Abstract
Here, we present the whole mitochondrial genome of commensal hydroid Eutima japonica McCrady 1859 (family Eirinidae); this is the first specimen of the family to have its mitogenome sequenced. The linear mitogenome is 15,315 bp in length and consists of 13 protein-coding genes (PCGs), large and small ribosomal subunits (rRNA), methionine and tryptophan transfer RNA (tRNA) genes (trnM and trnW), and a partial copy of cytochrome oxidase subunit I (cox1) pseudogene, as is typical for the class Hydrozoa. Nucleotide sequences of two cox1 genes at two ends of the linear mitogenome form a part of inverted terminal repeat. The overall genomic structure and gene arrangement of 13 PCGs were identical to the reported mitochondrial genomes of hydrozoans, except for the positions of two tRNA genes. Phylogenetic analysis of E. japonica 13 PCGs and other cnidarians recovers a closest relationship with the derived cluster of two hydrozoans, Laomedea flexuosa and Obelia longissimi within Leptothecata.
Keywords: Commensal hydroid, Eutima japonica, Leptothecata, linear mitogenome, phylogeny
Bivalve-inhabiting hydrozoans have lifestyles from simple epibiosis to mutualistic or commensalistic symbiosis with numerous aquatic animals (Piraino et al. 1994; Gili and Hughes 1995). Two genera of hydroids, Eutima McCrady, 1859 and Eugymnanthea Palombi, 1935 (e.g. Eutima japonica, E. sapinhoa, and Eugymnanthea japonica) as members in the family Eirenidae are generally observed inhabiting the bivalves via direct attachment to soft body parts (e.g. mantle) (Kubota 1992, 2012). Since controversy has occurred on the origin of Eirenidae and evolutionary relationships in hydrozoans, molecular phylogenetic approach has been conducted using several marker genes or transcriptome information (Leclère et al. 2009; Maronna et al. 2016; Kubota and Collins 2017; Kayal et al. 2018). Since partial genomic information such (e.g. 16S rRNA, 18S rRNA, and COI genes) has been available in Eutima and Eugymnanthea yet, their complete mitogenomes will provide a robust resource to understand phylogenetic relationship and evolutionary history of the bivalve-inhabiting Eirinidae species.
A specimen of E. japonica was collected in the mantle cavity of the Pacific oyster Crassostrea gigas from Southeast Sea in Korea (34°50′16.1′′N, 128°14′30.8′′W). The voucher specimen was registered in the Southeast Sea Fisheries Research Institute (Species ID: Cnidaria-01; Specimen ID: NIFS-SSFRI-01). Genomic DNA was isolated using a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany). A genomic library was constructed using a MGIEasy DNA library prep kit (MGI, Shenzhen, China) by MOAGEN (Pusan, Busan, South Korea), based on the manufacturer’s instructions. The quality of the library was checked using the Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA) and raw reads were obtained by MGI MGISEQ-2000 sequencing platform (paired-end, 150 bp read length, MGI). After the trimming process of 3′ adapter on the paired-end reads using Cutadapt version 1.9 (Martin 2011), the mitochondrial genome of E. japonica was recovered by direct mapping to the hydrozoan mitogenomes using Geneious version 11.1.3 (Kearse et al. 2012). Mitogenome feature and gene annotation were performed using MITOS2 (Bernt et al. 2013) and tRNAscan-SE version 2.0 (Lowe and Eddy 1997), and finally, the annotated gene was carefully confirmed using NCBI-BLAST (http://blast.ncbi.nlm.nih.gov).
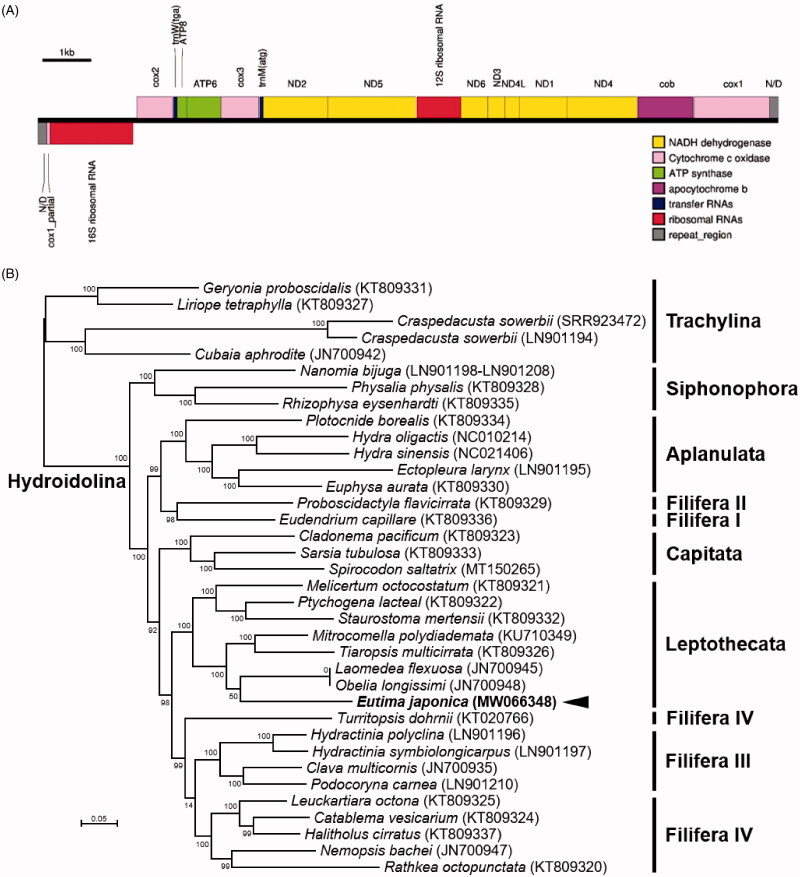
In this study, the complete linear mitochondrial genome of E. japonica was assembled and annotated as the first mitogenome of a member in the family Eirinidae. The mitogenome for E. japonica (GenBank accession no. MW066348) was 15,315 bp long and had a GC content of 32.8% (A: 35.5%; T: 31.7%; G: 12.7%; C: 20.1%). The E. japonica mitochondrion contained 13 PCGs, 22 tRNA genes (trnM and trnW), two rRNA genes, and a partial copy of cox1 pseudogene (Figure 1((A)). The order of 13 PCGs was identical to the reported 34 mitochondrial genomes of hydrozoans, except for the positions of two tRNA genes. The trnM gene was located between the cox3 and ND2, and the position of trnW gene was between cox2 and ATP8. Genomic positions of trnM and trnW were analyzed between cox1 pseudogene-16S rRNA and 16S rRNA-cox2 in the genus Hydra (Kayal and Lavrov 2008; Pan et al. 2014).
