Summary
RNA in situ hybridization can be time-consuming and difficult to troubleshoot. Here, we provide an optimized protocol for maize leaf tissue, though it can be applied to other plant tissues such as shoot apical meristems, embryos, and floral organs. We generate three >100 bp unique antisense probes for each gene of interest and hybridize them to tissue sections.
For complete details on the use and execution of this protocol, please refer to Bezrutczyk et al. (2021).
Subject areas: Cell Biology, Molecular Biology, In Situ Hybridization
Graphical Abstract

Highlights
-
•
Optimized for monocot tissues and tested on maize, sorghum, and teosinte
-
•
Clear results with low background and high resolution
-
•
Three unique samples for each gene of interest increase specificity
-
•
Comprehensive protocol with detailed explanation of all necessary steps
RNA in situ hybridization can be time-consuming and difficult to troubleshoot. Here, we provide an optimized protocol for maize leaf tissue, though it can be applied to other plant tissues such as shoot apical meristems, embryos, and floral organs. We generate three >100 bp unique antisense probes for each gene of interest and hybridize them to tissue sections.
Before you begin
This protocol is optimized for monocot tissue based on protocols from Jackson and Kellogg labs (Jackson, 1991; Jackson et al., 1994; Malcomber and Kellogg, 2004). The results are clearer, have less background and the protocol is therefore easier to troubleshoot, which was very complicated before (Stahl and Simon, 2010). It was primarily applied to the most prominent model organism among the monocots, Zea mays. Besides, the application to other organisms, such as Sorghum bicolor and Euchlaena mexicana, have successfully been performed (unpublished). The authors see no reason to doubt the suitability of this protocol for other monocots.
Solutions that can be prepared in advance
Timing: ∼1 day
Specific recipes for each reagent (Table 1) are found under ‘material and equipment’.
Table 1.
Solutions that can be prepared in advance and information about their treatment to achieve sterile and/or RNAse-free conditions
| REAGENT | Volume | Sterilization method | RNAse-free |
|---|---|---|---|
| 1 M Tris-HCl pH 6.8 | 1 L | Autoclave | DEPC water |
| 1 M Tris-HCl pH 7.5 | 1 L | Autoclave | DEPC water |
| 1 M Tris-HCl pH 9.5 | 1 L | Autoclave | DEPC water |
| 0.5 M EDTA pH 8 | 100 mL | Autoclave | DEPC water |
| 5 M NaCl | 500 mL | Autoclave | DEPC water |
| 1 M MgCl2 | 500 mL | n/a | n/a |
| 50% Dextran Sulfate | 50 mL | n/a | DEPC water |
| 100 mg/mL tRNA | 1 mL | n/a | DEPC water |
| 10% Acetic Acid | 20 mL | n/a | DEPC water |
| 10× in situ salts | 50 mL | Filter sterilize | DEPC water |
| 20× SSC pH 7 | 1 L | n/a | DEPC water |
| 10× PBS pH 7.4 | 1 L | Autoclave | n/a |
| 1× PBS pH 7.4 | 1 L | n/a | n/a |
| 50× TAE | 1 L | Autoclave | n/a |
| 1× TAE | 1 L | n/a | n/a |
| 1% (w/v) agarose in 1×TAE buffer | 500 mL | n/a | n/a |
| 50% Formamide | 50 mL | n/a | DEPC water |
| Hybridization solution | 40 mL | n/a | DEPC water |
| 15, 30, 50, 70, 80, 90% ethanol | 500 mL each | n/a | n/a |
| 25, 50, 75, 100% Histo-Clear in ethanol | 500 mL each | n/a | n/a |
| LB-Carbenicillin plates | n/a | n/a | n/a |
Specific recipes for each reagent are found under “materials and equipment”.
Solutions that have to be freshly prepared
Specific recipes for each reagent (Table 2) are found under “materials and equipment”.
Table 2.
Solutions that have to be prepared fresh and information about their treatment to achieve sterile and/or RNAse-free conditions
| REAGENT | Volume | Sterilization method | RNAse-free |
|---|---|---|---|
| 4% PFA-Triton in PBS pH 7.0 | 400 mL | n/a | n/a |
| 4% PFA in PBS pH 7.0 | 400 mL | n/a | n/a |
| Proteinase K buffer pH 7.5 at 37°C | 300 mL | n/a | n/a |
| 0.2% Glycine solution | 300 mL | n/a | n/a |
| 0.1 M Triethanolamine | 300 mL | n/a | n/a |
| 0.2× SSC 12 to 14 h at 55°C (for day 2) | 1 L | n/a | n/a |
| Block solution 1 | 50 mL | n/a | n/a |
| BSA block solution 2 | 300 mL | n/a | n/a |
| Buffer C | 100 mL | n/a | n/a |
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-digoxigenin AP-conjugated antibody | Merck/Roche | Cat#11333062910 |
| Bacterial and virus strains | ||
| One Shot TOP10 Chemically Competent E. coli | Thermo Fisher | Cat#1096176 |
| Chemicals, peptides, and recombinant proteins | ||
| 50× Denhardt's solution | Sigma-Aldrich | Cat#D2532 |
| Boehringer block reagent | Merck/Roche | Cat#1096176 |
| DEPC | Carl Roth | Cat#K028.3 |
| Proteinase K | Thermo Fisher | Cat#EO0491 |
| TRNA | Merck/Roche | Cat#10109517001 |
| Critical commercial assays | ||
| CloneJET PCR cloning kit | Thermo Fisher | Cat#K1232 |
| DIG labeling kit | Merck/Roche | Cat#11277073910 |
| NucleoSpin Gel and PCR Clean-up kit | Macherey-Nagel | Cat#740609.50 |
| PrimeSTAR GXL polymerase | Takara Bio | Cat#R050A |
| QuantiTect Reverse Transcription Kit | Qiagen | Cat#205311 |
| The MEGAscript SP6 Transcription kit | Thermo Fisher | Cat#Am1330 |
| Software and algorithms | ||
| Geneious R11 | N/A | www.geneious.com |
| Other | ||
| BCIP | Merck/Roche | Cat#11383221001 |
| Histo-Clear | Thermo Fisher | Cat#12358637 |
| Hot plate | VWR | Cat#442-0185 |
| Hybridization chamber | Merck | Cat#Z670146-1EA |
| HybriSlip hybridization coverslip | Thermo Fisher | Cat#10625983 |
| Microtome | Leica | RM 2255 replaced Cat#14051956472 |
| NBT | Merck/Roche | Cat#11383213001 |
| Paraplast | Carl Roth | Cat#X880.1 |
| ProbeOn Plus slides | Thermo Scientific (not available in Europe) | Cat#15-188-51 |
| RNase-ExitusPlus | PanReac AppliChem | Cat# A7153 |
| ROCKER 3D digital | IKA | Cat#0004001000 |
| Sectioning cartridge | Thermo Scientific | Cat#12740908100957 |
| Slide box for long-time storage | VWR | Cat#631-0735 |
| Slide dryer | Plano | Cat#12857-220-D |
| Slide rack with container | Merck | Cat#BR471800-5EA |
| SuperFrost Ultra Plus GOLD Adhesion slides | Thermo Scientific | Cat#11976299 |
| Water bath | Plano | Cat#28147-220 |
Alternatives: ProbeOn Plus slides are not available in Europe. The integrated spacer function allows the user to make slide sandwiches consisting of two slides facing each other rather than a single slide with a coverslip. This saves time and reagents because the same volume of probe and hybridization solution is used to treat twice as many slides. As an alternative, SuperFrost Ultra Plus GOLD Adhesion slides can be used in combination with HybriSlip Hybridization coverslips.
