Abstract
Background
Vertebrate teeth exhibit a wide range of regenerative systems. Many species, including most mammals, reptiles, and amphibians, form replacement teeth at a histologically distinct location called the successional dental lamina, while other species do not employ such a system. Notably, a ‘lamina-less’ tooth replacement condition is found in a paraphyletic array of ray-finned fishes, such as stickleback, trout, cod, medaka, and bichir. Furthermore, the position, renewal potential, and latency times appear to vary drastically across different vertebrate tooth regeneration systems. The progenitor cells underlying tooth regeneration thus present highly divergent arrangements and potentials. Given the spectrum of regeneration systems present in vertebrates, it is unclear if morphologically divergent tooth regeneration systems deploy an overlapping battery of genes in their naïve dental tissues.
Results
In the present work, we aimed to determine whether or not tooth progenitor epithelia could be composed of a conserved cell type between vertebrate dentitions with divergent regeneration systems. To address this question, we compared the pharyngeal tooth regeneration processes in two ray-finned fishes: zebrafish (Danio rerio) and threespine stickleback (Gasterosteus aculeatus). These two teleost species diverged approximately 250 million years ago and demonstrate some stark differences in dental morphology and regeneration. Here, we find that the naïve successional dental lamina in zebrafish expresses a battery of nine genes (bmpr1aa, bmp6, cd34, gli1, igfbp5a, lgr4, lgr6, nfatc1, and pitx2), while active Wnt signaling and Lef1 expression occur during early morphogenesis stages of tooth development. We also find that, despite the absence of a histologically distinct successional dental lamina in stickleback tooth fields, the same battery of nine genes (Bmpr1a, Bmp6, CD34, Gli1, Igfbp5a, Lgr4, Lgr6, Nfatc1, and Pitx2) are expressed in the basalmost endodermal cell layer, which is the region most closely associated with replacement tooth germs. Like zebrafish, stickleback replacement tooth germs additionally express Lef1 and exhibit active Wnt signaling. Thus, two fish systems that either have an organized successional dental lamina (zebrafish) or lack a morphologically distinct successional dental lamina (sticklebacks) deploy similar genetic programs during tooth regeneration.
Conclusions
We propose that the expression domains described here delineate a highly conserved “successional dental epithelium” (SDE). Furthermore, a set of orthologous genes is known to mark hair follicle epithelial stem cells in mice, suggesting that regenerative systems in other epithelial appendages may utilize a related epithelial progenitor cell type, despite the highly derived nature of the resulting functional organs.
Keywords: Successional dental lamina, Tooth regeneration, Odontode, Epithelial appendage
Background
Vertebrate teeth arose shortly after vertebrates themselves and have since greatly diversified. Vertebrate “odontodes”, which include teeth and denticles, are defined by a shared basic morphology: a dentine core derived from mesenchymal cells, and an enamel or enameloid tip sheath contributed (at least in part) by epithelial cells [1–4]. The first clear evidence of dentine and enameloid-bearing structures arose at least ~ 420 million years ago in fossils of placoderms, though an even earlier origin in conodonts has been debated [5–8]. Support for the homology of odontodes comes from developmental studies that document similar histogenesis, morphological development, and shared gene expression patterns between teeth and other odontodes across large phylogenetic distances within jawed vertebrates [9–12]. Despite these deeply conserved, fundamental aspects of tooth and odontode development, teeth have radiated vastly in their arrangement [13], shape [14], size [15], placement within the body plan [16], and the regeneration system they employ [17–21]. Thus, some aspects of tooth development are staunchly conserved, while others are wildly plastic in evolution. This variation in the conservation of different aspects of tooth development presents an interesting case study in the evolution of development: what genetic signatures, if any, are common to disparate dental morphologies? Here, we assess two divergent tooth regeneration strategies found in two different ray-finned fishes: zebrafish (Danio rerio) and threespine stickleback (Gasterosteus aculeatus).
The morphogenesis of individual tooth organs has been well-studied across living jawed vertebrates [9, 22]. Teeth form like other epithelial appendages, demonstrating an intricate epithelial–mesenchymal interaction and coordinated cell motions during their early development (Fig. 1) [23]. Primary teeth first appear as a placode (thickened epithelium) underlain by a mesenchymal condensation, followed by invagination, cap formation, and eventually deposition of dentine and enamel and/or enameloid, which are composed of highly concentrated hydroxyapatite (a hard, crystalline calcium phosphate mineral that gives bones their characteristic hardness) [3, 24]. Some vertebrates, such as mice, do not replace entire teeth during their lives. By contrast, in most known extant vertebrate dental systems, once a tooth position is established, tooth replacement via regeneration can occur anywhere between one (e.g., most mammalian teeth) to potentially one hundred times or more (e.g., some sharks) at a given tooth position [25–30]. In many vertebrates with tooth replacement, this process initiates at a site called the successional dental lamina (SDL). The SDL can be identified histologically as a deep extension of epithelium connected directly to the outer dental epithelium of a predecessor tooth [9, 31]. When present, during tooth replacement the SDL first thickens, similar to the placode stage of primary teeth [9]. Subsequently, this thickened epithelium undergoes the stereotypical tooth differentiation process outlined above (depicted in Fig. 1), and at all stages is tightly associated with underlying dental mesenchyme. The SDL was recently shown to be a key tissue in tooth regeneration: mouse molars, which do not naturally undergo regeneration, can be prompted to do so by stabilizing Wnt signaling in the rudimentary mouse SDL [32]. In most cases, a replacement tooth germ eventually prompts osteoclast activity to enzymatically dissociate the base of the predecessor tooth and remodel the underlying bone of attachment (if present) either before or during early eruption [33–37]. This osteoclast activity facilitates the shedding of the predecessor and makes space for the replacement tooth to finish mineralization and eruption, thus completing the regenerative process.
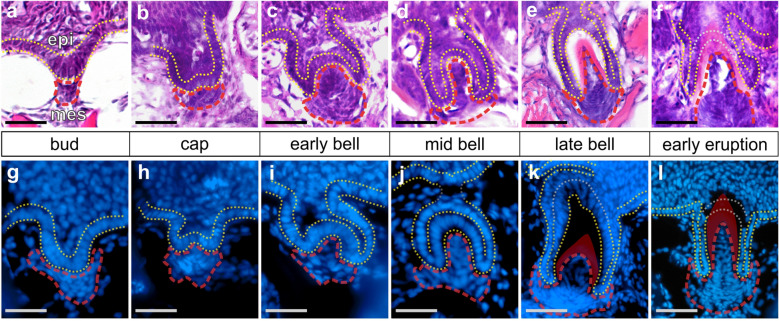
Fig. 1.
Stickleback pharyngeal tooth morphogenesis. All images show sagittal or transverse sections from sticklebacks (20–40 mm standard length). The basalmost layer of epithelium is outlined with yellow dashed lines in all panels, the mesenchyme is outlined in red. During late bell stages (e, k), the epithelium towards the apex of the forming tooth becomes thickened, no longer presents a clear bilayer, and will eventually be punctured during tooth eruption (gray outlines). a–f Staging series of hematoxylin and eosin-stained transverse sections through stickleback pharyngeal tooth germs at the stages indicated. Epithelium (“epi”) and mesenchyme (“mes”) are labeled in a. g–l Staging series of DAPI-stained sagittal sections through stickleback pharyngeal tooth germs at the stages indicated. Gray portions of dotted epithelium lines in J indicate the epithelium/tooth connection that was present on an adjacent section. Bone matrix is false-colored red in k and l. Scale bars = 20 μm
While many vertebrate tooth regeneration systems utilize an SDL, the arrangement of primary and replacement teeth is variable, both in modern species [9, 12, 17, 31, 38–44], as well as stem group jawed vertebrates [45, 46]. In some species (e.g., sharks), teeth are replaced in families from permanent, conspicuous regions in the oral jaw [12, 20]. By contrast, in other species, teeth are replaced from a relatively inconspicuous SDL that forms transiently (as in zebrafish) [42]. Some fishes such as bichir and salmonids present an even more abbreviated process, wherein the outer dental epithelium of the predecessor tooth directly relocalizes to a deeper region on the lingual side of the tooth, undergoes thickening, and eventually tooth differentiation, effectively ‘skipping’ the SDL phase [18, 47–49]. Cichlid oral teeth grossly undergo the same process, though interestingly their oral replacement teeth are derived from the opposite (labial) side of the predecessor [10, 50]. The patterning of tooth replacement can even vary throughout ontogeny of the same species: medaka fish initially have disorganized pharyngeal dentitions without visible tooth families, but later in life the teeth organize into highly patterned pharyngeal tooth plates with clear tooth families arranged in rows [38, 51]. Despite these clear tooth families, no discrete dental lamina was observed in medaka [38]. Together, these data show that much flexibility exists in how tooth progenitors can be arranged across dental regeneration systems [18, 21].
Despite high variation in the size, shape, and position of vertebrate teeth, gene expression and functional studies on tooth differentiation across vertebrates has revealed deeply conserved molecular processes at play [10, 21]. However, given the phylogenetic lability and morphological disparities of tooth regeneration strategies between some groups [45], it is unclear whether or not this process is homologous and uses shared cell types across vertebrates. Recent evidence suggests that not just teeth, but also other epithelial appendages (EAs) such as scales, feathers, and hair share some aspects of development, also raising the possibility of far-reaching EA homology among these organs [23, 52–59].
To further elucidate the developmental program underlying tooth regeneration, we compared the histology and gene expression patterns associated with the regenerative process in the pharyngeal teeth of two teleost fishes: zebrafish and threespine stickleback. These two species share a common ancestor that lived approximately 250 million years ago [60] and appear to demonstrate some stark differences in their dentitions. Sticklebacks are well documented as demonstrating differences in tooth number between populations, particularly between high-toothed freshwater populations and low-toothed marine populations [61–65]. These tooth number differences have a strong genetic basis, contributing to a significant difference in adult tooth number between certain populations, which is especially pronounced on the ventral pharyngeal jaw [63, 64]. We find that, unlike other vertebrates such as zebrafish, the threespine stickleback does not exhibit an SDL, nor does it regenerate teeth via the immediate relocalization of the deep outer dental epithelium of a predecessor tooth (as in salmonids and bichir). Furthermore, stickleback teeth appear to occasionally diverge from the common 1-for-1 mode of tooth replacement: some tooth germs appear to prompt osteoclast activity and shedding of more than one erupted tooth in concert. Despite these differences in modes of tooth regeneration, we find a suite of genes expressed in common between naïve dental epithelial cells in both fish species, suggesting that a conserved epithelial progenitor cell type underlies tooth regeneration, here referred to as the “successional dental epithelium” (SDE). We propose the use of this term to encompass not just the progenitor cells found in the histologically distinct successional dental lamina (SDL) of animals such as zebrafish, sharks, and tetrapods (where present), but also in less-pronounced epithelial regions in fishes like sticklebacks, which still undergo constant tooth regeneration across their tooth fields. Intriguingly, the genes we find localized to the SDE of both zebrafish and sticklebacks also mirror a cassette of gene expression that is well documented and functionally relevant in hair follicle epithelial stem cells (in the “bulge”) [66, 67], suggesting that such a genetic module may be common across not just teeth, but also other vertebrate epithelial appendages.
Results
A brief review and comparison of the gross morphologies of stickleback and zebrafish dentitions
Tooth field position and arrangement
Zebrafish and stickleback teeth are grossly similar in tooth shape and structure (Fig. 2a and b); both species’ teeth are unicuspid and mostly conical, with a slight bend or hook towards the posterior side. However, these two species differ drastically in tooth arrangement within the body plan. Zebrafish, like other members of Cypriniformes, lack oral teeth, and possess only a single (paired) tooth field found ventrally on the last branchial arch (on ceratobranchial 5) in the pharynx (see Fig. 2a, referred to here as the ventral tooth plate or VTP, also called the lower pharyngeal jaw or LPJ in some species). Sticklebacks, like most vertebrates, have oral teeth, which are located on their premaxilla and dentary bones. In the pharynx, sticklebacks also have a single paired VTP on ceratobranchial 5 (see Fig. 2b), as well as two paired dorsal tooth plates (DTPs) [68].
Fig. 2.
