Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is one of the most important swine viruses globally, including in Ontario, Canada. Understanding the evolution and relation of the various PRRSV genotypes in Ontario can provide insight into the epidemiology of the virus. The objectives of this study were to i) describe the variability of PRRSV genotypes in Ontario swine herds, and ii) evaluate possible groupings based on PRRSV genomic data. Virus open reading frame 5 (ORF-5) sequences collected from 2010 to 2018 were obtained from the Animal Health Laboratory, University of Guelph and Bayesian phylogenetic models were created from these. The PRRSV population of Ontario was then categorized into 10 distinct clades. Model comparisons indicated that the model with a constant population assumption fit the data best, which suggests that the net change in the PRRS virus variation of the entire population over the last decade was low. Nonetheless, viruses grouped into individual clades showed temporal clustering during distinct time intervals of the entire study period (P < 0.01).
Résumé
Le virus du syndrome reproducteur et respiratoire porcin (VSRRP) est l’un des virus porcins les plus importants au monde, y compris en Ontario, au Canada. Comprendre l’évolution et la relation des divers génotypes du VSRRP en Ontario peut donner un aperçu de l’épidémiologie du virus. Les objectifs de cette étude étaient de i) décrire la variabilité des génotypes du VSRRP dans les troupeaux de porcs de l’Ontario et ii) évaluer les regroupements possibles en fonction des données génomiques du VSRRP. Les séquences du cadre de lecture ouvert 5 du virus (ORF-5) recueillis de 2010 à 2018 ont été obtenues auprès du Laboratoire de santé animale de l’Université de Guelph et des modèles phylogénétiques bayésiens ont été créés à partir de ceux-ci. La population de VSRRP de l’Ontario a ensuite été classée en 10 clades distincts. Les comparaisons de modèles ont indiqué que le modèle avec une hypothèse de population constante correspondait le mieux aux données, ce qui suggère que le changement net de la variation du virus SRRP de l’ensemble de la population au cours de la dernière décennie était faible. Néanmoins, les virus regroupés en clades individuels ont montré un regroupement temporel à des intervalles de temps distincts de toute la période d’étude (P < 0,01).
(Traduit par Docteur Serge Messier)
Introduction
Porcine reproductive and respiratory syndrome, or PRRS, was first described in North America and Europe in the late 1980s to early 1990s (1–4). The disease is characterized by severe reproductive losses, increased mortality, respiratory disease, and decreased growth rate in pigs (1). Since its discovery, it has become an endemic disease with significant economic and animal health/welfare impact (5,6).
The PRRS virus (PRRSV) is an enveloped RNA virus belonging to the Arterivirus genus (order Nidovirales, family Arteriviridae) (7). It has been shown to have a high genetic variation, with rapid mutation rates and lineage diversification (1,4,8,9) due to the lack of proofreading done at the 3′ end during replication, which causes high rates of mutations by read errors (10,11). There are 2 main types of PRRSV, type 1 and type 2, which differ by roughly 44% of their genetic material (1). Most observed samples in North America belong to PRRSV type 2 (1). Currently, the type-2 genotypes are primarily classified using the open reading frame 5 (ORF-5) section of the genome, which encodes glycoprotein 5 on the surface of the virus. It is used for its high variability among isolated variants (12), with dissimilarity for type-2 PRRSV potentially exceeding 21% (1).
For practical purposes, PRRSV is frequently classified by using restriction fragment length polymorphism (RFLP) analysis (1,13), the original intention of which was to discriminate between field and vaccine strains (14). Subsequent use of this process has provided the swine industry with an efficient way to classify PRRSV strains and, in some situations, associate this classification with an expected clinical impact at the herd level (15). The RFLP classification system needs to be updated, however, due partly to the expanding variation of PRRSV (16–19).
Because of this high genetic variation, research has been conducted to establish categories of strains/variants. Recent work has used phylogenetic approaches to explore the variability of PRRSV within North America, acknowledging 9 distinct lineages (20). Additionally, the authors noted that due to pig flow, which means transportation patterns of pig exports/imports, strains within the United States and Canada appear to originate from 2 groups of PRRSV type-2 lineages (20). Acknowledging this, data from clinical samples collected from 2010 to 2018 will be used to investigate the variation of PRRSV diversity in Ontario, Canada. This will provide a focused analysis of PRRSV within a major pig-producing region and expand our knowledge of the evolution and diversification of the virus within Ontario.
Therefore, the specific objectives of this study were to i) describe the variability of PRRSV genotypes in Ontario swine herds, and ii) evaluate possible groupings based on PRRSV genomic data. The results of analysis stemming from such objectives would expand our understanding of PRRSV endemic circulation. Furthermore, accurate classification could provide a basis for understanding a possible link between discrete strains and their expected clinical impact in swine herds.
Materials and methods
Study population
The majority of sequence data was gathered from the Animal Health Laboratory (AHL) at the University of Guelph, with additional sequences gathered from the National Centre for Biotechnology Information (NCBI) (Supplementary table). The inclusion criteria for the AHL data were i) documented time of sample submission was from 2010 to 2018, and ii) sample origin was an Ontario swine herd. In addition, multiple ORF-5 sequences deposited in the NCBI GenBank were added to the AHL dataset. The AHL data totaled 939 ORF-5 PRRSV sequences. The GenBank data consisted of sequences originating from outside of Canada, which provides a geographical and temporal outgroup for subsequent phylogenetic analysis. This subgroup totaled 75 different sequences spanning 1990 to 2014, making the entire combined data set equal to 1014 ORF-5 PRRSV sequences.
The sequences from AHL were based on PRRSV-positive sample of tissue, sera, and oral fluid samples and represent a herd level status. The reasons for submission included disease investigation and monitoring of herd-level PRRSV. Total nucleic acids were extracted using MagMax Viral RNA Isolation Kit (catalog AM1836) in a magnetic particle processor (Mag-MAX Express-96; ThermoFisher Scientific, Waltham, Massachusetts, United States). Real-time polymerase chain reaction (RT-PCR) was done using the VetMAX PRRSV NA & EU Reagents (ThermoFisher Scientific).
Samples with cycle threshold (Ct) values of < 36 were considered positive and ≥ 36 were considered inconclusive. Samples that returned no Ct value were considered negative. Nucleic acids from PRRSV-positive samples, 603 base pairs long, were used to generate templates for sequencing of ORF-5 with a Qiagen One-Step RT-PCR Kit (Qiagen, Mississauga, Ontario). Nucleotide sequences of PCR products were determined at the University of Guelph Laboratory Services sequencing facility using the Sanger sequencing approach. The virtual RFLP patterns were determined from nucleotide sequences using established methodology (14) and the patterns were assigned using the list of RFLP patterns kindly provided by the University of Minnesota.
