Abstract
Background
Coronavirus Disease 2019 (COVID-19) is a viral pandemic disease that may induce severe pneumonia in humans. In this paper, we investigated the putative implication of 12 vaccines, including BCG, OPV and MMR in the protection against COVID-19. Sequences of the main antigenic proteins in the investigated vaccines and SARS-CoV-2 proteins were compared to identify similar patterns. The immunogenic effect of identified segments was, then, assessed using a combination of structural and antigenicity prediction tools.
Results
A total of 14 highly similar segments were identified in the investigated vaccines. Structural and antigenicity prediction analysis showed that, among the identified patterns, three segments in Hepatitis B, Tetanus, and Measles proteins presented antigenic properties that can induce putative protective effect against COVID-19.
Conclusions
Our results suggest a possible protective effect of HBV, Tetanus and Measles vaccines against COVID-19, which may explain the variation of the disease severity among regions.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12859-021-04045-3.
Keywords: SARS-CoV-2, BCG, OPV, MMR, Immunogenicity, Vaccine, Putative protection
Background
Since December 2019, an emerging coronavirus called Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has been spreading worldwide. This novel pathogen is responsible for the Coronavirus Disease 2019 (COVID-19), causing a worldwide pandemic, as declared by the World Health Organization (WHO) in March 2020 [1]. So far, SARS-CoV-2 has been responsible for more than 91 million confirmed cases and more than a million fatalities (from December 8, 2019 to January 15, 2021) [2].
SARS-CoV-2 is an enveloped, positive-sense single-stranded RNA virus. It belongs to the family of Coronaviridae, the subfamily of Orthocoronavirinae and the genus Betacoronavirus [3]. The viral genome is composed of approximately 29,903 nucleotides; it contains two untranslated regions (5′ and 3′) and eleven Open Reading Frames (ORF) encoding twelve proteins including the Spike (S) and the Nucleocapsid (N) proteins identified as the main antigenic proteins [4, 5].
The number of COVID-19 patients and death cases varied from a region to another [6]. In many countries, an important number of confirmed cases were reported, such as in the United States of America (USA), Brazil, India and Russia causing [6–9]. For instance, in the USA, more than eight million confirmed cases and two hundred thousand deaths were recorded until the 18th of October 2020 [6, 9]. However, in other regions, such as Madagascar, Sierra Leone, Nicaragua and Uruguay, the number of cases seems to be limited [6, 10, 11] and no more than three hundred deaths were notified [6]. The variation in the number of deaths and infections among countries can be explained by different factors, such as health infrastructure, mitigation strategies and also cultural behavior [12, 13]. The immunological background of the population, mainly due to the vaccination strategies used in those countries was also suggested [13–15]. Indeed, it was previously demonstrated that administration of attenuated vaccines such as OPV (Oral Poliovirus Vaccine), MMR (Measles, Mumps and Rubella vaccines) and BCG (Bacillus Calmette-Guérin) vaccines could improve the innate immune response to fight different pathogens [13–15]. Furthermore, it was suggested that the adoption of a universal and long-standing BCG policy may have a protective effect against COVID-19 [13]. However, and to date, no comprehensive fundamental evidence showed a relationship between regular vaccination and the acquisition of immunity to SARS-CoV-2. In recent epidemiological study, based on a large cohort of patients, no links between the administration of BCG vaccine and COVID-19 severity was found [16]; but, after refining the epidemiological study, a strong correlation was reported [17]. The protective potential of the MMR vaccine was also investigated based on bioinformatic analysis of the S protein [18]. However, no similarity with the crystal structure of S protein, in the Wuhan-Hu-1 isolate (MN908947.3), has been reported [18].
In this paper, we investigated the putative protective role against COVID-19 of three live attenuated vaccines (BCG, OPV and MMR) and nine inactivated vaccines (Tetanus, Corynebacterium diphtheriae, Bordetella pertussis, Hepatitis B, Hepatitis A, Haemophilus influenzae type B (Hib) and Streptococcus pneumoniae vaccines (PCV10)). Our aim was to identify similar amino-acid patterns in all SARS-CoV-2 proteins and the main antigenic proteins of the above-mentioned vaccines and to predict their immunogenicity, using a combination of bioinformatic tools. The in silico identified patterns may be the target of cross-reactive antibodies against their specific pathogen and SARS-CoV-2 and/or may induce cellular immunity.
Results
Amino acid sequence alignment and hot spot analysis
The global amino acid identity between the main antigenic protein of investigated vaccines and SARS-CoV-2 proteins does not exceed 63%. For structural proteins, it varied between 21 and 55% (identity levels for the S and M proteins respectively with the Polyprotein E1/E2 of the Rubella virus and the HAV VP1 protein). For non-structural proteins, identity levels varied between 21 and 63% (identity rates of ORF1a and ORF3a proteins respectively with HBsAg-adr protein of Hepatitis B virus and Tetanus Toxin protein) (Additional file 1).
Similar segments with main vaccine antigenic proteins were identified along with structural and non-structural proteins of SARS-CoV-2. The majority were shorter than five consecutive amino-acids for all SARS-CoV-2 proteins (Additional file 2). Nevertheless, a total of twelve patterns of six to eight similar consecutive amino-acids were identified in comparison with the main antigenic proteins of Poliovirus, Measles, Streptococcus pneumoniae, Tetanus, Mumps, Hepatitis B, Hib and BCG vaccines (Table1). Two similar segments were identified through comparison of Poliovirus, Measles, PCV10 and Hib proteins and SARS-CoV-2 structural proteins (S and N) and also non-structural proteins (ORF 1a, ORF 6 and ORF 8). In contrast, Tetanus, Mumps, Hepatitis B and BCG antigenic proteins showed no more than one similar segment with SARS-CoV-2 proteins (Table1). Among the described peptides, seven were similar to others in the S protein of SARS-CoV-2 and were identified in the antigenic proteins in poliovirus Sabin 3, S pneumoniae, tetanus, Mumps, Hepatitis B and Hib vaccines. The pattern’s length varied between six and seven amino acids. Also, one peptide of eight amino acids (GTSPARMA), detected in the Poliovirus VP1 sequence, matched with the N protein of the SARS-CoV-2.