Alternatives: PrimeSTAR GXL polymerase gives consistently good results but can be replaced by your preferred polymerase.
Alternatives: Geneious R11 can be replaced by any other software supporting cloning, primer design and sequencing analysis.
Materials and equipment
50% Dextran sulfate
| Reagent | Final concentration | Amount |
|---|---|---|
| Dextran sulfate | 50% w/v | 25 g |
| RNAse-free water | n/a | to 50 mL |
| Total | 50 mL |
Note: Dissolve by gently rocking over night at 55°C. Centrifuge to remove bubbles and mix for another 2 to 3 h. Store at −20°C for long-time storage.
tRNA
| Reagent | Final Concentration | Amount |
|---|---|---|
| tRNA | 100 mg/mL | 100 mg |
| RNAse-free water | n/a | 1 mL |
| Total | 1 mL |
Note: Make 2 aliquots of 500 μL. Store at −20°C for long-time storage.
0.5 M EDTA pH 8.0
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA | 500 mM | 14.6 g |
| DEPC water | n/a | to 100 mL |
| Total | 100 mL |
CRITICAL: For any solutions that contain EDTA, the DEPC water should be prepared first in bulk and then used to prepare the solutions, because EDTA chelates DEPC.
Note: Stir the solution for 2 h at 37°C. Adjust pH with HCl. Store at 22°C ± 2°C for long-time storage.
10× insitu salts
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 3000 mM | 8.78 g |
| NaH2PO4·H2O | 50 mM | 0.39 g |
| Na2HPO4 | 50 mM | 0.355 g |
| Tris-HCl pH 6.8 (1 M) | 100 mM | 5 mL |
| EDTA pH 8 (0.5 M) | 50 mM | 5 mL |
| RNAse-free water | n/a | to 50 mL |
| Total | 50 mL |
Note: Store at −20°C for long-time storage .
10% Acetic Acid
| Reagent | Final concentration | Amount |
|---|---|---|
| Glacial acetic acid | 10% v/v | 2 mL |
| RNAse-free water | n/a | to 20 mL |
| Total | 20 mL |
Note: Store at 22°C ± 2°C for long-time storage.
Hybridization solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× in situ salts | n/a | 5 mL |
| Formamide | 20 mM | 20 mL |
| 50% Dextran sulfate | n/a | 10 mL |
| tRNA 100 mg/mL | n/a | 0.5 mL |
| 50× Denhardt’s solution | n/a | 1 mL |
| RNAse-free water | n/a | to 40 mL |
| Total | 40 mL |
Note: Keep solution RNAse-free and store at −20°C for long-time storage. Details to 50× Denhardt’s solution can be found in key resource table.
CRITICAL: Handle formamide in a fume hood.
10× PBS pH 7.4
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 1750 mM | 102.2 g |
| Na2HPO4 | 84.1 mM | 11.94 g |
| NaH2PO4·H2O | 18.6 mM | 2.56 g |
| ddH2O | n/a | to 1 L |
| Total | 1 L |
Note: Adjust pH with HCl. Store at 22°C ± 2°C for long-time storage.
20× SSC pH 7.0
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 3000 mM | 175.3 g |
| Sodium citrate | 340 mM | 88.2 g |
| ddH2O | n/a | to 1 L |
| Total | 1 L |
Note: Adjust pH with HCl. Store at 22°C ± 2°C for long-time storage.
1 M Tris-HCl pH 6.8
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 1000 mM | 121 g |
| ddH2O | n/a | to 1 L |
| DEPC | n/a | 1 mL |
| Total | 1 L |
Note: Stir the solution for 2 h at 37°C and adjust pH with HCl. Autoclave afterward to inactivate DEPC. Store at 22°C ± 2°C for long-time storage.
1 M Tris-HCl pH 7.5
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 1000 mM | 121 g |
| ddH2O | n/a | to 1 L |
| DEPC | n/a | 1 mL |
| Total | 1 L |
Note: Stir the solution for 2 h at 37°C and adjust pH with HCl. Autoclave afterward to inactivate DEPC. Store at 22°C ± 2°C for long-time storage.
1 M Tris-HCl pH 9.5
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 1000 mM | 121 g |
| ddH2O | n/a | to 1 L |
| DEPC | n/a | 1 mL |
| Total | 1 L |
Note: Stir the solution for 2 h at 37°C and adjust pH with HCl. Autoclave afterward to inactivate DEPC. Store at 22°C ± 2°C for long-time storage.
5 M NaCl
| Reagent | Final concentration | Amount |
|---|---|---|
| NaCl | 5 M | 292 g |
| ddH2O | n/a | to 1 L |
| Total | 1 L |
Note: Store at 22°C ± 2°C for long-time storage.
1 M MgCl2
| Reagent | Final concentration | Amount |
|---|---|---|
| MgCl2 | 1 M | 95 g |
| ddH2O | n/a | to 1 L |
| Total | 1 L |
CRITICAL: Paraformaldehyde is harmful to health. Work in a fume hood when handling paraformaldehyde solutions and wear a protective mask when weighing PFA powder.
Note: Store at 22°C ± 2°C for long-time storage.
4% Paraformaldehyde-Triton pH 7.0
| Reagent | Final concentration | Amount |
|---|---|---|
| PFA | 200 mM | 16 g |
| 30% Triton-X100 | 0.03% | 400 μL |
| 1× PBS pH 7.4 | n/a | to 400 mL |
| Total | 400 mL |
Note: Add 3 NaOH pellets to dissolve PFA in 350 mL, then adjust pH to 7.0 with HCL; top up with PBS to 400 mL. Needs to be prepared freshly.
Note: Prepare 4% Paraformaldehyde (without triton) likewise, but without Triton-X100.
Proteinase K buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 7.5 (1 M) | 100 mM | 30 mL |
| EDTA pH 8 (0.5 M) | 10 mM | 6 mL |
| ddH2O | n/a | to 300 mL |
| Total | 300 mL |
Note: Needs to be prepared freshly.
0.2% Glycine
| Reagent | Final concentration | Amount |
|---|---|---|
| Glycine | 0.2% w/v | 0.6 g |
| 1× PBS pH 7.4 | n/a | to 300 mL |
| Total | 300 mL |
Note: The solution can be reused four to five times. Store at 22°C ± 2°C for this period.
0.1 M Triethanolamine pH 8.0
| Reagent | Final concentration | Amount |
|---|---|---|
| Triethanolamine | 100 mM | 4.125 mL |
| ddH2O | n/a | to 300 mL |
| Total | 300 mL |
Note: Triethanolamine is very viscous: pipette slowly and carefully. Needs to be prepared freshly. Adjust pH to 8.0 with HCl.
