Abstract
Interferons (IFN) are antiviral cytokines that mitigate the effects of invading viruses early on during the infection process. SARS-CoV and MERS induce weak IFN responses; hence, the clinical trials which included recombinant IFN accompanied with other antiviral drugs exhibited improved results in terms of shortening the duration of illness. The aim of the present study was to evaluate the type I IFN response in COVID-19 patients to determine whether it is sufficient to eliminate or reduce the severity of the infection, and whether it can be recommended as a potential therapy. Total RNA samples were converted to cDNA and used as templates to evaluate the gene expression levels of IFN regulatory factor (IRF)3 and IFN-β in COVID-19 patients or control. The results showed that IRF3 gene expression was upregulated ~250-fold compared with the negative samples. In contrast, IFN-β expression increased slightly in COVID-19 patients. Consistent with other coronaviruses, such as SARS-CoV and MERS, COVID-19 infection does not induce an efficient IFN response to reduce the severity of the virus. This may be attributed to an incomplete response of IRF3 in activating the IFN-β promoter in the infected patients. The results suggest IFN-β or α may be used as potential treatments.
Keywords: COVID-19, interferon, interferon regulatory factor, type I interferon, innate immune response
Introduction
The new coronavirus, termed COVID-19, emerged in Wuhan, China in late 2019. Although the newly emerged virus has a mutation in the sequence of the spike protein, its binding affinity for the angiotensin-converting enzyme 2 (ACE2) receptor is identical to that of the severe acute respiratory syndrome (SARS-CoV)-1(1). The cytokine storm is as a group of inflammatory responses, which includes interleukin (IL)-I, IL-2, IL-6, IL-10 and interferon (IFN)-γ (2), is a serious complication of COVID-19 infection (3-6).
Different types of vaccines are being developed to assist in limiting the spread of the virus, and reduce mortality rates going forward, some of which have been approved by the regulatory bodies of several countries, and are being widely distributed. Various companies are currently developing a vaccine introducing mRNA to produce viral proteins, specifically the spike protein, by the host cells. A more stable DNA vaccine is another option to prevent infection with SARS-CoV-2 using adenovirus plasmids encoding the SARS-CoV-2 spike protein (7,8). Another alternative is to use other viral proteins, usually by recombinant DNA. The inactivated whole SARS-CoV-2 vaccine is also a candidate being assessed in preclinical trials. However, using a SARS-CoV-2 live attenuated vaccine carries potential risks, such as the reactivation or the virulence of SARS-CoV-2 in immunocompromised patients (9).
IFNs were named initially due to their role in interfering with viral infections. Influenza-infected chick cells mounted antiviral resistance states by producing secreted cytokines, which were later termed IFNs (10). IFNs are cytokines that are implicated in antiviral responses, immune induction and regulating cell division (11). The gene expression of type I IFN is primarily regulated at the transcriptional stage, and in the absence of stimulators, such as double-stranded RNA, IFNs are not translated. The lack of the IFN-β gene through gene knockout makes mice vulnerable to infection with viruses, such as vaccinia virus and blocks the IFN-α response (12).
To induce the innate immune response during viral infection, it is essential to stimulate the IFN response. The absence of IFN regulatory factor (IRF)3 or defective IRF7 function decreases the gene expression of IFN-α/β, making mice more sensitive to viral infection (13). IRF3 modulates the innate antiviral response that is triggered by the invading virus. IRF3 is primarily modified by hyperphosphorylation when the virus begins replication (14).
IRF3 and IRF7 are the most common regulators of IFN-β. They replace IRF2, serving a key role in type I IFN responses (15-17). IRF3 and IRF7 have specific binding properties that allow them to bind to the type I IFN promoters, and their ratio to the bound elements modulates the IFN type I response during viral infection (18). However, IRF3 degradation has been reported to repress IFN-β rather than the activation of the transcriptional repressors. The lack of transcriptionally active IRF3 abolishes the activation of IFN-β to the Sendai virus in mouse embryonic fibroblasts (19).
