Abstract
Metabolomics is the study of profiles of small molecules in biological fluids, cells, or organs. These profiles can be thought of as the “fingerprints” left behind from chemical processes occurring in biological systems. Because of its potential for groundbreaking applications in disease diagnostics, biomarker discovery, and systems biology, metabolomics has emerged as a rapidly growing area of research. Metabolomics investigations often, but not always, involve the identification and quantification of endogenous and exogenous metabolites in biological samples. Software tools and databases play a crucial role in advancing the rigor, robustness, reproducibility, and validation of these studies. Specifically, the establishment of a robust library of spectral signatures with unique compound descriptors and atom identities plays a key role in profiling studies based on data from nuclear magnetic resonance (NMR) spectroscopy. Here, we discuss developments leading to a rigorous basis for unique identification of compounds, reproducible numbering of atoms, the compact representation of NMR spectra of metabolites and small molecules, tools for improved compound identification, quantification and visualization, and approaches toward the goal of rigorous analysis of metabolomics data.
Keywords: NMR, Metabolomics, Identification, Quantification, Numbering of atoms
1. Introduction
Owing to their close proximity to the functional endpoints that govern an organism’s phenotype, metabolites are highly informative about functional states. As a result, the metabolome, which is the collection of small molecules associated with an organism, has become the target of a rapidly growing body of research over the past decade. Metabolomics data have been utilized for systems biology, disease diagnostics, biomarker discovery, and the broader characterization of small molecules in mixtures. Information acquired from nuclear magnetic resonance (NMR) spectroscopy, mass spectrometry (MS), databases, and published literature are combined to produce an interpretation of available data [1, 2]. Although NMR has lower sensitivity when compared to MS, the utility of NMR as a rapid and nondestructive method is validated by its decades-long use in identifying and quantifying products and impurities in complex reaction mixtures [3]. NMR spectra are used routinely to identify and characterize molecules and molecular interactions in a wide range of applications [4–13]. The relatively short time needed to acquire a one-dimensional (1D) 1H NMR spectrum makes this experimental modality ideal for routine, high-throughput profiling and screening procedures.
Study design, biological sample collection, and sample preparation are key considerations prior to NMR data collection. Protocols and procedures for sample collection and preparation are often designed based on the goals of the study and the specific tissue under investigation. Protocols for sample collection from human participants in a clinical study, or from animals in the field, are varied and often require additional validation steps when compared to samples collected in the laboratory setting. Samples for NMR analysis typically require minimal preparation. NMR data can be collected at different field strengths, using a diverse set of pulse sequences and experimental parameters, including delays and number of scans. Data analysis and data interpretation practices are also diverse, and one is faced with choosing from a variety of possible workflows with potentially different outcomes [14]. For metabolic flux analysis, and for studies involving stable isotope enrichment, detailed tracing of peaks and their correspondence to moieties is required. Identification and quantification is not the only modality in which NMR data are utilized for metabolomics studies. Approaches that focus on global changes in spectral data using methods such as principal component analysis (PCA), or similar multivariate methods, are also popular. These methods also require a range of mathematical transformations during data processing and specific statistical approaches for classification and discrimination [15]. As the field develops, suggested protocols are beginning to emerge from individual laboratories [16]. As more studies and protocols develop, more process-oriented databases are likely to emerge that will make the sharing of data about the findings of studies, the detailed workflow, and the computational processes more practical. Toward this goal, tools and methodologies that improve the interpretation and validation of NMR metabolomics studies are critically needed.
This chapter focuses on metabolite identification and quantification. We examine challenging elements of the workflow, including data processing, analysis, interpretation, and deposition. We discuss tools that we have developed to address known challenges. We conclude with future approaches to address needs of the field, including our plans for new tools.
2. A Brief Overview of NMR-Based Metabolomics
The peak positions and intensities from a reference NMR spectrum generally serve as the identifying signature for a compound. Reference spectra normally are collected under specific conditions of pH, temperature, and magnetic field strength, because changes in conditions can distort the identifying signatures of compounds. The general approach to NMR-based metabolomics profiling utilizes sets of chemical shifts and intensities from reference spectra, which constitute the “fingerprint” used to detect the presence of a particular compound in the mixture of small molecules and to estimate its concentration [17]. The process of identifying and quantifying a metabolite from an 1D-1H NMR spectrum requires that NMR peaks from the experiment be matched against those of one- and two-dimensional reference spectra: for example, the experimental spectra archived at the Biological Magnetic Resonance Data Bank (BMRB) [18] or the Human Metabolome Database (HMDB) [19–21]. Full utilization of the reference NMR spectra in metabolic profiling requires the assignment of spectral transitions to the atoms of the molecule. In addition, effects of magnetic field strength on the positions and intensities of NMR peaks need to be taken into account (see Fig. 1).
