Abstract
Extracellular vesicles (EVs) can represent a novel source of disease biomarkers, and are under intensive study for their clinical potential. Most EV-based cancer diagnostic studies have focused on establishing EV assays that detect increased expression of a single cancer-associated marker or marker signatures based on multiplex detection of these biomarkers. EV biomarker readouts can be obscured by high background signal leading to false positives, and may markedly differ between analyses due to variation in sample purity during EV isolation. This can obstruct the comparisons among studies and lead to conflicting conclusions. This work reports that the nucleic acid to protein UV absorption ratio of an EV is a cell-specific EV characteristic. This EV collective attribute can be measured at low-cost to discriminate EVs derived from malignant and non-malignant cells rather than employing single markers that may be cancer- or subtype-specific. Our work also highlighted the application for accessing purity in EV preparations irrelevant to EV yield. It can be employed to distinguish from patients with and without malignant disease upon analysis of EVs isolated from their serum samples.
Keywords: Extracellular vesicles, Quality assessment, Collective attribute, PDAC
1. Introduction
Extracellular vesicles (EVs) carry biomolecules that play important roles in cell-to-cell communication, including tumor initiation, progression, and metastasis (Brinton et al., 2015; Kondratov et al., 2017; Minciacchi et al., 2015). Since EVs shuttle factors that are characteristic of their cell or tissue origins (de Candia et al., 2013; Muralidharan-Chari et al., 2010; van der Pol et al., 2012), circulating EVs have strong potential as novel biomarkers for minimally invasive cancer diagnosis. Tumor-derived EVs present in peripheral blood have demonstrated a strong association with clinical diagnosis and prognosis (Coumans et al., 2010; Enciso-Martinez et al., 2020). Most EV-based cancer diagnostic studies have focused on establishing EV assays that detect increased expression of a single cancer-associated marker (protein, RNA or miRNA, or mutant DNA allele), or marker signatures based on multiplex detection of single biomarker types. However, such single marker approaches or limited biomarker panels may miss a significant fraction of cancers that exhibit genetic or phenotypic heterogeneity. EVs are usually characterized by their membrane factors (Melo et al., 2015) or their cargoes (Taylor and Gercel-Taylor, 2008) when analyzed by molecular methods, and most translational studies have focused on the analysis of EV surface proteins (Castro-Marrero et al., 2018; Hoshino et al., 2015; Liang et al., 2017; Liu et al., 2020; Melo et al., 2015; Rupp et al., 2011; Sun et al., 2020; Sun and Hu, 2018) or intravesicular nucleotide cargoes (Chevillet et al., 2014; Donovan et al., 2015; Kahlert et al., 2014; Lässer et al., 2011; Miranda et al., 2010; Pefanis et al., 2015; Shao et al., 2015; Valadi et al., 2007). Distinctive properties of intact EVs derived from nonmalignant versus malignant cells have not been employed as diagnostic biomarkers, although more than 14,000 EV-related studies were published in 2019.
One major issue hindering the development of clinical assays is that most preliminary studies employ variable EV isolation procedures that can produce EV samples that produce conflicting results. Standardized protocols that yield EV samples with consistent purity, quality, and yield are essential for the development of clinical applications. Colorimetric assays (Zendrini et al., 2020), nanoflow cytometry (Tian et al., 2020), nanomechanical microscopy (Ridolfi et al., 2019; Whitehead et al., 2015), Fourier-transform infrared spectroscopy (Paolini et al., 2020), Raman spectroscopy (Enciso-Martinez et al., 2020; Gualerzi et al., 2019), and MALDI (Zhu et al., 2019) studies attempted to provide solutions, but are constrained by either complicated labeling procedure or delicate equipment that would limit their use in clinical settings.
In this study, we describe a simple approach to assess EV purity and discrimination different EV populations based on the characteristic nucleic acid-to-protein ratio (NPr) of specific EVs, as assessed by UV absorbance at 260 nm and 280 nm, respectively. Since nucleic acids and amino acids with aromatic rings exhibit absorbance maxima at these two wavelengths, while phospholipids, the major component of the EV membrane, exhibit low UV absorbance (McHowat et al., 1996), this approach permits dye-free analysis of EV composition, with research and clinical implications. We demonstrate that spectrophotometric EV NPr values, as a cell line-specific collective attribute rather than single factors, can evaluate EV purity, and differentiate EVs derived malignant and non-malignant cells when analyzing cell culture and patient samples. The findings indicate that EV NPr values evaluated by standard UV spectroscopy can function as a simple and inexpensive measure of EV purity, and provide proof-of-concept data that EV NPr values may have clinical utility for noninvasive cancer diagnosis improvement.
2. Material and methods
2.1. EV isolation from culture media
EV samples were isolated from independent replicate cultures maintained under the same conditions. Briefly, cells were grown in culture media supplemented with serum to a culture density of 107 cells, washed three times with pH 7.0 phosphate-buffered saline (PBS), and then cultured for 48 h in serum-free media to collect EVs during cell starvation. To collect EVs from cells not under nonstarvation conditions, the cells were PBS washed, and cultured for 48 h in media supplemented with 10% EV-depleted FBS (Thermo Scientific, US). After 48 h, culture supernatants were collected and centrifuged at 400 g for 15 min to pellet cells, centrifuged at 8,000 g for 40 min to remove cell debris, concentrated with 100 kDa centrifugal filter units (Merck Millipore Ltd) by centrifugation at 21,000 g for 60 min. Concentrated supernatants were then centrifuged at 110,000 g for 120 min, and resulting EV precipitates were collected, dissolved in 100 μL PBS (pH 7.4), and stored at 4 °C until analysis within the same day. TEM, nanoparticle tracking analysis, and Western blot analyses were used to validate the characteristics of isolated EV samples (Fig. S1).
2.2. EV isolation from serum
Cryopreserved human serum samples (600 μL) were thawed and mixed by gentle vortexing, centrifuged at 135 g for 15 min, supernatants were centrifuged for 45 min at 9400 g, after which supernatant was carefully isolated to avoid the separated lipid layers and debris pellets and then centrifuged supernatant for 3 h at 100,000 g at 4 °C, or for 9 h. EV pellets were resuspended in 100 μL PBS (pH 7.0) and resuspended EVs were stored at 4 °C until analysis within the same day. EVs were isolated from 600 μL to achieve stable EV NPr readouts (Fig. S2).
