Abstract
The mTOR complex 1 (mTORC1) kinase regulates cell growth by setting the balance between anabolic and catabolic processes. To be active, mTORC1 requires the environmental presence of amino acids and glucose. While a mechanistic understanding of amino acid sensing by mTORC1 is emerging, how glucose activates mTORC1 remains mysterious. Here, we used metabolically engineered human cells lacking the canonical energy sensor AMPK to identify glucose-derived metabolites required to activate mTORC1 independent of energetic stress. We show that mTORC1 senses a metabolite downstream of the aldolase and upstream of the glyceraldehyde 3-phosphate dehydrogenase steps of glycolysis and pinpoint dihydroxyacetone phosphate (DHAP) as the key molecule. In cells expressing a triose kinase, the synthesis of DHAP from dihydroxyacetone is sufficient to activate mTORC1 even in the absence of glucose. DHAP is a precursor for lipid synthesis, a process under the control of mTORC1, which provides a potential rationale for the sensing of DHAP by mTORC1.
How organisms sense and adapt to the availability of nutrients in the environment is incompletely understood. One key pathway for doing so is the signaling system anchored by the mTOR Complex 1 (mTORC1) kinase, which regulates cell growth and metabolism in response to nutrients, such as amino acids and glucose1. Aberrant mTORC1 signaling is implicated in many diseases, including several associated with nutrient overload, such as diabetes and non-alcoholic steatohepatitis (NASH)2–5. In contrast to the mechanistic understanding beginning to emerge for how mTORC1 senses amino acids, much less is known about how it detects glucose.
Glucose deprivation can inhibit mTORC1 through a pathway that requires the energy-sensing kinase AMPK6–8, which is activated by a rise in the AMP to ATP ratio. However, there is also significant evidence that mTORC1 can sense glucose through an AMPK-independent route9–12. While potential models for this route have been proposed, several involving glycolytic intermediates and their cognate enzymes, many are mutually incompatible, and no consensus has emerged on either the mechanisms involved or the molecule, if not glucose, that is sensed13–19. Here, we metabolically engineered cells to probe the role of glucose-derived metabolites in the regulation of mTORC1. We find that a metabolite previously not connected to mTORC1 plays a key role in its activation by glucose.
Results
Various sugars can activate mTORC1 independently of AMPK
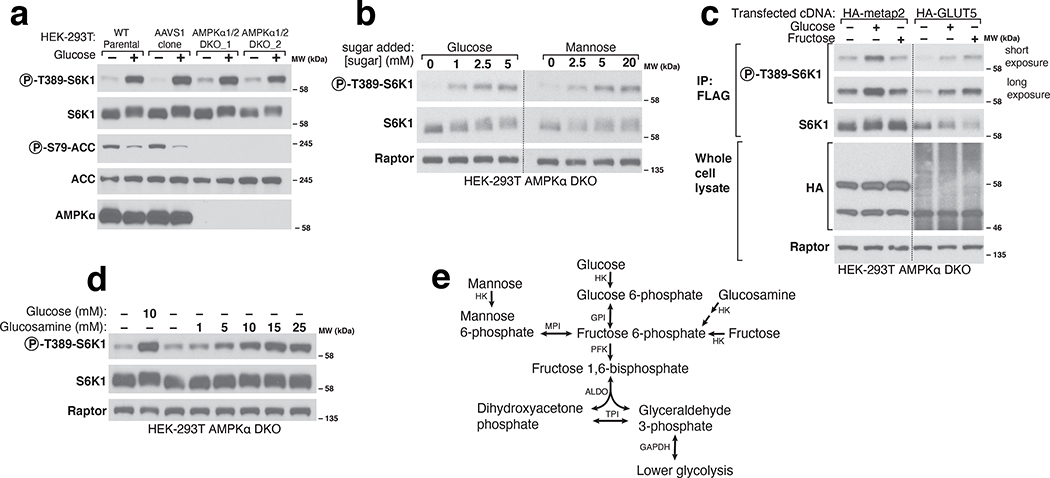
To examine in our cell system the role of AMPK in glucose-sensing by mTORC1, we generated AMPKα1 and AMPKα2 double knockout (DKO) HEK-293T cells. As expected, in these cells glucose starvation did not increase the phosphorylation of the canonical AMPK substrate Acetyl-CoA Carboxylase (ACC)20. Despite the absence of AMPK activity, glucose starvation still inhibited the phosphorylation of the mTORC1 substrate S6K1, albeit to a lesser degree than in wild-type cells (Fig. 1a). These results support the existing notion that mTORC1 can sense glucose through at least two pathways, one AMPK-dependent and the other not. In order to focus on the AMPK-independent mechanism, we undertook all further experiments in the AMPK DKO cells.
Figure 1. Various sugars can activate mTORC1 independently of AMPK.
a) AMPK is not necessary for glucose to regulate mTORC1. Wildtype parental HEK-293T cells, a control cell line generated by expressing Cas9 and an sgRNA targeting the AAVS1 locus (AAVS1 clone), or two AMPKα double knockout (DKO) cell lines generated with sgRNAs targeting both genes encoding the AMPKα subunit of AMPK (PRKAA1 and PRKAA2), were starved of glucose for 1 hour or starved and then re-stimulated with 10mM glucose for 10 minutes. Whole cell lysates were prepared and analyzed by immunoblotting using the indicated antibodies. b) In the absence of glucose, mannose activates mTORC1 in AMPKα DKO HEK-293T cells. Cells were incubated in media containing the indicated concentrations of glucose or mannose for 1 hour prior to the preparation and analysis of cell lysates. c) Fructose, in the absence of glucose, activates mTORC1 in cells expressing the fructose transporter GLUT5. Cells were incubated for an hour in either glucose or fructose for 1 hour. FLAG-immunoprecipitates were prepared from cells co-expressing FLAG-S6K1 and either a control protein (HA-metap2) or HA-GLUT5. Immunoprecipitates and cell lysates were analyzed by immunoblotting for the phosphorylation state or levels of the indicated proteins. d) Glucosamine, in the absence of glucose, activates mTORC1 in AMPKα DKO HEK-293T cells. Cells were incubated for 1 hour in the indicated concentrations of glucose or glucosamine. Whole cell lysates were analyzed by immunoblotting. E) Schematic demonstrating how hexoses feed into glycolysis according to the KEGG pathway (HK: hexokinase; GPI: glucose 6-phosphate isomerase; PFK: phosphofructokinase; ALDO: Aldolase; TPI: triosephosphate isomerase; GAPDH: glyceraldehyde 3-phosphate dehydrogenase; MPI: mannose 6-phosphate isomerase). Dashed lines separate lanes that were run on different gels (b) or where irrelevant intervening lanes were spliced out (c).
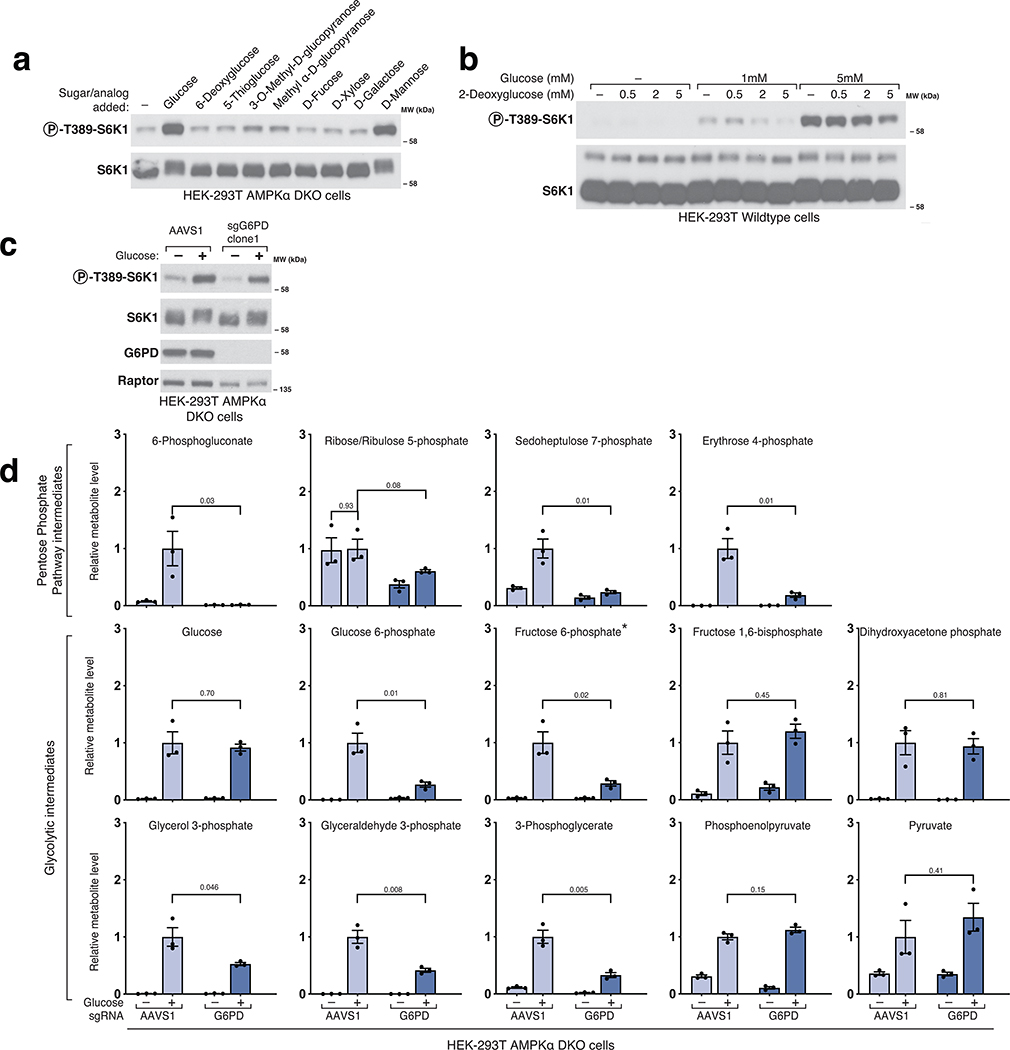
To probe the features of glucose necessary for it to activate mTORC1, we asked if glucose analogs as well as related sugars can substitute for glucose. While non-metabolizable (5-thio-glucose, 6-deoxyglucose, 2-deoxyglucose) and methylated (methyl α-D-glucopyranoside, 3-O-methyl-D-glucopyranose) glucose analogs could not (Extended Fig. 1a–b), the metabolizable sugars21 mannose, glucosamine, and fructose readily re-activated mTORC1 in glucose-starved cells (Fig. 1b–e). These results suggest that mTORC1 responds to a glucose-derived metabolite rather than to glucose itself, and that the pathway that generates the sensed molecule can be fed by fructose, mannose, or glucosamine. Given this, and that we found that the pentose phosphate pathway (PPP) is not required for glucose to activate mTORC1 (Extended Fig. 1c–d), we focused on the metabolism of glucose via glycolysis.
mTORC1 senses a metabolite downstream of aldolase and upstream of GAPDH
To pinpoint metabolites important for mTORC1 activation, we developed a genetic approach for inhibiting glycolytic enzymes because, other than for GAPDH22,23, there are few specific small molecule inhibitors targeting them. There are two key challenges to doing so: (1) glucose metabolism is essential in proliferating cells, and (2) many steps in glycolysis are catalyzed by enzymes with redundant paralogues. To overcome these issues, we built conditional knockout cell lines (‘dox-off’ cells) that lack all paralogues for a particular enzyme and express a complementing cDNA that is repressible by doxycycline.