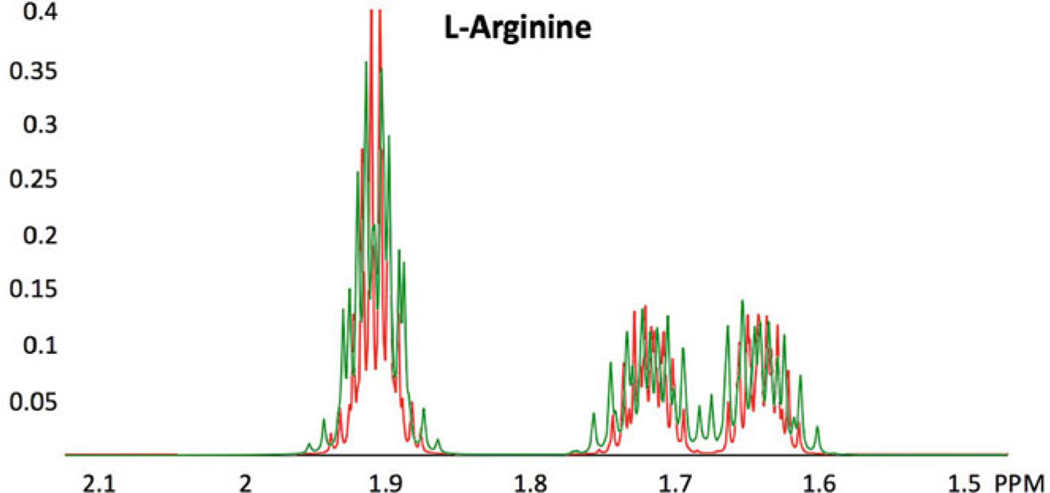
Fig. 1.
Effect of magnetic field strength on a spectrum. Comparison of simulated 1H NMR spectra of the 2.1–1.5 ppm region of L-arginine at 600 MHz (green) and 900 MHz (red). Aside from the improved resolution at 900 MHz, several peaks observed at 600 MHz are not present at 900 MHz, for example, those around 1.68 ppm. GISSMO software [22] was used in determining the spin system matrix for L-arginine by fitting the 500 MHz NMR spectrum of the compound from the BioMagResBank (BMRB) small molecule database [18]; the resulting spin system matrix was used subsequently to simulate the spectra at the two higher magnetic field strengths
Databases are being constructed to archive data from metabolomics experiments. MetabolomeXchange (http://www.metabolomexchange.org/site/) aggregates experimental data from four such repositories: Metabolites, Metabolomic Repository Bordeaux, Metabolomics Workbench, and Metabolonote. The National Institutes of Health (NIH) Common Fund Metabolomics Program has supported the establishment of a Data Repository and Coordinating Center (DRCC). The DRCC initiative is a response to the burgeoning development of databases. Proper identification of metabolites is key to establishing the identity of metabolites and the ability to cross-reference these small molecules across databases. Recent progress in establishing a unique naming and labeling convention that is entirely based on the InChI standard [23] has led to some remediation in a few databases, but much work remains to be done in this arena.
As an analytical method, NMR experimental data are highly reproducible; however, their interpretation often is subject to variability and uncertainty. A robust and reliable interpretation of data must include the following: (a) a rigorous quantification of uncertainty, (b) reliable small sample performance, and (c) wide applicability to different biological samples and sample conditions. For example, rigorous quantification must enable the comparison of uncertainty for results of experiments replicated in different laboratories. Moreover, because most exploratory data are obtained for a small number of samples, estimates of metabolite concentration and related uncertainties must be reliable despite the small number of samples.
3. What Would Constitute Optimal Tools for NMR-Based Metabolomics?
Ideally, metabolomics tools should be capable of harnessing information available on small molecules of interest in biological systems federated from a wide range of databases and other sources at all stages of an investigation. For example, in the early stages of sample preparation, information about sample collection and preparation methods and applicable data from prior studies should be easily accessible. Relevant information would include lists of compounds expected to be present in different biological fluids and tissue/organ extracts, including metabolites, drugs, and environmental compounds. NMR and MS signatures of these compounds are required, as is quantum chemical information on compounds for use in studies of molecular interactions. The keys to federating such information would be a system of easily derived unique descriptors for each compound and unique and reproducible designators for each atom present in each compound.
