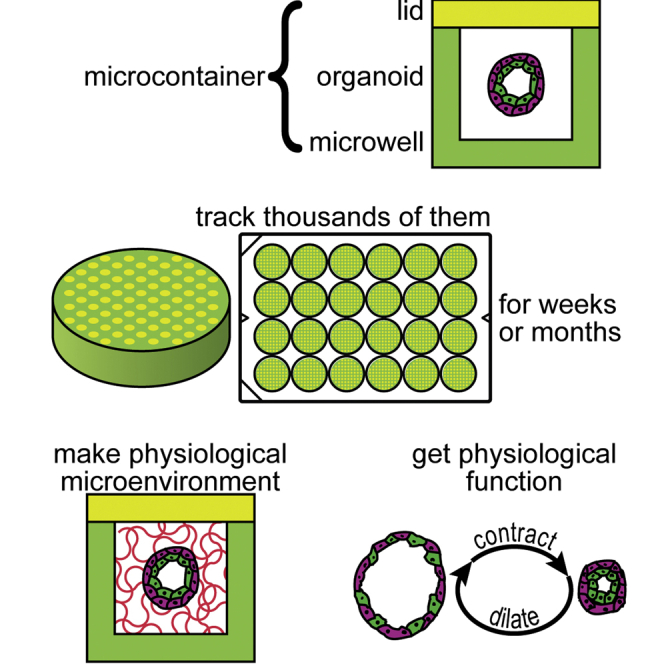
Summary
A long-standing constraint on organoid culture is the need to add exogenous substances to provide hydrogel matrix, which limits the study of fully human or fully native organoids. This paper introduces an approach to culture reconstituted mammary organoids without the impediment of exogenous matrix. We enclose organoids in nanoliter-scale, topologically enclosed, fluid compartments surrounded by agar. Organoids cultured in these “microcontainers” appear to secrete enough extracellular matrix to yield a self-sufficient microenvironment without exogenous supplements. In microcontainers, mammary organoids exhibit contractility and a high-level, physiological, myoepithelial (MEP) behavior that has not been previously reported in reconstituted organoids. The presence of contractility suggests that microcontainers elicit MEP functional differentiation, an important milestone. Microcontainers yield thousands of substantially identical and individually trackable organoids within a single culture vessel, enabling longitudinal studies and statistically powerful experiments, such as the evaluation of small effect sizes. Microcontainers open new doors for researchers who rely on organoid models.
Subject areas: Cell Biology, Stem Cells Research, Bioengineering, Tissue Engineering
Graphical abstract
Highlights
-
•
Microcontainers are volume-constrained microwells with hydrogel lids
-
•
Microcontainers enable statistically robust experimental design with organoids
-
•
Organoids produce their own extracellular matrix within microcontainers
-
•
Myoepithelial cells in mammary organoids achieve fully functional differentiation
Cell Biology; Stem Cells Research; Bioengineering; Tissue Engineering
Introduction
Organoid culture is a leading approach to obtain physiological data from human cells in the laboratory setting. Organoids provide their constituent cells with the microenvironment cues necessary to elicit native structure and function. Consequently, organoids can be better physiological models of biological tissue than cells grown on plastic, which has made organoid culture the subject of much research and development (Sachs et al., 2018). Nevertheless, organoid culture faces limitations to widespread and useful deployment. One of the most important limitations of organoid culture is its need for exogenous extracellular matrix scaffolds.
Exogenous scaffolds, ranging from semi-synthetic polymer-peptide conjugates (Cruz-Acuña and García, 2017) to jellied secretions from cancer cells (Lutolf and Gjorevski, 2018), are typically required for cells in three-dimensional culture to survive and develop into organoids. For mammary organoids, the typical scaffold is laminin-rich extracellular matrix (lrECM), such as Matrigel, which contains components of the epithelial basement membrane, as well as stromal components (Hansen et al., 2009). Although lrECM permits mammary organoids to recapitulate various physiological behaviors (Cerchiari et al., 2015; Todhunter et al., 2015), it has well-known limitations, including lot-to-lot variability, high cost, and discrepancies in both composition (Hansen et al., 2009) and structure (Kleinman et al., 1986) from bona fide basement membrane. These problems have driven a market for lrECM substitutes, but no substitute has yet been devised that emulates all aspects of physiological matrix. Scaffold-free cultures can bypass these issues for some cell types but are infeasible for anchorage-dependent cells, including human mammary epithelial cells (HMECs), which undergo anoikis under scaffold-free conditions (Hindupur et al., 2014).
Cultured HMECs express extracellular matrix components (Stampfer et al, 1981, 1993), making it puzzling that exogenous scaffolds are necessary for HMEC organoids. One plausible explanation is that cells do not express sufficient quantities of matrix for their culture systems, as the concentration of matrix polymers must exceed a minimum threshold in order to gel into a network (Yurchenco et al., 1985). In systems such as hanging droplets (Djomehri et al., 2019) or ultra-low-attachment plates (Keller et al., 2019), matrix secretions are diluted into a relatively large volume of culture media, an issue present even in systems such as droplet microfluidics (Yu et al., 2010), and dilution may prevent this gelation threshold from being reached.
To maximize the concentration of the endogenous, secreted matrix, we developed the microcontainer culture system. Within microcontainers, mammary organoids can be reconstituted from HMECs in the absence of lrECM. Microcontainer culture produces multiple arrays of 103-104 individually addressable organoids, meeting or exceeding the throughput of state-of-the-art techniques such as micropocket culture (Zhao et al., 2019). Mammary organoids in microcontainers demonstrate self-organization, polarization, and functional differentiation, including pulsatile myoepithelial (MEP) contractility (Mroue et al., 2015), a physiological behavior before now unseen in reconstituted organoids.
Results
Microcontainers enable lrECM-free organoid culture
Mammary epithelial organoids are traditionally grown in lrECM, which is typically derived from a non-human source, e.g. rodent Engelbreth-Holm-Swarm tumor cells (Hassell et al., 1980), or less often from a non-mammary human source (Okoh et al., 2013). It has been demonstrated that lrECM provides essential cues for maintaining proper organization and polarity in epithelial organoids and that culturing HMECs in suspension or low attachment cultures does not efficiently yield polarized acinar morphologies (Chanson et al., 2011). Previous work (Streuli and Bissell, 1990) has shown that mammary epithelial cells secrete matrix components under some culture conditions. We hypothesized that a confined volume could concentrate these secreted components and allow establishment of polarity cues.