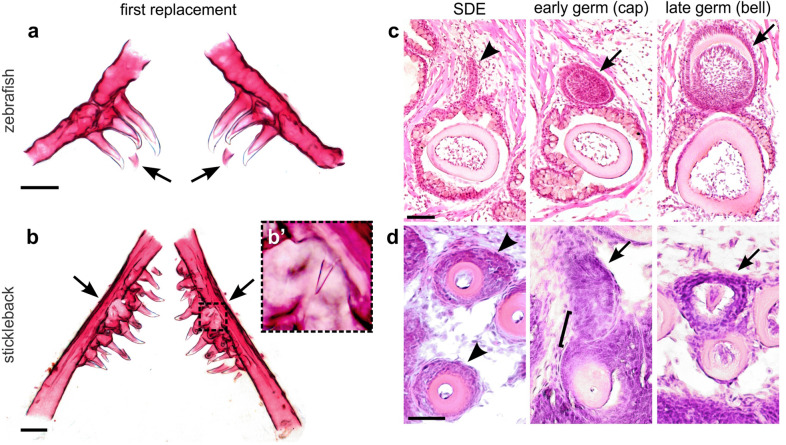
A comparison of zebrafish and stickleback pharyngeal tooth replacement. a, b Zebrafish and stickleback first tooth replacement events, respectively. Alizarin stained and dissected ceratobranchial 5 elements and their teeth from both species at 4 dpf zebrafish (a) and 30 dpf stickleback (b). Anterior to top. Arrows indicate first replacement tooth germs (bilaterally paired). Note that the zebrafish germs form on the ventral/medial side of the predecessor pioneer tooth (which in this case is the middle of 3 the ankylosed teeth on each side, tooth 4v1). Inset box in b magnified in b’ shows first replacement tooth. c, d Replacement histology on the coronal (d) and oblique coronal (c) axes (see Methods). Arrowheads mark the putative successional dental epithelium (SDE) in each image. In zebrafish, this tissue takes the form of a true successional dental lamina (SDL). In sticklebacks, there is only a subtle, non-elongated epithelial thickening that surrounds the tooth shaft like a collar, immediately beneath the rest of the nearby epithelial tissues (see Additional file 1: Fig. S1 for sagittal view). Arrows mark tooth germs at the stages indicated. A bracket marks the tight epithelial association between an early tooth germ and the position of the presumed SDE. Scale bars in a and b = 100 μm; c and d = 20 μm
Tooth number variation
Sticklebacks and zebrafish also have major differences in tooth number. In lab-reared zebrafish, each VTP typically hosts 11 tooth positions [69]. As in other cyprinids, this number is known to vary occasionally in zebrafish, occasionally observed as missing or bearing an extra tooth position [70]. Since replacement tooth mineral deposition occurs prior to the shedding of a predecessor tooth during the tooth regeneration process, an adult zebrafish typically hosts 15–17 mineralized teeth per VTP at any given point in time (i.e., there are typically 4–6 late bell stage replacement tooth germs per VTP that have not yet dislodged their predecessor). Given this relatively consistent dental arcade, zebrafish tooth positions have been named based on their location [69].
Threespine sticklebacks, on the other hand, can have up to ~ 200–300 + total teeth once fully grown, thus displaying both an order of magnitude more teeth than zebrafish, as well as higher within-species variation in adult tooth number. Tooth number in sticklebacks has a strong genetic basis, and varies significantly by population, with freshwater localities generally possessing more teeth than their marine counterparts [61, 63–65].
Early tooth replacement events
Sticklebacks and zebrafish both appear to replace their earliest primary teeth in a similar order, with those teeth that differentiate first also being replaced first [68, 69]. Interestingly, the tooth germs responsible for these early replacement events are essentially locationally opposite between the zebrafish and stickleback VTPs (arrows in Fig. 2a and b): zebrafish replacement tooth germs form on the medial/ventral side of each erupted tooth, towards the midline [69], while the first stickleback replacement tooth germs form on the lateral edge of the tooth plate, opposite the midline [68]. In zebrafish, this stereotypical polarized replacement direction (Fig. 2a) remains into adult stages. Conversely, as they mature, stickleback pharyngeal tooth fields do not appear to strictly adhere to the ‘lateral to medial’ replacement direction found in their earliest replacement events, which we detail below.
Replacement tooth histology
The histology of zebrafish tooth regeneration has been described previously in detail [42, 69]. Zebrafish tooth regeneration initiates from an SDL. In zebrafish, this region forms transiently, as an outpocketing on the ventro-medial side of each tooth, first visible as a thin projection of naïve epithelial cells stemming from the outer dental epithelium of a predecessor tooth (arrowhead in Fig. 2c) [42], protruding from the mineralized tooth and its associated epithelia deep in the tooth field. Subsequently, the SDL expands, forming a replacement tooth germ (arrows) while maintaining a close epithelial association with the predecessor tooth socket.
Stickleback pharyngeal replacement teeth arise from a more superficial region of the dentition than replacement teeth in zebrafish. After morphogenesis (Fig. 1), once ankylosis and eruption are complete, stickleback teeth do not retain a histologically distinct inner and outer dental epithelium (Additional file 1: Fig. S1). Instead, mature stickleback pharyngeal teeth retain a shallow “collar” of epithelial cells surrounding the site of eruption (arrowheads in Fig. 2d and Additional file 1: Fig. S1) that does not appear to be arranged in a clear bilayer. This collar of cells is contiguous with the basalmost epithelial sheet overlying the tooth field, juxtaposed to the pharyngeal epithelium (towards the lumen) and the basement membrane overlying mesenchymal cells (away from the lumen). It is from this collar of cells that the epithelial components of both oral and pharyngeal replacement teeth appear to be derived in sticklebacks (arrows in Fig. 2d), suggested by the tight epithelial association between early germs and the position of the putative SDE (bracket in Fig. 2d). Thus, a histologically distinct SDL is not present during oral or pharyngeal tooth regeneration in sticklebacks, a condition also reported in trout [49], the oral teeth of salmon [48], bichir [18], and cod [71], as well as pharyngeal teeth in medaka [38].
Stickleback pharyngeal teeth do not exhibit a tightly regulated 1-for-1 replacement scheme
Unlike zebrafish, stickleback tooth fields do not maintain any obvious arrangement of individual tooth positions across the entirety of each tooth field into adulthood, even within single populations. This observation initially led us to hypothesize that the stickleback tooth replacement process may introduce some spatial stochasticity of tooth position and spacing, similar to the condition in cod oral teeth, where their adult arrangement appears “haphazard” [71]. We thus sought to test the hypothesis that stickleback tooth replacement can diverge from a strict 1-for-1 tooth replacement system. We did this by analyzing H&E-stained serial sections cut in the sagittal, transverse, and coronal planes through pharyngeal tooth fields from subadult and adult fish (25–40 mm standard length; see Additional file 1: Fig. S1 for section plane orientation). Using these preparations, we identified every putative pharyngeal and oral replacement tooth germ that was in a mid- to late bell stage (the stages that coincide with bone remodeling during the regenerative cycle), allowing us to observe how many and which teeth were being actively dissociated during the replacement process. Of the few oral tooth germs we observed (n = 8 total observed from two separate fish) we found that stickleback oral replacement teeth formed at the labial side of each presumed predecessor tooth, and were not observed to be clearly diverging from a 1-for-1 replacement scheme (n = 0/8 tooth germs) (Fig. 3a and b). In the pharynx, most stickleback replacement tooth germs appeared to be anchoring themselves solely in the place of a single presumed predecessor tooth, which was usually the tooth to which they showed the nearest epithelial association (Fig. 3c). However, in ~ 25% of cases (17/67 tooth germs) a single replacement tooth was found to be abutting two erupted teeth that were both displaying interrupted mineralization and apparent concomitant osteoclast activity at their bases (Fig. 3d). We interpret these to be possible “1-for-2” tooth replacement events in the stickleback pharynx. These observations suggest that stickleback pharyngeal teeth deviate from a strict 1-for-1 mode of tooth replacement up to 25% of the time.
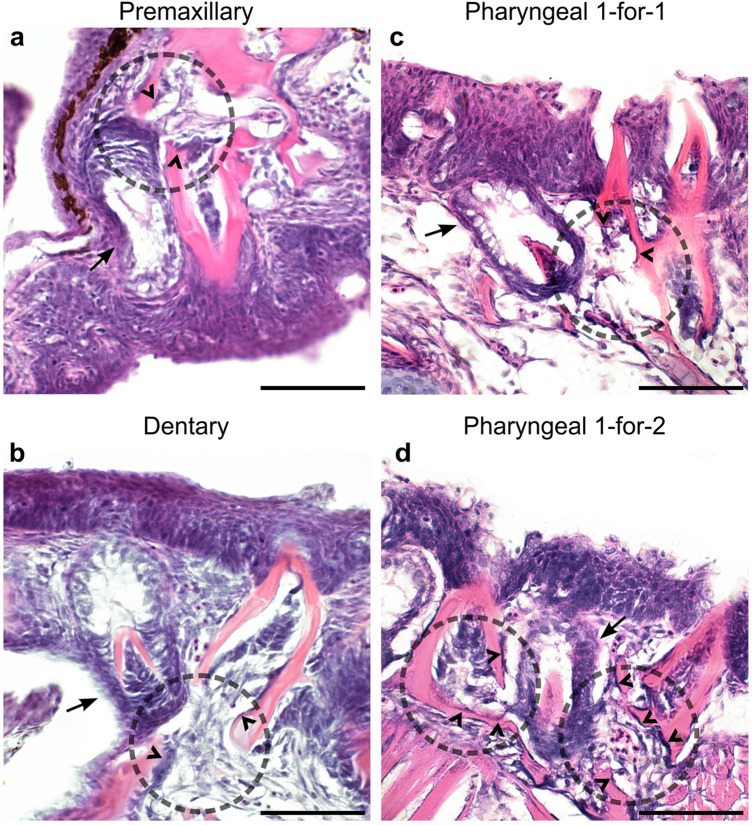
Fig. 3.
Oral and pharyngeal tooth replacement histology in stickleback. In all panels, dotted circles mark the general location of erupted tooth dislodgement adjacent to a presumed replacement tooth germ, arrows mark tooth germs, and carets indicate likely osteoclasts. All panels show sagittal (a, b) or transverse (c, d) sections and are oriented with the dorsal side facing upwards. a, b Oral tooth fields on the premaxilla and dentary bones were only observed as replacing via a 1-for-1 mechanism. c Pharyngeal replacement tooth germs were typically observed as dislodging only one other erupted pharyngeal tooth in 74.6% of cases (n = 50/67, see Methods). d By contrast, some pharyngeal replacement tooth germs appeared to be dislodging two erupted teeth simultaneously in 25.4% of cases (n = 17/67, see Methods). All scale bars = 20 μm
Gene expression in stickleback and zebrafish tooth germs and progenitors
Wnt signaling activation in zebrafish and stickleback replacement tooth germs
The role of Wnt signaling in tooth morphogenesis and differentiation has been well-established in mouse and zebrafish [72–76], and indicted in multiple human tooth phenotypes [77–84]. Using in situ hybridizations (ISHs) on thin sections through zebrafish and stickleback tooth plates, we compared the expression of Wnt10a and Lef1 in zebrafish and sticklebacks (Fig. 4a–d). We found highly conserved patterns of gene expression in tooth germs for both genes, with the expression of both Wnt10a and Lef1 upregulated in the future inner dental epithelia (especially at the central epithelial cells of the placode, black arrows in Fig. 4a–d) and early condensing mesenchyme of bud-stage tooth germs (white arrows in Fig. 4a–d).
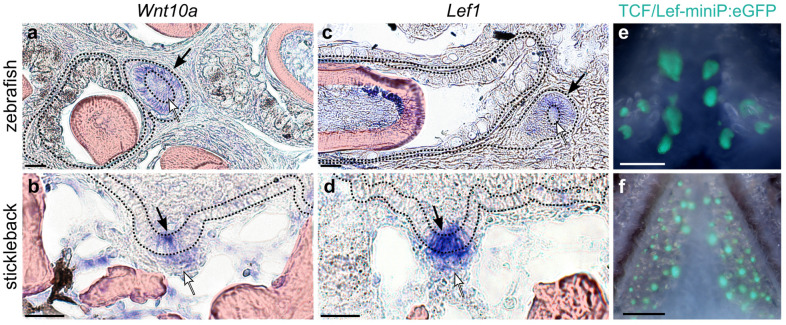
Fig. 4.