Libraries of known metabolites must be “dynamic” in the sense that templates for identification of metabolites must account for varying conditions such as field strength, pH, relaxation, and other effects. The tools that use library templates for analyzing the metabolomics data should integrate the information about the uncertainty of identification and quantification in their output. These tools should also identify gaps in the underlying information, such as compounds not present or improperly represented in particular databases, and assist in their remediation. In addition, data and workflow should be constructed in a way that streamlines data deposition and experimental reproducibility.
4. Naming and Numbering of Compounds for NMR (ALATIS)
Studies of small molecules rely on the unique and reproducible identification of each individual compound. The problem is that compound naming is not always unique (same descriptor can be used for more than one compound or multiple descriptors are used for a single compound). What uniquely discriminates one chemical compound from another is its three-dimensional chemical structure, which can distinguish isomers, enantiomers, and isotopomers. For this reason, we have advocated the adoption of InChI strings created from three-dimensional structure files as unique and reproducible compound identifiers. We have created an algorithm called ALATIS (Atom Label Assignment Tool using InChI String) and have incorporated it into software that produces a valid InChI string from a structure file in multiple standard formats (mol, sdf, cdx, pdb) [23]. In addition to creating the InChI string, ALATIS produces unique and reproducible numbering of all atoms in the compound. The universal ALATIS atom designators can be cross-referenced to specific atom designators in use in different scientific domains. The ALATIS approach enables the construction of validated cross-references among all small molecule databases that include 3D structures. In addition, the universal atom designators provide a mechanism for consolidating atom-specific information from these databases, such as NMR chemical shifts and coupling constants. ALATIS is embodied in a publicly available webserver located at (http://alatis.nmrfam.wisc.edu/). The ALATIS naming system has been fully implemented in the BMRB small molecule database and has been adopted by the NMReData initiative [24]. Through the application of ALATIS technology, we have been developing a federated database of information on metabolites and other small molecules that now includes more than 91,600,000 entries from PubChem and more than 400,000 entries from other databases: (http://gateway.nmrfam.wisc.edu/). Although most of these databases utilize InChI strings to represent individual compounds, our analysis has revealed widespread inconsistencies in their application that suggest the need for remediation [23].
5. NMR Templates for Metabolites and Other Compounds of Interest (GISSMO)
A spin system matrix, which parameterizes relationships among transitions, provides a much richer feature set for a compound than a spectral signature based on peak positions and intensities [25, 26]. Spin system matrices expand the applicability of NMR spectral libraries beyond the specific condition under which data were collected. In addition to their capability of simulating spectra at any field strength, spin system matrix parameters can be adjusted to systematically explore alterations in chemical shift patterns due to variations in other experimental conditions, such as pH or temperature.
Several simulation software packages have been developed for the purpose of predicting NMR experimental spectra (for a list see http://www.east-nmr.eu/index.php/databases-and-links). Among the nonproprietary software packages, NMRdb [27], GAMMA [28], and Spinach [29] are those more commonly used. The focus of these software packages is to produce an accurate approximation of the experimental data based on empirical or quantum mechanical computations—they are not designed to build spin system matrices by matching experimental spectra. A few methods have been introduced whose goal has been to automate fitting peak shapes to experimental spectra [17, 30–32]. We have developed an approach called GISSMO (Guided Ideographic Spin System Model Optimization) [22, 33] that facilitates the derivation of NMR spin system matrices by optimizing the fit between an experimental one-dimensional 1H NMR spectrum and the theoretical spin system matrix. GISSMO provides a graphical user interface (GUI) with several semi-automatic optimization modules. The GUI enables the splitting of spin system matrices so that portions can be optimized prior to merging. The GUI also allows for the consideration of couplings between 1H nuclei and other types of nuclei (e.g., 31P), and it supports the use of auxiliary two-dimensional spectra in refining chemical shifts and couplings. GISSMO utilizes ALATIS-derived compound identifiers and atom nomenclature [23].
The initial release of the GISSMO GUI reported spin system matrices for about 400 compounds from the BMRB archive [33]. In the current release, the number of entries from BMRB has increased to 511. In addition, the library now contains optimized spin system matrices for 666 molecular fragments from the Maybridge Ro3 fragment library (https://www.maybridge.com/), which are routinely utilized in NMR-based ligand screening for drug development. For every GISSMO entry in the library, we utilized ALATIS to identify its corresponding BMRB entry and applied an in-house text-processing module to extract the associated bio-locations of the compounds. The result, which is available on the GISSMO website, indicates that 338 out of 511 (66%) of the compounds have been observed in at least one cellular or tissue location. This library of optimized spin system matrices is publicly available in XML, NMR-STAR, and NMReData formats from GISSMO website. Figure 2 displays a histogram showing compounds as a function of the number of NMR spins analyzed and a histogram with a validation of the optimized spin system matrices in terms of the normalized RMSD100 between the experimental and fitted 1D-1H NMR spectra [34].