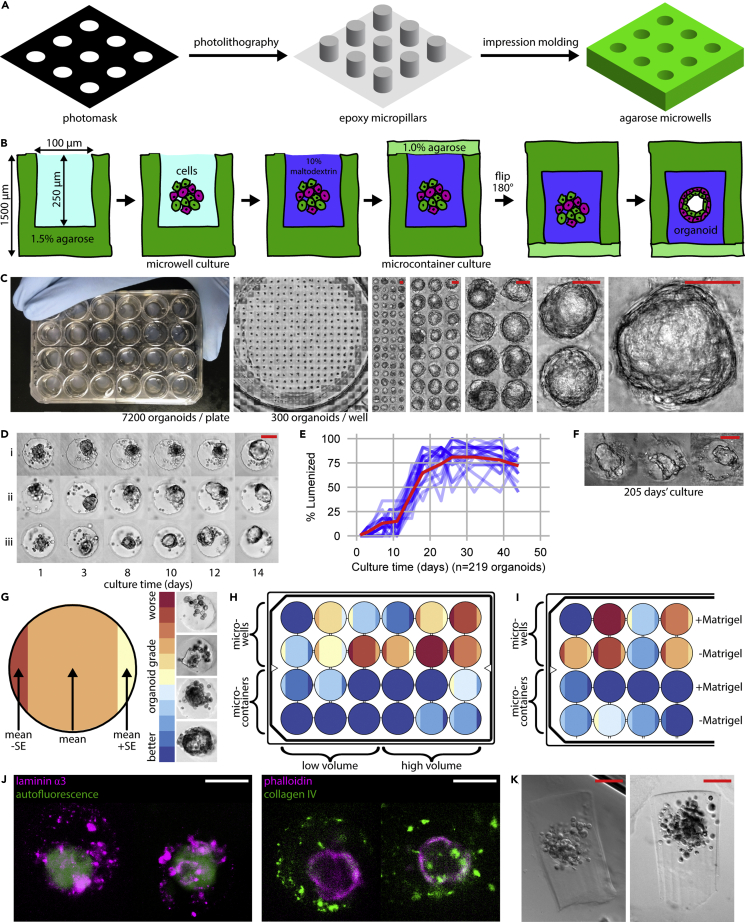
A central technical innovation in this paper is the development of the “microcontainer.” A microcontainer is a microwell made with standard photolithography techniques (Figure 1A) that, after being loaded with cells, is sealed shut with a hydrogel lid (Figure 1B), producing an enclosed liquid chamber instead of an open-topped microwell. To form the lids, microwells are first filled with a buffer brought to a high density (1.1 g/mL) with a soluble biocompatible solute such as maltodextrin or albumin (Table S1). Next, the lower density (1.0 g/mL) lid hydrogel is flowed over the tops of the microwells and allowed to gel, sealing the microwells. Buoyancy prevents the lid hydrogel from flowing into the microwells, forming sealed pockets of dilute solution (i.e., microcontainers) underneath the gelled lid. A cylindrical microcontainer with 100-μm diameter and 200-μm depth confines a population of 20-100 HMECs to a 1.6 nL volume. A hydrogel lid composed of 1% agarose has an expected pore size on the order of 100 nm (Righetti et al., 1981), slowing the diffusion of proteins (Boyer and Hsu, 1992) from the microcontainer and preventing the escape of larger macromolecular aggregates. This theory is corroborated by the ability of 15-nm quantum dots loaded into microcontainers to freely diffuse out, whereas fluorescently labeled Matrigel cannot (Figure S1A).
Figure 1.
Microcontainers enable lrECM-free organoid culture
(A) Microwells are produced by photolithography and impression molding.
(B) A novel microwell-based design encloses each organoid within a “microcontainer” of culture media.
(C) Seven thousand two hundred individually addressable organoids are grown in a single 24-well plate, shown here with a series of twofold magnifications.
(D and E) Organoids grown in microcontainer format reach stable morphology within 14 days, shown by (D) tracking individual organoids and (E) classifying the number of organoids with lumens, with each blue line showing the percent lumenization of a subgroup of 10 organoids (220 organoids total) and the red line showing the grand mean.
(F) Organoids can survive in microcontainers for months.
(G) Schematic of graphics for (H) and (I). Each well is summarized by mean +/− standard error (95% confidence interval).
(H) Qualitative assessment of organoid morphology shows differences between microcontainers and traditional microwells. n = 36 organoids/well.
(I) Microcontainers permit the growth of high-grade organoids in the absence of Matrigel. n = 36 organoids/well.
(J) Immunostaining showing laminin α3 and collagen IV secretion from organoids within microcontainers.
(K) Gelation of microcontainer contents visible upon removal from agarose. Scale bars are 50 μm.
Arrays of microcontainers provide a throughput of as many as 7200 organoids in a 24-well plate with microcontainers spaced on a grid of 500-μm pitch (Figure 1C), yielding about 300 organoids within each of the 24 wells. On a grid, each organoid is individually addressable by automated microscopy (Video S1), enabling each organoid to be tracked over months (Video S2). With this sample size, a Student's t-test can evaluate an effect size of 0.1 at the 0.005 significance level with power >0.9, and a 24-category ANOVA can evaluate an effect size of 0.1 at the 0.05 significance level with power >0.9 (Table 1).
Table 1.
Power analysis
| Power analysis, t test | |||
|---|---|---|---|
| Sample size n |
Effect size (Cohen's d) | Significance α |
Power 1 − β |
| 300 | 0.4 | 0.05 | 0.998 |
| 300 | 0.4 | 0.005 | 0.981 |
| 3600 | 0.1 | 0.05 | 0.989 |
| 3600 | 0.1 | 0.005 | 0.924 |
| Power analysis, ANOVA | ||||
|---|---|---|---|---|
| Groups | sample size n |
effect size f |
Significance α |
Power 1 − β |
| 24 | 300 | 0.09 | 0.01 | 0.997 |
| 12 | 600 | 0.08 | 0.01 | 0.996 |
| 6 | 1200 | 0.07 | 0.01 | 0.993 |
| 4 | 1800 | 0.06 | 0.01 | 0.975 |
Numbers assume a single 24-well plate of microcontainer organoids.
2880 organoids were sampled from a single experiment for this movie. Various magnifications are shown. Note that organoids were sampled with replacement, and certain organoids may appear multiple times.
All organoids in a 24-well plate are tracked across 39 days. At left, an overview of the entire well-plate. At right, a magnified view showing a region of a single well. Note that at day 26, two wells were lost.
Mammary organoid morphology develops across a two-week period. Each microcontainer is initially loaded with 20–100 individual HMECs. Within 12–48 hr, these HMECs agglomerate into a single spheroidal mass. The exact size of the spheroids depends on how many HMECs are loaded, but 100-μm diameter microcontainers readily yield spheroids with a cross-sectional area of ∼4,800 μm2 (∼78 μm diameter) (Figure S1B). Self-organization of luminal epithelial (LEP) and MEP cells forms a bilayered structure, and, over the ensuing days, a lumen forms and grows within the organoid, as measured by confocal microscopy (Figure S1C). Tracking individual organoids shows progressive lumen formation that typically reaches maximum size by 14 days of culture (Figure 1D). Applying binary classification to brightfield images of organoids allows the percentage of organoids with lumen to be non-destructively measured at each time point (Figure 1E) with reasonable sensitivity and precision (Figure S1D). Organoids can be maintained with stable lumens for many months in microcontainer formats (Figure 1F).
Initially, we observed that organoids with healthy morphology could be obtained in microcontainers containing Matrigel or collagen I at concentrations either above or below their respective gelation thresholds (Figure S1E). This was notable, as hydrogels at concentrations below their gelation threshold do not provide a substrate for cell anchorage. Next, we compared microcontainer cultures to standard microwell culture (Napolitano et al., 2007) by assessing organoid morphology in both formats. Our metric for organoid morphology was the fraction of non-squamous organoids with lumens, as assessed by brightfield microscopy (Figure 1G). Organoids were cultured for one week in media supplemented with 1 mg/mL Matrigel, a concentration below Matrigel's gelation threshold (∼3 mg/mL) (Corning, n.d.) and below the 3–10 mg/mL typically used for hydrogel-embedded organoids (Corning, n.d.). Under these culture conditions, organoids grown in microcontainers showed a greater fraction of non-squamous organoids with lumens than organoids grown in microwells (Figure 1H). We speculated that this effect may have been due to microcontainers limiting the dilution of survival-promoting factors such as growth factors or matrix components into the culture media reservoir. To test this hypothesis, microcontainers and microwells were assessed across two media volumes: either the highest volume of culture media permissible by plasticware geometry (∼1500 μL) or the lowest volume of culture media that would not desiccate the microwells (200 μL) (Figure 1H). We observed more non-squamous organoids with lumens in low-media microwells, consistent with the dilution hypothesis. In a follow-up experiment, we compared microwells with high or low media volumes to a third condition with high media volume provided by conditioned media from low media volume microwells. Conditioned high-volume microwells performed intermediate between high- and low-volume microwells, also consistent with the dilution hypothesis (Figure S1F).
Our sub-gelation Matrigel findings gave us confidence to explore Matrigel-free conditions. The organoid-culturing experiments in the above paragraph were repeated but this time entirely omitting Matrigel from the culture media. After seven days of culture, non-squamous organoids with lumens were present in microcontainers, even in the absence of Matrigel (Figure 1I). The fraction of non-squamous organoids with lumens was higher when 1 mg/mL Matrigel was present (mean = 97%) than when no Matrigel was present (mean = 73%), which suggests that although Matrigel-free conditions are feasible, the presence of Matrigel does provide some benefit to the quality of the culture.