Wnt signaling in zebrafish and stickleback replacement tooth germs. Black arrows mark replacement tooth germ epithelium, white arrows mark replacement tooth germ mesenchyme. a wnt10a expression in a cap-stage zebrafish replacement tooth germ. b Wnt10a expression in a bud-stage stickleback replacement tooth germ. Note concentrated expression in the distalmost epithelium (arrow). c lef1 expression in a cap-stage zebrafish replacement tooth germ. d Lef1 expression in a bud-stage stickleback replacement tooth germ. e, f TCF/Lef synthetic Wnt activity reporter is active in adult replacement tooth germs. Scale bars in a–d = 20 μm; e and f = 500 μm
One major output of Wnt signaling is upregulated transcriptional activity via β-catenin and TCF/Lef transcription factors [85]. A synthetic TCF/Lef reporter construct [86] was found to be expressed in a small number of cells in the young primary teeth of zebrafish, but expression was not detected in the first replacement tooth [76]. This result raised the possibility of a Wnt signaling disparity between primary teeth and replacement teeth. The construct used in these assays employed a destabilized enhanced GFP (dGFP) coding region, which adds temporal resolution to the reporter construct output by increasing the degradation rate of eGFP. To address this potential primary vs. replacement tooth difference, we first compared the activity of the previously tested TCF/Lef-miniP:dGFP reporter construct [86] in zebrafish and stickleback adults by generating novel transgenic lines in each species. We confirmed previous observations by Shim et al. (2019) that post-larval zebrafish teeth do not display detectable dGFP fluorescence (n = 4/4 juvenile zebrafish). Similarly, we also did not detect dGFP by fluorescence microscopy in stickleback teeth at any stage. Given our in situ data for Wnt10a and Lef1 expression in juvenile and adult fish (Fig. 4a–d), we suspected that downstream TCF/Lef transcriptional activity was also occurring in replacement tooth germs, but perhaps at a slightly reduced relative intensity and/or with faster turnover or signal dilution compared to the earliest zebrafish tooth germs. Thus, we re-stabilized the dGFP reporter cassette by removing the destabilization signal (see Methods) and again derived stable transgenic lines in zebrafish and sticklebacks. This change from dGFP to eGFP allowed the new TCF/Lef-miniP:eGFP cassette to be robustly detected in all developing young tooth germs in adult fish, especially tooth epithelium, and throughout the entire process of tooth morphogenesis (Fig. 3e and f). These data suggest that active Wnt signals are indeed transduced in replacement teeth of all ages in zebrafish and sticklebacks.
Expression in naïve epithelial tissues
As outlined above, sticklebacks lack a histologically defined SDL (per [31]) in their pharyngeal tooth fields. Despite this, sticklebacks must still possess epithelial replacement tooth progenitors in the pharynx, given that pharyngeal replacement teeth form in part from the endoderm adjacent to erupted teeth (Fig. 1). Here we refer to these putative epithelial progenitors collectively as the “successional dental epithelium” (SDE). We use this general term to encompass both the cell types found in permanent or transient SDLs of other vertebrates, as well as the putative epithelial tooth progenitors of less-organized systems such as stickleback pharyngeal teeth. Given our prediction regarding the location of the stickleback SDE based on histology data (arrowheads in Fig. 2d and Additional file 1: Fig. S1), we sought to assay detailed gene expression patterns in this tissue.
Previous expression assays revealed that β-catenin, Plakoglobin, and E-Cadherin are expressed in the zebrafish SDL [87, 88]. We aimed to bolster our understanding of gene expression in naïve dental epithelia by determining whether the putative stickleback SDE expresses gene orthologs that mark the zebrafish SDL. Thus, we first sought to identify a group of genes that were expressed in the putative stickleback SDE and subsequently asked if these genes also mark the zebrafish SDL by performing (ISH) on thin sections through tooth plates of both fish species (Fig. 5). To these ends, we selected twelve “SDE candidate gene” groups based on previously published data supporting expression or function in tooth or hair regeneration, namely in the SDL or hair follicle stem cells: Bambi [67], Bmi1 [50, 89], Bmp6 [62, 90], Bmpr1a [91], CD34 [92–97], Dkk3b [41], Gli1 [50, 66, 98–100], Igfbp5 [41, 50, 96, 99], Lgr4 [50, 101], Lgr6 [101], Nfatc1 [95, 102], and Pitx2 [11, 12, 47, 50, 99, 103, 104].
Fig. 5.
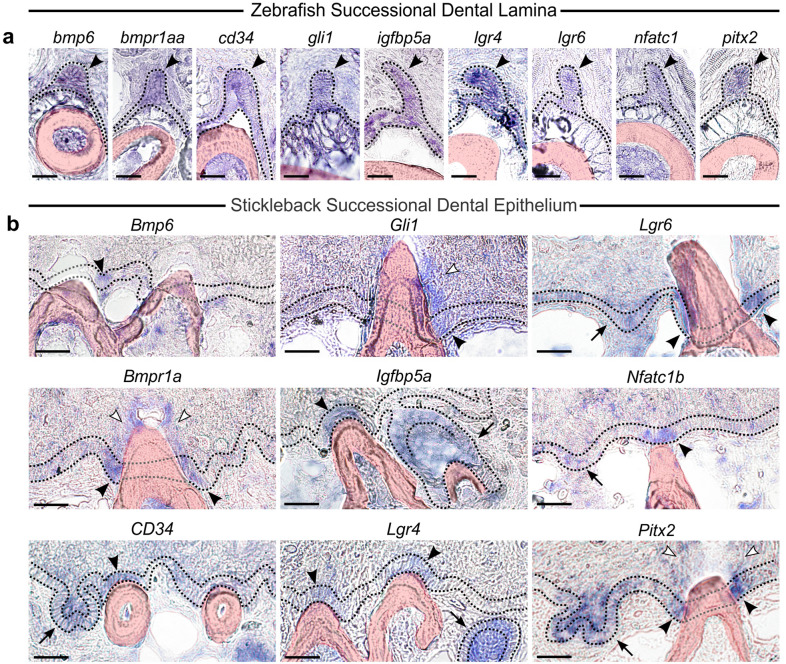
Successional dental epithelium (SDE) expression in zebrafish and stickleback. In all panels, a dotted line demarcates the basalmost pharyngeal epithelial layer, which is made gray when missing from a particular section (e.g., when it is interrupted by an erupted tooth). a Expression of bmp6, bmpr1aa, cd34, gli1, igfbp5a, nfatc1, lgr4, lgr6, pitx2 was assayed by in situ hybridization (ISH) in zebrafish. Expression of all 9 genes was detected in the successional dental lamina (SDL—black arrowheads). b Expression of Bmp6, Bmpr1a, CD34, Gli1, Igfbp5a, Lgr4, Lgr6, Nfatc1b, and Pitx2 was assayed by ISH in stickleback. Expression of all nine genes was detected in the proposed “successional dental epithelium” (SDE) of stickleback. Black arrowheads mark the SDE. Bmpr1a and Gli1 were additionally expressed in the superficial oral epithelium surrounding erupted teeth (white arrowheads). Tooth germs are indicated with a black arrow. All scale bars = 20 μm
In stickleback, 10 of these 12 gene groups were found to have at least one ortholog gene expressed in the putative SDE, i.e., the basalmost endodermal epithelium overlying pharyngeal tooth fields, specifically marking the deep “collar” epithelial cells immediately juxtaposed to some, but not all erupted teeth, occasionally on flanking contralateral sides of a tooth (black arrowheads in Fig. 5b; see Additional file 2: Fig. S2 for DAPI counterstain of nuclei of selected genes). Those genes we detected in the putative stickleback SDE were: Bmp6, Bmpr1a, CD34, Dkk3b, Gli1, Igfbp5a, Igfbp5b, Lgr4, Lgr6, Nfatc1b, and Pitx2 (black arrowheads in Fig. 5b and Additional file 3: Fig. S3). Of these 11 genes, Dkk3b and Igfbp5b were the least specific SDE markers, strongly marking many or most epithelial and mesenchymal cells in surrounding tissues (Additional file 3: Fig. S3), whereas the other nine genes exhibited more restricted patterns of expression (Fig. 5b). Importantly, these nine gene expression patterns in sticklebacks did not appear to collectively delineate any other cell type in the pharynx, other than early tooth germ epithelium (arrows in Fig. 5b), suggesting that this proposed “SDE genetic battery” is not common to other cell types or tissues in the pharynx. Five of these nine genes’ expression patterns appeared locally restricted to the SDE (Bmp6, Gli1, Igfbp5a, Lgr4, and Nfatc1b), while the rest were found in more expansive stretches of the basalmost epithelium (CD34, Bmpr1a, Lgr6, and Pitx2). Three of these genes (Bmpr1a, Gli1, and Pitx2) had consistent detectable expression in the superficial pharyngeal epithelium surrounding erupted teeth (white arrowheads in Fig. 5). Those candidate genes not detected in the stickleback SDE were Bmi1a, which we did not detect in any tissue, and Bambia, which was expressed only in epithelium and mesenchyme of differentiating tooth germs, and mesenchyme of erupted teeth (Additional file 3: Fig. S3).
In zebrafish, we performed ISH for nearest-neighbor orthologs of the nine “SDE genetic battery” genes we observed as most specifically marking the putative stickleback SDE (bmp6, bmpr1aa, cd34, gli1, igfbp5a, nfatc1, lgr4, lgr6, and pitx2). All nine of these genes were expressed in the naïve, transiently forming SDL during its earliest stages, when it consists only of a single epithelial bilayer (Fig. 5a). The naïve SDL and its presumed daughter cells in young tooth germs were the only observed pharyngeal cell types exhibiting expression of all nine of these genes (presumably due to the maintenance of this genetic battery, consistent with our observations in sticklebacks). We did not observe igfbp5b expression in the zebrafish SDL, and instead detected it in a subset of dental mesenchyme (Additional file 4: Fig. S4). We did not assay bambia, bmi1a, or dkk3b expression in zebrafish.
Dental mesenchymal expression in differentiating and erupted zebrafish and stickleback odontoblasts was also observed for eight of the nine “SDE marker” genes, to the exclusion of Pitx2, which was not detected in pharyngeal dental mesenchyme (Additional file 5: Fig. S5). Zebrafish mesenchymal expression of these eight genes appeared widespread, strongly marking most or all odontoblasts in tooth germs and erupted teeth, with the exception of Lgr6 which did not strongly mark mature tooth mesenchyme. Stickleback teeth showed slightly more dynamic mesenchymal expression of these genes: Bmp6 sparsely marked odontoblasts, while Igfbp5a and Lgr4 only marked younger tooth germs (Additional file 5: Fig. S5).