2.3. Protein/DNA/RNA extraction, EV quantification, and NPr measurement
Aliquots of isolated EVs (30 μL each from the 100 μL EV stock sample isolated from 107 cells) were incubated with M-PER mammalian protein extraction reagent (Thermo Scientific) for 30 min in an ice bath and protein lysate concentrations were measured by bicinchoninic acid (BCA) assay (micro BCA Kit, Thermo Scientific). Total EV DNA and RNA was isolated using the AllPrep DNA/RNA Mini Kit (Qiagen, USA) by suspending 40 μL of isolated EVs in 200 μL of RLT-Plus buffer, homogenizing samples at maximum power for 1 min, loading lysates onto isolation columns, and eluting RNA and DNA with 50 μL EB buffer and 30 μL of nuclease-free water, respectively. DNA/RNA concentrations were quantified by light absorption using a Nanodrop 8000 Spectrophotometer (Thermo Fisher Scientific). Concentrations and size distributions of EVs were assessed by nanoparticle tracking analysis using a NanoSight NS300 (NanoSight Ltd., Amesbury, UK) to analyze replicate EVs samples diluted 200-fold in PBS. Five PBS blank samples were measured between each EV sample to avoid cross-contamination, and five repeat NPr readings were recorded and calculated for each 2 μL EV sample from a 100 μL EV stock sample isolated from 107 cells or 600 μL of human serum or plasma using a Nanodrop 8000 Spectrophotometer.
2.4. mtDNA/nDNA quantification
mtDNA content was determined by quantitative polymerase chain reaction (qPCR) of MT-ND1(Taqman™ assay ID: Hs02596873_s1) and normalized against the nuclear GAPDH gene (TaqMan™ assay ID: Hs02786624_g1). DNA (5 ng) was amplified in technical triplicates in 10 μL reactions using TaqMan™ Universal Master Mix II on the Applied Biosystems (Foster City, CA) Quantstudio 6 real-time PCR system. Final Ct values were corrected and assessed for reaction efficiency based on standard curves of both sequence products (Fig. S3). The PCR products were confirmed by gel electrophoresis and subsequently were extracted from agarose gels using the QIAquick Gel Extraction Kit (Qiagen, Chatsworth, CA) and further confirmed by microcapillary gel electrophoresis on an Agilent 2100 Bioanalyzer system using the ‘high-sensitivity’ DNA chips (Agilent). mtDNA/nDNA was evaluated as 2(CtND1—CtGAPDH+1)
3. Result and discussion
3.1. UV nucleic acid to protein absorption ratios of EVs are cell-specific
EVs derived from cancer cells are reported to contain more DNA than those from non-malignant cells (Thakur et al., 2014). We analyzed EVs isolated from four pancreatic cell lines: a pancreatic cell line derived by immortalization of primary ductal pancreatic cells (HPNE, non-malignant), and three representative primary pancreatic ductal adenocarcinoma (PDAC) cell lines with different genotypes (PANC-1, BxPC-3, MIA PaCa-2). Analysis of the DNA and protein content of these EVs was performed by separate isolation and quantification procedures, which determined that the EV DNA-to-protein ratio ranged from 1:99 in HPNE EVs to 1:9 in MIA PaCa-2 EVs with PANC-1 and BxPC-3 demonstrating intermediate values of 1:49 and 1:12, respectively (Fig. 1a). Such DNA-to-protein ratio measurements require isolation and quantification steps that are subject to significant variation (Fig. S4). We therefore examined whether we could distinguish EVs isolated from malignant and non-malignant pancreatic cell lines using a simple, dye-free spectrophotometric assay. This approach analyzed the maximum 260 nm-280 nm absorbance ratio of an EV sample to evaluate its relative nucleic acid-to-protein ratio (NPr), since nucleic acids and proteins primarily absorb near 260 nm and 280 nm, respectively.
Fig. 1. EV NPr values distinguish EVs of malignant and non-malignant cells of the same lineage.
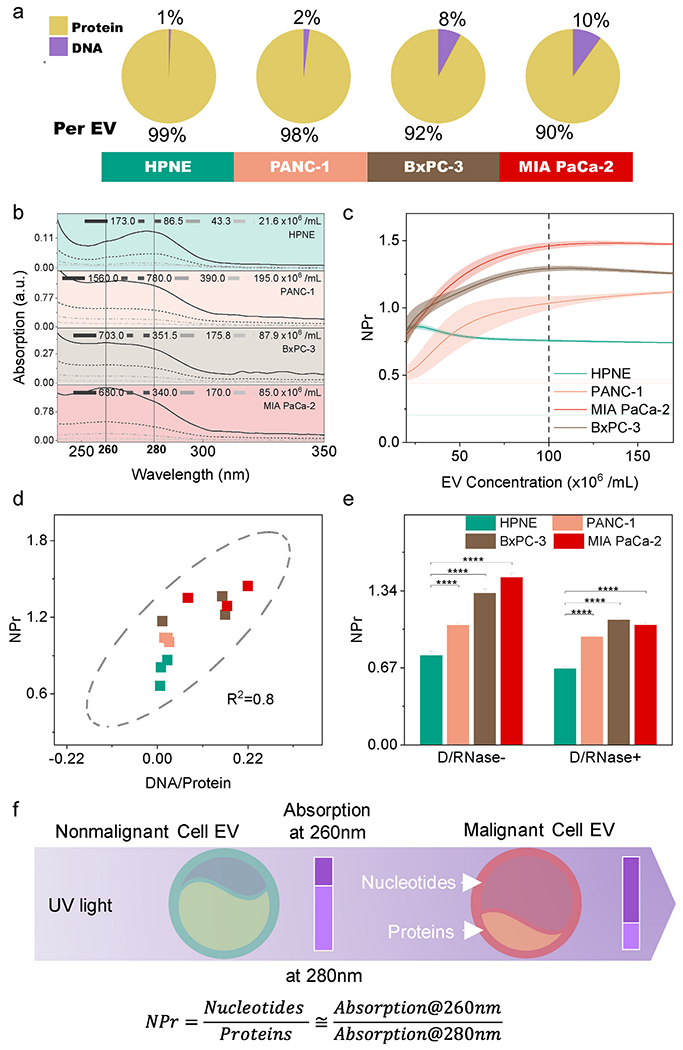
(a) DNA and protein composition of isolated EVs (n = 3/group). (b) The absorption spectrum of diluted EVs. The spectrum is EV concentration-dependent. (c) Mean (solid line) and SE (shaded area) of NPr values relative to EV concentration (n = 5/group). Cell-specific EV NPr proximate to constant when EV concentration > 108/mL. (d) Correlation of EV DNA/Protein mass ratio and NPr values, which is the rationale to use NPr for relative EV nucleotide quantification. (e) NPr values detected with and without DNase/RNase treatment to remove extravesicular nucleic acids (n = 5/group). Extra-EV nucleotides contamination does not mask out EV NPr difference between malignant and non-malignant cells. (f) Scheme of spectrophotometric NPr value for EV discrimination. Data points represent mean ± SE.