Table 1.
Description of similar patterns of more than five amino-acids obtained in vaccine antigenic proteins and SARS-CoV-2 proteins
| Vaccine | No of similar segment | Vaccine protein | SARS-CoV-2 protein | ||||
|---|---|---|---|---|---|---|---|
| Designation | Segment | Position | Designation | Segment | Position | ||
| Poliovirus | 2 | VP1 protein (Sabin 1) | GTAPARIS | 188–195 | N | GTSPARMA | 203–211 |
| VP1 protein (Sabin 3) | LDPLSE | 289–295 | S | LDPLSE | 293–299 | ||
| Measles | 2 | Fusion protein | QECLRG | 359–364 | ORF6 | QECVRG | 21–27 |
| IQVGSRR | 433–440 | ORF8 | IRVGARK | 47–54 | |||
| Streptococcus pneumoniae | 2 | Capsular polysaccharide biosynthesis protein (serotype19F, 18C, 14, 7, 4, 1) | IGFLAGVI | 182–190 | S | LGFIAGLI | 1218–1226 |
| Capsular polysaccharide biosynthesis protein (serotype19F, 18C, 14, 5) | SSVAFA | 33–39 | S | NSVAYS | 703–708 | ||
| Hib | 2 | Capsular polysaccharide biosynthesis protein | KNINDS | 210–215 | S | KNLNES | 1191–1196 |
| FILNKKI | 73–79 | ORF1a | FLLNKEM | 3183–3189 | |||
| BCG | 1 | Immunogenic protein MPB64 | IFMLVT | 5–11 | E | VFLLVT | 25–31 |
| Tetanus | 1 | Toxin protein | NILMQY | 84–90 | S | NLLLQY | 751–756 |
| Mumps virus | 1 | Fusion protein | DISTEL | 448–454 | S | DISTEI | 467–473 |
| Hepatitis B | 1 | HBs Ag-adr | PGTSTTS | 111–117 | S | PGTNTSN | 600–606 |
We also identified two discontinuous patterns of 10 amino-acids each, DISGFNSSVI and MSLSLLDLYL, in the tetanus toxin and the hemagglutinin Measles virus proteins which had 90% and 80% similarity with matching segments, DISGINASVV (1168-1177aa), IELSLIDFYL (2-11aa), in the S and ORF7b proteins of SARS-CoV-2 respectively.
Immunogenicity prediction
First, we focused on characterizing the immunogenicity of the matching sequences with S and N proteins for their involvement in modulating the immune response of the host [19, 20].
Regarding the pattern GTAPARIS matching with N protein sequence (GTSPARMA), it did not map to the structure of the N protein from SARS-CoV-2. Moreover, no significant match with CMH-I predicted epitope was distinguished. The prediction of the B-cell epitope using the N protein sequence showed a potential antigenic peptide of 51 amino acids (165–216) that harbors the pattern GTSPARMA identified from our similarity search.
Among the seven patterns identified in the SARS-CoV-2 S protein, four segments (LDPLSE, NSVAYS, NLLLQY, PGTNTSN) from Polio, PCV10, Tetanus and HBV vaccines, respectively, have been mapped on the structure of the spike protein S1 subunit (Fig. 1A). We were also able to map one other pattern, KNLNE, on the structure of the six-helical bundle fusion core solved independently (S2 subunit) from the rest of the ectodomain. The two other patterns (LGFIAGLI, and DISTEI) were not solved by the electron density map from the Cryo-EM structures. Among the five retained patterns, the segments PGTNTSN and LGFIAGLI showed a putative interaction with one of the MHC-I receptors predicted by IEDB analysis resource NetMHCpan. Furthermore, the prediction for these two peptides showed a weak peptide score of 0.07 and 0.02, respectively (0 indicates no MHC-I capacity, and 1 indicates a high probability). The segment PGTNTSN, existing in the Hbs Ag of Hepatitis B virus adr strain, is located in a turn region.
Fig. 1.
Structural mapping in S protein of the segments that match the antigenic proteins from different pathogens. A The location of the segments on the structure is marked by yellow patches. Different chains are represented in different colors. The S1 and S2 subunits have been solved independently. B B-cell epitope prediction from the sequence of SARS-CoV-2 protein. The sequences identified from the similarity analysis are marked in blue. Segments in which amino acid scores are above 0.5 are putative epitope sites. C Cumulative SASA measures for each of the putative antigenic sites calculated using different probe radii
On the other hand, the prediction of epitopes for B-cell response using Bepipred 2.0 from the IEDB analysis resource showed the implication of four putative patterns from the total set of the seven segments, namely LDPLSE, NSVAYS, DISTEI and PGTNTSN. These segments match the predicted epitopes LDPL, YTMSLGAENSVAYSNN, NLDSKVGGNYNYLYRLFRKSNLKPFERDISTEIYQAGSTPCNGVEGFNCYFPLQSYGFQPTN and TNTSN (Fig. 1B). The sequence KNLNES does not fall in a putative B-cell epitope region.
We also calculated the Solvent Accessible Surface Area (SASA) using different probe radii to allow better insight into the possible interaction of antibody Complementarity-Determining Regions (CDRs) to the predicted epitopes (Fig. 1C). Our results show that exposure to both water molecules and the antibody paratope is only preserved for the segment "PGTNTSN". Consequently, the SASA values at probe radii of 1.4 Å, 5 Å, and 10 Å are 528.69 Å2, 497.6 Å2, and 305.38 Å2, respectively.