Upon activation, IRF3 molecules translocate to the nucleus after phosphorylation and bind to the ornithine cyclodeaminase or P300 to form complexes in the IFN sensitive response element region (20). Most RNA viruses elicit a type I IFN response in toll-like receptors (TLRs) or independent mechanisms (cytosolic recognition system) through retinoic acid-inducible genes (RIG-1), which sense the viral RNA molecules (21). Moreover, RIG-1 is central to the stimulation of the type I IFN response to RNA virus infection via activation of IRF3 through kinases in fibroblasts and dendritic cells (22). Synthetic or natural dsRNAs are differentially recognised by RIG-1 and melanoma differentiation-associated protein 5 (MAD5), as the former induces production of IFNs to paramyxoviruses, the influenza virus and Japanese encephalitis virus, whereas picornavirus is detected by MAD5(23). As a coronavirus model, the mouse hepatitis virus antagonises the type I IFN through the Nucleocapsid protein (24). Previous outbreaks with SARS and MERS revealed that the virus does not increase the expression of IFN-β or its promoter activity. Therefore, treatments with recombinant interferons were used to boost the effects of antiviral drugs (25,26). Infection with respiratory viruses activates the TLR signalling pathways, and eventually leads to the induction of the type I IFN response. The SARS-CoV-2 infection stimulates the TLR downstream pathway to produce mature-IL-1β. An increase in IL-1β causes lung fibrosis and fever. The virus is more highly infectious in adults than children, which may be explained by the high expression levels of aryl hydrocarbon receptors in children compared with the relatively lower expression levels in adults (27).
The present study evaluated the gene expression of IFN and IRF3 in COVID-19-infected patients compared with the control, suggesting a mechanism for the induction of IFNs, and highlighting IFNs as a therapeutic option for treating COVID-19 patients in clinical trials.
Materials and methods
Sample collection, RNA extraction and reverse transcription quantitative PCR
RNA samples were collected from 30 patients suspected of infection with COVID-19 between February and April 2020 at the Public Health Laboratory in Basrah, Iraq. The age range of the patients was 25-55 years old, whereas that of the non-COVID-19 infected individuals was 28-60 years old. The infected patients included 8 females and 12 males whereas the non-infected individuals consisted of were 3 females and 7 males.
Infection was diagnosed using a LightMix SarbecoV E-gene plus EAV control (cat. no. 40-0776-96). The control samples were negative for COVID-19 and were diagnosed with either the common cold or influenza. The present study was approved by the Public Health Department, Basrah Health Directorate (approval no. F112020). All patients provided signed consent to participate in the present study.
The RNA from nasal swabs (total RNA) was extracted using an easy spin™ total RNA extraction kit (Intron; cat. no. 17221) according to the manufacturer's protocol, and used to evaluate the gene expression of IFN-β and IRF3 in COVID-19-infected or uninfected samples using gene-specific primers. An intron HiSenScript™ (RH-)RT-PCR PreMix transcription kit was used to reverse transcribe the RNA, according to the manufacturer's protocol. Quantitative PCR was performed using SYBR-Green MasterMix, according to the manufacturer's protocol (Bioneer; cat. no. K-6210). The reaction consisted of a mixture of 10 µl SYBR-Green, 3 µl cDNA template, 1 µl each of both the forward and reverse primers against IFN-β (forward, CAACTTGCTTGGATTCCTACAAAG and reverse primer, TATTCAAGCCTCCCATTCAATTG); IRF3 (forward, CGGAAAGAAGTGTTGCGGTTAG and reverse primer, TTTGCCATTGGTGTCAGGAGAG); and β-actin (forward, CCTGGCACCCAGCACAAT and reverse primer, GCCGATCCACACGGAGATCT), and 5 µl free deionised diethylpyrocarbonate D.W. to a final volume of 20 µl. The sequences of the primers are based on previous studies (28,29).
