Abstract
HMG-CoA reductase (HMGCR) is the rate-limiting enzyme in cholesterol biosynthesis and the target for cholesterol-lowering therapy. Acetaldehyde dehydrogenase 2 (ALDH2) is primarily responsible for detoxifying ethanol-derived acetaldehyde and endogenous lipid aldehydes derived from lipid peroxidation. Epidemiological and Genome Wide Association Studies (GWAS) have linked an inactive ALDH2 rs671 variant, responsible for alcohol flush in nearly 8% world population and 40% of Asians, with cholesterol levels and higher risk of cardiovascular disease (CVD) but the underlying mechanism remains elusive. Here we find that the cholesterol levels in the serum and liver of ALDH2 knockout (AKO) and ALDH2 rs671 knock-in (AKI) mice are significantly increased, consistent with the increase of intermediates in the cholesterol biosynthetic pathways. Mechanistically, mitochondrial ALDH2 translocates to the endoplasmic reticulum to promote the formation of GP78/Insig1/HMGCR complex to increase HMGCR degradation through ubiquitination. Conversely, ALDH2 mutant or ALDH2 deficiency in AKI or AKO mice stabilizes HMGCR, resulting in enhanced cholesterol synthesis, which can be reversed by Lovastatin. Moreover, ALDH2-regulated cholesterol synthesis is linked to the formation of mitochondria-associated endoplasmic reticulum membranes (MAMs). Together, our study has identified that ALDH2 is a novel regulator of cholesterol synthesis, which may play an important role in CVD.
Keywords: Cholesterol, ALDH2, HMGCR, MAM
Graphical abstract
Highlights
-
•
ALDH2 regulates HMGCR ubiquitination and degradation.
-
•
Mitochondrial ALDH2 translocates to ER to interact with HMGCR.
-
•
ALDH2 KO or rs671 KI mice have increased cholesterol levels.
-
•
Statins inhibit increased cholesterol in AKO and AKI mice.
1. Introduction
Cardiovascular disease (CVD) including myocardial infarction and stroke, remains the leading cause of deaths worldwide [1,2]. Among many CVDs, ischemic heart disease, caused by the rupture of atherosclerosis (AS) plaque, accounts for over 40% of mortality [3]. It is well-documented that oxidative stress-induced lipid peroxidation (LPO) has been linked to atherosclerosis and CVD [4,5]. Bioactive lipid aldehydes, such as malondialdehyde (MDA), 4-hydroxy-nonenal (4-HNE), generated from LPO, play an important role in regulating inflammatory response and mitochondrial functions in atherosclerosis [[6], [7], [8]]. Our recent study found that LPO is a prominent feature in the plasma of CVD patients and oxidized cholesterol esters in LDL elevated cholesterol levels by inhibiting cholesterol uptake in the liver and macrophages [[9], [10], [11]]. Acetaldehyde Dehydrogenase 2 (ALDH2) is an aldehyde dehydrogenase in mitochondria and its primary function in the liver is to catalyze ethanol-derived acetaldehyde and lipid aldehydes generated from LPO to acetate and other nontoxic metabolites [12]. Previous studies showed that ALDH2 protects against CVDs through detoxifying endogenous aldehydes and inhibiting inflammation [13,14]. Importantly, about 8% world population and 40% Asians carry an ALDH2 single-nucleotide polymorphism (SNP) rs671 (ALDH2*2). In addition to alcohol flush in these ALDH2*2 carriers, epidemiological studies have linked ALDH2*1/*2 and ALDH2*2 mutants to increased risk for CVD compared to ALDH2*1 subjects, but the underlying mechanisms beyond alcohol consumption remain poorly defined [[15], [16], [17]]. Our recent study found that ALDH2 interacts with LDLR and AMPK to regulate macrophage foam cell formation, which may increase the risk of CVD in ALDH2 SNP carriers [18]. However, ALDH2 may also mediate phenotype switch of vascular smooth muscle cells, suggesting that ALDH2*2 carriers have lower risk for aortic dissection and aneurysm (AAD) [19]. Interestingly, emerging human studies have associated ALDH2*1/*2 and ALDH2*2 mutants with increased serum cholesterol levels [20,21], but roles of ALDH2 in cholesterol metabolism have yet been examined.
An elevated cholesterol level in the circulation, especially low-density lipoprotein (LDL)-cholesterol, is one of the most important risk factors for atherosclerotic cardiovascular diseases (ASCVDs) [[22], [23], [24]]. Liver is the important organ for maintaining cholesterol homeostasis in the body, including absorption, uptake, synthesis, efflux and degradation. Cholesterol synthesis is delicately regulated in the liver [25]. The endoplasmic reticulum (ER) enzyme HMG-CoA reductase (HMGCR), the target of statins, catalyzes the rate-limiting step in cholesterol de novo synthesis [26]. HMGCR is regulated by cholesterol levels. Insulin-induced gene 1 (Insig1) and E3 ligase, GP78, are the major regulators of HMGCR protein levels [27]. When sterols are accumulated, Insig1 binds to HMGCR and recruits GP78 and an ATPase-Valosin-containing protein (VCP) to promote the ubiquitination and degradation of HMGCR [27]. In this process, sterol responsive element binding protein (SREBP) cleavage-activating protein (SCAP) can also bind to Insig1, which protects Insig1 from being ubiquitinated by GP78 [28]. On the other hand, when the cholesterol level drops, Insig1 is ubiquitinated by GP78 and HMGCR is stabilized [29,30], which initiates the cholesterol synthesis. Interestingly, a recent study identified a de-ubiquitylase ubiquitin-specific peptidase 20 (USP20) stabilizes HMGCR in the feeding state [31]. Statins are the main therapies for lowering cholesterol in patients with CVDs [[32], [33], [34]].
As the major subcellular site for cholesterol synthesis, ER has many membrane contact sites (MCSs) with other subcellular organelles, such as mitochondria, Golgi and peroxisomes. Emerging studies have found that these contact sites have their own specific biological functions [35]. For example, mitochondria-associated endoplasmic reticulum membranes (MAMs) can influence mitochondrial dynamics, Ca2+ signaling, and lipid transportation [36,37]. Studies with isolated MAMs from rat liver showed that proteins responsible for lipid biosynthesis, such as acyl-CoA synthetase, cholesterol acyltransferase and diacylglycerol acyltransferase, are enriched in MAMs [38,39]. However, it remains poorly defined for the roles of MAMs in maintaining cholesterol homeostasis, including biosynthesis and transportation of cholesterol between ER and mitochondria [[39], [40], [41], [42]].
In this study, we set out to investigate the mechanisms by which ALDH2 regulates cholesterol metabolism using ALDH2 knockout (AKO) and ALDH2 rs671 knock-in (AKI) mice, focusing on the roles of ALDH2 on HMGCR stability and cholesterol biosynthesis in the context of CVD.
2. Methods
2.1. Animals
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences. All mice were housed in a temperature-control and pathogen-free room under a 12 h/12 h-light/dark cycle. All mice used in this study is C57BL/6J background. WT and ALDH2−/− mice were obtained after crossing ALDH2+/− with ALDH2+/− mice. WT, ALDH2*1/*2, ALDH2*2 mice were obtained after crossing ALDH2*1/*2 with ALDH2*1/*2 mice [18,43]. For genotyping of ALDH2E487K mice (ALDH2*2), genomic DNA extracted from tails were used for PCR with the following primers are showed in Supplemental Table 1.
WT and AKO male mice were fed with chow diet for 12 weeks and sacrificed at 12th week. For statin experiments, 6-week-old WT, AKO and ALDH2*2 mice (male) were fed with western diet (Research Diets, catalog D12079B) and lovastatin (30 mg/kg/day) or vehicle (1% carboxymethylcellulose sodium salt) by intragastric administration for 7 weeks. For Adeno-associated virus (AAV) experiments, 6-week-old mice (mice) were injected with knocking down PCSK9 and control AAV through tail vein and fed western diet for 3 weeks.
2.2. Histological analyzes
After sacrifice, mouse liver tissues were fixed with 4% formaldehyde overnight at 4 °C. Liver tissues were embedded in paraffin or in optimal cutting temperature (OCT) compound (Tissue-Tek ® OCTTM Compound, Sakura Finetek USA). Liver tissues in OCT compound were stored in −80 °C. For H&E staining, liver tissues in paraffin were divided into 5 μm sections and subjected to hematoxylin and eosin (H & E) staining. For Oil Red O staining, tissues in OCT compound were cut with a freezing microtome (Leica CM1950) into sequential slices of 10 μm. Frozen tissues were stained with Oil Red O (catalog O0625, Sigma, St. Louis, MO) and counterstained with Mayer's hematoxylin to visualize intracellular lipid droplets. All sections were obtained with a light microscope (Vectra 2, Perkin Elmer, USA) and analyzed with Image J for quantitative measurements.
2.3. Cell culture
7702 cell lines and 293T cell lines were cultured in DMEM (Hyclone) containing 10% fetal bovine serum (FBS, Hyclone) and 1% penicillin-streptomycin (PS, Gibico).
Primary hepatocytes were isolated from mice, purified and cultured as previously described [44]. Briefly, mice (6–8 weeks old, chow diet) were anesthetized by intraperitoneal injection of 6% chloral hydrate (10 μL/g) and perfused the liver with perfusion buffer and enzyme buffer (collagenase type1, 0.6 mg/mL, Worthington, NJ, USA). Finally, we separated mice hepatocytes by using percoll (GE Healthcare) and cultured these hepatocytes in DMEM with 10% FBS and 1% PS.
2.4. Western blotting
Cells were incubated in cell lysis for 30 min at 4 °C and tissues were homogenized in 500 μL tissue lysis buffer. Proteins were separated by SDS-PAGE and transferred onto PVDF membranes (Millipore). Antibodies against ALDH2 (catalog 15310-1-AP), LDLR (catalog 10785-1-AP), MFN2 (catalog 12186-1-AP) and GAPDH (catalog 60004-1-Ig) were purchased from Proteintech. Antibodies against Myc-Tag (catalog 2276S), AMFR-GP78 (catalog 9590S) and normal rabbit IgG (catalog 2729S) were purchased from Cell Signaling Technology. Antibodies against PCSK9 (catalog ab181142), INSIG1 (catalog ab70784), HMGCR (catalog ab174830) and SCAP (catalog ab125186) were purchased from Abcam. IP3R–II (catalog sc-398434) was purchased from Santa Cruz Biotechnology.
