation [33]. To examine these signal transduction pathways in our HCC and CCA models, we analyzed the gene expression in livers after CCl4 injection using quantitative RT-PCR. Our transcriptional analysis showed strong upregulation of c-Myc and cyclinD1 (Wnt/ß-catenin pathway), Jagged1 (Notch pathway), and CTGF and Cyr61 (Hippo/YAP pathway) after CCl4 treatment (Fig. S1). Since the levels of their upregulation are similar in control, ndPTEN, p53-hKO, and ndPTEN::p53-hKO mice, the loss of nuclear PTEN and p53 does not affect these signal transduction mechanism upon oxidative stress.
4. Discussion
Liver cancer accounts for 8% of all cancer mortality in the world [34]. HCC is fast-growing and represents up to 90% of all primary liver cancers [35]. In contrast, CCA represents a relatively small percentage of liver cancer; however, CCA is very aggressive, and most patients die within 5 years [36]. There are currently no effective treatments for HCC and CCA. Therefore, there is an urgent need to decipher their disease mechanisms and develop therapeutic interventions. In our current study, we report a new preclinical model of both HCC and CCA using hepatocyte-specific KO of p53, which is deficient in essentially all human liver cancers. In this mouse model, the combination of the carcinogen DEN and hepatotoxin CCl4 with p53 loss robustly produces HCC and CCA. Additional loss of nuclear PTEN accelerated HCC and CCA in p53-hKO mice. Interestingly, a single loss of nuclear PTEN only induced HCC but not CCA. These data suggest that nuclear PTEN and p53 mitigate liver cancers through distinct tumor suppression mechanisms. While nuclear PTEN is important for protection from oxidative-stress induced DNA damage in hepatocytes, p53’s contribution to this protection process appears to be small. Further investigation of these mouse models would advance our understanding of both types of liver cancer.
HCC is derived from injured hepatocytes under stressed conditions, such as viral infection, toxic stimuli, and metabolic diseases [35]. CCA has been thought to originate from cholangiocytes (biliary epithelial cells); however, recent studies have shown that mature hepatocytes dedifferentiate into liver progenitor cells for cholangiocytes or transdifferentiate into cholangiocytes under stress conditions [37,38]. Our data showing that hepatocyte-specific loss of p53 leads to CCA suggests that p53 plays an important role in suppressing the transformation of hepatocytes to cholangiocytes and then CCA. Supporting this novel role of p53, we found giant cells in non-tumor regions in the liver of p53-hKO mice after DEN and CCl4 treatment (Fig. 3G, arrowheads). While giant cells are observed in the neonatal period, the transformation of mature hepatocytes into giant cells can also occur in adults after liver damage [39,40]. Therefore, in healthy livers, p53 likely inhibits the transformation of hepatocytes into giant cells, which could become precursors for CCA when oxidative stress and tissue injuries occur. It would be of great interest to uncover the molecular mechanisms underlying the new role of p53 in the suppression of liver cancers in future studies using our mouse models.
Supplementary Material
Highlights.
Nuclear PTEN deficiency upregulates p53 expression upon oxidative stress.
Loss of p53 potentiates stress-induced accumulation of PTEN in the nucleus.
Nuclear PTEN deficient mice increase hepatocellular carcinoma under oxidative stress.
Hepatic p53 KO enhances hepatocellular carcinoma and intrahepatic cholangiocarcinoma.
Acknowledgments
We thank past and present members of the Iijima and Sesaki labs for helpful discussions and technical assistance. This work was supported by NIH grants to MI (GM131768 and NS114458) and HS (GM123266).
Biochemical and Biophysical Research Communications
Abbreviation:
- (PTEN)
Phosphatase and tensin homolog deleted on chromosome ten
- (CCl4)
Carbon tetrachloride
- (phospho-H2AX)
Phosphorylated histone 2AX
- (DEN)
N-nitrosodiethylamine
- (HCC)
hepatocellular carcinoma
- (CCA)
cholangiocarcinoma
Footnotes
Declaration of competing interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Please note that all Biochemical and Biophysical Research Communications authors are required to report the following potential conflicts of interest with each submission. If applicable to your manuscript, please provide the necessary declaration in the box above.
(1) All third-party financial support for the work in the submitted manuscript.
(2) All financial relationships with any entities that could be viewed as relevant to the general area of the submitted manuscript.
(3) All sources of revenue with relevance to the submitted work who made payments to you, or to your institution on your behalf, in the 36 months prior to submission.
(4) Any other interactions with the sponsor of outside of the submitted work should also be reported. (5) Any relevant patents or copyrights (planned, pending, or issued).