Buffer C
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris-HCl pH 9.5 (1 M) | 100 mM | 10 mL |
| NaCl (5 M) | 100 mM | 2 mL |
| MgCl2 (1 M) | 50 mM | 5 mL |
| ddH2O | n/a | to 100 mL |
| Total | 100 mL |
Note: Needs to be prepared freshly.
Block solution 1
| Reagent | Final concentration | Amount |
|---|---|---|
| Boehringer Blocking reagent | n/a | 0.5 g |
| Tris-HCl pH 7.5 (1 M) | 100 mM | 5 mL |
| NaCl (5 M) | 150 mM | 1.5 mL |
| ddH2O | n/a | to 50 mL |
| Total | 50 mL |
Note: Dissolve at 60°C–70°C for at least 1 h. Details to Boehringer Blocking reagent can be found in key resource table. Needs to be prepared freshly.
BSA block solution 2
| Reagent | Final concentration | Amount |
|---|---|---|
| Bovine Serum Albumin | n/a | 3 g |
| Tris-HCl pH 7.5 (1 M) | 100 mM | 30 mL |
| NaCl (5 M) | 150 mM | 9 mL |
| 30% Triton-X-100 | 0.09% | 0.9 mL |
| ddH2O | n/a | to 300 mL |
| Total | 300 mL |
Note: Needs to be prepared freshly.
Step-by-step method details
Tissue preparation
Timing: 5 days
In this section plant samples are harvested and prepared for sectioning. The goal of this step is to create fixed, cleared samples that are embedded in Paraplast.
Harvesting and fixation
Note: Perform the steps under a fume hood.
-
1.
Add roughly 10 mL of PFA-Triton-X into a glass scintillation vial and keep on ice.
Alternatives: Any other small container which allows the tissue to lie flat can be used.
-
2.Cut a piece of the leaf tissue from a healthy plant and directly after cutting, place it into the fixative.
-
a.The size of the segment is relevant for penetration of the fixative and clearing agents.
-
b.One dimension of the tissue has to be less than 1 mm which is the leaf thickness in this case. We recommend a size of (LxWxH) 1 cm x 1 cm x 1 mm.
-
c.Each vial can carry up to 4 pieces of leaf tissue.
-
a.
-
3.
Place the scintillation vial on ice under a vacuum bell and pull a vacuum for 10 min.
-
4.
Carefully release the vacuum. Repeat the step two to five times until sinking of the tissue is observed.
-
5.
Replace with fresh fixative. Leave 12 to 14 h at 4°C.
Pause point: The tissue may be stored in 70% or 100% ethanol for longer periods of time at 4°C.
Dehydration
Note: Perform the steps on a rocker at 30 rpm.
-
6.
Incubate in 1× PBS for 1 h on ice.
-
7.
Incubate in 1× PBS for 1 h on ice.
-
8.
Incubate in 15% ethanol for 1 h on ice.
-
9.
Incubate in 30% ethanol for 1 h on ice.
-
10.
Incubate in 50% ethanol for 1 h on ice.
-
11.
Incubate in 70% ethanol for 1 h on ice.
-
12.
Incubate in 80% ethanol for 1.5 h on ice.
-
13.
Incubate in 90% ethanol for 1.5 h on ice.
-
14.
Incubate in 100% ethanol for 1.5 h on ice.
-
15.
Incubate in 100% ethanol for 12 to 14 h at 4°C.
Note: The tissue should slowly de-stain and chlorophyll should be visible in the ethanol.
Pause point: Dehydrated tissue may be stored in 100% ethanol at 4°C for 1 to 3 days.
Clearing
Note: Perform the steps on a rocker under a fume hood. Details to Histo-Clear can be found in key resource table.
-
16.
Incubate in 100% ethanol for 2 h at 22°C ± 2°C.
-
17.
Incubate in 25% Histo-Clear/75% EtOH for 1 h at 22°C ± 2°C .
-
18.
Incubate in 50% Histo-Clear/50% EtOH for 1 h at 22°C ± 2°C .
-
19.
Incubate in 75% Histo-Clear/25% EtOH for 1 h at 22°C ± 2°C .
-
20.
Incubate in 100% Histo-Clear for 1 h at 22°C ± 2°C .
-
21.
Incubate in 100% Histo-Clear for 1 h at 22°C ± 2°C .
-
22.
Incubate in 100% Histo-Clear for 1 h at 22°C ± 2°C .
Embedding and plate pouring
CRITICAL: Paraplast consists of paraffin wax (see key resource table), plastic polymers and DMSO. Paraplast solidifies between 56°C–57°C but can degrade above 62°C (Costa, 2018). Be careful to not heat Paraplast higher than 60°C and avoid re-melting, if possible (Jackson, 1991).
-
23.
Add Paraplast chips by half of the vial and fill it up with Histo-Clear. Leave it 12 to 14 h at 60°C.
-
24.
Also, place a 500 mL bottle fully filled with Paraplast chips to 60°C and leave it 12 to 14 h to melt.
-
25.
For three days in a row replace the vial content with freshly molten Paraplast in the morning and evening and leave at 60°C in between.
-
26.
Quickly pour the complete contents of the scintillation vial into a plastic petri dish.
-
27.
The wax will solidify before the samples can be arranged. Move the dish to a hot plate (<60°C) and let the Paraplast melt completely.
-
28.
Using preheated forceps, gently remove any air bubbles from above and underneath the plant tissue.
-
29.
Arrange the sections in the petri dish with maximum distance between them (Figure 1A).
-
30.
Remove the dish from the hot plate and let the Paraplast solidify at 22°C ± 2°C for ∼1 h.
Note: Solidified Paraplast easily breaks in an uncontrolled manner. Mark predetermined breaking points at the surface by scoring it with a razor blade (Figure 1A).
Figure 1.
Sample mounting and sectioning
(A) Embedded tissue in liquid Paraplast is poured into a petri dish, where a wax block is cut out after arranging and solidifying and mounted on a sample cartridge with fresh Paraplast as support structure.
(B) The mounted wax block is trimmed into a trapezoidal prism shape using a razor blade.
(C) Ribbons are produced by microtome sectioning the wax block, which is arranged with the long side touching the blade first. A good-looking ribbon is transferred to the surface of a water bath by detaching it from the blade also with a paint brush.
(D) Three ribbons with the length of the slide height are aligned close to each other to fill as much of the slide as possible.
Sectioning
Timing: 2 days
Sample mounting
Note: We prefer to mount the sample in two steps to make selecting the correct orientation of the final sections easier. The orientation is highly dependent on the experiment and should be considered carefully.
-
31.
Warp the petri dish to carefully loosen the solidified plate of wax out of the plastic. The sides can be separated from the dish by a razor blade.
-
32.
Cut out one single plant tissue as a wax block in a rectangular shape (Figure 1A).
-
33.
It is important not to cut directly at the tissue but to keep a surrounding wax layer of 2–5 mm.
-
34.
In a separate dish, melt Paraplast on the hot plate (<60°C).
Note: Molten Paraplast can be handled with disposable transfer pipets.
-
35.
Place the wax block in the desired orientation on a sectioning cartridge and add molten Paraplast to the sample holder and onto the sample sides building surrounding support structure. The mount will be stronger if Paraplast is added to the back of the cartridge as well (Figure 1A). The sample can also be poured into a mold, but we find it is easier to obtain the correct orientation with this method.