Under Matrigel-free conditions, human extracellular matrix proteins were detected within microcontainers occupied by HMEC organoids. Immunofluorescence microscopy of whole microcontainers showed both laminin α3 and collagen IV surrounding organoids (Figure 1J). The fluorescence signal was only visible within the microcontainers, with no signal in the agarose surrounding the microcontainers. This could be explained by laminin and collagen forming macromolecular aggregates sufficiently large to retard diffusion out of the microcontainers, consistent with their known polymerization behavior (Yurchenco et al., 1985). The presence of laminin α1 (Hansen et al., 2009) and laminin α3 is notable because they are key components of the mammary basement membrane. However, Matrigel does not contain laminin α3, only laminin α1 (Giannelli et al., 1999). The presence of laminin α3 could be explained by its production by the HMECs in microcontainers. The quantity of secreted protein is substantial: a plug of hydrogel that is visible by brightfield can be removed when a microcontainer is pried open (Figure 1K). These plugs stain negative with Lugol's iodine (Figure S1G), suggesting that they are not composed of the agarose used to construct the microcontainers. Furthermore, the plugs dissolve under collagenase treatment, suggesting that the matrix protein substantially comprises them.
Microcontainer organoids show hallmarks of structure and composition
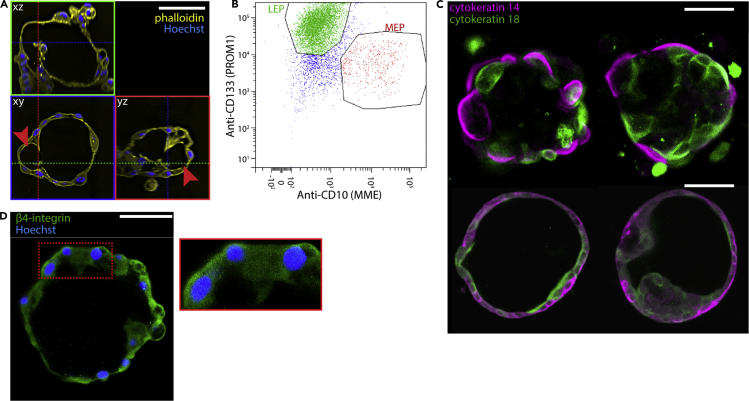
We sought to verify that HMEC organoids grown in microcontainers without lrECM conform to generally accepted hallmarks of mammary structure (Lategan, n.d.). First, microcontainer organoids reliably show lumens by 14 days of culture, apparent by confocal microscopy (Figure 2A). LEP and MEP cells were verified by both flow cytometry using antibodies against lineage-specific surface markers (CD133 for LEP cells and CD10 for MEP cells) (Figure 2B) and immunostaining using antibodies against lineage-specific cytokeratins (KRT18 for LEP cells and KRT14 for MEP cells) (Figure 2C). The abundance of MEP cells was apparently somewhat depleted relative to primary specimens (Figure S2A), but this depletion was not evident when staining for cytokeratins in intact organoids (Video S3) and may be associated with the difficulty dissociating MEP cells from organoids for flow analysis. Confocal sectioning of immunostained organoids showed self-organization of LEP cells to the apical layer and MEP cells to the basal layer (Figure 2C) (Figure S2B), and immunostaining for integrin β4 showed basal polarization (Figure 2D), as expected for mammary organoids. The establishment and maintenance of normal polarity and multi-lineage bilayers generally requires the extracellular matrix, supporting the claim that HMEC organoids in microcontainers secrete their own basement membrane components.
Figure 2.
Microcontainer organoids show hallmarks of structure and composition
(A) Organoid lumenization shown via orthogonal projection. Arrowheads denote secondary chamber within lumen.
(B) Flow cytometry plot showing sorted luminal and myoepithelial populations.
(C) Immunostaining showing bilayered organization of K14+ and K18 + cells. Organoids in the top row used cells from a 66-year-old specimen; organoids in the bottom row were from a different experiment using cells from a 19-year-old specimen.
(D) Immunostaining showing basal polarization of integrin. Zoomed inset at right. Scale bars are 50 μm.
Cytokeratin 18 (LEP lineage) pseudocolored as green and cytokeratin 14 (MEP lineage) pseudocolored as red. ∼150 organoids were sampled from an immunofluorescent staining experiment for this movie. Various magnifications are shown. Note that organoids were sampled with replacement, and certain organoids may appear multiple times.
High-level functional differentiation of myoepithelial cells
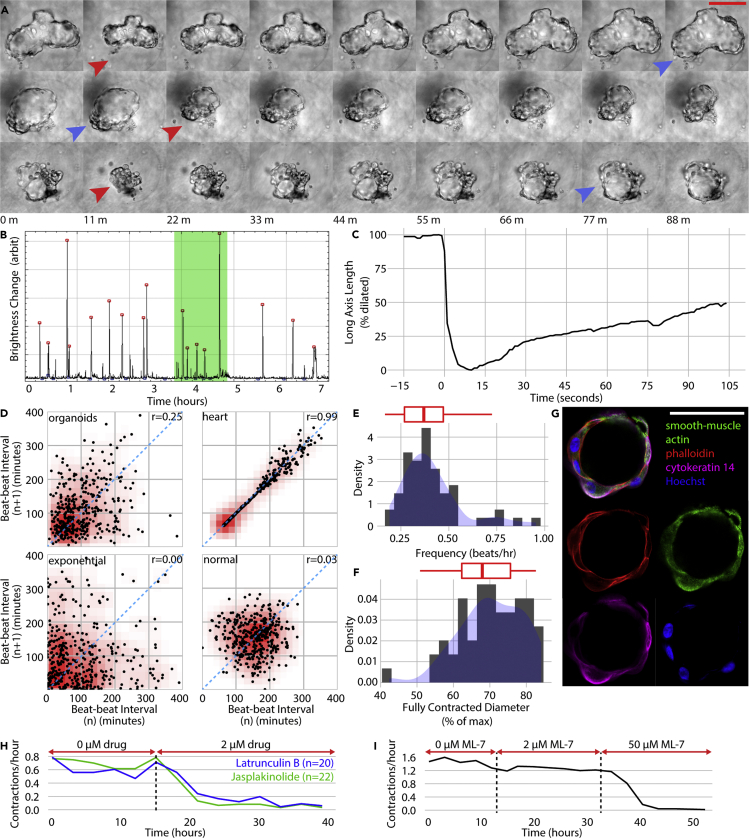
An unexpected finding in our microcontainer system is that organoids exhibit contractility. Although contractility is a known functional behavior of mammary tissue and has been observed in mouse mammary explants (Mroue et al., 2015; Sumbal et al., 2020), it has not been observed in reconstituted mammary organoids. Time-lapse microscopy showed that organoids in microcontainers gradually dilate across a span of one or more hours and then rapidly contract (Figure 3A). The dynamics of contractility vary by specimen and by experiment, but contractions typically have a frequency of ∼0.1/hr (Figure 3B), with individual contractions each spanning about 10s (Figure 3C) (Video S4). We found that the frequency of successive contractions is often correlated, as shown in Figure 3D via Poincaré plot. Poincaré plots, often used in cardiology, characterize the dynamics of pulsatile systems (Guzik et al., 2006). The temporal correlation is shown as a Pearson correlation. In the absence of temporal correlation, data points have a wide distribution and a near-zero Pearson correlation, as shown in the exponential and normal plots using simulated data. In the presence of a periodic rhythm, data points are centered elliptically on the identity line and have a high Pearson correlation, as shown in the cardiac plot. Mammary organoids show a correlation of r = 0.25, suggesting a temporally correlated but non-periodic contraction pattern. One possible explanation for such a pattern would be pulse trains (e.g., muscle clonus [Hidler and Rymer, 2000]), which can indeed be observed in organoid contractions, such as the highlighted region of Figure 3B. Physical fluidic models of lumen contractility have been proposed (Duclut et al., 2019; Ruiz-Herrero et al., 2017) but are not sufficient to predict pulse train or other temporally correlated behavior, suggesting that the contractility phenomenon is biomechanical.