(6) Any other relationships or affiliations that may be perceived by readers to have influenced, or give the appearance of potentially influencing, what you wrote in the submitted work. As a general guideline, it is usually better to disclose a relationship than not.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Baker SJ, PTEN enters the nuclear age, Cell 128 (2007) 25–28. [DOI] [PubMed] [Google Scholar]
- [2].Kreis P, Leondaritis G, Lieberam I, Eickholt BJ, Subcellular targeting and dynamic regulation of PTEN: implications for neuronal cells and neurological disorders, Front Mol Neurosci 7 (2014) 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Leslie NR, Kriplani N, Hermida MA, Alvarez-Garcia V, Wise HM, The PTEN protein: cellular localization and post-translational regulation, Biochem Soc Trans 44 (2016) 273–278. [DOI] [PubMed] [Google Scholar]
- [4].Iijima M, Devreotes P, Tumor suppressor PTEN mediates sensing of chemoattractant gradients, Cell 109 (2002) 599–610. [DOI] [PubMed] [Google Scholar]
- [5].Fruman DA, Chiu H, Hopkins BD, Bagrodia S, Cantley LC, Abraham RT, The PI3K Pathway in Human Disease, Cell 170 (2017) 605–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Iijima M, Huang YE, Devreotes P, Temporal and spatial regulation of chemotaxis, Dev Cell 3 (2002) 469–478. [DOI] [PubMed] [Google Scholar]
- [7].Song MS, Salmena L, Pandolfi PP, The functions and regulation of the PTEN tumour suppressor, Nature reviews. Molecular cell biology 13 (2012) 283–296. [DOI] [PubMed] [Google Scholar]
- [8].Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, Mak TW, Neel BG, Raught B, Stambolic V, Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress, Science 341 (2013) 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chung JH, Ostrowski MC, Romigh T, Minaguchi T, Waite KA, Eng C, The ERK1/2 pathway modulates nuclear PTEN-mediated cell cycle arrest by cyclin D1 transcriptional regulation, Hum Mol Genet 15 (2006) 2553–2559. [DOI] [PubMed] [Google Scholar]
- [10].Shen WH, Balajee AS, Wang J, Wu H, Eng C, Pandolfi PP, Yin Y, Essential role for nuclear PTEN in maintaining chromosomal integrity, Cell 128 (2007) 157–170. [DOI] [PubMed] [Google Scholar]
- [11].Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, Pandolfi PP, Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner, Cell 144 (2011) 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ozen C, Yildiz G, Dagcan AT, Cevik D, Ors A, Keles U, Topel H, Ozturk M, Genetics and epigenetics of liver cancer, N Biotechnol 30 (2013) 381–384. [DOI] [PubMed] [Google Scholar]
- [13].Lane DP, Cancer. p53, guardian of the genome, Nature 358 (1992) 15–16. [DOI] [PubMed] [Google Scholar]
- [14].Levine AJ, p53, the cellular gatekeeper for growth and division, Cell 88 (1997) 323–331. [DOI] [PubMed] [Google Scholar]
- [15].Vousden KH, Lane DP, p53 in health and disease, Nat Rev Mol Cell Biol 8 (2007) 275–283. [DOI] [PubMed] [Google Scholar]
- [16].Meek DW, Tumour suppression by p53: a role for the DNA damage response?, Nat Rev Cancer 9 (2009) 714–723. [DOI] [PubMed] [Google Scholar]
- [17].Mayo LD, Donner DB, A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus, Proc Natl Acad Sci U S A 98 (2001) 11598–11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mayo LD, Dixon JE, Durden DL, Tonks NK, Donner DB, PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy, J Biol Chem 277 (2002) 5484–5489. [DOI] [PubMed] [Google Scholar]
- [19].Freeman DJ, Li AG, Wei G, Li HH, Kertesz N, Lesche R, Whale AD, Martinez-Diaz H, Rozengurt N, Cardiff RD, Liu X, Wu H, PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms, Cancer Cell 3 (2003) 117–130. [DOI] [PubMed] [Google Scholar]
- [20].Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW, Regulation of PTEN transcription by p53, Mol Cell 8 (2001) 317–325. [DOI] [PubMed] [Google Scholar]
- [21].Eitel JA, Bijangi-Vishehsaraei K, Saadatzadeh MR, Bhavsar JR, Murphy MP, Pollok KE, Mayo LD, PTEN and p53 are required for hypoxia induced expression of maspin in glioblastoma cells, Cell Cycle 8 (2009) 896–901. [DOI] [PubMed] [Google Scholar]
- [22].Mao JH, Wu D, Perez-Losada J, Nagase H, DelRosario R, Balmain A, Genetic interactions between Pten and p53 in radiation-induced lymphoma development, Oncogene 22 (2003) 8379–8385. [DOI] [PubMed] [Google Scholar]