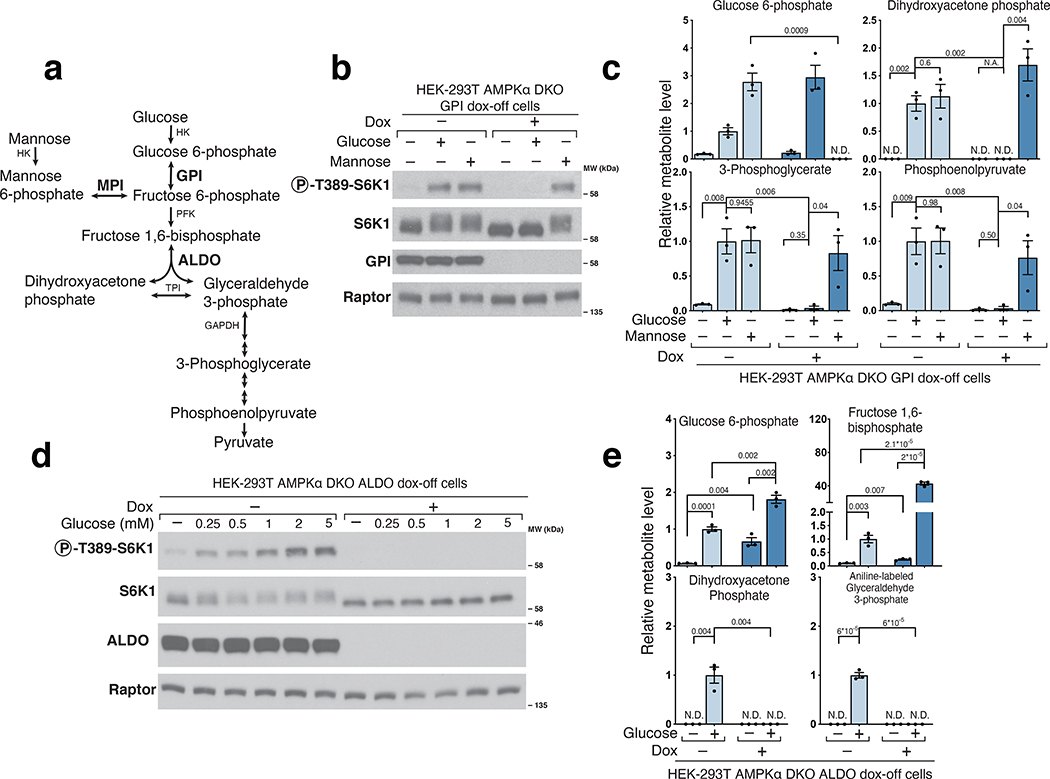
Hexokinase (HK) phosphorylates glucose and mannose to generate glucose 6-phosphate (G6P) and mannose 6-phosphate (M6P), respectively, which specific isomerases (Glucose 6-phosphate isomerase (GPI) and mannose 6-phosphate isomerase (MPI)) convert through a reversible reaction into fructose 6-phosphate (F6P) (Fig.2a)21. In GPI dox-off cells, doxycycline treatment eliminated GPI expression and prevented glucose, but not mannose, from activating mTORC1 (Fig.2b). Metabolite profiling confirmed that glucose was not metabolized further than G6P in cells lacking GPI. In contrast, the addition of mannose was able to restore the normal levels of glycolytic metabolites except for G6P (Fig. 2c). Collectively, these data indicate that the glucose-derived metabolite that leads to mTORC1 activation must be made downstream of GPI, and rule out glucose itself and G6P as candidate signaling molecules. Our results differ from previous work in cardiomyocytes proposing that G6P signals glucose levels to mTORC114,17–19, suggesting that the mechanisms underlying glucose sensing may vary between cell types.
Figure 2. Glucose 6-phosphate isomerase and Aldolase are upstream of the metabolite that signals glucose availability to mTORC1.
a) Diagram depicting glucose and mannose metabolism emphasizing the roles of Glucose 6-phosphate isomerase (GPI), Mannose 6-phosphate isomerase (MPI), and Aldolase (ALDO) in glycolysis. b) GPI is required for glucose, but not mannose, to activate mTORC1. AMPKα DKO GPI dox-off cells were treated with doxycycline (dox) for 10 days and placed for 2 hours in media with either no sugar, glucose, or mannose. Cell lysates were analyzed by immunoblotting for the phosphorylation state or levels of the indicated proteins. c) GPI is required for the metabolism of glucose beyond glucose 6-phosphate but not for that of mannose. Cells were treated as in (b) and metabolite extracts analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. N.D., peak not detected; N.A. statistical test not applicable. d) ALDO is required for the activation of mTORC1 by glucose. HEK-293T AMPKα DKO ALDO dox-off cells were treated with doxycycline for 5 days. Cells were then incubated in media containing the indicated concentrations of glucose for 3 hours. Cell lysates were analyzed by immunoblotting for the phosphorylation state and levels of the indicated proteins. e) Loss of ALDO expression leads to supraphysiological levels of Fructose 1,6-bisphosphate and prevents glucose from generating metabolites downstream of aldolase, including dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP). HEK-293T AMPKα DKO ALDO dox-off cells were incubated for 3 hours without glucose (-Gluc) or with 2 mM glucose (+Gluc). Metabolite extracts were analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. N.D., peak not detected.
After phosphofructokinase (PFK) converts the F6P made by GPI into fructose 1,6-bisphosphate (FBP), aldolase (ALDO), of which there are three paralogues (ALDOA, ALDOB, and ALDOC), cleaves it into two triose phosphates, dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (GAP) (Fig. 2a)21. ALDO dox-off cells were generated by targeting all three ALDOA/B/C paralogues using CRISPR/Cas9 and rescuing their loss with a doxycycline-repressible cDNA encoding ALDOA. In ALDO dox-off cells treated with doxycycline, glucose failed to activate mTORC1 (Fig. 2d) and did not generate GAP or DHAP while causing an accumulation of G6P and FBP above the amounts observed in normal cells (Fig.2e). These results suggest that a metabolite downstream of aldolase activates mTORC1 and eliminate F6P and FBP as candidate molecules.
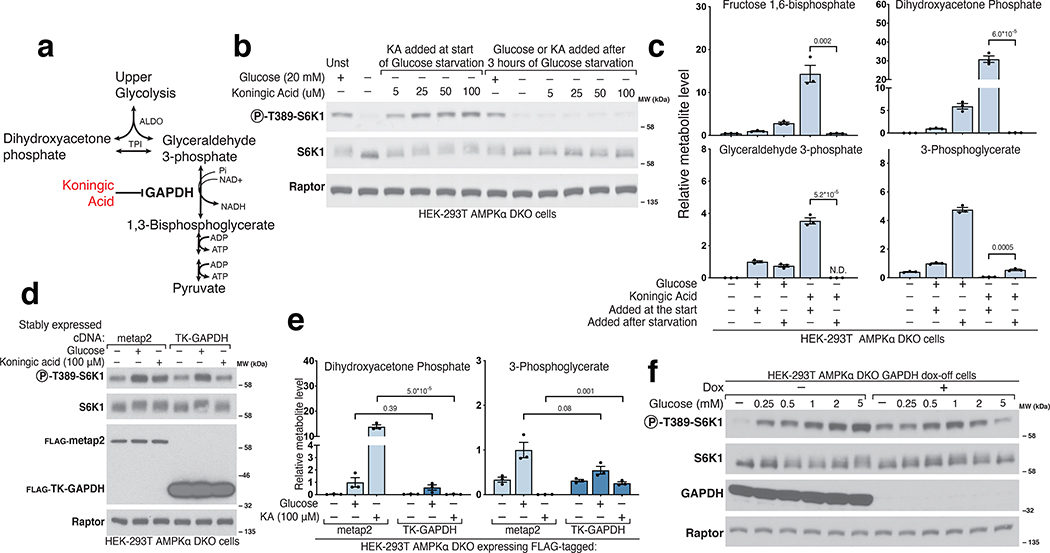
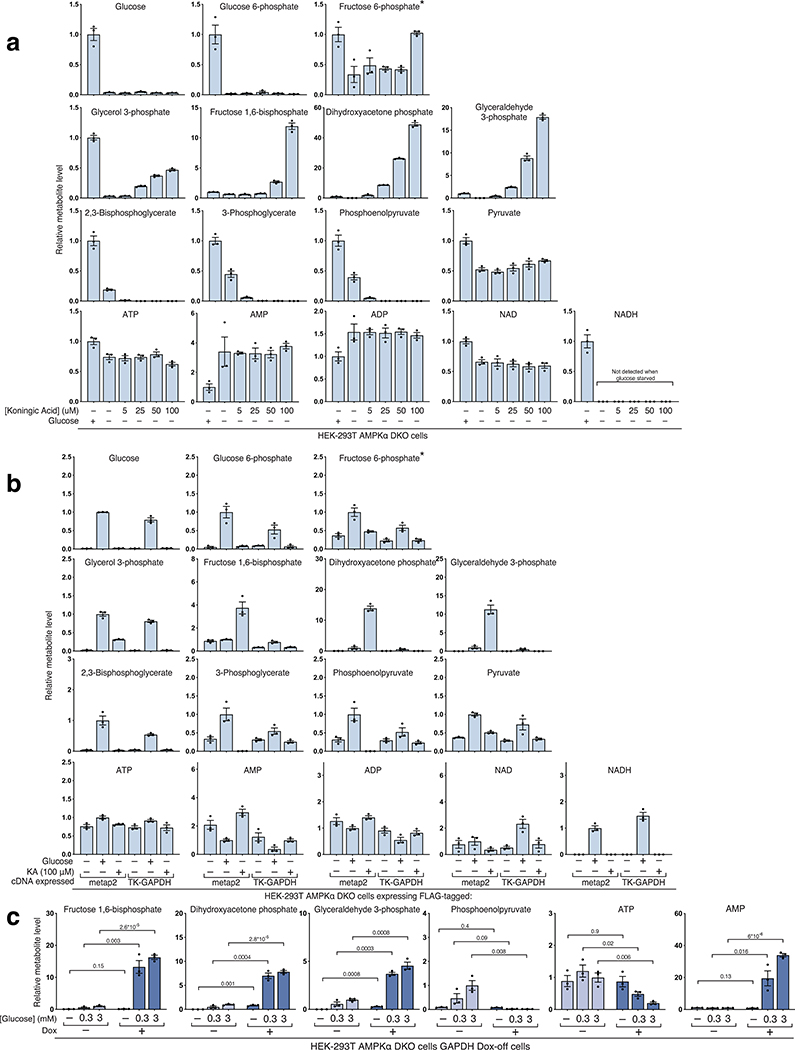
The next enzyme in glycolysis, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), couples the formation of NADH from NAD+ to the oxidation of GAP into 1,3-bisphosphoglycerate (1,3-BPG) (Fig.3a)21. To probe the role of GAPDH in mTORC1 activation, we placed cells in glucose-free media and simultaneously added Koningic acid (KA), a specific GAPDH inhibitor22,23. Interestingly, in the presence of KA, mTORC1 remained active even after cells were deprived of glucose for three hours (Fig.3b). Metabolite profiling showed that KA creates a dam so that even upon glucose-deprivation cells maintain high levels of metabolites upstream of GAPDH, such as GAP and DHAP, while depleting downstream metabolites (Fig.3c, Extended Fig. 2a). KA treatment of glucose-starved cells did not appear to effect ATP levels nor did it reverse the effects of glucose starvation on NAD+ or NADH (Extended Fig. 2a). Importantly, expression of a KA-resistant GAPDH from T. koningii (TK-GAPDH)22,23 reversed the increase in GAP and DHAP caused by KA and restored the mTORC1 inhibition normally caused by glucose starvation (Fig.3d–e, Extended Fig. 2b). KA prevented mTORC1 inhibition only if it was added at the beginning of the glucose withdrawal period, consistent with its impact on GAP and DHAP levels (Fig.3b–c). Doxycycline treatment of GAPDH dox-off cells had similar effects on metabolite levels and mTORC1 activity as KA, indicating that the GAPDH protein itself is not required for its inhibition to protect mTORC1 from glucose starvation (Fig.3f, Extended Fig. 2c).
