Abstract
Three-dimensional (3D) printing is a transformative manufacturing strategy, allowing rapid prototyping, customization, and flexible manipulation of structure-property relationships. Proteins are particularly appealing to formulate inks for 3D printing as they serve as essential structural components of living systems, provide a support presence in and around cells and for tissue functions, and also provide the basis for many essential ex vivo secreted structures in nature. Protein-based inks are beneficial in vivo due to their mechanics, chemical and physical match to the specific tissue, and full degradability, while also to promoting implant-host integration and serving as an interface between technology and biology. Exploiting the biological, chemical, and physical features of protein-based inks can provide key opportunities to meet the needs of tissue engineering and regenerative medicine. Despite these benefits, protein-based inks impose nontrivial challenges to 3D printing such as concentration and rheological features and reconstitution of the structural hierarchy observed in nature that is a source of the robust mechanics and functions of these materials. This review introduces photo-crosslinking mechanisms and rheological principles that underpins a variety of 3D printing techniques. The review also highlights recent advances in the design, development, and biomedical utility of monolithic and composite inks from a range of proteins, including collagen, silk, fibrinogen, and others. One particular focus throughout the review is to introduce unique material characteristics of proteins, including amino acid sequences, molecular assembly, and secondary conformations, which are useful for designing printing inks and for controlling the printed structures. Future perspectives of 3D printing with protein-based inks are also provided to support the promising spectrum of biomedical research accessible to these materials.
Keywords: Silk, Collagen, Fibrinogen, Rheology, Directed assembly, Photopolymerization, Simulation
Graphic abstract
1. Introduction
Three-dimensional (3D) printing/additive manufacturing is transforming the landscape of conventional manufacturing and is becoming a versatile and potent tool for a myriad of biomedical and clinical applications [1, 2]. 3D printing generally relies on adding materials in a layer-by-layer or voxel-by-voxel manner to generate 3D architectures. This additive strategy is in contrast to subtractive approaches such as mechanical machining, or formative ones such as molding/casting, and thus provides distinct manufacturing advantages. 3D printing enables rapid prototyping of structures customized in terms of composition and morphology, a feature desirable for precision medicine. 3D printing is also advantageous in spatially controlling the complex organization of multiple components to help recapitulate the heterogeneity of native tissues. Third, 3D printing enables tight control of structure-property relationships, providing tunable outcomes in terms of mechanical properties, as well as the degradation rate of the structures.
The key to 3D printing is printing ink [3, 4]. The performance and functionality of 3D printing are largely determined by the biochemical, rheological, and material properties of the ink. A broad spectrum of materials has been developed for 3D printing inks, including ceramics [5], metals [6, 7], glasses [8, 9], synthetic polymers [10, 11], and nature-origin polymers [12–14], consisting of polysaccharides and proteins. Among these materials, proteins represent the major building blocks for cells, extracellular matrices (ECM), and various biological structures. The prevalence of proteins in living systems makes protein-based inks particularly alluring to construct proteinaceous scaffolds that resemble or mimic the proteinaceous composition of native tissues (Fig. 1). In comparison to other materials, the use of these protein-based compositions is expected to minimize immune reactions and promote biomaterial-host integration, as well as to degrade fully over time, generating peptides and amino acids as nutrients for the cells.
Fig. 1.
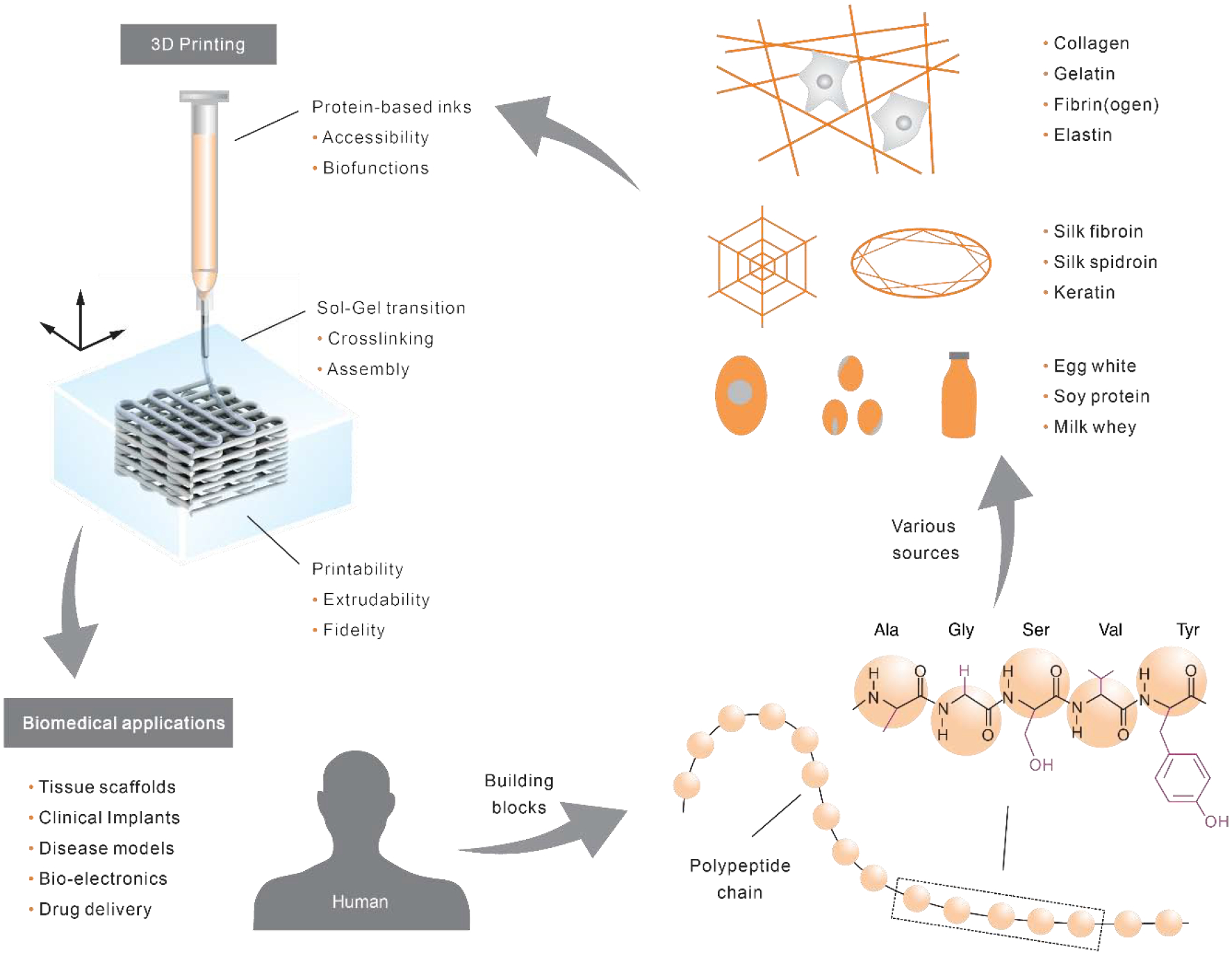
Overview of protein-constituted inks for 3D printing. Proteins, as the building blocks of living systems including humans, are linear macromolecules/polypeptide chains consisting of amino acids held together by peptide bonds. The chemical structure of a short model peptide, consisting of alanine (Ala), glycine (Gly), serine (Ser), valine (Val), and tyrosine (Tyr), is illustrated. The side chain groups of each amino acid are labeled in purple. Proteins can be derived from a variety of sources as well as genetic engineering techniques. Protein-constituted inks are printed, via a sol-gel transition, into 3D structures that hold promise to address critical challenges in biomedical and clinical applications.
Proteins are macromolecules consisting of amino acids [15], which have 20 standard types in eukaryotes yet result in an amazingly vast diversity of protein-based materials [16, 17]. Proteins can be roughly categorized as fibrous and globular proteins, depending on the capability to form fibrils. Most globular proteins form into singular entities; some globular proteins, such as lactoglobulin and insulin, can form fiber-like structures (e.g., amyloid fibrils) under certain conditions [18, 19]. Some fibrous proteins are derived from the decellularized native tissues or ECM [20], including collagens, fibronectin, elastin, fibrin, and cytokeratin [21–24]. These ECM proteins are tissue-specific and dynamically interact with cells via remodeling, degradation, and signal transduction [25], and help maintain tissue-specific functions and to support the differentiation of cells [26, 27]. Proteins, not from the decellularized tissues, include keratin, silk, soy, albumin, zein, and ovalbumin. The former two are fibrous while the rest are usually globular. These proteins are advantageous to be printing inks as they usually have a convenient and easy-scalable supply. For example, keratin and silk that can be harvested from the textile industrial production of wool and cocoons, respectively. Other food proteins are also available based on food infrastructure already in place.
Proteins are the material basis of tissues and structures with important mechanical properties, such as tendons from collagens and silks from fibroins and spidroins. For example, the specificenergy storage capacity of vertebrate tendons (~800 J kg−1) is higher than that of high-tensile spring steel (120 J kg−1) by six-fold [28]. Also, spider dragline silk is one of the toughest materials known; its maximum toughness (~350 MJ m−3) is higher than a range of high-performance polymers, including Kevlar 49 (50 MJ m−3) and nylon (80 MJ m−3) [29]. These mechanical feats are useful for regenerating native tissues and for developing new devices and systems where high-performance is desirable.
These exceptional material properties of proteins stem from a ubiquitous manufacturing strategy in biology, directed hierarchical molecular assembly. The protein molecules contain specific encoded designs, i.e., amino acid sequence information, which guides the assembly via weak interacting forces, such as hydrogen bonding and Van der Waals [28]. The molecular assembly process is dynamically regulated by cells, via rates of biosynthesis and cellular export, enzyme-cleavage, and environmental cues to form consistent, well-controlled structures across the hierarchical length scale under ambient and aqueous conditions. Translating these facile processing conditions to 3D printing with protein inks supports the integration of cells and bioactive molecules (e.g., cytokines and growth factors) into 3D printed proteinaceous structures, leading to hybrid living materials [30–32]. This biological manufacturing strategy is rich in information, including both molecular designs and the regulation of cell functions, in sharp contrast to the energy-intensive artificial manufacturing of polymers with organic solvents, thermal conditions or harsh chemical reactions. Such conditions usually compromise bioactivity.
Proteins have been printed into a host of 3D structures with substantial capability for tissue regeneration, disease models, and drug screening [33–35]. These exceptional studies mainly rely on conventional polymerization techniques, such as covalent crosslinking and organic solvents. However, the understanding of hierarchical macromolecular assembly can significantly facilitate the development of new 3D printing techniques to unleash the full potential of protein-based inks. Considerable progress has been made toward this goal, with the assembly of short peptides and polymers into functional structures [36–38]. Monolithic collagen and silk inks have been 3D printed via pH-induced assembly [39] and salt-directed assembly [40], respectively. Yet, there remains a largely untapped space of materials properties to be achieved via further understanding, and then the exploitation of proteins in 3D printed matrices for a wide range of utilities.
According to ASTM [41], the 3D printing/additive manufacturing for polymer materials encompasses a broad range of seven technical categories, including directed energy deposition, sheet lamination, binder jetting, material extrusion, material jetting, vat polymerization. Because not all these techniques apply to protein-based inks, we only focus on rheological behavior and photo-crosslinking approaches, underpinning these distinct 3D printing techniques. Viscoelastic behavior and rheological characterization of protein inks play a central role in the process of extrusion and jetting; photo-crosslinking determines printing efficiency and the structure and mechanical performance of protein hydrogels. We also cover the recent progress of computational simulation on predicting and understanding protein assembly, as it is promising for assisting 3D printing with protein inks. Then, we review several representative protein-based inks for 3D printing, including those based on collagen, silk, fibrinogen, keratin, and several food proteins. Some protein materials have been less exploited for 3D printing than others yet demonstrate unique value in biofunctions and accessibility. The inclusion of these less-investigated protein materials adds to the comprehensive nature of this review and motivates more investigations in the future. Finally, we briefly envision several promising future directions of 3D printing with protein-based inks. Throughout the review, the emphasis is placed on monolithic inks (single types of proteins), sequence-process-property relationships, and methods to improve structural integrity and to expand functions of protein prints.
2. Technical consideration for 3D printing
2.1. Rheological characterization
Several 3D printing techniques, such as material extrusion and jetting, involves the flow of ink. For example, the protein-based ink, often loaded in a movable syringe is extruded by external mechanical forces through a nozzle to deposit on a surface. During the extrusion, the ink is expected to flow as well as maintain shape after deposition [42–45]. The printing performance, such as extrudability, filament fidelity, and sol-gel transition, are largely determined by the viscoelastic properties of the ink that are characterized by a set of steady or dynamic rheological tests.
2.1.1. Extrudability
Extrudability is defined as how efficient an ink is extruded from a dispensing nozzle (Fig. 2A). The primary measure of extrudability is viscosity, which indicates the resistance of a fluid to deform under external forces, and at the molecular level, the resistance to the sliding between molecules. Specific to protein-based inks, viscosity is relevant to concentration, molecular weight, and importantly, inter- and intra-molecular interactions that are associated with several physical factors, including temperature, ion composition/concentrations, and pH. The molecular weight and concentration of proteins are proportional to molecular entanglement, which impedes molecular sliding and thus increases viscosity. The viscosity of collagen and regenerated silk fibroin solutions is associated with concentration [46] and molecular weight [47], respectively. Furthermore, the side chains of the amino acids endow a variety of secondary conformations and molecular interactions of proteins via covalent bonding, such as disulfide formation, as well as via weak bonding, including salt bridges, hydrogen bonding, and van der Waals forces. These molecular conformations and interactions also influence sliding between protein molecules and thus are relevant to the viscosity. For example, the viscosity of a partially hydrolyzed product of collagen, i.e., gelatin, is dependent on temperature because the hydrogen bonds that hold the triple-helix conformation of gelatin together are weakened by increased temperature, generally above 30°C [48]. According to previous definitions for injectable hydrogels [43], inks with a viscosity higher than 100 and less than 2,000 Pa·s may be called a paste; higher than 2,000 Pa·s may be called a putty. Depending on the viscosity of a protein-based ink, compressed air, pistons, and screw rods may be utilized to provide sufficient extrusion pressures.
Fig. 2.
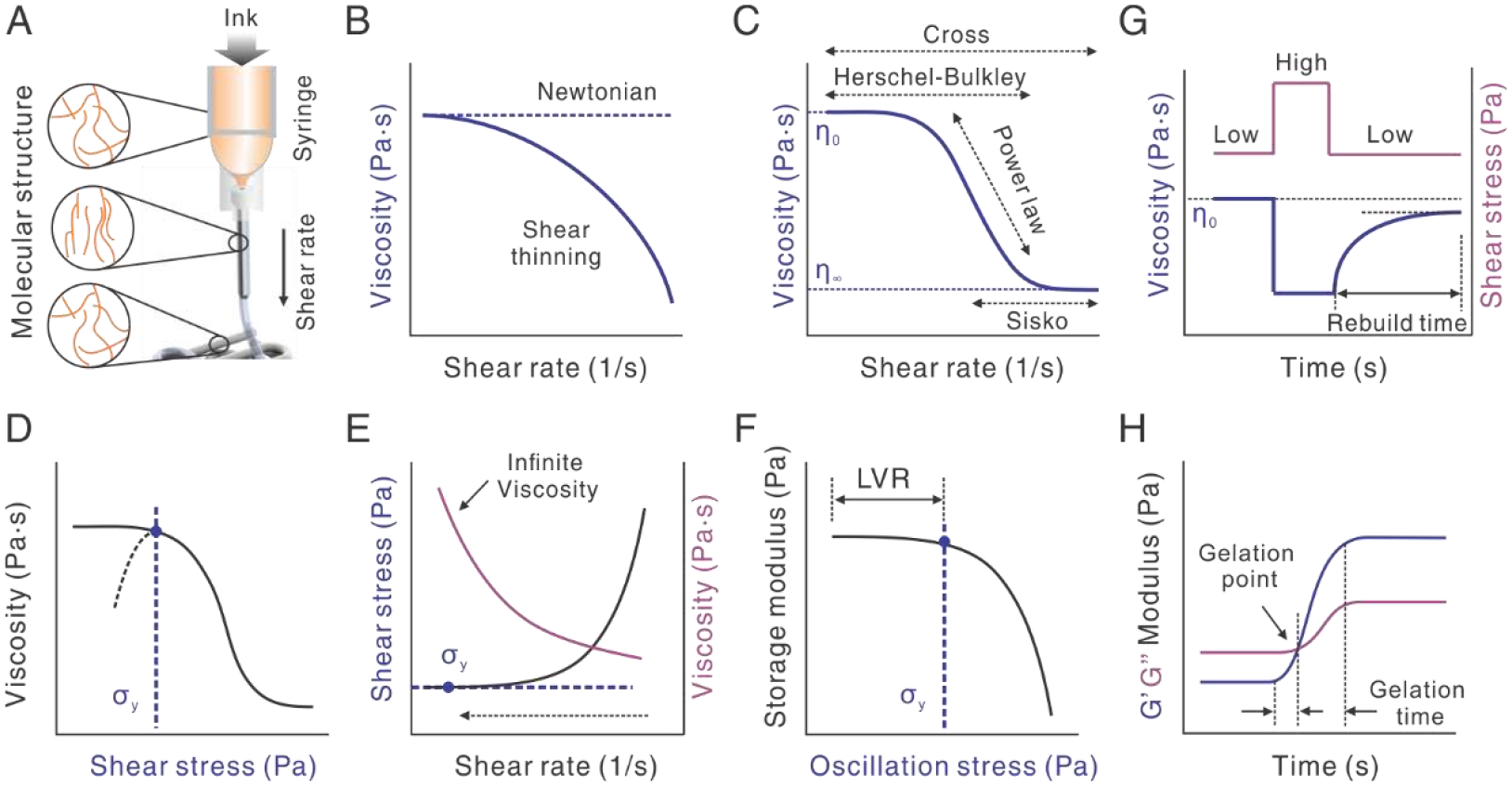
Rheological characterizations of viscoelastic protein inks for 3D printability. A. Illustration of the extrusion of ink and the stretching and rebuilding of molecular structure. B. Comparison between Newtonian and non-Newtonian shear-thinning behaviors. The flow curve is plotted in viscosity as a function of shear rate. C. Illustration of four models, including the Cross, Herschel-Bulkley (H-B), Power-law, and Sisko, for fitting the different viscoelastic behaviors of inks. η0 and η∞ indicate zero-shear and infinite-shear viscosity, respectively. D-F. Three approaches to measuring yield stress (σy). Shear stress equals shear rate times the viscosity. LVR indicates the linear viscoelastic region. The dashed arrow indicates the sweep starting from high to low shear rate. G. Thixotropy characterization by a three-step stress sweep. H. Crosslinking characterization by oscillatory time sweep. The gelation point is where storage modulus (G’) becomes equal to loss modulus (G”). The time between two equilibrated G’ values is gelation time. Sometimes, the time from the onset of G’ increase to the gelation point is also called gelation time.