Figure 3.
Mammary organoids grown in microcontainers exhibit contractility
(A) Filmstrips show single contractions of three organoids. Maximum dilation and contraction denoted with blue and red arrowheads, respectively.
(B) Overnight dynamics of a single organoid shows 17 contraction events. Green highlighting shows a sequence of high-frequency contractions.
(C) Dynamics of a single contraction.
(D) Poincaré plot embeds temporal dynamics for mammary organoids. Examples of heart, exponential, and normal distributions shown for comparison to organoid contractions. Density in red. n = 50 organoids.
(E and F) Contractility frequency (E) and magnitude (F) histograms for a population (n = 57) of organoids across 21 hr. Kernel density in blue, accompanying box-and-whisker plots in red.
(G) Immunostaining for smooth muscle actin, phalloidin, and cytokeratin 14.
(H) Latrunculin (2 μM) (n = 20 organoids) and jasplakinolide (2 μM) (n = 22 organoids) treatments, targeting actin, prevent contractions.
(I) ML-7 treatment, targeting smooth muscle myosin light-chain kinase, prevents contractions. n = 59 organoids. Scale bars are 50 μm. Code to construct (D), (H), and (I) is available in Data S1.
At left, brightfield timelapse microscopy. At right, analysis showing the first derivative of intensity with respect to time. Contractions of the organoid usually map to peaks on this plot and are denoted with red circles.
Organoid contractions exhibit high variation in frequency and magnitude, which we assessed via time-lapse microscopy on a set of 57 organoids across 48 hr (Figure 3E). Contraction magnitude was assessed by measuring the percent change of the long axis of organoids immediately before and after contractions (Figure 3F). We attempted various interventions to alter the contraction kinetics, including withdrawing cholera toxin (an activator of protein kinase A) and isoproterenol (an adrenoreceptor agonist) from the media, as well as changing the microenvironment by adding collagen I at 0 to 2 mg/mL, low-molecular-weight hyaluronan at 0 to 2 mg/mL, and Matrigel at 0 to 4 mg/mL. In all cases, no effect was observed (Table S2). Withdrawing oxytocin also showed no effect, which is notable considering that oxytocin is regarded as necessary for mammary contractions in vivo (Richardson, 1949).
Contractile function implies the presence of contractility-associated structural proteins. Staining organoids with phalloidin showed cortical actin and, more specifically, cytokeratin 14+ cells stained positive for alpha-smooth muscle actin (ASMA) (Figure 3G), a marker of the differentiated myoepithelium implicated in contractility. We tested the association between contractility and ASMA expression with inhibitor experiments. Contractile organoids were treated with either latrunculin B or jasplakinolide, inhibitors of cytoskeleton dynamics that either promote or inhibit actin polymerization, respectively. Both drugs were potent to inhibit contractions by >90% within 12 hr (Figure 3H), suggesting the necessity of actomyosin for contractions. Next, organoids were treated with ML-7, an inhibitor of the smooth muscle myosin light-chain kinase that interacts with ASMA. We observed partial inhibition of contractions with 2 μM ML-7 and >90% inhibition with 50 μM ML-7 (Figure 3I). This combination of experiments suggests that actomyosin in general and ASMA in particular are necessary for organoid contractions.
We did not detect luminal functional differentiation beyond the expression of keratin 19 and MUC1 expression and the observation of lumens. Immunostaining failed to show expression of the estrogen receptor, and prolactin treatment failed to show significant evidence of lactation: no morphological changes were induced (Figure S3A), with sudanophilia, a marker of lipophilic secretions (Dempsey et al., 1947), only evident in a single organoid (Figure S3B). Using previously described methods (Fridriksdottir et al., 2015; Meng et al., 2019), we attempted to induce estrogen receptor expression. Fourth-passage HMECs were cultured in the presence of 25 μM RepSox and 10 μM SB431542 until confluence and then incorporated into microcontainer organoids. Four conditions were tested: maintaining the drugs during microcontainer culture, withdrawing the drugs upon microcontainer culture, mixing the cells 1:10 with undrugged cells, and an undrugged negative control. We did not detect estrogen receptor expression by immunofluorescence under any of these conditions. Our evidence for increased functional differentiation is constrained to the MEP lineage.
Durability of differentiated states
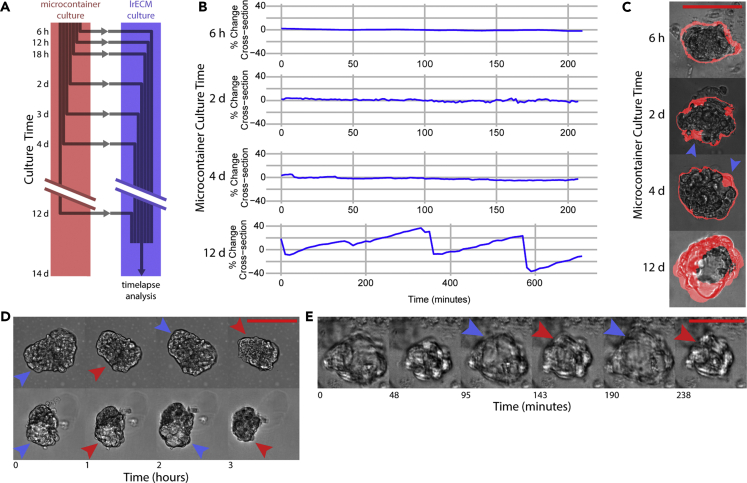
We investigated the stability of the contractile phenotype and its dependence on sustained microcontainer culture. To determine whether contractility could be sustained after organoids were removed from microcontainers, organoids were cultured in microcontainers and, after varied intervals, transferred out of the microcontainers and into embedded Matrigel culture (Figure 4A). After two weeks of culture, these organoids were imaged by time-lapse microscopy, and organoid contractility was assessed by cross-sectional area changes (Figure 4B). Organoids kept in microcontainers for less than two days showed no contractility, organoids kept in microcontainers for up to four days showed contractility localized to small regions, and organoids kept in microcontainers for at least twelve days showed global contractility (Figure 4C). In another experiment, organoids were cultured in microcontainers for two weeks and then transferred out of the microcontainers and into suspension culture, where they retained their contractility (Figure 4D). Contractility could reliably be observed in long-term cultures, with contractions observed in many organoids kept in microcontainers for 53 days (Video S5), as well as up to 157 days of sustained culture (Figure 4E). Together, this evidence suggests that the initial period of microcontainer culture causes durable changes that are sufficient to establish metastable organoid behavior.
Figure 4.
Durability of differentiated states
(A) Organoids begin culture in microcontainers and are subsequently transferred into lrECM culture for a total combined culture time of 14 days. Afterward, 24-hr time-lapse movies are taken.
(B) Time lapses are summarized in (B), graphing how the cross-sectional area of selected organoids changes over time. Alternate time scales of these data are available in Figure S4A.
(C)The spatial localization of cross-sectional changes is shown in (C), with regions of change highlighted in red. Organoids transferred at 2d or 4d show localized contractions (blue arrowheads), whereas organoids transferred at 12d show global contractions.
(D) Organoids transferred out of lrECM culture continue to show contractions.
(E) Contractile behavior persists in organoids cultured for 157 days. In (D) and (E), maximum dilation and contraction are denoted with blue and red arrowheads, respectively. Scale bars are 50 μm.
Brightfield timelapse microscopy was taken for 45 contracting organoids from a single experiment.