2.5. Immunoprecipitation and ubiquitination experiment
The immunoprecipitation experiment was conducted as in a previous report [18]. 7702 cells were transfected with target tagged protein by using Attractene Transfection Reagent (catalog 301005, Qiagen), harvested by using IP cell lysis buffer (50 mM Tris-HCL, 400 mM NaCl, 0.8% Triton-100X, PH7.5) and incubated with flag-beads or myc-beads (catalog B26301 for myc-beads, catalog B23102 for flag-beads, Biotool) overnight. Then, beads were separated and proteins were boiled with loading buffer at 95 °C for 5 min.
For ubiquitination experiment as described before [27], 7702 cells were transfected with HA-ub plasmids by using Transfection Kit (catalog L3000-015, Lipofectamine®3000), treated with MG132 (20 μM) and Bafilomycin A1 (50 μM) for 37 °C for 5 h. Then, cellular proteins were collected by using cell lysis buffer and then were incubated with HA-beads (catalog B26202) overnight at 4 °C. Proteins were boiled with loading buffer at 95 °C for 5 min and finally separated with HA-beads.
2.6. Real-time PCR
Total RNA was isolated from liver tissue or cells by using TRIzol reagent (Catalog 9109, Takara). The purity of extracted total RNA was determined by the A260/A280 ratio and the total RNA was reversely transcribed into cDNA by using Biosystems 7900 instrument. The fold change was calculated by using the comparative CT method and normalized to β-actin expression. The primer sequences are listed in Supplemental Table 1.
2.7. Total cholesterol, HDL-C, LDL-C and VLDL-C assays
Serum total cholesterol, HDL-C, VLDL-C, LDL-C, and triglyceride levels were enzymatically measured with the kits (Nanjing Jiancheng Bioengineering Institute, China). The total cholesterol levels in liver and cells were detected by GC-MS as in a previous study [11]. About 10 mg liver tissues were homogenized in ice-cold 1xPBS. Cholesterol was extracted with ions at m/z = 329, 368, 458 and internal standard (5α-cholestane) was extracted at m/z = 217, 357 in GC-MS.
2.8. The extraction of total and free cholesterol for GC-MS analysis
All samples were extracted according to our previous report [18]. In brief, cellular and liver cholesterol was extracted by using 2 mL hexane: isopropyl alcohol (IPA) (3:2, v/v) supplemented with 20 μL acetic acid and 6 μg internal standard (5α-cholestane). For free cholesterol, the dried residue was added 40 μL of pyridine and 40 μL of BSTFA and bathed at 90 °C for 60 min. For measuring total cholesterol, to the extract was added 200 μL 8 M KOH and incubated at 55 °C for 55 min. Then, the mixture was added HCl for adjusting pH to 3.0. The mixture was added 2 mL IPA: hexane: acetic acid (40:10:1, v/v/v), 1 mL hexane and 1 mL water. After vortex and centrifuged at 2000 rpm 3 min, the upper organic phase was collected and added 40 μL of pyridine and 40 μL of BSTFA for derivatization.
2.9. LC-MS analysis of metabolites in de novo cholesterol synthesis
Cells in 10-cm dish were cultured in a LPDS medium (DMEM with 5% lipoprotein deficient serum and 1% PS) for 12 h and labeled with 13C-acetate (2 mM, sigma) and 13C-glucose (2 g/L, Cambridge Isotope laboratory) for 24 h. The cells were washed with 1x PBS three times and then lysed with 500 μL EDTA-trypsin (Gibico). Cell lysates were collected after 3 min and the half of cell lysates were suspended in 400 μL methanol and 100 μL ddH2O (100 ng d3-mevalonate as the Internal standard). Other half of cell lysates were used for lanosterol and cholesterol extraction as described before. Lanosterol was extracted at m/z = 355,429,483. After vortex 1 min, samples were put in dry ice for 2 h and centrifuged at 14000 g for 15 min at 4 °C. Then the supernatant was transferred to a new 1.5 mL tube and the precipitation was re-suspended in 100 μL ACN: H2O = 1:1. After vortex and centrifugation, the supernatant was merged with previous supernatant. Samples were evaporated under a stream of N2 and dissolved in 0.1 mL of 25% ACN in water added with 5 mM ammonium acetate. Samples were analyzed using ACQUITY HSS T3 column (2.1 × 3x150 mm), 1.8 mm particle size (Waters, MA, USA) with an aqueous phase (A) of 25% ACN in water added with 5 mM ammonium acetate and a mobile phase (B) of 50% isopropanol, 45% ACN and 5% water with 5 mM ammonium acetate. The solvent gradient is the same as in a previous study [45]. The retention time and the m/z of cholesterol metabolites were determined by standards and were consistent with previous reports [46]. Peak areas of labeled cholesterol and cholesterol metabolites were normalized to that of internal standards and protein concentration (mg). The MS analysis was performed on a triple quadruple mass spectrometer TSQ Vantage (Thermo, San Jose, CA, USA) using multiple reaction monitoring.
2.10. Fluorescence microscopy
Cells were grown on glass coverslips in 6-well dishes and were cultured to 60–80% confluence. Next, Cells were washed with 1x PBS and fixed with 1 mL ice-cold methanol for 5 min at room temperature. They were rinsed twice with 1 mL 1x PBS for 5 min (room temperature) and then permeated with 1 mL of 1% Triton X-100 (diluted in PBS) for 5 min at room temperature. Cells were washed twice for 5 min with 1 mL 1x PBS, and were blocked with 1 mL 1% BSA for 30 min. Next, BMDMs were incubated with primary antibodies 30 min at 37 °C. The cells were then rinsed three times with 1 mL 1x PBS, and then incubated with Alexa-Fluor 488-conjugated anti-goat secondary antibody (Molecular Probes, A11058; diluted 1:100 in PBS) and Alexa-Fluor 594-conjugated anti-rat secondary antibody (Molecular Probes, cat. A21209; diluted 1:100 in PBS) or Alexa-Fluor 594-conjugated anti-mouse secondary antibody (Molecular Probes, cat. A11032; diluted 1:100 in PBS) for 30 min at 37 °C in the dark. They were washed four times with 1 mL PBS, and then mounted on slides using ProLong Diamond Antifade Mountant (Molecular Probes, cat. P36970). Cells were imaged under a Zeiss LSM 780. co-localization percentages (determined by Mander's overlap coefficient: the number of pixels from the red channel that overlap with pixels from the green channel divided by the total number of pixels detected in the red channel above the threshold value) were calculated by ZEN 2010 software (Zeiss). Thresholds were set automatically by the software as previously described [18].
2.11. Transmission electron microscopy (TEM)
Cells were grown on glass coverslips in 6-well dishes and were cultured to 60–80% confluence. Cultured 7702 cells (total medium or LPDS medium) were fixed with 2.5% glutaraldehyde in 0.1 mol/L sodium cacodylate buffer for 1 h at room temperature (RT). Cells were harvested with a cell scraper, and the aggregates were post-fixed with 1% osmium tetroxide in 0.1 mol/L cacodylate buffer for 1 h at RT, dehydrated in ethanol, and embedded in Epon 812 embedding resin. After polymerization, the plastic was removed and ultrathin sections were cut parallel and perpendicular to the flask surface. Thin sections were counterstained with uranyl acetate and lead nitrate, and examined with a FEI Tecnai G2 Spirit TEM.
2.12. Detection of interacting proteins with ALDH2 by using iTRAQ proteomics
After immunoprecipitation for ALDH2 in 7702 cells, we separated total cell lysate by using 10% SDS-PAGE for silver stain (catalog P0017S, Beyotime). The gel was extracted and the proteins were identified using iTRAQ proteomics by Shanghai Applied Protein Technology.
2.13. ChIP assay
7702 cells were cultured on 15-cm plates and transfected ALDH2 wild type and rs671 plasmid (2 μg) for 24 h. Metformin (2 mM) was used to treat cells for 24 h. ChIP assay was conducted using a ChIP Assay kit (catalog 17–295, Millipore). Eluted DNA was further purified using a PCR purification kit (Qiagen).
2.14. Luciferase assay
293T cells were transfected with pGL3.1 Insigs/pCMV-ATP6V0E2 (20:1) for 48 h. Metformin (2 mM) treated cells for 24 h. Luci-Insig2 plasmid is described in a previous publication [47]. Luciferase activity was measured using the Dual Luciferase Reporter Assay System (catalog E1960, Promega). The GL (firefly luciferase) activity was normalized to coex-pressed RL (renilla luciferase) activity. 293T cells were transfected with Lipofectamine 2000 Reagent (catalog 1854311, Invitrogen).