The viscosity of most protein-based inks becomes smaller as the shear rate increases, as shown in a rheological flow curve (Fig. 2B). This shear rate-dependent phenomenon is a non-Newtonian behavior, called shear-thinning, in comparison to shear-independent flow (Newtonian) and shear-thickening (another non-Newtonian flow behavior). This shear-thinning property of the ink is desired for 3D printing, as it lowers the pressure required to extrude the ink. Shear-thinning often results from the stretching or elongation of molecules under shear, which diminishes the molecular entanglements and facilitates the sliding between molecules, decreasing the viscosity (Fig. 2A). For example, a hydrogel of recombinant spider silks, eADF4(C16) is printable because of the shear-thinning behavior [49]. The complex viscosity decreased from 40 to 4 Pa·s with the increase of oscillatory frequency. Of note, frequency is used instead of the shear rate to describe the shear-thinning behavior, because the complex viscosity as a function of frequency approximates the viscosity as a function of shear rate, according to the Cox-Merz rule [50]. Another molecular mechanism leading to shear-thinning is host-guest interactions [51, 52], where intermolecular interactions between adamantane and β-cyclodextrin are interrupted under shear and restored upon deposition. The reversibility of the intermolecular interactions also makes the 3D prints self-healable.
The viscoelastic behavior of protein-based inks, especially shear-thinning, can be complex. Different inks can always be fitted by several models, including the Cross, Herschel-Bulkley (H-B), Power-law, and Sisko models (Fig. 2C and Table 1). The fitting parameters allow quantitative comparison between inks. The H-B model has long and widely been used to characterize the shear-thinning behavior of inks for extrusion-based 3D printing [53, 54]. In this model, the viscosity is a constant when the shear stress is smaller than the yield stress (σy); the viscosity starts to change when the shear stress is larger than σy. Yield stress implies the maximum strength of the molecular structures within the ink, and indicates the minimum shear stress to initiate the flow of ink, i.e., extrusion. Thus, for extrusion-based 3D printing, the extrusion stress needs to be higher than the yield stress of the ink. Yield stress is also important to filament fidelity and is discussed in the next section. In all models, when a factor, n approximates 1, the viscosity becomes more independent of the shear rate, indicating a more Newtonian behavior, i.e., shear-independence. Besides the viscoelastic properties of the ink, extrudability is relevant to the size and geometry of the nozzle, which contributes to the hydrodynamic resistance. Nozzles with a finer gauge, a longer length, and a cylinder shape, in comparison to a conical shape, lead to higher resistance and thus require a higher extrusion pressure.
Table 1.
Common flow models for extrusion-based 3D printing.
| Models | Equation a | Range |
|---|---|---|
| Cross | η0 → Shear-thinning → η∞ | |
| Herschel-Bulkley | η0 → Shear-thinning | |
| Power law | Shear-thinning | |
| Sisko | Shear-thinning → η∞ |
ηo is the zero-shear viscosity (Pa·s); η∞ is the infinite shear viscosity (Pa·s); is the shear rate (1/s); K and n are parameters related to the shape of the flow curve; σy is the yield stress (Pa).
2.1.2. Filament fidelity
Filament fidelity is the maintenance of the shape of the extruded filament upon deposition, which is associated with at least two viscoelastic properties, yield stress (σy) and thixotropy.
Insufficient yield stress may lead to collapse or sagging of printed filaments under its gravity and the weight of the above layers. The yield stress of the protein-based inks can be improved by adding thickeners and bulking agents, e.g., gellan [55]. Depending on the magnitude of the viscosity, “apparent” yield stress of the protein-based inks can be measured from a double logarithmic plot using three methods (Fig. 2D–F) [56]. The first one is a steady stress sweep for inks that have a medium viscosity and show a well-defined transition of viscosity (Fig. 2D). Depending on the equilibrium of the shear rate, the yield stress can be obtained from either the onset point of the decrease in viscosity or the maximum peak of the viscosity. The second one is a steady sweep from high to low shear rates (Fig. 2E). The approach is usually to utilize liquids with low viscosity and an infinity “apparent” viscosity. The yield stress is the plateau value at the low end of the shear rate and becomes independent of the share rate. The third approach is the oscillatory stress sweep (Fig. 2F). The yield stress is obtained from the onset point of the decrease in storage modulus. This approach is performed within the linear viscoelastic region (LVR), thus circumventing wall slippage issues and improving reproducibility. This approach is suitable for a wide range of viscosities.
Another viscoelastic property, relevant to filament fidelity, is thixotropy that describes a time-dependent process to rebuild a molecular structure after disruption of this structure by shear (Fig. 2A and G). Thixotropy can be experimentally characterized by a three-step shear sweep to indicate the recovered viscosity. The first step is to apply a low shear rate which corresponds to the slowly flowing ink in the syringe; then in the second step, the shear rate is dramatically increased, corresponding to the state of the ink flowing through the nozzle; in the third step, the shear rate returns to the low value, allowing the gradual building up of molecular structures and the recovery of viscosity. In Fig. 2G, the viscosity is recovered to 90% of the original value; the 10% gap may be permanent due to irreversible structural changes in the ink. For extrusion-based 3D printing, rapid rebuilding time and a large percentage of the recovered viscosity are favorable for filament fidelity. For a protein ink with collagen and gelatin nanoparticles [57], 75% storage modulus of the ink was recovered within one second. By contrast, an ink of alginate and methylcellulose showed recovery of ~56% viscosity after 30 seconds [58].
2.1.3. Sol-gel transition
The sol-gel transition in 3D printing, from liquid to solid phases, is often associated with the crosslinking of protein molecules. The crosslinking is characterized by the ratio between storage (G’) and loss (G”) moduli in an oscillatory test (Fig. 2H). G’ and G” describe the elastic (solid-like) and viscous (liquid-like) properties of the ink, respectively. When G’ becomes larger than G”, this signifies the sol-gel transition. Where G’ equals G” is usually called a gelation point in a sol-gel transition. An equilibrium value of G’ and the time required to reach the equilibrium can roughly indicate the density and rates of the crosslinking. A variety of crosslinking mechanisms have been used for making protein-based inks printable and for enhancing the printability. The mechanisms include chemical crosslinking via covalent bonding and molecular assembly via weak bonding. It should be noted that multiple crosslinking mechanisms are often used to enhance printability and strength [59, 60].
Chemical crosslinking of protein molecules has been achieved by using enzymes, amine-reactive chemicals, and light-based polymerization. Enzymes can crosslink amino acids side chains in the protein inks. Transglutaminase crosslinks lysine and glutamine [61, 62]; horseradish peroxidase can crosslink tyrosine groups [63]; microbial transglutaminase was used to partially crosslink gelatin inks with improved viscosity for optimizing printability [61]. In contrast to the three enzymes that form covalent bonds, thrombin cleaves fibrinogen to form fibrin that can form assembles for 3D printing [64]. For amine-reactive chemicals, glutaraldehyde [65] has been used to stabilize the prints of gelatin [66] and collagen [39] after printing. However, the toxicity of glutaraldehyde may be an issue for subsequent cell-involved applications [67]. Another chemical crosslinking mechanism for 3D printing is light-based polymerization. The detailed mechanisms of light-based polymerization will be discussed in section 2.2.
Molecular assembly via weak bonding is an appealing trait for printing protein-based inks. The secondary conformations of proteins, such as helices and β-sheets are both stabilized via hydrogen bonds, while also serving as physical crosslinking sites to connect molecules and to form gel networks (Fig. 3). The molecular basis of collagen to form α-helices is the sequence motif, XYG, where G is glycine at every third residue, while X and Y are most frequently the amino acids, proline and hydroxyproline[68]. Gelatin is a degraded product of collagen [69]. Below a critical temperature, gelatin molecules form a partial triple-helical conformation via hydrogen bonds, leading to a sol-gel transition; above this temperature, the hydrogen bonds are weakened, and the helices are lost, leading to a gel-sol transition. This well-defined property has made gelatin a versatile additive for formulating an array of composite protein inks. Similar to gelatin, collagen is able to assemble into gels under pH [70] and temperature [71].
Fig. 3.
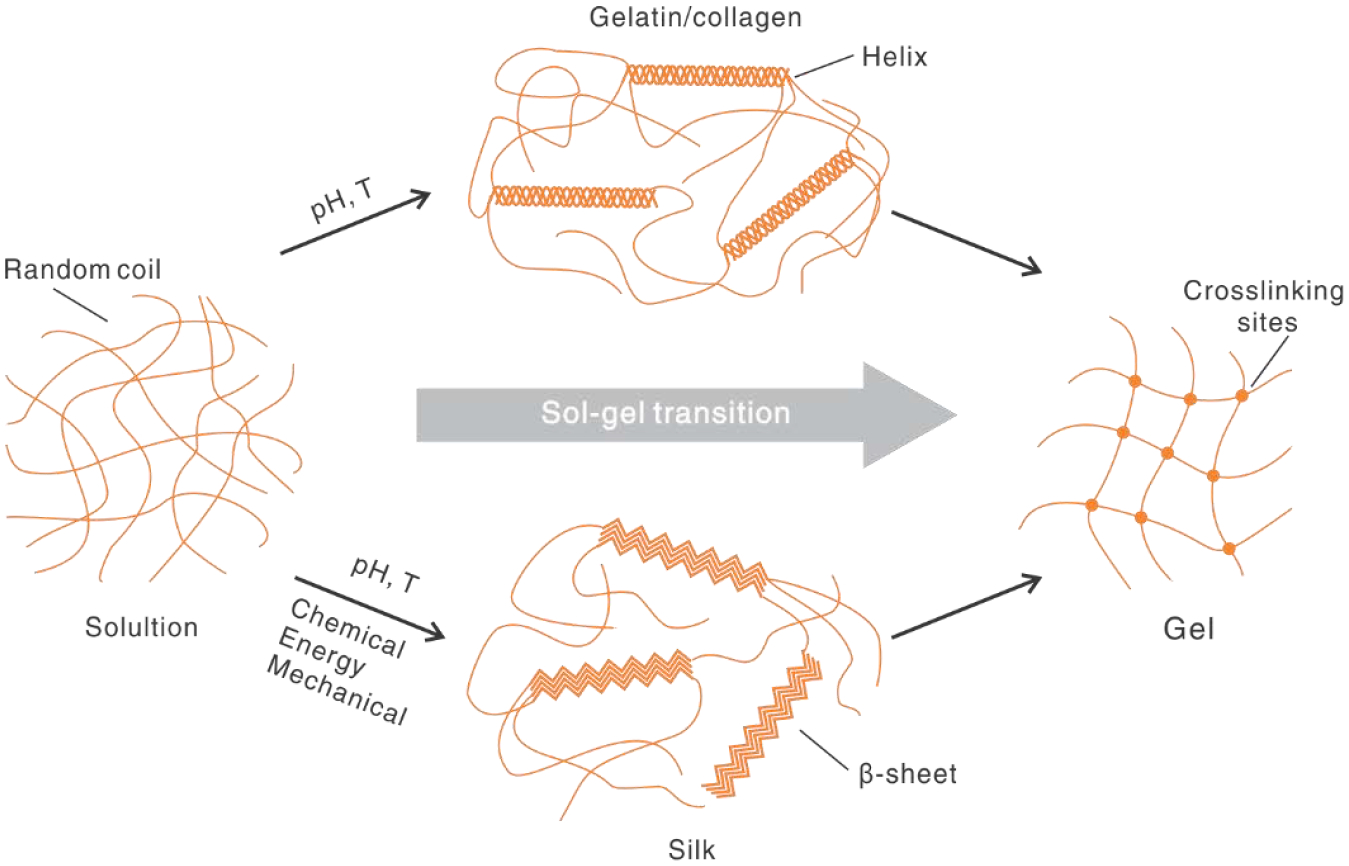
Crosslinking mechanisms of proteins via secondary conformation in a sol-gel transition. For gelatin/collagen, random coil is transitioned into helices by pH and temperature (T). For silk, random coil is transitioned to beta-sheets under several conditions, including chemical, energy, and mechanical environments.
It should be noted that the secondary conformation (here the helical conformation) is capable of initiating a sol-gel transition for 3D printing but often leads to mechanical weakness in comparison to that of native tissues. It is owing to insufficient control over the molecular assembly and, consequently, the lack of native structural hierarchy. For example, the compressive modulus of a meniscus print with 20 mg/ml collagen was around 30 kPa, less than half of the lower limit of the native meniscus [72]. A common way to improve the mechanical strength after molecular assembly is chemical crosslinking. The Young’s modulus of a collagen print is creased from around 10 kPa to around 100 kPa after glutaraldehyde treatment [39].
Besides gelatin/collagen, another example is silk proteins that form a β-sheet conformation [73–75] to crosslink molecular chains physically for a sol-gel transition (Fig. 3) [76]. The sequence motif of silk fibroin is GAGAGS that stacks together to form β-sheets. A range of processing conditions can induce the β-sheet conformation, including organic solvents [77], salts [78], heating [79], pH [80], mechanical shearing [81], and ultrasonication [82]. However, not all sol-gel transitions satisfy the dynamics required by 3D printing. The most widely used sol-gel transition for 3D printing with silk proteins is by organic solvents, such as methanol and isopropanol [83, 84]. Recently, a de novo aqueous salt bath has been developed for 3D printing with silk proteins [40]. This approach directs the assembly of silk fibroin into β-sheets, but also helps to align the molecular chains longitudinally for improved mechanical performance. This printing approach is discussed in section 3.2.
2.2. Photo-crosslinking approaches
Light-based polymerization usually allows high resolution and speed of 3D printing. The resolution is affected by multiple factors but theoretically can be as small as around the diffraction limit of the light used in the process. The scanning of the laser and the projecting of light are faster than the mechanical movement of dispensing nozzles. In addition, light-based 3D printing uses a reservoir to hold the photo-polymerizable ink during the printing, which can eliminate some technical issues in extrusion-based methods, such as nozzle clogging and shear stress as harmful to cells in inks [35, 85]. However, the configuration of an ink reservoir in light-based 3D printing usually restricts the use of multiple inks, as it is challenging to rinsing one ink and fixing the print before adding the next ink, despite some elegant works [86–89]. By contrast, extrusion-based 3D printing is advantageous in multi-material printing, due to the convenient and rapid exchange of inks between multiple printheads [90–92]. Light-assisted 3D printing can be roughly categorized as laser scanning and light projection, depending on the light source. The first category includes two-photon polymerization [93, 94] and stereolithography (SLA) [95], which employ lasers to scan protein-based inks in a voxel-by-voxel manner for (de)polymerization. The second one uses light projection via either a micro-mirror array or a dynamic mask to cure proteins in a layer-by-layer manner [96, 97], including digital light processing (DLP). In this section, we focus on three representative mechanisms of photo-polymerization of proteins [98–101], including acrylate free-radical polymerization, thiol-ene click reaction, and photo-oxidation, which have demonstrated utility with protein-based inks. Other photo-polymerization mechanisms suitable for protein inks are nitrogen radical reactions that were discussed elsewhere [102].
2.2.1. Acrylate
Free-radical photopolymerization of acrylate groups is widely used for 3D printing with protein-based inks, especially gelatin (meth)acryloyl (GelMA), due to the high efficiency of the reactions [103, 104]. Acrylate groups are not present in native proteins, so chemical immobilization is needed. In the presence of photo-generated radicals, (meth)acryloyl groups are polymerized with each other in a chain-growth manner (Fig. 4A). A cross-linker is not necessary for the polymerization, but its involvement may provide finer control over the photopolymerized molecular network of proteins. One drawback of the free-radical photopolymerization process is the inhibition of soluble oxygen, which can be addressed by using a nitrogen purge gas but may interfere with the encapsulation of cells if this is a consideration.
Fig. 4.
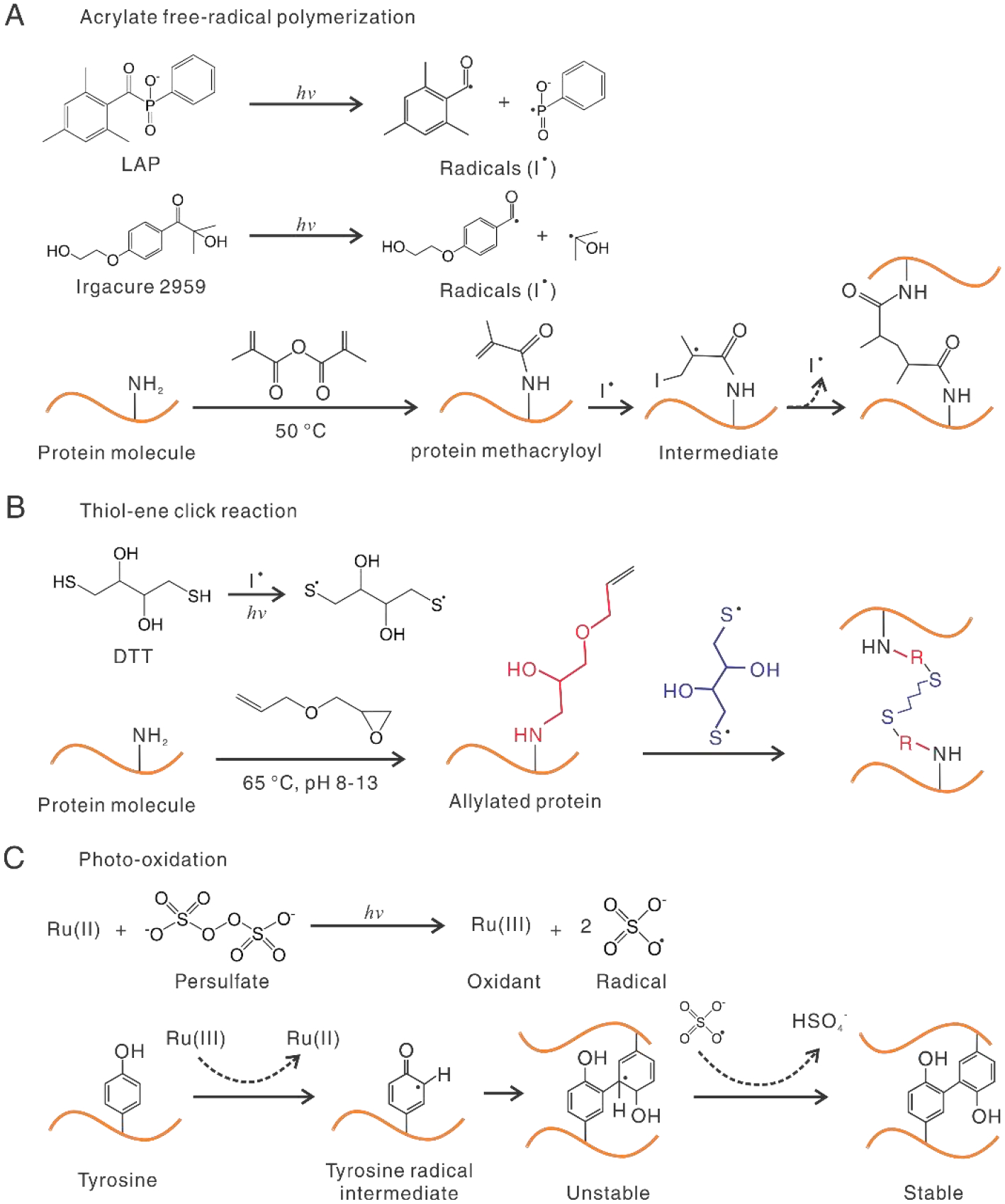
Three representative photo-crosslinking mechanisms for 3D printing with proteins. A. Acrylate free-radical photo-crosslinking. Photo-initiators, such as LAP and Irgacure 2959, produce free-radicals under UV or visible light. Protein molecules are modified with acrylate groups at primary amine and hydroxyl groups. Free radicals lead to chain polymerization between methacryloyl groups from one (intra-) or different (inter-) molecules. Chain propagation is not shown for clarity. B. 1,4-dithiothreitol (DTT) is activated by UV or photoinitiators and reacts with C-C bonds, i.e., “ene” groups, for crosslinking. R in red color represents the immobilized group in the previous step. C. ([Ru(II)(bpy)3]2+) /persulfate-based photo-oxidation. The unstable intermediate is fortified by removing a hydrogen atom.