Discussion
Successional dental epithelia
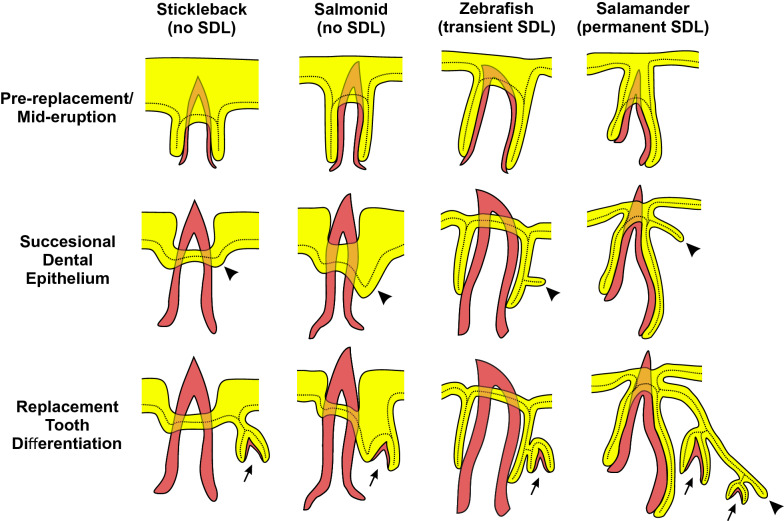
Across vertebrates, tooth replacement systems display variable arrangements, numbers of tooth replacement cycles, and levels of anatomical conspicuousness [9]. In some vertebrates, tooth replacement is morphologically well-defined and gives rise to many teeth per tooth position, while in other vertebrates, tooth replacement events are morphologically ill-defined (e.g., they lack an SDL), and/or temporally transient, in some cases only giving rise to a single tooth (e.g., humans and most other mammals that have only one wave of tooth replacement, a.k.a. diphyodonts) [9, 18, 42, 105]. Here we asked if two vertebrates with histologically distinct tooth regeneration systems exhibit similar localizations of orthologous gene expression consistent with the presence of a conserved tooth epithelial progenitor cell type. We hypothesized that, despite the morphological differences found across regenerative systems, vertebrate teeth might deploy a homologous cell type responsible for generating the epithelial portion of a replacement tooth, but that this cell type may not always be found sequestered within a deep dental lamina. Here we show that zebrafish and sticklebacks, which are around 250 million years diverged [60], express a conserved battery of nine genes in their newly emerging replacement tooth epithelia. We propose this battery of gene expression labels a homologous, conserved dental cell type: the successional dental epithelium (SDE). Most of these nine genes have required functions in vertebrate dentitions, including Bmp6, Bmpr1a, Igfbp5, Lgr4, Nfatc1, and Pitx2 [62, 91, 106–110], as well as other epithelial appendages (discussed below). Together, these data support a model wherein the successional dental epithelia (SDE) of these two species share a common evolutionary developmental history, despite the zebrafish SDE manifesting as a true histological SDL, while the stickleback SDE is histologically minute and is not arranged as an SDL (arrowheads in Fig. 2d and Additional file 1: Fig. S1). Overall, these data suggest a common cell type (the SDE) underlies tooth regeneration in divergent tooth regeneration systems, regardless of the presence of a morphologically obvious lamina or SDL (Fig. 6). We propose that SDE cells have thus retained a distinct cellular identity despite evolved changes in conformation, such as whether or not they participate in SDL formation. More work in other vertebrates will reveal the level of gene expression domain conservation across the SDE of different vertebrate groups.
Fig. 6.
Schematic of successional dental epithelia in different tooth regeneration systems. Each of the four columns of illustrations represents an idealized transverse cross-section of a tooth position from each of the polyphydont vertebrate groups indicated at the top. Each row of illustrations shows a different phase of the regeneration process as indicated on the left. Mineralized tissues are shown in red, and epithelia are shown in yellow. The continuous basalmost layer of epithelium that abuts the basement membrane is additionally delineated with a dotted line. Arrows indicate tooth germs, arrowheads indicate the successional dental epithelium (SDE) in each species as suggested by our model. The epithelia are depicted slightly off-plane (shallower on the z-plane) with respect to the erupted tooth (analogous to viewing a thick section). The left two columns (stickleback and salmonids) illustrate species that lack a successional dental lamina (SDL), whereas the right two columns illustrate species that do have an SDL; the zebrafish, which exhibits a transient SDL (note lack of SDL in the bottom row) vs. salamanders, which maintain a permanent SDL (arrowhead in bottom row). The stickleback drawings are representative of both oral and pharyngeal tooth regeneration and based on our own observations. The zebrafish pharyngeal tooth drawings are based on our own observations in conjunction with previous literature [42, 69]. The salmonid drawings are based on data from rainbow trout [47, 49] and Atlantic salmon [48, 136], and are based primarily on oral tooth histology (though pharyngeal teeth are described as behaving similarly in rainbow trout [49]). The salamander oral tooth drawings are based on data from the Iberian ribbed newt [137, 138] and axolotl [139].
Is the lack of a distinct successional dental lamina plesiomorphic, or derived?
Micro-CT studies found that tooth plate fossils can retain signatures of past replacement events. In the fossil species Andreolepis hedei, a stem bony vertebrate (osteichthyan), tooth replacement did not appear to have occurred in a static 1-for-1 fashion, and likely lacked a permanent SDL [45]. These fossil data suggest that early modes of tooth replacement in ancient bony vertebrates are grossly similar to the condition in stickleback and other bony fishes such as bichir, medaka, and cod [18, 47, 71]. While some have concluded that such a regenerative system is likely secondarily derived [31], it remains possible that these species have retained an ancient, less orderly tooth regeneration system (i.e., it is plesiomorphic), and that the presence of a successional dental lamina could be convergent in certain vertebrate groups, as recently proposed [45]. By analyzing the histology and gene expression of other ray-finned fishes’ dentitions, it could be possible to test if the tooth regeneration systems like those found in sticklebacks and other fishes are more likely a plesiomorphic or derived character.
Tooth number evolution in sticklebacks
We found that stickleback replacement tooth germs initiate possible “1-for-2” replacement events up to ~ 25% of the time. While our histological approach alone is unable to definitively determine the successional future or history at a given tooth position, our data leave open the possibility that adult tooth number variation in stickleback populations could be modulated by modifications to the replacement process, as previously posited [63]. Specifically, Ellis et al. (2015) found that marine and freshwater stickleback populations exhibited a consistent difference in adult (~ 40 mm standard length) bilateral ventral tooth plate number, with an average difference of around 25 teeth, an approximately 50% increase in tooth number in derived freshwater fish relative to ancestral marine fish. In this same study, tooth germ number was quantified on ventral tooth plates, and found to have an average difference of ~ 12 tooth germs in 40 mm adult fish, with freshwater fish from two different populations having more tooth germs than marine fish. As was previously addressed [63], these data suggest that the increased tooth numbers in freshwater stickleback populations relative to ancestral marine populations are likely not due to only differences in the appearance of new tooth germs; a difference in tooth shedding rate must also be at play. In line with these previous observations on inferred shedding rates, new histological data presented here suggest that the germ:shed ratio could be effectively doubled for some replacement events (1-for-1 vs 1-for-2) which conceivably could contribute to the overall tooth number differences seen between different populations. We hypothesize that up- or down-regulation of these “1-for-2” events may also contribute to these evolved tooth number differences in sticklebacks. Comparative studies on replacement events between multiple marine and freshwater stickleback populations could determine if “1-for-2” events differ in frequency between marine and freshwater fish, and if these differences could be related to genetic or developmental modifications to cell behavior or regeneration potential of the SDE.
Broader implications in epithelial appendage evolution and diversification
Ideas surrounding the homology of disparate epithelial appendages (EAs), like teeth and hair, have been proposed as early as Darwin’s On the Origin of Species [111], and were thereafter bolstered by morphological comparisons and pleiotropic phenotypes affecting teeth, hair, and other epithelial organs [112–114]. Some consensus regarding the homology or shared nature of the placode stage has been established [58, 59], while other work has indicated that larger-scale patterning mechanisms (e.g., concerted activity of Wnt, Eda, FGF, and SHH pathways) are at play across diverse EAs, including fish scales and amniote body coverings (reptile scales and feathers) [52, 115]. Distinguishing between uninterrupted ‘historical homology’ vs. deep homology [116, 117] of EAs like teeth and hair remains difficult to assay using only extant vertebrates; fossil evidence of intermediate forms could feasibly provide links between modernly disparate EAs. Although it is not clear whether teeth and hair are related by strict-sense historical homology or cooption, teeth, hair, and other epithelial appendages exhibit widespread similarities in their early development and regeneration [23, 57]. For example, in some rare instances, human dentitions form hair [118, 119], and Med1 mutant mouse dentitions form hair in the mouse incisor socket [120].
Here, we tested the hypothesis that two different tooth regeneration systems (zebrafish and stickleback) deploy similar patterns of gene expression (e.g., whether putative replacement epithelia have similar patterns of gene expression). Supporting this hypothesis, we found expression of a battery of nine candidate genes present in both the stickleback and zebrafish naïve dental epithelia, which we termed the SDE. Intriguingly, all nine of these genes are also expressed in mouse hair follicle stem cells, where most of these genes are known to play important roles in hair regeneration [66, 67, 90, 94, 101, 102, 104, 121–123]. This similarity in gene expression suggests that hair and teeth may regenerate using a shared epithelial cell type, namely one that responds to Bmp and Wnt signals during the regenerative cycle, with Bmps suppressing and Wnts facilitating differentiation. This work builds on previous studies on stickleback gene expression levels in ventral tooth plates of 254 known hair follicle stem cell gene orthologs, where this gene set was found to be reduced in Bmp6 mutants relative to wild type [62], and elevated in a high-toothed freshwater population relative to an ancestral low-toothed marine population [64]. Together, the expression profile similarities between hair follicle stem cells and dental tissues shown previously and presently will hopefully encourage further studies describing the shared and divergent developmental genetic mechanisms at play in these diverse organs. For example, the expression of the SDE markers reported here could be compared in other epithelial appendages such as amniote scales and feathers, where less is known about cellular and genetic bases of regeneration.
One curious feature of fish tooth regeneration addressed previously is the apparent lack of slow-cycling label-retaining cells (LRCs) in the bichir and salmon oral dentitions, which do not employ an SDL [18, 48]. Given that LRCs are present in other tooth regeneration systems [17, 41, 50, 124] and in the hair follicle stem cell niche [125–129], these findings could represent a major difference between some fish dentitions and other epithelial appendages, including other fish teeth [50]. Further studies on LRCs in other epithelial appendages, including other fish species’ teeth, will help delineate how common and/or consequential these changes in cell behavior might be. However, regardless of whether LRCs or a true epithelial stem cell niche exists across all fish dentitions, further elucidation of the shared gene expression and cellular processes between the SDE, hair follicle stem cells, and other epithelial appendages is warranted and ongoing, including defining functional responses to Bmp and Wnt signaling inputs, and testing function of genes that mark the SDE and/or mesenchymal tooth precursors during tooth regeneration.
Methods
Animal husbandry
Animals were raised following UC Berkeley IACUC protocol AUP-2015–01-7117. Zebrafish husbandry adhered to standard methods [130]. Sticklebacks were raised in 110-L aquaria in brackish water (3.5 g/L Instant Ocean salt, 0.0217 g/L sodium bicarbonate) at 17–18 °C in 8 h of light per day. Fish were fed live artemia as young fry, live artemia and frozen daphnia as young juveniles (1–2 cm standard length), frozen daphnia and frozen bloodworms as older juveniles (2–3 cm standard length), and frozen bloodworms and Mysis shrimp as sub-adults and adults (3 + cm standard length).
Generation of TCF/Lef-miniP stable transgenic lines
The TCF/Lef-miniP:dGFP plasmid was a gift from Tohru Ishitani [86]. To ‘restabilize’ GFP, we removed the destabilization signal by digesting the previously published plasmid with HindIII and ClaI, and filling this lesion with a short double-stranded piece of DNA that comprised two synthesized (Integrated DNA Technologies), phosphorylated, and annealed oligonucleotides. Forward oligo: 5′ AGCTTTAAAGCGACAT 3′, reverse oligo: 5′ CGATGTCGCTTTAA 3′. This change to the plasmid removed the majority of the destabilization signal, leaving just two extra amino acids (Lys, Leu) before a stop codon on the C terminus of eGFP. Zebrafish and stickleback transgenic lines were generated using both the unmodified and modified versions of the Tcf/Lef-miniP constructs according to standard methods in each species, using Tol2-driven transgenesis [131] by injecting zygotes at the one-cell stage [132, 133].
Alizarin Red staining
Alizarin Red was used to stain tooth plates as described previously [134]. In brief, whole fish were fixed in 10% neutrally buffered formalin overnight at room temperature and washed with tap water or 1 × phosphate-buffered saline with 0.1% Tween-20 (PBST) for 1 h at room temperature. Samples were then washed in a 0.008% Alizarin Red S solution in 1% KOH for 24 + hours. Once adequately stained, samples were transferred to 1% KOH for 1–3 days to continue clearing. Once cleared, tooth plates were dissected, further cleared in 50% glycerol for 1–5 days, flat-mounted, and imaged on a Leica DM 2500 compound microscope.