Model selection
Multiple nucleotide sequence alignment was carried out in MEGA 7 (Molecular Evolutionary Genetics Analysis version 7) (21), using ClustalW method. Recombination was tested using HyPhy software (22) and a single breakpoint test nonreversible model (23). From this alignment, 12 different Bayesian phylogenetic models were created through Bayesian evolutionary analysis by sampling trees (BEAST) software package version 1.8.4 (24).
Two different nucleotide replacement models, YANG96 (25) and SRD06 (26), 2 molecular clock priors, and 3 coalescent model priors (Table I) were considered. Each model was set to run for 500 000 000 states, with all effective sample size (ESS) values over 200, and was visualized through Tracer 1.6 (27). A burn in of 10% was used for posterior probability (PP) estimation. Final model selection was carried out by gathering an AICM score; a posterior simulation-based analogue of the Akaike’s information criterion (AIC). This comparison was done to provide the best estimate of virus evolution over time. The analogue for the AIC score is modified for the Markov’s chain Monte Carlo phylogeny and is interpreted in the same fashion (28). Model comparisons can be seen in Table I and node ages gathered from this final tree were used to estimate clade ages (Table II).
Table I.
Nucleotide replacement models and estimated priors’ description with posterior simulation-based analogue of Akaike’s information criterion (AICM), standard error (SE), and likelihood effective sample size (ESS).
| Codon model | Molecular clock | Population growth | AICM | SE | ESS |
|---|---|---|---|---|---|
| YANG96 | Loga | Constant | 78946.875 | 2.967 | 409.9691 |
| YANG96 | STRb | Constant | 78967.963 | 0.583 | 653.8571 |
| YANG96 | STRb | Skylinec | 78986.687 | 1.538 | 873.5631 |
| YANG96 | STRb | Exponential | 79117.035 | 1.394 | 463.2171 |
| SRD06 | STRb | Skylinec | 79182.723 | 1.069 | 1081.7194 |
| SRD06 | Loga | Exponential | 79193.505 | 1.38 | 370.2158 |
| YANG96 | Loga | Skylinec | 79198.16 | 2.468 | 379.3852 |
| SRD06 | Loga | Skylinec | 79287.803 | 1.098 | 782.3569 |
| SRD06 | STRb | Exponential | 79290.284 | 1.475 | 741.6301 |
| SRD06 | STRb | Constant | 79319.751 | 1.751 | 748.5941 |
| YANG96 | Loga | Exponential | 79337.422 | 3.305 | 444.8325 |
| SRD06 | Loga | Constant | 79517.949 | 2.171 | 358.1772 |
Lognormal relaxed clock.
Strict clock.
Bayesian skyline.
Table II.
Total count of restriction fragment length polymorphism (RFLP) type per phylogenetic clade from 2010 to 2018, top 11 RFLP designations displayed, other grouping made up of RFLP designations less than 20 cases in total. Clade descriptive measures included are node age (years ago), posterior probability (PP), and branch time (years).
| RFLP | 1-1-1 | 1-111-1 | 1-8-4 | 1-3-2 | 1-4-4 | 1-4-2 | 1-3-4 | 1-18-4 | 1-1-2 | 1-22-2 | 1-12-2 | 2-5-2 | Other | Total | Node age (95% HPD) | Posterior probability (PP) | Branch time (y) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clade | |||||||||||||||||
| 1 | 73 | 22 | 11 | 1 | 11 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 88 | 213 | 23.18 (19.26, 27.36) | 0.97 | 3.44 |
| 2 | 0 | 0 | 50 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 14 | 66 | 21.91 (18.03, 25.99) | 0.99 | 4.71 |
| 3 | 0 | 0 | 0 | 0 | 4 | 0 | 4 | 31 | 9 | 0 | 0 | 0 | 97 | 145 | 22.81 (16.86, 29.02) | 0.99 | 10.03 |
| 4 | 0 | 0 | 0 | 29 | 0 | 0 | 1 | 0 | 6 | 45 | 0 | 0 | 28 | 109 | 14.77 (12.24, 17.02) | 0.99 | 1.92 |
| 5 | 0 | 0 | 0 | 53 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 6 | 61 | 8.71 (8.12, 9.6) | 1 | 7.98 |
| 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 0 | 20 | 46 | 18.03 (14.53, 21.23) | 1 | 3.07 |
| 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 16 | 17 | 9.20 (8.29, 10.52) | 1 | 16.77 |
| 8 | 0 | 0 | 2 | 24 | 2 | 0 | 23 | 0 | 0 | 0 | 0 | 0 | 10 | 61 | 22.81 (18.99, 26.56) | 1 | 6.05 |
| 9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 120 | 105 | 226 | 38.52 (31.60, 44.03) | 0.88 | 5.86 |
| 10 | 0 | 0 | 0 | 2 | 0 | 14 | 0 | 0 | 5 | 0 | 0 | 0 | 9 | 30 | 30.55 (24.09, 37.69) | 0.98 | 13.83 |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 37 | 46.58 (36.70, 64.46) | 1 | 142.33 |
| Total | 73 | 22 | 63 | 109 | 19 | 17 | 32 | 31 | 23 | 45 | 27 | 120 | 430 | 1011 | |||
HPD — highest posterior density.
Tree interpretation and analysis
Final tree visualization was conducted using FigTree (29). Additional data management and analysis were conducted with R version 3.3.2 (30). Phylogenetic tree importation and analysis were conducted using ggplot2 and ggtree (31,32), along with ape and phytools (33,34). Additionally, clade groupings were selected based on visual interpretation of posterior probability (PP) and supported by a similarity matrix based on the ORF-5 genome sequences. Additional clade-specific population growth reconstructions were estimated from these clade designations. Historic population reconstruction was conducted via Bayesian skyline model plots and was based on the Bayesian skyline population prior (35), due to the methodological requirements. The clade subpopulation plots were constructed from new subtrees and root node ages were estimated from the best fit tree.
Total counts of the sequence data were summarized by year and clade to gather a yearly frequency count and proportion of total cases within a specific clade using functionalities in ggplot2 (31). In addition, frequency of RFLP types was visualized over time in an identical manner. The RFLP labeling was used for the communication of clade demographics. The similarity matrix was calculated from aligned sequences using raw similarities within the functionalities of ape (33).