The efficiency of the free-radical acrylate photopolymerization is associated with photo-initiators that produce free radicals in response to light. The photoinitiators for protein-based inks should be water-soluble and biocompatible [105–107]. Three broadly used photo-initiators with UV light absorbance are 2,2-azobis (2-methyl-N-2-hydroxyethyl) propionamide (VA-086) [67], 2-hydroxy-1-[4-(2-hydroxyethoxy) phenyl]-2-methyl-1-propanone (I2959), and lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) (Fig. 4A). LAP has a 10-fold faster gelation time than I2959 [105]. The photo-initiators with visible light absorbance include Rose Bengal [108], Eosin Y [109–111], and flavin-mononucleotide [112–115]. The use of visible light, in place of UV light, reduces potential damages to cells, especially to DNA [116]. Flavin-mononucleotide is a water-soluble variant of riboflavin and has been used with unmodified proteins, such as collagen [70, 117, 118] and silk [112]. Due to the absence of acrylate or other reactive groups, however the riboflavin-photocured protein structures are rather weak. For example, the storage modulus of 8 mg/ml collagen ink was about 20 Pa, while the pH-induced assembly was around 1,700 Pa [70].
2.2.2. Thiol-ene
Another photopolymerization mechanism is the thiol-ene reaction (Fig. 4B) [119]. This reaction couples two complementary groups, a C-C double bond/ene and a mercapto/thiol group. The thiol-ene reaction is a well-known example of click chemistry, which features fast kinetics and high efficiency [119]. Because of these benefits, photoinitiator concentration and the time of light exposure can be reduced, which favors cells in terms of compatibility and safety. The thiol-ene reaction, in comparison to the acrylates, is not limited by soluble oxygen and works in a step-growth manner, in contrast to the chain-growth manner, which is promising for more homogeneous polymer networks and a better filament fidelity.
The thiol-ene reaction may be beneficial for some proteins rich in cysteines with a thiol sidechain, such as soy proteins [120], because the chemical modification is not required. For other protein molecules without abandoned thiol side chains, chemical modification with “ene” groups is needed. Common ene groups include norbornene, vinyl esters, and allyl ethers [121, 122]. Norbornene provides the highest reaction rates, as ring strain leads to a “spring-relief” behavior [122, 123]. Less commonly, proteins were chemically modified with thiol groups and crosslinked with multi-dentate ‘ene’ crosslinkers [124, 125]. All photoinitiators, as discussed for the acrylate reactions, are applicable to the thiol-ene reactions. However, Deep-UV light, at a low wavelength of 254 nm, is able to activate the thiol group and carry out the thiol-ene reactions in the absence of photo-initiators (Fig. 4B) [126].
2.2.3. Photo-oxidation
A third mechanism is photo-oxidation, which crosslinks the side chain groups of native amino acids, such as tyrosine and tryptophan. This approach is particularly appealing to protein-based inks, as a wide range of proteins have considerable content of tyrosine, including recombinant resilin (~6.7%) [127], titin immunoglobulin domains (~6.7%) [128], fibrinogen (β chain, 4.9%) [129], gelatin (0.9%) [130], fibronectin (4%) [131], collagen (1%) [131] fibrin(~4%) [131], and silk fibroin (5.3%) [132, 133]. The percentage in the brackets indicates the content of tyrosine. Besides, the tyrosine side chain can be incorporated into proteins via chemical modification for higher tyrosine content [132, 134]. The higher content of tyrosine indicates a possible higher density of crosslinking, thus leading to more satisfied mechanical performance.
The photo-oxidation process was illustrated with an example of the widely used ruthenium (II) trisbipyridyl ([Ru(II)(bpy)3]2+)/persulfate system (Fig. 4C) [135]. [Ru(II)(bpy)3]2+ has a high molar attenuation coefficient of 14,600 M-1 cm-1 at 450 nm. Under visible light, Ru(II) and persulfate are photolyzed into Ru(III) and sulfate radicals, respectively. Ru(III) is an oxidant that can oxidize tyrosine residues to give rise to a tyrosine radical intermediate. The tyrosine intermediate can form arene to couple with nearby tyrosine groups, which is further stabilized by the persulfate radial and the removal of a hydrogen atom. In comparison to a free-radical acrylate reaction based on I2959, the Ru/persulfate photo-oxidation system showed less oxygen inhibition and maintained higher cell viability even with increased concentrations and light intensity. For example, with light intensity was increased from 3 to 100 mW/cm2, I2595 decreased cell viability from 90% to around 45%, while Ru/persulfate maintained cell viability at around 90% [136].
2.3. Computational simulation for protein assembly
Computational simulation has been widely used for designing tissue scaffolds and biomedical devices with regard to mechanics [137–139], mass transport [140–142], and shear stress [143]. The simulations can predict experimental results and thus reduce costly and lengthy experimental iterations [144, 145]. Computational simulation increasingly plays important roles in 3D printing to predict flow behavior within nozzles [40, 146, 147] and the morphology of the deposited inks [148]. Specific to optimizing protein-based inks, computational simulation is also a promising tool to elucidate the molecular basis for assembly, mechanical performance, and degradation [149].
The assembly of large proteins, instead of short peptides, is often challenging for traditional molecular dynamics (MD) simulations, owing to the long folding times on the scale of seconds. To address this issue, replica exchange MD (REMD) simulation, combined with experimental characterization, has been developed to simulate large proteins with 698 residues, tropoelastin (~60kDa) (Fig. 6A) [150]. This approach demonstrated intramolecular assembly in water from a fully extended chain of tropoelastin into elastic fibers. Three mutations (R515A, D72A, and G685D) were simulated to change molecular conformation as well as assembly propensity, providing important insight into sequence-structure relationships. Furthermore, coarse-grained MD (CGMD) suggested that the hierarchical assembly of tropoelastin was driven by specific hydrophobic domains [151]. The interaction between hydrophobic domains reorients surrounding water molecules, and in turn, gives rise to multi-molecule coacervation. While the simulated results were consistent with experimental observations, several co-factors in the assembly process, including integrins, glycosaminoglycans, and environmental cues, such as pH and salts, can be included in the future to further mimic in vivo conditions [151].
Fig. 6.
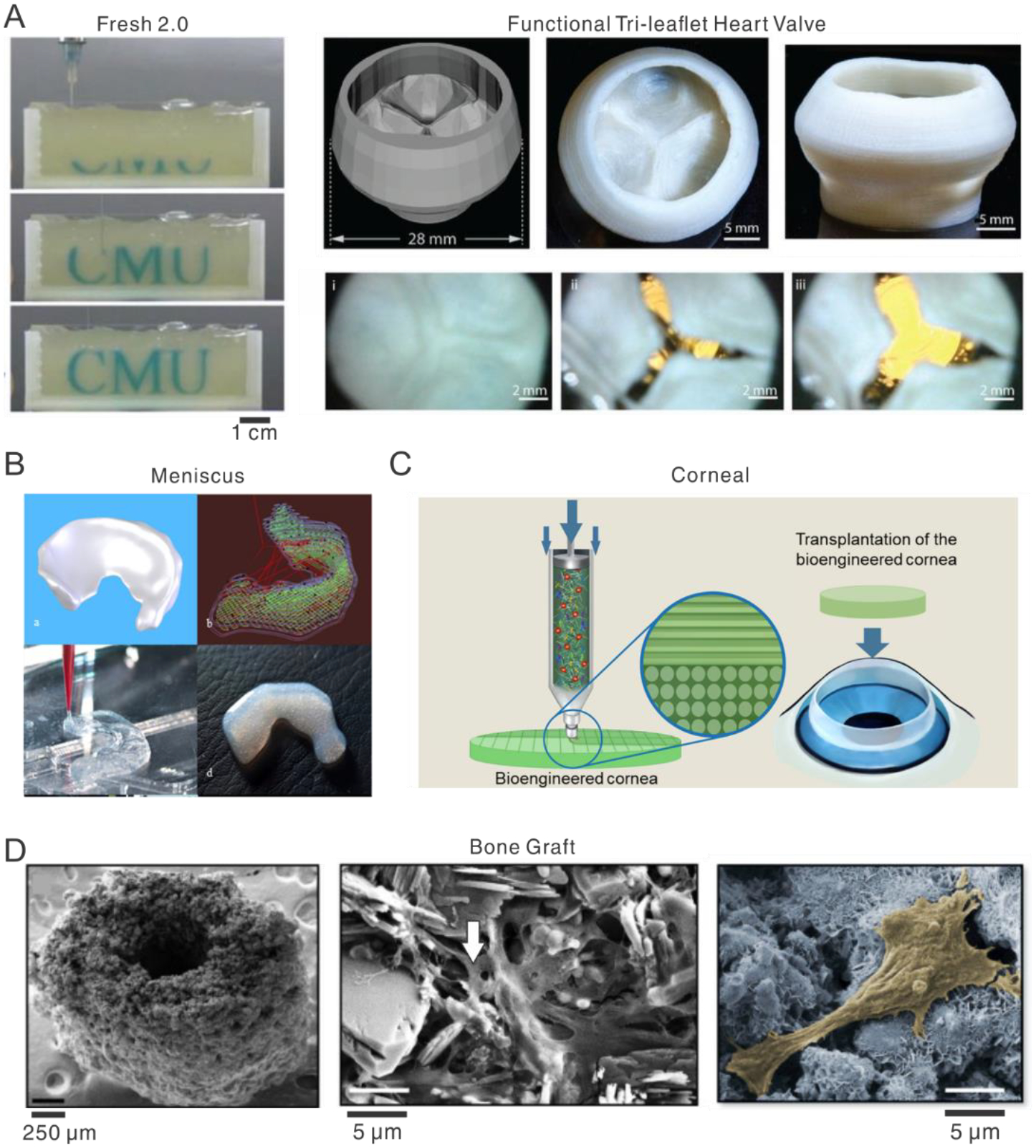
Representative 3D prints with collagen-based inks. A. Left, time-lapse images of the 3D printing of the letters “CMU” dyed in cyan in a creamy suspension of gelatin microparticles with diameters of 25 μm. FRESH means Freeform reversible embedding of suspended hydrogels. Right, a 3D printed functional tri-leaflet heart valve, mechanically enhanced by glutaraldehyde. The opening of the valve was observed underflow. [39], Copyright 2019. Reprinted with permission from the American Association for the Advancement of Science. B. Sheep meniscus analogy printed by using 17.5 mg/ml collagen and bovine meniscal fibrochondrocytes. Top left, CT scan of native meniscus; top right, print path; bottom, 3D printed meniscus. [72], Copyright 2016. Reprinted with permission from the American Chemical Soc. C. Illustration of 3D printing of transparent cornea with aligned collagen fibrils. The alignment results from extrusion-caused shear stress during printing. [177], Copyright 2019. Reprinted with permission from IOP publishing. D. Left, 3D printed bone graft represents the geometry of the murine femoral mid-diaphysis. Middle, collagen, indicated by a white arrow, dispersed among the bound calcium phosphate crystals in the 3D printed structure. Right, a representative cell spreading on the 3D printed structure is healthy. [179], Copyright 2014. Reproduced with permission from the Elsevier.
Dimers of amino-terminal domains play an important role in the assembly of spider silks. Previous experimental results suggested a high concentration of sodium chloride (up to 300 mM) stabilizes the monomer of the N-terminus of the silk during storage in the glands of the animals prior to spinning the web, while the absence of sodium chloride promotes the formation of dimers for molecular assembly and fibrillogenesis during web-spinning [152, 153]. This finding is consistent with the observation that along with the spinning gland, from silk storage to spinning, the concentration of sodium chloride decreases [154]. MD simulation has been used to support further the influence of salt on the N-terminus and these sol-gel transitions. A high concentration of sodium chloride, up to 0.5 M, weakened salt bridges between the N-terminal domains by 28.6%, resulting in stabilized monomers [155].
The amino acid sequences contain essential “information” that directs protein assembly. These sequence-assembly relationships have been investigated by CGMD [156], artificial intelligence (AI) [157, 158], and de novo folding calculations [159, 160]. CGMD predicted the size of aggregations of two recombinant silk peptides, H(AB)12 and H(AB)2, with different numbers of domains or blocks, i.e., polymer chain length [156]. The simulations showed that longer chain length led to larger micelle-like structures, which was experimentally confirmed by atomic force microscopy (AFM) and dynamic light scattering (DLS). H(AB)12 with a molecular weight of 43.7 kDa showed an average diameter of 25 nm; H(AB)2 with 11.6 kDa showed a diameter below 2 nm. Furthermore, MD simulations identified the roles of specific amino acids in the response of proteins to environmental stimuli, such as temperature [161]. In recombinant silk–elastin-like proteins (SELPs), the X in the sequence of (GVGVP)(GXGVP)(GVGVP) has been substituted with valine (V) with a hydrophobic methyl side chain and lysine (K) with a positively charged amino group. The former increases the temperature for the thermal transition with folding via hydrophobic interactions; the latter suppressed this structural transition and the response.
Besides simulating the assembly of proteins, computational simulation has elucidated the molecular basis of the mechanical strength of protein-based materials at the macroscale [162, 163]. All-atom MD simulations indicated that the strength of silk materials mainly results from the appropriate size of β-sheet nanocrystals (2–3 nm) (Fig. 6B) [164, 165]. β-sheet nanocrystals, longer than 3 nm, tend to bend, thus decreasing pulling strength. Mesoscale dissipative particle dynamics (DPD) simulations identified that intermediate hydrophobic/hydrophilic block ratios determined the optimal combination of protein solubility, size of assembled structures, and polymer network topology, which in turn leads to high-performance silk-based materials [166]. Simulations also showed the scale-up of nanoscale properties of silk repeat units to generate macroscopic silk fibers with outstanding mechanical properties, despite the presence of cavities, tears, and cracks [167].
3. Protein-based inks and applications
3.1. Collagens
Collagens are a family of around 29 variants. The most well-known fibrillar collagen is type I, which is accounts for ~90% of all collagens in the human body and constitutes the main structural component in most connective tissues, skin, and bone of mammals [168, 169]. Type I collagen is composed of three left-handed peptide chains that wind-up together to form a right-handed superhelix. The biological significance of type I collagen prompts the use of this protein as inks for 3D printing [170–172]. Type I collagen is most commonly harvested from rat tail, bovine skin, and tendons [173].
Monolithic collagen inks have been 3D printed for constructing a range of tissue equivalents, including heart, meniscus, and cornea (Fig. 6). One outstanding printing approach is the freeform reversible embedding of suspended hydrogels, or FRESH (Fig. 6A) [39, 174]. A suspension of gelatin microparticles was employed to support and maintain the shape of the extruded collagen filament until the gelation of collagen was complete. In FRESH 2.0, the size and distribution of gelatin microparticles were finely controlled, resulting in an improvement of printing resolution from ~200 μm to ~20 μm. Optimized pH (7.4), a high concentration (up to 24 mg/ml), and glutaraldehyde fixation [175] were also adopted to improve printing performance. For example, a 3D printed tri-leaflet heart valve showed Young’s modulus of around 110 kPa. The mechanical strength enabled cyclical opening and closing of the valve leaflets and a maximum transvalvular pressure of around 40 mmHg, above normal physiologic pressures for the tricuspid (~25 mmHg) and pulmonary valves (~10 mmHg)[39].
Collagen inks were also used to print a 3D meniscus by adopting a sharp transition of temperature that expedite the gelation rate (Fig. 6B) [72]. The temperature transition is achieved using an infrared heat lamp to keep the printing surface at 37°C and an ice pack to cool the printing nozzle and syringe. The sharp change of temperature expedites the gelation of collagen and avoids the collapse and spread of filaments. The heating of the printing surface, however, led to premature gelation and syneresis of 20 mg/ml collagen inks, thus limiting the highest concentration available for printing. Nevertheless, 17.5 mg/ml collagen inks led to a maximum modulus of 25 kPa, almost half of the native meniscus. A follow-up study explained the relationship between rheological behavior and the density of chondrocytes in the collagen inks [176]. The chondrocytes with a high cell density up to 100 million/ml remained viable through 14-day cultures.