Figure 1.
(A) Schematic diagram for the linear genomic structure of E. japonica mitogenome. (B) Maximum-likelihood (ML) phylogeny of 36 hydrozoans including E. japonica based on the concatenated nucleotide sequences of entire protein-coding genes (PCGs). Numbers at nodes represent ML bootstrap percentages (1000 replicates). DDBJ/EMBL/Genbank accession numbers for published sequences are incorporated. The black arrow indicates the Eutima sp. analyzed in this study.
A maximum-likelihood phylogenetic hypothesis was established using sequence data from the concatenated set of 13 PCGs of E. japonica mitogenome with including 35 published hydrozoan mitogenomes. JModelTest version 2.1.10 (Darriba et al. 2012) was used to select the best substitution model and a substitution model (HKY + G + I) was applied to perform a maximum-likelihood (ML) method in the PhyML version 2.4.5 (Guindon and Gascuel 2003) with 1000 bootstrap replicates. Hydrozoa consists of two main clades, Trachylina and Hydroidolina (Aplanulata, Capitata s.s., Filifera I–IV, Leptothecata, Siphonophorae) that comprises almost all hydrozoans except for Limnomedusae, which is part of Trachylina (Kayal et al. 2015). Based on the monophyly of Hydroidolina and Trachylina (Kayal et al. 2015), five members of Trachylina were set as an outgroup. Phylogenetic analysis using mitogenomic data resolved a close relationship of E. japonica with the cluster of Leptothecata species (Figure 1(B)).
Funding Statement
This research was supported by the National Institute of Fisheries Science under a grant [NIFS-R2020016].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.ni h.gov/bioproject/PRJNA684984, https://www.ncbi.nlm.nih.gov/sra/SRR13249693, and https://www.ncbi.nlm.nih.gov/biosample/SAMN17073642, respectively. The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MW066348.
References
- Bernt A, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF.. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gili JM, Hughes RG.. 1995. The ecology of marine benthic hydroids. Oceanogr Mar Biol Annu Rev. 33:351–426. [Google Scholar]
- Guindon S, Gascuel O.. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52(5):696–704. [DOI] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG.. 2015. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ. 3:e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Pankey MS, Ohdera AH, Medina M, Plachetzki DC, Collins AG, Ryan JF.. 2018. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol Biol. 18:68. [Google Scholar]
- Kayal E, Lavrov DV.. 2008. The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene. 410(1):177–186. [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S. 1992. Four bivalve-inhabiting hydrozoans in Japan differing in range and host preference. p. Sci Mar 56:149–159. [Google Scholar]
- Kubota S. 2012. The life cycle of a bivalve-inhabiting hydrozoan, Eutima sapinhoa (Cnidaria, Hydrozoa), from Florida, USA. Biogeography. 14:87–91. [Google Scholar]
- Kubota S, Collins A.. 2017. A single origin of bivalve-inhabiting hydrozoans (Cnidaria, Hydrozoa, Leptomedusae) in the family Eirenidae based on an analysis of 16S rRNA gene. Biogeography. 19:75–79. [Google Scholar]
- Leclère C, Schuchert P, Cruaud C, Couloux A, Manuel M.. 2009. Molecular phylogenetics of Thecata (Hydrozoa, Cnidaria) reveals long-term maintenance of life history traits despite high frequency of recent character changes. Syst Biol. 58(5):509–526. [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR.. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maronna MM, Miranda TP, Peña Cantero ÁL, Barbeitos MS, Marques AC.. 2016. Towards a phylogenetic classification of Leptothecata (Cnidaria, Hydrozoa). Sci Rep. 6:18075–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from highthroughput sequencing reads. EMBnet J. 17(1):10–12. [Google Scholar]
- Pan H, Qian X, Li P, Li X, Wang A.. 2014. The complete mitochondrial genome of Chinese green hydra, Hydra sinensis (Hydroida: Hydridae). Mitochondrial DNA. 25(1):44–45. [DOI] [PubMed] [Google Scholar]
- Piraino S, Todaro C, Geraci S, Boero F.. 1994. Ecology of the bivalve-inhabiting hydroid Eugymnanthea inquilina in the coastal sounds of Taranto (Ionian Sea, SE Italy. Mar Biol. 118(4):695–703. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
BioProject, SRA, and BioSample accession numbers are https://www.ncbi.nlm.ni h.gov/bioproject/PRJNA684984, https://www.ncbi.nlm.nih.gov/sra/SRR13249693, and https://www.ncbi.nlm.nih.gov/biosample/SAMN17073642, respectively. The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov, accession number MW066348.