Sectioning
Stickleback sub-adults (~ 2 cm) derived from marine (Rabbit Slough [RABS]) and/or freshwater (Cerrito Creek [CERC] or Fishtrap Creek [FTC]) populations, and AB strain zebrafish adults or sub-adults (1.5–2 cm) were euthanized, decapitated, and fixed overnight in 4% formaldehyde (Sigma P6148) in 1 × phosphate-buffered saline (PBS) at 4° C with heavy agitation, washed 3 × 20 min with PBST on a nutator, then decalcified for 5–7 days (sticklebacks) or 3–5 days (zebrafish) in 20% ethylenediaminetetraacetic acid (EDTA, pH 8.0) at room temperature on a nutator. Once decalcified, specimens were again washed 3 × 10 min in PBST, then gradually stepped into 100% EtOH via 15–60 min washes in 30, 50, 70, and 100% EtOH in RNAse-free H2O. Samples were sometimes washed into MeOH and stored at this stage for up to 12 weeks. Samples were then washed for 1–2 h in 50/50 EtOH/Hemo-De at room temperature, then 1 h + in 100% Hemo-De at room temperature, then 1–2 h in 50/50 Hemo-De/paraffin (Paraplast X-tra, Fisher) at 65 °C, then rinsed and washed overnight at 65 °C in 100% paraffin. Fish heads were embedded in plastic molds with 100% paraffin (heated to 65 °C), mounted, sectioned on the sagittal (stickleback), off-sagittal (~ 30 degree tilt on the coronal axis; zebrafish), coronal (both species) or transverse (stickleback) planes with a Microm HM 340 E microtome. Sections were captured on Superfrost Plus slides, sometimes stored for up to 3 weeks at room temperature prior to analysis. To prepare slides for histological staining or ISH, slides were de-paraffinized (5 min incubation at 65 °C, let cool, submerge for 5 then 10 min in 100% Hemo-De, 5 min 100% EtOH, 10 min 80% EtOH/H2O, 10 min 100% H2O).
Hematoxylin and eosin (H&E) staining on sections
H&E staining was performed on sectioned tooth material using the following series of washes: hematoxylin solution (VWR) for 3 min, tap water for 2 × 20 s, bluing solution (0.1% sodium bicarbonate) for 2 min, tap water for 20 s, acid alcohol (0.32% HCl in 95% EtOH) for 20 s, tap water for 20 s, eosin solution (VWR) for 30 s, 95% EtOH for 2 × 2 min, 100% EtOH for 2 × 2 min, then Hemo-De for 2 × 5 min. Slides were then immediately coverslipped with Permount (Fisher), allowed to dry overnight, and imaged on a Leica DM2500 compound microscope.
In situ hybridizations on sections
All in situ hybridization (ISH) steps were performed on freshwater (CERC) stickleback, or AB strain zebrafish sections. The following steps were performed in LockMailer microscope slide jars (Sigma-Aldrich) in a volume of 9–11 mL unless stated otherwise. To begin the in situ process, slides were washed for 5 min in PBST, 5 min in proteinase K solution (15 μg/mL in PBST), rinsed briefly with PBST, then re-fixed for 20 min at room temperature in 4% formaldehyde in PBS. Fixative was then washed out with one PBST rinse and 2 × 10 min PBST washes before pre-hybridization. Slides were washed 2 × 5 min at room temperature in hybridization buffer, followed a long incubation in hybridization buffer (no probe, “pre-hyb” step) for 1–4 h at 67° C in a rotating hybridization oven (hybridization buffer is 50% formamide, 5 × SSC, 0.1% Tween, 5 mg/mL CHAPS, 1 mg/mL yeast RNA, 0.1 mg/mL heparin, pH 6.0 with citric acid). Riboprobes were generated essentially as described previously [68] against gene orthologs of interest (see Additional file 6: Table S1). Riboprobe templates were created either by PCR cloning gene fragments from cDNA or genomic DNA, or by synthesizing whole or partial inferred coding sequences (Gene Universal, Delaware, USA). See Additional file 6: Table S1 for a complete list of transcript accession numbers and sequences corresponding to exact riboprobe templates used in this study. Stickleback Bmp6 and Pitx2 riboprobes were previously published [61]. The zebrafish pitx2 riboprobe was previously published [135]. Zebrafish and stickleback CD34 orthologs were identified using the Genomicus synteny browser v100.01 (www.genomicus.biologie.ens.fr), which supported orthology of mammalian CD34 to the fish genes assayed here (ENSGACG00000011016 and ENSDARG00000095268). The translated products of these genes appear to code for proteins containing a partially recognizable CD34 domain (pfam06365). Riboprobes were synthesized with digoxigenin-labeled UTP and added at a concentration of ~ 100–500 ng/mL in 10 mL of hybridization buffer and agitated overnight in a rotating hybridization oven at 67 °C. Riboprobes in hybridization solution were stored at − 80° C and reused in some cases. The following day, six pre-heated hybridization washes at 67 °C in a rotating hybridization oven were performed for 20–90 min each, totaling 5–6 h of total “hyb wash” time (hyb wash is the same recipe as hyb buffer, excluding CHAPS, RNA, and heparin). Slides were then rinsed and washed in pre-heated Maleic acid buffer with Tween (MABT) at 67 °C for 20 min, then washed in pre-heated MABT for 20 min at room temperature (to allow for slow cooling). Slides were then removed from slide jars, placed in a humidor (a sealed plastic container with wet paper towels for moisture and pieces of plastic to raise slides form the bottom), and blocked with 70–100 μl (enough to cover all tissue on slides) of 2% Boehringer Blocking Reagent (BBR), covered with parafilm for 1 to 3 h at room temperature. Following the block step, block was poured off each slide, and anti-digoxigenin alkaline phosphatase conjugated antibody (Roche SKU 11093274910) was added at a concentration of 1:2000 in 2% BBR, again using enough to submerge each piece of tissue on each slide (70–100 μl) beneath parafilm and incubated at 4 °C overnight, or 3 h at room temperature in the dark. Importantly, tissue was not allowed to dry out beneath the parafilm during the block or antibody steps above. The following day, we performed one MABT rinse and 5 × 20–50 min MABT washes (back into slide jars) over the course of 3–4 h, agitated at room temperature, to wash out residual antibody, usually followed with an overnight MABT wash unagitated at 4 °C. To begin the coloration process, slides were changed into NTMT (0.1 M Tris pH 9.5, 0.05 M MgCl2, 0.1 M NaCl, 0.1% Tween) via 3 × 5–10 min washes before removing the final NTMT wash and replacing it with 10 mL of coloration solution (NTMT with 25 μg/mL Nitro blue tetrazolium chloride [NBT] and 175 μg/mL 5-bromo 4-chloro 3-indolyl phosphate [BCIP]). Signal development was carried out for 2–30 h to visualize mRNA localization. Once adequately developed, slides were rinsed, then washed for 10 min in PBST, fixed in 4% formaldehyde in PBS for 1–5 days at 4 °C. Slides were then washed 3 × 5 min in PBST. At this point, some select slides were counterstained with 0.1 μg/mL DAPI in 1 × PBST for 5–30 min at room temperature (the same 10 mL of solution was used to stain all sections shown in this paper, stored at 4 °C). To prepare slides for imaging, they were rinsed then washed 3 × 5 + min with deionized H2O, coverslipped with deionized H2O, and imaged on a Leica DM2500 microscope.
Quantification of 1-for-1 vs 1-for-2 tooth replacement in sticklebacks
To address the possibility of 1-for-2 tooth replacement in sticklebacks, we H&E-stained serial sections (described above) of nine sticklebacks of various sizes (20–40 mm) from various backgrounds (5 × CERC, 1 × FTC, 2 × RABS, 1 × marine/freshwater hybrid). Across these preparations, we identified a total of 67 pharyngeal tooth germs for which we had complete histological profiles at the mid- to late bell stage, that were also present in the middle of the tooth plate (i.e., surrounded by erupted teeth), which represent presumed or potential replacement teeth. Of these 67 tooth germs, 17 (25.4%) were clearly observed to be abutting and connected to bone remodeling events at the bases of two erupted teeth. These associations also appeared to contain an abundance of likely osteoclasts surrounding the disrupted bone in the erupted teeth (carets in Fig. 4), which we interpret as evidence of possible shedding events (or at the very least, loosening events) in-progress at the time of tissue preparation. We similarly located all oral tooth germs present on these slides, though the oral tooth fields were only visible on two sectioned specimens. We did not observe oral tooth germs as diverging from a 1-for-1 replacement scheme (n = 0/8).
Supplementary information
Additional file 1: Fig. S1. Histological plane orientation and description in sticklebacks. Illustrations at top depict a tooth (red cone) and indicate section planes with dotted rectangles, which are pictured below for each respective plane of section (labeled at top). These planes of section roughly translate to whole-animal anatomical planes. Left and middle illustrations show the sagittal and/or transverse plane (because stickleback teeth are mostly conical, these two planes of section appear generally similar in section). On these planes of section, teeth often appear off their medial axis (left vs center illustration, see labels). Rightmost illustration depicts the coronal plane, where teeth appear as rings of bone. Bottom images are of H&E-stained sections representing each illustrated plane of section above. Left bottom image shows a pharyngeal tooth on the transverse plane. Center bottom image shows an oral tooth on the sagittal plane. Right bottom image shows a pharyngeal tooth sectioned on the coronal plane. An erupted tooth (“et”) is indicated in the center image of an off-medial tooth (suggested with a red dotted line) sectioned on the sagittal plane. In all H&E images, tooth germs are indicated with an arrow, and the SDE is indicated with arrowheads. All scale bars = 50 μm.
Additional file 2: Fig. S2. DAPI counterstaining in situ hybridized sections aids in distinguishing epithelium from mesenchyme. Three gene expression patterns are shown as examples, labeled at top of columns in figure. Each column shows the same section corresponding to each gene, top two images are in brightfield, bottom two show DAPI fluorescence. 1st row of brightfield images are shown without any markup. 2nd row of brightfield images outline the basalmost epithelium with black dotted lines where possible (gray segments suggest borders out of the plane of section); bone is false-colored red. Expression domains in tooth germs are indicated with black arrows. Expression in the SDE is indicated with black arrowheads. Superficial epithelium with Gli1 expression adjacent to erupted teeth is indicated with white arrowheads. 3rd row of images shows the DAPI counterstain of each brightfield image above it, with the same epithelium outlines overlain (white dotted lines). 4th row shows DAPI images without any markup. Scale bar = 50 μm and applies to all panels.
Additional file 3: Fig. S3. Stickleback Dkk3b, Igfbp5b, and Bambia ISH on sections through dental tissue. The stickleback SDE was positive for Dkk3b and Igfbp5b (black arrowheads in top images), though these genes marked many cell populations across the dental field and surrounding tissues. Bambia was detected in tooth germ epithelium (black arrows) and mesenchyme (white arrows), and additionally in mature tooth mesenchyme, but never in the SDE (gray arrowheads). Scale bar = 50 μm and applies to all panels.
Additional file 4: Fig. S4. Zebrafish igfbp5b expression in deep tooth mesenchyme. Top row shows brightfield images, bottom row shows the corresponding DAPI counterstains to each brightfield image above. The SDL of zebrafish did not show igfbp5b expression (gray arrowhead). Expression was detected in a subset of the deep dental mesenchyme (white arrows). Scale bar = 50 μm and applies to all panels.
Additional file 5: Fig. S5. Expression of Bmp6, Bmpr1a, CD34, Gli1, Igfbp5a, Lgr4, Lgr6, Nfatc1, and Pitx2, highlighting mesenchymal expression. All of these genes were found to mark the SDE, except for Pitx2, and are expressed in some portion of dental mesenchyme (white arrows). Species and gene assayed are indicated in the figure. All scale bars = 50 μm.
Additional file 6: Table S1. Riboprobe sequence information. The first column (A) designates the species for which each riboprobe was designed. The second column (B) shows the gene symbol corresponding to each riboprobe. The third column (C) is the NCBI or Ensembl accession number representing the closest deposited sequence to the riboprobe template used. The fourth column (D) shows the original publication, if any, where the riboprobe activity was previously documented. The fifth column (E) contains the nucleotide sequence of the entire gene-specific portion of each riboprobe template, shown in the sense orientation (i.e. each riboprobe RNA sequence was the reverse complement of the sequence shown in column D).
Acknowledgements
We thank Andrew Glazer, Priscilla Erickson, and Justin Roncaioli for generating a subset of riboprobe templates, James Hart for assistance and advice on stickleback expression data, Amy Shyer and Richard Harland for microtome assistance and access, Sophie Archambeault, Alyssa Bormann, and Mark Stepaniak for valuable feedback on the manuscript, and two anonymous reviewers for their constructive feedback on the manuscript.
Authors’ contributions
TAS and CTM conceived the study. TAS designed the experiments. TAS, SBS, and EJM performed the experiments and collected the data. TAS prepared the figures. TAS and CTM wrote the manuscript. All authors provided input on the contents of the final manuscript. All authors read and approved the final manuscript.