Besides water-soluble collagen molecules, a suspension of monolithic collagen fibrils was used for 3D printing of transparent cornea stroma (Fig. 6C) [177]. The collagen fibrils were obtained from decellularized corneal tissues (Co-dECM) and were around 50 nm in diameter and 2,000 nm in length. The shear-induced alignment was adopted primarily by manipulating the gauge of the printing nozzle to recapitulate the spatial organization of collagen fibrils in the cornea. The alignment of collagen fibrils increased with increased shear stress, resulting from the decrease of the nozzle diameter. However, a 25 G nozzle, in comparison to a finer 30 G nozzle, maintained the phenotype of keratocytes better, with fewer the formation of myofibroblasts. Myofibroblasts are problematic due to the loss of transparency [178]. The 3D printed cornea analogy exhibited densely packed organization of collagen fibrils and visual transmittance comparable to the human cornea and better than Co-dECM hydrogels.
Composite collagen inks have also been developed for enhancing structural integrity and for adding extra biofunctions. The common additives include calcium phosphate [179], gelatin [180–182], GelMA [67, 183], gold nanorods [184], and alginate [59, 185–187]. In addition, the inherent bioactivity of collagen, largely resulting from the collagen-specific amino acid sequence, was harnessed for bone regeneration [179]. Collagen at a low concentration of 1–2 wt% was blended with other components, including 0.25 wt% Tween 80 and calcium phosphate, to print bone graft substitutes with improved flexural strength and cell viability (Fig. 6D).
3.2. Silk proteins
Silk proteins have been appealing for 3D printing inks [84, 188–190], because of the strong analogy between native silk spinning and extrusion-based printing processes, as well as other material properties. Silk is spun from a gland via a duct, which is similar to the extrusion of printing inks from a syringe through a nozzle [188]. Furthermore, the spinning speed of silks is around 20 mm/s [191], comparable to that of extrusion-based 3D printing and much faster than the weeks- and months-long growth of other natural protein structures, such as wool and tendons. This analogy implies a natural source of inspiration for developing extrusion-based 3D printing techniques with ambient processing conditions and exceptional material performance.
The translation of silk-spinning into 3D printing requires the understanding of molecular mechanisms underlying silk-spinning [29, 192, 193], especially those governing molecular assembly and phase-transitions. Silks are spun by a variety of animals, including insects and spiders. However, all silk proteins, despite species-specific peculiarities, share similar general design principles at the amino acid sequence level. Silk protein consists of a long central segment, flanked by terminal domains; the central core includes short, repeat hydrophobic regions interlaced with hydrophilic domains [194]. For example, the representative repeated hydrophobic regions of spiders and silkworms are often AAAAAA and GAGAGS, respectively. Here, A is alanine, G glycine, and S serine. The hydrophobic regions of silk proteins are folded into crystalline β-sheets that underpin the high stiffness and strength of silk fibers [164]. The terminal domains regulate the formation of micelle-like structures, which enable the storage of silk proteins (>25 wt% in aqueous solution) in the glands prior to spinning [195, 196]. Furthermore, the spinning conditions, such as salts, pH, and water content, have critical roles in mediating the hierarchical molecular assembly that underlies the phase transition from water-soluble silk protein to insoluble fibers [197, 198]. All of these biochemical, mechanical, and processing properties of silk proteins are valuable sources of inspiration for the development of 3D printing techniques with silk proteins or silk-inspired proteins and polymers [40, 54].
Silk-based inks are often regenerated from silkworm cocoons [199, 200] or produced by recombinant DNA techniques with the spider or silkworm motifs [201, 202]. In particular, sericulture provides a massive supply of cocoons for textile industries, thus beneficial for scaling up silk-based inks from silk fibroin, the core protein of domestic B. mori silkworm cocoons. Recombinant DNA approaches provide flexibility to manipulate amino acid sequences based on silks to bring new functions to these proteins. The native silk obtained directly from the spinning gland may also be used as the ink for light-based patterning [203]. It should be noted that regenerated or recombinant silk proteins may possess different properties from that of native silks due to changes in molecular weight, concentration, amino acid sequence, and molecular conformation. These differences need to be considered in designing and developing 3D printing processes with regenerated and recombinant silk proteins.
Monolithic silk protein solutions have been generated for extrusion-based 3D printing. A concentrated solution of regenerated silk fibroin (28–30 wt%) was printed into a methanol bath (85%) for constructing bone scaffolds [83, 204]. However, the methanol bath aggregates silk molecules in a random manner, leading to the brittleness of 3D printed silk structures. Monolithic silk ink was also 3D printed into a bath consisting of synthetic nanoclay (Laponite) and polyethylene glycol (PEG) [205]. The bath mechanically supports the extruded silk ink and physically crosslinks silk molecules via electrostatic interactions and osmotic pressure-induced water removal [206]. The resulting mechanical properties, however, were weak. Hydrogels of recombinant spider silk protein, eADF4(C16) were used for 3D printing cell-laden lattice structures (Fig. 7A) [49, 207]. The hydrogels were formed by thermal gelation (at 37°C) of the eADF4(C16) via physical crosslinking. The cells encapsulated in the hydrogel remained viable for at least one week. RGD sequences can be added to the eADF4(C16) to improve cell adhesion, an advantage in recombinant silk proteins, although such peptides can also be appended onto native silk fibroin via chemical coupling [208]. The elastic modulus of eADF4(C16) is increased from 20 Pa to 200 Pa after adding RGD in the sequence.
Fig. 7.
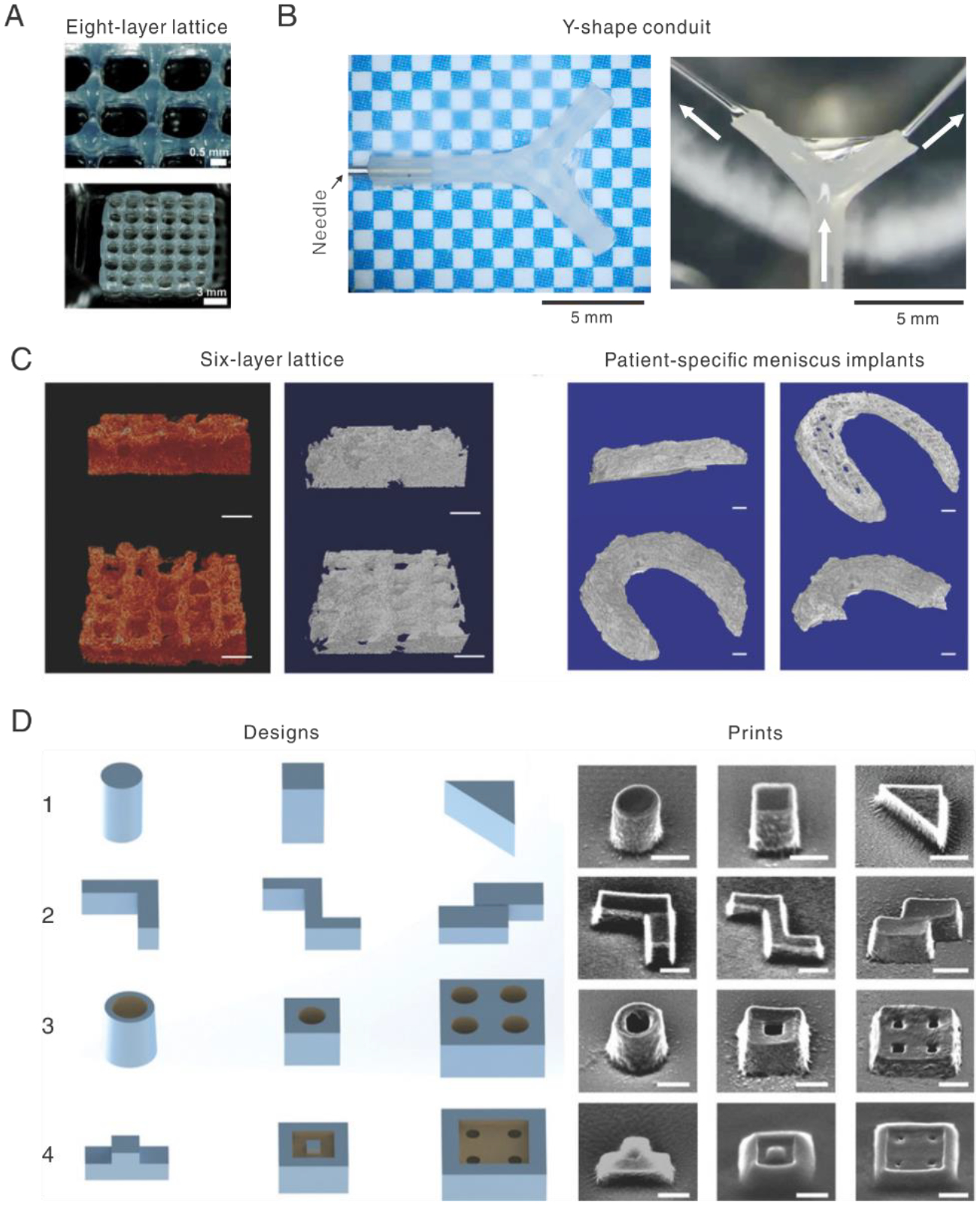
Typical 3D prints with silk-based inks. A. Eight-layer lattice scaffold printed with recombinant spider silk. [49], Copyright 2015. Reprinted with permission from the John Wiley and Sons. B. Y-shape conduit printed with regenerated silk fibroin without using sacrificial or supporting materials. Water jet from the silk conduit, indicated by white arrows, demonstrates the remarkable strength and perfusability. [40], Copyright 2020. Reprinted with permission from the John Wiley and Sons. C. Micro-CT images of enzyme-crosslinked silk prints after freeze-drying. Scale bars: left, 1 mm; right 2.5 mm. [211], Copyright 2017. Reprinted with permission from the John Wiley and Sons. D. Left, a set of designs; right, corresponding SEM images of silk structures by ion and electron beam lithography. The scale bar is 500 nm. [240], Copyright 2018. Reprinted with permission from the John Wiley and Sons.
A de novo aqueous salt bath for 3D printing with monolithic silk inks was developed to improve the mechanical strength of the structures while also avoiding the use of organic solvents in the process [40]. The bath consisted of 0.5 M dipotassium phosphate and 4 M sodium chloride to recapitulate solvent conditions found in the native spinning glands of both spiders and silkworms, such as ion composition and pH. In particular, the high osmolarity (around 9.5 M) of the bath removes water from the silk ink to mimic the decreased water content along the native spinning gland where the silk is pre-assembled prior to spinning into the air. By using the biomimetic salt bath, silk fibroin is directed to physically assemble at hierarchical length scales [40], which is supported by the longitudinal alignment of nanofibrils. The hierarchical assembly also improves the printability of the monolithic silk ink, so fine resolution (about 100 μm), complex geometry, and superior mechanical performance were achieved. Specifically, a Y-shape microfluidic channel with a burst pressure of about 300 kPa was printed in the absence of sacrificial and supporting materials (Fig. 7B). Also, an overhanging filament of monolithic silk fibroin was printed with a high aspect-ratio of ~375, in comparison to previous results of ~20 (polyelectrolyte ink) [53], ~33 (Carbomer-laden hydrogel) [209], and ~1 (silk fibroin) [63, 210]. The overhanging filament is only supported by its two ends (or by one end during printing), thus representing a measure of the printability. In addition, the mechanical performance of silk prints was dramatically improved. The tensile strength, toughness, and Young’s modulus of the 3D printed silk filaments in the wet state were around 39 MPa, 37 MJ m−3, and 117.37 MPa, respectively, superior to other 3D printed protein materials by orders of magnitude, and comparable to synthetic polymers [40].
Silk inks can be 3D printed using enzymatic reactions for a human meniscus scaffold [211] (Fig. 7C) and for microalgal-laden hydrogel structures [63]. Horseradish peroxidase (HRP) and hydrogen peroxide (H2O2) can crosslink silk via the formation of dityrosine bonds [212]. The enzymatically crosslinked silk meniscus scaffolds were post-processed via freeze-drying to be porous. Micro-CT images showed a porosity of about 59.2% and a pore size of about 224.4 μm (Fig. 7C). The meniscus silk scaffold also exhibited shape recovery after a compression strain of 80% and a storage modulus close to the native meniscus. These mechanical and morphological properties are desired for meniscus regeneration [213]. The cytocompatibility of enzymatic crosslinked silk prints was confirmed by the viability of human adipose-derived stem cells [211] and oxygen production by microalgae encapsulated in the 3D printed silk structures [63].
Besides monolithic silk inks, a range of functional additives has been used to formulate composite silk inks for extrusion-based 3D printing. A major portion of additives were thickeners or bulking agents to improve printability and structural integrity, which include agar [214], gelatin [214–221], hydroxypropyl methylcellulose [63, 222, 223], PEG [224, 225], glycerol [216], and Konjac gum [226]. Another category of additives is used to add specific biofunctions to silk inks, including tricalcium phosphate [219], extracellular matrix components [227, 228], hydroxyapatite [204, 229], and bioactive glass [230]. Some components have been used to formulate self-curing silk inks [231, 232], including hydroxypropyl methylcellulose [233], Eri silk/gelatin [218], and polyol/gelatin [214]. Silk was also used to enhance the mechanics of other printing inks, such as chitosan [234, 235] and soy protein isolate [236].
Light-based 3D printing with silk-based inks is mostly based on the photo-polymerization mechanism [95, 210, 237–243], as discussed in section 2.2. The polymorphic nature of silk proteins has also been exploited [244] as a new mechanism to form pseudo-3D and 3D structures via electron-beam and laser irradiation [245, 246]. In particular, irradiation can change the solubility of silks. A set of typical silk structures via the combination of ion and electron beam lithography is shown in Fig. 7D [240]. The first row shows simple geometries; the second row shows combinations of simple geometries; the third row shows both nanosintered (embossing) and nanosculptured (debossing) geometries; and the last row shows geometries with two levels of nanosintering and nanosculpturing, respectively.
3.3. Fibrinogen/Fibrin
Fibrinogen and fibrin are essential for hemostasis, play important roles in many pathophysiological processes, and are promising biomaterials in tissue engineering [247–249]. A unique advantage of fibrinogen, in comparison with other protein materials, is the autologous source [250]. Fibrinogen is produced in the liver and circulates through blood vessels, and thus can be derived from patients’ plasma albeit in limited quantities.
Fibrinogen consists of three pairs of polypeptide chains (Aα, Bβ, and γ) with a total molecular weight of about 340 kDa. Fibrinogen is cleaved by thrombin, a highly specific serine protease, to remove fibrinopeptides (A and B) to form fibrin monomer (α, β, and γ). The fibrin monomer has two complementary domains at the central region and chain ends, which are called “holes” and “knobs”, respectively. Via interactions between the holes and knobs, fibrin monomers form fibrin oligomers and protofibrils that further assemble, laterally and longitudinally, to form a network of branched and interwoven fibrin fibers, i.e., fibrin gels or clots. The chemical and mechanical stability of the fibrin gel are enhanced by calcium-mediated ionic binding, and a plasma transglutaminase, Factor XIIIa, via covalent crosslinking between glutamine and lysine [248].
The understanding of the molecular mechanisms of fibrinogen assembly/ polymerization underpins 3D printing with fibrinogen-based inks. For example, thrombin is used as a printing bath to trigger the phase-transition of fibrinogen solution after extrusion [64, 251, 252]. Factor XIIIa and calcium ions are also frequently used to enhance printability [253, 254]. However, it remains challenging to use fibrin(ogen) alone for printing, owing to inadequate structural stability, and poorly defined geometry. Thus, structural enhancements of fibrinogen-based inks, by adding thickeners and multiple crosslinking mechanisms, are often necessary.
Hyaluronic acid (HA) and polyvinyl alcohol (PVA) were added to fibrinogen ink to increase printability [254]. Moreover, extrusion-induced shear stress aligned fibrin fibers, which directs the growth of neurites and Schwann cells. Other thickeners for fibrinogen-based inks include 3 w/v% alginate, 9 w/v% methylcellulose [255], and gelatin [251, 256] for bone and vascular bioengineering. The printed fibrinogen-gelatin vascular graft, after two-month culture with primary neonatal human dermal fibroblasts, demonstrated a burst pressure of about 1,100 mmHg, roughly 52% of that of the human saphenous vein. A PEG-alginate supporting bath was used for the printed fibrinogen inks to make cartilage-like tissues with controlled mechanical properties[257]. The PEG-alginate formed an interpenetrating polymer network with similar mechanical properties to native cartilages; the fibrinogen prints, softer than PEG-alginate networks, supports the growth of human mesenchymal stem cell (hMSC) spheroids. Also, a blend of polycaprolactone (PCL) and polylactide-co-caprolactone (PLCL) was used, as an internal structural framework, to strengthen the fibrin gel to approach the mechanical properties of the rabbit urethra (Fig. 8A) [252]. Another interesting work blended fibrinogen with multiple additives, including alginate, genipin, and chitosan, and adopted multiple crosslinking mechanisms (Fig. 8B) [60]. This approach has been exploited to print a glioblastoma tumor model with the U87MG cell line for drug screening [258].
Fig. 8.
