Figure 1.
Experimental design and quantitative terminomics workflow for large-scale analysis of cleavage sites by metalloproteases
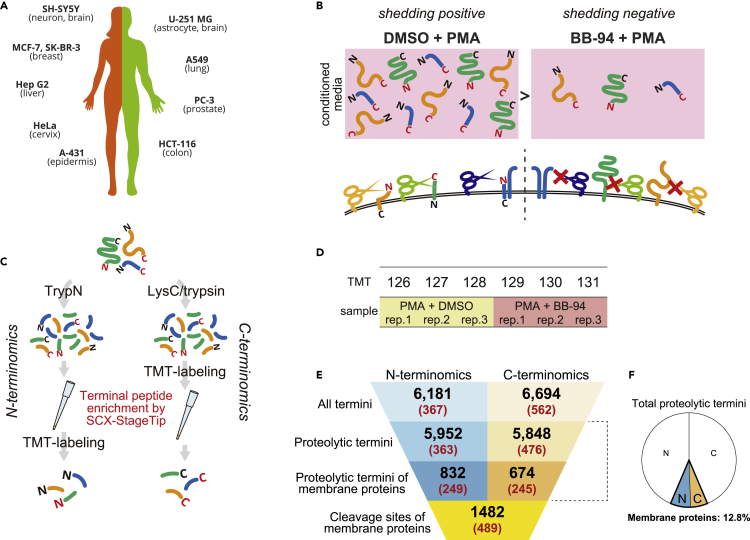
(A) List of investigated cultured cell lines (10 human cancer cell lines).
(B) Experimental design. Cells were treated with DMSO or BB-94 for 1 hr, followed by PMA treatment for 1 hr. Triplicate samples were prepared for each condition.
(C) Workflow of sample preparation. Proteins were digested with either TrypN or LysC and trypsin. Note that N-terminal peptides were TMT-labeled after terminal peptide enrichment, while C-terminal peptide enrichment was performed following TMT-labeling. See Figures S1C and S1D for details.
(D) Triplicate shedding-positive samples (PMA + DMSO) and shedding-negative samples (PMA + BB-94) were labeled as shown with TMT channels and form a 6-plexed TMT.
(E) Summary of the number of quantified termini and resulting cleavage sites. The parenthesized numbers show those significantly downregulated by BB-94. All termini: the total numbers of N- or C- termini (native protein termini and proteolytic termini). Proteolytic termini: the numbers of N- or C-termini that are presumably generated by endogenous proteases. Proteolytic termini on membrane proteins: the numbers of proteolytic N- or C-termini that are mapped on membrane proteins. Cleavage sites of membrane proteins: the total number of cleavage sites presented by proteolytic N- and C-termini on membrane proteins.
(F) Pie chart depicting the ratio of proteolytic termini mapped on membrane proteins in total proteolytic termini. The white areas show non-membrane proteins, and the areas highlighted with colors show membrane proteins. N, N-termini; C, C-termini.