Temporal cluster analysis of PRRSV cases classified into individual clades was conducted through SaTScan, software for the spatial, temporal, and space-time scan statistics (36). The analysis was retrospective and focused on temporal clustering using a multinomial model (37) with clade designations. Additional Poisson models for individual clades were also conducted (38). For all models, the emphasis was on detecting clusters of high incidence. The minimum number of cases for a cluster was 2 cases with a maximum restriction to 50% of the temporal time frame with 999 replications and 10 iterations.
Results
Population and recombination
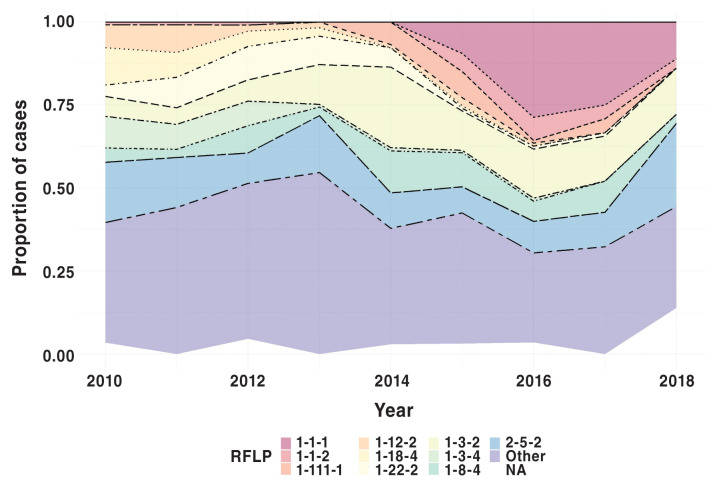
The aligned sequences were 618 base pairs long and showed no evidence of recombination based on the single breakpoint nonreversible model. The overall RFLP distribution from 2010 to 2018 as a proportion of all PRRS viruses in a given year is shown in Figure 1. Of note, RFLP 1-1-1, 1-1-2, and 1-111-1 were recently emerged strains in 2018, at the end of the study period. Other RFLP designations, specifically 2-5-2, 1-8-4, and 1-3-2, had a more consistent presence within the population, whereas other RFLP types, such as 1-18-4 and 1-22-2, were not detected near the end of the study period.
Figure 1.
Total PRRSV samples, including phylogenetic outgroup as a percentage of sampled cases by RFLP designation. Other category represents compilation of cases less than 20 samples total from 2010 to 2018.
Temporal clustering analysis
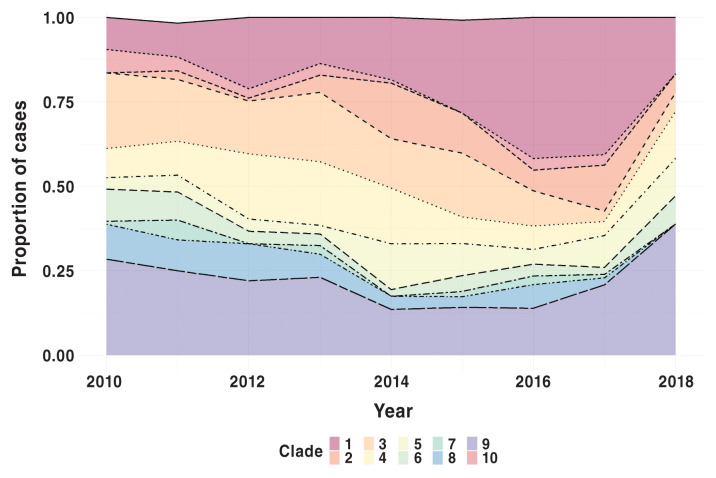
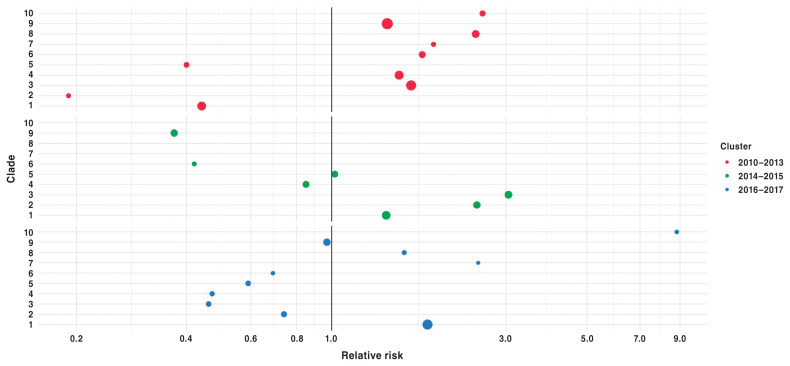
Results of scan statistics based on the Poisson model to determine primary temporal clusters for each clade separately are provided in Table III. The analysis included only Ontario-based sequences, (N = 939), with 3 sequences removed as they were not being grouped within a clade, ending with a total population of 936. Although present throughout the study period (Figure 3), PRRS viruses classified into clades 1 and 2 showed significant temporal clustering near the end of the study period (Table III, Figure 4). Contrary to this, other clades, e.g., clade 3 and 6, showed temporal clustering near the beginning of the study period (Table III). Similar findings were seen based on the multinomial model; specifically, clade 1 had a relative risk (RR) greater than 1 in the latter temporal clusters (Figure 4).
Table III.
Clade-specific primary temporal clusters of the SaTScan Poisson models displaying the time period when cluster was detected, ratio of observed and expected cases, relative risk, total number of cases in the primary cluster, and P-value.
| Clade | Temporal cluster | Observed/expected | Relative risk | Cases | P-value |
|---|---|---|---|---|---|
| 1 | 2015–2017 | 1.68 | 2.64 | 122 | 0.001 |
| 2 | 2014–2017 | 1.76 | 5.03 | 52 | 0.001 |
| 3 | 2010–2013 | 1.25 | 1.63 | 89 | 0.028 |
| 4 | 2012–2014 | 1.54 | 2.2 | 60 | 0.001 |
| 5 | 2014–2015 | 1.72 | 2.26 | 26 | 0.021 |
| 6 | 2010–2011 | 1.82 | 2.5 | 21 | 0.022 |
| 7 | 2011 | 3.26 | 4.85 | 7 | 0.033 |
| 8 | 2010–2013 | 1.43 | 2.46 | 43 | 0.008 |
| 9 | 2010–2013 | 1.17 | 1.41 | 114 | 0.138 |
| 10 | 2010–2011 | 1.85 | 2.58 | 13 | 0.128 |
Figure 3.