2.15. Statistics
Results are shown as mean ± SD or mean ± SEM. Statistical analysis was conducted using one or two-way ANOVA with appropriate post-hoc tests or an unpaired two tailed, Student's t-test. P < 0.05 is considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
3. Results
3.1. ALDH2 deficiency increases cholesterol levels in mouse serum and liver
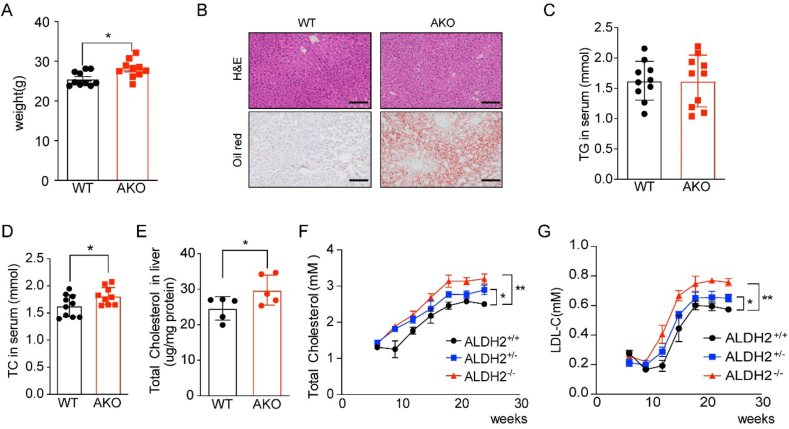
To determine the effect of ALDH2 on the cholesterol metabolism, we fed WT and AKO mice with chow diet and sacrificed these mice at 12 weeks old. ALDH2 knockout significantly increased mouse weight (Fig. 1A) and the lipid accumulation in the mouse liver without affecting the levels of triglycerides (TG) (Fig. 1B–C). Next, we measured total cholesterol levels in mouse serum and liver and found that total cholesterol (TC) in mouse serum and liver tissues was significantly increased in AKO mice (Fig. 1D–E). To further confirm the elevated cholesterol levels in AKO mice compared with WT mice, we collected mouse serum every three weeks for 24 weeks with chow diet and found that the levels of TC and LDL-C were significantly higher in homozygous AKO mice than those of heterozygous AKO and WT (Fig. 1F–G). Together, these results clearly show that ALDH2 affects cholesterol levels in mouse serum and liver, whereas ALDH2 deficiency leads to increased cholesterol levels in mice, suggesting hepatic ALDH2 plays an important role in cholesterol metabolism.
Fig. 1.
ALDH2 knockout increases cholesterol levels in mouse serum and liver even with chow diet feeding. (A) Mouse weights increased in AKO mice compared to WT at 12th week (chow diet, n = 10). (B) Representative H&E and oil red staining for liver, Scale bar: 400 μm. (C) Triglycerides (TG) in WT and AKO mouse serum at 12th week (chow diet, n = 10). (D) Total cholesterol in serum (chow diet, n = 10) and (E) liver of 12th week mice (chow diet, n = 5). (F–G) Total cholesterol and LDL-C in WT and AKO mouse serum from 5 to 28-week old (chow diet, n = 10). Statistical comparisons were made using a 2-tailed Student's t-test. All data are mean ± SD. *P < 0.05, **P < 0.01, WT: wild type; AKO: ALDH2 Knockout; ALDH2+/−, heterozygous; ALDH2−/−, homozygous. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.2. ALDH2 deficiency leads to increased de novo cholesterol synthesis in vivo and in liver cells in vitro
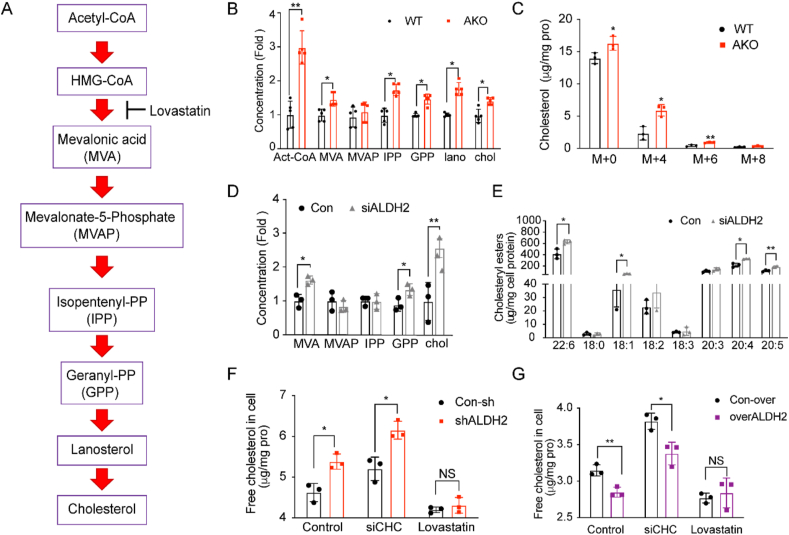
Cholesterol homeostasis, including cholesterol synthesis, uptake and degradation, is delicately controlled [32]. To investigate the underlying mechanisms regulating cholesterol levels in mouse liver by ALDH2, we first examined the mRNA levels of genes related to cholesterol synthesis (Hmgcs and Hmgcr), efflux (Abca1, Abcc1, and Abcg1), and bile acid metabolism (Abcg5, Abcg8, and Cyp7a1) in WT and AKO mouse liver tissues. Interestingly, none of these genes was significantly altered between WT and AKO mice (supplemental Fig. 1), suggesting that post-translational regulation of cholesterol homeostasis may play a more important role. Thus, we postulate that ALDH2 may regulates the cholesterol de novo synthesis in the liver through affecting protein stability of HMGCR. To test this hypothesis, we developed a liquid chromatography/tandem mass spectrometry (LC-MS/MS) method to measure the major intermediates of cholesterol synthesis pathways (Fig. 2A) and found that the levels of most intermediates in cholesterol synthesis were indeed elevated in AKO mouse liver (Fig. 2B), in addition to the increased cholesterol levels. Consistently, we treated mice hepatocytes with 13C-labeled acetate and found that the levels of labeled cholesterol (M+4, and M+6) were significantly increased in AKO compared to WT, as well as the major intermediates in the cholesterol synthesis pathways (Fig. 2C and Supplemental Fig. 2). To further confirm the role of ALDH2 in regulating cholesterol synthesis, we knocked down the ALDH2 expressions by transfecting human liver cell lines (7702) with ALDH2 siRNA and measured the levels of intermediates in cholesterol synthesis pathway, cholesterol and cholesterol esters. Consistently, knocking down ALDH2 significantly increased cholesterol synthesis (Fig. 2D) and cholesterol esters (Fig. 2E). Furthermore, down-regulation of ALDH2 in shALDH2 cells led to increased cholesterol levels, whereas over-expression of ALDH2 decreased cholesterol levels when the cells were incubated with lipoprotein deficient serum (LPDS) to stimulate cholesterol de novo synthesis; importantly, the differences of cholesterol levels with the manipulation of ALDH2 levels were diminished after lovastatin treatment (Supplemental Fig. 3). To provide further evidence that ALDH2 regulates hepatic cholesterol levels primarily through de novo synthesis instead of the uptake pathway by LDLR, we further measured the cholesterol levels in liver cells by knocking down the expression of Clathrin heavy chain (CHC) in the LDLR endocytosis pathway. Inhibiting cholesterol uptake by siCHC did not significantly affect the increased cholesterol levels in ALDH2 knockdown or decreased cholesterol levels in over-expression cells, whereas lovastatin treatment diminished the difference of cholesterol levels under both conditions (Fig. 2F–G). Taken together, ALDH2 primarily inhibits cholesterol de novo synthesis in mouse liver, whereas ALDH2 KO leads to elevated cholesterol levels through upregulating cholesterol de novo synthesis.
Fig. 2.
ALDH2 knockout increases cholesterol synthesis. (A) Schematics of the major intermediates in pathways of de novo cholesterol synthesis. (B) LC-MS analysis of de novo cholesterol synthesis in WT and AKO mouse liver at 12th week (chow diet, n = 10). (C) 13C-acetate incorporation into cholesterol in WT and AKO hepatocytes (n = 3). (D) LC-MS analysis of intermediates in de novo cholesterol synthesis in 7702 cells transfected with ALDH2 siRNA (n = 3). (E) LC-MS analysis of cholesterol esters in 7702 cells transfected with ALDH2 siRNA (n = 3). (F–G) GC-MS analysis of free cholesterol in control and overALDH2 7702 cells treated with siCHC and lovastatin (n = 3). Statistical comparisons were made using a 2-tailed Student's t-test. All data are mean ± SD. *P < 0.05, **P < 0.01, WT: wild type; AKO: ALDH2 Knockout; CHC, Clathrin heavy chain.
3.3. ALDH2 promotes the ubiquitination/degradation of HMGCR in the presence of cholesterol
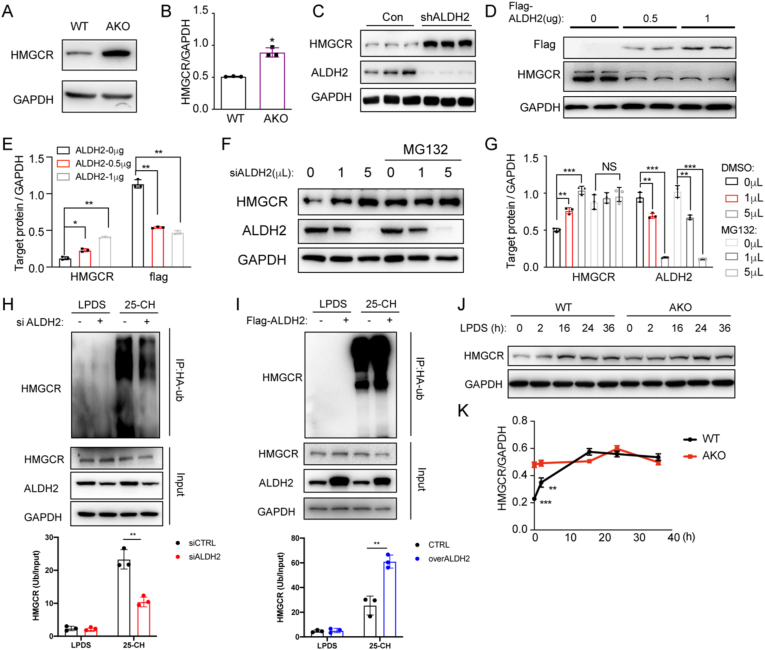
To investigate how ALDH2 regulates cholesterol synthesis, we focused on HMGCR, the rate-limiting enzyme of cholesterol synthesis and the target of statins. We found that HMGCR protein levels in AKO mouse liver were significantly increased compared to WT (Fig. 3A–B), consistent with the upregulation of cholesterol synthesis in AKO mouse liver (Fig. 2). Furthermore, ALDH2 knocking down in 7702 cells significantly increased HMGCR expression (Fig. 3C), whereas transient over-expression of ALDH2 dose-dependently decreased HMGCR protein level (Fig. 3D and E). Because the mRNA levels of HMGCR are not significantly different in AKO and WT mouse liver (Supplemental Fig. 1), we speculate that ALDH2 decreases HMGCR protein expression primarily through promoting the degradation of HMGCR in the proteasome after ubiquitination. To this end, we observed that HMGCR protein levels were dose-dependently increased when ALDH2 was knocked down by transfecting with increasing dose of siRNA in 7702 cells. Consistently, the increase of HMGCR levels were completely diminished with the treatment of a proteasome inhibitor, MG132 (Fig. 3F–G). Furthermore, knocking down ALDH2 decreased the ubiquitination of HMGCR (Fig. 3H), whereas over-expressing ALDH2 increased the ubiquitination of HMGCR (Fig. 3I), stimulated by 25-hydroxycholesterol (25-CH). Moreover, HMGCR expression was responsive to cholesterol levels when we treated WT and AKO hepatocytes with LPDS medium at different time points. The differences of HMGCR levels between WT and AKO hepatocytes disappeared after LPDS treatment for 16 h (Fig. 3J–K), suggesting that ALDH2 regulates HMGCR degradation in response to cholesterol levels. Taken together, all these data demonstrate that ALDH2 decreases cholesterol synthesis through increasing the ubiquitination and degradation of HMGCR in the presence of excess cholesterol.