Figure 3. GAPDH is downstream of the metabolite that signals glucose availability to mTORC1.
a) Diagram of glycolysis with an emphasis on the role of GAPDH and its inhibitor Koningic Acid (KA). b) Inhibition of GAPDH by KA prevents the suppression of mTORC1 normally caused by glucose starvation, but only if KA is added at the beginning of the starvation period. AMPKα DKO HEK-293T cells were incubated with glucose (Unst) or starved of it for 3 hours. Koningic acid was added to cells either at the beginning of the 3-hour glucose starvation period or for 15 minutes after a 3-hour starvation. Cell lysates were analyzed by immunoblotting for the phosphorylation state or levels of the indicated proteins. c) GAPDH inhibition by 50 μM KA prevents depletion of metabolites upstream of GAPDH upon glucose starvation but only if added at the beginning of the starvation period. Cells were treated as in (B), metabolite extracts were analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. N.D., peak not detected. d) Overexpression of the KA-resistant version of GAPDH from the fungus T. koningii (TK-GAPDH) eliminated the effects of KA on mTORC1 signaling. Cells stably expressing FLAG-metap2 or FLAG-TK-GAPDH were incubated under the indicated conditions. KA was added at the beginning of the starvation. Cell lysates were analyzed by immunoblotting for the phosphorylation state or levels of the indicated proteins. e) Overexpression of TK-GAPDH prevents the accumulation of metabolites upstream of GAPDH normally caused by KA treatment in glucose-starved cells. Cells were treated as in (d). Metabolite extracts were analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. f) Loss of GAPDH expression has the same phenotype as inhibition of GAPDH by KA. GAPDH dox-off cells treated with doxycycline maintain mTORC1 activity even after a 3-hour starvation of glucose.
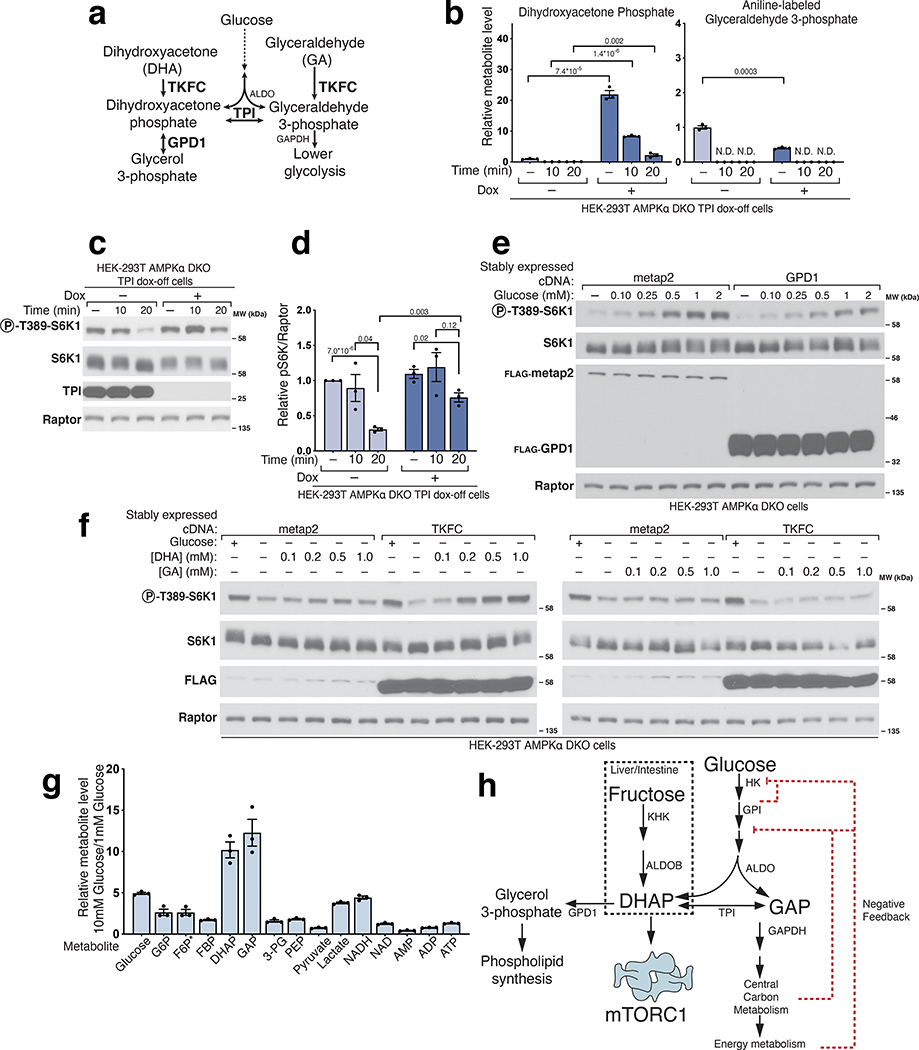
DHAP synthesis is sufficient to activate mTORC1 in glucose starved cells
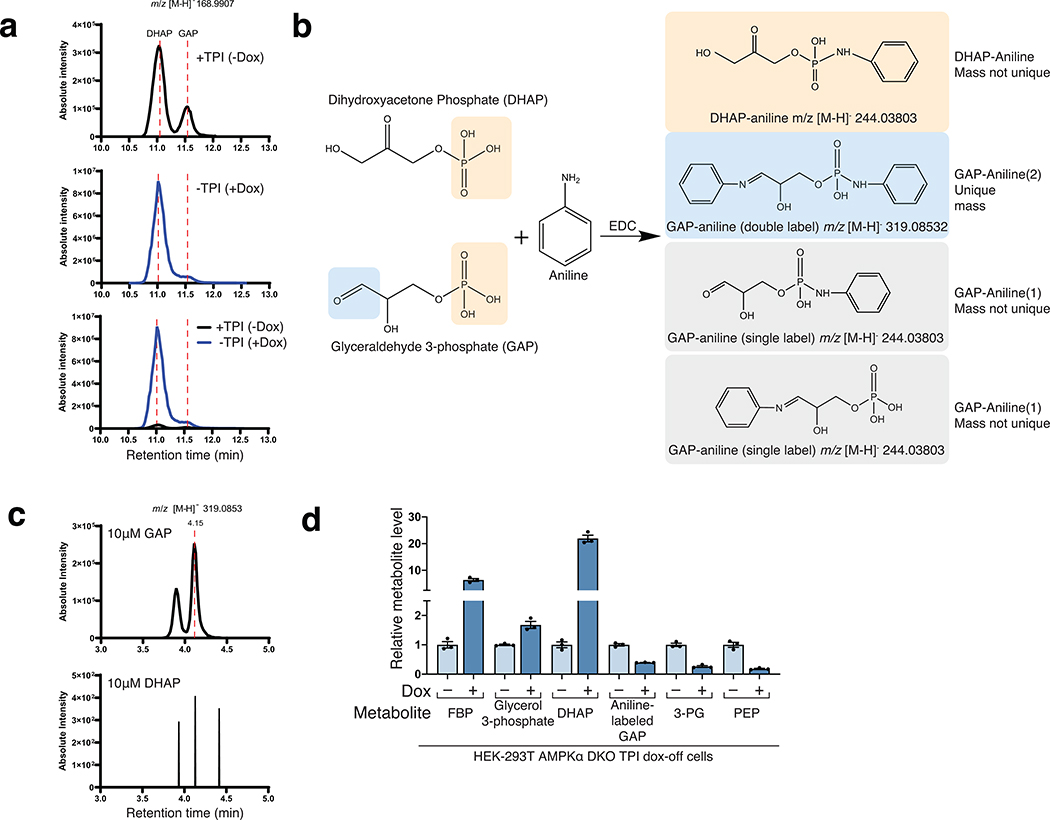
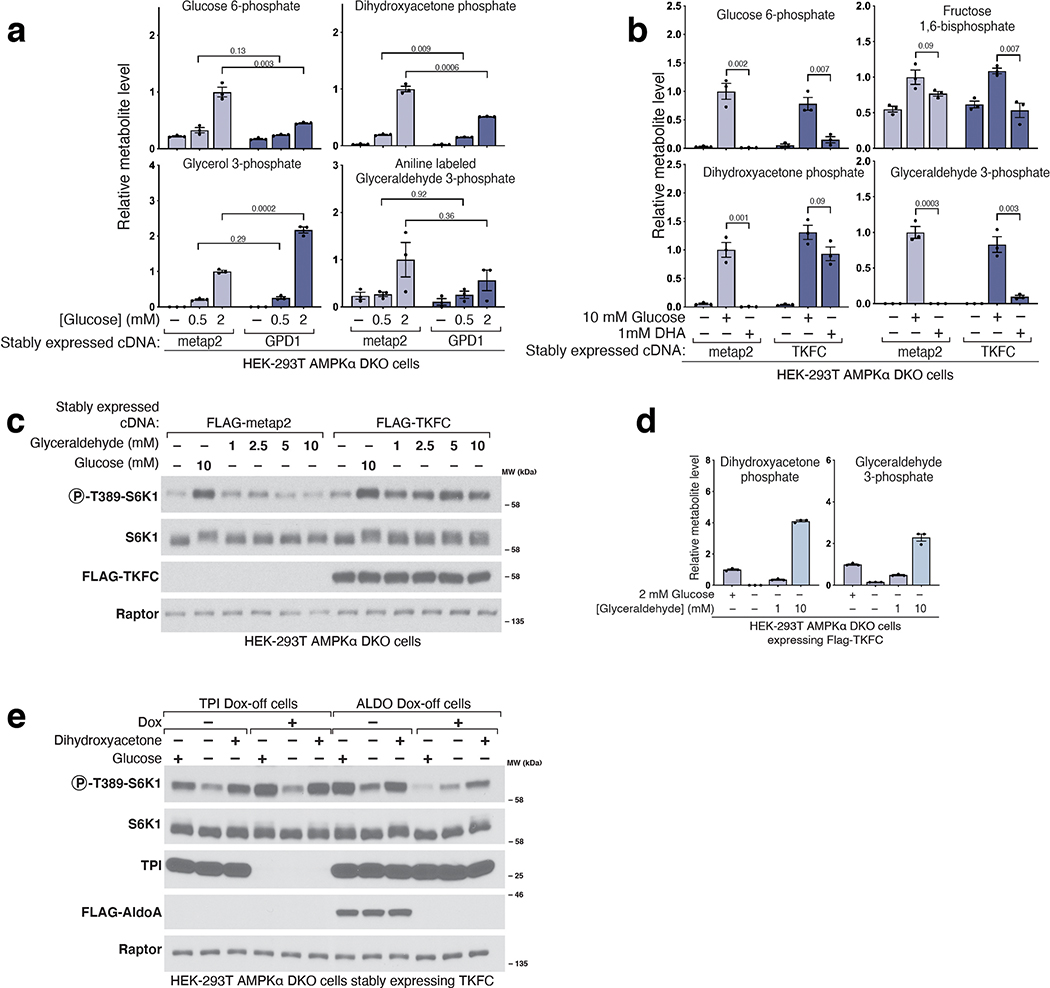
Collectively, these data indicate that a glycolytic intermediate downstream of ALDO but upstream of GAPDH is required for glucose to activate mTORC1. Candidate molecules include GAP, DHAP, or derivatives of either. While GAP and DHAP can spontaneously interconvert, the rate of DHAP conversion to GAP is negligible in the absence of triosephosphate isomerase (TPI)24 (Fig. 3a), so to differentiate between the two, we generated a TPI dox-off cell line and measured DHAP and GAP levels. In the cells lacking TPI, GAP was measured as its aniline adduct for the technical reasons explained in Extended Fig. 3. In glucose-starved cells, loss of TPI greatly slowed the catabolism of DHAP without seemingly impacting that of GAP, consistent with the direction of normal net flux being from DHAP to GAP (Fig. 4b). Given this, we reasoned that a kinetic analysis afforded a simple way to discern which molecule is most relevant to mTORC1 activation: if glucose starvation inhibited mTORC1 more slowly in cells lacking TPI, it would favor DHAP, while, if the timing of mTORC1 inhibition was unaffected, it would favor a model in which GAP has a key role. Quite revealingly, TPI loss markedly slowed the inhibition of mTORC1 caused by glucose starvation (Fig. 4c–d). Thus, TPI is downstream of the metabolite that activates mTORC1, which favors DHAP or a molecule derived from it (except GAP), as the preferred candidate.
Figure 4. Triose phosphate isomerase (TPI) is downstream of the metabolite that signals glucose availability to mTORC1 and DHAP synthesis is sufficient to activate mTORC1.