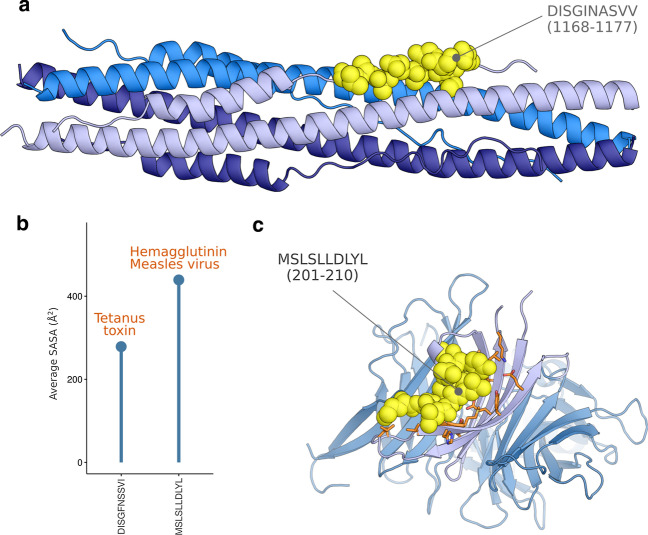
Second, we focused on a list of hits that belonged to the investigated vaccine sequences and that match any of the other proteins of SARS-CoV-2. All the patterns have been explored for their antigenic potential using IEDB Bepipred and IEDB NetMHCpan methods. None of the investigated patterns showed a significant putative B-cell antibody binding property. Discontinuous patterns with more than ten residues were discarded from the analysis as they showed low levels of similarity. Consequently, we have retained two segments from Tetanus toxin protein (DISGFNSSVI) and chain A hemagglutinin protein of the Measles virus (MSLSLLDLYL) that significantly matched SARS-CoV-2 Spike and ORF7b proteins, respectively. The segment DISGINASVV of the S protein (Fig. 2A) showed a putative interaction with the MHC-I receptor encoded by one of the corresponding HLA alleles. DISGINASVV and corresponding matching segment DISGFNSSVI showed high peptide scores of 0.88 and 0.76 for the SARS-CoV-2 S and the tetanus toxin proteins, respectively. The segment DISGINASVV is part of the six-helical bundle fusion core of the spike protein. It belongs to the HR2 domain as a random coil structure [21]. The peptide shows an extended conformation within its native environment stabilized by the residues of a small groove formed between two HR1 parallel helices from different monomers. The SASA value for DISGINASVV peptide is 504.88 Å2. In contrast, its matching sequence from Tetanus toxin DISGFNSSVI corresponds to a SASA value of 243.3 Å2 (Fig. 2B) and the Bepipred tool shows only a partial implication of the sub-string "DISGI" as an epitope in the context of B-cell response.
Fig. 2.
Structural properties of CMH-I putative epitopes resembling segments from Tetanus toxin protein and chain A of the Measles hemagglutinin Protein. A Location of DISGINASVV on the structure of the six-helical bundle fusion core from the spike protein. B Calculation of SASA for the vaccines DISGFNSSVI and MSLSLLDLYL segment using a probe of 1.4 Å. C Crystal structure of the Measles virus hemagglutinin [22]. The peptide (in yellow) shows putative T-cell immunogenicity with the interaction pocket residues (light purple)
Regarding the ORF7b and Measles hemagglutinin proteins, the identified similar segments overlap significantly with regions of putative T-cell antigenicity. The matching segment of the Measles hemagglutinin protein (Fig. 2C) corresponded to a random coil segment (MSLS) spanned by an alpha helix of six residues (LLDLYL) in the crystal structure of the hemagglutinin [22]. The segment also interacts with a large pocket formed mainly by four strands of a beta-sheet containing many aromatic amino acids. The pocket is similar to the groove of the MHC-I molecule (Fig. 2C and Additional file 3). Moreover, MSLSLLDLYL corresponds to a SASA measured at 439.19 Å2 (Fig. 2B). The NetMHCpan tool predicted an antigenicity score of 0.18 for the MSLSLLDLYL segment using the sequence of ORF7b. We also noticed that the matching segment of the Measles hemagglutinin Protein, i.e. “IELSLIDFYL” is represented by a substring “IELSLIDFY” that shows the highest antigenicity score of 0.59 among all the predicted epitopes.
Discussion
In this study, we investigated the potential protective effect against COVID-19 induced by regularly used vaccines. In the aim to assess their possible implication of in the immune response against SARS-CoV-2, we used a combination of sequence similarity analysis, structural and antigenicity prediction tools to evaluate main antigenic proteins in twelve commonly used vaccines including BCG, OPV and MMR vaccines.
In our study, we identified of similar patterns and found that most of the detected segments were shorter than five amino acids; therefore, they could not constitute a putative T-cell or B-cell epitopes [23–25].
Nevertheless, twelve patterns of six to eight amino-acids were found and further investigated. We think that PGTNTSN is the most putative to bind to endogenous antibodies among the four patterns that have been identified by the B-cell epitope prediction tool. Segments of less than 5 amino acids such as the LDPL, a substring of the LDPLSE, are rarely responsible for inducing humoral immunity response [25]. Moreover, NSVAYS and DISTEI segments are shorter with 10 and 56 amino acids less than the matching predicted epitopes using the entire sequence of the spike protein from SARS-CoV-2. In such a case, the sequence length would be a constraining factor in reproducing the immunological properties for the studied vaccines. That also applies to GTSPARMA segment which is a substring of 51 amino acid putative epitope from the N protein. On the other hand, the PGTNTSN segment of SARS-CoV-2 matches with the predicted epitope TNTSN which is only shorter by two amino acids, compared to both patterns identified for SARS-CoV-2 and its matching segment on HBs Ag-adr.