RNA-seq comparison of microcontainer organoids and primary cells
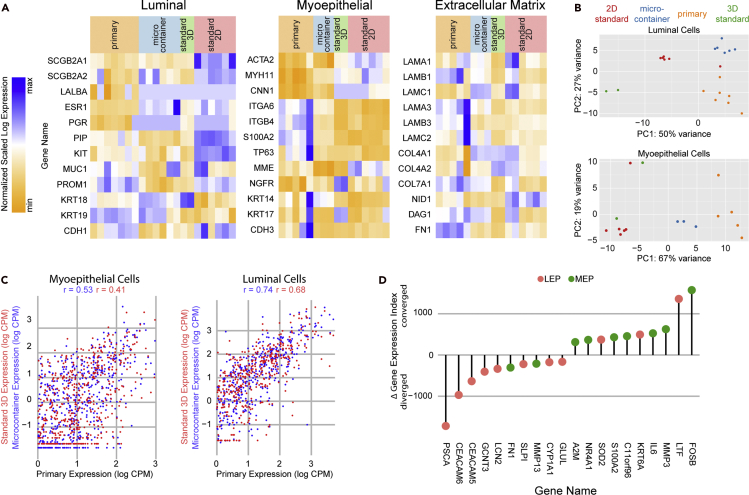
To better characterize the distinctiveness of microcontainer culture, RNA-seq (Data S2, S3, and S4) was performed on MEP cells and LEP cells across several specimens from primary tissue, microcontainer culture, best practice three-dimensional culture (using on-top Matrigel format, see transparent methods), and best practice two-dimensional cell culture (see transparent methods). About 9600 organoids were harvested from microcontainers for the RNA isolation of each specimen. Expression was evaluated for defined markers of the LEP and MEP lineages (Sayaman et al., 2021), as well as for extracellular matrix proteins, especially basement membrane components (Figures 5A and S5A). MEP cells from microcontainers show markedly increased expression of the contractility-associated genes ACTA2 and CNN1 (but not MYH11). MEP cells show reduced expression of integrins ITGA6 and ITGB4 and lineage-specific keratins KRT18 and KRT19, bringing their expression closer to the low levels seen in primary tissue. LEP cells from microcontainers show the same trend of reduced expression of lineage-specific keratin KRT14 (but not KRT17). These LEP cells also show markedly increased expression of the progenitor-associated gene KIT and mucous barrier gene MUC1. The hormone receptors ESR1 and PGR, found in the primary tissue, are absent in LEP cells from standard cell culture and remain absent in microcontainers. Major components of the basement membrane—LAMA1, LAMB1, LAMC1, LAMA3, LAMB3, LAMC2, LAMA5, COL4A1, and COL4A2—are expressed under all culture conditions analyzed, suggesting that, although microcontainers may uniquely allow the accumulation of the associated gene products, the expression of these genes is not unique to any culture condition. Moreover, principal component analysis on these curated genes (Figure 5B) showed clustering of microcontainer specimens proximal to primary specimens, especially for luminal cells.
Figure 5.
RNA-seq comparison of microcontainer organoids and primary cells
(A) Relative gene expression for characteristic genes of luminal cells, myoepithelial cells, and the extracellular matrix genes for standard two-dimensional culture, standard three-dimensional culture, microcontainer culture, and primary tissue specimens.
(B) Principal component analysis of specimens on the basis of the genes listed in (A), with specimens colored by culture method.
(C) Correlation analysis of lineage-specific genes across culture conditions, divided by lineage. Gene expression values were regularized and log-transformed as per the DESeq2 rlog function. Pearson coefficients are shown above.
(D) Lollipop plot showing the top ten genes whose expression levels most highly converged or diverged from that of primary tissue, as per the index described in Figure S5C.
We widened the scope of our RNA-seq analysis by calculating Pearson correlations on lineage-specific genes across primary tissue, microcontainer culture, and standard three-dimensional cell culture. To avoid spuriously inflating the Pearson correlation with housekeeping genes, we enriched for lineage-specific genes using differential expression (DE) analysis between MEP cells and LEP cells via DESeq2 (Love et al., 2014), constraining correlation analysis to the 5157 genes with at least two-fold DE between MEP cells and LEP cells across our set of primary tissue (Figure 5C). Although the resulting correlation coefficients appear to compare favorably with published Pearson-based analyses of mammary organoids with their tissues of origin (Rosenbluth et al., 2020), the absolute differences between Pearson correlation coefficients is small (r = 0.53 for microcontainers vs r = 0.41 for standard 3D for MEP cells and r = 0.74 for microcontainers vs r = 0.68 for standard 3D for LEP cells). However, this form of analysis may not be sensitive enough to detect meaningful differences in gene expression patterns, so we moved on to other techniques.
To further analyze differences across culture conditions, principal component and gene ontology analyses (Ashburner et al., 2000; The Gene Ontology Consortium, 2019) were performed on lineage-specific genes (as defined above). Principal component analysis showed modest clustering of samples on the basis of culture conditions (Figure S5B). To identify the genes with the greatest contributions to the principal components, varimax rotation was applied to the principal components, and the gene factors with the most extreme loadings for each rotated component were identified (Figure S5B). The second varimax-rotated component (RC2) identifies expressed genes shared by microcontainers and primary samples but not fourth-passage cells. Particularly notable among these genes, KCNQ1 stands out for its known role in apical ion transport into the mammary lumen (Abbott, 2014) and FDSCP stands out for its detectability in human milk (van Herwijnen et al., 2016) and casein homology (Kawasaki et al., 2011). As a complementary analysis to determine the most salient differences between standard 2D cell culture and microcontainer culture, genes were sorted by a gene expression distance index, calculated as the geometric mean of the additive and multiplicative gene expression differences versus primary tissue (Figure S5C). With this index, positive values denote genes whose expression in microcontainers converges on primary tissue, and negative values denote genes whose expression in microcontainers diverges from primary tissue. The top ten most converged and diverged genes are shown in Figure 5D. The same analysis was performed between standard 3D cell culture and microcontainer culture (Figure S5D), but the smaller number of standard 3D specimens limited the interpretability of this analysis. To get a broader view of gene expression changes, the 200 most converged and hundred most diverged genes were analyzed by gene set analysis using PANTHER (Mi et al., 2019), with the statistically significant (false discovery rate-adjusted p < 0.05) gene sets and fold enrichments shown in Table 2 and the breakdown of specific enrichments in Data S3. This analysis shows that microcontainer organoids approach primary tissue with respect to serine protease inhibitors, as well as the binding of growth factors, proteases, and carbohydrate derivatives. In other respects, microcontainer organoids do not resemble primary tissue, with the predominant differences being in ribosomal and rRNA genes, as well as certain extracellular-matrix-binding genes. Although microcontainers obviate the requirement of exogenous extracellular matrix in organoid cultures, these distance analyses highlight the challenges that still lie ahead.
Table 2.
Gene set enrichment analysis
| Gene set | Fold enrichment |
|---|---|
| Convergent gene expression | |
| Growth factor binding | 15 |
| Serine-type endopeptidase inhibitora | 10 |
| Protease bindinga | 7 |
| Carbohydrate derivative binding | 4 |
| Divergent gene expression | |
| rRNA bindingb | 28 |
| Structural constituent of ribosomeb | 25 |
| Extracellular matrix binding | 24 |
RNA-seq analysis indicates that the expression of particular gene sets in microcontainer-based HMEC organoids converges or diverges from the expression in primary tissue specimens. The PANTHER Go-Slim Molecular Function gene sets were chosen for this analysis. Detail in Data S5.
The majority of matched genes in these sets overlap.
The majority of matched genes in these sets overlap.
Discussion
Exogenous lrECM is generally regarded as necessary to sustain mammary epithelial organoids with normal apical-basal polarity and bilayered morphology . Microcontainers appear able to bypass this requirement, possibly due to allowing the secreted endogenous matrix to accumulate in a confined volume, allowing the matrix to provide microenvironment cues (Cerchiari et al., 2015; Chanson et al., 2011). Optimal conditions for mammary organoids in microcontainers include at least some exogenous matrix (1 mg/mL Matrigel), but Matrigel-free culture is viable to a much greater extent in microcontainers than in microwells. The profile of laminin genes expressed by organoids within microcontainers is distinct from the profile of laminins characterized in Matrigel, which suggests that the microcontainer microenvironment may provide cues that are elusive when using the exogenous matrix formulas typical to organoid culture.