a) Model depicting the metabolism of trioses and triose-phosphates. b-d) TPI loss causes supraphysiological levels of dihydroxyacetone phosphate (DHAP) and slower catabolism and slower inhibition of mTORC1 following glucose starvation. The levels of glyceraldehyde 3-phosphate (GAP) are decreased and consumed at a normal rate. AMPKα DKO TPI dox-off HEK-293T cells treated with doxycycline for ten days were starved of glucose for the indicated periods of time. Metabolite extracts were analyzed by LC/MS (b) and protein lysates by immunoblot (c). Data are shown in (b) as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-tests. N.D., peak not detected. Immunoblots for the phosphorylation state and levels of the indicated proteins (c) were quantified in (d). Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. e) Overexpression of GPD1 decreases the activation of mTORC1 by glucose. AMPKα DKO HEK-293T cells expressing the indicated cDNAs were starved of glucose for 3 hours or starved of glucose and re-stimulated with the indicated concentration of glucose for 10 minutes. f) DHAP synthesis is sufficient to activate mTORC1 in the absence of glucose. AMPKα DKO HEK-293T cells stably expressing the control protein FLAG-metap2 or FLAG-TKFC were starved of glucose for 3 hours or starved and stimulated for 10 minutes with glucose or the indicated concentrations of dihydroxyacetone (DHA) or glyceraldehyde (GA). g) The fold-change in DHAP and GAP levels between cells in low (1 mM) and high (10 mM) glucose is greater than that for any other glycolytic metabolite. AMPKα DKO HEK-293T cells were starved of glucose for 3 hours and then restimulated for 15 minutes with either 1 mM glucose or 10 mM glucose. Metabolite extracts were analyzed by LC/MS. Data are shown as mean ± s.e.m. for n = 3 biologically independent replicates. P-values were determined for two-sided Student’s t-test. * The peak used to quantify fructose 6-phosphate (F6P) levels may also contain glucose 1-phosphate. h) Diagram depicting the metabolism of glucose in most cells and of fructose in the liver and intestine.
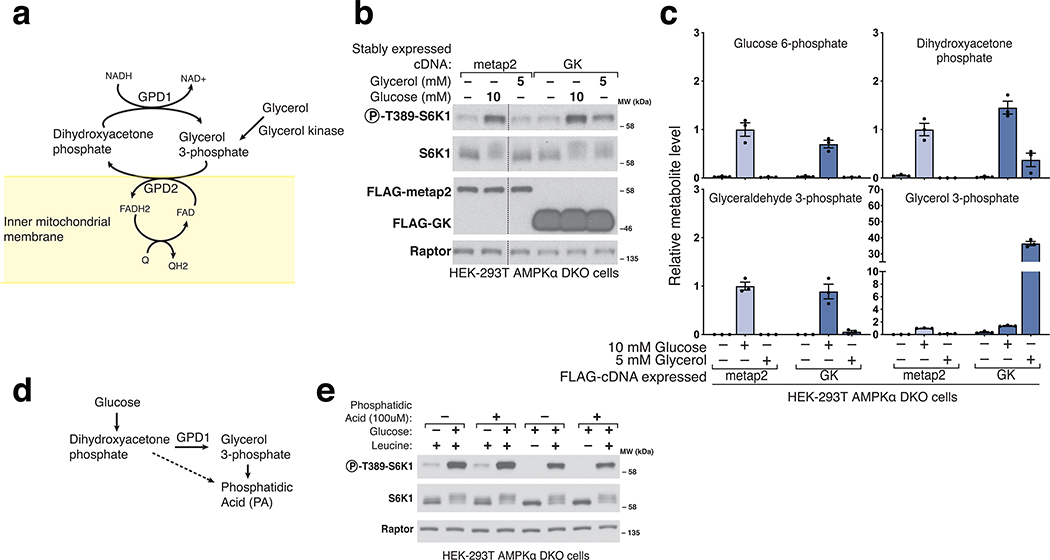
One such derivative is glycerol 3-phosphate (G3P), a precursor in the synthesis of lipids, which glycerol 3-phosphate dehydrogenase (GPD1/GPD1L) generates by the reduction of DHAP (Fig. 4a, Extended Fig. 4a)21. An alternative pathway for the generation of G3P is the phosphorylation of glycerol by glycerol kinase (GK) (Extended Fig. 4a), an enzyme not expressed in HEK-293T cells. We generated cells that stably express human GK and after glucose starvation added glycerol to them. Glycerol caused a partial reactivation of mTORC1 (Extended Fig. 4b) while raising G3P to supraphysiological levels and partially restoring those of DHAP (Extended Fig. 4c). These findings are not consistent with a model in which G3P mediates the activation of mTORC1 by glucose. Because G3P and DHAP are precursors in the biosynthesis of phosphatidic acid (PA) (Extended Fig. 4d), a lipid that has been shown to activate mTORC125,26, we tested if PA could re-activate mTORC1 in cells starved of glucose or leucine. However, this was not the case (Extended Fig. 4e), indicating that glucose sensing by mTORC1, like that of leucine, is not mediated by PA.
To characterize the role of GPD1 activity in the activation of mTORC1 by glucose, we generated cells that overexpress FLAG-GPD1. We found that overexpression of GPD1 suppressed the activation of mTORC1 by glucose (Fig.4e). Metabolite profiling of these cells showed decreased DHAP and boosted G3P levels (Extended Fig. 5a). As above, these results point to DHAP, but not G3P, as the glucose-derived metabolite necessary for transmitting the presence of glucose to mTORC1. We also conclude that the metabolite that drives mTORC1 activation in response to glucose is upstream of GPD1, again arguing against a key role for PA, which depends on GPD1 for its synthesis26.