This absorption spectroscopy approach, which is commonly used to characterize the molecular content of purified samples (Schmid, 2001), is sensitive and nondestructive, and requires only small amounts of material for analysis (1 μL). Samples primarily composed of nucleic acid or protein respectively exhibit absorption A260/A280 ratios (NPr) >1.5 and < 0.8, while those with mixed compositions exhibit intermediate values that reflect the relative composition of nucleic acid and protein in the specimen (Patterson and Mura, 2013). However, the absorption spectrum was not well accepted to identify EVs because of the quantity dependency (Fig. 1b). Since the major component of the EV membrane is low-UV-absorbing phospholipids, NPr values should primarily reflect the nucleic acid and protein composition of an EV sample. NPr values of EVs isolated from the four pancreatic cell lines exhibited differential variability at EV concentrations below 108 EVs/mL due to limitations of the detector, but were highly reproducible for EVs derived from each cell line when measured at higher EV concentrations, with EVs from the three tumor cells exhibiting higher NPr values (Fig. 1c). These NPr values thus have the potential to serve as an identifier that distinguishes EVs derived from each of these cells, and potentially other cells, due to their composition differences.
DNA has been reported to represent the bulk of EV nucleic acid (Thakur et al., 2014), and we found that the EV RNA content fell below the limit of detection in our analysis (UV absorption and bioanalyzer). EV NPr values and EV DNA/protein ratios determined with purified EV material demonstrated a strong linear correlation (R2 = 0.8, Fig. 1d), suggesting that NPr can reliably evaluate the relative amount of DNA to protein in isolated EVs. DNase/RNase digestion revealed these samples contained varying amounts of extravesicular nucleic acid, as indicated by NPr decreases following the digestion (Fig. 1e). However, relative NPr value differences among these cell lines persisted following digestion, indicating that most EV-associated nucleic acid detected in these samples was intravesicular EV cargo. Spectrophotometric NPr EV value thus appears to be a useful means for EV discrimination, given its minimal sample requirement and the concentration-independent stability of the detected values when analyses are performed above a minimum EV concentration threshold (Fig. 1f).
3.2. Mitochondria DNA contributes to EV NPr
NPr increases detected for EVs from malignant PDAC cell lines were associated with increased EV DNA-to-protein ratios, which may reflect changes in DNA contributions from the nucleus (nDNA) or mitochondria (mtDNA) (Sansone et al., 2017). Mitochondrial metabolism plays a central role in cancer development and progression, and increased mitochondrial abundance is a common characteristic of tumors that exhibit worse clinical outcomes (Carew and Huang, 2002; Vyas et al., 2016). Larger NPr values observed in PDAC-derived EVs could thus reflect increased mtDNA content, since all three PDAC cell lines revealed greater mitochondrial density than the non-malignant HPNE cell line when analyzed by microscopy (Fig. 2a and S5) and flow cytometry (Fig. 2b and S6). Similarly, mtDNA to nDNA ratios were increased 11 × and 5 × when comparing MIA PaCa-2 versus HPNE cells and EVs, respectively (Fig. 2c). Since relative mitochondrial protein content of MIA PaCa-2 cells and their EVs increased by 1.4 × and 1.75 × versus that of HPNE cells and their EVs (Fig. 2d), it is reasonable to postulate that mtDNA (compared with mitochondrial proteins) contributes to the high NPr value of the EVs from PDAC cells.
Fig. 2. Mitochondrial effects on EV composition and NPr.
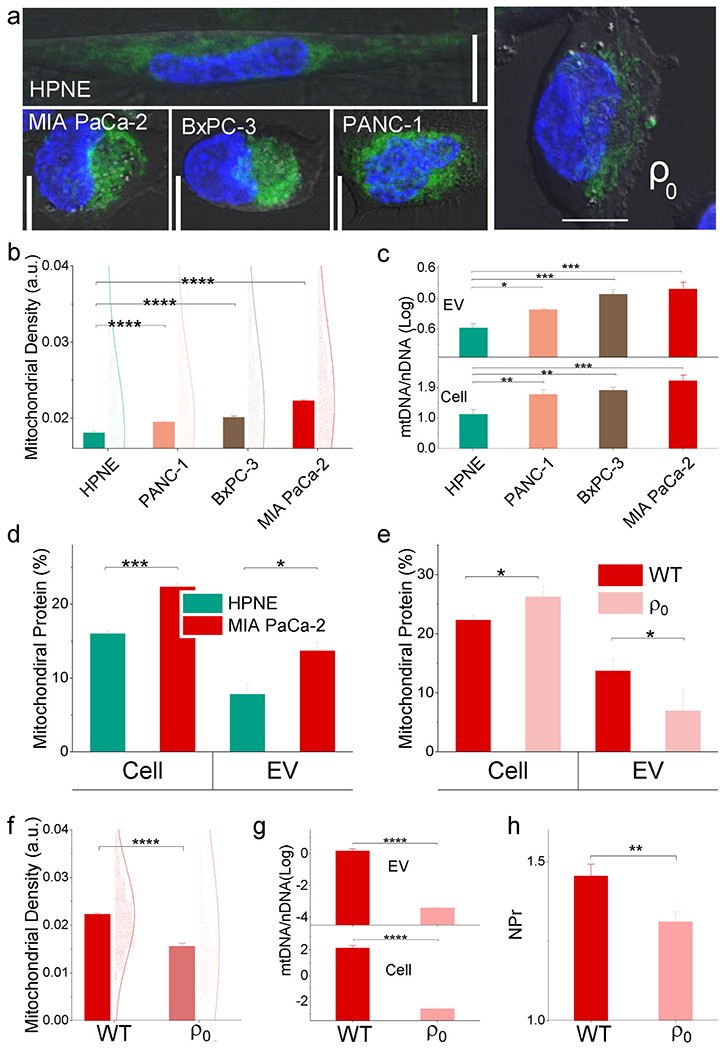
(a) Representative confocal images depicting mitochondria (green) and nuclei (blue) in the indicated cell types. Scale bar: 10 μm. The malignant cell is mitochondrial rich compared to the non-malignant cell. Wild type (WT) MIA PaCa-2 cell is mitochondrial rich compared to mitochondrial DNA depleted cells (ρ0). (b) and (f) Flow cytometry analysis of mitochondrial density normalized by cell size. (c) and (g) mtDNA/nDNA ratio and (d) and (e) mitochondrial protein composition of HPNE and MIA PaCa-2 cells and their EVs. (h) NPr comparison between WT and ρ0. Mitochondrial DNA contributes to EV NPr of malignant cells (mean ± SE; n = 4/group). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
We thus evaluated the contribution of mtDNA to EV NPr, by comparing mtDNA depleted MIA PaCa-2 cells (ρ0) with the wild type (WT). The mitochondrial mass density of ρ0 declined versus WT (Fig. 2a and f) with decreased cytoactivities (Fig. S7). The mtDNA of the ρ0 was depleted, but the mitochondrial protein was not (Fig. 2e and g). The lower NPr of po EVs compared with WT (Fig. 2h) suggested that mtDNA contributes to the high NPr of EVs from malignant cells.