The thermocycling conditions were: Initial denaturation at 94˚C for 5 min; followed by 40 cycles of 15 sec of denaturation at 95˚C, annealing at 58˚C for 30 sec, and an extension step at 72˚C for 45 sec; with a final extension at 72˚C for 5 min. The products were subjected to dissociation curve analysis. Using the 2-∆∆Cq method for analysis of mRNA expression, data were normalised to β-actin, which was used as a housekeeping gene (30).
Statistical analysis
Data are presented as the mean ± standard deviation of three technical repeats per patient. All data were analysed using a Student's t-test. Statistical analysis was performed using Microsoft Excel (Office 365; Microsoft Corporation). P<0.05 was considered to indicate a statistically significant difference.
Results
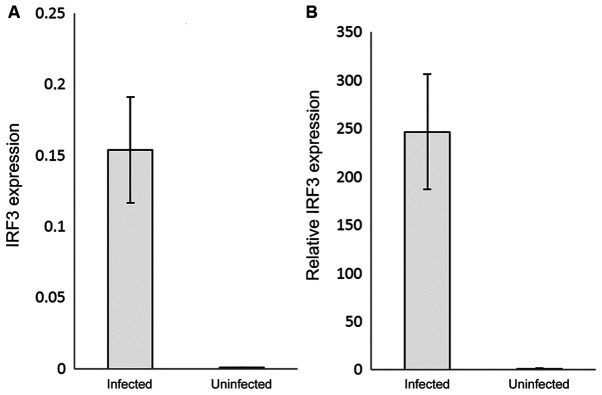
The RNAs of the COVID-19 positive and negative samples were extracted, reverse transcribed, used as the templates and mixed with IRF3 or IFN-β specific primers to assess their relative expression by qPCR. IRF3 gene expression was significantly (P<0.05) upregulated in the COVID-19-infected patients by ~250-fold compared with the control (uninfected samples) (Fig. 1A and B). Interestingly, IFN-β relative expression was ~1.5-fold higher in the COVID-19-infected samples compared with the control samples (P<0.05) (Fig. 2A and B). The 2-∆∆Cq analysis was used to detect the relative expression after subtracting the β-actin value from each sample, and the control was normalised to 1, to express the results as the fold change.
Figure 1.
IRF3 gene expression in COVID-19 infected individuals compared with uninfected controls. (A) IRF3 mRNA expression was quantified by reverse transcription-quantitative PCR. (B) Relative IRF3 mRNA expression levels. The results were normalised to the control values, and are presented as the mean ± standard deviation of the three technical replicates. β-actin was used as the internal control. The fluorescence was detected using SYBR-Green as the intercalating dye. IRF, interferon regulatory factor-3.
Figure 2.
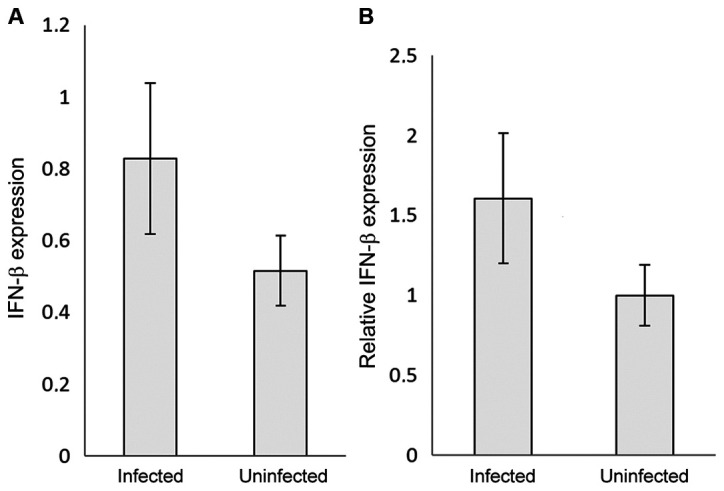
(A) IFN-β gene expression in COVID-19 infected individuals compared with uninfected controls. IFN-β mRNA expression was quantified by reverse transcription-quantitative PCR. (B) Relative IFN-β mRNA expression levels. The results were normalised to the control values, and are presented as the mean ± standard deviation of the three technical replicates. β-actin was used as the internal control. The fluorescence was detected using SYBR-Green as the intercalating dye. IFB-β, interferon-β.