To test the sufficiency of DHAP in the activation of mTORC1 by glucose, we needed a way to restore DHAP levels in glucose-starved cells. However, DHAP does not readily cross membranes and there is no known plasma membrane transporter for DHAP in eukaryotes. Thus, we took advantage of human triokinase (TKFC), which can phosphorylate the membrane-permeable trioses dihydroxyacetone (DHA) and glyceraldehyde (GA) to make DHAP and GAP, respectively (Fig. 4a)21. Expression of TKFC on its own had no effect on the regulation of mTORC1 signaling by glucose. However, in cells starved of glucose for 3 hours and expressing TKFC, but not a control protein, the addition of DHA for just ten-minutes was sufficient to reactivate mTORC1 (Fig. 4f). Metabolite profiling confirmed that DHA addition leads to DHAP synthesis only in the TKFC-expressing cells and only partially restored GAP levels, consistent with our previous observation suggesting DHAP is more important than GAP in the activation of mTORC1 (Extended Fig. 5b). Glyceraldehyde, when added at the same concentration as DHA in TKFC expressing cells, did not activate mTORC1, while at much higher doses (5–10 mM), it did (Extended Fig. 5c). However, this effect correlated with the production of DHAP, likely from the fact that commercially available glyceraldehyde is contaminated with DHA (Extended Fig. 5d). Lastly, DHA also restored mTORC1 activity in TPI- and ALDO-deficient cells expressing TKFC (Extended Fig. 5e). Therefore, we conclude that DHAP synthesis is sufficient to activate mTORC1.
We noticed that glucose starvation caused DHAP levels to drop below the level of detection in our experiments, an observation that was not true for most other glycolytic intermediates (Extended Fig. 2b). To accurately measure the fold-change in DHAP upon changes in glucose availability, we compared the levels of glycolytic metabolites in cells in media containing 1 or 10 mM glucose (Fig. 4g). The level of DHAP, along with that of GAP, changed about 10-fold between these two media glucose concentrations, the most of all other metabolites in glycolysis (Fig. 4g). Under the same conditions, we observed more subtle changes in the levels of adenine nucleotides (AMP, ADP, and ATP), which reflects the involvement of other nutrients besides glucose, such as glutamine and fatty acids, in energy metabolism (Fig. 4g).
Perturbations in the GATOR-Rag signaling axis abrogate glucose sensing by mTORC1
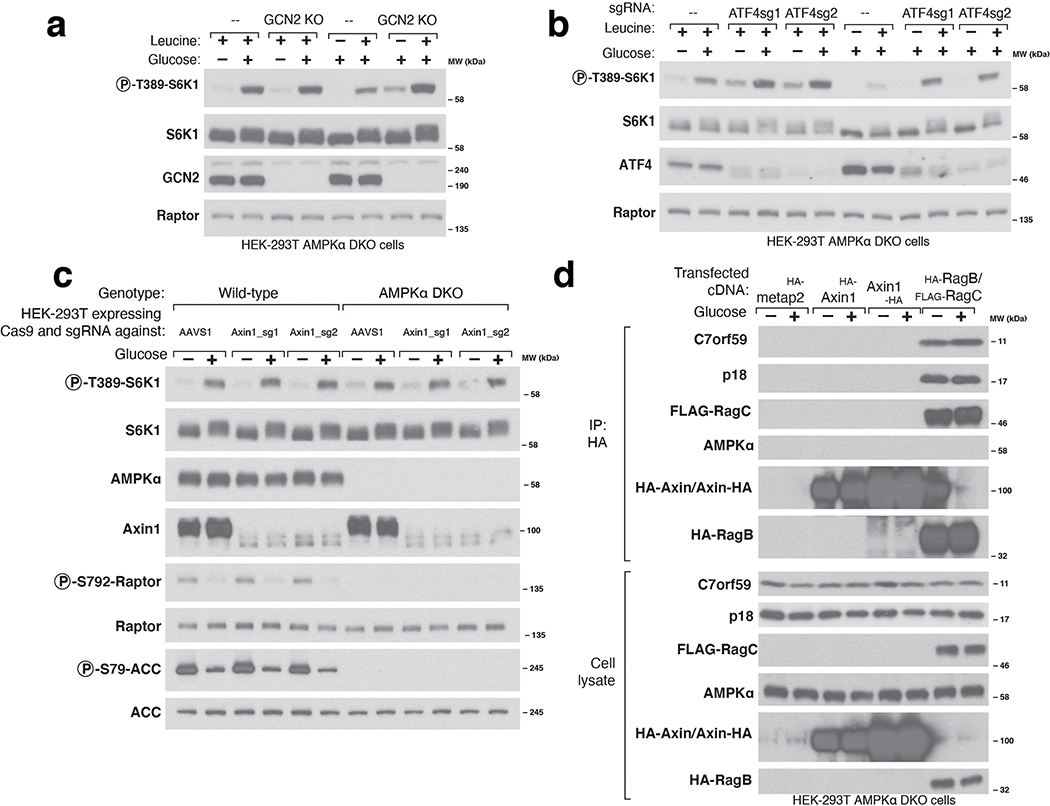
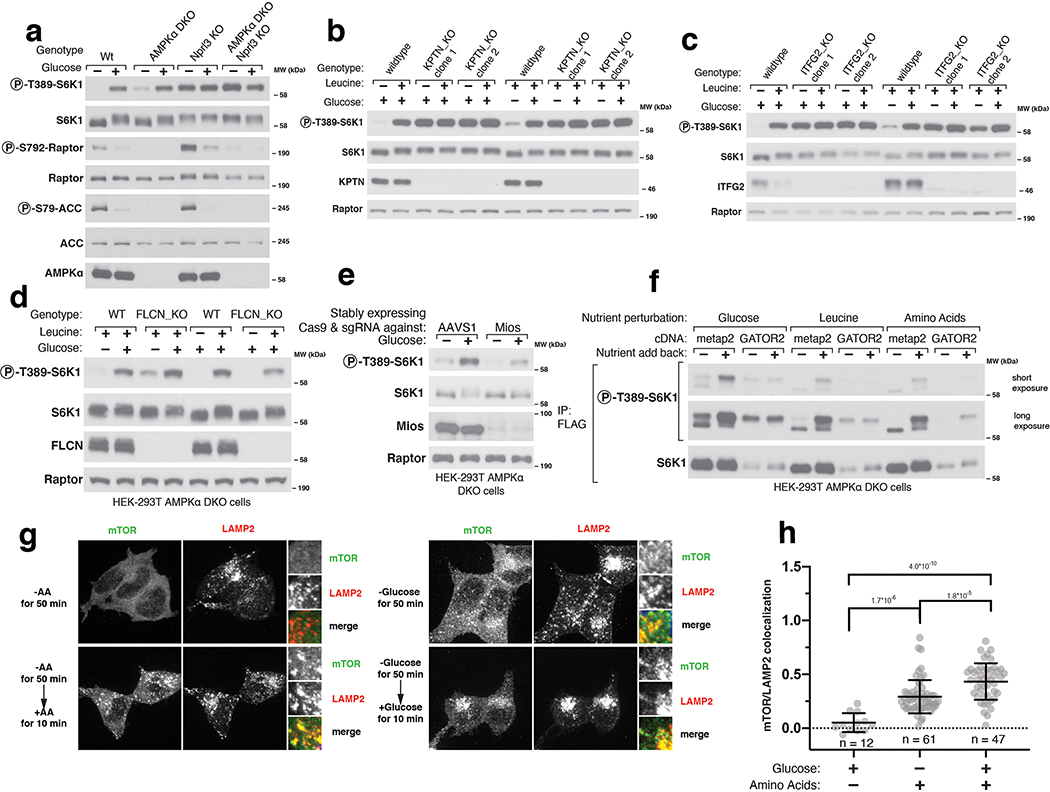
Many upstream inputs regulate mTORC1 activity, including the nutrient-sensing pathway anchored by the heterodimeric Rag GTPases that recruit mTORC1 to the lysosomal surface, its site of activation. The Rag GTPases and their positive (GATOR2, Ragulator, SLC38A9, FLCN-FNIP) and negative (GATOR1, KICSTOR) upstream regulators play key roles in the sensing of amino acids by mTORC11. In addition, several stress-sensing pathways activate the ATF4 transcription factor, which suppresses mTORC1 signaling, at least in part, by upregulating the Sestrin2 leucine sensor that acts through GATOR227. We found that neither loss of ATF4 nor its upstream activator, GCN2, prevented glucose sensing by mTORC1 in AMPK DKO cells (Extended Fig. 6A–B). Recently, the Wnt pathway component Axin1 has been implicated in glucose sensing upstream of AMPK via an interaction with the Ragulator complex15. In our cells, loss of Axin1 did not affect the capacity of mTORC1 or AMPK to be regulated by glucose and we could not detect Ragulator in Axin1-immunoprecipitates, suggesting that the role of Axin in detecting glucose might be cell-type dependent (Extended Fig. 6C–D).
Consistent with previous work implicating the Rag GTPases in the AMPK-independent sensing of glucose by mTORC19,10, loss of core components of GATOR1 or KICSTOR eliminated the capacity of glucose starvation to inhibit mTORC1 (Extended Fig. 7A–C). While loss of FLCN did not affect the ability of glucose to regulate mTORC1 (Extended Fig. 7D), loss of a GATOR2 component (Mios) prevents full activation of mTORC1 by glucose (Extended Fig. 7E). Moreover, in cells overexpressing all five components of GATOR2 (WDR59, WDR24, MIOS, SEH1L, and SEC13) glucose starvation no longer inhibited mTORC1 (Extended Fig. 7F). These data agree with the previous reports that glucose sensing in the mTORC1 pathway occurs upstream of the Rag-GTPases and specifically through the GATOR2-GATOR1-KICSTOR input. As previously seen9 and consistent with the known roles of the Rag GTPases in mTORC1 activation, starvation of glucose regulated the localization of mTORC1 to the lysosomal surface, albeit to a lesser degree than that of amino acids (Extended Fig. 7G–H).