The pattern PGTNTSN detected in HBsAg of Hepatitis B virus corresponded to an exposed site in the S protein and showed the highest values of accessible surface area compared to the segments identified in the S1 subunit. Additionally, the accessibility of PGTNTSN to the probing spheres mimicking the CDRs antibodies supports its implication in the B-cell mediated response. Thus, its structural properties were consistent with its putative neutralizing capacity. Naturally, the antibodies would be able to recognize the targeted epitope on the whole assembled structure of the virus, and therefore, the epitope must be accessible at the surface of the spike protein. On the other hand, in their recent attempt to establish the antigenicity map of SARS-CoV-2, Zhang et al. have found that a segment called IDh spanning residues 522–646 induces a positive B-cells reaction in sera of convalescent COVID-19 patients [20]. The pattern PGTNTSN was included in the IDh epitope and we were able to identify strong prediction metrics using the IEDB Bepipred tool. Therefore, the induced immunological reaction by this segment would be a humoral response. Furthermore, our results were in agreement with the work published by Tajiri et al. [26] who showed that two regions of HBsAg (residues 104–123 and 108–123) containing the epitope matching the PGTNTSN segment of SARS-CoV-2, were able to bind with two human monoclonal antibodies. This highlighted the immunogenic capability of these segments. There have been concerns about the antibody-dependent enhancement (ADE) of the SARS-CoV-2 infection due to the possible activation of effector functions [27]. The antibody repertoire is thought to be the main culprit for such an effect [28]. However, its magnitude still unknown and recent evidence suggests a non-significant or unclear contribution in enhancing the infectivity of SARS-CoV-2. For instance, the expression of Fcγ receptors through which the effector functions are triggered seems to be very low in alveolar, bronchial, and nasal-cavity epithelial cells (idem). Moreover, it is difficult to distinguish the contribution of the antibody-dependent enhancement of the infection from a severity due to other factors. Recently, in a detailed review, Arvin et al. have stated that current clinical experience is insufficient to implicate a role for ADE of disease, or immune enhancement by any other mechanism, in the severity of COVID-19 [28].
The segment PGTNTSN is located away from the RBD interaction site to ACE2, separated by an approximate distance of 75 Å. However, the putative antigen, is very close to the fusion peptide SFIEDLLFNKV (residues 816–826 on the PDB structure 7BYR) located at an approximate distance of 35 Å. Moreover, the same region includes the S21P2 segment that has been identified as the epitope for antibodies targeting protein S and enabling the neutralization of the SARS-CoV-2 pseudovirus infection [28]. Therefore, it would be possible to have the same scenario for the PGTNTSN predicted epitope. Furthermore, the location of the PGTNTSN segment overlaps with a putative interaction surface with TMPRSS2 which would impact the cleavage of S1/S2 and S2 sites required for the priming of the S protein [29, 30].
On the other hand, and considering the S protein conservation, which is constantly facing a selective pressure from the immune system, several studies demonstrated the existence of highly conserved domains in the S protein such as “SD2.1” (amino acids 589–605) which matches with the ‘PGTNTSN’ segment (600–606) [31–33]. Still, only, randomized controlled trials might provide evidence of induced protective effect against COVID-19. In many countries, the HBV vaccine is commonly recommended or mandatory for healthcare and wet lab workers. Therefore, it would be interesting to investigate the prevalence of SARS-CoV-2 and clinical manifestations of COVID-19 among HBV vaccinated health workers.
Interestingly, our analysis showed the presence of two segments of ten amino acids from the Tetanus toxin protein and the chain A of the Measles hemagglutinin protein, similar to others located in the S and ORF7b proteins of SARS-CoV-2. The segment DISGINASVV, matching with the toxin tetanus protein has been previously described to be part of an antigenic peptide in the S protein of SARS-CoV-2 [34]. Trigueiro-Louro et al. performed a structure-based strategy targeting highly conserved regions in the Spike domains and demonstrated that the domain “CD-HR2.1” (amino acids 1112–1232), that matches with the regions DISGINASVV, is a “highly conserved druggable regions” [14]. Regarding the segment matching with the ORF7b protein, which may have an accessory function and whose role is yet to be determined [35], we could not exclude its possible immunogenic role. On the other hand, we have also recorded a significant global identity level between the Measles fusion and hemagglutinin proteins and SARS-CoV-2 spike, envelope and matrix proteins (45–50%) (suppl mat. 1). Furthermore, another study using other Measles and Rubella sequences, different from Edmonston Measles and Wistar RA 27/3 Rubella vaccine strains, revealed similarity between the N terminal region of SARS-COV-2 Spike protein and the Fusion protein of Measles virus as well as the envelope protein of Rubella virus. Still, no similarity was obtained with the crystal structure [18]. It was previously demonstrated that live attenuated vaccines such as OPV, BCG and MMR could improve the innate immune response to other pathogens [36]. These non-specific effects of live vaccines involved the trained immunity which refers to the memory-like characteristics of innate immune cells [37]. Indeed, following exposure to a primary stimulus like a vaccine or a microbial component, innate immune cells, especially monocytes and NK-cells, undergo epigenetic reprogramming that subsequently regulates cytokine production and cell metabolism and it collectively enhances responsiveness to an unrelated secondary stimulus. In this line, observational studies reported a decrease in hospitalization rate and overall mortality among children immunized with live attenuated vaccines [14]. Furthermore, pediatric populations seem to be less vulnerable to COVID-19, especially in low and middle- income countries [14, 38, 39]. The long-term use of an attenuated vaccine, with high coverage rate, could, partially, explain the low symptomatic infection rate among children. Thus, epidemiological studies targeting a largely vaccinated population can help in assessing the protective effect of the MMR vaccine against COVID-19.
Conclusions
Since December 2019, the novel Coronavirus, SARS-CoV-2, spread all around the word causing a worldwide pandemic, and more than 91 million confirmed cases and a million fatalities.
Using an in silico strategy, this study suggests a possible protective effect of HBV, Tetanus and Measles vaccines against SARS-CoV-2 which should be confirmed by extensive epidemiological studies targeting large populations. This possible cross-protection may explain the variation of the disease severity among countries.
Material and methods
Investigated vaccines and sequences
Our study focused on twelve vaccines including live attenuated (BCG, OPV, MMR vaccines) and inactivated ones (Tetanus, Corynebacterium diphtheriae, Bordetella pertussis, Hepatitis B, Hepatitis A, Haemophilus influenzae type B (Hib) and Streptococcus pneumoniae vaccines (PCV10) (Table 2).