MEP contractility is a key physiological function of mammary epithelia that is attainable via microcontainer culture. MEP cells, along with fibroblasts, can deform the matrix, such as by contracting collagen (Nielsen et al., 2003), and it has been shown that such contractions may be mediated through MEP motility (Buchmann et al., 2019). The pulsatility of contractions seen in microcontainers appears to be novel. Relatedly, ASMA is a key clinical marker of MEP cells (Lategan, n.d.) whose expression is either entirely absent or rapidly declines (Taylor-Papadimitriou et al., 1989) during HMEC culture. Differentiation of MEP cells toward an ASMA-expressing phenotype is influenced by media composition (Fridriksdottir et al., 2017), and we show here that microcontainer culture has a similar differentiating effect. Achieving this level of differentiation may aid examination into the tumor-suppressive functions of MEP cells. We were less successful facilitating further differentiation of luminal cells, achieving expression of neither lactation products nor sex hormone receptors. In vitro differentiation of normal human luminal cells to milk-secreting states has been a persistent challenge, and maintenance of estrogen receptor expression has so far only been reported with high concentrations of transforming growth factor beta-receptor inhibitors (Fridriksdottir et al., 2015). We speculate that adding stromal tissue components and retaining hormone response genes, possibly through cyclic application of estrous hormones and inclusion of TGF-beta receptor inhibitors in the media, could significantly narrow the gap between organoid culture and primary tissue.
Although we emphasize the significance of microcontainers in enabling lrECM-free HMEC culture, microcontainers offer other distinct advantages. Microcontainer throughput is among the highest throughput of organoid culture techniques, rivaling microwells (Cerchiari et al., 2014), micropockets (Zhao et al., 2019), and droplet microfluidics (Yu et al., 2010). The relative simplicity of the approach lends itself to scalability. Although matrix components can accumulate in microcontainers, it is likely that microcontainers are in paracrine contact with one another due to diffusion of low-molecular-weight factors through the agarose. Microcontainer culture is useful for longitudinal tracking of organoids, due to microcontainers keeping organoids in defined locations for months. Pulsatile contractility, which stands out as a functional behavior, appears to be either inhibited, absent, or unobserved in other culture systems. Taken together, microcontainer culture provides the means to study statistically robust quantities of physiologically relevant organoids without the cost or confoundment of lrECM.
A prospect for microcontainers is producing microenvironments in vitro that cannot be produced using available exogenous scaffolds. The extracellular matrix consists of a wide variety of proteins, including at least twenty-eight distinct collagens (Ricard-Blum, 2011) and at least fourteen distinct laminins that are found in different combinations in different tissues. The scope of matrices commercially available is far narrower. However, microcontainers may be able to produce usable microenvironments from any combination of matrix proteins secreted by cells. In the case of HMECs, cells produce a microenvironment enriched in laminin-332, as opposed to the laminin-111 characteristic of Matrigel. In this manner, microcontainers may provide access to a broader range of more physiological microenvironments than otherwise attainable.
It is not trivial that cells would be capable of reconstructing their native microenvironments. The notion presupposes that cells retain lineage-specific information about their native matrix composition, presumably via epigenetic states; our previous work has shown that cultured primary HMECs retain lineage-specific DNA methylation patterns consistent with uncultured tissue for at least four passages (Miyano et al., 2017). Curiously, uncultured HMECs show less expression of the laminin-332 genes than do cultured HMECs. From the perspective of dynamic reciprocity (Roskelley and Bissell, 1995), one could reason that cells produce less matrix as the abundance of native matrix increases. Or viewing cell culture as a wound healing response, perhaps cells “repair” their microenvironment by producing their native matrix. It is tempting to speculate that cells retain the necessary information to reconstruct tissue-appropriate microenvironments within their epigenetic memories.
The tissue microenvironment is characterized not only by its biochemical composition but also by its mechanical characteristics, and the mechanical characteristics of microcontainer culture are yet to be determined. Extracellular matrix mechanics are of particular interest in the mammary gland (Chaudhuri et al., 2014; Pelissier et al., 2014; Schedin and Keely, 2011). Agarose mechanically isolates microcontainers from one another, as well as from the plastic cultureware, which should limit the stiffness experienced by the organoids. It is unclear how mechanically different native extracellular matrix would be from substances like Matrigel. Since plugs of the extracellular matrix can be extracted from mature microcontainers, future work could include mechanical characterization with atomic force microscopy or rheometry.
Although the specific findings of this paper are limited to mammary epithelial cells, microcontainer culture is probably useful for culturing other cell types. Microcontainers permit the buildup of endogenous secreted matrix, which could prove useful for cells that rely on lrECM for in vitro culture, including the epithelium such as the prostate and gut or liver hepatocytes. Furthermore, stem and progenitor cells, which are exquisitely sensitive to their microenvironment, would likely benefit from the endogenous matrix. In circumstances where cells of interest cannot secrete sufficient matrix on their own, it may be feasible to combine cell types, such as stromal cells with epithelial cells. By increasing the availability of tissue-specific, species-specific microenvironments, microcontainer culture may increase the physiological validity of 3D cell culture.
Limitations of the study
Regarding the characterization of mammary organoids in microcontainers, perhaps the greatest limitation of this study is the absence of evidence of luminal differentiation—we were unable to demonstrate hormone sensitivity and we were unable to elicit lactation. Second, the microcontainers as described in this work are too small to produce entire terminal ductal lobular units, which are generally regarded as the fundamental histological and functional subunit of the mammary gland. Third, this work does not describe the incorporation of stromal cells into microcontainer systems. An ideal mammary organoid system would overcome these three shortcomings.
Regarding the capacity of microcontainers to permit organoid culture without the exogenous matrix, our claims are limited to mammary epithelial organoids. Furthermore, this work does not thoroughly characterize the composition or properties of the matrix that HMECs produce within microcontainers. It is possible that the matrix produced by HMECs within microcontainers has important deviations from normal histology, despite its endogenous origins.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Mark LaBarge (mlabarge@coh.org).
Materials availability
HMEC strains used in this study are made available either by contacting the lead contact or by contacting Dr. Martha Stampfer at http://hmec.lbl.gov.
Data and code availability
The authors declare that all data and code supporting the findings of this study are available within the paper and its supplemental information files.
Methods
All methods can be found in the accompanying transparent methods supplemental file.
Acknowledgments
The authors thank the consistent input from patient advocates Susan Samson and Sany Preto and the Roberts Academy at COH for support of D.M. This work was supported by awards from the Department of Defense/Army Breast Cancer Era of Hope Scholar Award (BC141351, BC181737), from the National Institutes of Health (U01CA244109, R33AG059206, R01EB024989, R01CA237602), Circle 1500, City of Hope Center for Cancer and Aging to M.A.L., the American Cancer Society Postdoctoral Fellowship (131311-PF-18-188-01-TBG) to M.E.T., the National Cancer Institute (NCI) Cancer Metabolism Training Program Postdoctoral Fellowship (T32CA221709) to R.W.S., and the Dr. Arthur Riggs Predoctoral Fellowship to A.F. Research reported in this publication included work performed in the Analytical Cytometry and Integrative Genomics Cores supported by the National Cancer Institute of the National Institutes of Health under grant number P30CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
M.E.T. and M.A.L. conceived the study; M.E.T., M.M., and M.A.L. designed experiments; M.E.T., M.M., D.M., and A.F. performed experiments; M.E.T., D.M., A.F., and R.W.S. analyzed and interpreted the data; and M.E.T amd M.A.L. wrote the manuscript. All authors discussed and commented on the manuscript.
Declaration of interests
A provisional patent application has been filed on the basis of this work.
Published: April 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102253.
Supplemental information
R code sufficient to recapitulate certain graphics in Figure 3.
Contains the raw counts used for all RNA analysis in this paper. Reads were aligned to Homo sapiens reference genome hg19 and assigned to HGNC symbols. This data is suitable as input for Data S4.
Contains specimen ID, chronological age, lineage (MEP/LEP), culture type, and library type for the 37 samples analyzed in this paper. This data is suitable as input for Data S4.
R code sufficient to recapitulate all analysis in Figure 5.