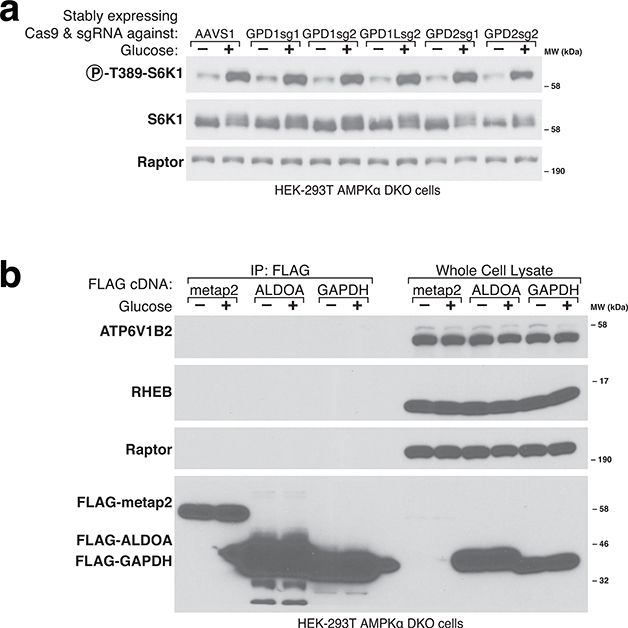
Our extensive attempts to identify DHAP sensor proteins akin to Sestrin2 for leucine28 and CASTOR1 for arginine29 have been unsuccessful. In addition, our existing data suggest that the enzymes TPI, ALDO, and GPD1/GPD1L, which are known to consume or generate DHAP, are not DHAP sensors for the mTORC1 pathway. TPI cannot be the sensor because while its loss slows the inhibition of mTORC1 upon glucose starvation, ultimately it is not necessary for glucose to regulate the pathway (Fig.4c–d, S5E). Likewise, none of the aldolase paralogues can be the sensor because DHA treatment of TKFC-expressing cells activates mTORC1 in cells lacking all of them (Extended Fig. 5E). Lastly, loss of GPD1 and its paralogue GPD1L does not prevent glucose regulation of mTORC1, ruling them out as sensors (Extended Fig. 8A). GAPDH was previously reported to bind the mTORC1-activating Rheb GTPase and to be a GAP sensor for the mTORC1 pathway16, but in our hands we have not detected an interaction between GAPDH and Rheb (Extended Fig. 8B), nor is it reported in the Bioplex network of protein-protein interactions30,31. Similarly, ALDOA was recently proposed to act as a glucose sensor to regulate the AMPK pathway via a nutrient-regulated interaction with the v-ATPase complex15. However, we are unable to detect an interaction between ALDOA and the v-ATPase component ATP6V1B2 in our cell system (Extended Fig. 8B).
Discussion
In this study we took a systematic approach to test the importance of specific steps in glycolysis in the activation of mTORC1 in response to glucose and we narrow in on the metabolite DHAP as playing a central role. Of course, we are unable to determine whether or not DHAP is the only glycolytic metabolite that signals to mTORC1, particularly in systems beyond the cultured human cells we have used, namely HEK-293T cells lacking the canonical energy sensor AMPK.
While confirmation that DHAP is a bona fide signaling molecule awaits the discovery and manipulation of the sensing mechanism, our current findings support the conclusion that DHAP has a key role in transmitting glucose availability to mTORC1: (i) the synthesis of DHAP is sufficient to activate mTORC1 in the absence of glucose; (ii) enzymes upstream of DHAP are necessary for the activation of mTORC1 by glucose (iii) and those downstream for its suppression by glucose starvation. Two key findings lead us to conclude that DHAP is more important than GAP for the sensing of glucose by mTORC1. First, loss of TPI, which slowed the turnover of DHAP but not of GAP, also slowed the inhibition of mTORC1 caused by glucose starvation. Second, the addition of DHA to glucose-starved cells expressing TKFC was sufficient to activate mTORC1 and restore DHAP levels but only had a partial effect on those of GAP.
An important question is why the mTORC1 pathway acquired the capacity to respond to glucose in a manner independent of the cellular energy status. Because mTORC1 increases the rate of glycolysis by regulating HIF1α32, it makes sense for it to sense glucose availability. However, this does not answer why, in particular, the cell uses DHAP to do so. In retrospect, there are many reasons why DHAP may be particularly well suited to play a role as a signaling molecule. First, DHAP levels, along with those for GAP, change more than any other glycolytic intermediate between high- and low-glucose conditions. Second, DHAP, in addition to being a glycolytic intermediate, is a precursor for the glycerol backbone used in the synthesis of triglycerides and phospholipids, processes that mTORC1 promotes when active33. Linking DHAP levels to mTORC1 activation ensures that lipid synthesis is only fully activated when the precursor metabolite DHAP is at acceptable levels. Therefore, DHAP sensing by the mTORC1 pathway could play a critical role in post-prandial de novo lipogenesis in adipose tissue. In agreement with this notion, a previous study demonstrated that GLUT4 overexpression in adipose tissue led to greater increases in insulin-stimulated mTORC1 activation suggesting that glucose uptake, in concert with insulin action, plays a key role in driving mTORC1-dependent anabolism34. Third, the position of DHAP in both glucose and fructose metabolism allows the mTORC1 pathway to sense both sugars, which are the major simple sugars in human diets. Moreover, because fructose metabolism to DHAP bypasses the steps of glycolysis that are subject to feedback inhibition35, fructose has the potential of activating mTORC1 without some of the intrinsic negative feedback in glycolysis (Fig.4h) in tissues that metabolize fructose, such as the small intestine and the liver36. In the latter, fructose stimulates lipogenesis and drives diet-induced fatty liver disease37. While much more work is necessary to determine the role of DHAP sensing by mTORC1 in vivo, it is tempting to speculate that the link between dietary fructose and lipogenesis might involve the activation of anabolic processes by mTORC1. Future work should also clarify the role in mTORC1 activation of other carbon sources such as lactate or glycerol, which under the right physiological contexts have the potential to activate mTORC1 by way of DHAP production.
Methods
Antibodies
Antibodies names, source, and catalogue numbers are described in Supplmentary Table 1.
Chemicals
Chemical names, source, and catalogue numbers are described in Supplmentary Table 2.
Plasmids used
Plasmid used with their Addgene numbers are described in Supplmentary Table 3.
Other materials
Anti-FLAG M2 affinity gel from Millipore Sigma; XtremeGene9 and Complete Protease Cocktail from Roche; Alexa 488, 568 and 647-conjugated secondary antibodies, and Inactivated Fetal Bovine Serum (IFS) from Invitrogen; leucine-free and amino acid-free RPMI from US Biologicals; and anti-HA magnetic beads, glucose-free RPMI (cat # 11879020) from Thermo Fisher Scientific. DMEM high glucose (catalog number: 11995040) and DMEM low glucose (catalogue number: 11885092) were purchased from Thermo Fisher Scientific. For LC/MS experiments, all solvents (including water) were purchased from Fisher and were Optima LC/MS grade. Formic acid 99% HPLC/MS Mobile Phase Component was purchased from Thermo Fisher Scientific.
Cell lines and tissue culture
HEK-293T cells obtained from ATCC were normally cultured in DMEM high-glucose (25 mM) with 10% IFS and supplemented with 2 mM glutamine. Cells were maintained at 37°C and 5% CO2. Cells were passaged every 48 hours and were not allowed to ever reach more than 90% confluency. For ALDO and GAPDH dox off experiments, cells were grown in DMEM with low glucose (5 mM).
cDNA Transfections
For the transfection of cDNA expression constructs into HEK-293T cells, 1.5 – 2 million cells were seeded in 10 cm dishes. Using the polyethylenimine method (39), cells were transfected 24 hours after seeding with the indicated pRK5-based expression vectors. Experiments were undertaken 36–48 hours after transfection. The total amount of DNA transfected was normalized to 5 μg with the empty pRK5 vector. The following amounts of cDNA were used in the indicated figures.
Fig1C: 2 ng of FLAG-S6K (pRK5) + 500 ng (HA-metap2 in pRK5 or HA-GLUT5 in pRK5)
Supp_Fig.7F: 2 ng of FLAG-S6K (pRK5) + either 2 ug HA-Metap2 (pRK5) or 800 ng each of HA-Mios, WDR24, and WDR59, and 1200 ng of SEC13 and SEH1L.
Lentiviral production and lentiviral infections
HEK-293T cells were seeded at a density of 750,000 cells per well of a 6-well plate in IMDM with 20% IFS. 24 hours after seeding, VSV-G envelope and CMV ΔVPR packaging plasmids were co-transfected with either pLJM60 containing cDNAs, pLentiCRISPRv2 with indicated guide sequences, or pCW57.1_tTA with the indicated cDNA, using XTremeGene 9 transfection reagent (Roche). 12 hours after transfection, the media was changed to fresh IMDM 20% IFS. 36 hours after the media change, the virus-containing supernatant was collected and passed through a 0.45 μm filter. Target cells were plated in 6-well plates with 8 μg/mL polybrene and incubated with virus-containing media. Cells were spinfected at 2200 rpm for 45 minutes at 37°C. Twelve hours later, the virus containing media was replaced with fresh DMEM media. 24–48 hours after spinfection, cells were passaged into puromycin containing media for pLJM60 or pLentiCRISPR or blasticidin containing media for pCW57.1_tTA.
Nutrient starvation experiments
Cells were seeded at 1 million cells per well of a fibronectin-coated 6-well plate the day before the experiment. Cells were washed 1x with 1 mL of the starvation media (-glucose RPMI, -leucine RPMI, or -AA RPMI) and starved in 1 mL of the same starvation media for 1–3 hours, as indicated in each figure legend, or starved for and restimulated for 10–20 minutes with glucose or the indicated nutrient. For glyceraldehyde (GA) and dihydroxyacetone (DHA) restimulations, 500 mM solutions of GA or DHA in water were prepared immediately before adding them to cells. Unless otherwise specified, glucose or other sugars were added at a final concentration of 10 mM. For the phosphatidic acid (PA) experiment, PA was prepared as previously reported 25,26,38,39. A 1 mM solution of PA in PBS was prepared by sonication, and added to starved cells to a final concentration of 100 μM.
Cell lysis and immunoprecipitations
Cells were rinsed with cold PBS and lysed in lysis buffer (1% Triton, 10 mM β-glycerol phosphate, 10 mM pyrophosphate, 40 mM Hepes pH 7.4, 2.5 mM MgCl2 and 1 tablet of EDTA-free protease inhibitor [Roche] (per 25 ml of buffer)). Cell lysates were cleared by centrifugation in a microcentrifuge (15,000 rpm for 10 minutes at 4°C). Cell lysate samples were prepared by addition of 5X sample buffer (0.242 M Tris, 10% SDS, 25% glycerol, 0.5 M DTT, and bromophenol blue), resolved by 8%−16% SDS-PAGE, and analyzed by immunoblotting.