Fig. 3.
ALDH2 destabilizes HMGCR through increasing its ubiquitination and proteasomal degradation in sterol-loaded cells. (A–B) HMGCR protein levels increased in AKO mouse hepatocytes compared to WT (n = 3). (C) HMGCR protein levels increased in ALDH2 knockdown 7702 cells (n = 3). (D–E) Representative western blot (D) and the quantification (E, n = 3) of HMGCR expression in 7702 cells transfected with different dose of flag-ALDH2 (WT) plasmids. (F–G) Representative western blot and the quantification of HMGCR expression in 7702 cells (treat with or without MG132) transfected with different dose of ALDH2 siRNA (n = 3). (H–I) The representative image and quantitation of ubiquitination of HMGCR in siALDH2 (H) and overexpressed ALDH2 7702 cells (I). (J–K) Representative western blot and the quantification of HMGCR expression in WT and AKO hepatocytes treated with lipoprotein deficient medium (LPDS) from 0 to 36 h (n = 3). Statistical comparisons were made using a 2-tailed Student's t-test or ANOVA. All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
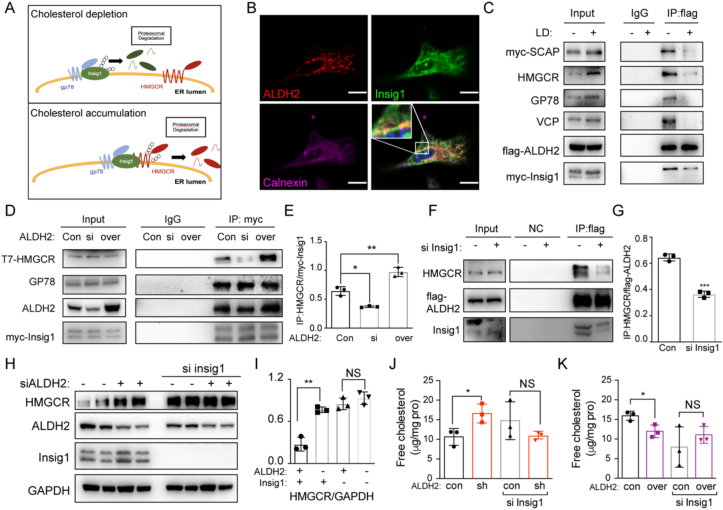
3.4. ALDH2 enhances the formation of Insig1/HMGCR/GP78 complex
It is well-documented that Insig1 and E3 ligase GP78 are key factors responsible for regulating HMGCR degradation, which represents one of the most important feedbacks for cholesterol metabolism in the liver [30]. In the absence of cholesterol, HMGCR is stabilized to increase the synthesis of cholesterol in ER when Insig1 is degraded after it binds to GP78 and ubiquitinated by GP78. On the other hand, in the accumulation of cholesterol, Insig1 binds to HMGCR and recruits GP78 to ubiquitinate HMGCR itself. The degradation of HMGCR decreases cholesterol synthesis (Fig. 4A). We performed proteomic analysis on ALDH2-enriched proteins to identify potential interacting proteins with ALDH2 and found that ALDH2 binds to many mitochondrial and ER proteins including Insig1 (Supplemental Table 2). Therefore, we hypothesized that ALDH2 increased the ubiquitination of HMGCR through promoting the formation of Insig1/HMGCR/GP78 complex. Consistently, we observed a co-localization of ALDH2 with Insig1 in the ER in immunofluorescent experiments (Fig. 4B). Due to the potential cross-reaction of ALDH2 antibody with other isoform of ALDHs, such as ALDH1A1 and ALDH1B1, we validated the specificity of ALDH2 using AKO tissues or shALDH2 cells and found that AKO or knockdown specifically downregulated ALDH2 expressions without significantly affecting the expression of ALDH1A1 and ALDH1B1 (Supplemental Fig. 4A-D). Furthermore, immunoprecipitation (IP) results showed that ALDH2 bound to Insig1/HMGCR/GP78 complex and this binding significantly decreased in the absence of cholesterol with LPDS treatment (Fig. 4C). Moreover, over-expressing ALDH2 increased the binding of Insig1 and HMGCR, whereas knockdown ALDH2 decreased the interaction of Insig1 and HMGCR (Fig. 4D–E). Consistently, knocking down Insig1 led to the decreased binding of HMGCR and ALDH2 (Fig. 4F–G). Furthermore, the increased HMGCR protein level due to ALDH2 knocking down disappeared after Insig1 knocking down (Fig. 4H–I). Consistently, the increased cholesterol levels in shALDH2 cells and decreased cholesterol levels in ALDH2 overexpression cells were diminished after Insig1 knockdown (Fig. 4J–K), suggesting that ALDH2 decreases HMGCR protein levels through interacting with Insig1. In summary, all these results demonstrate that ALDH2 potentiates the ubiquitination/degradation of HMGCR through promoting Insig1 binding to HMGCR and recruiting E3 ligase GP78, which in turn attenuates the de novo cholesterol synthesis.
Fig. 4.
ALDH2 promotes the interaction of HMGCR and Insig1 in sterol-loaded cells. (A) Schematics of the interactions of HMGCR, Insig1 and gp78 in the presence or absence of cholesterol. (B) Immunofluorescent results in 7702 cells (Red, ALDH2; Green, Insig1; Pink, Calnexin, ER marker); Scale bar: 5 μm. (C) Immunoprecipitation of ALDH2 and cholesterol synthesis-related proteins in the presence or absence of cholesterol. (D–E) Representative western blot and the quantification of immunoprecipitation of Insig1 and HMGCR, gp78 in si ALDH2 and overexpressed ALDH2 7702 cells (n = 3). (F–G) Immunoprecipitation of ALDH2 and HMGCR in control and si Insig1 7702 cells (F, representative image; G, quantification, n = 3). (H–I) Representative western blot and the quantification of HMGCR expression in 7702 cells treated with ALDH2 siRNA or Insig1 siRNA (n = 3). (J–K) Free cholesterol levels in control, shALDH2 (J) and overexpressed ALDH2 (K) 7702 cells treated with or without Insig1 siRNA (n = 3). Statistical comparisons were made using a 2-tailed Student's t-test or ANOVA. All data are mean ± SD. *P < 0.05, **P < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
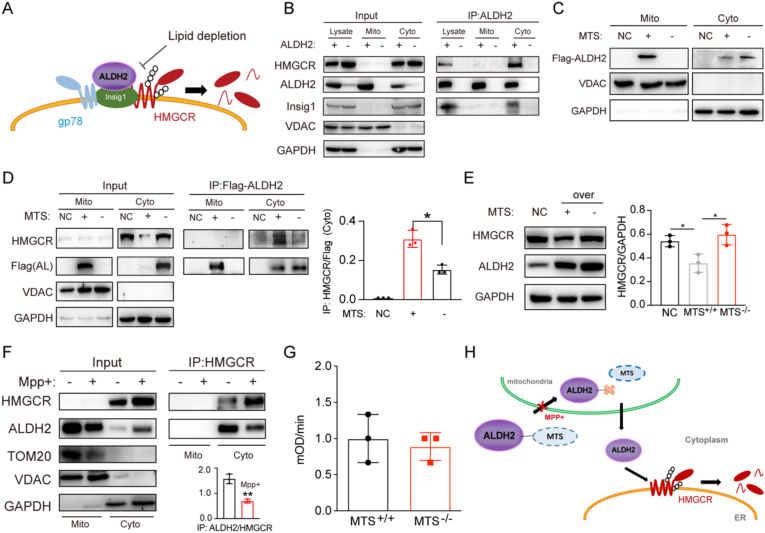
3.5. HMGCR stability in the ER membrane is regulated by ALDH2 after its exit from mitochondria
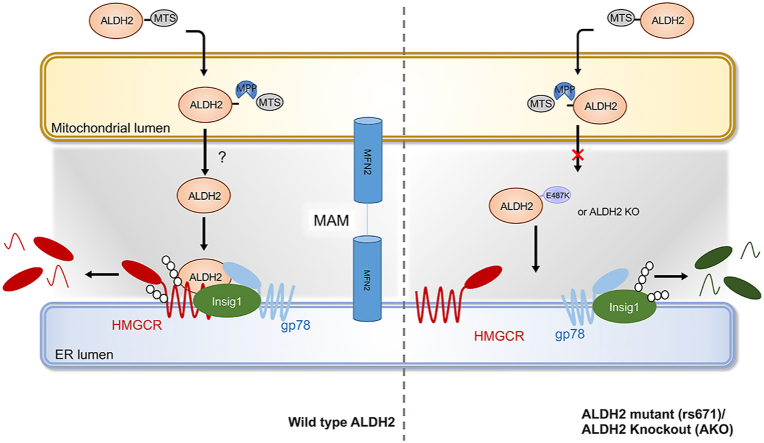
It is well-established that the cholesterol synthesis machinery, including HMGCR, Insig1 and gp78, resides in ER membrane and ALDH2 is primarily a mitochondrial protein. We next examined whether ALDH2 exits from mitochondria to interact with HMGCR and Insig1 in the ER (Fig. 5A). First, we performed IP experiments in cytosolic and mitochondrial fractions of AKO and WT liver tissues and found that ALDH2 indeed presented in both mitochondrial and cytosolic fractions, while only cytosolic ALDH2 pulled down Insig1 and HMGCR (Fig. 5B). Next, we overexpressed Flag-tagged ALDH2 in the presence or absence of mitochondrial targeting sequence (MTS) in liver cells and found that a majority of ALDH2 was localized in mitochondria and small fraction of ALDH2 appeared in cytosol with MTS, whereas MTS deletion resulted in the exclusion of ALDH2 from mitochondria and slightly increased presence of cytosolic ALDH2 (Fig. 5C). Furthermore, deletion of MTS significantly decreased the interaction of ALDH2 with HMGCR in the cytosolic fraction compared to the presence of MTS (Fig. 5D). Consistently, MTS deletion stabilized HMGCR, presumably through the decreased ubiquitination and proteasomal degradation of HMGCR (Fig. 5E). Moreover, previous study found that MPP+ (1-Methyl-4-phenylpyridinium) treatment blocked mitochondrial proteins transport [48] and we observed that MPP+ significantly decreased the ALDH2/HMGCR interaction and stabilized HMGCR in the cytosol (Fig. 5F), suggesting that ALDH2's entry and exit of mitochondria is essential for the interaction with HMGCR. Interestingly, expression of MTS did not significantly affect the enzymatic activity of ALDH2 (Fig. 5G), suggesting that the ALDH2 enzymatic activity plays a limited role in regulating cholesterol levels through interaction with HMGCR and ALDH2. Taken together, our data support the model that ALDH2 with MTS enters mitochondria and ALDH2 exits from mitochondria to interact with HMGCR/Insig1 to regulate HMGCR protein stability in the ER and subsequent cholesterol de novo synthesis (Fig. 5H).
