Abstract
Ramie (Boehmeria nivea L. Gaud) is a traditional fiber crop and important medicinal plant belonging to the family Urticaceae. In this study, we determine the complete chloroplast genome sequence of B. nivea. The assembled chloroplast genome is 156065 bp in length and shares the conserved quadripartite structure as other cp genomes in Boehmeria. The genome contains 131 genes, including 84 protein genes, 8 rRNA genes, 37 tRNA genes and 2 pseudo genes. There are 17 duplicated genes in the IR region. The overall GC content of B. nivea is 36.33%, with the highest GC content of 42.72% in IR region. A total of 67 simple sequence repeats are identified in the cp genome of B. nivea. Phylogenetic analysis demonstrated that B. nivea clustered together with B. tomentosa, further forming a monophyletic group with the species of Debregeasia and Pipturus. This work provides basic genetic resources for developing robust markers and investigating the population genetics diversities for B. nivea.
Keywords: Boehmeria nivea, complete chloroplast genome, phylogenetic analysis, Urticaceae family
Ramie (Boehmeria nivea), also known as China grass, is a perennial herb with high economic values due to its production of natural fiber. It is a widely planted species in south of China. The ramie leaf was also utilized as feed supplement and livestock forage grass since the high content of protein and nutrients (Mu et al. 2020). Furthermore, the leaves and roots of B. nivea have been used as traditional Chinese medicine that possesses a variety of pharmacological properties. The ethanol extract of B. nivea suppressed mast cell mediated allergic inflammation, indicating its potential application in the treatment of various allergic disorders (Lim et al. 2020). In addition, the leaf of B. nivea exhibited significant anti-hepatitis B virus activity (Wei et al. 2014). It is also reported that B. nivea is rich in active polyphenol compounds and exerts potent laxative and antioxidant effects in model rats with loperamide-induced constipation (Lee et al. 2020). The complete chloroplast genome information would contribute to the cultivation research and species identification of this important plant. Here, we report the complete chloroplast genome of B. nivea to provide a genomic resource for molecular marker development and clarify the phylogenetic relationship in family Urticaceae.
The sample of Boehmeria nivea was collected from Fuyang area of Zhejiang Province (30°05′2.4″N, 119°53′20.4″E). The specimen is deposited at Medicinal Herbarium Center of Zhejiang Chinese Medical University, Hangzhou, China (Voucher Identifying Number ZM-1963). Total genomic DNA was extracted and sequenced using the Illumina Hiseq Platform according to the previous reprot (Gao et al. 2020; Wang et al. 2020). The chloroplast genome of B. nivea was assembled by metaSPAdes with the chloroplast sequence of B. umbrosa as reference (Nurk et al. 2017). The chloroplast was annotated using GeSeq and further confirmed by BLAST (Tillich et al. 2017). The complete cp genome of B. nivea was submitted to GenBank with the accession number of MW057777.
The length of the complete chloroplast genome sequence of B. nivea was 156065 bp, with a large single copy (LSC) region of 85901 bp, a small single copy (SSC) region of 18972 bp, and two separated inverted repeated (IR) regions of 25596 bp each. A total of 131 genes were identified in the cp of B. nivea, containing 84 protein-coding genes, 37 tRNA genes, 8 rRNA genes and 2 putative pseudo genes. The overall GC content was 36.33%, and the corresponding GC contents for LSC, SSC and IR regions were 34%, 29.61% and 42.72%, respectively. The genome included 17 duplicated genes in the IR region and exhibited 50.23% protein-coding sequences. Moreover, a total of 67 small single repeats (SSR) are identified in the cp of B. nivea, ranging from 10 bp to 123 bp.