-
36.
Let the mounted sample harden at 22°C ± 2°C for ∼1 h.
Pause point: Embedded tissue in petri dishes or on sample holders may be stored at 22°C ± 2°C for several years.
Microtome sectioning
Note: Paraffin sectioning takes time and patience to learn. To prepare high quality sections, a calm state of mental clarity is essential.
-
37.
Heat the slide dryer and water bath to 37°C–42°C.
-
38.
Place the mounted sample on ice until use.
-
39.
Use a razor blade to trim the sample before securing it to the microtome sample holder. A trapezoidal prism shape is best, with the long side striking the blade first; this will facilitate the formation of a continuous ribbon of sections for easy handling (Figure 1B).
-
40.
Use the handwheel to bring the sample to the same height as the blade. Carefully position the blade as close as possible to the sample and fix the blade position.
-
41.
Orient the sample parallel to the blade with the setscrews (Figure 1C).
-
42.To remove the first layer of paraffin without any sample and to generate an even sample surface, trim it by choosing a cutting thickness of ∼60 μm.
-
a.Thick sections such as these will form rolls of Paraplast.
-
b.Trimming can be stopped when the paraffin rolls have a uniform width at both sides.
-
a.
-
43.Change the operating mode from trimming to sectioning.
-
a.Set a sectioning of 10 μm.
-
a.
-
44.Cut Paraplast ribbons of your sample (Figure 1C). Troubleshooting 1
-
a.The ribbon should have the length of the slide width.
-
b.The number of sections per ribbon therefore depends on the single section thickness and changes with every sample (Figure 1D).
-
a.
-
45.
Loosen the ribbon from the blade by carefully detaching it with a paint brush (Figure 1C).
-
46.Use the paint brush to transfer the ribbon to the water surface. Troubleshooting 1.
-
a.This will flatten the section.
-
b.If the paraffin sticks to the brush, a graphite pencil tip can help to detach.
-
a.
Note: Use ProbeOn Plus slides if possible (see key resource table).
-
47.
Hold the slide at the labeling end with one hand and dip it at an angle into the water bath. The slide should be nearly completely under water.
-
48.
Use the other hand to bring one section by the other to the slide with a paint brush. Arrange it with the brush on the water surface over the slide in the desired orientation and carefully lift the slide to fix there.
-
49.Continue with all remaining ribbons.
-
a.Try to move as many ribbons onto the slide as possible for maximal space usage (Figure 1D).
-
a.
-
50.
Drain off excess water with a Kim wipe from underneath the slide and place them on the slide dryer for 12 to 14 h.
Pause point: If slides are not immediately needed, they may be stored in a slide box for several weeks at 4°C.
Probe preparation
Timing: 7–10 days
Depending on the probe design, probes are generated from either gDNA or cDNA. In general probes can be cloned from gDNA, which can be prepared using standard DNA extraction protocols. If probes target cDNA regions spanning introns or if specific probes targeting isoforms of a gene are necessary, it is recommended to clone probes from cDNA. In this section leaf RNA is extracted and reverse transcribed into cDNA. RNA sense and antisense probes are designed and produced. Here, this protocol differs from other protocols in that we recommend you generate several unique and specific probes for each gene.
Probe design
Note: For each gene two to three probes will be designed to cover different regions of the gene. The specification of those as antisense and sense probe will occur later during transcription.
-
51.
We recommend designing three independent probes of approximately 100 bp each in the 5′-UTR, 3′-UTR, and in at least one other location (Figures 2B and 2C).
-
52.
As a general point, it is important to consider whether there are multiple isoforms of your gene of interest, which should be aligned first to ensure that the probe binds to all transcripts, unless the purpose of the experiment is to target a specific isoform of a gene.
-
53.
If there are many genes with high sequence homology to your gene of interest, the probe template regions must be carefully selected to avoid designing a non-specific probe, and this may limit the number of probes per gene if three unique 100 bp regions are not available.
-
54.
We recommend using one sense probe for each antisense probe as a negative control, as these have identical GC content and length to the antisense probes, and may be required by journal editors or reviewers for publication. Alternatively, one could generate random sequence controls.
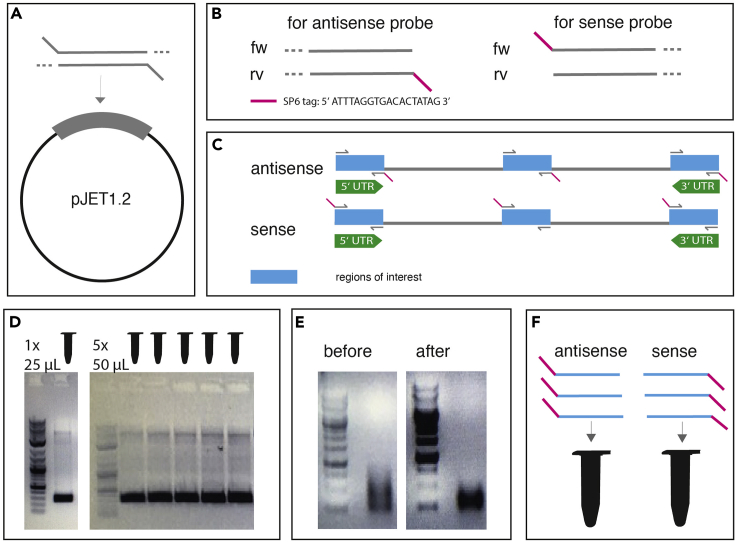
Figure 2.
Probe design, cloning, and generation
(A) Ligation of the gene of interest into the pJET1.2 vector using the CloneJet PCR cloning kit.
(B) Schematic design of SP6-tagged probe template primers.
(C) Schematic design of hybridization probes from three different regions of interest. One region is located in the 5′-UTR, one in the middle and one in the 3′-UTR.
(D) Assembly of small and big probe template PCRs and their product purification from an agarose gel after agarose gel electrophoresis. GeneRuler 1 kb serves as DNA ladder.
(E) After SP6 transcription over night the products of sense and antisense probes can be differentiated by their location of the tag. The removement of the remaining DNA from the DNA template is supervised by two agarose gel electrophoreses containing DNase un- and treated transcription product. GeneRuler 1 kb serves as DNA ladder.
(F) The 5′-UTR, coding region, and 3′-UTR RNA probes can be mixed in equal amounts to generate sense and antisense probe mixes.
RNA extraction for probe production
Alternatives: The procedures for performing RNA extraction are included for completeness. We recommend this protocol, however, any established RNA extraction protocol can be used.
CRITICAL: Maintain RNAse-free conditions.
CRITICAL: Phenol and chloroform are toxic. Work under a fume hood and wear a lab coat, safety goggles and appropriate gloves.
-
55.
Prepare RNA buffer with 100 mM sodium acetate, 1 mM sodium-EDTA and 4% (w/v) SDS.
-
56.
Prepare acid (pH 4.7) phenol:chloroform:isoamylalcohol 25:24:1. This mixture must sit until fully cleared, which can take up to an hour.
-
57.
Mix RNA buffer with phenol:chloroform:isoamylalcohol in 1:1 (v/v) ratio to prepare the RNA extraction solution.
-
58.