Fig. 5.
ALDH2 regulates HMGCR after its exit from the mitochondria. (A) Schematics of ALDH2 promoting the interaction of Insig1 and HMGCR. (B) ALDH2 interacts with HMGCR in the cytosol instead of the mitochondria. (C) The mitochondrial targeting sequence (MTS) is important for the cellular localization of ALDH2. (D) The interaction of ALDH2 with HMGCR is affected by MTS (right, quantification, n = 3). (E) HMGCR expression in mouse hepatocytes transfected vector (NC), total ALDH2 and ALDH2 without MTS (right, quantification, n = 3). (F) Blocking the entry into mitochondria by MPP+ stabilizes HMGCR (n = 3). (G) MPP+ treatment does not significantly affect total ALDH2 enzymatic activity. (H) Schematics of ALDH2 decreasing HMGCR after entering into mitochondrion. Statistical comparisons were made using a 2-tailed Student's t-test or ANOVA. All data are mean ± SD. *P < 0.05. Mito: mitochondrion; Cyto: cytosol.
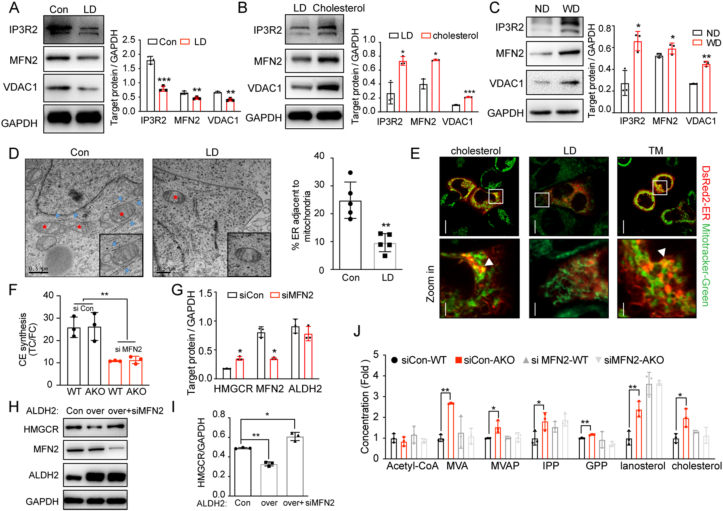
3.6. Mitochondria-associated membrane with ER (MAM) is involved in ALDH2 interaction with HMGCR
Because ALDH2 is a mitochondrial matrix protein and HMGCR and Insig1 reside in the ER membrane, we hypothesize that ALDH2 binds to Insig1 potentially through the formation of MAMs in response to cholesterol levels. Our proteomics results showed that ALDH2 bound to several MAM proteins, such as Voltage-dependent anion channel 2 (VDAC2) and ER chaperone protein Bip (Supplemental Table 2). We further found that cholesterol depletion significantly decreased the expression of makers of MAMs: Inositol 1,4,5-trisphosphate receptor type 2 (IP3R2), mitofusin 2 (MFN2) and Voltage-dependent anion-selective channel 1 (VDAC1) (Fig. 5, Fig. 6). On the other hand, treatment with cholesterol increased the makers of MAMs formation (Fig. 5, Fig. 6B). Consistently, Western diet feeding also increased IP3R2, MFN2 and VDAC1 expressions in WT mouse liver compared to chow diet (Fig. 5, Fig. 6C). Moreover, transmission electron microscopy (TEM) experiments and immunofluorescent experiments showed that the contact of mitochondria and ER decreased after cholesterol depletion, suggesting that cholesterol modulates the formation of MAMs (Fig. 6D–E). Next, to investigate whether the formation of MAMs affects ALDH2-regulated cholesterol ester (CE) synthesis through interactions with HMGCR, we decreased MAM formation by knocking down MFN236 and observed that knocking down MFN2 in WT and AKO hepatocytes decreased CE synthesis by measuring the ratios of TC/FC (Fig. 5, Fig. 6F). Consistently, knocking down MFN2 in WT mouse hepatocytes stabilized HMGCR without affection ALDH2 expressions (Supplemental Fig. 5A and Fig. 5, Fig. 6). Furthermore, the destabilization effects of HMGCR by over-expressing ALDH2 were reversed after knocking down MFN2 (Fig. 5, Fig. 6H–I). Consistently, the increased cholesterol synthesis in AKO hepatocytes also diminished by knocking down MFN2 (Fig. 5, Fig. 6J). Taken together, all these results strongly suggest that MAMs are involved in ALDH2-regulated cholesterol synthesis through interaction with HMGCR/Insig1 in the ER membrane.
Fig. 6.
Mitochondria-associated membrane with ER (MAM) is involved in ALDH2 interaction with HMGCR. (A–B) Markers of MAM in 7702 cells treated with lipoprotein deficient (LD) medium for 12 h (A, n = 3) and re-added cholesterol in lipoprotein deficient medium (B, n = 3). (C) Representative western blot (left) and the quantification (right, n = 3) of makers of MAM formation in 12-week old mouse liver fed with chow diet and 6-week western diet, respectively. (D) Transmission Electron Microscopy (TEM) results of MAM in 7702 cells treated with or without lipoprotein deficient medium (left, representative image; right, quantification, n = 5). (E) Immunofluorescence results of MAM in 7702 cells treated with total medium, LD and cholesterol. (F) Cholesterol esters synthesis in WT and AKO hepatocytes treated with si control or MFN2 siRNA (n = 3). (G) Western blot quantification of expression of HMGCR in WT hepatocytes treated with si control or siMFN2 RNA (n = 3). (H–I) Representative western blot (H) and the quantification (I, n = 3) of HMGCR expression in control and overexpressed ALDH2 7702 cells treated with or without MFN2 siRNA. (J) LC-MS analysis of de novo cholesterol synthesis in WT and AKO hepatocytes treated with or without MFN2 siRNA (n = 3). Statistical comparisons were made using a 2-tailed Student's t-test. All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. WT: wild type; AKO: ALDH2 Knockout.
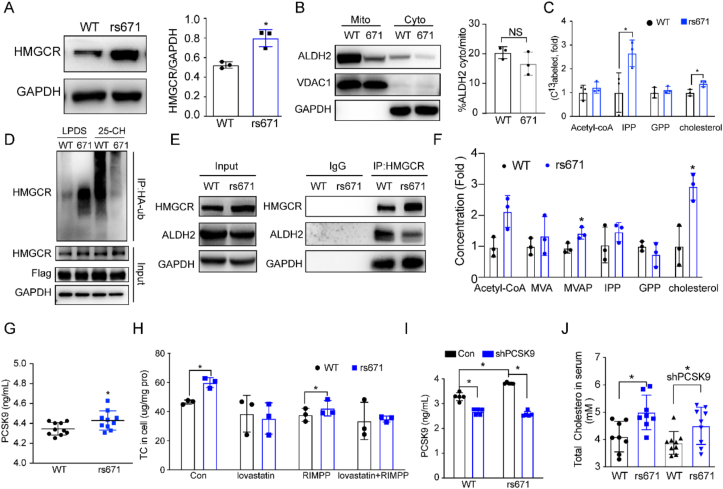
3.7. ALDH2*2 (rs671) mutant increases cholesterol synthesis through stabilizing HMGCR
Previous studies found that Asians with ALDH2*2 mutant have higher total cholesterol and LDL-C levels compared with ALDH2*1 counterparts [21], suggesting that similar mechanism operates in ALDH2*2 mutant as that in AKO mice in the context of regulating cholesterol synthesis. Consistently, we observed increased TC, LDL-C levels in ALDH2*2 mice compared to the WT (Supplemental Fig. 5B and C), similar to the AKO mice. To study the role of ALDH2*2 mutant on cholesterol synthesis, we transfected 7702 cells with WT and ALDH2*2 mutant plasmids and showed that ALDH2*2 mutant significantly stabilized HMGCR expression (Supplemental Fig. 6A). Furthermore, mutant ALDH2 increased the binding to HMGCR but decreased the binding to Insig1 (Supplemental Fig. 6B), suggesting that ALDH2*2 mutant stabilized HMGCR through decreasing its binding to Insig1, which is responsible for recruiting E3 ligase GP78. Consistently, the protein levels of HMGCR in ALDH2*2 mouse hepatocytes are higher than the WT (Fig. 7A). Furthermore, the expression levels of mutant ALDH2 were both decreased in the mitochondria and cytosol compared to WT although more ALDH2 rs671 resides in mitochondria than ER (Fig. 7B). Moreover, metabolite analysis and metabolic flux experiments in WT and ALDH2*2 mutant hepatocytes and 7702 cells showed that ALDH2*2 mutant increased cholesterol synthesis in vitro (Fig. 7C, supplemental Fig. 6C and D). Similar to ALDH2 KO, ALDH2 ubiquitination was significantly attenuated in ALDH2 rs671 with the stimulation of 25-hydoxycholesterol (Fig. 7D). Importantly, HMGCR expression is much higher in human liver tissues with ALDH2 rs671 compared to WT, so as the intermediates in cholesterol synthesis and cholesterol levels (Fig. 7E–F).