Contains details of gene ontology analysis. All gene hits considered converged or diverged from primary tissue are listed here, as per the methodology described in the transparent methods. The specific genes found in each of the putative gene sets as well as the full report from geneontology.org.
References
- Abbott G.W. Biology of the KCNQ1 potassium channel. New J. Sci. 2014;2014 [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Michael Cherry J., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer P.M., Hsu J.T. Experimental studies of restricted protein diffusion in an agarose matrix. Aiche J. 1992;38:259–272. [Google Scholar]
- Buchmann B., Meixner L.K., Fernandez P., Hutterer F.P., Raich M.K., Scheel C.H., Bausch A.R. Mechanical plasticity of the ECM directs invasive branching morphogenesis in human mammary gland organoids. bioRxiv. 2019 doi: 10.1101/860015. [DOI] [Google Scholar]
- Cerchiari A.E., Garbe J.C., Jee N.Y., Todhunter M.E., Broaders K.E., Peehl D.M., Desai T.A., LaBarge M.A., Thomson M., Gartner Z.J. A strategy for tissue self-organization that is robust to cellular heterogeneity and plasticity. Proc. Natl. Acad. Sci. U S A. 2015;112:2287–2292. doi: 10.1073/pnas.1410776112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerchiari A., Garbe J.C., Todhunter M.E., Jee N.Y., Pinney J.R., LaBarge M.A., Desai T.A., Gartner Z.J. Formation of spatially and geometrically controlled three-dimensional tissues in soft gels by sacrificial micromolding. Tissue Eng. Part C Methods. 2014;21:541–547. doi: 10.1089/ten.tec.2014.0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanson L., Brownfield D., Garbe J.C., Kuhn I., Stampfer M.R., Bissell M.J., LaBarge M.A. Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proc. Natl. Acad. Sci. U S A. 2011;108:3264–3269. doi: 10.1073/pnas.1019556108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri O., Koshy S.T., Branco da Cunha C., Shin J.-W., Verbeke C.S., Allison K.H., Mooney D.J. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater. 2014;13:970–978. doi: 10.1038/nmat4009. [DOI] [PubMed] [Google Scholar]
- Corning. (n.d.). Corning Matrigel Matrix Frequently Asked Questions. https://www.corning.com/catalog/cls/documents/faqs/CLS-DL-CC-026.pdf.
- Cruz-Acuña R., García A.J. Synthetic hydrogels mimicking basement membrane matrices to promote cell-matrix interactions. Matrix Biol. 2017;57-58:324–333. doi: 10.1016/j.matbio.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey E.W., Bunting H., Wislocki G.B. Observations on the chemical cytology of the mammary gland. Am. J. Anat. 1947;81:309–341. doi: 10.1002/aja.1000810302. [DOI] [PubMed] [Google Scholar]
- Djomehri S.I., Burman B., Gonzalez M.E., Takayama S., Kleer C.G. A reproducible scaffold-free 3D organoid model to study neoplastic progression in breast cancer. J. Cell Commun. Signal. 2019;13:129–143. doi: 10.1007/s12079-018-0498-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclut C., Sarkar N., Prost J., Jülicher F. Fluid pumping and active flexoelectricity can promote lumen nucleation in cell assemblies. Proc. Natl. Acad. Sci. U S A. 2019;116:19264–19273. doi: 10.1073/pnas.1908481116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridriksdottir A.J., Kim J., Villadsen R., Klitgaard M.C., Hopkinson B.M., Petersen O.W., Rønnov-Jessen L. Propagation of oestrogen receptor-positive and oestrogen-responsive normal human breast cells in culture. Nat. Commun. 2015;6:8786. doi: 10.1038/ncomms9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridriksdottir A.J., Villadsen R., Morsing M., Klitgaard M.C., Kim J., Petersen O.W., Rønnov-Jessen L. Proof of region-specific multipotent progenitors in human breast epithelia. Proc. Natl. Acad. Sci. U S A. 2017;114:E10102–E10111. doi: 10.1073/pnas.1714063114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli G., Pozzi A., Stetler-Stevenson W.G., Gardner H.A., Quaranta V. Expression of matrix metalloprotease-2-cleaved laminin-5 in breast remodeling stimulated by sex steroids. Am. J. Pathol. 1999;154:1193–1201. doi: 10.1016/S0002-9440(10)65371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik P., Piskorski J., Krauze T., Wykretowicz A., Wysocki H. Heart rate asymmetry by Poincaré plots of RR intervals. Biomed. Tech. 2006;51:272–275. doi: 10.1515/BMT.2006.054. [DOI] [PubMed] [Google Scholar]
- Hansen K.C., Kiemele L., Maller O., O’Brien J., Shankar A., Fornetti J., Schedin P. An in-solution ultrasonication-assisted digestion method for improved extracellular matrix proteome coverage. Mol. Cell. Proteomics. 2009;8:1648–1657. doi: 10.1074/mcp.M900039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassell J.R., Robey P.G., Barrach H.J., Wilczek J., Rennard S.I., Martin G.R. Isolation of a heparan sulfate-containing proteoglycan from basement membrane. Proc. Natl. Acad. Sci. U S A. 1980;77:4494–4498. doi: 10.1073/pnas.77.8.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidler J.M., Rymer W.Z. Limit cycle behavior in spasticity: analysis and evaluation. IEEE Trans. Biomed. Eng. 2000;47:1565–1575. doi: 10.1109/10.887937. [DOI] [PubMed] [Google Scholar]
- Hindupur S.K., Balaji S.A., Saxena M., Pandey S., Sravan G.S., Heda N., Kumar M.V., Mukherjee G., Dey D., Rangarajan A. Identification of a novel AMPK-PEA15 axis in the anoikis-resistant growth of mammary cells. Breast Cancer Res. 2014;16:420. doi: 10.1186/s13058-014-0420-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K., Lafont A.-G., Sire J.-Y. The evolution of milk casein genes from tooth genes before the origin of mammals. Mol. Biol. Evol. 2011;28:2053–2061. doi: 10.1093/molbev/msr020. [DOI] [PubMed] [Google Scholar]
- Keller F., Rudolf R., Hafner M. Towards optimized breast cancer 3D spheroid mono- and co-culture models for pharmacological research and screening. J. Cell Biotechnol. 2019;5:89–101. [Google Scholar]
- Kleinman H.K., McGarvey M.L., Hassell J.R., Star V.L., Cannon F.B., Laurie G.W., Martin G.R. Basement membrane complexes with biological activity. Biochemistry. 1986;25:312–318. doi: 10.1021/bi00350a005. [DOI] [PubMed] [Google Scholar]
- Lategan, B. (n.d.). Normal histology of breast [WWW Document]. PathologyOutlines.com. http://www.pathologyoutlines.com/topic/breastnormal.html (accessed 4.20.20).
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutolf, M., Gjorevski, N. 2018. Three dimensional hydrogels for culturing organoids. US Patent. 20180258403:A1.