3.3. NPr functions as a cell-specific EV quality assessment measure
EV content is expected to reflect the composition of its cell at the time of its secretion, and variations in the physiologic state of a cell can thus affect EV cargoes. However, variation in the collection conditions and isolation procedures can significantly impact the EV sample compositions, and thus the analysis results based on these EVs. The current practice of EV quality assessment and integrity checking is by accessing individual markers, but it is impossible to propose universal markers of one or the other type of EVs(Théry et al., 2018). We hence examine the possibility of using NPr as a practical quality assessment to ensure consistent EV production in research.
We analyzed NPr response to the collection conditions affecting EV constitute. Most EVs isolated from cultured cells are isolated under serum starvation to avoid contaminating secreted EVs with serum-derived EVs. Serum starvation altered EV NPr values, which rapidly decreased and then stabilized at lower levels (Fig. 3a), perhaps reflecting changes in cytosolic DNA levels or sorting in these cells. EVs isolated from actively growing HPNE and PDAC cells cultured with EV-depleted serum (Fig. 3b), however, demonstrated NPr relationships similar to those observed in EVs isolated under starvation conditions (Fig. 1e), indicating that cell starvation-induced NPr changes were similar among all these cells. NPr values for EVs derived from HPNE and PDAC cell cultures exhibited low variation when EVs were isolated from the same cell lines under equivalent culture conditions and using a consistent protocol (starvation or nonstarvation). However, variation in EV isolation procedures could significantly alter the composition of EV fractions, by differentially enriching certain EV subtypes or altering the amount and type of extravesicular contaminants present in these fractions. Since NPr values for EVs of a given cell line are stable when EVs are collected under constant growth conditions, NPr changes for purified EV fractions from these cells could reveal the degree and type of EV contamination. Reducing extravesicular nucleic acid or protein contamination by DNase/RNase digestion or washing steps, respectively reduced (Fig. 1e) or increased (Fig. 3c) absolute NPr values of EV samples. NPr values could therefore assess sample purity and the type and relative degree of sample contamination for EVs derived from a single cell type. Spiking EV fractions with known amounts of nucleic acid or protein also resulted in predictable effects on NPr values (Fig. 3d), where the amount of contamination by the dominant material could be predicted by Albert’s law (see Supporting Information). A similar modeling approach (see Supporting Information) was also be employed to determine the relative proportions of two EV populations (e.g. malignant and nonmalignant cell EVs) with different NPr values in a mixed population (Fig. 3e). Thus, in a sample with single EV population, NPr can access purity of the EVs by indicating nucleic acid and protein contaminations. Based on this data, NPr may serve to evaluate the purity and relative protein or nucleic acid contamination of EVs isolated from cell culture experiments and could evaluate the relative contribution of EVs derived from defined cell cultures.
Fig. 3. NPr as a measure of EV purity.
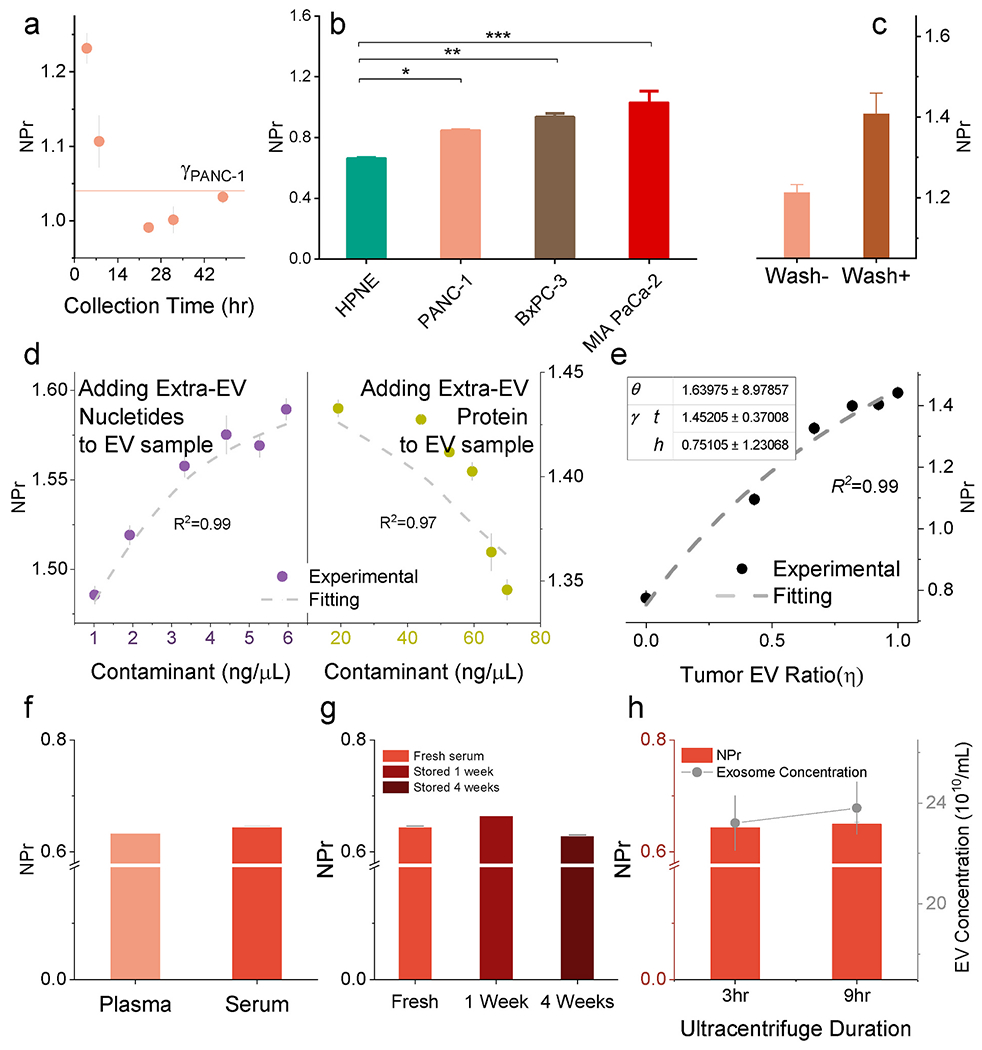
(a) The collection condition affects EV NPr value. PANC-1 NPr decreased in relation to starvation time. (b) NPr difference between malignant and non-malignant EV was not masked out by changing the collection condition. NPr values of EVs isolated from cells maintained under nonstarvation conditions (n = 5). (c) NPr values were increased by wash steps corresponding to reduced protein contamination. (d) EV NPr affected by adding contaminants (nucleotides and protein) following the nonlinear model as reflected by Alber’s law (Supporting Information). (e) NPr reflected the relative abundance of MIA PaCa-2 EVs in mixed EV samples containing MIA PaCa-2 and HPNE EVs. NPr indicates the relative purity of a specific EV population in a mix. (f) NPr values of EVs purified from healthy human serum and plasma. (n = 6). (g) Stability of NPr values in relation to storage time at 4 °C for weeks (n = 5). (h) NPr values in response to ultracentrifugation time during EV isolation (n = 5). Increasing ultracentrifugation time escalated EV yield but not NPr. Data points represent mean ± SE.