Place a bottle of 8M LiCl and bottle of 80% ethanol at −20°C.
CRITICAL: Plant tissue must remain frozen. Pre-cool spatulas, mortar and pestles, and tubes with liquid nitrogen.
CRITICAL: Working with liquid nitrogen has to be performed carefully while wearing a lab coat, safety goggles and appropriate gloves.
-
59.
Homogenize plant tissue using mortar and pestle in liquid nitrogen. Transfer about 300 mg of powder to a pre-cooled 2 mL tube.
-
60.
Aliquot 1.5 mL RNA extraction solution, add ground tissue powder, and vortex immediately.
-
61.
Incubate at 58°C (< 60°C) and vortex every few minutes during the 30 min incubation time.
CRITICAL: During the incubation chloroform evaporation may cause the tube to explode if heated above 58°C: handle with care and wear PPE.
-
62.
Centrifuge samples for 10 min at 14.000 × g and 22°C ± 2°C.
-
63.
Transfer the top phase to a new tube and add 950 μL phenol:chloroform:isoamylalcohol (pH 4.7). Vortex under a fume hood.
-
64.
Centrifuge samples for 5 min at 14.000 × g and 22°C ± 2°C.
-
65.
Transfer the top phase to a new tube and add 950 μL phenol:chloroform:isoamylalcohol (pH 4.7). Vortex under a fume hood.
-
66.
Centrifuge samples for 5 min at 14.000 × g and 22°C ± 2°C.
-
67.
Carefully transfer the top phase into a new 2 mL tube.
-
68.
Centrifuge samples for 5 min at 14.000 × g and 22°C ± 2°C.
-
69.
Carefully transfer the top phase into a new 2 mL tube and add 0.5 volumes of ice-cold 8M LiCl.
-
70.
Leave RNA at −20°C to precipitate for at least 2 h; 12 to 14 h results in better yield.
-
71.
Centrifuge samples for 15 min at 14.000 × g and 4°C.
-
72.
Remove the supernatant.
-
73.
Add 1 mL of ice-cold 80% ethanol to the tube and invert; pellet becomes opaque.
-
74.
Centrifuge samples for 2 min at 14.000 × g and 22°C ± 2°C.
-
75.
Remove the supernatant.
-
76.
Add 1 mL of ice-cold 80% ethanol to the tube and invert.
-
77.
Centrifuge samples for 2 min at 14.000 × g and 22°C ± 2°C.
-
78.
Carefully remove as much ethanol as possible and air dry the pellet under a sterile bench until the pellet edges become clear, but no longer than 5 min.
-
79.
Dissolve the pellet in 20–40 μL RNA elution buffer.
-
80.
Either store RNA at −80°C, leave at 4°C for analysis or keep on ice for cDNA synthesis.
cDNA synthesis and cloning
-
81.
Synthesize cDNA from your isolated RNA using the QuantiTect Reverse Transcription kit and its enclosed random hexamers primer mix (see key resource table).
-
82.
Amplify the gene of interest from cDNA with Takara PrimeSTAR GXL DNA Polymerase.
-
83.
Ligate the PCR product into pJET1.2 following instructions of CloneJet PCR cloning kit (Figure 2A).
-
84.
Transform chemically competent TOP10 cells with the ligation product and plate on carbenicillin selection plates.
-
85.
Perform a colony PCR with a standard Taq polymerase on at least 5 colonies.
-
86.
Isolate plasmids containing the gene of interest with the NucleoSpin Plasmid kit.
-
87.
Sequence the pJET plasmids containing your gene of interest and continue with the protocol if results match expectations.
Template PCR
-
88.
Amplify the regions of interest you selected using PrimeSTAR GXL DNA polymerase to generate a DNA template for RNA probe synthesis.
-
89.
Perform one small test probe template PCR (preliminary PCR) (1× 25 μL) in advance to verify that PCR settings are successful, and the reaction is specific (Figure 2D).
-
90.Perform two large (we recommend 5× 50 μL) probe template PCR reactions for each region of interest, one for the antisense and the other for the sense probe:
-
91.If several DNA fragments visible in the preliminary PCR:
-
a.Load total PCR product (5× 50 μL) onto a 1% agarose gel prepared with 1× TAE buffer. Separate DNA fragments at 100 V and 400 mA for 40 min (Figure 2D).
-
b.Excise the fragment of interest and purify with a dedicated NucleoSpin Gel and PCR Clean-up kit for RNAse-sensitive experiments.
-
c.Use preheated (70˚C) RNAse-free water to elute and maintain RNAse free environment from this step forward.
-
a.
-
92.If one DNA fragment visible in the preliminary PCR:
-
a.Purify total PCR product (5× 50 μL) with NucleoSpin Gel and PCR Clean-up kit.
-
b.Use preheated RNAse-free water to elute and maintain RNAse free environment from this step forward.
-
a.
SP6 transcription
-
93.
Estimate the dsDNA concentration of the purified probe template using a spectrophotometer.
-
94.
Calculate the final volume needed to reach a concentration of 500 ng/μL.
-
95.
Place the open tube under a sterile bench and evaporate until the desired volume and concentration is reached. This can take up to several hours, depending on the initial concentration.
-
96.
Perform a transcription reaction using the concentrated probe template as described in the manual of the MEGAscript SP6 transcription kit, using DIG RNA Labeling Mix (1:3) with kit-supplied rNTPs (see key resource table). This reaction is performed 12 to 14 h at 37°C (Figure 2E).
Note: Do not use a heat block for this reaction, as the small reaction volume may result in complete evaporation and condensation on the lid within a 1 mL tube. Place the entire tube in a 37°C oven or chamber.
Probe purification
-
97.
Save 0.2 μL in a separate tube to visualize on an agarose gel, then add 1 μL TURBO DNase (included in the MEGAscript SP6 transcription kit) to the remaining transcription RNA probe.
-
98.
Incubate for 20 min at 37°C.
-
99.
To stop the DNA digestion, add 24.6 μL RNAse-free water and 0.4 μL 0.5 M EDTA. Incubate for 10 min at 75°C.
-
100.
Again, save 0.2 μL of TURBO DNAsetreated RNA probe for agarose gel imaging.
-
101.
Perform a quick agarose gel electrophoresis with both samples: 1% agarose gel, 5 min, 100 V and 400 mA (Figure 2E).
-
102.Fragments should appear as discrete bands of the expected size in both gels: if so, continue with purification. If not, repeat SP6 transcription. To purify, treat your sample with the following solutions:
-
a.2 μL 2 mg/ml glycogen
-
b.4 μL 10% acetic acid
-
c.4 μL 3 M NaOAc
-
d.96 μL 100% ethanol
-
a.
Note: It is possible that the RNA forms secondary and tertiary structures, preventing efficient migration through the gel. This is typically visualized by multiple bands or a smear above the expected discrete band.
-
103.
Centrifuge the reaction tube for 30 min at 20.000 × g and 4°C.
-
104.
Carefully remove the supernatant and wash the pellet with 70% ethanol in RNAse-free water at 22°C ± 2°C.
-
105.
Decant and dry pellet for 5 min under sterile hood.
CRITICAL: Handle formamide in a fume hood.
-
106.
Resuspend the pellet in 25 μL RNAse-free 10 mM TE buffer pH 8 and 25 μL formamide.