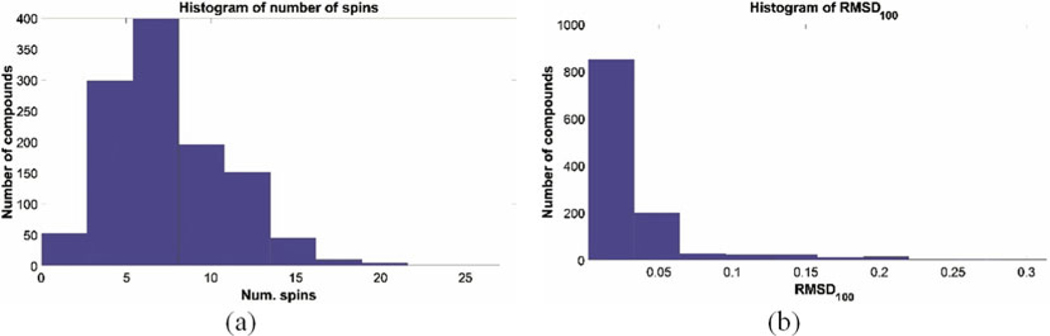
Fig. 2.
Histograms showing the number of spins and normalized RMSD of the entries in the current release of GISSMO library. (a) This histogram shows the number of spins (x-axis) versus the number of compounds (y-axis). (b) Normalized RMSD between simulated and experimental spectra. While the majority of simulated spectra are accurate representations of the experimental data (RMSD <0.1), there are cases where RMSD values are higher than 0.1 owing to relaxation effects or low signal-to-noise ratio of the experimental spectra
Because the 1H NMR spectra of many compounds are not strictly first order, a major challenge of databases containing reference NMR data is how to deal with their dependence on field strength. One approach is to collect reference spectra at a single field strength and require that this field strength be used for collecting experimental spectra. Alternatively, some reference databases contain data collected at more than one field strength. A more general solution to this challenge is to derive parametric representations of 1H NMR spectra in terms of spin system matrices, which can be utilized to generate spectra at any desired magnetic field strength. We have taken advantage of this feature of spin system matrices to produce spectra of all compounds in the GISSMO database at 1H resonance frequencies of 40, 60, 80, 90, 100, 200, 300, 400, 500, 600, 700, 750, 800, 900, 950, 1000, and 1300 MHz. These spectra, which cover the range of magnetic fields used in NMR spectroscopy and MRI, can be accessed and downloaded from the GISSMO website. To display the spectra, we utilize the open source graphing library Plotly (https://plot.ly/), which provides interactive zooming and pan features for visual investigation of the spectra. The downloadable spectra are formatted in two columns (ppm and amplitude) as a comma-separated values (CSV) file. These files can be loaded into NMR software programs such as Mestrelab Mnova (http://mestrelab.com) and NMRFx [35], which are accessible through the NMRbox project [36] (https://www.nmrbox.org/). In addition, these files can be easily loaded using any scripting and programming languages.
6. Advantages of Spectra Simulated from Spin System Matrices as References in Spectral Profiling
Spectral peak pattern matching is a common approach for profiling of small molecules. This approach involves peak picking the NMR spectra and subsequently using the resulting chemical shifts to search for matching peak patterns in a small molecule reference database. This process of identification relies strongly on the spectral peak lists, especially those archived in the reference databases. Because these databases utilize peak picking programs on their archived experimental spectra, the accuracy and reliability of the reference spectral peaks depend on a variety of factors including the correct identification of the reference compound, the presence of impurities, spectral artifacts from water signal suppression or other sources, and the signal-to-noise ratio of the experimental spectra. By contrast, the reference spectra generated from parameterized spin system matrices are noise-free, contain no impurity peaks, and are free from spectral artifacts. In addition, they serve to validate the identity of the reference compound.
6.1. Peak Lists and Searching
Generating peak lists from these highly refined spectra can be readily achieved by standard peak picking approaches. We utilized the peak picking modules of the Mnova program, under both the “Standard” and “Global Spectral Deconvolution (GSD)” options, to generate peak lists from the entire library of compounds parameterized by GISSMO at all aforementioned magnetic field strengths. We used these to generate interactive lists of chemical shifts and peak amplitudes (standard or GSD) for each compound at a selectable magnetic field strength, which are available on the GISSMO website. Clicking on a chemical shift brings up a region of the 1H NMR spectrum with the corresponding peak identified.