Funding
TAS was supported by NIH Fellowship F32-DE027871 to TAS and CTM; TAS, SBS, EJM, and CTM were supported by NIH Grant R01-DE021475 to CTM.
Availability of data and materials
The datasets used and analyzed during the current study are available from the first author on reasonable request.
Ethics approval and consent to participate
Animals were raised following UC Berkeley IACUC protocol AUP-2015-01-7117.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tyler A. Square, Email: square@berkeley.edu
Craig T. Miller, Email: ctmiller@berkeley.edu
Supplementary information
The online version contains supplementary material available at 10.1186/s13227-021-00172-3.
References
- 1.Huysseune A, Sire J-Y. Evolution of patterns and processes in teeth and tooth-related tissues in non-mammalian vertebrates. Eur J Oral Sci. 1998;106:437–481. doi: 10.1111/j.1600-0722.1998.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 2.Oralová V, Rosa JT, Larionova D, Witten PE, Huysseune A. Multiple epithelia are required to develop teeth deep inside the pharynx. Proc Natl Acad Sci USA. 2020;117:11503–11512. doi: 10.1073/pnas.2000279117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sasagawa I, Ishiyama M, Yokosuka H, Mikami M, Uchida T. Tooth enamel and enameloid in actinopterygian fish. Front Mater Sci China. 2009;3:174. doi: 10.1007/s11706-009-0030-3. [DOI] [Google Scholar]
- 4.Smith MM, Hall BK. A developmental model for evolution of the vertebrate exoskeleton and teeth. In: Hecht MK, MacIntyre RJ, Clegg MT, editors. Evolutionary biology. Boston: Springer; 1993. pp. 387–448. [Google Scholar]
- 5.Aldridge RJ, Purnell MA. The conodont controversies. Trends Ecol Evol. 1996;11:463–468. doi: 10.1016/0169-5347(96)10048-3. [DOI] [PubMed] [Google Scholar]
- 6.Donoghue PCJ. Growth and patterning in the conodont skeleton. Philos Trans: Biolo Sci. 1998;353:633–666. doi: 10.1098/rstb.1998.0231. [DOI] [Google Scholar]
- 7.McCollum M, Sharpe PT. Evolution and development of teeth. J Anat. 2001;199:153–159. doi: 10.1046/j.1469-7580.2001.19910153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rücklin M, Donoghue PCJ. Romundina and the evolutionary origin of teeth. Biol Let. 2015;11:20150326. doi: 10.1098/rsbl.2015.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkovitz B, Shellis P. Chapter 10-tooth replacement and ontogeny of the dentition. In: Berkovitz B, Shellis P, editors. The teeth of non-mammalian vertebrates. Cambridge: Academic Press; 2017. pp. 255–289. [Google Scholar]
- 10.Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An ancient gene network is co-opted for teeth on old and new jaws. PLoS Biol. 2009;7:e1000031. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin KJ, Rasch LJ, Cooper RL, Metscher BD, Johanson Z, Fraser GJ. Sox2+ progenitors in sharks link taste development with the evolution of regenerative teeth from denticles. PNAS. 2016;113:14769–14774. doi: 10.1073/pnas.1612354113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rasch LJ, Martin KJ, Cooper RL, Metscher BD, Underwood CJ, Fraser GJ. An ancient dental gene set governs development and continuous regeneration of teeth in sharks. Dev Biol. 2016;415:347–370. doi: 10.1016/j.ydbio.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 13.Vandewalle P, Parmentier E, Chardon M. The branchial basket in Teleost feeding. Cybium. 2000;24:319–342. [Google Scholar]
- 14.Jernvall J, Thesleff I. Tooth shape formation and tooth renewal: evolving with the same signals. Development. 2012;139:3487–3497. doi: 10.1242/dev.085084. [DOI] [PubMed] [Google Scholar]
- 15.Carlson SJ. Vertebrate dental structures. Skelet Biominer Patt Process Evol Trends. 2013;1:531–556. [Google Scholar]
- 16.Stock DW. The genetic basis of modularity in the development and evolution of the vertebrate dentition. Phil Trans R Soc Lond B. 2001;356:1633–1653. doi: 10.1098/rstb.2001.0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salomies L, Eymann J, Khan I, Di-Poï N. The alternative regenerative strategy of bearded dragon unveils the key processes underlying vertebrate tooth renewal. eLife. 2019;8:e47702. doi: 10.7554/eLife.47702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenplas S, De Clercq A, Huysseune A. Tooth replacement without a dental lamina: the search for epithelial stem cells in Polypterus senegalus. J Exp Zool B Mol Dev Evol. 2014;322:281–293. doi: 10.1002/jez.b.22577. [DOI] [PubMed] [Google Scholar]
- 19.Fraser G, Thiery A. Evolution, Development, and Regeneration of Fish Dentitions. Cambridge University Press 978–1–107–17944–8 -Evolution and Development of Fishes. 2020.
- 20.Fraser GJ, Standing A, Underwood C, Thiery AP. The dental lamina: an essential structure for perpetual tooth regeneration in Sharks. Integr Comp Biol. 2020;60:644–655. doi: 10.1093/icb/icaa102. [DOI] [PubMed] [Google Scholar]
- 21.Tucker AS, Fraser GJ. Evolution and developmental diversity of tooth regeneration. Semin Cell Dev Biol. 2014;25–26:71–80. doi: 10.1016/j.semcdb.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- 23.Chuong C-M, Randall VA, Widelitz RB, Wu P, Jiang T-X. Physiological regeneration of skin appendages and implications for regenerative medicine. Physiology. 2012;27:61–72. doi: 10.1152/physiol.00028.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Von Euw S, Wang Y, Laurent G, Drouet C, Babonneau F, Nassif N, et al. Bone mineral: new insights into its chemical composition. Sci Rep. 2019;9:8456. doi: 10.1038/s41598-019-44620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Botella H, Valenzuela-Ríos JI, Martínez-Pérez C. Tooth replacement rates in early chondrichthyans: a qualitative approach. Lethaia. 2009;42:365–376. doi: 10.1111/j.1502-3931.2009.00152.x. [DOI] [Google Scholar]
- 26.Breder CM. The shedding of teeth by Carcharias littoralis (Mitchill) Copeia. 1942;1942:42–44. doi: 10.2307/1437945. [DOI] [Google Scholar]
- 27.Correia JP. Tooth loss rate from two captive sandtiger sharks (Carcharias taurus) Zoo Biol. 1999;18:313–317. doi: 10.1002/(SICI)1098-2361(1999)18:4<313::AID-ZOO5>3.0.CO;2-D. [DOI] [Google Scholar]
- 28.Luer CA, Blum PC, Gilbert PW. Rate of tooth replacement in the nurse shark, Ginglymostoma cirratum. Copeia. 1990;1990:182–191. doi: 10.2307/1445834. [DOI] [Google Scholar]
- 29.Overstrom NA. Estimated tooth replacement rate in captive sand tiger sharks (Carcharias taurus Rafinesque, 1810) Copeia. 1991;1991:525. doi: 10.2307/1446602. [DOI] [Google Scholar]
- 30.Wass RC. Size, growth, and reproduction of the Sandbar Shark, Carcharhinus milberti, in Hawaii. Hawaii: University of Hawaii Press; 1973. [Google Scholar]
- 31.Reif W-E. Evolution of dermal skeleton and dentition in vertebrates. In: Hecht MK, Wallace B, Prance GT, editors. Evolutionary biology. Boston: Springe; 1982. [Google Scholar]
- 32.Popa EM, Buchtova M, Tucker AS. Revitalising the rudimentary replacement dentition in the mouse. Development; 2019. [DOI] [PubMed]
- 33.Kague E, Witten PE, Soenens M, Campos CL, Lubiana T, Fisher S, et al. Zebrafish sp7 mutants show tooth cycling independent of attachment, eruption and poor differentiation of teeth. Dev Biol. 2018;435:176–184. doi: 10.1016/j.ydbio.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 34.Kamat M, Puranik R, Vanaki S, Kamat S. An insight into the regulatory mechanisms of cells involved in resorption of dental hard tissues. J Oral Maxillofac Pathol. 2013;17:228–233. doi: 10.4103/0973-029X.119736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantoku A, Chatani M, Aono K, Inohaya K, Kudo A. Osteoblast and osteoclast behaviors in the turnover of attachment bones during medaka tooth replacement. Dev Biol. 2016;409:370–381. doi: 10.1016/j.ydbio.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 36.To TT, Witten PE, Huysseune A, Winkler C. An adult osteopetrosis model in medaka reveals the importance of osteoclast function for bone remodeling in teleost fish. Comp Biochem Physiol C: Toxicol Pharmacol. 2015;178:68–75. doi: 10.1016/j.cbpc.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 37.Witten PE, Huysseune A. A comparative view on mechanisms and functions of skeletal remodelling in teleost fish, with special emphasis on osteoclasts and their function. Biol Rev. 2009;84:315–346. doi: 10.1111/j.1469-185X.2009.00077.x. [DOI] [PubMed] [Google Scholar]
- 38.Abduweli D, Baba O, Tabata MJ, Higuchi K, Mitani H, Takano Y. Tooth replacement and putative odontogenic stem cell niches in pharyngeal dentition of medaka (Oryzias latipes) Microscopy. 2014;63:141–153. doi: 10.1093/jmicro/dft085. [DOI] [PubMed] [Google Scholar]
- 39.Gaete M, Tucker AS. Organized emergence of multiple-generations of teeth in snakes is dysregulated by activation of Wnt/Beta-catenin signalling. PLoS ONE. 2013;8:e74484. doi: 10.1371/journal.pone.0074484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graver HT. The polarity of the dental lamina in the regenerating salamander jaw. Dev. 1973;30:635–646. [PubMed] [Google Scholar]
- 41.Handrigan GR, Leung KJ, Richman JM. Identification of putative dental epithelial stem cells in a lizard with life-long tooth replacement. Development. 2010;137:3545–3549. doi: 10.1242/dev.052415. [DOI] [PubMed] [Google Scholar]
- 42.Huysseune A. Formation of a successional dental lamina in the zebrafish (Danio rerio): support for a local control of replacement tooth initiation. Int J Dev Biol. 2006;50:637–643. doi: 10.1387/ijdb.052098ah. [DOI] [PubMed] [Google Scholar]
- 43.Moriyama K, Watanabe S, Iida M, Sahara N. Plate-like permanent dental laminae of upper jaw dentition in adult gobiid fish, Sicyopterus japonicus. Cell Tissue Res. 2010;340:189–200. doi: 10.1007/s00441-010-0935-2. [DOI] [PubMed] [Google Scholar]
- 44.Popa EM, Anthwal N, Tucker AS. Complex patterns of tooth replacement revealed in the fruit bat (Eidolon helvum) J Anat. 2016;229:847–856. doi: 10.1111/joa.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen D, Blom H, Sanchez S, Tafforeau P, Ahlberg PE. The stem osteichthyan Andreolepis and the origin of tooth replacement. Nature. 2016;539:237–241. doi: 10.1038/nature19812. [DOI] [PubMed] [Google Scholar]
- 46.Vaškaninová V, Chen D, Tafforeau P, Johanson Z, Ekrt B, Blom H, et al. Marginal dentition and multiple dermal jawbones as the ancestral condition of jawed vertebrates. Science. 2020;369:211–216. doi: 10.1126/science.aaz9431. [DOI] [PubMed] [Google Scholar]
- 47.Fraser GJ, Berkovitz BK, Graham A, Smith MM. Gene deployment for tooth replacement in the rainbow trout (Oncorhynchus mykiss): a developmental model for evolution of the osteichthyan dentition. Evol Dev. 2006;8:446–457. doi: 10.1111/j.1525-142X.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- 48.Vandenplas S, Willems M, Witten PE, Hansen T, Fjelldal PG, Huysseune A. Epithelial label-retaining cells are absent during tooth cycling in Salmo salar and Polypterus senegalus. PLoS ONE. 2016;11:e0152870. doi: 10.1371/journal.pone.0152870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser GJ, Graham A, Smith MM. Conserved deployment of genes during odontogenesis across osteichthyans. Proc R Soc Lond B. 2004;271:2311–2317. doi: 10.1098/rspb.2004.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bloomquist RF, Fowler TE, An Z, Yu TY, Abdilleh K, Fraser GJ, et al. Developmental plasticity of epithelial stem cells in tooth and taste bud renewal. Natl Acad Sci. 2019;116:17858–17866. doi: 10.1073/pnas.1821202116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Debiais-Thibaud M, Borday-Birraux V, Germon I, Bourrat F, Metcalfe CJ, Casane D, et al. Development of oral and pharyngeal teeth in the medaka (Oryzias latipes): comparison of morphology and expression of eve1 gene. J Exp Zool B Mol Dev Evol. 2007;308:693–708. doi: 10.1002/jez.b.21183. [DOI] [PubMed] [Google Scholar]
- 52.Aman AJ, Fulbright AN, Parichy DM. Wnt/β-catenin regulates an ancient signaling network during zebrafish scale development. ELife. 2018;7:e37001. doi: 10.7554/eLife.37001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Debiais-Thibaud M, Oulion S, Bourrat F, Laurenti P, Casane D, Borday-Birraux V. The homology of odontodes in gnathostomes: insights from Dlx gene expression in the dogfish Scyliorhinus canicula. BMC Evol Biol. 2011;11:307. doi: 10.1186/1471-2148-11-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Di-Poï N, Milinkovitch MC. The anatomical placode in reptile scale morphogenesis indicates shared ancestry among skin appendages in amniotes. Sci Adv. 2016;2(6):e1600708. doi: 10.1126/sciadv.1600708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mikkola ML. Genetic basis of skin appendage development. Semin Cell Dev Biol. 2007;18:225–236. doi: 10.1016/j.semcdb.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Moss ML. Comparative anatomy of vertebrate dermal bone and teeth. I. The epidermal co-participation hypothesis. Acta Anat. 1968;71:178–208. doi: 10.1159/000143185. [DOI] [PubMed] [Google Scholar]