-
107.
Store RNAse safe at −20°C.
In situ hybridization
Timing: min 2 days
In this step, your sectioned tissue is first pretreated to make it accessible for the RNA probes. Then the hybridization is performed and the detection process will be initiated. At the end of this step your sample will be ready to image.
Note: It is paramount to maintain an RNAse-free environment. We recommend cleaning the bench, pipettes, tip boxes etc. with Rnase-ExitusPlus or other similar treatment. Wear gloves and change them frequently. Wear a dedicated lab coat that is only for RNA in situ hybridization. The solutions used in this step should be exclusively for RNA in situ hybridization. Most can be reused several times, except for the 100% ethanol, PFA, proteinase K buffer, PBS and Histoclear.
Note: To treat the sections on the slides we recommend placing 18 or any other even number of the slides into the slide rack (see key resource table). Use empty, clean 1000 μL tip boxes, which fit the slide rack. Move the rack from box to box. Use a volume of solution that covers the slides completely: we recommend 300 mL. Incubations can be performed on an orbital shaker. The given incubation times are minimums, except for the proteinase K, which should not incubate longer than 10 min. Perform all steps at 22°C ± 2°C unless otherwise stated.
Deparaffinization and rehydration
-
108.
Incubate in Histo-Clear for 10 min.
-
109.
Incubate in Histo-Clear for 10 min.
-
110.
Incubate in 100% ethanol for 30 s.
-
111.
Incubate in 100% ethanol for 30 s.
-
112.
Incubate in 90% ethanol for 30 s.
-
113.
Incubate in 80% ethanol for 30 s.
-
114.
Incubate in 70% ethanol for 30 s.
-
115.
Incubate in 50% ethanol for 30 s.
-
116.
Incubate in 30% ethanol for 30 s.
-
117.
Incubate in H2O for 30 s.
-
118.
Incubate in 1× PBS for 2 min.
Permeabilization
-
119.
Add 30 μL proteinase K (20 mg/mL) to 300 mL pre-warmed proteinase K buffer.
-
120.
Incubate slides for 10 min in the proteinase K-containing buffer at 37˚C.
-
121.
Incubate in 0.2% glycine solution for 2 min.
-
122.
Incubate in 1× PBS for 2 min.
Refixation
-
123.
Incubate in 4% PFA for 10 min.
-
124.
Incubate in 1× PBS for 2 min.
Acetylation
CRITICAL: This is a unique but critical step for this in situ hybridization protocol. Acetylation reduces non-specific binding of the probe to the tissue and reduces background.
-
125.
Incubate in 0.1 M triethanolamine while dribbling 1.5 mL acetic anhydride over the slides.
-
126.
If possible, suspend the slide rack above a stir bar and place its container on a stirring plate, so that the solutions can be constantly mixed.
-
127.
Incubate for 10 min.
-
128.
Incubate in 1× PBS for 2 min.
Dehydration
-
129.
Incubate in 30% ethanol for 30 s.
-
130.
Incubate in 50% ethanol for 30 s.
-
131.
Incubate in 70% ethanol for 30 s.
-
132.
Incubate in 80% ethanol for 30 s.
-
133.
Incubate in 90% ethanol for 30 s.
-
134.
Incubate in 100% ethanol for 1 min.
-
135.
Incubate in 100% ethanol for 2 min.
Pause point: At this point it is possible to store the slides at 4°C for up to several days. Therefore, keep the slide rack in the tip box filled with a small amount of ethanol to cover the bottom and seal the box with Parafilm.
Hybridization
Note: Decide on the probe concentration for the hybridization solution. For new probe, use multiple concentrations ranging from 0.5× to 2×.
-
136.
Thaw the hybridization solution at 22°C ± 2°C, set an oven to 55°C, and a heat block to 98°C.
CRITICAL: Handle formamide in a fume hood.
-
137.
Thaw all RNA sense and antisense probes on ice and fill hybridization chamber (see key resource table) with 100 mL formamide and 100 mL H2O.
-
138.
Remove slides from ethanol by arranging them on a fresh paper towel. Place slide pairs next to each other, whereby a slide pair is considered to be two slides that will be treated with the same probe. If you have an uneven number of slides, use a HybriSlip Hybridization coverslip for the final slide. If you are not using ProbeOn Plus slides with which you can make ‘slide sandwiches’ because of the raised tabs, you will treat each slide individually and use flexible coverslips, instead.
CRITICAL: Handle formamide in a fume hood.
-
139.When the probes are thawed, dilute the hybridization probes with formamide.
-
a.Mix equal volumes of the different probes for a given gene of interest (i.e., 5′-UTR antisense probe + central antisense probe +3′ antisense probe; Figure 2F).
-
b.For a 1× concentrated probe mix: Add 1 μL of RNA probe mix to 50 μL 50% formamide in RNAse-free water for 1 slide pair.
-
a.
-
140.Denature the diluted probes for 3 min at 98°C. Immediately, place them on ice. Further dilute hybridization probes with hybridization solution.
-
a.For 1 slide pair: Add 200 μL of hybridization solution.
-
a.
-
141.
Pipette 220 μL of prepared hybridization probe on one slide of a slide pair
-
142.
Lay one slide onto the other, sections facing each other, and make sure that no bubbles remain. Place the slide sandwich on the rubber-lined risers in the hybridization chamber. Avoid touching the slides, which could cause cross-contamination. It is also recommended to change the gloves between samples with different probes.
Alternatives: If slide sandwiches are not feasible, add 200 μL of hybridization probe on a SuperFrost Ultra Plus GOLD Adhesion slide and cover it with a HybriSlip Hybridization coverslip.
-
143.
Close the lid of the hybridization chamber and seal with parafilm.
-
144.
Carefully place the chamber in the oven at 55°C.
-
145.
Place a beaker filled with 500 mL H2O in the oven for 12 to 14 h to maintain humidity, as well as the prepared 0.2× SSC to pre-warm it.
Washing
-
146.
Set incubator to 55°C.
-
147.
0.2× SSC can be microwaved to reach 55°C if not pre-warmed.
-
148.
Carefully, remove hybridization chamber from the oven.
-
149.
Separate the slide pairs by dipping them into pre-warmed 0.2× SSC solution.
Alternatives: If slide sandwiches are not feasible, remove the coverslip by dipping the slide into pre-warmed 0.2× SSC solution.
-
150.
Place separated slides into the slide rack.
-
151.
Place the slide rack into the slide container and fill it with pre-warmed 0.2× SSC solution.
Note: Be careful not to pour SSC directly on the sections, but on the labeling end of the slide. Alternatively, remove the slides, add the SSC and replace the slides.
-
152.
Place the slide container in the incubator and gently shake at 55°C for 1 h.
-
153.
Replace 0.2× SSC solution and incubate gently shaking at 55°C for 1 h.
-
154.
Again, replace 0.2× SSC solution and incubate gently shaking at 55°C for 1 h.
Blocking
-
155.
Wash slides in 1× PBS on an orbital shaker for 5 min at 22°C ± 2°C.
-
156.
Move slides into a plastic box, where slides can be arranged facing up.
-
157.Incubate slides with 50 mL block solution 1 on an orbital shaker for 45 min at 22°C ± 2°C.