Percentage of PRRSV samples per clade, including phylogenetic outgroup, from 2010 to 2018, n = 1011 as 3 samples were not within any grouping.
Figure 4.
Clade specific relative risks from primary (2010–2013), secondary (2016–2017) and tertiary (2014–2015) temporal cluster, SaTScan multinomial model. Node size is proportional to number of detected cases within the cluster for a specific clade.
The full multinomial model determined 3 temporal clusters, 2010–2013, 2014–2015, and 2016–2017 (Figure 4). The primary temporal cluster was found in the period from 2010–2013. Subsequent secondary and tertiary clusters followed within 2016–2017 and 2014–2015. The primary and secondary clusters were deemed significant (Figure 4; P < 0.05), whereas the tertiary temporal cluster (2014–2015) was not (Figure 4; P = 0.068). Clade 10 had the largest RR (8.82) in magnitude compared to other clades within the 2016–2017 secondary cluster. In comparison to the number of cases (symbol size), however, clade 10 had the lowest number of observed cases (n = 7) when compared to other clades during that same cluster time frame, e.g., clade 1 (n = 87). Additionally, no clade had an RR greater than 1 across all the temporal clusters. However, specific clades were seen to have RRs greater than 1 in 2 of the temporal clusters, specifically clade 1 within the secondary and tertiary clusters, clade 3 in the primary and tertiary clusters, and clade 10 in the primary and secondary clusters. With some exceptions, the overall trend visually demonstrates that more recently emerged clades have RR values greater than 1 within the latter temporal clusters (Figure 4).
Comparison of clade designation and RFLP patterns
Total RFLP count per clade designation is shown in Table II. The “other” category represents all RFLP designations that had less than 20 samples over the course of the 9 y of sampling. The top 6 RFLP designations within the “other” category were: 1-12-4 [17], 1-30-4 [16], 1-8-2 [16], 1-16-2 [15], 1-26-4 [15], and 1-5-2 [15]. Type 2-5-2 RFLP, which represented 120 of the total samples or 12.7% of the Ontario-based samples, was most prevalent. This was followed by 1-3-2, representing 109 samples, then 1-1-1 and 1-8-4. Clade 9 had the greatest number of sequences within it (226, 22.3%), followed by clade 1 (213, 21.1%), and clade 3 (145, 14.3%).
Additionally, clade 9 contained all sequences that were categorized as RFLP type 2-5-2, a known RFLP designation for a specific vaccine strain of PRRSV. Similarly, clade 1 contained all sequences that were categorized as RFLP patterns 1-1-1 and 1-111-1, clade 3 contained all sequences from RFLP 1-18-4, and clade 4 contained all sequences with the RFLP designation of 1-22-2. Conversely, 1-8-4, 1-3-2, 1-4-4, and 1-1-2 were classified to more than 1 clade, with most viruses from these RFLP patterns classified to clades 2, 5, 1, and 3, respectively.
Clade statistics and similarity
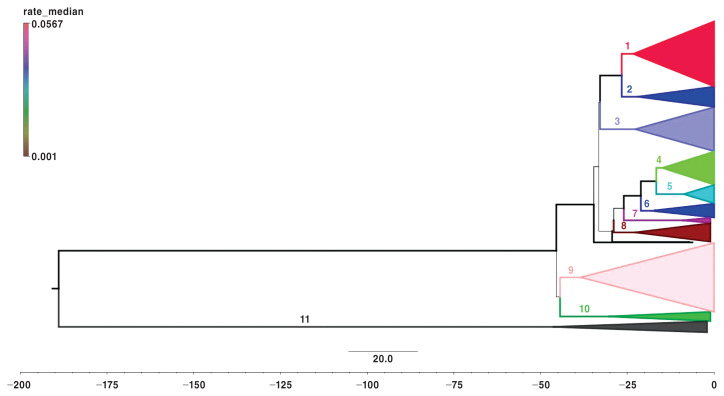
The estimated age of clades varied greatly, from as early as ~9 y ago to as late as ~47 y ago. The average node age was 23.37 y with a standard deviation (SD) of 11.04 y. Clade-specific node ages, with corresponding 95% highest posterior density (HPD), are shown in Table II. The posterior probability (PP) for each clade ranges from 0.88 to 1.0 with a mean of 0.98 (SD = 0.03). Clade-specific values and branch time estimates can also be found in Table II. Excluding clade 11, clade branch times range from 1.92 y (clade 4) to 16.77 y (clade 7).
From their recent common ancestral split, it was estimated that clades 1 and 2 evolved roughly over the same time (clade 1 = 3.44 y, clade 2 = 4.71 y). Similar evolutionary branch times can be seen with clades 9, 8, 6, and 5, however, with clades 4 and 7 being the extremes, as mentioned previously. Specifically, within clade 1, an expansion of subclade-containing viruses designated as RFLP 1-1-1 and 1-111-1 can be seen approximately 6 y ago (5.9 y ago, HPD = 5.3, 6.73; PP = 0.91). Additionally, the ancestral split between clades 1 and 2 is estimated at 26.63 y ago (HPD = 21.80, 31.64; PP = 1) and within clade 2, a subclade containing most of the viruses designated as RFLP 1-8-4, emerged within Ontario approximately 10.47 y ago (HPD 8.76, 12.5; PP = 1).