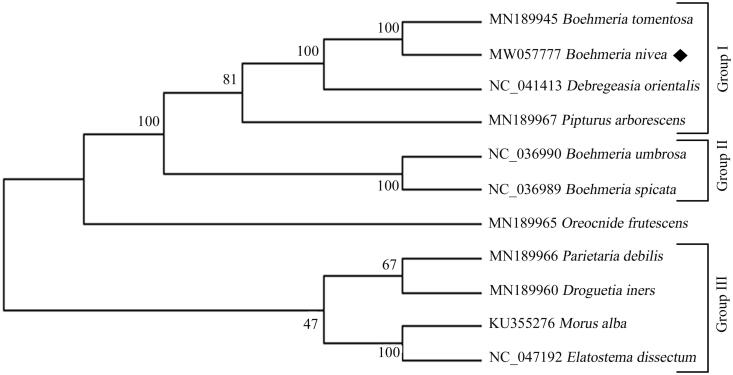
The complete genome sequences of B. nivea and other 10 representative Urticaceae species were analyzed using MEGA 7.0 by maximum-likelihood (ML) method to confirm its phylogenetic position. The result demonstrated a sister relationship between B. nivea and B. tomentosa, indicating a close genetic relationship between the two species (Figure 1). The monophyletic group of B. nivea and B. tomentosa clustered with the combined clade of species from genuses Debregeasia and Pipturus, further forming the Group I in the the family Urticaceae (Figure 1). Furthermore, the species of B. umbrosa and B. spicata formed Group II, but did not display sister relationship with the monophyletic group of B. nivea and B. tomentosa, indicating further investigation and classification revision in the genus Boehmeria. These results would contribute the development of molecular markers and understanding of evolutionary history and cultivation strategy for Boehmeria nivea.
Figure 1.
ML phylogenetic tree analysis of Boehmeria nivea and other representative Urticaceae species based on the complete chloroplast genome sequences. Numbers on the nodes are bootstrap values from 100 replicates. The GenBank accession numbers were listed before the species name.
Funding Statement
This work was supported by the Opening Project of Zhejiang Provincial Preponderant and Characteristic Subject of Key University (Traditional Chinese Pharmacology), Zhejiang Chinese Medical University [No. ZYAOX2018033] and Project of Quality Guarantee System of Chinese Herbal Medicines [Grant No. 201507002-4].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW057777. The associated BioProject, SRA, and BioSample numbers are PRJNA689705, SRR13357339 and SAMN17215053, respectively.
References
- Gao C, Wang Q, Ying Z, Ge Y, Cheng R.. 2020. Molecular structure and phylogenetic analysis of complete chloroplast genomes of medicinal species Paeonia lactiflora from Zhejiang Province. Mitochondrial DNA B Resour. 5(1):1077–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Choi EJ, Park S, Lee JJ.. 2020. Laxative and antioxidant effects of ramie (Boehmeria nivea L.) leaf extract in experimental constipated rats. Food Sci Nutr. 8(7):3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JY, Lee JH, Lee BR, Kim MA, Lee YM, Kim DK, Choi JK.. 2020. Extract of Boehmeria nivea suppresses mast cell-mediated allergic inflammation by inhibiting mitogen-activated protein kinase and nuclear factor-κB. Molecules. 25(18):4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L, Cai M, Wang Z, Liu J, Liu T, Wanapat M, Huang B.. 2020. Assessment of ramie leaf (Boehmeria nivea L. gaud) as an animal feed supplement in P.R. China. Trop Anim Health Prod. 52(1):115–121. [DOI] [PubMed] [Google Scholar]
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA.. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Yu S, Gao C, Ge Y, Cheng R.. 2020. The complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Rubus chingii Hu. Mitochondrial DNA Part B. 5(2):1307–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Lin L, Su X, Qin S, Xu Q, Tang Z, Deng Y, Zhou Y, He S.. 2014. Anti-hepatitis B virus activity of Boehmeria nivea leaf extracts in human HepG2.2.15 cells. Biomed Rep. 2(1):147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW057777. The associated BioProject, SRA, and BioSample numbers are PRJNA689705, SRR13357339 and SAMN17215053, respectively.