EVs present in blood samples derive from multiple cell types and represent a more heterogeneous mixture and background for EV diagnostics. Notably, though, NPr values differed by only 1.79% between matched serum and plasma EV samples (Fig. 3f). Serum EVs also revealed low within- and between-run coefficients of variation across a broad EV dilution range, varying from 0.06 to 0.69 at the high end to 2.74 and 5.56 at the lowest limit of detection (Supporting Information, Table S1). NPr values varied more significantly when EVs were isolated from serum stored at 4 °C for a week – a common limit for clinical analysis – exhibiting a 2.99% maximum within-run variation (Fig. 3g). Centrifugation time can be a major source of variation for EVs isolated by ultracentrifugation, but NPr values were not markedly altered by extended ultracentrifugation time (Fig. 3h). Taken together, these data suggest that NPr values for serum- or plasma-derived EV samples are sufficiently reproducible to allow their use in clinical applications.
3.4. NPr can identify serum containing PDAC-derived EVs
Since NPr values of EVs isolated from PDAC cells and serum are stable and reproducible, we analyzed the NPr values of EV samples isolated from the serum of PDAC patients or individuals with nonmalignant conditions that have similar symptom profiles (disease controls) to determine if NPr values could distinguish these groups. Mean NPr values of serum EVs derived from these two groups did not significantly differ (Fig. 4b, lower panel). However, since PDAC-derived EVs are expected to constitute a small fraction of the total serum EV population, the NPr signal contributed by these EVs could be masked by heterogenous NPr signals arising from the bulk of circulating that derive from non-malignant cells. Serum EV isolates were thus immunoprecipitated to enrich for EVs expressing EpCAM (Fig. 4a), an epithelial cell marker that is overexpressed on most PDAC tumors (Costa-Silva et al., 2015; Hoshino et al., 2015), finding that the change in NPr value pre-versus post-enrichment (∆NPr%) was significantly larger in serum EVs isolated from the PDAC group (Fig. 4b, upper panel). The ∆NPr% values observed in the PDAC and disease control groups exhibited substantial overlap, however, which could reflect the heterogeneity of disease in these two populations that exhibit similar symptoms, and may be further complicated by other factors that might influence NPr values and circulating EV composition, including age, gender, BMI, hypertension and other physiologic parameters.
Fig. 4. Serum NPr may serve as biomarker for PDAC diagnosis.
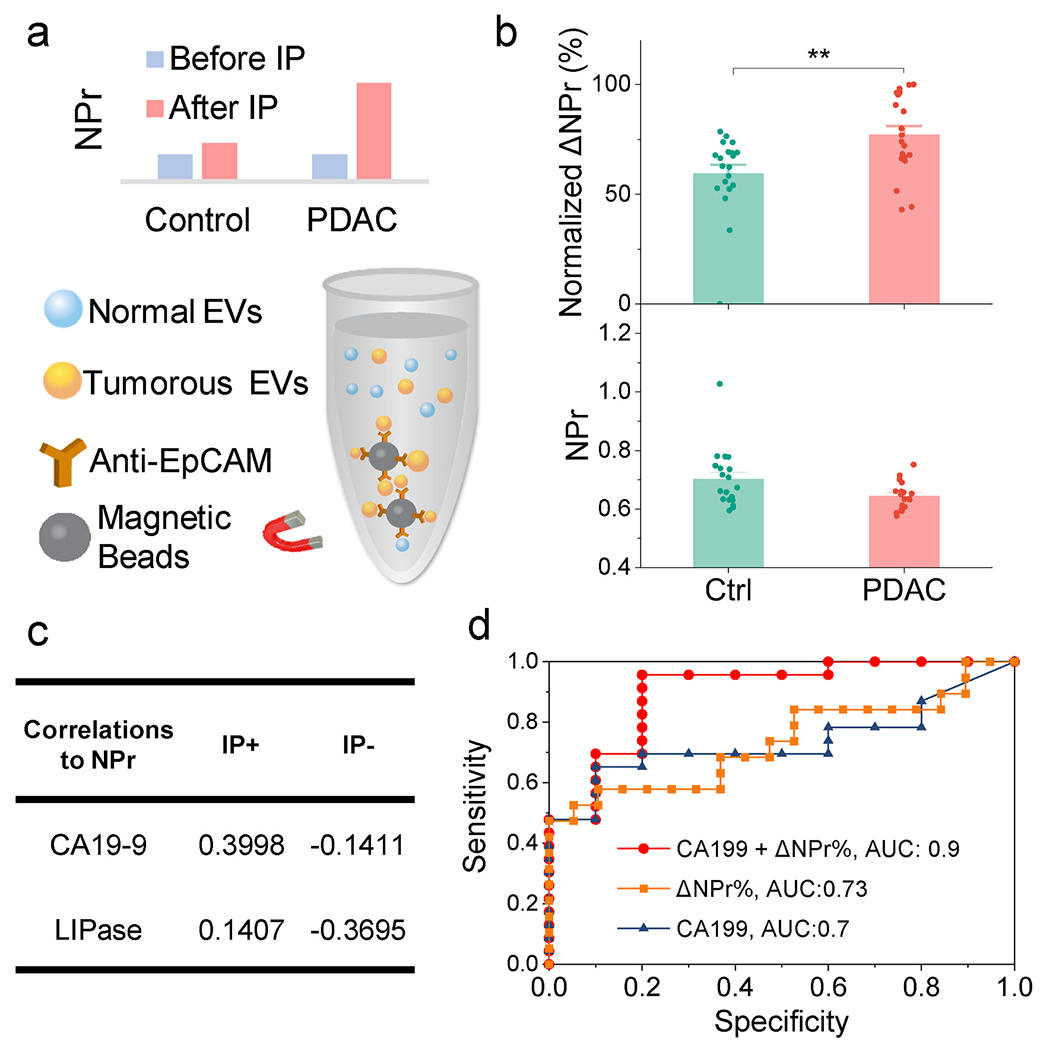
(a) Schematic of EV enrichment by immunoprecipitation (IP). Tumorous EV was enriched and detected after IP. (b) NPr values of total serum EVs isolated from PDAC patients (n = 19) and disease controls (Ctrl, n = 19), and the change in NPr (∆NPr%) after enrichment for EpCAM + EVs. Data points represent mean ± SE. **, p < 0.005 by two-tail Studenťs t-test. (c) Correlation of the PDAC associated markers CA19-9 and LIPase with ∆NPr% (IP+) and NPr (IP−), (d) Receiver-operating characteristic (ROC) curve analysis for the ability of the indicated factors to distinguish serum of PDAC cases from that of disease controls.
The correlation of serum ∆NPr% with two PDAC biomarkers (CA19-9 and LIPase) in this cohort also improved after enrichment for EpCAM + EVs, although these correlations remained low (Fig. 4c). The low correlations raised the potential for a complementary noninvasive diagnostic method using NPr of circulating tumorous EVs with IP enrichment. Evaluation of the ability of serum ∆NPr% and CA19-9 values to distinguish individuals with PDAC from a disease control population, found that both factors exhibited similar diagnostic performance when employed alone, but enhanced performance when used in combination (Fig. 4d).
4. Conclusion
Our results indicate that the nucleic acid to protein ratio of isolated EVs, as evaluated by their 260 nm/280 nm absorbance ratio, is a stable characteristic of specific cell populations when EV samples are collected under constant conditions. This measure has the potential to serve as a means of evaluating the purity of EV samples isolated from cell lines, and as a cancer screening approach pending large scale validation studies.
Supplementary Material
Acknowledgments
This work was financially supported by grants from the National Institutes of Health (R01AI122932, R01HD090927, R01AI113725, R21AI126361, R03CA252783), an Arizona Board Research Grant (ABRC/ADHS18-198856), the Weatherhead presidential endowment fund, and National Science Foundation under NSF EPSCoR Track-1 Cooperative Agreement (OIA #1355466).
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2021.113058.
References