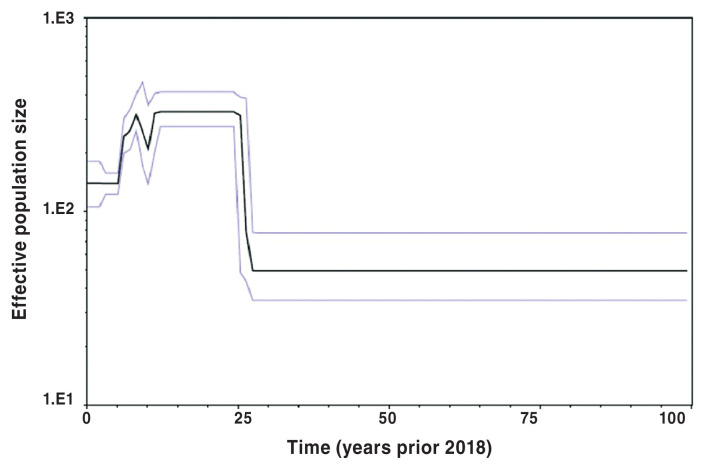
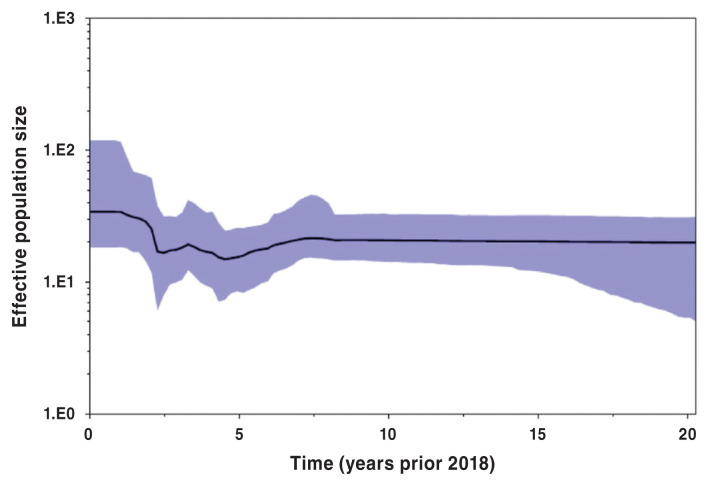
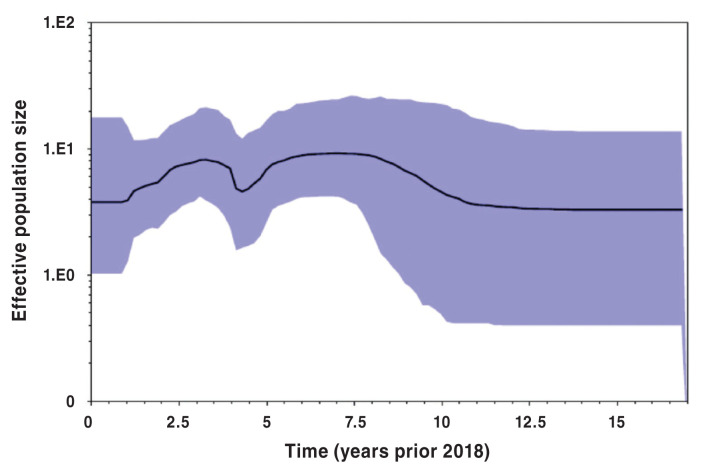
Bayesian skyline population reconstructions, which were based on the Yang96 Bayesian skyline model, with a lognormal relaxed molecular clock, are shown in Figures 5, 6, and 7. The clade subpopulation plots were constructed from new subtrees using a Bayesian skyline prior and node ages from the best-fit tree with the constant population prior. The effective population size (Y-axis) remained constant for 5 y prior to 2018 (Figure 5), when all PRRS viruses were considered. When subpopulations were investigated, i.e., clades 1 and 2, however, the population dynamics become more variable, displaying a non-linear nature. This is shown in Figures 6 and 7.
Figure 5.
Bayesian skyline plot of full phylogenetic tree based on Yang96 lognormal relaxed Bayesian skyline model. Y-axis displays effective population size at time (t) in the past prior to 2018 (t = 0).
Figure 6.
Clade 1 Bayesian skyline plot. Y-axis displays effective population size at time (t) in the past prior to 2018 (t = 0).
Figure 7.
Clade 21 Bayesian skyline plot. Y-axis displays effective population size at time (t) in the past prior to 2018 (t = 0).
Within-clade genomic similarity can be seen in Table IV. Within-clade similarity ranged on average from 88.28 to 97.73%, with a maximum sequence similarity of 100% within all clades. Conversely, between-clade mean dissimilarity is shown in Table V, with the diagonal representing within-clade dissimilarity. Note that all Ontario-based clades (1–10) had the highest dissimilarity with the geographical outgroup, clade 11. Clades 1 and 10 had the highest mean dissimilarity between Ontario sequences with 15.82%, followed by clades 1 and 9 (15.18%), and clades 8 and 10 (14.93%). Clades 1 and 2, which are predominantly made up of 3, currently well-known RFLP types in Ontario, 1-1-1, 1-111-1, and 1-8-4, have an average dissimilarity of 12.32%. The overall minimum average dissimilarity between the viruses from 2 clades was 7.29% between clades 4 and 5 (Table V).
Table IV.
Within-clade percent similarity of open reading frame 5 (ORF-5) nucleotide sequences of Ontario PRRSV collected from 2010 to 2018, N = 1014, outgroup included (clade 11). Mean, median, minimum, maximum, standard deviation (SD), and interquartile range (IQR) displayed.
| Clade | Mean | Minimum | Maximum | SD | Median | IQR |
|---|---|---|---|---|---|---|
| 1 | 92.05% | 82.62% | 100.00% | 4.14% | 91.43% | 7.14% |
| 2 | 97.13% | 91.67% | 100.00% | 1.65% | 97.14% | 2.38% |
| 3 | 90.83% | 25.24% | 100.00% | 10.85% | 92.38% | 3.10% |
| 4 | 95.49% | 88.81% | 100.00% | 1.98% | 95.48% | 2.86% |
| 5 | 96.56% | 92.38% | 100.00% | 1.93% | 96.67% | 3.10% |
| 6 | 93.20% | 88.57% | 100.00% | 2.94% | 92.38% | 3.81% |
| 7 | 97.73% | 95.71% | 100.00% | 1.22% | 97.62% | 1.90% |
| 8 | 90.65% | 84.76% | 100.00% | 3.79% | 89.76% | 4.29% |
| 9 | 97.61% | 85.24% | 100.00% | 2.49% | 98.57% | 2.62% |
| 10 | 94.50% | 87.86% | 100.00% | 3.07% | 94.76% | 4.76% |
| 11 | 88.28% | 81.90% | 100.00% | 3.97% | 87.38% | 4.76% |
| NG | 92.22% | 88.33% | 100.00% | 6.02% | 88.33% | 8.75% |
NG — No group, sequences not belonging to any clade.
Table V.