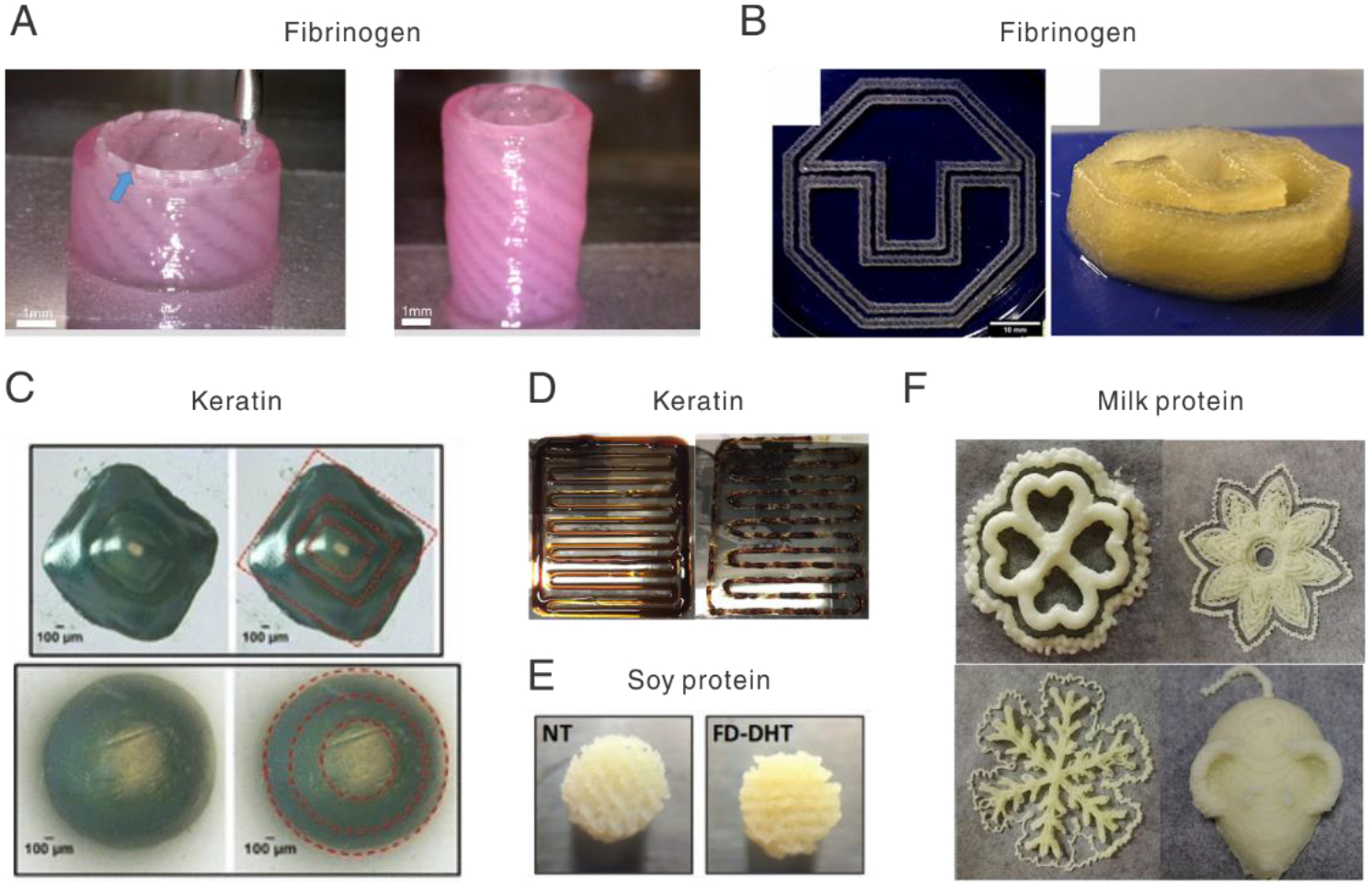
Representative 3D prints of fibrinogen, keratin, and food proteins. A. Printed urethra with fibrinogen-based inks. Blue arrow indicates the PCL/PLCL blend. [252], Copyright 2017. Reproduced with permission from the Elsevier. B. Printed 3D logo with a composite ink of fibrinogen, alginate, and methylcellulose. [255], Copyright 2020. Reproduced with permission from the American Chemical Soc. C. Pyramid structures in keratin were printed by light-based 3D printing. The red dash line outlines the boundary of each layer in cube or cylinder shapes. [113], Copyright 2017. Reprinted with permission from the Springer Nature. D. Printing results of a keratin-lignin (4:1) composite ink. The left and right traces were printed at 60 and 95 min after ink preparation. [268], Copyright 2020. Reprinted with permission from the Elsevier. E. Porous scaffolds printed with soy protein. NT means non-treatment with a diameter of 6.5 ± 0.19mm; FD-DHT means freeze-dried and dehydrothermal treatment with a diameter of 6.57 ± 0.14mm. [273], Copyright 2013. Reproduced with permission from the Elsevier. F. A variety of patterns and structures printed with milk proteins doped with whey protein isolate and other additives. [274], Copyright 2018. Reprinted with permission from the Elsevier.
3.4. Keratin
Keratin, commonly derived from wool, hair, and feathers, has been used for tissue scaffolds and drug delivery [259–261]. One benefit of keratin is the presence of cell-binding amino acid motifs, including leucine-aspartic acid-valine (LDV), glutamic acid-aspartic acid-serine (EDS), and arginine-glycine-aspartic acid (RGD) [262, 263], which promote cell adhesion and proliferation. Keratin has two pentapeptide repeats, CCXPX and CCXS(T)S(T), where C is cysteine, P is proline, S is serine, and T is threonine[264]. The sequence motif enables keratin to form α-helix and coiled-coil architectures that transform to β-sheets in either an irreversible or reversible way. The reversible transformation between coiled-coil and β-sheets for 3D printing shape-memory Keratin structures was recently reported[265].
Keratin was oxidized to reduce the formation of disulfate bonds for preparing inks for light-based 3D printing (Fig. 8C) [113, 266]. 3D printed keratin scaffolds have been used to load a drug, halofuginone, to inhibit contraction for the treatment of dermal burn wounds [267]. The post-processing of the keratin prints, such as lyophilization and sterilization, was investigated toward scalable production and shelf-storage. Besides formulating photo-curable inks, keratin was blended with lignin and other additives, such as Guar gum, for extrusion-based 3D printing (Fig. 8D) [268]. The blending of keratin and lignin forms copolymers via a range of molecular interactions, for example, between keratin amido and lignin aryl hydroxyls, evidenced by solubility and thermal stability.
3.5. Food proteins
Soy, egg white, and milk proteins are common food ingredients and have been formulated into composite inks for 3D printing [269]. Soy protein-based ink has been developed by blending with thickeners, such as polyurethane [270], gelatin [271], and glycerol [272]. An inhibiting reagent for disulfide bond, dithiothreitol, was also added into the soy ink to improve extrudability and printability [272]. The 3D printed soy scaffolds, when used as in vivo implants, showed immune-compatibility and macrophage-mediated degradability (Fig. 8E) [273]. Milk protein concentrate and egg white protein were supplemented with a range of additives, including gum and glycerol [274], gelatin [275, 276], cornstarch [275, 276], sucrose [275, 276], egg yok [277], and rice flour [277]. Several representative 3D prints of milk proteins and whey protein isolate (5:2) are shown in Fig. 8F. It should be noted that these additives adjust rheological behaviors for printing while also optimize the texture and the oral processing of the prints that are edible. For example, the addition of whey protein isolate into the milk protein inks altered microstructures of the 3D printed structures, and thus increased the springiness, cohesiveness, and chewiness, but decreased the gumminess [275]. Furthermore, egg white protein has been developed as a highly transparent and conductive liquid, which was used as an ink for 3D printing for bioelectronics and as a triboelectric nanogenerator [278]. In this study, strong alkaline solutions were used to hydrolyze the egg white proteins, leading to higher conductivity (20.4 S/m). Because both egg white and milk proteins have been useful for 3D cultures of cells and angiogenesis assays [279–283], the ink based on these proteins holds promise for 3D printed tissue scaffolds.
4. Conclusions and perspectives
3D printing with protein-based inks takes advantage of both the material benefits of proteins and the versatile fabrication capability of 3D printing, which has shown the potential to change the landscape of biomedical and clinical research. A wide range of protein materials, nature-derived and recombinant, have been used as essential components of inks for 3D printing with considerable biomedical and clinical interests. 3D printing with protein-based inks brings a new level of the capability to construct structures that underpin disease pathogenesis and contribute to regenerative medicine, as well as to support drug delivery and screening. Despite the substantial promise and rapid progress in recent years, 3D printing with protein-based inks is still in its infancy, with many challenges that require interdisciplinary endeavors between polymer science, molecular biology, rheology, physical chemistry, computational simulations, microfluidics, and biotechniques.
In particular, one major challenge is the weak mechanical strength of protein prints [287], especially for monolithic protein inks in comparison to their natural counterparts. This issue of weak strength also limits structural integrity and shape complexity, thus restricting the broad utility of protein prints. Except for the 3D printing of silk fibroin using bioinspired aqueous salt solution [40], monolithic protein inks are rare to print overhanging filaments and perfusable conduits. Some techniques have been developed to address this issue, including covalent crosslinking [39], suspension baths [39, 174], and the addition of structural enhancing materials [22, 288]. However, non-protein materials and potentially toxic conditions, even approved by the FDA, may impair the merits of protein materials.
Thus, it is reasonable to understand the fundamental molecular mechanisms of high-performance protein tissues for better 3D printing with protein inks. These mechanisms embrace the encoded sequence information, hierarchical structures of protein tissues, and more importantly, the processing principles to govern macromolecular assembly. To recapitulate the assembly process in nature, but here in a 3D printing process, is a route to capture the unique and important mechanical properties of protein tissues.
Besides their unique assembly properties as macromolecular building blocks, they possess other polymeric properties which are valuable to exploit for 3D printing. For example, several proteins, including silk [75], suckerins [289], zein [290, 291], and keratin [268], are thermoplastic. These proteins can be extruded and molded with appropriate thermal and pressure processes. The thermoplastic properties of these proteins represent a promising mechanism for fused deposition modelling (FDM) widely used for 3D printing with traditional thermoplastics [292] and more recently, liquid crystal polymers [10, 293]. The benefits of the thermoplastic approach include the long shelf-storage of dry protein-based inks and the streamlined printing process for scale-up.
The functionality of 3D prints has been growing in emphasis [294], with protein-based inks particularly promising toward this goal. Protein inks, instead of synthetic materials, such as PEG, Polydimethylsiloxane (PDMS), and PCL, bring new opportunities to responsive actuators [295, 296], in vivo 3D printing [297, 298], as well as to the construction of microphysiological systems and tissue-chips [299]. Moreover, 3D printed protein structures have potential in bio-optoelectronics with improved capacity to interface between biology and devices, especially for on-skin applications and in vivo implantation [300–303].
Fig. 5.
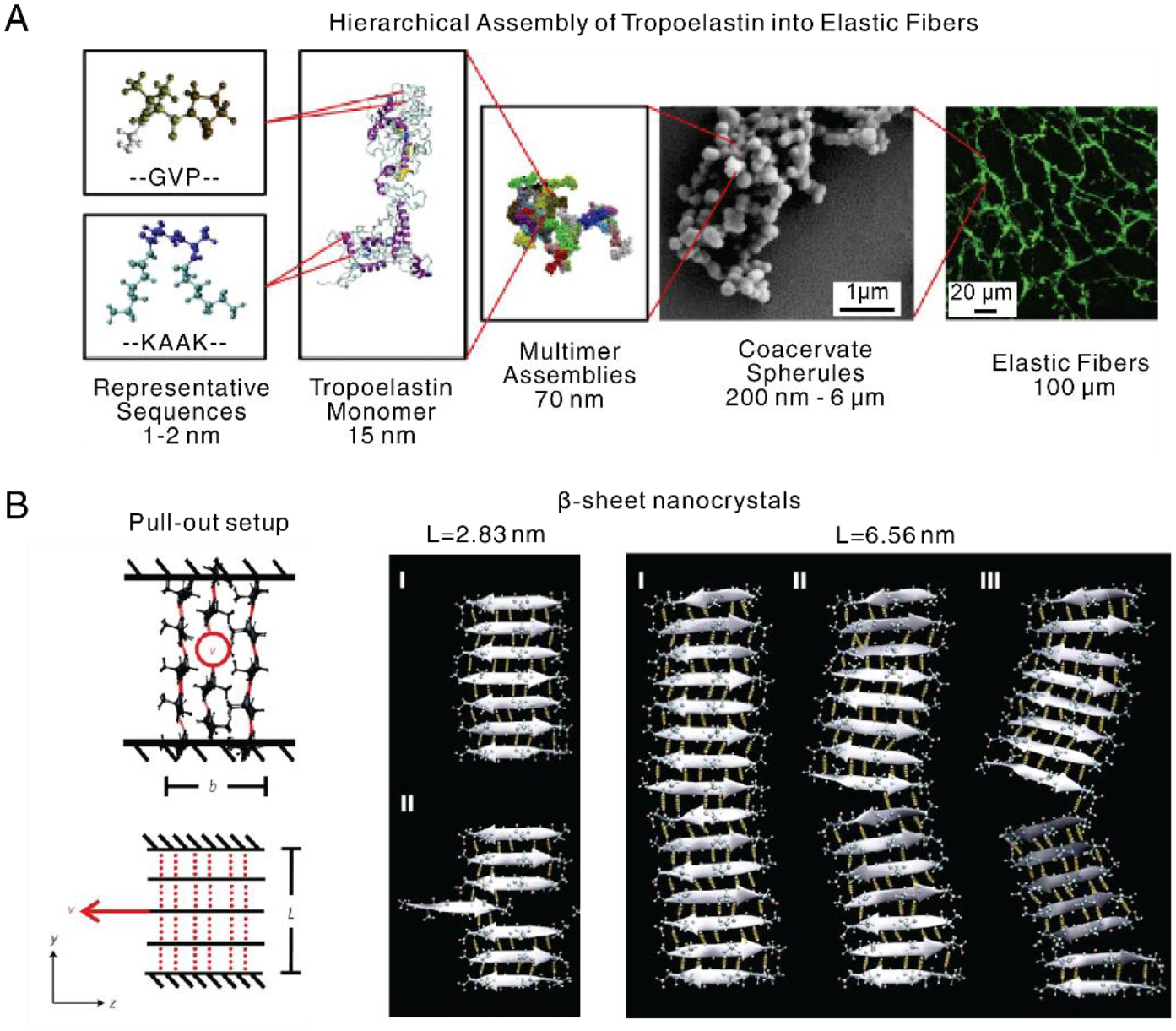
Computational simulations for protein’ structures. A. Tropoelastin, as a basic unit of elastic fibers, consists of alternating GVP-rich hydrophobic domains and hydrophilic K-containing cross-linking domains. Tropoelastin interacts to form multimer assemblies, spherules, and eventually microfibers. [151], Copyright 2019. Reproduced with permission from the Elsevier. B. Pull-out configuration to characterize fracture resistance of nanocrystals, where the central strand of the middle sheet is pulled out with constant velocity (indicated by the red arrow), and the top and bottom strands are restrained. Red dash line and black line indicate hydrogen bonding and amino acid chain, respectively. Nanocrystal with short size (L=2.83 nm) shows a stick-slip motion, due to uniform shear loading, preserving high strength; long size (L=6.56 nm) leads to a crack-like flaw and significant bending, thus decreasing the strength. [164], Copyright 2019. Reprinted with permission from the Springer Nature.
Table 2.
Summary of protein-based inks for 3D printed tissue scaffolds
| Proteins | Ink composition | Methods | Crosslinking | Auxiliary | Young’s modulus | Tissues | References |
|---|---|---|---|---|---|---|---|
| Collagen | Monolithic (up to 24 mg/ml) | Extrusion | Physical | Supporting bath | 10 kPa; 100 kPa (GLU crosslinking) | Heart | [39] |
| Monolithic (up to 17.5 mg/ml) | Extrusion | Physical | Heating | ~30 kPa | Meniscus | [72] | |
| Monolithic (up to 12 mg/ml) | Extrusion | Physical | pH | NA | cartilage | [70] | |
| Collagen fibrils | Extrusion | Physical | Shear-induced fiber alignment | NA | Corneal | [177] | |
| Composite (Gelatin; hyaluronic acid) | Extrusion; Light | Physical; Covalent | Rheology additives | NA | Liver | [57] | |
| Composite (gold nanorods) | Light | Physical | Near-infrared femtosecond laser | NA | Vessel | [184] | |
| Composite (Pluronic® F-127) | Extrusion | Physical | Rheology additives | 7 kPa | Bone marrow | [71] | |
| Composite (Alginate) | Extrusion | Physical | Rheology additives | NA | Bone; liver | [284] | |
| Gelatin | Gelatin methacrylamide (up to 20 w/v%) | Extrusion; Light | Covalent | Photopolymerization | NA | Liver | [67] |
| Gelatin methacrylamide (10 w/v%) | Extrusion | Covalent | Structural enhancement | ~130 kPa | Cartilage | [285] | |
| Gelatin methacryloyl (up to 20 w/v%; Gellan) | Extrusion | Physical | Rheology additives | ~ 186 kPa | Cartilage | [55] | |
| Gelatin-methacryloyl (10 w/v%; 0.6% Collagen) | Extrusion | Covalent | Photopolymerization | NA | Cartilage | [136] | |
| Gelatin methacrylamide (10 w/v%) | Extrusion; Light | Covalent | Enzymatic reactions; Photopolymerization | NA | Fibroblast | [61] | |
| Silk | Monolithic (up to ~30 wt%) | Extrusion | Physical | Aqueous salt bath | 117 MPa (wet) 2 GPa (dry) | Vessel | [40] |
| Monolithic (up to ~30 wt%) | Extrusion | Physical | Methanol bath | 5.6 GPa | Bone | [83] | |
| Monolithic (16 wt%) | Extrusion | Covalent | Enzymatic reactions | 150 – 320 kPa | Meniscus | [211] | |
| Methacryloyl-modified (up to 30% w/v) | Light | Covalent | Digital micromirror device | 15 kPa (Tensile); 40 kPa (Compressive, 20% strain) | Brain; cartilage | [237] | |
| Monolithic (up to 2 wt%); Composite (Gelatin) | Extrusion; Light | Covalent | Photopolymerization; Rheology additives; Sacrificial templates | ~400 kPa | Cartilage | [239] | |
| Monolithic (Hydrogel) | Extrusion | Physical | Amino acid sequence | 0.21 kPa | Fibroblast | [49] | |
| Composite (PEG) | Extrusion | Physical | Rheology additives | NA | Fibroblast | [225] | |
| Composite (glycerol; Gelatin) | Extrusion | Physical | Rheology additives | 1.4 kPa | Bone | [216] | |
| Composite (GelMA; PEGDMA) | Extrusion; Light | Physical; Covalent | Photopolymerization; Enzymatic reactions | ~40 kPa | Heart | [243] | |
| Composite (Fibrin/Gelatin) | Extrusion | Physical; Covalent | Rheology additives | ~16 kPa | Cartilage | [217] | |
| Composite (Gelatin) | Extrusion | Physical | Rheology additives | ~70 kPa (Compressive, 20% strain) | Cartilage | [218] | |
| Fibrin(ogen) | Monolithic (ca. 1mg/ml) | Extrusion | Physical | Ionic bonding | NA | Skin | [253] |
| Composite (Alginate; Methylcellulose) | Extrusion | Physical | Rheology additives | NA | Bone | [255] | |
| Composite (PCL; PLCL) | Extrusion | Physical | Structural enhancement | 1 MPa | Urethra | [252] | |
| Composite (Hyaluronic acid; Polyvinyl alcohol) | Extrusion | Physical; Covalent | Rheology additives; Enzymatic reactions | NA | Nerve | [254] | |
| Composite (Furfuryl-gelatin) | Extrusion; Light | Covalent | Photopolymerization; Rheology additives; Enzymatic reactions | NA | Heart | [286] | |
| Composite (PEG; alginate) | Extrusion | Physical; Covalent | Ionic bonding; Enzymatic reactions; Photopolymerization | Ca. 0.9 – 3 MPa | Cartilage | [257] | |
| Composite (Gelatin) | Extrusion | Physical; Covalent | Heating; Enzymatic reactions | 1 kPa | Vessel | [251] | |
| Keratin | Monolithic (up to 6% w/v) | Light | Covalent | Photopolymerization | ~15 kPa | Fibroblast | [113] |
| Composite (Lignin; guar gum; glycerol) | Extrusion | Physical | Rheology additives | NA | NA | [268] | |
| Soy | Composite (glycerol; dithiothreitol) | Extrusion | Covalent | Rheology additives | 1 kPa ~4 kPa (EDC crosslinking) | NA | [272] |
| Composite (polyurethane) | Extrusion | Physical; Covalent | Rheology additives | NA | Fibroblast | [270] |
GLU: glutaraldehyde; EDC: 1-ethyl-3-(3 dimethylaminopropyl) carbodiimide; PEGDMA, Poly(ethylene glycol) diacrylate.