A “Peak Search” module is now available on the GISSMO website. The search is linked to a PostgreSQL (https://www.postgresql.org/) database containing all curated spectral peak lists. The user can query the database by specifying one or more peak positions in ppm (standard or GSD) with a specified tolerance at a selected magnetic field strength. The result is a list of compounds associated with the queried peaks within the specified matching tolerance. The compounds returned from the query are sorted based on the minimum differences between the queried peaks and those archived in the database. Users can investigate the resulting compounds by browsing the corresponding GISSMO webpages, downloading the GISSMO entries, or downloading the output in CSV format. Additional details regarding the results are provided on the website.
6.2. Simulated Spectra of Compound Mixtures
1H NMR is widely used to identify and quantify metabolites in biological fluids or tissue extracts. We are developing tools based on spin system matrix parameterizations for use in NMR-based metabolomics. As a first step, we have created a module for simulating 1H NMR spectra of compound mixtures [33]. This module utilizes the archived spin system matrices in the GISSMO library that have been manually optimized against the reference spectra. As its input, the module accepts a list of these compounds and their corresponding concentrations. Users can upload a CSV file or provide them through an interactive webpage on GISSMO’s website. The spectrum of the simulated mixture can be downloaded as a two-column CSV file. A user-controlled slider enables adjustments of the components in the simulated spectrum to achieve the best match to an experimental spectrum containing the same components. The mixture simulation module uses the Plotly library to display the spectra. As an example, Fig. 3 shows 1H NMR spectra of blood plasma simulated at two magnetic resonance fields.
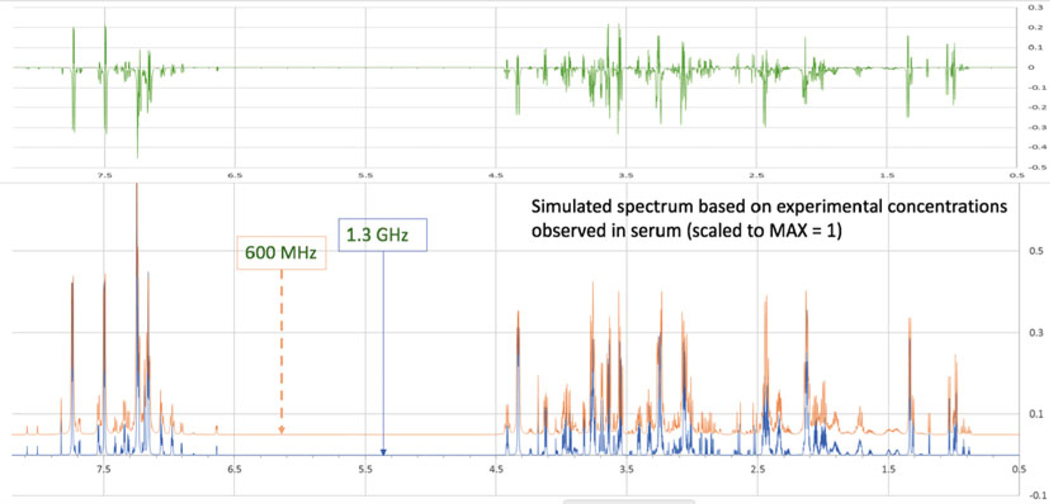
Fig. 3.
Simulated spectrum of a biological mixture created from GISSMO spectral templates. Simulated 1H NMR spectra at 600 MHz (red) and 1.3 GHz (blue) of the 34 most abundant metabolites of blood plasma at their relative concentrations. The difference spectrum (green) shown above documents the additional level of detail available at the higher magnetic field
The mixture module [33] accepts an optional experimental spectrum in CSV format and displays an overlay of the uploaded experimental and simulated mixtures on the website. Comparisons of simulated spectra with experimental spectra of mixtures can be used to validate prior analyses of compounds present and their relative concentrations. If experimental spectra of mixtures have been collected at multiple fields, the simulation can provide an additional layer of validation. Additional effects from pH differences and molecular interactions can, in principle, be incorporated into the ideographs representing the spin system matrix of each compound. We are currently working to expand this first step of processing mixture spectra by developing ways of optimizing spin system matrices against experimental spectra of biological samples.