Between-clade mean percent dissimilarity of open reading frame 5 (ORF-5) nucleotide sequences of Ontario PRRSV collected from 2010 to 2018, N = 1014, outgroup included (clade 11).
| Clade | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | NG |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7.95% | |||||||||||
| 2 | 12.32% | 2.87% | ||||||||||
| 3 | 14.46% | 13.83% | 9.17% | |||||||||
| 4 | 13.74% | 13.07% | 14.10% | 4.51% | ||||||||
| 5 | 12.32% | 11.16% | 13.11% | 7.29% | 3.44% | |||||||
| 6 | 13.18% | 10.45% | 12.79% | 10.63% | 9.04% | 6.80% | ||||||
| 7 | 11.61% | 11.74% | 12.59% | 10.86% | 9.91% | 10.14% | 2.27% | |||||
| 8 | 14.38% | 13.46% | 14.28% | 13.07% | 11.92% | 13.11% | 13.79% | 9.35% | ||||
| 9 | 15.18% | 13.86% | 13.88% | 13.47% | 11.37% | 12.52% | 13.33% | 14.58% | 2.39% | |||
| 10 | 15.82% | 14.69% | 14.52% | 14.21% | 12.62% | 12.82% | 14.29% | 14.93% | 11.07% | 5.50% | ||
| 11 | 37.01% | 35.90% | 37.73% | 36.67% | 36.57% | 36.57% | 36.22% | 37.13% | 35.78% | 35.78% | 11.72% | |
| NG | 12.45% | 11.72% | 12.97% | 11.32% | 10.53% | 10.64% | 10.49% | 12.97% | 13.57% | 13.92% | 36.48% | 7.78% |
NG — No group, sequences not belonging to any clade.
Discussion
Model summary and population dynamics
With the known limitations associated with RFLP, alternative classification of PRRSV genotypes has become an important topic of research (8,13,18,20) and recent research has created new classifications for PRRSV (20,39). The most recent research has focused on Canadian variants and used maximum likelihood tree estimations to identify clusters of sequences, with support from bootstrap scores to define those clusters (39). Our research differs from this study through the base methodology, as we used Bayesian methods on a smaller population of PRRSV samples. The purpose of using the Bayesian methodology was based on the capability to gather internal validation statistics quickly and the ability to compare models, and by extension, the parameters within them. Bayesian analysis through BEAST can provide insights into the evolutionary history of PRRSV by model comparison and the selection of parameters.
Our defined clades were based on the Yang96 model, assuming constant population size and a lognormal relaxed molecular clock. A constant population size was determined to best fit our population of samples compared to the Bayesian skyline prior used in Shi et al and 2010 (20,40). As overall model fit along with prior selection are based on the specific population and its evolutionary history, it is difficult to compare models among different populations of the same virus or different viruses. Comparing models based on a single population, however, can provide insight into a population and its evolutionary history. This important step has been taken in a limited capacity for PRRSV research.
A constant population size prior used in this study suggests that the net viral population number has not changed over the 9 years included in this study. A previous study has suggested that the inherent variation of PRRSV strains is more attributable to the changes in the practices of the swine industry than to the individual host immunological selection pressures (8). The data used in this study were geographically and temporally constrained, i.e., Ontario, Canada and 2010 to 2018, respectively. As we have a localized data set of PRRSV sequences, this population dynamic may be representative of this constraint.
Previous research using data from 1998 to 2016 investigated spatial patterns of PRRSV cases and their phylogenetic relatedness within the United States (41). Their findings suggested that endemic strains demonstrated a slower population growth and dissemination rate when compared to emerging strains (41). Specifically, individual clades better fit different population growth assumptions. These results are comparable to results obtained in this study through formal temporal clustering analysis, visualization of patterns, and Bayesian skyline population reconstructions of viruses from 2 clades. More research is warranted to further explore these findings, however, taking longer temporal and wider geographical context into consideration.
The Bayesian population dynamic was needed to produce graphical representation of the historic population growth. Using the methodology, the effective population size of the entire population and subpopulations was estimated. The effective population size is a measure of the historic size of a population that must be replicated in order to give the total population at that time. Based on this, there is evidence supporting a constant population size over the last 5 y of our study period, although individual clade dynamics varied from this pattern. This can be illustrated by the increase of effective population size in clade 1 during the last 5 y of the study period. As such, the results from Bayesian skyline plots were in general agreement with the results of temporal scan cluster analysis. It is possible that the net change in the overall population of Ontario PRRS virus remains constant, but distinct viral subpopulations, represented by individual clades, may vary greatly in their frequency of occurrence with respect to time.
In this study, Poisson temporal scan models for individual clades and multinomial temporal scan models for all clades were used to investigate temporal clustering and the findings were in agreement. Although 3 temporal clusters were identified over the study period, when evaluated using the multinomial temporal scan statistics, no single clade of virus present at consistently high frequencies across all 3 clusters. In addition, viruses detected from clades 7, 8, and 10 in the secondary cluster seemed to reemerge near the end of the study period, although their overall detection frequency was low. Clades that were present at low frequencies in the primary cluster (clades 1, 2, and 5) were then detected at higher than expected levels in the tertiary and subsequently less than expected in the secondary cluster, with the exception of clade 1. This finding suggests temporal patterns with regard to clade-specific outbreaks in which new clades emerge over time, but viruses from older clades could still circulate and reemerge.
Previous research has indicated that there were temporal and geographic factors associated with clusters or clades of PRRSV (41). It would be interesting to investigate whether distinct viruses from these broadly defined clades reemerged in the same segment, e.g., region or production system, of the swine industry or a different one. Such an investigation would require a similar approach to an outbreak investigation using molecular data. It would be of particular interest given the ability of the virus to be present for an extended period of time in individual animals and perhaps in entire populations (42). Overall, we concur with the conclusions of the other research projects, specifically that investigations of PRRS virus populations should include larger geographical and temporal scales, as well as data on animal movement (20,41).
Clade demographics and similarity
In this study, PRRS virus clades were compared to RFLP patterns as an additional reference tool for veterinary practitioners who continue to use RFLP patterns and other molecular diagnostic data when investigating PRRSV outbreaks and planning interventions. It has been well-established that RFLP designations related to PRRSV genotyping must be continuously updated (16–18).
In this study, there were limited RFLP groupings that were grouped entirely into distinct PRRSV clades, with the exceptions of 1-1-1 (clade 1), 1-22-2 (clade 4), 1-18-4 (clade 3), and 2-5-2 (clade 9). However, these RFLP designations were not the sole RFLP pattern within their respective clades. It was shown that RFLP strain 2-5-2, which is a known vaccine/vaccine-like strain (43,44), was grouped in clade 9 with known RFLP patterns 2-6-2, 2-1-2, and 1-5-2, along with other vaccine-like strains (1-4-4) and other potential “gray” strains, i.e., 1-5-4, 1-1-2, and 1-2-4 (44). Some of these RFLP designations could represent possible new vaccine-like genotypes, as suggested in previous studies (20,44).
With further investigation of temporal clustering, clades 9 and 10 should also be considered differently than other clades identified in this study. Vaccine strains are introduced to populations as part of control measures and any temporal clustering could be a consequence of the greater need for vaccination or a deliberate modification in their use by a substantial number of farms in the source population.