- Brinton LT, Sloane HS, Kester M, Kelly KA, 2015. Formation and role of exosomes in cancer. Cell. Mol. Life Sci 10.1007/s00018-014-1764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carew JS, Huang P, 2002. Mitochondrial defects in cancer. Mol. Canc 1, 1–12. 10.1186/1476-4598-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Marrero J, Serrano-Pertierra E, Oliveira-Rodríguez M, Zaragozá MC, Martínez-Martínez A, Blanco-López M, del C, Alegre J, 2018. Circulating extracellular vesicles as potential biomarkers in chronic fatigue syndrome/myalgic encephalomyelitis: an exploratory pilot study. J. Extracell. Vesicles 7, 1453730. 10.1080/20013078.2018.1453730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet JR, Kang Q, Ruf IK, Briggs H.a., Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M, 2014. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. Unit. States Am 111, 14888–14893. 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK, Becker A, Hoshino A, Mark MT, Molina H, Xiang J, Zhang T, Theilen TM, García-Santos G, Williams C, Ararso Y, Huang Y, Rodrigues G, Shen TL, Labori KJ, Lothe IMB, Kure EH, Hernandez J, Doussot A, Ebbesen SH, Grandgenett PM, Hollingsworth MA, Jain M, Mallya K, Batra SK, Jarnagin WR, Schwartz RE, Matei I, Peinado H, Stanger BZ, Bromberg J, Lyden D, 2015. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol 17, 816–826. 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coumans FAW, Doggen CJM, Attard G, de Bono JS, Terstappen LWMM, 2010. All circulating EpCAM+CK+CD45-objects predict overall survival in castration-resistant prostate cancer. Ann. Oncol 21, 1851–1857. 10.1093/annonc/mdq030. [DOI] [PubMed] [Google Scholar]
- de Candia P, Torri A, Gorletta T, Fedeli M, Bulgheroni E, Cheroni C, Marabita F, Crosti M, Moro M, Pariani E, Romanò L, Esposito S, Mosca F, Rossetti G, Rossi RL, Geginat J, Casorati G, Dellabona P, Pagani M, Abrignani S, 2013. Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PloS One 8, 75348. 10.1371/journal.pone.0075348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan MJ, Noerholm M, Bentink S, Belzer S, Skog J, O’Neill V, Cochran JS, Brown G. a. , 2015. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis 1–6. 10.1038/pcan.2015.40. [DOI] [PubMed] [Google Scholar]
- Enciso-Martinez A, Van Der Pol E, Hau CM, Nieuwland R, Van Leeuwen TG, Terstappen LWMM, Otto C, 2020. Label-free identification and chemical characterisation of single extracellular vesicles and lipoproteins by synchronous Rayleigh and Raman scattering. J. Extracell. Vesicles 9. 10.1080/20013078.2020.1730134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi A, Kooijmans SAA, Niada S, Picciolini S, Brini AT, Camussi G, Bedoni M, 2019. Raman spectroscopy as a quick tool to assess purity of extracellular vesicle preparations and predict their functionality. J. Extracell. Vesicles 8. 10.1080/20013078.2019.1568780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Jørgen Labori K, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, 2015. Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. 10.1038/naturel5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert C, Melo SA, Protopopov A, Tang J, Seth S, Koch O, Zhang J, Weitz J, Chin L, Futreal A, Kalluri R, 2014. Identification of doublestranded genomic dna spanning all chromosomes with mutated KRAS and P53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem 289, 3869–3875. 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov KA, Petrova TA, Mikhailovskii VY, Ivanova AN, Kostareva AA, Fedorov AV, 2017. A study of extracellular vesicles isolated from blood plasma conducted by low-voltage scanning electron microscopy. Cell Tissue Biol 11, 181–190. 10.1134/S1990519X17030051. [DOI] [PubMed] [Google Scholar]
- Lässer C, Seyed Alikhani V, Ekström K, Eldh M, Torregrosa Paredes P, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H, 2011. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J. Transl. Med 9, 9. 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z, Hu Y, 2017. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng 1, 0021 10.1038/s41551016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Fan J, Xu T, Ahmadinejad N, Hess K, Lin SH, Zhang J, Liu X, Liu L, Ning B, Liao Z, Hu TY, 2020. Extracellular vesicle tetraspanin-8 level predicts distant metastasis in non-small cell lung cancer after concurrent chemoradiation. Sci. Adv 6, 6162–6173. 10.1126/sciadv.aaz6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHowat J, Jones JH, Creer MH, 1996. Quantitation of individual phospholipid molecular species by UV absorption measurements. J. Lipid Res 37, 2450–2460. [PubMed] [Google Scholar]
- Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R, 2015. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 523, 177–182. 10.1038/naturel4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minciacchi VR, Freeman MR, Di Vizio D, 2015. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, Da Silva N, Brown D, Russo LM, 2010. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78, 191–199. 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C, 2010. Microvesicles: mediators of extracellular communication during cancer progression. J. Cell Sci 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolini L, Federici S, Consoli G, Arceri D, Radeghieri A, Alessandri I, Bergese P, 2020. Fourier-transform Infrared (FT-IR) spectroscopy fingerprints subpopulations of extracellular vesicles of different sizes and cellular origin. J. Extracell. Vesicles 9. 10.1080/20013078.2020.1741174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson J, Mura C, 2013. Rapid colorimetric assays to qualitatively distinguish RNA and DNA in biomolecular samples. JoVE e50225. 10.3791/50225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu Z-P, Economides AN, Bradner JE, Rabadan R, Basu U, 2015. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 161, 774–789. 10.1016/j.cell.2015.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridolfi A, Brucale M, Montis C, Caselli L, Paolini L, Borup A, Boysen A, Loria F, van Herwijnen M, Kleinjan M, Nejsum P, Zarovni N, Wauben M, Berti D, Bergese P, Valle F, 2019. AFM-based high-throughput nano mechanical screening of single extracellular vesicles. bioRxiv, 854539. 10.1101/854539. [DOI] [PubMed] [Google Scholar]
- Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, Moldenhauer G, Marm F, Sltmann H, Altevogt P, 2011. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol. Oncol 122, 437–446. 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- Sansone P, Savini C, Kurelac I, Chang Q, Amato LB, Strillacci A, Stepanova A, Iommarini L, Mastroleo C, Daly L, Galkin A, Thakur BK, Soplop N, Uryu K, Hoshino A, Norton L, Bonafé M, Cricca M, Gasparre G, Lyden D, Bromberg J, Sansone P, Kurelac I, Chang Q, Amato LB, Strillacci A, Stepanova A, Iommarini L, Daly L, Galkin A, Thakur BK, Soplop N, Uryu K, Hoshinob A, Norton L, Bonafe M, Cricca M, Gasparre G, Sansone P, Savini C, Kurelac I, Chang Q, Benedetta L, Strillacci A, 2017. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer 201718630, Proc. Natl. Acad. Sci. Unit. States Am, 1704862114. 10.1073/pnas.1718630114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F-X, 2001. Biological macromolecules: UV-visible spectrophotometry. In: Encyclopedia of Life Sciences. John Wiley & Sons, Ltd, Chichester, UK. 10.1038/npg.els.0003142. [DOI] [Google Scholar]
- Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R, 2015. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat. Commun. 6, 6999. 10.1038/ncomms7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Hu TY, 2018. A low cost mobile phone dark-field microscope for nanoparticle-based quantitative studies. Biosens. Bioelectron. 99, 513–518. 10.1016/j.bios.2017.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Yang L, Lyon CJ, Hu T, 2020. Simulation-directed amplifiable nanoparticle enhanced quantitative scattering assay under low magnification dark field microscopy. J. Mater. Chem. B 10.1039/d0tb00350f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C, 2008. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol 110, 13–21. 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez-Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova-Todorova K, Welte K, Bromberg J, Peinado H, Lyden D, 2014. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24, 766–769. 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, Ayre DC, Bach JM, Bachurski D, Baharvand H, Balaj L, Baldacchino S, Bauer NN, Baxter AA, Bebawy M, Beckham C, Bedina Zavec A, Benmoussa A, Berardi AC, Bergese P, Bielska E, Blenkiron C, Bobis-Wozowicz S, Boilard E, Boireau W, Bongiovanni A, Borràs FE, Bosch S, Boulanger CM, Breakefield X, Breglio AM, Brennan M, Brigstock DR, Brisson A, Broekman MLD, Bromberg JF, Bryl-Górecka P, Buch S, Buck AH, Burger D, Busatto S, Buschmann D, Bussolati B, Buzás EI, Byrd JB, Camussi G, Carter DRF, Caruso S, Chamley LW, Chang YT, Chaudhuri AD, Chen C, Chen S, Cheng L, Chin AR, Clayton A, Clerici SP, Cocks A, Cocucci E, Coffey RJ, Cordeiro-da-Silva A, Couch Y, Coumans FAW, Coyle B, Crescitelli R, Criado MF, D’Souza-Schorey C, Das S, de Candia P, De Santana EF, De Wever O, del Portillo HA, Demaret T, Deville S, Devitt A, Dhondt B, Di Vizio D, Dieterich LC, Dolo V, Dominguez Rubio AP, Dominici M, Dourado MR, Driedonks TAP, Duarte FV, Duncan HM, Eichenberger RM, Ekström K, EL Andaloussi S, Elie-Caille C, Erdbrügger U, Falcón-Pérez JM, Fatima F, Fish JE, Flores-Bellver M, Försönits A, Frelet-Barrand A, Fricke F, Fuhrmann G, Gabrielsson S, Gámez-Valero A, Gardiner C, Gärtner K, Gaudin R, Gho YS, Giebel B, Gilbert C, Gimona M, Giusti