Acknowledgments
We thank the AFOSR (FA9550-17-1-0333), the NIH (P41EB002520, R01EB021264, R01NS092847, U01EB014976), NSF through DMR-1608125 for the support of the research work as a foundation for this review.
Abbreviation:
- 3D
Three-dimensional
- ECM
extracellular matrices
- Ala
alanine
- Gly
glycine
- Ser
serine
- Val
valine
- Tyr
tyrosine
- H-B
Herschel-Bulkley
- σy
yield stress
- LVR
linear viscoelastic region
- G’
storage modulus
- G”
loss modulus
- T
temperature
- SLA
stereolithography
- DLP
digital light processing
- VA-086
2,2-azobis (2-methyl-N-2-hydroxyethyl) propionamide (VA-086)
- I2959
2-hydroxy-1-[4-(2-hydroxyethoxy) phenyl]-2-methyl-1-propanone
- LAP
lithium phenyl-2,4,6-trimethylbenzoylphosphinate
- [Ru(II)(bpy)3]2+
ruthenium (II) trisbipyridyl
- MD
molecular dynamics
- REMD
replica exchange MD
- CGMD
coarse-grained MD
- DLS
dynamic light scattering
- AFM
atomic force microscopy
- AI
artificial intelligence
- SELPs
silk-elastin-like proteins
- DPD
dissipative particle dynamics
- GelMA
gelatin (meth)acryloyl
- HA
Hyaluronic acid
- PVA
polyvinyl alcohol
- FDM
fused deposition modelling
- GLU
glutaraldehyde
- EDC
1-ethyl-3-(3 dimethylaminopropyl) carbodiimide
- PEGDMA
Poly(ethylene glycol) diacrylate
- FRESH
Freeform reversible embedding of suspended hydrogels
- RGD
arginine-glycine-aspartate
- HRP
Horseradish peroxidase
- H2O2
hydrogen peroxide
- PCL
polycaprolactone
- PLCL
polylactide-co-caprolactone
- LDV
leucine-aspartic acid-valine
- EDS
glutamic acid-aspartic acid-serine
- PDMS
polydimethylsiloxane
- hMSC
human mesenchymal stem cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
References
- [1].Ligon SC, Liska R, Stampfl Jr, Gurr M, Mülhaupt R. Polymers for 3D printing and customized additive manufacturing. Chem Rev 2017;117:10212–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Truby RL, Lewis JA. Printing soft matter in three dimensions. Nature 2016;540:371–8. [DOI] [PubMed] [Google Scholar]
- [3].Valot L, Martinez J, Mehdi A, Subra G. Chemical insights into bioinks for 3D printing. Chem Soc Rev 2019;48:4049–86. [DOI] [PubMed] [Google Scholar]
- [4].Gopinathan J, Noh I. Recent trends in bioinks for 3D printing. Biomater Res 2018;22:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu G, Zhao Y, Wu G, Lu J. Origami and 4D printing of elastomer-derived ceramic structures. Sci Adv 2018;4:eaat0641.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Martin JH, Yahata BD, Hundley JM, Mayer JA, Schaedler TA, Pollock TM. 3D printing of high-strength aluminium alloys. Nature 2017;549:365–9. [DOI] [PubMed] [Google Scholar]
- [7].Zhang D, Qiu D, Gibson MA, Zheng Y, Fraser HL, StJohn DH, et al. Additive manufacturing of ultrafine-grained high-strength titanium alloys. Nature 2019;576:91–5. [DOI] [PubMed] [Google Scholar]
- [8].Kotz F, Arnold K, Bauer W, Schild D, Keller N, Sachsenheimer K, et al. Three-dimensional printing of transparent fused silica glass. Nature 2017;544:337–9. [DOI] [PubMed] [Google Scholar]
- [9].Klein J, Stern M, Franchin G, Kayser M, Inamura C, Dave S, et al. Additive manufacturing of optically transparent glass. 3D Print Addit Manuf 2015;2:92–105. [Google Scholar]
- [10].Gantenbein S, Masania K, Woigk W, Sesseg JP, Tervoort TA, Studart AR. Three-dimensional printing of hierarchical liquid-crystal-polymer structures. Nature 2018;561:226. [DOI] [PubMed] [Google Scholar]
- [11].Grigoryan B, Paulsen SJ, Corbett DC, Sazer DW, Fortin CL, Zaita AJ, et al. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 2019;364:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Malafaya PB, Silva GA, Reis RL. Natural–origin polymers as carriers and scaffolds for biomolecules and cell delivery in tissue engineering applications. Adv Drug Delivery Rev 2007;59:207–33. [DOI] [PubMed] [Google Scholar]
- [13].Guvendiren M, Molde J, Soares RMD, Kohn J. Designing Biomaterials for 3D Printing. ACS Biomater Sci Eng 2016;2:1679–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Feinberg AW, Miller JS. Progress in three-dimensional bioprinting. Mrs Bull 2017;42:557–62. [Google Scholar]
- [15].Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, et al. Silk-based biomaterials. Biomaterials 2003;24:401–16. [DOI] [PubMed] [Google Scholar]
- [16].Abascal NC, Regan L. The past, present and future of protein-based materials. Open Biol 2018;8:180113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hu X, Cebe P, Weiss AS, Omenetto F, Kaplan DL. Protein-based composite materials. Materials Today 2012;15:208–15. [Google Scholar]
- [18].Adamcik J, Mezzenga R. Proteins fibrils from a polymer physics perspective. Macromolecules 2012;45:1137–50. [Google Scholar]
- [19].Knowles TP, Mezzenga R. Amyloid fibrils as building blocks for natural and artificial functional materials. Adv Mater 2016;28:6546–61. [DOI] [PubMed] [Google Scholar]
- [20].Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials 2011;32:3233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Jang J, Park H-J, Kim S-W, Kim H, Park JY, Na SJ, et al. 3D printed complex tissue construct using stem cell-laden decellularized extracellular matrix bioinks for cardiac repair. Biomaterials 2017;112:264–74. [DOI] [PubMed] [Google Scholar]
- [22].Pati F, Jang J, Ha D-H, Kim SW, Rhie J-W, Shim J-H, et al. Printing three-dimensional tissue analogues with decellularized extracellular matrix bioink. Nat Commun 2014;5:3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hussey GS, Dziki JL, Badylak SF. Extracellular matrix-based materials for regenerative medicine. Nat Rev Mater 2018;3:159–73. [Google Scholar]
- [24].Da Silva K, Kumar P, Choonara YE, du Toit LC, Pillay V. 3D Printing of Extracellular Matrix (ECM) - Mimicking Scaffolds: A Critical Review of the Current ECM Materials. J Biomed Mater Res, Part A 2020:;in press. 10.1002/jbm.a.36981 (27 pp). [DOI] [PubMed] [Google Scholar]
- [25].Hynes RO. The Extracellular Matrix: Not Just Pretty Fibrils. Science 2009;326:1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Adams JC, Watt FM. Regulation of development and differentiation by the extracellular matrix. Development 1993;117:1183–98. [DOI] [PubMed] [Google Scholar]
- [27].Muncie JM, Weaver VM. The physical and biochemical properties of the extracellular matrix regulate cell fate. Curr Top Dev Biol 2018;130:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gosline JM. The Structural Design of Collagen Tendons and Ligaments. Mechanical Design of Structural Materials in Animals: Princeton University Press; 2018. p. 103–51. [Google Scholar]
- [29].Guo C, Li C, Mu X, Kaplan DL. Engineering silk materials: From natural spinning to artificial processing. Appl Phys Rev 2020;7:011313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Smith RSH, Bader C, Sharma S, Kolb D, Tang T-C, Hosny A, et al. Hybrid Living Materials: Digital Design and Fabrication of 3D Multimaterial Structures with Programmable Biohybrid Surfaces. Adv Funct Mater 2020;30:1907401. [Google Scholar]
- [31].Schaffner M, Rühs PA, Coulter F, Kilcher S, Studart AR. 3D printing of bacteria into functional complex materials. Sci Adv 2017;3:eaao6804.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Marelli B, Patel N, Duggan T, Perotto G, Shirman E, Li C, et al. Programming function into mechanical forms by directed assembly of silk bulk materials. Proc Natl Acad Sci 2017;114:451–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Placone JK, Engler AJ. Recent Advances in Extrusion - Based 3D Printing for Biomedical Applications. Adv Healthcare Mater 2018;7:1701161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jose RR, Rodriguez MJ, Dixon TA, Omenetto F, Kaplan DL. Evolution of bioinks and additive manufacturing technologies for 3D bioprinting. ACS Biomater Sci Eng 2016;2:1662–78. [DOI] [PubMed] [Google Scholar]
- [35].Heinrich MA, Liu W, Jimenez A, Yang J, Akpek A, Liu X, et al. Bioprinting: 3D Bioprinting: from Benches to Translational Applications. Small 2019;15:1970126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Furst EM. Directed self-assembly. Soft Matter 2013;9:9039–45. [Google Scholar]
- [37].Freeman R, Han M, Álvarez Z, Lewis JA, Wester JR, Stephanopoulos N, et al. Reversible self-assembly of superstructured networks. Science 2018;362:808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Qiu H, Hudson ZM, Winnik MA, Manners I. Multidimensional hierarchical self-assembly of amphiphilic cylindrical block comicelles. Science 2015;347:1329–32. [DOI] [PubMed] [Google Scholar]
- [39].Lee A, Hudson A, Shiwarski D, Tashman J, Hinton T, Yerneni S, et al. 3D bioprinting of collagen to rebuild components of the human heart. Science 2019;365:482–7. [DOI] [PubMed] [Google Scholar]
- [40].Mu X, Wang Y, Guo C, Li Y, Ling S, Huang W, et al. 3D Printing of Silk Protein Structures by Aqueous Solvent-Directed Molecular Assembly. Macromol Biosci 2020;20:1900191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].International A ISO/ASTM52900–15 Standard Terminology for Additive Manufacturing - General Principles - Terminology. West Conshohocken, PA: ASTM International; 2015. p. 1–9. [Google Scholar]
- [42].Malda J, Visser J, Melchels FP, Jüngst T, Hennink WE, Dhert WJ, et al. 25th anniversary article: engineering hydrogels for biofabrication. Adv Mater 2013;25:5011–28. [DOI] [PubMed] [Google Scholar]
- [43].Townsend JM, Beck EC, Gehrke SH, Berkland CJ, Detamore MS. Flow behavior prior to crosslinking: The need for precursor rheology for placement of hydrogels in medical applications and for 3D bioprinting. Prog Polym Sci 2019;91:126–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gao T, Gillispie GJ, Copus JS, PR AK, Seol Y-J, Atala A, et al. Optimization of gelatin–alginate composite bioink printability using rheological parameters: a systematic approach. Biofabrication 2018;10:034106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Loebel C, Rodell CB, Chen MH, Burdick JA. Shear-thinning and self-healing hydrogels as injectable therapeutics and for 3D-printing. Nat Protoc 2017;12:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Lai G, Li Y, Li G. Effect of concentration and temperature on the rheological behavior of collagen solution. Int J Biol Macromol 2008;42:285–91. [DOI] [PubMed] [Google Scholar]
- [47].Pritchard EM, Hu X, Finley V, Kuo CK, Kaplan DL. Effect of Silk Protein Processing on Drug Delivery from Silk Films. Macromol Biosci 2013;13:311–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bohidar HB, Jena SS. Kinetics of sol–gel transition in thermoreversible gelation of gelatin. J Chem Phys 1993;98:8970–7. [Google Scholar]
- [49].Schacht K, Jüngst T, Schweinlin M, Ewald A, Groll J, Scheibel T. Biofabrication of cell-loaded 3D spider silk constructs. Angew Chem, Int Ed 2015;54:2816–20. [DOI] [PubMed] [Google Scholar]
- [50].Cox W, Merz E. Correlation of dynamic and steady flow viscosities. J Polym Sci 1958;28:619–22. [Google Scholar]
- [51].Ouyang L, Highley CB, Rodell CB, Sun W, Burdick JA. 3D Printing of Shear-Thinning Hyaluronic Acid Hydrogels with Secondary Cross-Linking. ACS Biomater Sci Eng 2016;2:1743–51. [DOI] [PubMed] [Google Scholar]
- [52].Highley CB, Rodell CB, Burdick JA. Direct 3D printing of shear-thinning hydrogels into self-healing hydrogels. Adv Mater 2015;27:5075–9. [DOI] [PubMed] [Google Scholar]
- [53].Smay JE, Cesarano J, Lewis JA. Colloidal inks for directed assembly of 3-D periodic structures. Langmuir 2002;18:5429–37. [Google Scholar]
- [54].Lewis JA. Direct ink writing of 3D functional materials. Adv Funct Mater 2006;16:2193–204. [Google Scholar]
- [55].Mouser VH, Melchels FP, Visser J, Dhert WJ, Gawlitta D, Malda J. Yield stress determines bioprintability of hydrogels based on gelatin-methacryloyl and gellan gum for cartilage bioprinting. Biofabrication 2016;8:035003/1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chen T Rheological techniques for yield Stress analysis. TA Instruments Applications Note, AAN017 2017. [Google Scholar]
- [57].Clark CC, Aleman J, Mutkus L, Skardal A. A mechanically robust thixotropic collagen and hyaluronic acid bioink supplemented with gelatin nanoparticles. Bioprinting 2019;16:e00058. [Google Scholar]
- [58].Li H, Tan YJ, Leong KF, Li L. 3D bioprinting of highly thixotropic alginate/methylcellulose hydrogel with strong interface bonding. ACS Appl Mater Interfaces 2017;9:20086–97. [DOI] [PubMed] [Google Scholar]
- [59].Zhu K, Chen N, Liu X, Mu X, Zhang W, Wang C, et al. A General Strategy for Extrusion Bioprinting of Bio-Macromolecular Bioinks through Alginate-Templated Dual-Stage Crosslinking. Macromol Biosci 2018;18:1800127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Abelseth E, Abelseth L, De la Vega L, Beyer ST, Wadsworth SJ, Willerth SM. 3D printing of neural tissues derived from human induced pluripotent stem cells using a fibrin-based bioink. ACS Biomater Sci Eng 2018;5:234–43. [DOI] [PubMed] [Google Scholar]
- [61].Zhou M, Lee BH, Tan YJ, Tan LP. Microbial transglutaminase induced controlled crosslinking of gelatin methacryloyl to tailor rheological properties for 3D printing. Biofabrication 2019;11:025011. [DOI] [PubMed] [Google Scholar]
- [62].Wolberg AS. Fibrinogen and Factor XIII: Newly-Recognized Roles in Venous Thrombosis Formation and Composition. Curr Opin Hematol 2018;25:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Zhao S, Guo C, Kumarasena A, Omenetto FG, Kaplan DL. 3D printing of functional microalgal silk structures for environmental applications. ACS Biomater Sci Eng 2019;5:4808–16. [DOI] [PubMed] [Google Scholar]
- [64].Cui X, Boland T. Human microvasculature fabrication using thermal inkjet printing technology. Biomaterials 2009;30:6221–7. [DOI] [PubMed] [Google Scholar]
- [65].Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques 2004;37:790–802. [DOI] [PubMed] [Google Scholar]
- [66].Wang X, Yan Y, Pan Y, Xiong Z, Liu H, Cheng J, et al. Generation of three-dimensional hepatocyte/gelatin structures with rapid prototyping system. Tissue Eng 2006;12:83–90. [DOI] [PubMed] [Google Scholar]
- [67].Billiet T, Gevaert E, De Schryver T, Cornelissen M, Dubruel P. The 3D printing of gelatin methacrylamide cell-laden tissue-engineered constructs with high cell viability. Biomaterials 2014;35:49–62. [DOI] [PubMed] [Google Scholar]
- [68].Kramer RZ, Bella J, Mayville P, Brodsky B, Berman HM. Sequence dependent conformational variations of collagen triple-helical structure. Nature Structural Biology 1999;6:454–7. [DOI] [PubMed] [Google Scholar]
- [69].Jeong B, Kim SW, Bae YH. Thermosensitive sol–gel reversible hydrogels. Adv Drug Delivery Rev 2012;64:154–62. [DOI] [PubMed] [Google Scholar]
- [70].Diamantides N, Wang L, Pruiksma T, Siemiatkoski J, Dugopolski C, Shortkroff S, et al. Correlating rheological properties and printability of collagen bioinks: the effects of riboflavin photocrosslinking and pH. Biofabrication 2017;9:034102. [DOI] [PubMed] [Google Scholar]
- [71].Moncal KK, Ozbolat V, Datta P, Heo DN, Ozbolat IT. Thermally-controlled extrusion-based bioprinting of collagen. J Mater Sci: Mater Med 2019;30:55. [DOI] [PubMed] [Google Scholar]
- [72].Rhee S, Puetzer JL, Mason BN, Reinhart-King CA, Bonassar LJ. 3D bioprinting of spatially heterogeneous collagen constructs for cartilage tissue engineering. ACS Biomater Sci Eng 2016;2:1800–5. [DOI] [PubMed] [Google Scholar]
- [73].Cebe P, Hu X, Kaplan DL, Zhuravlev E, Wurm A, Arbeiter D, et al. Beating the Heat - Fast Scanning Melts Silk Beta Sheet Crystals. Sci Rep 2013;3:1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Koh L-D, Cheng Y, Teng C-P, Khin Y-W, Loh X-J, Tee S-Y, et al. Structures, mechanical properties and applications of silk fibroin materials. Prog Polym Sci 2015;46:86–110. [Google Scholar]
- [75].Guo C, Li C, Vu HV, Hanna P, Lechtig A, Qiu Y, et al. Thermoplastic moulding of regenerated silk. Nat Mater 2020;19:102–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Matsumoto A, Chen J, Collette AL, Kim U-J, Altman GH, Cebe P, et al. Mechanisms of Silk Fibroin Sol–Gel Transitions. J Phys Chem B 2006;110:21630–8. [DOI] [PubMed] [Google Scholar]