7. Dynamic Metabolite Libraries: Archiving, Maintenance, and Retrieval of Data
The establishment of permanent and dynamic data records is a key requisite for robust scientific progress in the developing and expanding field of metabolomics. Owing to its flexible data model, the NMR-STAR ontology at the BMRB has evolved gracefully over time to include tags that accommodate almost all requirements for the archiving of metabolite library records [37]. The database contains more than 1400 entries that correspond to different experimental conditions (sample conditions and magnetic field strengths) of 1250 small molecules. For the majority of these entries, BMRB archives 10 different NMR spectra that can be used in metabolomics profiling procedures. Permanence and identity of records for small molecules has been achieved through the adoption of the ALATIS identifier, and entries with a corresponding GISSMO spin system matrix are cross-linked to GISSMO website. Data records for each metabolite contain cross-references to multiple relevant databases, including PubChem, KEGG, BioCyc, PDB, literature records, and other sources. In addition to references to the GISSMO record, the entries provide detailed data relevant to sample conditions. Additional tags for connecting additional experimental records, for example from MS, provide a path to full data integration. Because the NMR-STAR format is self-defining, it is a dynamic data record that can evolve over time in order to satisfy new demands on additional content relevant to each metabolite. Users can retrieve data from the NMR-STAR archive by means of query interfaces available on the BMRB website. The instant search on BMRB website provides a fast pattern matching tool to look up small molecule names. In addition, BMRB’s application program interface (API) (https://github.com/uwbmrb/BMRB-API) and BMRB’s FTP access (http://www.bmrb.wisc.edu/ftp/pub/bmrb/) facilitate batch download of the entries and their corresponding NMR experimental spectra. The PyNMRSTAR program (https://github.com/uwbmrb/PyNMRSTAR) has been developed for easy access and processing of NMR-STAR files. This program can be used for seamless extraction of every information archived in the corresponding NMR-STAR file of the compounds (e.g., compound name, sample condition, assigned chemical shifts, and list of experimental data associated). The reference entries of the BMRB database are constantly being expanded to cover the small molecules routinely studied in biomedical research.
8. Discussion
A number of initiatives to standardize metabolomics data formats are underway, and these efforts are expected to face the same growing pains as those in other omics fields. In the EU, the “Coordination of Standards in Metabolomics” (COSMOS) project is developing infrastructure and exchange standards for metabolomics within the European metabolomics community [38]. Along with standardization, national and international funding agencies are increasingly requesting publicly funded research data and software to become open access. In practice, changes in standards are the result of social, technical, economic, political, and legal activities, which include strategic decisions that can have a dramatic effect on day-to-day operations of the research enterprise. Moreover, the impact felt by changing practices is not homogeneous. For example, defining a new data exchange protocol has a significant impact on technology developers, but may have little or no impact on technology users. On the other hand, the use of standard vocabularies and protocols within defined laboratory management systems has its highest impact on technology users. Nonetheless, apt technological innovations can find balanced solutions that address the tension between standardization and the scientific endeavor.
For example, decoupling data exchange, data storage, and compute-cycle delivery would enable continued scientific innovation at the computational level, while, at the same time, meeting the goals of standardization and data preservation. One approach to decoupling data exchange and storage is to adopt a flexible data model. The Self-Defining Text Archive and Retrieval (STAR) specification [39, 40] has served as the model for the mmCIF format used by the Protein Data Bank [41] as well as the NMR-STAR format used by the BioMagResBank (BMRB)[37,42].NMR-STAR is a backward compatible self-defining format in which every data item has attributes that describe its features, including explicit definitions of relationships among data items. It is human-readable, and no domain knowledge is required to read the files. The format is easily converted to XML; it supports the requisite RDF (Resource Description Framework); and its flexibility provides a strong hedge against the potential of future disruptions due to format. The decoupling of compute-cycle delivery can be achieved by using a virtual machine as the technology delivery vehicle. Moreover, the use of virtual machines simplifies the long-term preservation of data and procedures. More broadly, we can improve the odds for success by adopting newer design methodologies and architectures.
We envision the use of these tools in constructing a federated database containing a vast reservoir of knowledge of relevance to metabolomics (see Fig. 4) [34]. Rigor within this database will be ensured through unique identification of compounds through standard InChI strings and unique and reproducible ALATIS atom numbering [23]. And we are developing an automated probabilistic approach to the analysis of NMR spectra of metabolite mixtures based on GISSMO-derived templates for individual compounds tuned to experimental conditions.
Fig. 4.
Proposed federation of information from multiple databases on the basis of standard InChI strings and the ALATIS universal atom numbering system [23]. The wheel lists various databases from different scientific domains that can be integrated. The inset indicates a compound with ALATIS atom numbering and arrows indicate1H NMR signals associated with particular atoms. This information can then be used in analyzing NMR spectra of compound mixtures
9. Conclusions
To enable the precise reproduction of results, it is important to define reporting requirements associated with experimental design, data acquisition, data processing, and downstream statistical analysis and interpretation. Ideally, this will include a complete description of the workflow and access to the versions of the software used. The scientific enterprise is an increasingly interconnected activity in which data exchange and data preservation are both essential requirements.