Table 2.
vaccines and corresponding antigenic proteins investigated in this study
| Vaccine | Protein | Accession N° | Reference |
|---|---|---|---|
| Tetanus | Toxin protein | AAA23282.1 | [40] |
| Corynebacterium diphtheriae | Toxin protein | CAA00374.1 | [40] |
| Hepatitis B | HBsAg-adw2 | AAW65557.1 | [41] |
| HBsAg-adr | AAW65588.1 | ||
| Bordetella pertussis | Toxin protein | AQW64178.1 | [40] |
| Measles | Hemagglutinin protein | AAF85705.1 | |
| Fusion protein | AAF85704.1 | [42] | |
| Rubella | Polyprotein E1/E2 | ACN50046.1 | [42] |
| Mumps | Fusion protein | ACN50030.1 | |
| Hemagglutinin/neuraminidase protein | ACN50032.1 | [42] | |
| Hepatitis A | VP1 protein | AAA45466.1 | |
| VP3 protein | AAA45466.1 | [43, 44] | |
| Bacillus Calmette-Guérin (BCG) | Immunogenic protein MPB83 | BAA11027.1 | |
| Immunogenic protein MPB70 | BAA07402.1 | [45–47] | |
| Immunogenic protein MPB64 | AIC33023.1 | ||
| Hemophilus influenzae serotype B (Hib) | Capsulation protein | CWW30252.1 | [40] |
| Capsular polysaccharide biosynthesis protein | WP_015702013.1 | ||
| Poliovirus | VP1 protein (Sabin 1 strain) | AAL89597.1 | [48] |
| VP1 protein (Sabin 2 strain) | AAL92486.1 | ||
| VP1 protein (Sabin 3 strain) | AAL89592.1 | ||
| Streptococcus pneumoniae (PCV10) | Capsular polysaccharide biosynthesis protein [serotype 19F] | AEO88919.1 | [49] |
| Capsular polysaccharide biosynthesis protein [serotype 23F] | AAC69522.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 18C] | CAI33577.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 14] | CAI33319.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 9 V] | CAI33023.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 7F] | CAI32924.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 6B] | AAK20683.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 5] | CAI32793.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 1] | COS99248.1 | ||
| Capsular polysaccharide biosynthesis protein [serotype 4] | AAK20668.1 |
The full amino-acid sequences of the main antigenic proteins (n = 30) corresponding to the 12 investigated vaccines were obtained from NCBI Genbank database (https://www.ncbi.nlm.nih.gov). Accession numbers are listed in Table 2. In addition, the amino-acid sequences of the structural proteins (Spike (S), Envelope (E), Membrane glycoprotein (M), Nucleocapsid (N) and non-structural proteins (ORF1ab, ORF1a, ORF3a, ORF6, ORF7a, ORF7ab, ORF8 and ORF10) of SARS-CoV-2 Wuhan reference strain (NC_045512) were obtained from NCBI.
Amino acid sequence alignment and hot spot analysis
Identification of similar segments, including identical amino-acids and/or similar amino-acids (with similar biochemical properties), was assessed using Blastp homology search by querying the protein sequences of SARS-CoV-2 over the set of antigenic sequences of the vaccines [50]. Blast 2 sequences tool was used with an Expect threshold (E-value) of 10, in order to see shorter alignments, according to the stochastic model of Karlin and Altschul (1990) [51]. Pairwise alignments obtained from Blastp were explored and analyzed using BioEdit software, version 7.2.5 (http://www.mybiosoftware.com/bioedit-7-0-9-biological-sequence-alignment-editor.html).
Structural analysis and antigenicity prediction
The structure of the SARS-CoV-2 spike protein was obtained from PDB entries 7BYR [52] and 6LXT [21] corresponding to the structure of S1 and S2 subunits respectively. Both structures showed a respective sequence identity of 99.6 and 100% compared to the reference sequence of the S protein from the Wuhan-Hu-1 isolate of SARS-CoV-2 (accession number YP_009724390.1 for the spike protein). The segments matching one of the sequences of S and N proteins were mapped on the structure. The Solvent Accessible Surface Area (SASA) per residue was calculated using freesasa [52]. The B-cell and T-cell epitope predictions were conducted using IEDB analysis resource Bepipred 2.0 [29] and the IEDB analysis resource NetMHCpan [53] methods by uploading the primary structure of SARS-CoV-2 protein; considering all the possible human HLA alleles for MHC class I. These correspond to HLA genes A, B, C, E, and G and cover 134 alleles from different allele groups. A list of these alleles is provided in Additional file 4. The length of the predicted peptides was set to a default value of 8–11 residues, with respect to the proteasomal processing mechanism [54]. A pattern is retained if it shows a good quality local alignment with no indels and no more than two successive dissimilar residues. The matching pattern of the query has to show significant antigenicity prediction, at least with one of the methods, IEDB Bepipred or IEDB NetMHCpan. A cutoff of peptide score no less than 0.1 was used. At this level, the sensitivity and specificity values would be above 0.9, according to the evaluation by Jutz et al.[55]. For IEDB Bepipred, a putative epitope has to show a score above 0.5 for all its constructing amino acids.
The Solvent Accessible Surface Area (SASA) was calculated residue wise. Three probing radii were used including one that mimics the solvent molecules (1.4 Å) and two other (5 and 10 Å) to access the accessibility of the Antibody Complementarity-Determining region (CDR) to the putative B-cell epitope [56].
Supplementary Information
Additional file 1: Global amino-acid identities between structural protein sequences of SARS-CoV-2 and main antigenic proteins of investigated vaccines.
Additional file 2: Similar patterns identified between SARS-CoV-2 proteins and antigenic proteins in investigated vaccines.
Additional file 3: Structure of MHC class I heavy chain in complex with Vesicular stomatitis virus nucleoprotein (PDB code 2VAA) [57]. The binding groove floor is composed of 5 strands beta-sheet, resembling the stabilizing beta-sheet from of MSLSLLDLYL peptide within the Measles hemagglutinin Protein.