- [77].Tsukada M, Gotoh Y, Nagura M, Minoura N, Kasai N, Freddi G. Structural changes of silk fibroin membranes induced by immersion in methanol aqueous solutions. J Polym Sci, Part B: Polym Phys 1994;32:961–8. [Google Scholar]
- [78].Kim U-J, Park J, Kim HJ, Wada M, Kaplan DL. Three-dimensional aqueous-derived biomaterial scaffolds from silk fibroin. Biomaterials 2005;26:2775–85. [DOI] [PubMed] [Google Scholar]
- [79].Drummy LF, Phillips DM, Stone MO, Farmer B, Naik RR. Thermally induced α-helix to β-sheet transition in regenerated silk fibers and films. Biomacromolecules 2005;6:3328–33. [DOI] [PubMed] [Google Scholar]
- [80].Foo CWP, Bini E, Hensman J, Knight D, Lewis R, Kaplan D. Role of pH and charge on silk protein assembly in insects and spiders. Appl Phys A 2006;82:223–33. [Google Scholar]
- [81].Knight D, Knight M, Vollrath F. Beta transition and stress-induced phase separation in the spinning of spider dragline silk. Int J Biol Macromol 2000;27:205–10. [DOI] [PubMed] [Google Scholar]
- [82].Wang X, Kluge JA, Leisk GG, Kaplan DL. Sonication-induced gelation of silk fibroin for cell encapsulation. Biomaterials 2008;29:1054–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Ghosh S, Parker ST, Wang X, Kaplan DL, Lewis JA. Direct - Write Assembly of Microperiodic Silk Fibroin Scaffolds for Tissue Engineering Applications. Adv Funct Mater 2008;18:1883–9. [Google Scholar]
- [84].Chawla S, Midha S, Sharma A, Ghosh S. Silk‐based bioinks for 3D bioprinting. Adv Healthcare Mater 2018;7:1701204. [DOI] [PubMed] [Google Scholar]
- [85].Shen Y, Tang H, Huang X, Hang R, Zhang X, Wang Y, et al. DLP printing photocurable chitosan to build bio-constructs for tissue engineering. Carbohydr Polym 2020;235:115970. [DOI] [PubMed] [Google Scholar]
- [86].Ma X, Yu C, Wang P, Xu W, Wan X, Lai CSE, et al. Rapid 3D bioprinting of decellularized extracellular matrix with regionally varied mechanical properties and biomimetic microarchitecture. Biomaterials 2018;185:310–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Ma X, Qu X, Zhu W, Li Y-S, Yuan S, Zhang H, et al. Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting. Proc Natl Acad Sci 2016;113:2206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Ge Q, Sakhaei AH, Lee H, Dunn CK, Fang NX, Dunn ML. Multimaterial 4D printing with tailorable shape memory polymers. Sci Rep 2016;6:31110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wang P, Berry D, Moran A, He F, Tam T, Chen L, et al. Controlled Growth Factor Release in 3D-Printed Hydrogels. Adv Healthcare Mater 2020:1900977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Skylar-Scott MA, Mueller J, Visser CW, Lewis JA. Voxelated soft matter via multimaterial multinozzle 3D printing. Nature 2019;575:330–5. [DOI] [PubMed] [Google Scholar]
- [91].Bader C, Kolb D, Weaver JC, Sharma S, Hosny A, Costa J, et al. Making data matter: Voxel printing for the digital fabrication of data across scales and domains. Sci Adv 2018;4:eaas8652.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Shaegh SAM, et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci 2017;114:E2293–E302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Xing J-F, Zheng M-L, Duan X-M. Two-photon polymerization microfabrication of hydrogels: an advanced 3D printing technology for tissue engineering and drug delivery. Chem Soc Rev 2015;44:5031–9. [DOI] [PubMed] [Google Scholar]
- [94].Worthington KS, Do AV, Smith R, Tucker BA, Salem AK. Two-Photon Polymerization as a Tool for Studying 3D Printed Topography-Induced Stem Cell Fate. Macromol Biosci 2019;19:1800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Sun Y-L, Li Q, Sun S-M, Huang J-C, Zheng B-Y, Chen Q-D, et al. Aqueous multiphoton lithography with multifunctional silk-centred bio-resists. Nat Commun 2015;6:8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Mu Q, Wang L, Dunn CK, Kuang X, Duan F, Zhang Z, et al. Digital light processing 3D printing of conductive complex structures. Addit Manuf 2017;18:74–83. [Google Scholar]
- [97].Lu Y, Mapili G, Suhali G, Chen S, Roy K. A digital micro-mirror device-based system for the microfabrication of complex, spatially patterned tissue engineering scaffolds. J Biomed Mater Res, Part A 2006;77:396–405. [DOI] [PubMed] [Google Scholar]
- [98].Hughes T, Simon GP, Saito K. Chemistries and capabilities of photo-formable and photoreversible crosslinked polymer networks. Mater Horiz 2019;6:1762–73. [Google Scholar]
- [99].Lim KS, Galarraga JH, Cui X, Lindberg GC, Burdick JA, Woodfield TB. Fundamentals and Applications of Photo-Cross-Linking in Bioprinting. Chem Rev 2020;120:10662–94. [DOI] [PubMed] [Google Scholar]
- [100].Knowlton S, Yenilmez B, Anand S, Tasoglu S. Photocrosslinking-based bioprinting: Examining crosslinking schemes. Bioprinting 2017;5:10–8. [Google Scholar]
- [101].Yu C, Schimelman J, Wang P, Miller KL, Ma X, You S, et al. Photopolymerizable Biomaterials and Light-Based 3D Printing Strategies for Biomedical Applications. Chem Rev 2020;120:10695–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Zhang X, Jiang C, Cheng M, Zhou Y, Zhu X, Nie J, et al. Facile Method for the Fabrication of Robust Polyelectrolyte Multilayers by Post-Photo-Cross-Linking of Azido Groups. Langmuir 2012;28:7096–100. [DOI] [PubMed] [Google Scholar]
- [103].Van Den Bulcke AI, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules 2000;1:31–8. [DOI] [PubMed] [Google Scholar]
- [104].Yue K, Trujillo-de Santiago G, Alvarez MM, Tamayol A, Annabi N, Khademhosseini A. Synthesis, properties, and biomedical applications of gelatin methacryloyl (GelMA) hydrogels. Biomaterials 2015;73:254–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials 2009;30:6702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Nguyen AK, Goering PL, Reipa V, Narayan RJ. Toxicity and photosensitizing assessment of gelatin methacryloyl-based hydrogels photoinitiated with lithium phenyl-2,4,6-trimethylbenzoylphosphinate in human primary renal proximal tubule epithelial cells. Biointerphases 2019;14:021007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Williams CG, Malik AN, Kim TK, Manson PN, Elisseeff JH. Variable cytocompatibility of six cell lines with photoinitiators used for polymerizing hydrogels and cell encapsulation. Biomaterials 2005;26:1211–8. [DOI] [PubMed] [Google Scholar]
- [108].Son TI, Sakuragi M, Takahashi S, Obuse S, Kang J, Fujishiro M, et al. Visible light-induced crosslinkable gelatin. Acta Biomater 2010;6:4005–10. [DOI] [PubMed] [Google Scholar]
- [109].Bahney C, Lujan T, Hsu C, Bottlang M, West J, Johnstone B. Visible light photoinitiation of mesenchymal stem cell-laden bioresponsive hydrogels. Eur Cells Mater 2011;22:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Wang Z, Abdulla R, Parker B, Samanipour R, Ghosh S, Kim K. A simple and high-resolution stereolithography-based 3D bioprinting system using visible light crosslinkable bioinks. Biofabrication 2015;7:045009. [DOI] [PubMed] [Google Scholar]
- [111].Noshadi I, Hong S, Sullivan KE, Sani ES, Portillo-Lara R, Tamayol A, et al. In vitro and in vivo analysis of visible light crosslinkable gelatin methacryloyl (GelMA) hydrogels. Biomater Sci 2017;5:2093–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Applegate MB, Partlow BP, Coburn J, Marelli B, Pirie C, Pineda R, et al. Photocrosslinking of Silk Fibroin Using Riboflavin for Ocular Prostheses. Adv Mater 2016;28:2417–20. [DOI] [PubMed] [Google Scholar]
- [113].Placone JK, Navarro J, Laslo GW, Lerman MJ, Gabard AR, Herendeen GJ, et al. Development and characterization of a 3D printed, keratin-based hydrogel. Ann Biomed Eng 2017;45:237–48. [DOI] [PubMed] [Google Scholar]
- [114].Nguyen AK, Gittard SD, Koroleva A, Schlie S, Gaidukeviciute A, Chichkov BN, et al. Two-photon polymerization of polyethylene glycol diacrylate scaffolds with riboflavin and triethanolamine used as a water-soluble photoinitiator. Regener Med 2013;8:725–38. [DOI] [PubMed] [Google Scholar]
- [115].Bell A, Kofron M, Nistor V. Multiphoton crosslinking for biocompatible 3D printing of type I collagen. Biofabrication 2015;7:035007. [DOI] [PubMed] [Google Scholar]
- [116].Lewis JB, Wataha JC, Messer RL, Caughman GB, Yamamoto T, Hsu SD. Blue light differentially alters cellular redox properties. J Biomed Mater Res, Part B 2005;72:223–9. [DOI] [PubMed] [Google Scholar]
- [117].Subasinghe SK, Ogbuehi KC, Dias GJ. Current perspectives on corneal collagen crosslinking (CXL). Graefe’s Arch Clin Exp Ophthalmol 2018;256:1363–84. [DOI] [PubMed] [Google Scholar]
- [118].Tirella A, Liberto T, Ahluwalia A. Riboflavin and collagen: New crosslinking methods to tailor the stiffness of hydrogels. Mater Lett 2012;74:58–61. [Google Scholar]
- [119].Hoyle CE, Bowman CN. Thiol–ene click chemistry. Angew Chem, Int Ed 2010;49:1540–73. [DOI] [PubMed] [Google Scholar]
- [120].Tansaz S, Boccaccini AR. Biomedical applications of soy protein: A brief overview. J Biomed Mater Res, Part A 2016;104:553–69. [DOI] [PubMed] [Google Scholar]
- [121].Bertlein S, Brown G, Lim KS, Jungst T, Boeck T, Blunk T, et al. Thiol–ene clickable gelatin: a platform bioink for multiple 3D biofabrication technologies. Adv Mater 2017;29:1703404. [DOI] [PubMed] [Google Scholar]
- [122].Mũnoz Z, Shih H, Lin C-C. Gelatin hydrogels formed by orthogonal thiol–norbornene photochemistry for cell encapsulation. Biomater Sci 2014;2:1063–72. [DOI] [PubMed] [Google Scholar]
- [123].Van Hoorick J, Gruber P, Markovic M, Rollot M, Graulus G-J, Vagenende M, et al. Highly Reactive Thiol-Norbornene Photo-Click Hydrogels: Toward Improved Processability. Macromol Rapid Commun 2018;39:1800181. [DOI] [PubMed] [Google Scholar]
- [124].Xu K, Fu Y, Chung W, Zheng X, Cui Y, Hsu IC, et al. Thiol–ene-based biological/synthetic hybrid biomatrix for 3-D living cell culture. Acta Biomater 2012;8:2504–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Bao Z, Gao M, Fan X, Cui Y, Yang J, Peng X, et al. Development and characterization of a photo-cross-linked functionalized type-I collagen (Oreochromis niloticus) and polyethylene glycol diacrylate hydrogel. Int J Biol Macromol 2020;155:163–73. [DOI] [PubMed] [Google Scholar]
- [126].Cramer NB, Scott JP, Bowman CN. Photopolymerizations of Thiol–Ene Polymers without Photoinitiators. Macromolecules 2002;35:5361–5. [Google Scholar]
- [127].Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, et al. Synthesis and properties of crosslinked recombinant pro-resilin. Nature 2005;437:999–1002. [DOI] [PubMed] [Google Scholar]
- [128].Lv S, Dudek DM, Cao Y, Balamurali M, Gosline J, Li H. Designed biomaterials to mimic the mechanical properties of muscles. Nature 2010;465:69–73. [DOI] [PubMed] [Google Scholar]
- [129].Elvin CM, Brownlee AG, Huson MG, Tebb TA, Kim M, Lyons RE, et al. The development of photochemically crosslinked native fibrinogen as a rapidly formed and mechanically strong surgical tissue sealant. Biomaterials 2009;30:2059–65. [DOI] [PubMed] [Google Scholar]
- [130].Elvin CM, Vuocolo T, Brownlee AG, Sando L, Huson MG, Liyou NE, et al. A highly elastic tissue sealant based on photopolymerised gelatin. Biomaterials 2010;31:8323–31. [DOI] [PubMed] [Google Scholar]
- [131].Bjork JW, Johnson SL, Tranquillo RT. Ruthenium-catalyzed photo cross-linking of fibrin-based engineered tissue. Biomaterials 2011;32:2479–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Murphy AR, Kaplan DL. Biomedical applications of chemically-modified silk fibroin. J Mater Chem 2009;19:6443–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Whittaker JL, Choudhury NR, Dutta NK, Zannettino A. Facile and rapid ruthenium mediated photo-crosslinking of Bombyx mori silk fibroin. J Mater Chem B 2014;2:6259–70. [DOI] [PubMed] [Google Scholar]
- [134].Heichel DL, Burke KA. Dual-Mode Cross-Linking Enhances Adhesion of Silk Fibroin Hydrogels to Intestinal Tissue. ACS Biomater Sci Eng 2019;5:3246–59. [DOI] [PubMed] [Google Scholar]
- [135].Fancy DA, Kodadek T. Chemistry for the analysis of protein–protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc Natl Acad Sci 1999;96:6020–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Lim KS, Schon BS, Mekhileri NV, Brown GC, Chia CM, Prabakar S, et al. New visible-light photoinitiating system for improved print fidelity in gelatin-based bioinks. ACS Biomater Sci Eng 2016;2:1752–62. [DOI] [PubMed] [Google Scholar]
- [137].Zhang S, Vijayavenkataraman S, Lu WF, Fuh JY. A review on the use of computational methods to characterize, design, and optimize tissue engineering scaffolds, with a potential in 3D printing fabrication. J Biomed Mater Res, Part B 2018;107B:1329–51. [DOI] [PubMed] [Google Scholar]
- [138].Adachi T, Osako Y, Tanaka M, Hojo M, Hollister SJ. Framework for optimal design of porous scaffold microstructure by computational simulation of bone regeneration. Biomaterials 2006;27:3964–72. [DOI] [PubMed] [Google Scholar]
- [139].Hollister SJ, Lin CY. Computational design of tissue engineering scaffolds. COMPUT METHOD APPL M 2007;196:2991–8. [Google Scholar]
- [140].Tozzi L, Laurent P-A, Di Buduo CA, Mu X, Massaro A, Bretherton R, et al. Multi-channel silk sponge mimicking bone marrow vascular niche for platelet production. Biomaterials 2018;178:122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Mu X, Kaplan DL. Modeling of Mass Transport in 3D Vascularized Porous Tissue Scaffolds. Comsol Conference. Boston 2019. [Google Scholar]
- [142].Kuo C-Y, Eranki A, Placone JK, Rhodes KR, Aranda-Espinoza H, Fernandes R, et al. Development of a 3D Printed, Bioengineered Placenta Model to Evaluate the Role of Trophoblast Migration in Preeclampsia. ACS Biomater Sci Eng 2016;2:1817–26. [DOI] [PubMed] [Google Scholar]
- [143].Sun H, Chan C-W, Wang Y, Yao X, Mu X, Lu X, et al. Reliable and reusable whole polypropylene plastic microfluidic devices for a rapid, low-cost antimicrobial susceptibility test. Lab Chip 2019;19:2915–24. [DOI] [PubMed] [Google Scholar]
- [144].Dimas LS, Bratzel GH, Eylon I, Buehler MJ. Tough Composites Inspired by Mineralized Natural Materials: Computation, 3D printing, and Testing. Adv Funct Mater 2013;23:4629–38. [Google Scholar]
- [145].To AC. Preface. Addit Manuf 2016;12:143-. [Google Scholar]
- [146].Emmermacher J, Spura D, Cziommer J, Kilian D, Wollborn T, Fritsching U, et al. Engineering considerations on extrusion-based bioprinting: interactions of material behavior, mechanical forces and cells in the printing needle. Biofabrication 2020;12:025022. [DOI] [PubMed] [Google Scholar]
- [147].Yang F, Guo C, Zhang M, Bhandari B, Liu Y. Improving 3D printing process of lemon juice gel based on fluid flow numerical simulation. Lwt 2019;102:89–99. [Google Scholar]
- [148].Göhl J, Markstedt K, Mark A, Håkansson K, Gatenholm P, Edelvik F. Simulations of 3D bioprinting: predicting bioprintability of nanofibrillar inks. Biofabrication 2018;10:034105. [DOI] [PubMed] [Google Scholar]
- [149].Dinjaski N, Ebrahimi D, Qin Z, Giordano JE, Ling S, Buehler MJ, et al. Predicting rates of in vivo degradation of recombinant spider silk proteins. J Tissue Eng Regener Med 2018;12:e97–e105. [DOI] [PubMed] [Google Scholar]
- [150].Tarakanova A, Yeo GC, Baldock C, Weiss AS, Buehler MJ. Molecular model of human tropoelastin and implications of associated mutations. Proc Natl Acad Sci 2018;115:7338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Tarakanova A, Ozsvar J, Weiss A, Buehler M. Coarse-grained model of tropoelastin self-assembly into nascent fibrils. Materials Today Bio 2019;3:100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Hagn F, Thamm C, Scheibel T, Kessler H. pH-Dependent Dimerization and Salt – Dependent Stabilization of the N-terminal Domain of Spider Dragline Silk-Implications for Fiber Formation. Angew Chem, Int Ed 2011;50:310–3. [DOI] [PubMed] [Google Scholar]
- [153].Askarieh G, Nordling K, Saenz A, Casals C, Rising A, Johansson J, et al. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 2010;465:236. [DOI] [PubMed] [Google Scholar]
- [154].Knight DP, Vollrath F. Changes in element composition along the spinning duct in a Nephila spider. Naturwissenschaften 2001;88:179–82. [DOI] [PubMed] [Google Scholar]