-
a.Ensure that the slides do not slide over each other to avoid damaging the sections.
-
a.
-
158.
Remove block solution 1 from the slides using a serological pipette.
-
159.
Incubate slides with 50 mL BSA block solution 2 on an orbital shaker for 45 min at 22°C ± 2°C.
Antibody
-
160.
Prepare the antibody solution before the incubation time is over. Dilute anti-Dig alkaline phosphatase conjugated antibody (see key resource table) 1:1250 in BSA block solution 2. Mix 4 μL antibody with 5 mL BSA block solution 2.
-
161.
Remove slides from the plastic box by arranging them on a fresh paper towel.
-
162.
Place the slide pairs next to each other and fill the hybridization chamber with 200 mL H2O.
-
163.Pour the antibody dilution into a weigh boat and treat slides with antibody dilution (Figure 3):
-
a.Make slide sandwiches with respective slide pairs with sections facing towards each other.
-
b.Place the short edge of the slide sandwich on a stack of paper towels to wick out any solution.
-
c.If all liquid is wicked away, dip the slide sandwich edge into the antibody solution to wick it away until all sections are covered.
-
d.Press the slide sandwich edge to the paper towels to wick away the antibody dilution, and then dip slide sandwich edge into the antibody solution again, until all sections are covered and avoid bubbles.
-
e.Dry the outside of the slide sandwich with a paper towel and place it in the hybridization chamber.
-
f.Close the lid and seal it with Parafilm.
-
g.Incubate for 2 h at 22°C ± 2°C.
-
a.
Alternatives: If slide sandwiches are not feasible, add 200 μL of antibody dilution on a SuperFrost Ultra Plus GOLD Adhesion slide and cover it with a HybriSlip Hybridization coverslip.
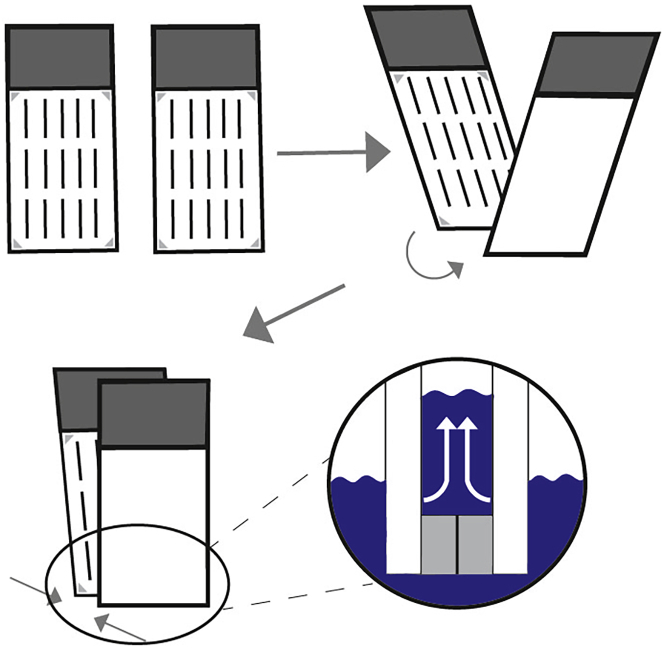
Figure 3.
Handling of slide sandwiches
Move the bottoms of two slides in the treatment solution against each other. Carefully bring the rest of the slides together until the tops are touching. The sections will be protected by spacers on ProbeOn Plus slides. The treatment solution will be wicked away by capillary forces.
Washing
-
164.
Move the slides out of the hybridization chamber and separate them carefully.
-
165.
Place single slides back into the plastic box to lay them tissue facing up. Place the box on an orbital shaker.
-
166.
Wash slides with 50 mL BSA blocking solution 2 for 15 min at 22°C ± 2°C.
-
167.
Wash slides with 50 mL BSA blocking solution 2 for 15 min at 22°C ± 2°C.
-
168.
Wash slides with 50 mL BSA blocking solution 2 for 15 min at 22°C ± 2°C.
-
169.
Wash slides with 50 mL BSA blocking solution 2 for 15 min at 22°C ± 2°C.
-
170.
Wash slides with 50 mL buffer C for 15 min at 22°C ± 2°C.
Detection
Note: Though there are several possible detection methods for in situ hybridization, we find that the reaction of alkaline phosphatase with BCIP/NBT (see key resource table) gives a reproducible result for various genes on maize leaf sections. The detection reagent BCIP is hydrolyzed by alkaline phosphatase conjugated to the secondary antibody to form an intermediate leucoindigo compound, which is oxidized by NBT to form 5, 5′-dibromo-4,4′-dichloro indigo (BCI). NBT is reduced to insoluble purple diformazan (DF) and forms NBT-DF. The combination of NBT-DF and BCI results in a dark purple colored precipitate. The color of the stain may vary from blue to purple to brown.
-
171.
Remove slides from the plastic box. Arrange pairs next to each other on a fresh paper towel.
-
172.
Fill the hybridization chamber with 200 mL H2O.
-
173.
Add 50 μL NBT and 37.5 μL BCIP to 10 mL buffer C.
-
174.
Pour the solution into a weigh boat and keep protected from bright light.
-
175.
Wick NBT-BCIP detection solution into the slide sandwiches as described for antibody treatment (Figure 3).
-
176.
Place them in the hybridization chamber, close the lid and seal it with Parafilm.
-
177.Incubate slides with detection solution for several hours to 3 days.
-
a.The duration depends on several factors such as abundance of mRNA and probe quality, but the staining process can be checked periodically.
-
b.Typically, in situ hybridization results are detectable as a purple precipitate after one night.
-
a.
-
178.
After checking the staining progress of one slide, apply fresh detection solution if staining has not reached the desired intensity or after 24 h of staining.
-
179.
Stop further incubation when purple precipitate can be observed but background noise is still low.
Note: The detection step is light sensitive. Keep light exposure times as short as possible.
Note: To stop the detection, an ethanol series is performed to reduce background staining. Notice that the stain is also slightly ethanol soluble and longer incubation periods will reduce the intensity of staining. This is particularly important for very faint signals, as the signal can be washed away by the ethanol steps.
Stop detection
-
180.
Separate slide sandwiches by carefully placing them in water, allowing them to fall apart.
-
181.
Place separated slides into the slide rack.
-
182.
Incubate in water for 3 min.
-
183.
Incubate in 30% ethanol for 30 s.
-
184.
Incubate in 50% ethanol for 30 s.
-
185.
Incubate in 70% ethanol for 30 s.
-
186.
Incubate in 80% ethanol for 30 s.
-
187.
Incubate in 90% ethanol for 30 s.
-
188.
Incubate in 100% ethanol for 30 s.
-
189.
Incubate in 100% ethanol for 30 s.
-
190.
Incubate in Histo-Clear for 5 min.
-
191.
Incubate in Histo-Clear for 5 min.
CRITICAL: To mount the slides, work under a fume hood and let your slides dry there, as well.
Mounting
-
192.
Move slides on a fresh paper towel. Troubleshooting 5
-
193.
Add a line of 120 μL Eukitt on the slide and cover it with a coverslip. Avoid bubbles.
-
194.
Let the mounting medium solidify for 12 to 14 h.