Discussion
Studies have shown that IRF7 is expressed at a very low level physiologically, and requires activation of a type I interferon response for its induction (31,32). Both MERS and SARS trigger a low level of interferon response (33,34). IRF3 is a key regulator of type I IFN, which triggers the host response against the invading viruses. IRF3 also implicated in unwanted inflammatory responses and septic shock response (35-37). Thus, in the present study, the effects of COVID-19 on an innate immune response were determined.
The results showed that the gene expression levels of IRF3 were notably increased by ~250-fold compared with its expression in the virus-free samples. The increase in the IFN-β levels were not consistent with the increase in the expression of its primary regulator. The results agree with a study concerning coronavirus infection and IFN responses; infection with SARS-CoV does not induce IFN-β production or its promoter activity (38). A lower IFN response was detected in the COVID-19-infected lung tissue compared with SARS, which makes the former virus more sensitive to treatment with a type I IFN (22,39). However, in SARS infections, IRF3 is shown to translocate to the nucleus, independent of nay phosphorylation, dimerization or binding to cAMP response element-binding protein (CREB) binding protein. The SARS-CoV virus may block IRF3 hyperphosphorylation-mediated homodimerization CREB after transport of IRF3 to the nucleus (38).
Another hypothesis suggests that coronaviruses use the IFN-inducible transmembrane proteins (IFITM) to enter the cell, and the IFITM structural motifs required for entry inhibit the entry of other viruses. The IFITM theory explains how the virus can invade the lower respiratory tract (40). Coronaviruses, such as SARS, avoid the inhibitory effects of type I IFNs either through induction of double-membrane vesicles to physically hide the viral RNA intermediates from pattern recognition receptors or by expressing open reading frame (ORF)3, ORF6, ORF7, nucleocapsid protein and non-structural RNA binding protein 1, which when combined, abolish the IRF-3-dependent IFN-β pathway (41,42).
Based on the mechanism by which SARS inhibits the IFN response, recombinant IFNs were used to treat SARS-infected patients. The treatment of human corona Erasmus medical centre (HcoV-EMC) human-infected tissues with the type I or III IFN, 1 h post-infection, decreased the replication of the virus (43). In vitro, treating SARS-CoV-infected Vero and Caco2 cells with human recombinant IFN-β inhibited viral replication in the Caco2 cells by ~5 times compared with the Vero cells (44). Replication of HcoV-EMC was notably reduced when treated with type I or type III IFN in the human airway epithelium culture (43,44).
A delay in the induction of the type I IFN response enables SARS-CoV to replicate efficiently in mice and augments the accumulation of inflammatory monocyte-macrophages (45). A lack of type I and type III IFN responses in signal transducer and activator transcription-1 knockout mice resulted in uncontrolled SARS-CoV replication with both liver and neurological consequences (46). Treatment of MERS-CoV-infected patients with IFN-α2a improved the survival rates to a maximum of 14 days (43). The type I IFN and TLR3 agonist were the most effective combined drugs for SARS/MERS CoV treatment (26).
Regarding COVID-19 infections, a clinical trial showed that treating hospitalised patients with IFN-α2b, either alone or in combination with arbidol, shortened the time of detectable viral presence in upper respiratory infections and reduced the IL-6 and C-reactive protein levels (47). Addition of IFNs to the national regime of treating COVID-19 patients reduced the 28-day mortality rate (48). The antiviral effect was augmented when Lopinavir-ritonavir was administered to mild or moderate cases of COVID-19 in combination with IFN-β 1b, and the enhancing effect was associated with a reduction in symptoms, the length of stay in hospital and viral shedding (49). In terms of COVID-19 infections and IFN responses, it was revealed that the reduced type I IFN levels in the peripheral blood system increased the expression of IL-6 and tumour necrosis factor (50). A limited type I IFN response was detected concomitantly with a large chemokine response, including production of IL-6, in the transcriptomes of SARS-CoV2 infected cells (51). In contrast, increased type I IFN and interferon stimulatory gene responses were reported in COVID-19 hospitalised patients. Several factors may underlie these contradictory results, such as the individual immune systems of patients, duration between initial infection and when the samples were obtained, and the severity of the infection (52).