Additional file 4: List of the HLA alleles used for the prediction of CMH-I binding using IEDB analysis resource.
Acknowledgements
This study was funded by the Tunisian ministry of higher education and Scientific Research (Research laboratory: Virus, Vectors and Hosts). It was also partially supported by the European project PHINDaccess: Strengthening Omics data analysis capacities in pathogen-host interaction (Grant Agreement ID: 811034).
Authors' contributions
SH-B, HO and KG designed the study, SH-B and HO wrote the main text, HO, RT, KA and ML contributed to carry out analysis and to prepare figures, SH-B, IBM, MK and HT validated the study. All authors read and approved the final manuscript.
Authors' Information
Sondes Haddad-Boubaker is an assistant professor in Virology from the Faculty of Sciences of Tunis. She is an Assistant Professor in the Laboratory of Clinical Virology, at Pasteur Institute of Tunis, which acts as the WHO Regional Reference Laboratory for Poliomyelitis and Measles in the EMR. She is also a Professor of Clinical Virology and the coordinator of the Microbiology section in the High Institute of Health Techniques in Tunis, Tunisia. Her research interest includes molecular characterization and omics data analysis of Human viruses, especially poliovirus, enteric viruses and SARS-CoV2.
Houcemeddine Othman is a bioinformatician at the Sydney Brenner Institute for Molecular Bioscience at the University of the Witwatersrand. His main interests are pharmacogenomics, Molecular modeling, and data science. He is an avid supporter of reproducible research practices and data sharing trying to increase awareness about these issues in bioinformatics and genomics fields.
Rabeb Touati obtained a PhD in electrical engineering from the National Engineering School of Tunisia (ENIT). Currently, she has a Postdoctoral position at the Laboratory of Human Genetics (LR99ES10) at the Faculty of Medicine of Tunis (FMT). Her research interest includes genomic signal processing, bioinformatics, pattern recognition and machine learning.
Kaouther Ayouni is a PhD student in the Laboratory of Clinical Virology, at Pasteur Institute of Tunis, Tunisia.
Marwa Lakhal is a PhD student in Human Genetics at the Faculty of Medicine of Tunis, Tunisia.
Imen Ben Mustapha is a Professor of Immunology at the Faculty of Medicine of Tunis and the head of a research team at the Laboratory of Transmission, Control and Immunobiology of Infections at Institut Pasteur de Tunis, Tunisia.
Kais Ghedira is an assistant professor in Bioinformatics at the Institut Pasteur de Tunis. He is head of the research team at the Laboratory of Biomathematics, Biomathematics and Biostatistics in Institut Pasteur de Tunis, Tunisia. His research interests are mainly focused on OMICS data integration and functional genomics.
Maher Kharrat is a Professor in Human Genetics from the Faculty of Medicine of Tunis (FMT). He is a Professor of Human Genetics in 2006 at the Faculty of Medicine of Tunis (FMT). He is the head of the Genetic Human laboratory (LR99ES10) at the Faculty of Medicine of Tunis (FMT). His research interests include Human Genetics.
Henda Triki is a Professor in Virology since 2006 at the Faculty of Medicine of Tunis (FMT). She is the head of the Laboratory of Clinical Virology, which acts as the WHO Regional Reference Laboratory for Poliomyelitis and Measles in the EMR region. In 2018, she was assigned as Director of the Clinical Investigation Center entitled: “Transmissible diseases: Natural history and innovative tools for diagnostic, prevention and treatment” in Pasteur Institute of Tunis. Currently, she is a member of the National COVID-19 vaccination comity.
Funding
This study was funded by the Tunisian Ministry of Higher Education and Scientific Research (Research laboratory: Virus, Vectors and Hosts; LR20IPT10). It was also partially supported by the European project PHINDaccess: Strengthening Omics data analysis capacities in pathogen-host interaction (Grant Agreement ID: 811034).
Availability of data and materials
All data generated or analyzed as part of this study are included in this published article and its supplementary information files. Accession numbers of sequences used in this study are indicated in Table2, in the Material and Methods section of the article. All data generated are available in a public repository https://figshare.com/articles/dataset/Comparative_study_of_SARS-CoV-2_proteins_and_antigenic_proteins_in_BCG_OPV_MMR_and_other_vaccines_evidence_of_possible_putative_protective_effect/13220762
Declarations
Ethics approval and consent to participate
This study did not include Human participants or Patient data. Hence no ethical approval and consent to participate is required.
Consent for publication
Not applicable. This study did not include patients.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References:
- 1.Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19(3):149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 2.WHO. WHO Coronavirus Disease (COVID-19) Dashboard 2020 [15 January 2020]. https://covid19.who.int/.
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of V. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5(4):536–44. [DOI] [PMC free article] [PubMed]
- 4.Touati R, Haddad-Boubaker S, Ferchichi I, Messaoudi I, Ouesleti AE, Triki H, et al. Comparative genomic signature representations of the emerging COVID-19 coronavirus and other coronaviruses: high identity and possible recombination between Bat and Pangolin coronaviruses. Genomics. 2020. [DOI] [PMC free article] [PubMed]
- 5.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO. Coronavirus disease (COVID-19) weekly epidemiological update 2020 [22 October 2020]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports.