- [23].Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, Cordon-Cardo C, Pandolfi PP, Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis, Nature 436 (2005) 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Guijarro MV, Dahiya S, Danielson LS, Segura MF, Vales-Lara FM, Menendez S, Popiolek D, Mittal K, Wei JJ, Zavadil J, Cordon-Cardo C, Pandolfi PP, Hernando E, Dual Pten/Tp53 suppression promotes sarcoma progression by activating Notch signaling, Am J Pathol 182 (2013) 2015–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Igarashi A, Itoh K, Yamada T, Adachi Y, Kato T, Murata D, Sesaki H, Iijima M, Nuclear PTEN deficiency causes microcephaly with decreased neuronal soma size and increased seizure susceptibility, J Biol Chem 293 (2018) 9292–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kato T, Yamada T, Nakamura H, Igarashi A, Anders RA, Sesaki H, Iijima M, The Loss of Nuclear PTEN Increases Tumorigenesis in a Preclinical Mouse Model for Hepatocellular Carcinoma, iScience 23 (2020) 101548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Heindryckx F, Colle I, Van Vlierberghe H, Experimental mouse models for hepatocellular carcinoma research, Int J Exp Pathol 90 (2009) 367–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Uehara T, Pogribny IP, Rusyn I, The DEN and CCl4 -Induced Mouse Model of Fibrosis and Inflammation-Associated Hepatocellular Carcinoma, Curr Protoc Pharmacol 66 (2014) 14.30.1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Recknagel RO, Glende EA Jr., Dolak JA, Waller RL, Mechanisms of carbon tetrachloride toxicity, Pharmacol Ther 43 (1989) 139–154. [DOI] [PubMed] [Google Scholar]
- [30].Perugorria MJ, Olaizola P, Labiano I, Esparza-Baquer A, Marzioni M, Marin JJG, Bujanda L, Banales JM, Wnt-beta-catenin signalling in liver development, health and disease, Nat Rev Gastroenterol Hepatol 16 (2019) 121–136. [DOI] [PubMed] [Google Scholar]
- [31].Dimri M, Satyanarayana A, Molecular Signaling Pathways and Therapeutic Targets in Hepatocellular Carcinoma, Cancers (Basel) 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rauff B, Malik A, Bhatti YA, Chudhary SA, Qadri I, Rafiq S, Notch signalling pathway in development of cholangiocarcinoma, World J Gastrointest Oncol 12 (2020) 957–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim W, Khan SK, Gvozdenovic-Jeremic J, Kim Y, Dahlman J, Kim H, Park O, Ishitani T, Jho EH, Gao B, Yang Y, Hippo signaling interactions with Wnt/beta-catenin and Notch signaling repress liver tumorigenesis, J Clin Invest 127 (2017) 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA Cancer J Clin 68 (2018) 394–424. [DOI] [PubMed] [Google Scholar]
- [35].Craig AJ, von Felden J, Garcia-Lezana T, Sarcognato S, Villanueva A, Tumour evolution in hepatocellular carcinoma, Nat Rev Gastroenterol Hepatol 17 (2020) 139–152. [DOI] [PubMed] [Google Scholar]
- [36].Banales JM, Cardinale V, Carpino G, Marzioni M, Andersen JB, Invernizzi P, Lind GE, Folseraas T, Forbes SJ, Fouassier L, Geier A, Calvisi DF, Mertens JC, Trauner M, Benedetti A, Maroni L, Vaquero J, Macias RI, Raggi C, Perugorria MJ, Gaudio E, Boberg KM, Marin JJ, Alvaro D, Expert consensus document: Cholangiocarcinoma: current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA), Nat Rev Gastroenterol Hepatol 13 (2016) 261–280. [DOI] [PubMed] [Google Scholar]
- [37].Chen Y, Wong PP, Sjeklocha L, Steer CJ, Sahin MB, Mature hepatocytes exhibit unexpected plasticity by direct dedifferentiation into liver progenitor cells in culture, Hepatology 55 (2012) 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Tarlow BD, Pelz C, Naugler WE, Wakefield L, Wilson EM, Finegold MJ, Grompe M, Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes, Cell Stem Cell 15 (2014) 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Maggiore G, Sciveres M, Fabre M, Gori L, Pacifico L, Resti M, Choulot JJ, Jacquemin E, Bernard O, Giant cell hepatitis with autoimmune hemolytic anemia in early childhood: long-term outcome in 16 children, J Pediatr 159 (2011) 127–132.e1. [DOI] [PubMed] [Google Scholar]
- [40].Estradas J, Pascual-Ramos V, Martinez B, Uribe M, Torre A, Autoimmune hepatitis with giant-cell transformation, Ann Hepatol 8 (2009) 68–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.