Based on the similarities between the results of the present study and previous studies regarding the pattern of IFN responses, it is hypothesized that IFNs may be used as a potential treatment for management of COVID-19 infections. However, the present study has some limitations. The data assessed was done so irrespective of the severity of infections. Additionally, clinical trials will be required to assess both the safety and efficacy of IFN in managing COVID-19 infections.
In conclusion, increases in the gene expression of the key regulator of type I interferon was not shown to be effective and efficient in mounting an interferon response.
Acknowledgements
Not applicable.
Funding Statement
Funding:No funding was received.
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
AAS and MHW completed the RNA extraction and SARS-CoV-2 diagnosis. AAA-A and ZWA achieved the gene expression of the target gene and data analysis. The writing of the study was mainly conducted by ZWA. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Health Directorate (approval no. F112020) and according to an application that was made by the authors. All patients provided signed consent to participate in the present study and gave their written consent to publish any corresponding data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Docea AO, Tsatsakis A, Albulescu D, Cristea O, Zlatian O, Vinceti M, Moschos SA, Tsoukalas D, Goumenou M, Drakoulis N, et al. A new threat from an old enemy: Re emergence of coronavirus (Review) Int J Mol Med. 2020;45:1631–1643. doi: 10.3892/ijmm.2020.4555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsatsakis A, Petrakis D, Nikolouzakis TK, Docea AO, Calina D, Vinceti M, Goumenou M, Kostoff RN, Mamoulakis C, Aschner M, et al. COVID-19, an opportunity to reevaluate the correlation between long-term effects of anthropogenic pollutants on viral epidemic/pandemic events and prevalence. Food Chem Toxicol. 2020;141(111418) doi: 10.1016/j.fct.2020.111418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conti P, Caraffa A, Gallenga CE, Ross R, Kritas SK, Frydas I, Younes A, Ronconi G. Coronavirus-19 (SARS-CoV-2) induces acute severe lung inflammation via IL-1 causing cytokine storm in COVID-19: A promising inhibitory strategy. J Biol Regul Homeost Agents. 2020;34:1971–1975. doi: 10.23812/20-1-E. [DOI] [PubMed] [Google Scholar]
- 4.Shi H, Wang W, Yin J, Ouyang Y, Pang L, Feng Y, Qiao L, Guo X, Shi H, Jin R, et al. The inhibition of IL-2/IL-2R gives rise to CD8+ T cell and lymphocyte decrease through JAK1-STAT5 in critical patients with COVID-19 pneumonia. Cell Death Dis. 2020;11(429) doi: 10.1038/s41419-020-2636-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han H, Ma Q, Li C, Liu R, Zhao L, Wang W, Zhang P, Liu X, Gao G, Liu F, et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–1130. doi: 10.1080/22221751.2020.1770129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, Li JX, Yang BF, Wang L, Wang WJ, et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calina D, Docea AO, Petrakis D, Egorov AM, Ishmukhametov AA, Gabibov AG, Shtilman MI, Kostoff R, Carvalho F, Vinceti M, et al. Towards effective COVID-19 vaccines: Updates, perspectives and challenges (Review) Int J Mol Med. 2020;46:3–16. doi: 10.3892/ijmm.2020.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaacs A, Lindenmann J. Virus interference. I. The interferon. Proc R Soc Lond B Biol Sci. 1957;147:258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 11.Vilcek J, Sen GC. Interferons and other cytokines. In: Fields Virology. Fields BN, Knipe DM and Howley PM (eds). 3rd edition. Lippincott-Raven Publishers, Philadelphia, PA, pp375-399, 1996. [Google Scholar]