Fig. 7.
ALDH2 rs671 (ALDH2*2) mutant increases cholesterol synthesis primarily through decreasing HMGCR degradation. (A) Representative image and quantification of HMGCR expression in wild type ALDH2 (WT) and ALDH2*2 (671) primary mouse hepatocytes (n = 3). (B) Wild type ALDH2 (WT) and ALDH2*2 (671) expression in mitochondria and cytoplasm (n = 3). (C) 13C-acetate incorporation into cholesterol in WT and ALDH2*2 hepatocytes (n = 3). (D) HMGCR ubiquitination is diminished in rs671 cells stimulated with 25-HC compared to WT. (E) HMGCR is stabilized in rs671 human liver tissues (n = 3). (F) Major intermediates in de novo cholesterol synthesis in WT and ALDH2 rs671 human liver tissues (n = 5). (G) PCSK9 concentrations in 13-week old mouse serum after Western diet feeding for 7 weeks (n = 10). (H) GC-MS analysis of total cholesterol in hepatocytes treated with RIMPP or lovastatin (n = 3). (I) PCSK9 levels in mice injected with control or shPCSK9 AAV (n = 9). (J) Total cholesterol concentration in mouse serum after Western diet feeding for 9 weeks (n = 8–9). Statistical comparisons were made using a 2-tailed Student's t-test or ANOVA. All data are mean ± SD. *P < 0.05, **P < 0.01.
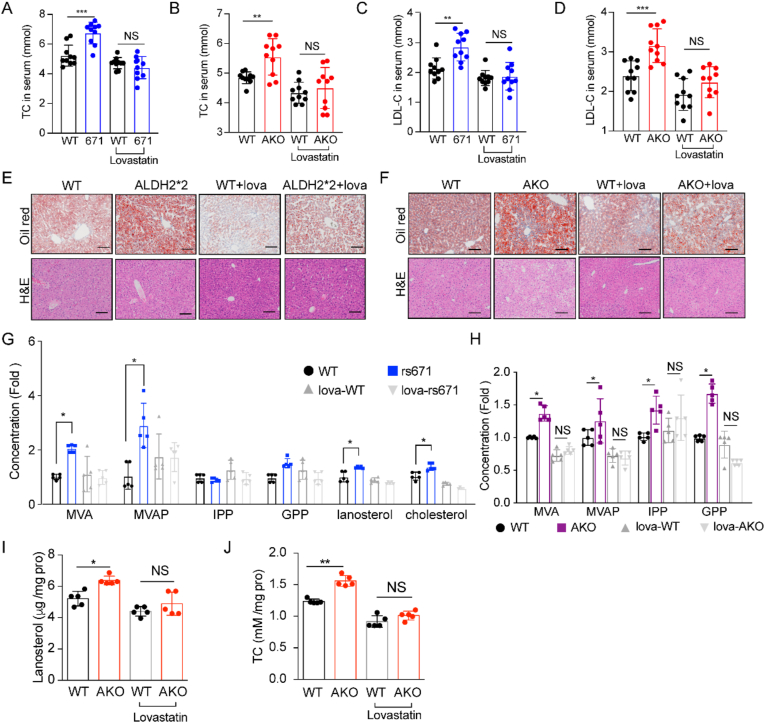
To examine whether ALDH2 regulates cholesterol levels through cholesterol uptake by PCSK9, we found that protein levels of PCSK9 in hepatocytes and serum of ALDH2*2 mouse were increased compared to WT, whereas the LDLR levels were decreased in ALDH2*2 mouse liver (Fig. 7G, supplemental Fig. 7A-B). Intriguingly, the mRNA levels of PCSK9 have no difference but the transcription of LDLR, downstream of PCSK9, surprisingly increased in ALDH2*2 mouse liver (Supplemental Fig. 7C). However, protein levels of LDLR decreased and PCSK9 increased in ALDH2*2 mutant cells (Supplemental Fig. 7D and E) and this phenotype disappeared after inhibiting the function of lysosome through treatment with Bafilomycin A1 (Baf-A1) (Supplemental Fig. 7F and G), suggesting that ALDH2*2 mutant increases PCSK9 through decreasing its lysosomal degradation. However, inhibition of PCSK9 expression by treating WT and ALDH2*2 mutant hepatocytes with RIMPP, an inhibitor of PCSK9 transcription, did not affect the increased cholesterol levels in ALDH2*2 cells (Fig. 7H–J). Furthermore, knocking down PCSK9 in mice by the tail vein injection of AAV showed that knocking down PCSK9 failed to attenuate the increased cholesterol levels in mice serum and liver although the protein levels of PCSK9 were successfully decreased (Fig. 7I–J, supplemental Fig. 7H). Fortunately, statin treatment efficiently decreased the elevated cholesterol levels in ALDH2*2 mice (Fig. 7H, supplemental Fig. 7H).
Taken together, all these data demonstrate that increased cholesterol levels in ALDH2 mutant carrier are primarily due to the upregulation of de novo cholesterol synthesis through stabilizing HMGCR (Supplemental Fig. 6E); although ALDH2 rs671 increases protein levels of PCSK9 in the mouse liver and serum compared to WT, knocking down PCSK9 in ALDH2*2 by AAV fails to lower cholesterol level, implying that PCSK9 antibody or siRNA therapy for lowing cholesterol may fail for ALDH2*2 carriers if validated in humans.
3.8. Lovastatin inhibits the increased cholesterol levels in AKO and AKI mice
To support our hypothesis that ALDH2 decreases cholesterol synthesis through increasing the degradation of HMGCR in the context of CVD, we treated 6-week-old WT, AKO and AKI mice with Lovastatin (30 mg/kg) and western diet (WD) for 7 weeks (Supplemental Fig. 8A-F). WD feeding significantly increased the serum levels of TC, LDL-C in AKO and AKI mice, which were completed diminished after Lovastatin treatment (Fig. 8A–D). Consistently, lipid accumulation in the liver was also decreased after Lovastatin treatment (Fig. 8E–F). Interestingly, Lovastatin treatment efficiently decreased the liver weights in AKO and AKI mice, while the increased weight of WAT in AKO mice remained elevated (supplemental Fig. 8B). Moreover, the metabolite analysis showed that increased levels of cholesterol, lanosterol, and major intermediates in de novo cholesterol synthesis pathways in AKO and AKI mouse liver also disappeared with lovastatin treatment (Fig. 8G–J). Together, these data clearly demonstrate that ALDH2 deficiency or mutation increases cholesterol synthesis, which can be inhibited by targeting HMGCR with statins.
Fig. 8.
Lovastatin inhibits the increased cholesterol levels in ALDH2 KO and rs671 mouse serum and liver. A-D. Total cholesterol and LDL-C in rs671 (A) and AKO (B) mouse serum; LDL-C levels in rs671 (C) and AKO (D) mouse serum. (E–F) Lovastatin treatment diminished lipid accumulation in ALDH2 2*2 and ALDH2 KO mouse liver (n = 10). (G–H) LC-MS analysis of major intermediates in cholesterol synthesis in rs 671 and AKO mouse liver (n = 5). (I–J) GC-MS analysis of lanosterol and total cholesterol in mice liver (n = 5). Statistical comparisons were made using an ANOVA. All data are mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. WT: wild type; AKO: ALDH2 Knockout.
4. Discussion
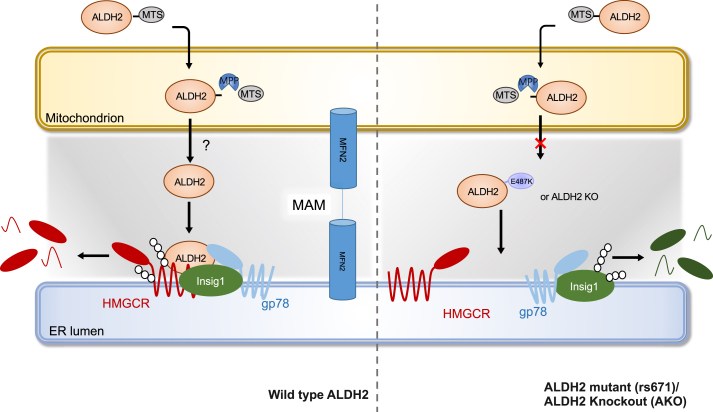
Elevated LDL cholesterol levels in the circulation is one of the most important risk factors for atherosclerotic CVDs. It's been estimated that a 1 mmol reduction in LDL-C in middle-aged individuals for 5 years leads to a 20% reduction in CVD risk [49]. Thus, understanding key factors regulating cholesterol metabolism is instrumental for developing safer and more efficient therapies for the prevention and treatment of CVDs. The mitochondrial ALDH2 is primarily responsible for detoxifying acetaldehyde derived from alcohol in the liver. Around 40% of Asians carry ALDH2 rs671 SNP and also have higher risk of CVD [50,51]. Epidemiological studies have identified an association of cholesterol levels in human subjects with ALDH2 rs671 SNP [20,51]. Moreover, studies showed that alcohol intake is related to cholesterol level in human serum [21,52], suggesting that ALDH2 is possibly involved in the regulation of cholesterol metabolism. However, mechanisms underlying cholesterol metabolism and ALDH2 remain elusive. In this study, we have discovered a mechanism by which mitochondrial ALDH2 regulates cholesterol synthesis through promoting the interaction of HMGCR and Insig1/gp78 in ER membrane (Fig. 9): when cholesterol is accumulated, mitochondrial ALDH2 transfers to ER, presumably through MAM, to promote the interaction of HMGCR with Insig1 and gp78, which leads to the ubiquitination and degradation of HMGCR. On the other hand, HMGCR is stabilized in ALDH2 KO or mutant liver due to the attenuated interaction with Insig1 and HMGCR, which is primarily responsible for the elevated levels of cholesterol in AKO and ALDH2*2 mice. Taken together, our data demonstrate, for the first time, that ALDH2 is a novel regulator for hepatic cholesterol synthesis.