- 7.Hallal PC, Hartwig FP, Horta BL, Silveira MF, Struchiner CJ, Vidaletti LP, et al. SARS-CoV-2 antibody prevalence in Brazil: results from two successive nationwide serological household surveys. The Lancet Global health. 2020;8(11):e1390–e1398. doi: 10.1016/S2214-109X(20)30387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lauxmann MA, Santucci NE, Autran-Gomez AM. The SARS-CoV-2 Coronavirus and the COVID-19 Outbreak. Int Braz J Urol. 2020;46(Suppl 1):6–18. doi: 10.1590/s1677-5538.ibju.2020.s101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martines RB, Ritter JM, Matkovic E, Gary J, Bollweg BC, Bullock H, et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease. US Emerg Infect Dis. 2020;26(9):2005–2015. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haider N, Osman AY, Gadzekpo A, Akipede GO, Asogun D, Ansumana R, et al. Lockdown measures in response to COVID-19 in nine sub-Saharan African countries. BMJ Global Health. 2020;5(10). [DOI] [PMC free article] [PubMed]
- 11.Post LA, Argaw ST, Jones C, Moss CB, Resnick D, Singh LN, et al. A SARS-CoV-2 surveillance system in Sub-Saharan Africa: modeling study for persistence and transmission to inform policy. J Med Internet Res. 2020;22(11):e24248. doi: 10.2196/24248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riggioni C, Comberiati P, Giovannini M, Agache I, Akdis M, Alves-Correia M, et al. A compendium answering 150 questions on COVID-19 and SARS-CoV-2. Allergy. 2020;75(10):2503–2541. doi: 10.1111/all.14449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis N, Sparrow A, Ghebreyesus TA, Netea MG. Considering BCG vaccination to reduce the impact of COVID-19. Lancet. 2020;395(10236):1545–1546. doi: 10.1016/S0140-6736(20)31025-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Neill LAJ, Netea MG. BCG-induced trained immunity: can it offer protection against COVID-19? Nat Rev Immunol. 2020;20(6):335–337. doi: 10.1038/s41577-020-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vojtek I, Buchy P, Doherty TM, Hoet B. Would immunization be the same without cross-reactivity? Vaccine. 2019;37(4):539–549. doi: 10.1016/j.vaccine.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Hamiel U, Kozer E, Youngster I. SARS-CoV-2 rates in BCG-vaccinated and unvaccinated young adults. JAMA. 2020. [DOI] [PMC free article] [PubMed]
- 17.Escobar LE, Molina-Cruz A, Barillas-Mury C. BCG vaccine protection from severe coronavirus disease 2019 (COVID-19) Proc Natl Acad Sci USA. 2020;117(30):17720–17726. doi: 10.1073/pnas.2008410117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidiq KR, Sabir DK, Ali SM, Kodzius R. Does Early Childhood Vaccination Protect Against COVID-19? Front Mol Biosci. 2020;7:120. doi: 10.3389/fmolb.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Li J, Heck S, Lustigman S, Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J Virol. 2006;80(12):5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang BZ, Hu YF, Chen LL, Yau T, Tong YG, Hu JC, et al. Mining of epitopes on spike protein of SARS-CoV-2 from COVID-19 patients. Cell Res. 2020;30(8):702–704. doi: 10.1038/s41422-020-0366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S, Liu M, Wang C, Xu W, Lan Q, Feng S, et al. Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343–355. doi: 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, et al. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc Natl Acad Sci USA. 2007;104(49):19535–19540. doi: 10.1073/pnas.0707830104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dougan DA, Malby RL, Gruen LC, Kortt AA, Hudson PJ. Effects of substitutions in the binding surface of an antibody on antigen affinity. Protein Eng. 1998;11(1):65–74. doi: 10.1093/protein/11.1.65. [DOI] [PubMed] [Google Scholar]
- 24.Frankild S, de Boer RJ, Lund O, Nielsen M, Kesmir C. Amino acid similarity accounts for T cell cross-reactivity and for "holes" in the T cell repertoire. PLoS ONE. 2008;3(3):e1831. doi: 10.1371/journal.pone.0001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh H, Ansari HR, Raghava GP. Improved method for linear B-cell epitope prediction using antigen's primary sequence. PLoS ONE. 2013;8(5):e62216. doi: 10.1371/journal.pone.0062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tajiri K, Ozawa T, Jin A, Tokimitsu Y, Minemura M, Kishi H, et al. Analysis of the epitope and neutralizing capacity of human monoclonal antibodies induced by hepatitis B vaccine. Antiviral Res. 2010;87(1):40–49. doi: 10.1016/j.antiviral.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Lu LL, Suscovich TJ, Fortune SM, Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat Rev Immunol. 2018;18(1):46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arvin AM, Fink K, Schmid MA, Cathcart A, Spreafico R, Havenar-Daughton C, et al. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584(7821):353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 29.Baughn LB, Sharma N, Elhaik E, Sekulic A, Bryce AH, Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 Infection. Mayo Clin Proc. 2020;95(9):1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poh CM, Carissimo G, Wang B, Amrun SN, Lee CY, Chee RS, et al. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun. 2020;11(1):2806. doi: 10.1038/s41467-020-16638-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cagliani R, Forni D, Clerici M, Sironi M. Coding potential and sequence conservation of SARS-CoV-2 and related animal viruses. Infect Genetics Evol. 2020;83:104353. doi: 10.1016/j.meegid.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trigueiro-Louro J, Correia V, Figueiredo-Nunes I, Giria M, Rebelo-de-Andrade H. Unlocking COVID therapeutic targets: a structure-based rationale against SARS-CoV-2, SARS-CoV and MERS-CoV Spike. Comput Struct Biotechnol J. 2020;18:2117–2131. doi: 10.1016/j.csbj.2020.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–92 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y, Zhou Y, Wu H, Luo B, Chen J, Li W, et al. Identification of immunodominant sites on the spike protein of severe acute respiratory syndrome (SARS) coronavirus: implication for developing SARS diagnostics and vaccines. J Immunol. 2004;173(6):4050–4057. doi: 10.4049/jimmunol.173.6.4050. [DOI] [PubMed] [Google Scholar]