For anti-FLAG immunoprecipitations, anti-FLAG M2 Affinity Gel (Sigma A2220) was washed with lysis buffer three times and then resuspended to a ratio of 50:50 affinity gel to lysis buffer. 25 μL of a well-mixed slurry was added to cleared lysates and incubated at 4°C with shaking for 90–120 minutes. Immunoprecipitates were then washed three times, once with lysis buffer, and twice with lysis buffer with 500 mM NaCl. Immunoprecipitated proteins were denatured by addition of 50 μL of SDS-containing sample buffer (0.121 M Tris, 5% SDS, 12.5% glycerol, 0.25 M DTT, and bromophenol blue) and heated in boiling water for 5 minutes. Denatured samples were resolved by 8%−12% SDS-PAGE, and analyzed by immunoblotting.
Quantification of Western Blots
The intensities of western blot bands were measured using Fiji software. Average pixel intensity was taken over boxes of equal area that covered each band. A nearby area on the blot without a band was used to obtain a background measure. The difference in average pixel intensity between each band and the background was obtained. The relative change between each band and a control lane was computed by taking the ratio of these two values, for Figure 4C,D the first lane was taken as the control lane. The relative differences in the p-S6K1 band were normalized to the differences observed in a loading control, Raptor. Raptor was chosen as the loading control to normalize to instead of S6K1 because the S6K1 protein runs with apparent changes in its molecular weight as a function of its phosphorylation state, the very variable we are trying to quantify. The mobility changes to S6K could potentially confound the normalization process.
Generation of knock-out cell lines using CRISPR-Cas9 technology
To generate CRISPR/Cas9-mediated gene knockouts in HEK-293T cells, we used the guide sequences described in Supplmentary Table 4. For clonal KOs, guides were cloned into pX330 and for stable overexpression into pLentiCRISPR.
On day one, 2 million HEK-293T cells were seeded in a 10 cm plate. Twenty-four hours after seeding, each well was transfected with 1 μg shGFP pLKO, 1 μg of the pX330 guide construct and 3 μg of empty pRK5 using XtremeGene9. Two days after transfection, cells were moved to a new 10 cm plate in puromycin-containing media. Forty-eight hours after selection, the media was switched to media not containing puromycin. Cells were allowed to recover for 1 week after selection prior to single-cell sorting with a flow cytometer into the wells of a 96-well plate containing 150 μl of DMEM supplemented with 30% IFS.
Generation of Dox-off cell lines
cDNAs for human GPI, ALDOA, TPI, or GAPDH were cloned from HEK-293T cDNA. The cDNAs were made resistant for their respective sgRNA as follows. GPI: sgRNA sequence <CAACCATGGGCATATCCTGG> was used; the PAM sequence in the cDNA was mutated by mutating the codon for valine 53 from GTG>GTA using overhang extension PCR. ALDOA: sgRNA sequence < CATTGGCACCGAGAACACCG> was used; the PAM sequence in the cDNA was removed by mutating the codon for glutamate 72 from GAG>GAA. TPI: Based on molecular weight, HEK-293T cells express isoform 2 (Uniprot: P60174–1) of TPI1. The sgRNA < TGTCTTTGGGGAGTCAGATG> was used, the cDNA was naturally resistant to this guide because the PAM sequence is partially in an intron, therefore no modification was made for this cDNA. GAPDH: sgRNA sequence < TGCTGGCGCTGAGTACGTCG> was used; the PAM sequence in the cDNA was removed by mutating the codon for valine 96 from GTG>GTA.
Cells were transduced with lentivirus produced from the pCW57 vector encoding the dox-off cDNA and a blasticidin resistance gene and were selected for 7 days with blasticidin. For GPI, TPI, and GAPDH dox-off cells, they were transduced to express stable Cas9 and the respective sgRNA along with a puromycin resistance gene. For ALDO dox off cells, guides against ALDOA, ALDOB, and ALDOC encoded in the pX330 plasmid were co-transfected along with an shGFP pLKO vector encoding a puromycin resistance gene. Next, cells were puromycin selected and allowed to expand for an additional 7 days. Cells were single-cell sorted and screened by western blot for the appropriate dox-off status.
Doxycycline Treatment
Doxycycline was prepared as a 30 ug/mL stock solution and aliquots were stored at −80°C. Aliquots are good for 2–3 months at −80°C. Cells were treated with 30 ng/mL doxycycline and cultured in 5 mM glucose DMEM for 5 days for the GAPDH and ALDOA experiments. For GPI and TPI, cells were treated for at least 9 days prior to the experiment.
Notably, when >3 mM glucose was added to cells lacking GAPDH expression, it caused an 80% decrease in ATP levels and a 30-fold increase in AMP levels. This is likely due to the fact that glucose is phosphorylated twice in upper glycolysis, but the phosphates are not liberated as this occurs after the GAPDH step of glycolysis. Therefore, when GAPDH is absent, glucose acts as an ATP-sink. Thus, it is hard to interpret the signaling status of cells in the presence of >3 mM glucose and lacking GAPDH expression because large decreases in ATP levels are known to decrease mTORC1 activity, potentially because of a relatively high Km for ATP40.
Koningic Acid Treatment
Koningic acid was ordered in a 250 ug size and resuspended in 178 uL of sterile water to make a 5 mM stock solution. The stock solution can be aliquoted and stored at −80° C for 2 weeks at most. Cells were starved of glucose and KA was added as indicated. If added to glucose replete cells, KA was very toxic, so it was only added to glucose-starved cells where it was well tolerated during the 3-hour incubation period. Similar to cells lacking GAPDH expression, glucose addition to KA treated cells caused a marked decrease in ATP levels and in mTORC1 signaling. This effect is likely similar to the phenomenon observed in cells lacking GAPDH expression and to that observed by Dennis et al40.
Immunofluorescence assays
For the experiment in Extended Fig. 7G–H, 400,000 HEK-293T cells were plated on fibronectin-coated glass coverslips in 6-well tissue culture plates. After 24 hours, the slides were rinsed once with PBS and fixed with 4% paraformaldehyde in PBS for 15 minutes at room temperature. The slides were then rinsed three times with PBS and the cells permeabilized with 0.05% Triton X-100 in PBS for 5 minutes at room temperature. The slides were rinsed three times with PBS and then blocked for 1 hour in Odyssey blocking buffer at room temperature. The slides were incubated with primary antibody in Odyssey blocking buffer at 4°C overnight, rinsed three times with PBS, and incubated with secondary antibodies produced in donkey (diluted 1:1000 in Odyssey blocking buffer) for 50 minutes at room temperature in the dark, and washed three times with PBS. The primary antibodies used were directed against mTOR (CST; 1:100 dilution), LAMP2 (Santa Cruz Biotechnology; 1:300 dilution). Slides were mounted on glass coverslips using Vectashield (Vector Laboratories) containing DAPI. Images were acquired on a Zeiss AxioVert200M microscope with a 63X oil immersion objective and a Yokogawa CSU-22 spinning disk confocal head with a Borealis modification (Spectral Applied Research/Andor) and a Hamamatsu ORCA-ER CCD camera. The MetaMorph software package (Molecular Devices) was used to control the hardware and image acquisition. The excitation lasers used to capture the images were 488 nm (LAMP2) and 561 nm (mTOR). Lysosomal enrichment was quantified as previously described10. Raw image files were opened in the Fiji software and a maximum-intensity projection of a Z stack of ~6–8 contiguous focal planes (∼0.5μm each) was used. In each cell analyzed, a cytoplasmic region of interest containing lysosomes was chosen by finding a punctum of high LAMP2 signal and in this area the mean fluorescence intensities (MFIs) of the 488 nm (LAMP2) and 561 nm channels (mTOR) were measured. In the same cell an equivalently sized area in a region of the cytoplasm not containing lysosomes with low LAMP2 signal was chosen and the MFIs of the 488 and 561 nm channels were also measured in this area. For each channel, the MFI of the non-lysosomal area was subtracted from that of the lysosomal area. The value obtained for the 561 nm channel was then divided by the analogous value for the 488 nm channel to give the lysosomal enrichment factor shown in the bar graphs in the figures. A lysosomal enrichment factor near 1 indicates that the mTOR (561 nm) signal was enriched in a region of the cell containing lysosomes over one that does not. A lysosomal enrichment factor closer to 0 or below zero indicates that the mTOR (561 nm) signal was not enriched at the lysosomes, indicating no specific co-localization with the LAMP2 signal (488 nm).
Extraction of metabolites for LC/MS analyses
Cells were seeded in fibronectin coated 6 well plates at a density of 1 million cells per well the day before the experiment. On the day of the experiment, cells were starved or starved and restimulated as indicated. At the time of lysis, the media was aspirated, cells were washed once with 1mL of cold saline, and metabolites were extracted by adding 800 uL of cold 80% methanol containing 500 nM isotope-labeled internal standards (Cambridge Isotope Laboratories, Inc.; MSK-A2–1.2). Methanol extracts were moved to pre-chilled Eppendorf tubes and samples were moved immediately to dry ice. Samples were briefly vortexed for 1 min, spun 15,000 rpm at 4°C for 10 minutes. The supernatant was dried by vacuum centrifugation and stored at −80°C. Just before LC/MS analysis, samples were resuspended in 50uL of LC/MS grade water, cleared of any insoluble debris by centrifugation at 15,000rpm.
Hydrophilic interaction chromatography (HILIC) LC/MS
Polar metabolite profiling was conducted on a QExactive benchtop orbitrap mass spectrometer equipped with an Ion Max source and a HESI II probe, which was coupled to a Dionex UltiMate 3000 HPLC system (Thermo Fisher Scientific). External mass calibration was performed using the standard calibration mixture every 7 days. Typically, samples were reconstituted in 100 uL water and 2 uL were injected onto a SeQuant® ZIC®-pHILIC 150 × 2.1 mm analytical column equipped with a 2.1 × 20 mm guard column (both 5 mm particle size; EMD Millipore). Buffer A was 20 mM ammonium carbonate, 0.1% ammonium hydroxide; Buffer B was acetonitrile. The column oven and autosampler tray were held at 25°C and 4°C, respectively. The chromatographic gradient was run at a flow rate of 0.150 mL/min as follows: 0–20 min: linear gradient from 80–20% B; 20–20.5 min: linear gradient from 20–80% B; 20.5–28 min: hold at 80% B. The mass spectrometer was operated in full-scan, polarity-switching mode, with the spray voltage set to 3.0 kV, the heated capillary held at 275°C, and the HESI probe held at 350°C. The sheath gas flow was set to 40 units, the auxiliary gas flow was set to 15 units, and the sweep gas flow was set to 1 unit. MS data acquisition was performed in a range of m/z = 70–1000, with the resolution set at 70,000, the AGC target at 1×106, and the maximum injection time (Max IT) at 20 msec. To increase sensitivity to DHAP/GAP, a targeted selected ion monitoring (tSIM) scan in negative mode was included. The isolation window was set at 1.0 m/z and tSIM scans were centered at m/z = 168.99080. Relative quantitation of polar metabolites was performed with XCalibur QuanBrowser 2.2 and TraceFinder 4.1 (both Thermo Fisher Scientific) using a 5 ppm mass tolerance and referencing an in-house library of chemical standards.