Fig. 9.
The working model of ALDH2 in regulating HMGCR stability and cholesterol synthesis. When cholesterol is accumulated, mitochondrial ALDH2 translocates to ER (presumably through MAM formation) to promote the interaction of Insig1/gp78 with HMGCR, which leads to the ubiquitination and degradation of HMGCR. However, in ALDH2 KO or ALDH2*2 liver, HMGCR is stabilized due to the attenuated interaction of Insig1 and HMGCR, resulting in increased cholesterol de novo synthesis. When cholesterol is depleted, ALDH2 predominantly resides in mitochondrial due to the decreased MAM formation, which decreases the interaction of HMGCR and Insig1, ultimately leading to stabilized HMGCR to initiate cholesterol synthesis.
It is well-documented that liver plays an important role in cholesterol homeostasis by regulating cholesterol synthesis, uptake, esterification, and metabolism [25,53]. As the rate-limiting enzyme and target for cholesterol-lowering therapies, HMGCR is one of most important factors in cholesterol synthesis, which is tightly controlled at the transcriptional and post-translational levels in response to cholesterol levels. Interestingly, a recent study showed that a deubiquitylase ubiquitin-specific peptidase 20 (USP20) stabilizes HMG-CoA reductase (HMGCR) in the feeding state in response to post-prandial increase in insulin and glucose [31]. Our study has significant implications in understanding the roles of mitochondrial protein ALDH2 in regulating HMGCR stability through interaction with Insig1/gp78 in the ER membrane. Mitochondrial ALDH2 has been shown to play protective roles in heart failure after myocardial infarction via suppression of cytosolic JNK/p53 pathway [54]. Furthermore, p53 represses the cholesterol synthesis to mediate its tumor suppression in the liver [45]. It remains to be studied whether p53 is involved in the ALDH2-regulated hepatic cholesterol synthesis.
Our study shows that ALDH2 is such a dynamic protein with diverse functions in the context of CVD, in addition to the detoxification of acetaldehyde and bioactive lipid aldehydes, such as 4-HNE, derived from lipid peroxidation (LPO) [6,55,56]. Previous study identified that LPO is a prominent feature in human plasma with CVD patients [9]. Oxidation products in the LDL particles, such as oxidized cholesterol esters, may increase the cholesterol levels by inhibiting the cholesterol uptake by hepatocytes and macrophages through LXRα-LDLR pathways [10,11]. In addition, a recent study identified that ALDH2 interactions with LDLR and AMPK play an important role in regulating macrophage foam cell formation [18]. Interestingly, ALDH2 also mediates vascular smooth muscle cell phenotype switch, which may be implicated in the lower risk of aortic aneurysm/dissection (AAD) with ALDH2 SNP carriers [19]. Nonetheless, the factors that governs cellular distributions of ALDH2 are poorly understood although limited studies suggest that phosphorylation by AMPK or PKA may play a role [18,57].
Since cholesterol-lowering therapies are important in CVD prevention and treatment, our study has significant clinical implication in our understanding of the responses to various cholesterol-lowering therapies in ALDH2 rs671 populations. Statins and PCSK9 antibody are the main therapies for lowing cholesterol, especially LDL-C, in the treatment and prevention of ASCVD [32]. Previous studies showed that further lowering LDL-C by PCSK9 antibody reduces CVD risk even with initial LDL levels of 1.8 mmol/L [58]. However, therapeutic effects of statins and PCSK9 antibodies can be different with different genetic background. For example, PCSK9 antibody has no effect in humans carrying a loss of function PCSK9 mutant (rs11591147) [59]. With the high cost of PCSK9 antibody, it is important to identify human genetic variations on the responses to PCSK9 antibody. In this study, we found that levels of PCSK9 was increased in ALDH2*2 mutant mice. Treatment with Baf-A1 diminished the increase of PCSK9 in ALDH2*2 mutant cells, suggesting that ALDH2*2 increased PCSK9 through decreasing the lysosomal degradation. However, in ALDH2*2 mice, knocking down PCSK9 through AAV injection failed to decrease cholesterol level, suggesting that patients carrying ALDH2 mutant may have poor response to PCSK9 antibody once validate in future human studies.
We have provided compelling evidence that MAMs are most likely involved in regulation of HMGCR and cholesterol synthesis by mitochondrial ALDH2 (Fig. 6). There are many proteins involved in the formation and biological function of MAMs. For examples, MFN2 is reported to be responsible for the formation of MAMs [36]. 1,4,5-Triphosphate receptor (IP3R) and the voltage-dependent anion-selective channel (VDAC) are responsible for Ca2+ transporting between ER and mitochondria [35]. Interestingly, previous studies showed that contact sites between mitochondrial and ER enriched many proteins related to cholesterol transfer and synthesis [35]. Since cholesterol levels in mitochondria are approximately 4.5-fold lower than those in the ER, emerging studies indicated that cholesterol transfer occurs through MAMs [60]. It remains to be better defined, however, whether MAMs are involved in cholesterol synthesis. In this study, we showed that MAMs play an important role in the regulation of cholesterol synthesis by ALDH2.
In summary, our study defines a novel mechanism by which ALDH2 regulates cholesterol synthesis through interactions with HMGCR and MAMs. Importantly, this study sheds new light on how individuals carrying ALDH2*2 mutant may respond to various cholesterol-lowering therapies including statins and PCSK9 antibodies.
Declaration of competing interest
The authors claim no conflict of interests.
Acknowledgements
The authors would like to acknowledge Drs. Jun Ren and Aijun Sun from Shanghai Institute of Cardiovascular Diseases, Zhongshan Hospital affiliated with Fudan University, Shanghai, China, for providing ALDH2−/− mice. A constructive discussion with Dr. Dawei Zhang at University of Alberta, Canada, Dr. Ming-Hui Zou from Georgia State University, USA, and Dr. Bao-liang Song from Wuhan University, China, is also greatly appreciated. This work was financially supported by the National Natural Science Foundation of China (32030053, 91857112), National Key R&D Program of China administered by Chinese Ministry of Science and Technology (MOST) (2018YFA0800301). We thank molecular biology/biochemistry/cell technology platform, experimental animal platform, mass spectrometry core lab, and biological sample pathology analysis platform in Shanghai Institute of Nutrition and Health, Chinese Academy of Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.101919.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Arnett D.K. ACC/AHA guideline on the primary prevention of cardiovascular disease: executive summary. A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. 2019;74:1376–1414. doi: 10.1016/j.jacc.2019.03.009. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Celermajer D.S., Chow C.K., Marijon E., Anstey N.M., Woo K.S. Cardiovascular disease in the developing world: prevalences, patterns, and the potential of early disease detection. J. Am. Coll. Cardiol. 2012;60:1207–1216. doi: 10.1016/j.jacc.2012.03.074. [DOI] [PubMed] [Google Scholar]
- 3.Vergallo R., Crea F. Atherosclerotic plaque healing. N. Engl. J. Med. 2020;383:846–857. doi: 10.1056/NEJMra2000317. [DOI] [PubMed] [Google Scholar]
- 4.Yin H., Xu L., Porter N.A. Free radical lipid peroxidation: mechanisms and analysis. Chem. Rev. 2011;111:5944–5972. doi: 10.1021/cr200084z. [DOI] [PubMed] [Google Scholar]
- 5.Chen X. Ferroptosis and cardiovascular disease: role of free radical-induced lipid peroxidation. Free Radic. Res. 2021:1–11. doi: 10.1080/10715762.2021.1876856. [DOI] [PubMed] [Google Scholar]
- 6.Zhong H., Lu J., Xia L., Zhu M., Yin H. Formation of electrophilic oxidation products from mitochondrial cardiolipin in vitro and in vivo in the context of apoptosis and atherosclerosis. Redox Biol. 2014;2:878–883. doi: 10.1016/j.redox.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao M., Zhong H., Xia L., Tao Y., Yin H. Pathophysiology of mitochondrial lipid oxidation: role of 4-hydroxynonenal (4-HNE) and other bioactive lipids in mitochondria. Free Radic. Biol. Med. 2017;111:316–327. doi: 10.1016/j.freeradbiomed.2017.04.363. [DOI] [PubMed] [Google Scholar]
- 8.Wei X., Yin H. Covalent modification of DNA by α, β-unsaturated aldehydes derived from lipid peroxidation: recent progress and challenges. Free Radic. Res. 2015;49:905–917. doi: 10.3109/10715762.2015.1040009. [DOI] [PubMed] [Google Scholar]
- 9.Lu J. Comprehensive metabolomics identified lipid peroxidation as a prominent feature in human plasma of patients with coronary heart diseases. Redox Biol. 2017;12:899–907. doi: 10.1016/j.redox.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo S., Li L., Yin H. Cholesterol homeostasis and liver X receptor (LXR) in atherosclerosis. Cardiovasc. Haematol. Disord. - Drug Targets. 2018;18:27–33. doi: 10.2174/1871529X18666180302113713. [DOI] [PubMed] [Google Scholar]