The variability in the mean intra-clade similarity, as well as further examination of clades from the Bayesian analysis, confirms that the clades proposed in the current study represent a mixture of groups in terms of diversity, some with diverse viruses, e.g., clade 3 and clade 8, and some that show lower diversity, e.g., clades 2, 5, 7, and 9. Nonetheless, results of average between-clade similarity coincide with previous research findings that suggest that between-lineage difference was greater than 10% (12,40). The clades proposed in this study could, therefore, potentially be considered as candidates for groupings during epidemiological investigations on a regional level or as inputs for prognostic models.
As a final note, the similarity analysis was done in conjunction with the phylogenetic analysis and should not be interpreted alone. The intention was to provide more tangible quantitative values to visualize the variability within and among the clades described in this study. Although other more complex methodologies were attempted, due to the inclusion of the geographic and temporal outgroup, the similarity was less than 75% for some sequences. This caused the creation of non-integer values using these more intricate methods.
Limitations and conclusion
The data are a subset of Ontario’s PRRSV population, which would be inherently biased due to the potential differences for submitting diagnostic specimens mentioned previously or the reasons for requesting sequencing on a positive specimen. In addition, phylogenetic classification would ideally be based on the wholegenome sequence of PRRSV isolates. Such data were not available, however, whole-genome sequencing of PRRSV rarely occurs for the purposes of disease surveillance and control. In addition, sequencing of a larger number of positive specimens from the same submission could provide insight into the frequency of infections with more than 1 PRRSV strain and could provide greater insight into PRRSV epidemiology, both at the farm and provincial level.
The best fit tree had a constant population assumption and, using that criterion alone, the skyline plots may be considered unnecessary in some respect. Estimated skyline plots require Bayesian skyline modeling before being constructed, and the resulting plots, in combination with the temporal scan model, provide additional insight into the population dynamics of the entire Ontario source population and key PRRSV subpopulations, which contain strains important to the swine industry.
The limited number of tests to detect possible recombinant viruses was another possible limitation of this study. There are other approaches that should be considered in the future (45). Similarly, other approaches to classification could have been used, such as maximum likelihood methods and phylogenetic methods based on amino acid sequences. This was not reported in this study because it would have added another level of complexity to this work, which already has an additional classification method, i.e., RFLP.
In conclusion, this research has provided an in-depth phylogenetic description of the PRRSV population within Ontario on the basis of ORF-5 sequences obtained from regular monitoring and diagnostic investigations. Our analysis indicates that PRRS viruses detected in Ontario from 2010 to 2018 could be grouped into 10 broad clades of type-2 PRRS viruses. Distinct PRRSV clades demonstrated temporal clustering, which suggests that specific PRRSV strains spread in epidemic manner and show peak frequency in different time periods.
When applied to the entire study population, the Bayesian model assuming the constant population size of PRRS viruses was most consistent with the observed sequence data, using modified Akaike information criterion (AICM) for model comparison. Thus, despite high overall variability, the net change in the variation of the PRRSV strains in the population over time was negligible in this study population. This suggests that there could be minimal change in the overall variation of PRRSV strains and the overall population in Ontario could be stable. This is a novel finding that needs to be evaluated in similar populations.
Model comparisons related to Bayesian phylogenetic models have been done in a limited manner for PRRSV and should become a more common practice. Furthermore, consistent with previous findings, RFLP typing showed poor concordance with the broad genetic classification of PRRSV established in this study.
Despite its popularity and widespread use among veterinary practitioners, RFLP typing cannot be recommended to make conclusions about the spread of PRRSV among herds, particularly for RFLP types that have been present over longer periods of time. Interestingly, distinct PRRSV genotypes could re-emerge, although mechanisms responsible for their re-emergence could not be deducted from the data. From a practical standpoint, the clades established in this study could be used for epidemiological investigations of the spread of PRRS virus in target populations.
Figure 2.
Phylogenetic tree of 1014 PRRSV samples from 2010 to 2018 from Yang96 model assuming constant population growth and a lognormal relaxed clock. Node labels represent clade designations and line thickness corresponds to size of the posterior probability (PP).
Acknowledgments
This research was funded by the Ontario Ministry of Agriculture Food and Rural Affairs (OMAFRA) and a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (400558).
References
- 1.Zimmerman JJ, Dee SA, Holtkamp DJ, et al. Porcine reproductive and respiratory syndrome viruses (porcine arteriviruses) In: Zimmerman JJ, Karriker LA, Ramirez A, et al., editors. Diseases of Swine. 11th ed. Hoboken, New Jersey: John Wiley & Sons; 2019. pp. 685–708. [Google Scholar]
- 2.Loula T. Mystery pig disease. Agri-Practice. 1991;12:23–34. [Google Scholar]
- 3.Wensvoort G. Lelystad virus and the porcine epidemic abortion and respiratory syndrome. Vet Res. 1993;24:117–124. [PubMed] [Google Scholar]
- 4.Wensvoort G, Terpstra C, Pol JM, et al. Mystery swine disease in The Netherlands: The isolation of Lelystad virus. Vet Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 5.Cho JG, Dee SA. Porcine reproductive and respiratory syndrome virus. Theriogenology. 2006;66:655–662. doi: 10.1016/j.theriogenology.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 6.Morin M, Carpenter J, Poljack Z, et al. Production and economic aspects of pig production sites involved in PRRS area regional control projects in Canada. AASV 45th Annu Meet; 2014; Dallas, Texas. [Google Scholar]
- 7.Meulenberg JJ, Hulst MM, de Meijer EJ, et al. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murtaugh MP, Stadejek T, Abrahante JE, Lam TTY, Leung FCC. The ever-expanding diversity of porcine reproductive and respiratory syndrome virus. Virus Res. 2010;154:18–30. doi: 10.1016/j.virusres.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 9.Snijder EJ, Meulenberg JJ. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg R, Oleksiewicz MB, Petersen AM, Hein J, Bøtner A, Storgaard T. A molecular clock dates the common ancestor of European-type porcine reproductive and respiratory syndrome virus at more than 10 years before the emergence of disease. Virology. 2001;289:174–179. doi: 10.1006/viro.2001.1102. [DOI] [PubMed] [Google Scholar]