- Meng P., Vaapil M., Tagmount A., Loguinov A., Vulpe C., Yaswen P. Propagation of functional estrogen receptor positive normal human breast cells in 3D cultures. Breast Cancer Res. Treat. 2019;176:131–140. doi: 10.1007/s10549-019-05229-5. [DOI] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyano M., Sayaman R.W., Stoiber M.H., Lin C.-H., Stampfer M.R., Brown J.B., LaBarge M.A. Age-related gene expression in luminal epithelial cells is driven by a microenvironment made from myoepithelial cells. Aging. 2017;9:2026–2051. doi: 10.18632/aging.101298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroue R., Inman J., Mott J., Budunova I., Bissell M.J. Asymmetric expression of connexins between luminal epithelial- and myoepithelial- cells is essential for contractile function of the mammary gland. Dev. Biol. 2015;399:15–26. doi: 10.1016/j.ydbio.2014.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano A.P., Dean D.M., Man A.J., Youssef J., Ho D.N., Rago A.P., Lech M.P., Morgan J.R. Scaffold-free three-dimensional cell culture utilizing micromolded nonadhesive hydrogels. Biotechniques. 2007;43:494, 496–500. doi: 10.2144/000112591. [DOI] [PubMed] [Google Scholar]
- Nielsen H.L., Gudjonsson T., Villadsen R., Rønnov-Jessen L., Petersen O.W. Collagen gel contraction serves to rapidly distinguish epithelial-and mesenchymal-derived cells irrespective of α-smooth muscle actin expression. In Vitro Cell. Dev. Biol. Anim. 2003;39:297–303. doi: 10.1290/1543-706X(2003)039<0297:CGCSTR>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Okoh V.O., Felty Q., Parkash J., Poppiti R., Roy D. Reactive oxygen species via redox signaling to PI3K/AKT pathway contribute to the malignant growth of 4-hydroxy estradiol-transformed mammary epithelial cells. PLoS One. 2013;8:e54206. doi: 10.1371/journal.pone.0054206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelissier F.A., Garbe J.C., Ananthanarayanan B., Miyano M., Lin C., Jokela T., Kumar S., Stampfer M.R., Lorens J.B., LaBarge M.A. Age-related dysfunction in mechanotransduction impairs differentiation of human mammary epithelial progenitors. Cell Rep. 2014;7:1926–1939. doi: 10.1016/j.celrep.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard-Blum S. The collagen family. Cold Spring Harb. Perspect. Biol. 2011;3:a004978. doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson K.C. Contractile tissues in the mammary gland, with special reference to myoepithelium in the goat. Proc. R. Soc. Lond. B Biol. Sci. 1949;136:30–45. doi: 10.1098/rspb.1949.0004. [DOI] [PubMed] [Google Scholar]
- Righetti P.G., Brost B.C.W., Snyder R.S. On the limiting pore size of hydrophilic gels for electrophoresis and isoelectric focussing. J. Biochem. Biophys. Methods. 1981;4:347–363. doi: 10.1016/0165-022x(81)90075-0. [DOI] [PubMed] [Google Scholar]
- Rosenbluth J.M., Schackmann R.C.J., Gray G.K., Selfors L.M., Li C.M.-C., Boedicker M., Kuiken H.J., Richardson A., Brock J., Garber J. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat. Commun. 2020;11:1711. doi: 10.1038/s41467-020-15548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskelley C.D., Bissell M.J. Dynamic reciprocity revisited: a continuous, bidirectional flow of information between cells and the extracellular matrix regulates mammary epithelial cell function. Biochem. Cell Biol. 1995;73:391–397. doi: 10.1139/o95-046. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrero T., Alessandri K., Gurchenkov B.V., Nassoy P., Mahadevan L. Organ size control via hydraulically gated oscillations. Development. 2017;144:4422–4427. doi: 10.1242/dev.153056. [DOI] [PubMed] [Google Scholar]
- Sachs N., de Ligt J., Kopper O., Gogola E., Bounova G., Weeber F., Balgobind A.V., Wind K., Gracanin A., Begthel H. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172:373–386.e10. doi: 10.1016/j.cell.2017.11.010. [DOI] [PubMed] [Google Scholar]
- Sayaman R.W., Miyano M., Senapati P., Shalabi S., Zirbes A., Todhunter M.E., Seewaldt V., Neuhausen S.L., Stampfer M.R., Schones D.E., LaBarge M.A. Epigenetic changes with age primes mammary luminal epithelia for cancer initiation. bioRxiv. 2021 doi: 10.1101/2021.02.12.430777. [DOI] [Google Scholar]
- Schedin P., Keely P.J. Mammary gland ECM remodeling, stiffness, and mechanosignaling in normal development and tumor progression. Cold Spring Harb. Perspect. Biol. 2011;3:a003228. doi: 10.1101/cshperspect.a003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfer M.R., Vlodavsky I., Smith H.S., Ford R., Becker F.F., Riggs J. Fibronectin production by human mammary cells. J. Natl. Cancer Inst. 1981;67:253–261. [PubMed] [Google Scholar]
- Stampfer M.R., Yaswen P., Alhadeff M., Hosoda J. TGF beta induction of extracellular matrix associated proteins in normal and transformed human mammary epithelial cells in culture is independent of growth effects. J. Cell. Physiol. 1993;155:210–221. doi: 10.1002/jcp.1041550127. [DOI] [PubMed] [Google Scholar]
- Streuli C.H., Bissell M.J. Expression of extracellular matrix components is regulated by substratum. J. Cell Biol. 1990;110:1405–1415. doi: 10.1083/jcb.110.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumbal J., Chiche A., Charifou E., Koledova Z., Li H. Primary mammary organoid model of lactation and involution. Front. Cell Dev. Biol. 2020;8:68. doi: 10.3389/fcell.2020.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Papadimitriou J., Stampfer M., Bartek J., Lewis A., Boshell M., Lane E.B., Leigh I.M. Keratin expression in human mammary epithelial cells cultured from normal and malignant tissue: relation to in vivo phenotypes and influence of medium. J. Cell Sci. 1989;94(Pt 3):403–413. doi: 10.1242/jcs.94.3.403. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todhunter M.E., Jee N.Y., Hughes A.J., Coyle M.C., Cerchiari A., Farlow J., Garbe J.C., LaBarge M.A., Desai T.A., Gartner Z.J. Programmed synthesis of three-dimensional tissues. Nat. Methods. 2015;12:975–981. doi: 10.1038/nmeth.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herwijnen M.J.C., Zonneveld M.I., Goerdayal S., Nolte-’t Hoen E.N.M., Garssen J., Stahl B., Maarten Altelaar A.F., Redegeld F.A., Wauben M.H.M. Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol. Cell. Proteomics. 2016;15:3412–3423. doi: 10.1074/mcp.M116.060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L., Chen M.C.W., Cheung K.C. Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing. Lab Chip. 2010;10:2424–2432. doi: 10.1039/c004590j. [DOI] [PubMed] [Google Scholar]
- Yurchenco P.D., Tsilibary E.C., Charonis A.S., Furthmayr H. Laminin polymerization in vitro. Evidence for a two-step assembly with domain specificity. J. Biol. Chem. 1985;260:7636–7644. [PubMed] [Google Scholar]
- Zhao L., Mok S., Moraes C. Micropocket hydrogel devices for all-in-one formation, assembly, and analysis of aggregate-based tissues. Biofabrication. 2019;11:045013. doi: 10.1088/1758-5090/ab30b4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2880 organoids were sampled from a single experiment for this movie. Various magnifications are shown. Note that organoids were sampled with replacement, and certain organoids may appear multiple times.
All organoids in a 24-well plate are tracked across 39 days. At left, an overview of the entire well-plate. At right, a magnified view showing a region of a single well. Note that at day 26, two wells were lost.
Cytokeratin 18 (LEP lineage) pseudocolored as green and cytokeratin 14 (MEP lineage) pseudocolored as red. ∼150 organoids were sampled from an immunofluorescent staining experiment for this movie. Various magnifications are shown. Note that organoids were sampled with replacement, and certain organoids may appear multiple times.
At left, brightfield timelapse microscopy. At right, analysis showing the first derivative of intensity with respect to time. Contractions of the organoid usually map to peaks on this plot and are denoted with red circles.
Brightfield timelapse microscopy was taken for 45 contracting organoids from a single experiment.
R code sufficient to recapitulate certain graphics in Figure 3.
Contains the raw counts used for all RNA analysis in this paper. Reads were aligned to Homo sapiens reference genome hg19 and assigned to HGNC symbols. This data is suitable as input for Data S4.
Contains specimen ID, chronological age, lineage (MEP/LEP), culture type, and library type for the 37 samples analyzed in this paper. This data is suitable as input for Data S4.
R code sufficient to recapitulate all analysis in Figure 5.
Contains details of gene ontology analysis. All gene hits considered converged or diverged from primary tissue are listed here, as per the methodology described in the transparent methods. The specific genes found in each of the putative gene sets as well as the full report from geneontology.org.
Data Availability Statement
The authors declare that all data and code supporting the findings of this study are available within the paper and its supplemental information files.