- 12.Deonarain R, Alcami A, Alexiou M, Dallman MJ, Gewert DR, Porter AC. Impaired antiviral response and alpha/β interferon induction in mice lacking beta interferon. J Virol. 2000;74:3404–3409. doi: 10.1128/jvi.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, et al. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 14.Collins SE, Noyce RS, Mossman KL. Innate cellular response to virus particle entry requires IRF3 but not virus replication. J Virol. 2004;78:1706–1717. doi: 10.1128/jvi.78.4.1706-1717.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 16.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, ARoulston A, Lacoste J, Hiscott J. Viral induction of the human beta interferon promoter: modulation of transcription by NF-kappaB/rel proteins and interferon regulatory factors. J Virol. 1994;68:4707–4715. doi: 10.1128/JVI.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Génin P, Vaccaro A, Civas A. The role of differential expression of human interferon - a genes in antiviral immunity. Cytokine Growth Factor Rev. 2009;20:283–295. doi: 10.1016/j.cytogfr.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Ye J, Maniatis T. Negative regulation of interferon-beta gene expression during acute and persistent virus infections. PLoS One. 2011;6(e20681) doi: 10.1371/journal.pone.0020681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weaver BK, Kumar KP, Reich NC. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi O, Akira S. MDA5/RIG-I and virus recognition. Curr Opin Immunol. 2008;20:17–22. doi: 10.1016/j.coi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, et al. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19–28. doi: 10.1016/j.immuni.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 23.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 24.Ye Y, Hauns K, Langland JO, Jacobs BL, Hogue BG. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J Virol. 2007;81:2554–2563. doi: 10.1128/JVI.01634-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Z, Tunnacliffe A. Response of human cells to desiccation: comparison with hyperosmotic stress response. J Physiol. 2004;558:181–191. doi: 10.1113/jphysiol.2004.065540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strayer DR, Dickey R, Carter WA. Sensitivity of SARS/MERS CoV to interferons and other drugs based on achievable serum concentrations in humans. Infect Disord Drug Targets. 2014;14:37–43. doi: 10.2174/1871526514666140713152858. [DOI] [PubMed] [Google Scholar]
- 27.Tsatsakis A, Calina D, Falzone L, Petrakis D, Mitrut R, Siokas V, Pennisi M, Lanza G, Libra M, Doukas SG, et al. SARS-CoV-2 pathophysiology and its clinical implications: An integrative overview of the pharmacotherapeutic management of COVID-19. Food Chem Toxicol. 2020;146(111769) doi: 10.1016/j.fct.2020.111769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura M, Funami K, Komori A, Yokoyama T, Aiba Y, Araki A, Takii Y, Ito M, Matsuyama M, Koyabu M, et al. Increased expression of Toll-like receptor 3 in intrahepatic biliary epithelial cells at sites of ductular reaction in diseased livers. Hepatol Int. 2008;2:222–230. doi: 10.1007/s12072-008-9055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu LM, Tu WJ, Zhu T, Wang XT, Tan ZL, Zhong H, Gao DY, Liang DY. IRF3 is an important molecule in the UII/UT system and mediates immune inflammatory injury in acute liver failure. Oncotarget. 2016;7:49027–49041. doi: 10.18632/oncotarget.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu RH, Schuster DM, Lee JE, Smith M, Potter J, Dhariwal G, Rosenthal K, Nathan M, Gerard GF, Rashtchian A. One-step analysis and quantification of RNA by RT-PCR: Using high-temperature reverse transcription. Focus. 2000;22:3–5. [Google Scholar]