Imaging
Timing: 1 h
In this section the slides will be observed under a light microscope. Purple precipitate will locate the mRNA of the gene of interest.
Microscopy
-
195.Observe sections on a standard bright-field microscope. Troubleshooting 2, 3, and 4
-
a.For bundle sheath cells of maize plants, a 20× magnification is optimal.
-
a.
Expected outcomes
Sections hybridized with an antisense probe should show purple stain in the region or cell type where the mRNAs of interest are located. Sections treated with sense probes should not have any stain. NBT-BCIP incubation of corresponding sense and antisense probes should be stopped together to maintain comparable conditions (Figures 4A and 4B).
Figure 4.
Expected outcome
(A) In situ hybridization of ME1. Rank-2 intermediate vein from leaf section hybridized with antisense probe shows mRNA localization specifically to bundle sheath cells. Scale bar, 50 μm.
(B) In situ hybridization of CUP-SHAPED COTYLEDON2 (CUC2) in the maize shoot apical meristem. CUC2 marks the organ boundaries and specific expression is observed at the edges of the leaf primordium and at the boundaries between stem and leaves. Scale bar, 100 μm.
Limitations
The protocol can be applied to detection of mRNAs in monocot leaves and other tissues with similar properties. The use of Paraplast as embedding medium is optimal for mature leaves but may not be suitable for tissues which are softer, such as Arabidopsis leaves, or harder, such as maize kernels. In this case, another embedding medium such as LR white resin may be preferable.
Troubleshooting
Problem 1
Paraffin ribbons rip during sectioning e.g., during microtome sectioning.
Potential solution 1
-
1.
Change the blade angle. We usually position the blade at an angle of 7.5 degrees respective to the tissue.
-
2.
If the room temperature is too hot or too cold, it can affect the consistency of the wax. If possible, keep it at 22°C ± 2°C.
-
3.
If the sample has been at 22°C, place it back on ice and let it cool again.
-
4.
Wipe the edges of the section with a wet brush, occasionally.
-
5.
Use a fresh blade.
-
6.
Trim the sample with a razor blade back to a trapezoidal shape.
-
7.
Rearrange the sample to the blade. It should be parallel.
-
8.
Pause and come back when a calm state of mental clarity is reached.
-
9.
Tissue is not suitable for paraffin embedding.
Problem 2
No stain can be detected while imaging.
Potential solution 2
-
1.
Use a positive control probe, for example, in leaf tissue: RuBisCO small chain homologs (RBCS1 or RBCS2) have high levels of mRNA in bundle sheath cells. In the shoot apical meristem working probes have been published for a multitude of genes such CUP-SHAPED COTYLEDON2 (Johnston et al., 2014) (Figure 4B).
-
2.
RNA is degraded. Perform in situ hybridization steps absolutely RNAse-free.
-
3.
Antibody detection incubation time is too short. Incubate in NBT/BCIP longer.
-
4.
Permeabilization step was too long.
-
5.
Permeabilization was not successful. It is crucial for sample penetration.
-
6.
NBT/BCIP has expired. Use fresh solutions and reduce light exposure to a minimum.
-
7.
Ethanol de-staining washes were too intensive and reduced the stain intensity accordingly. Reduce ethanol incubation time.
-
8.
Probe did not bind to transcripts. Make new probes in a different region.
Problem 3
Paraffin sections under the microscope are not intact or have disintegrated.
Potential solution 3
-
1.
The tissue was not completely fixed. Be careful that the tissue sinks after pulling a vacuum. If FAA was used instead of PFA the incubation time has to be adjusted. In our experience 1 h fixation in FAA is insufficient.
-
2.
We highly recommend ProbeOn Plus Slides (see key resource table). They feature small spacers so that you can make slide sandwiches without removing the samples. If these are not available (as in Europe), it is not possible to make slide sandwiches and these steps must be performed with flexible coverslips.
-
3.
Replace the blade of the microtome. If the blade is blunted or notched it can damage the tissue.
Problem 4
During imaging, staining is detected in cells above and below major and/or rank-1 intermediate veins of maize leaves, even in sense probe-hybridized sections.
Potential solution 4
These are hypodermal sclerenchyma cells, and staining of these cells is considered unspecific since also seen when using sense control probes in maize leaves.
Problem 5
I want to use a counterstain to see cell walls more clearly e.g., when mounting.
Potential solution 5
In our experience, counterstaining is not necessary to visualize cell walls. However, several stains are compatible with this protocol. Classical counterstains such as Eosin-Y require xylene-containing mounting media, such as Eukitt, for visualization. Though NBT/BCIP may form crystals when mounted with xylene-containing media, we have not encountered this problem. In principle, Eukitt mounting medium and Eosin-Y counterstain are compatible with this protocol. As an alternative, Xylene-free mounting media such as Vectamount can be used instead, with a compatible counterstain such as Vector Methyl Green or Vector Nuclear Fast Red.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Dr. Wolf B. Frommer (frommew@hhu.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
This research was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy-EXC-2048/1-project ID 390686111 and SFB 1208-Project-ID 267205415, as well as the Alexander von Humboldt Professorship to W.B.F. The research at PSB-VIB UGent was performed by R.L., an FWO-predoctoral fellow (application number 1S10817N). We would also like to thank Clinton Whipple for sharing his experience and teaching us how to perform in situ hybridizations at PSB.
Author contributions
Conceptualization, N.R.Z., M.B., and R.L.; methodology and investigation, N.R.Z., M.B., and R.L.; resources and supervision, H.N., W.B.F., and R.S.; and writing, N.R.Z., M.B., and W.B.F.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Nora R. Zöllner, Email: nora.zoellner@hhu.de.
Wolf B. Frommer, Email: frommew@hhu.de.
References
- Bezrutczyk M., Zöllner N.R., Kruse C.P.S., Hartwig T., Lautwein T., Köhrer K., Frommer W.B., Kim J.-Y. Evidence for phloem loading via the abaxial bundle sheath cells in maize leaves. Plant Cell. 2021:koaa055. doi: 10.1093/plcell/koaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa P.M. Chapter 3 - Sample Preparation. In: Costa P.M., editor. The Handbook of Histopathological Practices in Aquatic Environments. Academic Press; 2018. pp. 51–81. [Google Scholar]
- Jackson D. In situ hybridization in plants. In: Gurr S.J., McPherson M.J., Bowles D.J., editors. Molecular Plant Pathology. A Practical Approach. Oxford University Press; 1991. pp. 163–174. [Google Scholar]
- Jackson D., Veit B., Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405. [Google Scholar]
- Johnston R., Wang M., Sun Q., Sylvester A.W., Hake S., Scanlon M.J. Transcriptomic analyses indicate that maize ligule development recapitulates gene expression patterns that occur during lateral organ initiation. Plant Cell. 2014;26:4718. doi: 10.1105/tpc.114.132688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcomber S.T., Kellogg E.A. Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell. 2004;16:1692. doi: 10.1105/tpc.021576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl Y., Simon R. mRNA detection by whole mount in situ hybridization (WISH) or sectioned tissue in situ hybridization (SISH) in Arabidopsis. Methods Mol. Biol. 2010;655:239–251. doi: 10.1007/978-1-60761-765-5_16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.