- 35.Schaecher SR, Mackenzie JM, Pekosz A. The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles. J Virol. 2007;81(2):718–731. doi: 10.1128/JVI.01691-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uthayakumar D, Paris S, Chapat L, Freyburger L, Poulet H, De Luca K. Non-specific effects of vaccines illustrated through the BCG example: from observations to demonstrations. Front Immunol. 2018;9:2869. doi: 10.3389/fimmu.2018.02869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Netea MG, Dominguez-Andres J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, et al. Defining trained immunity and its role in health and disease. Nat Rev Immunol. 2020;20(6):375–388. doi: 10.1038/s41577-020-0285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel NA. Pediatric COVID-19: Systematic review of the literature. Am J Otolaryngol. 2020;41(5):102573. doi: 10.1016/j.amjoto.2020.102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoldas MA, Yoldas H. Pediatric COVID-19 disease: a review of the recent literature. Pediatr Ann. 2020;49(7):e319–e325. doi: 10.3928/19382359-20200615-01. [DOI] [PubMed] [Google Scholar]
- 40.Dhillon S. DTPa-HBV-IPV/Hib vaccine (Infanrix hexa): a review of its use as primary and booster vaccination. Drugs. 2010;70(8):1021–1058. doi: 10.2165/11204830-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 41.Heijtink RA, Bergen P, Melber K, Janowicz ZA, Osterhaus AD. Hepatitis B surface antigen (HBsAg) derived from yeast cells (Hansenula polymorpha) used to establish an influence of antigenic subtype (adw2, adr, ayw3) in measuring the immune response after vaccination. Vaccine. 2002;20(17–18):2191–2196. doi: 10.1016/S0264-410X(02)00145-7. [DOI] [PubMed] [Google Scholar]
- 42.Tillieux SL, Halsey WS, Sathe GM, Vassilev V. Comparative analysis of the complete nucleotide sequences of measles, mumps, and rubella strain genomes contained in Priorix-Tetra and ProQuad live attenuated combined vaccines. Vaccine. 2009;27(16):2265–2273. doi: 10.1016/j.vaccine.2009.01.112. [DOI] [PubMed] [Google Scholar]
- 43.Haro I, Perez S, Garcia M, Chan WC, Ercilla G. Liposome entrapment and immunogenic studies of a synthetic lipophilic multiple antigenic peptide bearing VP1 and VP3 domains of the hepatitis A virus: a robust method for vaccine design. FEBS Lett. 2003;540(1–3):133–140. doi: 10.1016/S0014-5793(03)00249-7. [DOI] [PubMed] [Google Scholar]
- 44.Ping LH, Jansen RW, Stapleton JT, Cohen JI, Lemon SM. Identification of an immunodominant antigenic site involving the capsid protein VP3 of hepatitis A virus. Proc Natl Acad Sci USA. 1988;85(21):8281–8285. doi: 10.1073/pnas.85.21.8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harboe M, Nagai S, Patarroyo ME, Torres ML, Ramirez C, Cruz N. Properties of proteins MPB64, MPB70, and MPB80 of Mycobacterium bovis BCG. Infect Immun. 1986;52(1):293–302. doi: 10.1128/IAI.52.1.293-302.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seki M, Honda I, Fujita I, Yano I, Yamamoto S, Koyama A. Whole genome sequence analysis of Mycobacterium bovis bacillus Calmette-Guerin (BCG) Tokyo 172: a comparative study of BCG vaccine substrains. Vaccine. 2009;27(11):1710–1716. doi: 10.1016/j.vaccine.2009.01.034. [DOI] [PubMed] [Google Scholar]
- 47.Wiker HG, Nagai S, Hewinson RG, Russell WP, Harboe M. Heterogenous expression of the related MPB70 and MPB83 proteins distinguish various substrains of Mycobacterium bovis BCG and Mycobacterium tuberculosis H37Rv. Scand J Immunol. 1996;43(4):374–380. doi: 10.1046/j.1365-3083.1996.d01-61.x. [DOI] [PubMed] [Google Scholar]
- 48.Katz SL. From culture to vaccine–Salk and Sabin. N Engl J Med. 2004;351(15):1485–1487. doi: 10.1056/NEJMp048217. [DOI] [PubMed] [Google Scholar]
- 49.Croxtall JD, Keating GM. Pneumococcal polysaccharide protein D-conjugate vaccine (Synflorix; PHiD-CV) Paediatr Drugs. 2009;11(5):349–357. doi: 10.2165/11202760-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 51.Karlin S, Altschul SF. Methods for assessing the statistical significance of molecular sequence features by using general scoring schemes. Proc Natl Acad Sci USA. 1990;87(6):2264–2268. doi: 10.1073/pnas.87.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Y, Su B, Guo X, Sun W, Deng Y, Bao L, et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells. Cell. 2020;182(1):73–84 e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucl Acids Res. 2020;48(W1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kisselev AF, Akopian TN, Woo KM, Goldberg AL. The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. Implications for understanding the degradative mechanism and antigen presentation. J Biol Chem. 1999;274(6):3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 55.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Urbanowicz RA, Wang R, Schiel JE, Keck ZY, Kerzic MC, Lau P, et al. Antigenicity and immunogenicity of differentially glycosylated hepatitis C virus E2 envelope proteins expressed in mammalian and insect cells. J Virol. 2019;93(7). [DOI] [PMC free article] [PubMed]
- 57.Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Global amino-acid identities between structural protein sequences of SARS-CoV-2 and main antigenic proteins of investigated vaccines.
Additional file 2: Similar patterns identified between SARS-CoV-2 proteins and antigenic proteins in investigated vaccines.
Additional file 3: Structure of MHC class I heavy chain in complex with Vesicular stomatitis virus nucleoprotein (PDB code 2VAA) [57]. The binding groove floor is composed of 5 strands beta-sheet, resembling the stabilizing beta-sheet from of MSLSLLDLYL peptide within the Measles hemagglutinin Protein.
Additional file 4: List of the HLA alleles used for the prediction of CMH-I binding using IEDB analysis resource.
Data Availability Statement
All data generated or analyzed as part of this study are included in this published article and its supplementary information files. Accession numbers of sequences used in this study are indicated in Table2, in the Material and Methods section of the article. All data generated are available in a public repository https://figshare.com/articles/dataset/Comparative_study_of_SARS-CoV-2_proteins_and_antigenic_proteins_in_BCG_OPV_MMR_and_other_vaccines_evidence_of_possible_putative_protective_effect/13220762