- 11.Guo S. Endogenous cholesterol ester hydroperoxides modulate cholesterol levels and inhibit cholesterol uptake in hepatocytes and macrophages. Redox Biol. 2019;21:101069. doi: 10.1016/j.redox.2018.101069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C.H., Sun L., Mochly-Rosen D. Mitochondrial aldehyde dehydrogenase and cardiac diseases. Cardiovasc. Res. 2010;88:51–57. doi: 10.1093/cvr/cvq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo J.M. ALDH2 protects against stroke by clearing 4-HNE. Cell Res. 2013;23:915–930. doi: 10.1038/cr.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan C. Aldehyde dehydrogenase 2 inhibits inflammatory response and regulates atherosclerotic plaque. Oncotarget. 2016;7:35562–35576. doi: 10.18632/oncotarget.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu X. Exome chip meta-analysis identifies novel loci and East Asian–specific coding variants that contribute to lipid levels and coronary artery disease. Nat. Genet. 2017;49:1722. doi: 10.1038/ng.3978. https://www.nature.com/articles/ng.3978#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato N. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat. Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ebert A.D. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci. Transl. Med. 2014;6:255ra130. doi: 10.1126/scitranslmed.3009027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong S. Acetaldehyde dehydrogenase 2 interactions with LDLR and AMPK regulate foam cell formation. J. Clin. Invest. 2019;129:252–267. doi: 10.1172/JCI122064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang K. Prevention of aortic dissection and aneurysm via an ALDH2-mediated switch in vascular smooth muscle cell phenotype. Eur. Heart J. 2020;41:2442–2453. doi: 10.1093/eurheartj/ehaa352. [DOI] [PubMed] [Google Scholar]
- 20.Lu X. Exome chip meta-analysis identifies novel loci and East Asian-specific coding variants that contribute to lipid levels and coronary artery disease. Nat. Genet. 2017;49:1722–1730. doi: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasakabe T. Modification of the associations of alcohol intake with serum low-density lipoprotein cholesterol and triglycerides by ALDH2 and ADH1B polymorphisms in Japanese men. J. Epidemiol. 2018;28:185–193. doi: 10.2188/jea.JE20160189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joseph P. Reducing the global burden of cardiovascular disease, Part 1: the epidemiology and risk factors. Circ. Res. 2017;121:677–694. doi: 10.1161/CIRCRESAHA.117.308903. [DOI] [PubMed] [Google Scholar]
- 23.Goldberg I.J. Deciphering the role of lipid droplets in cardiovascular disease: a report from the 2017 national heart, lung, and blood Institute workshop. Circulation. 2018;138:305–315. doi: 10.1161/CIRCULATIONAHA.118.033704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paynter N.P., Ridker P.M., Chasman D.I. Are genetic tests for atherosclerosis ready for routine clinical use? Circ. Res. 2016;118:607–619. doi: 10.1161/circresaha.115.306360. [DOI] [PubMed] [Google Scholar]
- 25.Luo, J., Yang, H. & Song, B.-L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21, 225-245, doi:10.1038/s41580-019-0190-7 (2020). [DOI] [PubMed]
- 26.Leong D.P. Reducing the global burden of cardiovascular disease, Part 2: prevention and treatment of cardiovascular disease. Circ. Res. 2017;121:695–710. doi: 10.1161/circresaha.117.311849. [DOI] [PubMed] [Google Scholar]
- 27.Song B.L., Sever N., DeBose-Boyd R.A. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol. Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Gong Y. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metabol. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 29.Lee J.N., Zhang X., Feramisco J.D., Gong Y., Ye J. Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J. Biol. Chem. 2008;283:33772–33783. doi: 10.1074/jbc.M806108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.N., Song B., DeBose-Boyd R.A., Ye J. Sterol-regulated degradation of Insig-1 mediated by the membrane-bound ubiquitin ligase gp78. J. Biol. Chem. 2006;281:39308–39315. doi: 10.1074/jbc.M608999200. [DOI] [PubMed] [Google Scholar]
- 31.Lu X.-Y. Feeding induces cholesterol biosynthesis via the mTORC1–USP20–HMGCR axis. Nature. 2020 doi: 10.1038/s41586-020-2928-y. [DOI] [PubMed] [Google Scholar]
- 32.Michos E.D., McEvoy J.W., Blumenthal R.S. Lipid management for the prevention of atherosclerotic cardiovascular disease. N. Engl. J. Med. 2019;381:1557–1567. doi: 10.1056/NEJMra1806939. [DOI] [PubMed] [Google Scholar]
- 33.Rosenson Robert S., Hegele Robert A., Koenig W. Cholesterol-lowering agents. Circ. Res. 2019;124:364–385. doi: 10.1161/CIRCRESAHA.118.313238. [DOI] [PubMed] [Google Scholar]
- 34.Amarenco P. A comparison of two LDL cholesterol targets after ischemic stroke. N. Engl. J. Med. 2019;382:9–19. doi: 10.1056/NEJMoa1910355. [DOI] [PubMed] [Google Scholar]
- 35.Wu H., Carvalho P., Voeltz G.K. Here, there, and everywhere: the importance of ER membrane contact sites. Science. 2018;361 doi: 10.1126/science.aan5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Brito O.M., Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y. Mitofusin 2-containing mitochondrial-reticular microdomains direct rapid cardiomyocyte bioenergetic responses via interorganelle Ca(2+) crosstalk. Circ. Res. 2012;111:863–875. doi: 10.1161/circresaha.112.266585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vance J.E. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 39.Rusiñol A.E., Cui Z., Chen M.H., Vance J.E. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 1994;269:27494–27502. [PubMed] [Google Scholar]
- 40.Prasad M. Mitochondria-associated endoplasmic reticulum membrane (MAM) regulates steroidogenic activity via steroidogenic acute regulatory protein (StAR)-voltage-dependent anion channel 2 (VDAC2) interaction. J. Biol. Chem. 2015;290:2604–2616. doi: 10.1074/jbc.M114.605808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galmes R. ORP5/ORP8 localize to endoplasmic reticulum-mitochondria contacts and are involved in mitochondrial function. EMBO Rep. 2016;17:800–810. doi: 10.15252/embr.201541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L. p38 MAP kinase-dependent phosphorylation of the Gp78 E3 ubiquitin ligase controls ER-mitochondria association and mitochondria motility. Mol. Biol. Cell. 2015;26:3828–3840. doi: 10.1091/mbc.E15-02-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin S. ALDH2(E487K) mutation increases protein turnover and promotes murine hepatocarcinogenesis. Proc. Natl. Acad. Sci. U. S. A. 2015;112:9088–9093. doi: 10.1073/pnas.1510757112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei L. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat. Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 45.Moon S.H. p53 represses the mevalonate pathway to mediate tumor suppression. Cell. 2019;176 doi: 10.1016/j.cell.2018.11.011. 564-580.e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Henneman L., van Cruchten A.G., Kulik W., Waterham H.R. Inhibition of the isoprenoid biosynthesis pathway; detection of intermediates by UPLC-MS/MS. Biochim. Biophys. Acta. 2011;1811:227–233. doi: 10.1016/j.bbalip.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Zhang F. Hepatic CREBZF couples insulin to lipogenesis by inhibiting insig activity and contributes to hepatic steatosis in diet-induced insulin-resistant mice. Hepatology. 2018;68:1361–1375. doi: 10.1002/hep.29926. [DOI] [PubMed] [Google Scholar]
- 48.Franco-Iborra S. Defective mitochondrial protein import contributes to complex I-induced mitochondrial dysfunction and neurodegeneration in Parkinson's disease. Cell Death Dis. 2018;9:1122. doi: 10.1038/s41419-018-1154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins R. Interpretation of the evidence for the efficacy and safety of statin therapy. Lancet. 2016;388:2532–2561. doi: 10.1016/s0140-6736(16)31357-5. [DOI] [PubMed] [Google Scholar]
- 50.Sung Y.F. Homozygous ALDH2*2 is an independent risk factor for ischemic stroke in Taiwanese men. Stroke. 2016;47:2174–2179. doi: 10.1161/strokeaha.116.013204. [DOI] [PubMed] [Google Scholar]
- 51.Hao P.P. Association between aldehyde dehydrogenase 2 genetic polymorphism and serum lipids or lipoproteins: a meta-analysis of seven East Asian populations. Atherosclerosis. 2010;212:213–216. doi: 10.1016/j.atherosclerosis.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Mizuno Y. East asian variant of aldehyde dehydrogenase 2 is associated with coronary spastic angina: possible roles of reactive aldehydes and implications of alcohol flushing syndrome. Circulation. 2015;131:1665–1673. doi: 10.1161/circulationaha.114.013120. [DOI] [PubMed] [Google Scholar]
- 53.Chen L. Regulation of glucose and lipid metabolism in health and disease. Sci. China Life Sci. 2019;62:1420–1458. doi: 10.1007/s11427-019-1563-3. [DOI] [PubMed] [Google Scholar]
- 54.Sun A. Mitochondrial aldehyde dehydrogenase 2 plays protective roles in heart failure after myocardial infarction via suppression of the cytosolic JNK/p53 pathway in mice. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000779. e000779-e000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong H. Mitochondrial control of apoptosis through modulation of cardiolipin oxidation in hepatocellular carcinoma: a novel link between oxidative stress and cancer. Free Radic. Biol. Med. 2017;102:67–76. doi: 10.1016/j.freeradbiomed.2016.10.494. [DOI] [PubMed] [Google Scholar]
- 56.Zhong H., Yin H. Role of lipid peroxidation derived 4-hydroxynonenal (4-HNE) in cancer: focusing on mitochondria. Redox Biol. 2015;4:193–199. doi: 10.1016/j.redox.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen C.-H., Sun L., Mochly-Rosen D. Mitochondrial aldehyde dehydrogenase and cardiac diseases. Cardiovasc. Res. 2010;88:51–57. doi: 10.1093/cvr/cvq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabatine M.S. Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 59.Feng Q. The effect of genetic variation in PCSK9 on the LDL-cholesterol response to statin therapy. Pharmacogenomics J. 2017;17:204–208. doi: 10.1038/tpj.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin L.A., Kennedy B.E., Karten B. Mitochondrial cholesterol: mechanisms of import and effects on mitochondrial function. J. Bioenerg. Biomembr. 2016;48:137–151. doi: 10.1007/s10863-014-9592-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.