Abstract
Aster pekinensis is a perennial herb that distributes widely in China, Korea, and Eeastern Russia. The complete plastome of A. pekinensis is reported here. It is a circular molecular of 152,815 bp in length and consists of a large single-copy region (LSC: 84,530 bp), a small single-copy region (SSC: 18,219 bp), and two inverted repeats (IR: 25,033 bp) regions. GC content is 37.3%. This plastome encodes 113 unique genes, including 79 protein-coding genes, 30 tRNAs, and 4 rRNAs. Phylogenomic analysis of 17 plastomes within Aster and closely related genera revealed that A. pekinensis was sister to the clade comprising A. flaccidus and A. altaicus.
Keywords: Aster pekinensis, plastome, phylogeny
Aster is a genus of the family Asteraceae. This genus has 152 species, 80% of which are distributed in China. A. pekinensis is a widespread perennial herb growing on forest margins, thickets, mountain slopes, riverbanks, and roadsides (Chen et al. 2011). Due to its good quality and palatability, it can be used as a kind of forage grass (Yang and Li 2003). Based on combined sequences of ITS, ETS, and trnL-F, the recent phylogenetic tree demonstrated that A. pekinensis was sister to A. indicus (Zhang et al. 2019). In this study, we reported the complete plastome of A. pekinensis, which will be helpful for species identification and the phylogenetic analysis of the genus Aster.
Silica-dried leaves of A. pekinensis were collected from Zibo, Shandong, China (118°6′10.01″E, 36°23′32.16″N). Voucher specimen (No.198-2) was deposited at School of Life Sciences, Shandong Normal University. Total genomic DNA was isolated using a modified CTAB-based protocol (Wang et al. 2013). DNA library preparation and sequencing were conducted by Illumina Novaseq platform at Novogene (Beijing, China). After obtaining sequencing data, we used Organelle Genome Assembler (OGA, https://github.com/quxiaojian/OGA) to assemble plastome (Qu, Fan, et al. 2019). Annotation was performed with Plastid Genome Annotator (PGA, https://github.com/quxiaojian/PGA) (Qu, Moore, et al. 2019). Referring to previous published studies (Qu 2019; Wang et al. 2019), we used Geneious v9.1.4 to do manual correction (https://www.geneious.com). All 79 protein-coding genes were selected to construct the maximum likelihood (ML) tree by RAxML v8.2.10 (Stamatakis 2014), using 1000 bootstrap replicates with GTRCAT model after alignment using MAFFT v7.313 (Katoh and Standley 2013).
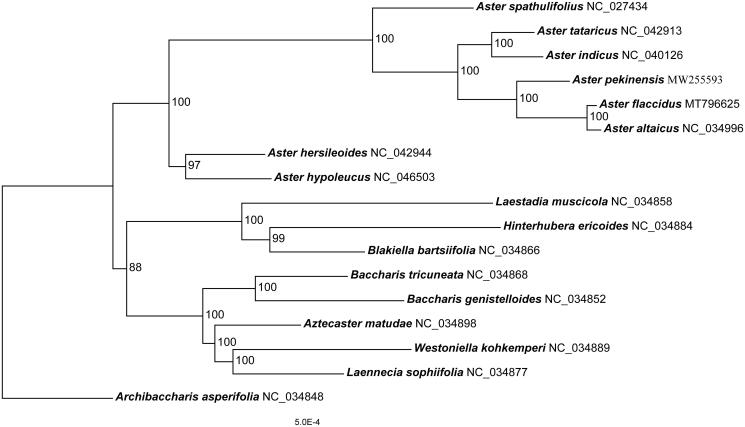
The complete plastome of A. pekinensis (GenBank accession number: MW255593) is a circular molecular of 152,815 bp in length. It consists of a large single-copy region (LSC: 84,530 bp), a small single-copy region (SSC: 18,219 bp), and two inverted repeats regions (IRs: 25,033 bp). GC content is 37.3%. This plastome encodes 113 unique genes, including 79 protein-coding genes, 30 tRNAs, and 4 rRNAs. There are genes with two copies, including rps12, ycf2, rrn23, ndhB, rpl2, rrn16, trnA-UGC, trnI-GAU, rps7, rpl23, rrn5, rrn4.5, trnL-CAA, trnI-CAU, trnR-ACG, trnV-GAC, and trnN-GUU gene. Phylogenomic analysis of 17 plastomes within Aster and its related genera revealed that A. pekinensis was sister to the clade comprising A. flaccidus and A. altaicus (Figure 1).
Figure 1.
A maximum likelihood (ML) phylogenetic tree based on 17 Asteraceae species is shown. Bootstrap support values are shown as numbers next to branches.
Funding Statement
The study was financially supported by Shandong Agricultural Science and Technology Fund Project [2019LY002], Subsidy for the Enhancement of Medical Services and Security Capability [2019,39], and large instrument Open Fundation of Shandong Normal University [KFJJ2021002].
Disclosure statement
No potential conflict of interest was reported by the authors.
Data avaliability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MW255593. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA679197, SUB8572223, and SAMN16825934, respectively.
References
- Chen YL, Chen YS, Brouillet L, Semple JC.. 2011. Aster L. In: Wu CY, Raven PH, Hong DY, editors. FOC. VOI. 20-21. Beijing and Missouri: Science Press; St. Louis: Botanical Garden Press. p. 574–632. [Google Scholar]
- Katoh K, Standley DM.. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ. 2019. Complete plastome sequence of an endangered species, Calocedrus rupestris (Cupressaceae). Mitochondrial DNA B. 4(1):762–763. [Google Scholar]
- Qu XJ, Fan SJ, Wicke S, Yi TS.. 2019. Plastome reduction in the only parasitic gymnosperm Parasitaxus is due to losses of photosynthesis but not housekeeping genes and apparently involves the secondary gain of a large Inverted repeat. Genome Biol Evol. 11(10):2789–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XJ, Moore MJ, Li DZ, Yi TS.. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, , Jiang DF, , Huang YH, , Wang PM, , Li T. 2013. Study on the phylogeny of Nephroma helveticum and allied species. Mycotaxon. 125(1):263–275. [Google Scholar]
- Wang R, Wang QJ, Qu XJ, Fan SJ.. 2019. Characterization of the complete plastome of Alopecurus aequalis (Poaceae), a widespread weed. Mitochondrial DNA B Resour. 4(2):4216–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YF, Li JD.. 2003. Growth strategies of different age classes of ramets in Kalimeris integrifolia population at the Songnen Plains of China. Ying Yong Sheng Tai Xue Bao. 14(12):2171–2175. [PubMed] [Google Scholar]
- Zhang GJ, Hu HH, Gao TG, Gilbert MG, Jin XF.. 2019. Convergent origin of the narrowly lanceolate leaf in the genus Aster with special reference to an unexpected discovery of a new Aster species from East China. PeerJ. 7:e6288. [DOI] [PMC free article] [PubMed] [Google Scholar]