Acknowledgments
This work received funding from the National Institutes of Health (NIH). Grants from the NIH National Institute of General Medical Science provided support to the National Magnetic Resonance Facility at Madison (P41 GM103399) and the Biological Magnetic Resonance Data Bank (R01 GM109046) and partial support for HRE, HD, and JRW (P41 GM111135 to the National Center for Biomolecular NMR Data Processing and Analysis). H.D. currently is supported by National Heart, Lung, and Blood Institute grant T32 HL007575.
References
- 1.Misra BB (2018) New tools and resources in metabolomics: 2016–2017. Electrophoresis 39 (7):909–923 [DOI] [PubMed] [Google Scholar]
- 2.Misra BB, Mohapatra S (2018) Tools and resources for metabolomics research community: A 2017–2018 update. Electrophoresis. doi: 10.1002/elps.201800428. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 3.Halabalaki M, Vougogiannopoulou K, Mikros E, Skaltsounis AL (2014) Recent advances and new strategies in the NMR-based identification of natural products. Curr Opin Biotechnol 25:1–7 [DOI] [PubMed] [Google Scholar]
- 4.Ravanbakhsh S, Liu P, Bjorndahl TC, Mandal R, Grant JR, Wilson M et al. (2015) Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS One 10(5): e0124219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke CJ, Haselden JN (2008) Metabolic profiling as a tool for understanding mechanisms of toxicity. Toxicol Pathol 36(1):140–147 [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Liu L, Steffen D, Ye T, Raftery D(2012) Metabolic profiling of gender: head-space-SPME/GC–MS and 1H NMR analysis of urine. Metabolomics 8(2):323–334 [Google Scholar]
- 7.Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC et al. (2007) Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2(11):2692–2703 [DOI] [PubMed] [Google Scholar]
- 8.Jupin M, Michiels PJ, Girard FC, Spraul M, Wijmenga SS (2014) NMR metabolomics profiling of blood plasma mimics shows that medium- and long-chain fatty acids differently release metabolites from human serum albumin. J Magn Reson 239:34–43 [DOI] [PubMed] [Google Scholar]
- 9.Larive CK, Barding GA, Dinges MM (2015) NMR spectroscopy for metabolomics and metabolic profiling. Anal Chem 87(1):133–146 [DOI] [PubMed] [Google Scholar]
- 10.Elmsjo A, Rosqvist F, Engskog MK, Haglof J, Kullberg J, Iggman D et al. (2015) NMR-based metabolic profiling in healthy individuals overfed different types of fat: links to changes in liver fat accumulation and lean tissue mass. Nutr Diabetes 5:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palma M, Hernandez-Castellano LE, Castro N, Arguello A, Capote J, Matzapetakis M et al. (2016) NMR-metabolomics profiling of mammary gland secretory tissue and milk serum in two goat breeds with different levels of tolerance to seasonal weight loss. Mol BioSyst 12(7):2094–2107 [DOI] [PubMed] [Google Scholar]
- 12.Vermathen M, Paul LEH, Diserens G, Vermathen P, Furrer J (2015) 1H HR-MAS NMR based metabolic profiling of cells in response to treatment with a Hexacationic ruthenium Metallaprism as potential anticancer drug. PLoS One 10(5):e0128478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whigham LD, Butz DE, Dashti H, Tonelli M,Johnson LK, Cook ME et al. (2014) Metabolic evidence of diminished lipid oxidation in women with polycystic ovary syndrome. Curr Metabolomics 2(4):269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bingol K (2018) Recent advances in targeted and untargeted metabolomics by NMR and MS/NMR methods. High Throughput 7(2): E9. 10.3390/ht7020009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Worley B, Powers R (2014) MVAPACK: a complete data handling package for NMR metabolomics. ACS Chem Biol 9 (5):1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vignoli A, Ghini V, Meoni G, Licari C, TakisPG, Tenori L et al. (2018) High-throughput metabolomics by 1D NMR. Angew Chem Int Ed Engl. 10.1002/anie.201804736e. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Napolitano JG, Lankin DC, McAlpine JB,Niemitz M, Korhonen S-P, Chen S-N et al. (2013) Proton fingerprints portray molecular structures: enhanced description of the 1H NMR spectra of small molecules. J Org Chem 78(19):9963–9968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J et al. (2008) BioMagResBank. Nucl Acids Res 36(suppl 1):402–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y et al. (2012) HMDB 3.0--the human metabolome database in 2013. Nucl Acids Res 41(Database issue):D801–D807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wishart DS, Knox C, Guo AC, Eisner R,Young N, Gautam B et al. (2008) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37(Database