- 11.Forsberg R. Divergence time of porcine reproductive and respiratory syndrome virus subtypes. Mol Biol Evol. 2005;22:2131–2134. doi: 10.1093/molbev/msi208. [DOI] [PubMed] [Google Scholar]
- 12.Delisle B, Gagnon CA, Lambert MÈ, D’Allaire S. Porcine reproductive and respiratory syndrome virus diversity of Eastern Canada swine herds in a large sequence dataset reveals two hypervariable regions under positive selection. Infect Genet Evol. 2012;12:1111–1119. doi: 10.1016/j.meegid.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 13.Shi M, Lam TT, Hon CC, et al. Molecular epidemiology of PRRSV: A phylogenetic perspective. Virus Res. 2010;154:7–17. doi: 10.1016/j.virusres.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 14.Wesley RD, Mengeling WL, Lager KM, Clouser DF, Landgraf JG, Frey ML. Differentiation of a porcine reproductive and respiratory syndrome virus vaccine strain from North American field strains by restriction fragment length polymorphism analysis of ORF 5. J Vet Diagn Invest. 1998;10:140–144. doi: 10.1177/104063879801000204. [DOI] [PubMed] [Google Scholar]
- 15.Rosendal T, Dewey C, Friendship R, Wootton S, Young B, Poljak Z. Association between the genetic similarity of the open reading frame 5 sequence of porcine reproductive and respiratory syndrome virus and the similarity in clinical signs of porcine reproductive and respiratory syndrome in Ontario swine herds. Can J Vet Res. 2014;78:250–259. [PMC free article] [PubMed] [Google Scholar]
- 16.Larochelle R, D’Allaire S, Magar R. Molecular epidemiology of porcine reproductive and respiratory syndrome virus (PRRSV) in Quebec. Virus Res. 2003;96:3–14. doi: 10.1016/s0168-1702(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 17.Cha SH, Chang CC, Yoon KJ. Instability of the restriction instability of the restriction fragment length polymorphism pattern of open reading frame 5 of porcine reproductive and respiratory syndrome virus during sequential pig-to-pig passages. J Clin Microbiol. 2004;42:4462–4467. doi: 10.1128/JCM.42.10.4462-4467.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brar MS, Shi M, Ge L, Carman S, Murtaugh MP, Leung FC. Porcine reproductive and respiratory syndrome virus in Ontario, Canada 1999 to 2010: Genetic diversity and restriction fragment length polymorphisms. J Gen Virol. 2011;92:1391–1397. doi: 10.1099/vir.0.030155-0. [DOI] [PubMed] [Google Scholar]
- 19.Lambert MÈ, Delisle B, Arsenault J, Poljak Z, D’Allaire S. Positioning Quebec ORF5 sequences of porcine reproductive and respiratory syndrome virus (PRRSV) within Canada and worldwide diversity. Infect Genet Evol. 2019;74:103999. doi: 10.1016/j.meegid.2019.103999. [DOI] [PubMed] [Google Scholar]
- 20.Shi M, Lemey P, Singh Brar M, et al. The spread of type 2 porcine reproductive and respiratory syndrome virus (PRRSV) in North America: A phylogeographic approach. Virology. 2013;447:146–154. doi: 10.1016/j.virol.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosakovsky Pond SL, Frost SD, Muse SV. HyPhy: Hypothesis testing using phylogenics. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 23.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 24.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z. Maximum-likelihood models for combined analyses of multiple sequence data. J Mol Evol. 1996;42:587–596. doi: 10.1007/BF02352289. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro B, Rambaut A, Drummond AJ. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol. 2006;23:7–9. doi: 10.1093/molbev/msj021. [DOI] [PubMed] [Google Scholar]
- 27.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018;67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raftery AE, Newton MA, Satagopan JM, Krivitsky PN. Estimating the integrated likelihood via posterior simulation using the harmonic mean identity. Bayesian Stat. 2007;8:1–45. [Google Scholar]
- 29.Rambaut A. [Last accessed February 12, 2021];FigTree version 1.4.3 [Internet] 2016 Available from: http://tree.bio.ed.ac.uk/software/figtree.
- 30.R Core Team. R: A language and environment for statistical computing [Internet] Vienna, Austria: R Foundation for Statistical Computing; 2019. [Last accessed February 12, 2021]. Available from: https://www.r-project.org. [Google Scholar]
- 31.Wickham H. ggplot2: Elegant graphics for data analysis [Internet] New York: Springer-Verlag; 2016. [Last accessed February 12, 2021]. Available from: https://ggplot2.tidyverse.org. [Google Scholar]
- 32.Yu G, Smith D, Zhu H, Guan Y, Lam TTY. ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 33.Paradis E, Schliep K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 34.Revell LJ. Phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2011;3:217–223. [Google Scholar]
- 35.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 36.SaTScan. [Last accessed February 12, 2021];Software for the spatial, temporal, and space-time scan statistics [Internet] 2005 Available from: https://www.satscan.org.
- 37.Jung I, Kulldorff M, Otukei JR. A spatial scan statistic for multinomial data. Stat Med. 2010;29:1910–1918. doi: 10.1002/sim.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulldorff M. A spactial scan statistic. Commun Stat Theory Methods. 1997;26:1481–1496. [Google Scholar]
- 39.Lambert MÈ, Arsenault J, Audet P, Delisle B, D’Allaire S. Evaluating an automated clustering approach in a perspective of ongoing surveillance of porcine reproductive and respiratory syndrome virus (PRRSV) field strains. Infect Genet Evol. 2019;73:295–305. doi: 10.1016/j.meegid.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Shi M, Lam TTY, Hon CC, et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. 2010;84:8700–8711. doi: 10.1128/JVI.02551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alkhamis MA, Arruda AG, Morrison RB, Perez AM. Novel approaches for spatial and molecular surveillance of porcine reproductive and respiratory syndrome virus (PRRSv) in the United States. Sci Rep. 2017;7:4343. doi: 10.1038/s41598-017-04628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goyal SM. Porcine reproductive and respiratory syndrome. J Vet Diagn Invest. 1993;5:656–664. doi: 10.1177/104063879300500435. [DOI] [PubMed] [Google Scholar]
- 43.Rosendal T, Dewey C, Friendship R, Wootton S, Young B, Poljak Z. Spatial and temporal patterns of porcine reproductive and respiratory syndrome virus (PRRSV) genotypes in Ontario, Canada, 2004–2007. BMC Vet Res. 2014;10:83. doi: 10.1186/1746-6148-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christopher-Hennings J, Faaberg KS, Murtaugh MP, et al. Porcine reproductive and respiratory syndrome (PRRS) diagnostics: Interpretation and limitations. J Swine Health Prod. 2002;10:213–218. [Google Scholar]
- 45.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. RDP3: A flexible and fast computer program for analyzing recombination. Bioinformatics. 2010;26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]