- 57.Naveau A, Seidel K, Klein OD. Tooth, hair and claw: comparing epithelial stem cell niches of ectodermal appendages. Exp Cell Res. 2014;325:96–103. doi: 10.1016/j.yexcr.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/S0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 59.Sharpe PT. Fish scale development: hair today, teeth and scales yesterday? Curr Biol. 2001;11:R751–R752. doi: 10.1016/S0960-9822(01)00438-9. [DOI] [PubMed] [Google Scholar]
- 60.Near TJ, Eytan RI, Dornburg A, Kuhn KL, Moore JA, Davis MP, et al. Resolution of ray-finned fish phylogeny and timing of diversification. Natl Acad Sci. 2012;109:13698–13703. doi: 10.1073/pnas.1206625109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cleves PA, Ellis NA, Jimenez MT, Nunez SM, Schluter D, Kingsley DM, et al. Evolved tooth gain in sticklebacks is associated with a cis-regulatory allele of Bmp6. Natl Acad Sci. 2014;111:13912–13917. doi: 10.1073/pnas.1407567111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cleves PA, Hart JC, Agoglia RM, Jimenez MT, Erickson PA, Gai L, et al. An intronic enhancer of Bmp6 underlies evolved tooth gain in sticklebacks. PLoS Genet. 2018;14:e1007449. doi: 10.1371/journal.pgen.1007449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ellis NA, Glazer AM, Donde NN, Cleves PA, Agoglia RM, Miller CT. Distinct developmental genetic mechanisms underlie convergently evolved tooth gain in sticklebacks. Development. 2015;142:2442–2451. doi: 10.1242/dev.124248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hart JC, Ellis NA, Eisen MB, Miller CT. Convergent evolution of gene expression in two high-toothed stickleback populations. PLoS Genet. 2018;14:e1007443. doi: 10.1371/journal.pgen.1007443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller CT, Glazer AM, Summers BR, Blackman BK, Norman AR, Shapiro MD, et al. Modular skeletal evolution in sticklebacks is controlled by additive and clustered quantitative trait loci. Genetics. 2014;197:405–420. doi: 10.1534/genetics.114.162420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abe Y, Tanaka N. Roles of the Hedgehog Signaling Pathway in Epidermal and Hair Follicle Development, Homeostasis, and Cancer. J Dev Biol [Internet]. 2017 [cited 2020 Jul 6];5. [DOI] [PMC free article] [PubMed]
- 67.Kandyba E, Leung Y, Chen Y-B, Widelitz R, Chuong C-M, Kobielak K. Competitive balance of intrabulge BMP/Wnt signaling reveals a robust gene network ruling stem cell homeostasis and cyclic activation. Natl Acad Sci. 2013;110:1351–1356. doi: 10.1073/pnas.1121312110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ellis NA, Donde NN, Miller CT. Early development and replacement of the stickleback dentition. J Morphol. 2016;277:1072–1083. doi: 10.1002/jmor.20557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van der Heyden C, Huysseune A. Dynamics of tooth formation and replacement in the zebrafish (Danio rerio) (Teleostei, Cyprinidae) Dev Dyn. 2000;219:486–496. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1069>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 70.Pasco-Viel E, Charles C, Chevret P, Semon M, Tafforeau P, Viriot L, et al. Evolutionary trends of the pharyngeal dentition in Cypriniformes (Actinopterygii: Ostariophysi) PLoS ONE. 2010;5:e11293. doi: 10.1371/journal.pone.0011293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holmbakken N, Fosse G. Tooth replacement in Gadus callarias. Z Anat Entwicklungsgesch. 1973;143:65–79. doi: 10.1007/BF00519911. [DOI] [PubMed] [Google Scholar]
- 72.Chen J, Lan Y, Baek J-A, Gao Y, Jiang R. Wnt/beta-catenin signaling plays an essential role in activation of odontogenic mesenchyme during early tooth development. Dev Biol. 2009;334:174–185. doi: 10.1016/j.ydbio.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huysseune A, Soenens M, Elderweirdt F. Wnt signaling during tooth replacement in zebrafish (Danio rerio): pitfalls and perspectives. Front Physiol. 2014;5:386. doi: 10.3389/fphys.2014.00386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Järvinen E, Salazar-Ciudad I, Birchmeier W, Taketo MM, Jernvall J, Thesleff I. Continuous tooth generation in mouse is induced by activated epithelial Wnt/β-catenin signaling. Natl Acad Sci. 2006;103:18627–18632. doi: 10.1073/pnas.0607289103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu F, Millar SE. Wnt/β-catenin signaling in oral tissue development and disease. J Dent Res. 2010;89:318–330. doi: 10.1177/0022034510363373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shim JS, Kim B, Park HC, Ryu JJ. Temporal control of WNT activity regulates tooth number in fish. J Dent Res. 2019;98:339–346. doi: 10.1177/0022034518811322. [DOI] [PubMed] [Google Scholar]
- 77.Callahan N, Modesto A, Meira R, Seymen F, Patir A, Vieira AR. Axis inhibition protein 2 (AXIN2) polymorphisms and tooth agenesis. Arch Oral Biol. 2009;54:45–49. doi: 10.1016/j.archoralbio.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He H, Han D, Feng H, Qu H, Song S, Bai B, et al. Involvement of and Interaction between WNT10A and EDA Mutations in Tooth Agenesis Cases in the Chinese Population. PLOS ONE. Public Library of Science; 2013;8:e80393. [DOI] [PMC free article] [PubMed]
- 79.Kantaputra PN, Kapoor S, Verma P, Kaewgahya M, Kawasaki K, Ohazama A, et al. Al-Awadi-Raas-Rothschild syndrome with dental anomalies and a novel WNT7A mutation. European Journal of Medical Genetics. 2017;60:695–700. doi: 10.1016/j.ejmg.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 80.Lammi L, Arte S, Somer M, Järvinen H, Lahermo P, Thesleff I, et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet. 2004;74:1043–1050. doi: 10.1086/386293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nawaz S, Klar J, Wajid M, Aslam M, Tariq M, Schuster J, et al. WNT10A missense mutation associated with a complete odonto-onycho-dermal dysplasia syndrome. Eur J Hum Genet. 2009;17:1600–1605. doi: 10.1038/ejhg.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ockeloen CW, Khandelwal KD, Dreesen K, Ludwig KU, Sullivan R, van Rooij IALM, et al. Novel mutations in LRP6 highlight the role of WNT signaling in tooth agenesis. Genet Med. 2016;18:1158–1162. doi: 10.1038/gim.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang B, Li H, Liu Y, Lin X, Lin Y, Wang Y, et al. Expression patterns of WNT/β-CATENIN signaling molecules during human tooth development. J Mol Hist. 2014;45:487–496. doi: 10.1007/s10735-014-9572-5. [DOI] [PubMed] [Google Scholar]
- 84.Xu M, Horrell J, Snitow M, Cui J, Gochnauer H, Syrett CM, et al. WNT10A mutation causes ectodermal dysplasia by impairing progenitor cell proliferation and KLF4-mediated differentiation. Nat Commun. 2017;8:15397. doi: 10.1038/ncomms15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiese KE, Nusse R, van Amerongen R. Wnt signalling: conquering complexity. Development. 2018;145:165902. doi: 10.1242/dev.165902. [DOI] [PubMed] [Google Scholar]
- 86.Shimizu N, Kawakami K, Ishitani T. Visualization and exploration of Tcf/Lef function using a highly responsive Wnt/β-catenin signaling-reporter transgenic zebrafish. Dev Biol. 2012;370:71–85. doi: 10.1016/j.ydbio.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 87.Verstraeten B, Sanders E, van Hengel J, Huysseune A. Zebrafish teeth as a model for repetitive epithelial morphogenesis: dynamics of E-cadherin expression. BMC Dev Biol. 2010;10:58. doi: 10.1186/1471-213X-10-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verstraeten B, van Hengel J, Huysseune A. Beta-catenin and plakoglobin expression during zebrafish tooth development and replacement. PLoS ONE. 2016;11:e0148114. doi: 10.1371/journal.pone.0148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Biehs B, Hu JK, Strauli NB, Sangiorgi E, Jung H, Heber R-P, et al. Bmi1 represses Ink4a/Arf and Hox genes to regulate stem cells in the rodent incisor. Nat Cell Biol. 2013;15:846–852. doi: 10.1038/ncb2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu P, Zhang Y, Xing Y, Xu W, Guo H, Deng F, et al. The balance of Bmp6 and Wnt10b regulates the telogen–anagen transition of hair follicles. Cell Commun Signal. 2019;17:16. doi: 10.1186/s12964-019-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andl T, Ahn K, Kairo A, Chu EY, Wine-Lee L, Reddy ST, et al. Epithelial Bmpr1a regulates differentiation and proliferation in postnatal hair follicles and is essential for tooth development. Development. 2004;131:2257–2268. doi: 10.1242/dev.01125. [DOI] [PubMed] [Google Scholar]
- 92.Lorz C, García-Escudero R, Segrelles C, Garín MI, Ariza JM, Santos M, et al. A Functional role of RB-dependent pathway in the control of quiescence in adult epidermal stem cells revealed by genomic profiling. Stem Cell Rev. 2010;6:162–177. doi: 10.1007/s12015-010-9139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martínez-Martínez E, Galván-Hernández CI, Toscano-Márquez B, Gutiérrez-Ospina G. Modulatory role of sensory innervation on hair follicle stem cell progeny during wound healing of the rat skin. PLoS ONE. 2012;7:e36421. doi: 10.1371/journal.pone.0036421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trempus CS, Morris RJ, Ehinger M, Elmore A, Bortner CD, Ito M, et al. CD34 Expression by hair follicle stem cells is required for skin tumor development in mice. Cancer Res. 2007;67:4173–4181. doi: 10.1158/0008-5472.CAN-06-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-Renewal, multipotency, and the existence of two cell populations within an epithelial stem cell Niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 97.Trempus CS, Morris RJ, Bortner CD, Cotsarelis G, Faircloth RS, Reece JM, et al. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J Invest Dermatol. 2003;120:501–511. doi: 10.1046/j.1523-1747.2003.12088.x. [DOI] [PubMed] [Google Scholar]
- 98.Peterson SC, Eberl M, Vagnozzi AN, Belkadi A, Veniaminova NA, Verhaegen ME, et al. Basal cell carcinoma preferentially arises from stem cells within hair follicle and mechanosensory niches. Cell Stem Cell. 2015;16:400–412. doi: 10.1016/j.stem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, et al. Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. eLife. 2017;6:e24712. doi: 10.7554/eLife.24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Walker JV, Zhuang H, Singer D, Illsley CS, Kok WL, Sivaraj KK, et al. Transit amplifying cells coordinate mouse incisor mesenchymal stem cell activation. Nat Commun. 2019;10:3596. doi: 10.1038/s41467-019-11611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327:1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 102.Keyes BE, Segal JP, Heller E, Lien W-H, Chang C-Y, Guo X, et al. Nfatc1 orchestrates aging in hair follicle stem cells. Proc Natl Acad Sci USA. 2013;110:E4950–4959. doi: 10.1073/pnas.1320301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sun Z, Yu W, Navarro MS, Sweat M, Eliason S, Sharp T, et al. Sox2 and Lef-1 interact with Pitx2 to regulate incisor development and stem cell renewal. Development. 2016;143:4115–4126. doi: 10.1242/dev.138883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sohn K-C, Shi G, Jang S, Choi D-K, Lee Y, Yoon T-J, et al. Pitx2, a β-catenin-regulated transcription factor, regulates the differentiation of outer root sheath cells cultured in vitro. J Dermatol Sci. 2009;54:6–11. doi: 10.1016/j.jdermsci.2008.11.006. [DOI] [PubMed] [Google Scholar]
- 105.Bertonnier-Brouty L, Viriot L, Joly T, Charles C. Morphological features of tooth development and replacement in the rabbit Oryctolagus cuniculus. Arch Oral Biol. 2020;109:104576. doi: 10.1016/j.archoralbio.2019.104576. [DOI] [PubMed] [Google Scholar]
- 106.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, et al. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118:3775–3789. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ji Y, Buel SM, Amack JD. Mutations in zebrafish pitx2 model congenital malformations in Axenfeld-Rieger syndrome but do not disrupt left-right placement of visceral organs. Dev Biol. 2016;416:69–81. doi: 10.1016/j.ydbio.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, et al. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 109.Saito K, Ohshima H. The putative role of insulin-like growth factor (IGF)-binding protein 5 independent of IGF in the maintenance of pulpal homeostasis in mice. Regen Ther. 2019;11:217–224. doi: 10.1016/j.reth.2019.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamakami Y, Kohashi K, Oyama K, Mohri Y, Hidema S, Nishimori K. LGR4 is required for sequential molar development. Biochem Biophys Rep. 2016;8:174–183. doi: 10.1016/j.bbrep.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Darwin C. On the origin of species by means of natural selection, or the Preservation of Favoured Races in the Struggle for Life. Darwin C. On the origin of species by means of natuLondon/Die Entstehung der Arten durch natürliche Zuchtwahl, Leipzig oJ; 1859.