Aniline labeling of metabolite extracts for metabolomics
We adapted a previously published method in order to measure aniline-GAP (glyceraldehyde 3-phosphate) adducts41,42. Following methanol extraction, 300 uL of the ~800 uL samples were used in the aniline reaction. Both aniline and EDC solutions were prepared fresh in water. For the aniline solution, 777.5 mg/mL solution (6 M) was prepared in water and 6 uL of 10 M NaOH was added per 100 uL of the solution to increase pH to 4.5. To 300 uL of methanol extract, 30 uL of 6M Aniline-HCl and 30 uL of 200 mg/mL EDC solution were added. Samples were gently vortexed for 2 hours at room temperature. At the end of the two hours 5 uL of triethylamine was added to stop the reaction. Samples were then dried by vacuum centrifugation and stored at −80°C. Just before LC/MS analysis, samples were resuspended in 50 uL of LC/MS grade water with 0.1% FA/acetonitrile with 0.1% FA (50:50 v/v), cleared of any insoluble debris by centrifugation at 15,000 rpm.
Reverse Phase LC/MS
The LC and mass spectrometer general settings were as described above. Typically, 10 uL of a sample was injected onto a Kinetex C8 2.6 μm, 2.1 × 30 mm column (Phenomenex). Mobile Phase A was 0.1% formic acid in water and Mobile Phase B was 0.1% formic acid in acetonitrile. The column oven was set to 25°C and the flow rate was 0.250 mL/min. The gradient was as follows: 0–1 min: hold at 5% B; 1–6 min: linear gradient 5–70% B; 6–8 min: linear gradient 70–100% B; 8.1–10 min: hold at 5% B. The data were acquired in negative ion mode with a scan range of m/z = 140–380. A tSIM was included for the doubly labeled GAP-aniline adduct and was centered on m/z = 319.08420. Relative quantification of GAP-aniline was performed with XCalibur QuanBrowser 2.2 and TraceFinder 4.1 (both Thermo Fisher Scientific) using a 5 ppm mass tolerance.
Statistics and Reproducibility
Two-tailed t-tests were used for comparisons between groups. Reported p-values were not adjusted for multiple hypothesis testing. Therefore, comparisons with p-values <0.05 that are interpreted to be significantly different may reflect type I error. The Reporting Summary includes additional information.
All metabolite profiling experiments were conducted with 3 biological replicates. Signaling experiments shown as western blots are representative of at least two biologically independent experiments. For the immunofluorescence experiments, we show representative images of 3 biologically independent experiments.
Data Availability
The data supporting the plots within this paper are available from the corresponding author upon reasonable request.
Extended Data
Extended Fig. 1.
Extended Fig. 2.
Extended Fig. 3.
Extended Fig. 4.
Extended Fig. 5.
Extended Fig. 6.
Extended Fig. 7.
Extended Fig. 8.
Supplementary Material
Acknowledgements
The authors would like to thank Max Valenstein, Jessica Spinelli, and all the current members of the Sabatini lab for helpful insights. This work was supported by grants from the NIH (R01 CA103866, R01 CA129105, and R37 AI047389). J.M.O. was supported by a fellowship grant F30CA210373 from the National Cancer Institute and the Harvard-MIT MSTP training grant T32GM007753 from the National Institute of General Medical Sciences. P.A.K. was supported by a scholarship from Santander Universidades Mobility Fund granted by Adam Mickiewicz University in Poznan, Poland. A.L.C. was supported by a fellowship grant F31DK113665 from the National Institute of Diabetes and Digestive and Kidney Diseases. D.M.S. is an investigator of the Howard Hughes Medical Institute and an American Cancer Society Research Professor.
Footnotes
Competing Interests Statement
DMS is a founder, shareholder, and a member of the scientific advisory board for Navitor Pharmaceuticals, which is targeting the mTORC1 pathway for therapeutic benefit. J.M.O. is a shareholder of Navitor Pharmaceuticals. The other authors declare no competing intersts.
References
- 1.Saxton RA & Sabatini DM mTOR Signaling in Growth, Metabolism, and Disease. Cell 168, 960–976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kubrusly MS et al. A role for mammalian target of rapamycin (mTOR) pathway in non alcoholic steatohepatitis related-cirrhosis. Histol. Histopathol 25, 1123–1131 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Guillén C & Benito M mTORC1 Overactivation as a Key Aging Factor in the Progression to Type 2 Diabetes Mellitus. Front. Endocrinol 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ardestani A, Lupse B, Kido Y, Leibowitz G & Maedler K mTORC1 Signaling: A Double-Edged Sword in Diabetic β Cells. Cell Metab 27, 314–331 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Lee PL, Tang Y, Li H & Guertin DA Raptor/mTORC1 loss in adipocytes causes progressive lipodystrophy and fatty liver disease. Mol. Metab 5, 422–432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoki K, Zhu T & Guan K-L TSC2 Mediates Cellular Energy Response to Control Cell Growth and Survival. Cell 115, 577–590 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Shaw RJ et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6, 91–99 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Gwinn DM & Shaw RJ 3 - AMPK Control of mTOR Signaling and Growth. in The Enzymes (eds. Tamanoi F & Hall MN) vol. 28 49–75 (Academic Press, 2010). [Google Scholar]
- 9.Efeyan A et al. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfson RL et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 543, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalender A et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab 11, 390–401 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buller CL, Heilig CW & Brosius FC GLUT1 enhances mTOR activity independently of TSC2 and AMPK. Am. J. Physiol.-Ren. Physiol 301, F588–F596 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almacellas E et al. Phosphofructokinases Axis Controls Glucose-Dependent mTORC1 Activation Driven by E2F1. iScience 20, 434–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts DJ, Tan-Sah VP, Ding EY, Smith JM & Miyamoto S Hexokinase-II positively regulates glucose starvation induced autophagy through TORC1 inhibition. Mol. Cell 53, 521–533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C-S et al. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature 548, 112–116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee MN et al. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol. Cell. Biol 29, 3991–4001 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S, Guthrie PH, Chan SS, Haq S & Taegtmeyer H Glucose phosphorylation is required for insulin-dependent mTOR signalling in the heart. Cardiovasc. Res 76, 71–80 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anja Karlstaedt, Radhika Khanna, Manoj Thangam & Heinrich Taegtmeyer. Glucose 6-Phosphate Accumulates via Phosphoglucose Isomerase Inhibition in Heart Muscle. Circ. Res 126, 60–74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiraj Sen et al. Glucose Regulation of Load‐Induced mTOR Signaling and ER Stress in Mammalian Heart. J. Am. Heart Assoc 2, e004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hardie DG, Ross FA & Hawley SA AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol 13, 251–262 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.KEGG PATHWAY Database. https://www.genome.jp/kegg/pathway.html#metabolism.
- 22.Sakai K, Hasumi K & Endo A Two glyceraldehyde-3-phosphate dehydrogenase isozymes from the koningic acid (heptelidic acid) producer Trichoderma koningii. Eur. J. Biochem 193, 195–202 (1990). [DOI] [PubMed] [Google Scholar]
- 23.Liberti MV et al. A Predictive Model for Selective Targeting of the Warburg Effect through GAPDH Inhibition with a Natural Product. Cell Metab 26, 648–659.e8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rieder SV & Rose IA The Mechanism of the Triosephosphate Isomerase Reaction. J. Biol. Chem 234, 1007–1010 (1959). [PubMed] [Google Scholar]
- 25.Fang Y, Vilella-Bach M, Bachmann R, Flanigan A & Chen J Phosphatidic Acid-Mediated Mitogenic Activation of mTOR Signaling. Science 294, 1942–1945 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Menon D et al. Lipid sensing by mTOR complexes via de novo synthesis of phosphatidic acid. J. Biol. Chem 292, 6303–6311 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye J et al. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev 29, 2331–2336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfson RL et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 351, 43–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chantranupong L et al. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huttlin EL et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huttlin EL et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Düvel K et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson TR et al. mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman MA, She P, Peroni OD, Lynch CJ & Kahn BB Adipose Tissue Branched Chain Amino Acid (BCAA) Metabolism Modulates Circulating BCAA Levels. J. Biol. Chem 285, 11348–11356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson DL & Cox MM Lehninger Principles of Biochemistry. (W.H. Freeman, 2012). [Google Scholar]
- 36.Jang C et al. The Small Intestine Converts Dietary Fructose into Glucose and Organic Acids. Cell Metab 27, 351–361.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herman MA & Samuel VT The sweet path to metabolic demise: fructose and lipid synthesis. Trends Endocrinol. Metab. TEM 27, 719–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joy JM et al. Phosphatidic acid enhances mTOR signaling and resistance exercise induced hypertrophy. Nutr. Metab 11, 29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frias MA et al. Phosphatidic acid drives mTORC1 lysosomal translocation in the absence of amino acids. J. Biol. Chem 295, 263–274 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dennis PB et al. Mammalian TOR: A Homeostatic ATP Sensor. Science 294, 1102–1105 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Yang W-C et al. Simultaneous Quantification of Metabolites Involved in Central Carbon and Energy Metabolism Using Reversed-Phase Liquid Chromatography−Mass Spectrometry and in Vitro 13 C Labeling. Anal. Chem 80, 9508–9516 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Jannasch A, Sedlak M & Adamec J Quantification of Pentose Phosphate Pathway (PPP) Metabolites by Liquid Chromatography-Mass Spectrometry (LC-MS). in Metabolic Profiling: Methods and Protocols (ed. Metz TO) 159–171 (Humana Press, 2011). doi: 10.1007/978-1-61737-985-7_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the plots within this paper are available from the corresponding author upon reasonable request.