issue): D603–D610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N et al. (2007) HMDB: the human Metabolome database. Nucleic Acids Res 35 (Database issue):D521–D526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dashti H, Westler WM, Tonelli M, Wedell JR, Markley JL, Eghbalnia HR (2007) Spin system modeling of nuclear magnetic resonance spectra for applications in metabolomics and small molecule screening. Anal Chem 89 (22):12201–12208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dashti H, Westler WM, Markley JL, Eghbalnia HR (2017) Unique identifiers for small molecules enable rigorous labeling of their atoms. Sci Data 4:170073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pupier M, Nuzillard JM, Wist J, Schlorer Nils E, Kuhn S, Erdelyi M et al. (2018) NMRe-DATA, a standard to report the NMR assignment and parameters of organic compounds. Magn Reson Chem 56(8):703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson WA, McConnell HM (1957) Analysis of high-resolution NMR spectra. J Chem Phys 26:1496 [Google Scholar]
- 26.Corio PL (1960) The analysis of nuclear magnetic resonance spectra. Chem Rev 60:363–429 [Google Scholar]
- 27.Binev Y, Marques MMB, Aires-de-Sousa J(2007) Prediction of 1H NMR coupling constants with associative neural networks trained for chemical shifts. J Chem Inform Modeling 47(6):2089–2097 [DOI] [PubMed] [Google Scholar]
- 28.Smith SA, Levante TO, Meier BH, Ernst RR(1994) Computer simulations in magnetic resonance. An object-oriented programming approach. J Magn Reson Series A 106 (1):75–105 [Google Scholar]
- 29.Goodwin DL, Kuprov I (2015) Auxiliarymatrix formalism for interaction representation transformations, optimal control, and spin relaxation theories. J Chem Phys 143 (8):084113 [DOI] [PubMed] [Google Scholar]
- 30.Cheshkov DA, Sinitsyn DO, Sheberstov KF, Chertkov VA (2016) Total lineshape analysis of high-resolution NMR spectra powered by simulated annealing. J Magn Reson 272:10–19 [DOI] [PubMed] [Google Scholar]
- 31.Laatikainen R, Niemitz M, Weber U, Sundelin J, Hassinen T, Vepsalainen J (1996) General strategies for total-Lineshape-type spectral analysis of NMR spectra using integral-transform iterator. J Mag Reson Series A 120(1):1–10 [Google Scholar]
- 32.Stephenson DS, Binsch G (1980) Automatedanalysis of high-resolution NMR spectra. I. Principles and computational strategy. JMagReson (1969) 37(3):395–407 [Google Scholar]
- 33.Dashti H, Wedell JR, Westler WM, Tonelli M,Aceti D, Amarasinghe GK et al. (2018) Applications of parametrized NMR spin systems of small molecules. Anal Chem 90 (18):10646–10649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dashti H, Wedell JR, Cornilescu G, Schwieters C, Westler WM, Markley JL et al. (2018) Robust nomenclature and software for enhanced reproducibility in molecular modeling of small molecules. bioRxiv:429530. 10.1101/429530 [DOI] [Google Scholar]
- 35.Norris M, Fetler B, Marchant J, Johnson BA(2016) NMRFx processor: a cross-platform NMR data processing program. J Biomol NMR 65(3–4):205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maciejewski MW, Schuyler AD, Gryk MR,Moraru I, Romero PR, Ulrich ER et al. (2017) NMRbox: a resource for biomolecular NMR computation. Biophy J 112 (8):1529–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulrich EL, Baskaran K, Dashti H, Ioannidis YE, Livny M, Romero PR et al. (2018) NMR-STAR: comprehensive ontology for representing, archiving and exchanging data from nuclear magnetic resonance spectroscopic experiments. J Biomol NMR. 10.1007/s10858-018-0220-3. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salek RM, Neumann S, Schober D, Hummel J, Billiau K, Kopka J et al. (2015) Coordination of standards in metabolomics (COSMOS): facilitating integrated metabolomics data access. Metabolomics 11(6):1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall SR (1991) The STAR file: a new format for electronic data transfer and archiving. J Chem Inf Comput Sci 31:326–333 [Google Scholar]
- 40.Hall SR, Cook APF (1995) STAR dictionary definition language: initial specification. J Chem Inf Comput Sci 35:819–825 [Google Scholar]
- 41.Fitzgerald PMD, Westbrook JD, Bourne PE, McMahon B, Watenpaugh KD, Berman HM (2005) 4.5 macromolecular dictionary (mmCIF). In: International tables for crystallography G definition and exchange of crystallographic dataedited by Hall SR, McMahon B. Springer, Dordrecht, pp 295–443 [Google Scholar]
- 42.Ulrich EL, Argentar D, Klimowicz A, Markley JL (1996) STAR/CIF macromolecular NMR data dictionaries and data file formats. Acta Crystallogr A 52(a1):C577–C577 [Google Scholar]