I, Goberdhan DCI, Görgens A, Gorski SM, Greening DW, Gross JC, Gualerzi A, Gupta GN, Gustafson D, Handberg A, Haraszti RA, Harrison P, Hegyesi H, Hendrix A, Hill AF, Hochberg FH, Hoffmann KF, Holder B, Holthofer H, Hosseinkhani B, Hu G, Huang Y, Huber V, Hunt S, Ibrahim AGE, Ikezu T, Inal JM, Isin M, Ivanova A, Jackson HK, Jacobsen S, Jay SM, Jayachandran M, Jenster G, Jiang L, Johnson SM, Jones JC, Jong A, Jovanovic-Talisman T, Jung S, Kalluri R, Kano S ichi, Kaur S, Kawamura Y, Keller ET, Khamari D, Khomyakova E, Khvorova A, Kierulf P, Kim KP, Kislinger T, Klingeborn M, Klinke DJ, Kornek M, Kosanović MM, Kovács ÁF, Krämer-Albers EM, Krasemann S, Krause M, Kurochkin IV, Kusuma GD, Kuypers S, Laitinen S, Langevin SM, Languino LR, Lannigan J, Lässer C, Laurent LC, Lavieu G, Lázaro-Ibáñez E, Le Lay S, Lee MS, Lee YXF, Lemos DS, Lenassi M, Leszczynska A, Li ITS, Liao K, Libregts SF, Ligeti E, Lim R, Lim SK, Linē A, Linnemannstöns K, Llorente A, Lombard CA, Lorenowicz MJ, Lörincz ÁM, Lötvall J, Lovett J, Lowry MC, Loyer X, Lu Q, Lukomska B, Lunavat TR, Maas SLN, Malhi H, Marcilla A, Mariani J, Mariscal J, Martens-Uzunova ES, Martin-Jaular L, Martinez MC, Martins VR, Mathieu M, Mathivanan S, Maugeri M, McGinnis LK, McVey MJ, Meckes DG, Meehan KL, Mertens I, Minciacchi VR, Möller A, Møller Jørgensen M, Morales-Kastresana A, Morhayim J, Mullier F, Muraca M, Musante L, Mussack V, Muth DC, Myburgh KH, Najrana T, Nawaz M, Nazarenko I, Nejsum P, Neri C, Neri T, Nieuwland R, Nimrichter L, Nolan JP, Nolte-’t Hoen ENM, Noren Hooten N, O’Driscoll L, O’Grady T, O’Loghlen A, Ochiya T, Olivier M, Ortiz A, Ortiz LA, Osteikoetxea X, Ostegaard O, Ostrowski M, Park J, Pegtel DM, Peinado H, Perut F, Pfaffl MW, Phinney DG, Pieters BCH, Pink RC, Pisetsky DS, Pogge von Strandmann E, Polakovicova I, Poon IKH, Powell BH, Prada I, Pulliam L, Quesenberry P, Radeghieri A, Raffai RL, Raimondo S, Rak J, Ramirez MI, Raposo G, Rayyan MS, Regev-Rudzki N, Ricklefs FL, Robbins PD, Roberts DD, Rodrigues SC, Rohde E, Rome S, Rouschop KMA, Rughetti A, Russell AE, Saá P, Sahoo S, Salas-Huenuleo E, Sánchez C, Saugstad JA, Saul MJ, Schiffelers RM, Schneider R, Schøyen TH, Scott A, Shahaj E, Sharma S, Shatnyeva O, Shekari F, Shelke GV, Shetty AK, Shiba K, Siljander PRM, Silva AM, Skowronek A, Snyder OL, Soares RP, Sódar BW, Soekmadji C, Sotillo J, Stahl PD, Stoorvogel W, Stott SL, Strasser EF, Swift S, Tahara H, Tewari M, Timms K, Tiwari S, Tixeira R, Tkach M, Toh WS, Tomasini R, Torrecilhas AC, Tosar JP, Toxavidis V, Urbanelli L, Vader P, van Balkom BWM, van der Grein SG., Van Deun J, van Herwijnen MJC, Van Keuren-Jensen K, van Niel G, van Royen ME, Wijnen AJ, Vasconcelos MH, Vechetti IJ, Veit TD, Vella LJ, Velot É, Verweij FJ, Vestad B, Viñas JL, Visnovitz T, Vukman KV, Wahlgren J, Watson DC, Wauben MHM, Weaver A, Webber JP, Weber V, Wehman AM, Weiss DJ, Welsh JA, Wendt S, Wheelock AM, Wiener Z, Witte L, Wolfram J, Xagorari A, Xander P, Xu J, Yan X, Yáñez-Mó M, Yin H, Yuana Y, Zappulli V, Zarubova J, Žėekas V, Zhang J. ye, Zhao Z, Zheng L, Zheutlin AR, Zickler AM, Zimmermann P, Zivkovic AM, Zocco D, Zuba-Surma EK, 2018. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7. 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Gong M, Hu Y, Liu H, Zhang W, Zhang M, Hu X, Aubert D, Zhu S, Wu L, Yan X, 2020. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 9, 1697028. 10.1080/20013078.2019.1697028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO, 2007. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9, 654–659. 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R, 2012. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev 64, 676–705. 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- Vyas S, Zaganjor E, Haigis MC, 2016. Mitochondria and cancer. Cell 166, 555–566. 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead B, Wu LP, Hvam ML, Aslan H, Dong M, DyrskjØt L, Ostenfeld MS, Moghimi SM, Howard KA, 2015. Tumour exosomes display differential mechanical and complement activation properties dependent on malignant state: implications in endothelial leakiness. J. Extracell. Vesicles, 10.3402/jev.v4.29685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendrini A, Paolini L, Busatto S, Radeghieri A, Romano M, Wauben MHM, van Herwijnen MJC, Nejsum P, Borup A, Ridolfi A, Montis C, Bergese P, 2020. Augmented Colorimetric NANoplasmonic (CONAN) method for grading purity and determine concentration of EV microliter volume solutions. Front. Bioeng. Biotechnol 7, 452. 10.3389/fbioe.2019.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pick H, Gasilova N, Li X, Lin TE, Laeubli HP, Zippelius A, Ho PC, Girault HH, 2019. MALDI detection of exosomes: a potential tool for cancer studies. Inside Chem 5, 1318–1336. 10.1016/j.chempr.2019.04.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