- 31.Génin P, Morin P, Civas A. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J Virol. 2003;77:7113–7119. doi: 10.1128/jvi.77.12.7113-7119.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Lang A, Baas T, Smits SL, Katze MG, Osterhaus AD, Haagmans BL. Unraveling the complexities of the interferon response during SARS-CoV infection. Future Virol. 2009;4:71–78. doi: 10.2217/17460794.4.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan RW, Chan MC, Agnihothram S, Chan LL, Kuok DI, Fong JH, Guan Y, Poon LL, Baric RS, Nicholls JM, et al. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J Virol. 2013;87:6604–6614. doi: 10.1128/JVI.00009-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang BS, Chan KH, Cheng VC, Woo PC, Lau SK, Lam CC, Chan TL, Wu AK, Hung IF, Leung SY, et al. Comparative host gene transcription by microarray analysis early after infection of the Huh7 cell line by severe acute respiratory syndrome coronavirus and human coronavirus 229E. J Virol. 2005;79:6180–6193. doi: 10.1128/JVI.79.10.6180-6193.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Au WC, Moore PA, Lowther W, Juang YT, Pitha PM. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atwan Z, Wright J, Woodman A, Leppard KN. Promyelocytic leukemia protein isoform II inhibits infection by human adenovirus type 5 through effects on HSP70 and the interferon response. J Gen Virol. 2016;97:1955–1967. doi: 10.1099/jgv.0.000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker WE, Bozzi AT, Goldstein DR. IRF3 contributes to sepsis pathogenesis in the mouse cecal ligation and puncture model. J Leukoc Biol. 2012;92:1261–1268. doi: 10.1189/jlb.0312138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiegel M, Pichlmair A, Martínez-Sobrido L, Cros J, García-Sastre A, Haller O, Weber F. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol. 2005;79:2079–2086. doi: 10.1128/JVI.79.4.2079-2086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loukugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, Rajsbaum R, Menachery VD. SARS-CoV-2 sensitive to type I interferon pretreatment. bioRxiv: Apr 9, 2020 (Epub ahead of print). doi: 10.1101/2020.03.07.982264. [Google Scholar]
- 40.Zhao X, Guo F, Liu F, Cuconati A, Chang J, Block TM, Guo JT. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc Natl Acad Sci USA. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stertz S, Reichelt M, Spiegel M, Kuri T, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Koch G. The intracellular sites of early replication and budding of SARS-coronaviru. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cinatl J, Morgenstern B, Bauer G, Chandra P, Rabenau H, Doerr HW. Treatment of SARS with human interferons. Lancet. 2003;362:293–294. doi: 10.1016/s0140-6736(03)13973-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahlakõiv T, Ritz D, Mordstein M, DeDiego ML, Enjuanes L, Müller MA, Drosten C, Staeheli P. Combined action of type I and type III interferon restricts initial replication of severe acute respiratory syndrome coronavirus in the lung but fails to inhibit systemic virus spread. J Gen Virol. 2012;93:2601–2605. doi: 10.1099/vir.0.046284-0. [DOI] [PubMed] [Google Scholar]
- 45.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Q, Chen V, Shannon CP, Wei XS, Xiang X, Wang X, Wang ZH, Tebbutt SJ, Kollmann TR, Fish EN. Interferon-α2b Treatment for COVID-19. Front Immunol. 2020;11(1061) doi: 10.3390/v13010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davoudi-Monfared E, Rahmani H, Khalili H, Hajiabdolbaghi M, Salehi M, Abbasian L, Kazemzadeh H, Yekaninejad MS. A randomized clinical trial of the efficacy and safety of interferon beta-1a in treatment of severe COVID-19. Antimicrob Agents Chemother. 2020;64:e01061–e20. doi: 10.1016/j.intimp.2020.106903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hung IF, Lung KC, Tso EY, Liu R, Chung TW, Chu MY, Ng YY, Lo J, Chan J, Tam AR, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, Péré H, Charbit B, Bondet V, Chenevier-Gobeaux C, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, Jordan TX, Oishi K, Panis M, Sachs D, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JS, Shin EC. The type I interferon response in COVID-19: Implications for treatment. Nat Rev Immunol. 2020;20:585–586. doi: 10.1038/s41577-020-00429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author on reasonable request.