- 112.Danforth CH. Hair in its relation to questions of homology and phylogeny. Am J Anatomy. 1925;36:47–68. doi: 10.1002/aja.1000360103. [DOI] [Google Scholar]
- 113.El-Mangoury MH. Homology between teeth and hair. J Egypt Med Assoc. 1954;37:676–679. [PubMed] [Google Scholar]
- 114.Guilford SH. The Teeth and Hair. Am J Dent Sci. 1894;27:471–474. [PMC free article] [PubMed] [Google Scholar]
- 115.McKinnell IW, Turmaine M, Patel K. Sonic Hedgehog functions by localizing the region of proliferation in early developing feather buds. Dev Biol. 2004;272:76–88. doi: 10.1016/j.ydbio.2004.04.019. [DOI] [PubMed] [Google Scholar]
- 116.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 117.Tschopp P, Tabin CJ. Deep homology in the age of next-generation sequencing. Philos Trans Royal Soci B: Biol Sci. 2017;372:20150475. doi: 10.1098/rstb.2015.0475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Agha-Hosseini F, Etesam F, Rohani B. A boy with oral hair: case report. Med Oral Patol Oral Cir Bucal. 2007;12:E357–359. [PubMed] [Google Scholar]
- 119.Zhurakivska K, Toni G, Laino G, Franco R, Troiano G, Laino L, et al. An unusual case of recurrent gingival hirsutism. Oral Surg Oral Med Oral Pathol Oral Radiol. 2020;129:e200–e203. doi: 10.1016/j.oooo.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 120.Yoshizaki K, Hu L, Nguyen T, Sakai K, He B, Fong C, et al. Ablation of coactivator Med1 switches the cell fate of dental epithelia to that generating hair. PLoS ONE. 2014;9:e99991. doi: 10.1371/journal.pone.0099991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Brownell I, Guevara E, Bai CB, Loomis CA, Joyner AL. Nerve-derived Sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell. 2011;8:552–565. doi: 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. PNAS Natl Acad Sci. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mignone JL, Roig-Lopez JL, Fedtsova N, Schones DE, Manganas LN, Maletic-Savatic M, et al. Neural potential of a stem cell population in the hair follicle. Cell Cycle. 2007;6:2161–2170. doi: 10.4161/cc.6.17.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu P, Wu X, Jiang T-X, Elsey RM, Temple BL, Divers SJ, et al. Specialized stem cell niche enables repetitive renewal of alligator teeth. PNAS Natl Acad Sci. 2013;110:E2009–E2018. doi: 10.1073/pnas.1213202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Braun KM, Niemann C, Jensen UB, Sundberg JP, Silva-Vargas V, Watt FM. Manipulation of stem cell proliferation and lineage commitment: visualisation of label-retaining cells in wholemounts of mouse epidermis. Development The Company of Biologists Ltd. 2003;130:5241–5255. doi: 10.1242/dev.00703. [DOI] [PubMed] [Google Scholar]
- 126.Cotsarelis G, Sun T-T, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-C. [DOI] [PubMed] [Google Scholar]
- 127.Morris RJ, Potten CS. Highly persistent label-retaining cells in the hair follicles of mice and their fate following induction of anagen. J Investig Dermatol. 1999;112:470–475. doi: 10.1046/j.1523-1747.1999.00537.x. [DOI] [PubMed] [Google Scholar]
- 128.Rodriguez CN, Nguyen H. Identifying quiescent stem cells in hair follicles. Methods Mol Biol. 2018;1686:137–147. doi: 10.1007/978-1-4939-7371-2_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.da Silva-Diz V, Solé-Sánchez S, Valdés-Gutiérrez A, Urpí M, Riba-Artés D, Penin RM, et al. Progeny of Lgr5-expressing hair follicle stem cell contributes to papillomavirus-induced tumor development in epidermis. Oncogene. 2013;32:3732–3743. doi: 10.1038/onc.2012.375. [DOI] [PubMed] [Google Scholar]
- 130.Sprague J, Doerry E, Douglas S, Westerfield M. The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Res. 2001;29:87–90. doi: 10.1093/nar/29.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 132.Erickson PA, Cleves PA, Ellis NA, Schwalbach KT, Hart JC, Miller CT. A 190 base pair, TGF-β responsive tooth and fin enhancer is required for stickleback Bmp6 expression. Dev Biol. 2015;401:310–323. doi: 10.1016/j.ydbio.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- 134.Ellis NA, Miller CT. Dissection and Flat-mounting of the Threespine Stickleback Branchial Skeleton. JoVE (Journal of Visualized Experiments). 2016;e54056. [DOI] [PMC free article] [PubMed]
- 135.Essner JJ, Branford WW, Zhang J, Yost HJ. Mesendoderm and left-right brain, heart and gut development are differentially regulated by pitx2 isoforms. Development The Company of Biologists Ltd. 2000;127:1081–1093. doi: 10.1242/dev.127.5.1081. [DOI] [PubMed] [Google Scholar]
- 136.Huysseune A, Sire J-Y, Witten PE. Evolutionary and developmental origins of the vertebrate dentition. J Anat. 2009;214:465–476. doi: 10.1111/j.1469-7580.2009.01053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Davit-Béal T, Allizard F, Sire J-Y. Morphological variations in a tooth family through ontogeny in Pleurodeles waltl (Lissamphibia, Caudata) J Morphol. 2006;267:1048–1065. doi: 10.1002/jmor.10455. [DOI] [PubMed] [Google Scholar]
- 138.Berkovitz B, Shellis P. Chapter 9 - Tooth Formation. In: Berkovitz B, Shellis P, editors. The Teeth of Non-Mammalian Vertebrates [Internet]. Academic Press; 2017.
- 139.Fraser GJ, Graham A, Smith MM. Developmental and evolutionary origins of the vertebrate dentition: molecular controls for spatio-temporal organisation of tooth sites in osteichthyans. J Exp Zool B Mol Dev Evol. 2006;306:183–203. doi: 10.1002/jez.b.21097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Histological plane orientation and description in sticklebacks. Illustrations at top depict a tooth (red cone) and indicate section planes with dotted rectangles, which are pictured below for each respective plane of section (labeled at top). These planes of section roughly translate to whole-animal anatomical planes. Left and middle illustrations show the sagittal and/or transverse plane (because stickleback teeth are mostly conical, these two planes of section appear generally similar in section). On these planes of section, teeth often appear off their medial axis (left vs center illustration, see labels). Rightmost illustration depicts the coronal plane, where teeth appear as rings of bone. Bottom images are of H&E-stained sections representing each illustrated plane of section above. Left bottom image shows a pharyngeal tooth on the transverse plane. Center bottom image shows an oral tooth on the sagittal plane. Right bottom image shows a pharyngeal tooth sectioned on the coronal plane. An erupted tooth (“et”) is indicated in the center image of an off-medial tooth (suggested with a red dotted line) sectioned on the sagittal plane. In all H&E images, tooth germs are indicated with an arrow, and the SDE is indicated with arrowheads. All scale bars = 50 μm.
Additional file 2: Fig. S2. DAPI counterstaining in situ hybridized sections aids in distinguishing epithelium from mesenchyme. Three gene expression patterns are shown as examples, labeled at top of columns in figure. Each column shows the same section corresponding to each gene, top two images are in brightfield, bottom two show DAPI fluorescence. 1st row of brightfield images are shown without any markup. 2nd row of brightfield images outline the basalmost epithelium with black dotted lines where possible (gray segments suggest borders out of the plane of section); bone is false-colored red. Expression domains in tooth germs are indicated with black arrows. Expression in the SDE is indicated with black arrowheads. Superficial epithelium with Gli1 expression adjacent to erupted teeth is indicated with white arrowheads. 3rd row of images shows the DAPI counterstain of each brightfield image above it, with the same epithelium outlines overlain (white dotted lines). 4th row shows DAPI images without any markup. Scale bar = 50 μm and applies to all panels.
Additional file 3: Fig. S3. Stickleback Dkk3b, Igfbp5b, and Bambia ISH on sections through dental tissue. The stickleback SDE was positive for Dkk3b and Igfbp5b (black arrowheads in top images), though these genes marked many cell populations across the dental field and surrounding tissues. Bambia was detected in tooth germ epithelium (black arrows) and mesenchyme (white arrows), and additionally in mature tooth mesenchyme, but never in the SDE (gray arrowheads). Scale bar = 50 μm and applies to all panels.
Additional file 4: Fig. S4. Zebrafish igfbp5b expression in deep tooth mesenchyme. Top row shows brightfield images, bottom row shows the corresponding DAPI counterstains to each brightfield image above. The SDL of zebrafish did not show igfbp5b expression (gray arrowhead). Expression was detected in a subset of the deep dental mesenchyme (white arrows). Scale bar = 50 μm and applies to all panels.
Additional file 5: Fig. S5. Expression of Bmp6, Bmpr1a, CD34, Gli1, Igfbp5a, Lgr4, Lgr6, Nfatc1, and Pitx2, highlighting mesenchymal expression. All of these genes were found to mark the SDE, except for Pitx2, and are expressed in some portion of dental mesenchyme (white arrows). Species and gene assayed are indicated in the figure. All scale bars = 50 μm.
Additional file 6: Table S1. Riboprobe sequence information. The first column (A) designates the species for which each riboprobe was designed. The second column (B) shows the gene symbol corresponding to each riboprobe. The third column (C) is the NCBI or Ensembl accession number representing the closest deposited sequence to the riboprobe template used. The fourth column (D) shows the original publication, if any, where the riboprobe activity was previously documented. The fifth column (E) contains the nucleotide sequence of the entire gene-specific portion of each riboprobe template, shown in the sense orientation (i.e. each riboprobe RNA sequence was the reverse complement of the sequence shown in column D).
Data Availability Statement
The datasets used and analyzed during the current study are available from the first author on reasonable request.