- [155].Gronau G, Qin Z, Buehler MJ. Effect of sodium chloride on the structure and stability of spider silk’s N-terminal protein domain. Biomater Sci 2013;1:276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Tokareva OS, Lin S, Jacobsen MM, Huang W, Rizzo D, Li D, et al. Effect of sequence features on assembly of spider silk block copolymers. J Struct Biol 2014;186:412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Yu C-H, Qin Z, Martin-Martinez FJ, Buehler MJ. A Self-Consistent Sonification Method to Translate Amino Acid Sequences into Musical Compositions and Application in Protein Design Using Artificial Intelligence. ACS Nano 2019;13:7471–82. [DOI] [PubMed] [Google Scholar]
- [158].Qin Z, Wu L, Sun H, Huo S, Ma T, Lim E, et al. Artificial intelligence method to design and fold alpha-helical structural proteins from the primary amino acid sequence. Extreme Mech Lett 2020;36:100652. [Google Scholar]
- [159].Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods in enzymology: Elsevier; 2004. p. 66–93. [DOI] [PubMed] [Google Scholar]
- [160].Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, et al. Principles for designing ideal protein structures. Nature 2012;491:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Huang W, Tarakanova A, Dinjaski N, Wang Q, Xia X, Chen Y, et al. Design of Multistimuli Responsive Hydrogels Using Integrated Modeling and Genetically Engineered Silk – Elastin - Like Proteins. Adv Funct Mater 2016;26:4113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Su I, Jung GS, Narayanan N, Buehler MJ. Perspectives on 3D printing of self-assembling materials and structures. Cur Opini Biomed Eng 2020;15:59–67. [Google Scholar]
- [163].López Barreiro D, Yeo J, Tarakanova A, Martin - Martinez FJ, Buehler MJ. Multiscale Modeling of Silk and Silk - Based Biomaterials—A Review. Macromolecular bioscience 2019;19:1800253. [DOI] [PubMed] [Google Scholar]
- [164].Keten S, Xu Z, Ihle B, Buehler MJ. Nanoconfinement controls stiffness, strength and mechanical toughness of β-sheet crystals in silk. Nat Mater 2010;9:359. [DOI] [PubMed] [Google Scholar]
- [165].Cheng Y, Koh L-D, Li D, Ji B, Han M-Y, Zhang Y-W. On the strength of β-sheet crystallites of Bombyx mori silk fibroin. Journal of the Royal Society Interface 2014;11:20140305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Lin S, Ryu S, Tokareva O, Gronau G, Jacobsen MM, Huang W, et al. Predictive modelling-based design and experiments for synthesis and spinning of bioinspired silk fibres. Nat Commun 2015;6:6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Giesa T, Arslan M, Pugno NM, Buehler MJ. Nanoconfinement of Spider Silk Fibrils Begets Superior Strength, Extensibility, and Toughness. Nano Lett 2011;11:5038–46. [DOI] [PubMed] [Google Scholar]
- [168].Fratzl P Collagen: structure and mechanics, an introduction. Collagen: Springer; 2008. p. 1–13. [Google Scholar]
- [169].Sorushanova A, Delgado LM, Wu Z, Shologu N, Kshirsagar A, Raghunath R, et al. The Collagen Suprafamily: From Biosynthesis to Advanced Biomaterial Development. Adv Mater 2019;31:1801651. [DOI] [PubMed] [Google Scholar]
- [170].Marques C, Diogo G, Pina S, Oliveira J, Silva T, Reis R. Collagen-based bioinks for hard tissue engineering applications: a comprehensive review. J Mater Sci: Mater Med 2019;30:32. [DOI] [PubMed] [Google Scholar]
- [171].Osidak EO, Kozhukhov VI, Osidak MS, Domogatskiy SP. Collagen as Bioink for Bioprinting: A Comprehensive Review. Int J Bioprint 2020;6:17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Osidak EO, Karalkin PA, Osidak MS, Parfenov VA, Sivogrivov DE, Pereira FDAS, et al. Viscoll collagen solution as a novel bioink for direct 3D bioprinting. J Mater Sci: Mater Med 2019;30:31. [DOI] [PubMed] [Google Scholar]
- [173].Cen L, Liu W, Cui L, Zhang W, Cao Y. Collagen tissue engineering: development of novel biomaterials and applications. Pediatr Res 2008;63:492–6. [DOI] [PubMed] [Google Scholar]
- [174].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, et al. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci Adv 2015;1:e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Lim H-G, Kim GB, Jeong S, Kim YJ. Development of a next-generation tissue valve using a glutaraldehyde-fixed porcine aortic valve treated with decellularization, α-galactosidase, space filler, organic solvent and detoxification. Eur J Cardiothorac Surg 2015;48:104–13. [DOI] [PubMed] [Google Scholar]
- [176].Diamantides N, Dugopolski C, Blahut E, Kennedy S, Bonassar LJ. High density cell seeding affects the rheology and printability of collagen bioinks. Biofabrication 2019;11:045016. [DOI] [PubMed] [Google Scholar]
- [177].Kim H, Jang J, Park J, Lee K-P, Lee S, Lee D-M, et al. Shear-induced alignment of collagen fibrils using 3D cell printing for corneal stroma tissue engineering. Biofabrication 2019;11:035017. [DOI] [PubMed] [Google Scholar]
- [178].Torricelli AA, Wilson SE. Cellular and extracellular matrix modulation of corneal stromal opacity. Exp Eye Res 2014;129:151–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Inzana JA, Olvera D, Fuller SM, Kelly JP, Graeve OA, Schwarz EM, et al. 3D printing of composite calcium phosphate and collagen scaffolds for bone regeneration. Biomaterials 2014;35:4026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo S-S, et al. Creating perfused functional vascular channels using 3D bio-printing technology. Biomaterials 2014;35:8092–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Lee W, Lee V, Polio S, Keegan P, Lee JH, Fischer K, et al. On - demand three - dimensional freeform fabrication of multi - layered hydrogel scaffold with fluidic channels. Biotechnol Bioeng 2010;105:1178–86. [DOI] [PubMed] [Google Scholar]
- [182].Kuttappan S, Mathew D, Nair MB. Biomimetic composite scaffolds containing bioceramics and collagen/gelatin for bone tissue engineering-A mini review. Int J Biol Macromol 2016;93:1390–401. [DOI] [PubMed] [Google Scholar]
- [183].Stratesteffen H, Köpf M, Kreimendahl F, Blaeser A, Jockenhoevel S, Fischer H. GelMA-collagen blends enable drop-on-demand 3D printablility and promote angiogenesis. Biofabrication 2017;9:045002. [DOI] [PubMed] [Google Scholar]
- [184].Hribar KC, Meggs K, Liu J, Zhu W, Qu X, Chen S. Three-dimensional direct cell patterning in collagen hydrogels with near-infrared femtosecond laser. Sci Rep 2015;5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Yeo M, Lee J-S, Chun W, Kim GH. An innovative collagen-based cell-printing method for obtaining human adipose stem cell-laden structures consisting of core–sheath structures for tissue engineering. Biomacromolecules 2016;17:1365–75. [DOI] [PubMed] [Google Scholar]
- [186].Mahou R, Vlahos AE, Shulman A, Sefton MV. Interpenetrating Alginate-Collagen Polymer Network Microspheres for Modular Tissue Engineering. ACS Biomater Sci Eng 2018;4:3704–12. [DOI] [PubMed] [Google Scholar]
- [187].Yang X, Lu Z, Wu H, Li W, Zheng L, Zhao J. Collagen-alginate as bioink for three-dimensional (3D) cell printing based cartilage tissue engineering. Mater Sci Eng C 2018;83:195–201. [DOI] [PubMed] [Google Scholar]
- [188].Mu X, Fitzpatrick V, Kaplan DL. From Silk Spinning to 3D Printing: Polymer Manufacturing using Directed Hierarchical Molecular Assembly. Adv Healthcare Mater 2020:1901552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Wang Q, Han G, Yan S, Zhang Q. 3D printing of silk fibroin for biomedical applications. Materials 2019;12:504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].DeBari MK, Keyser MN, Bai MA, Abbott RD. 3D printing with silk: considerations and applications. Connect Tissue Res 2018;61:163–73. [DOI] [PubMed] [Google Scholar]
- [191].Shao Z, Vollrath F. Surprising strength of silkworm silk. Nature 2002;418:741. [DOI] [PubMed] [Google Scholar]
- [192].Liu Y, Ren J, Ling S. Bioinspired and biomimetic silk spinning. Compos Commun 2019;13:85–96. [Google Scholar]
- [193].Rising A, Johansson J. Toward spinning artificial spider silk. Nat Chem Biol 2015;11:309. [DOI] [PubMed] [Google Scholar]
- [194].Bini E, Knight DP, Kaplan DL. Mapping domain structures in silks from insects and spiders related to protein assembly. J Mol Biol 2004;335:27–40. [DOI] [PubMed] [Google Scholar]
- [195].Jin H-J, Kaplan DL. Mechanism of silk processing in insects and spiders. Nature 2003;424:1057. [DOI] [PubMed] [Google Scholar]
- [196].Hagn F, Eisoldt L, Hardy JG, Vendrely C, Coles M, Scheibel T, et al. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 2010;465:239–42. [DOI] [PubMed] [Google Scholar]
- [197].Heim M, Keerl D, Scheibel T. Spider silk: from soluble protein to extraordinary fiber. Angew Chem, Int Ed 2009;48:3584–96. [DOI] [PubMed] [Google Scholar]
- [198].Eisoldt L, Hardy JG, Heim M, Scheibel TR. The role of salt and shear on the storage and assembly of spider silk proteins. J Struct Biol 2010;170:413–9. [DOI] [PubMed] [Google Scholar]
- [199].Holland C, Numata K, Rnjak - Kovacina J, Seib FP. The biomedical use of silk: past, present, future. Adv Healthcare Mater 2019;8:1800465. [DOI] [PubMed] [Google Scholar]
- [200].Rockwood DN, Preda RC, Yücel T, Wang X, Lovett ML, Kaplan DL. Materials fabrication from Bombyx mori silk fibroin. Nat Protoc 2011;6:1612–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Teulé F, Cooper AR, Furin WA, Bittencourt D, Rech EL, Brooks A, et al. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat Protoc 2009;4:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Heidebrecht A, Eisoldt L, Diehl J, Schmidt A, Geffers M, Lang G, et al. Biomimetic fibers made of recombinant spidroins with the same toughness as natural spider silk. Adv Mater 2015;27:2189–94. [DOI] [PubMed] [Google Scholar]
- [203].Brif A, Laity P, Claeyssens F, Holland C. Dynamic Photo-cross-linking of Native Silk Enables Macroscale Patterning at a Microscale Resolution. ACS Biomater Sci Eng 2020;6:705–14. [DOI] [PubMed] [Google Scholar]
- [204].Sun L, Parker ST, Syoji D, Wang X, Lewis JA, Kaplan DL. Direct - Write Assembly of 3D Silk/Hydroxyapatite Scaffolds for Bone Co‐Cultures. Adv Healthcare Mater 2012;1:729–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Rodriguez MJ, Dixon TA, Cohen E, Huang W, Omenetto FG, Kaplan DL. 3D freeform printing of silk fibroin. Acta Biomater 2018;71:379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Dang Q, Lu S, Yu S, Sun P, Yuan Z. Silk Fibroin/Montmorillonite Nanocomposites: Effect of pH on the Conformational Transition and Clay Dispersion. Biomacromolecules 2010;11:1796–801. [DOI] [PubMed] [Google Scholar]
- [207].DeSimone E, Schacht K, Pellert A, Scheibel T. Recombinant spider silk-based bioinks. Biofabrication 2017;9:044104. [DOI] [PubMed] [Google Scholar]
- [208].Sofia S, McCarthy MB, Gronowicz G, Kaplan DL. Functionalized silk-based biomaterials for bone formation. J Biomed Mater Res 2001;54:139–48. [DOI] [PubMed] [Google Scholar]
- [209].Chen Z, Zhao D, Liu B, Nian G, Li X, Yin J, et al. 3D Printing of Multifunctional Hydrogels. Adv Funct Mater 2019;29:1900971/1–8. [Google Scholar]
- [210].Dickerson MB, Dennis PB, Tondiglia VP, Nadeau LJ, Singh KM, Drummy LF, et al. 3D Printing of Regenerated Silk Fibroin and Antibody-Containing Microstructures via Multiphoton Lithography. ACS Biomater Sci Eng 2017;3:2064–75. [DOI] [PubMed] [Google Scholar]
- [211].Costa JB, Silva - Correia J, Oliveira JM, Reis RL. Fast setting silk fibroin bioink for bioprinting of patient - specific memory - shape implants. Adv Healthcare Mater 2017;6:1701021. [DOI] [PubMed] [Google Scholar]
- [212].McGill M, Coburn JM, Partlow BP, Mu X, Kaplan DL. Molecular and macro-scale analysis of enzyme-crosslinked silk hydrogels for rational biomaterial design. Acta Biomater 2017;63:76–84. [DOI] [PubMed] [Google Scholar]
- [213].Zhang Z-Z, Jiang D, Ding J-X, Wang S-J, Zhang L, Zhang J-Y, et al. Role of scaffold mean pore size in meniscus regeneration. Acta Biomater 2016;43:314–26. [DOI] [PubMed] [Google Scholar]
- [214].Jose RR, Brown JE, Polido KE, Omenetto FG, Kaplan DL. Polyol-silk bioink formulations as two-part room-temperature curable materials for 3D printing. ACS Biomater Sci Eng 2015;1:780–8. [DOI] [PubMed] [Google Scholar]
- [215].Xiong S, Zhang X, Lu P, Wu Y, Wang Q, Sun H, et al. A gelatin-sulfonated silk composite scaffold based on 3D printing technology enhances skin regeneration by stimulating epidermal growth and dermal neovascularization. Sci Rep 2017;7:4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Rodriguez MJ, Brown J, Giordano J, Lin SJ, Omenetto FG, Kaplan DL. Silk based bioinks for soft tissue reconstruction using 3-dimensional (3D) printing with in vitro and in vivo assessments. Biomaterials 2017;117:105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].The journal of physical chemistry B Shi W, Sun M, Hu, Ren B, Cheng J, Li C, et al. Structurally and functionally optimized silk-fibroin-gelatin scaffold using 3D printing to repair cartilage injury in vitro and in vivo. Adv Mater 2017;29:1701089. [DOI] [PubMed] [Google Scholar]
- [218].Singh YP, Bandyopadhyay A, Mandal BB. 3D Bioprinting Using Cross-Linker-Free Silk–Gelatin Bioink for Cartilage Tissue Engineering. ACS Appl Mater Interfaces 2019;11:33684–96. [DOI] [PubMed] [Google Scholar]
- [219].Wei L, Wu S, Kuss M, Jiang X, Sun R, Reid P, et al. 3D printing of silk fibroin-based hybrid scaffold treated with platelet rich plasma for bone tissue engineering. Bioact Mater 2019;4:256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Bandyopadhyay A, Mandal BB. A three-dimensional printed silk-based biomimetic trilayered meniscus for potential patient-specific implantation. Biofabrication 2019;12:015003. [DOI] [PubMed] [Google Scholar]
- [221].Chawla S, Kumar A, Admane P, Bandyopadhyay A, Ghosh S. Elucidating role of silk-gelatin bioink to recapitulate articular cartilage differentiation in 3D bioprinted constructs. Bioprinting 2017;7:1–13. [Google Scholar]
- [222].Dong T, Mi R, Wu M, Zhong N, Zhao X, Chen X, et al. The regenerated silk fibroin hydrogel with designed architecture bioprinted by its microhydrogel. J Mater Chem B 2019;7:4328–37. [Google Scholar]
- [223].Zhong N, Dong T, Chen Z, Guo Y, Shao Z, Zhao X. A novel 3D-printed silk fibroin-based scaffold facilitates tracheal epithelium proliferation in vitro. J Biomater Appl 2019;34:3–11. [DOI] [PubMed] [Google Scholar]
- [224].Kwak H, Shin S, Lee H, Hyun J. Formation of a keratin layer with silk fibroin-polyethylene glycol composite hydrogel fabricated by digital light processing 3D printing. J Ind Eng Chem 2019;72:232–40. [Google Scholar]
- [225].Zheng Z, Wu J, Liu M, Wang H, Li C, Rodriguez MJ, et al. 3D bioprinting of self - standing silk - based bioink. Adv Healthcare Mater 2018;7:1701026. [DOI] [PubMed] [Google Scholar]
- [226].Sommer MR, Schaffner M, Carnelli D, Studart AR. 3D printing of hierarchical silk fibroin structures. ACS Appl Mater Interfaces 2016;8:34677–85. [DOI] [PubMed] [Google Scholar]
- [227].Zhang X, Zhai C, Fei H, Liu Y, Wang Z, Luo C, et al. Composite Silk-Extracellular Matrix Scaffolds for Enhanced Chondrogenesis of Mesenchymal Stem Cells. Tissue Eng, Part C 2018;24:645–58. [DOI] [PubMed] [Google Scholar]
- [228].Lee H, Yang GH, Kim M, Lee J, Huh J, Kim G. Fabrication of micro/nanoporous collagen/dECM/silk-fibroin biocomposite scaffolds using a low temperature 3D printing process for bone tissue regeneration. Mater Sci Eng C 2018;84:140–7. [DOI] [PubMed] [Google Scholar]
- [229].Huang T, Fan C, Zhu M, Zhu Y, Zhang W, Li L. 3D-printed scaffolds of biomineralized hydroxyapatite nanocomposite on silk fibroin for improving bone regeneration. Appl Surf Sci 2019;467:345–53. [Google Scholar]
- [230].Du X, Wei D, Huang L, Zhu M, Zhang Y, Zhu Y. 3D printing of mesoporous bioactive glass/silk fibroin composite scaffolds for bone tissue engineering. Mater Sci Eng C 2019;103:109731. [DOI] [PubMed] [Google Scholar]
- [231].Bhunia BK, Mandal BB. Exploring Gelation and Physicochemical Behavior of in Situ Bioresponsive Silk Hydrogels for Disc Degeneration Therapy. ACS Biomater Sci Eng 2019;5:870–86. [DOI] [PubMed] [Google Scholar]
- [232].Meng L, Shao C, Cui C, Xu F, Lei J, Yang J. Autonomous Self-Healing Silk Fibroin Injectable Hydrogels Formed via Surfactant-Free Hydrophobic Association. ACS Appl Mater Interfaces 2020;12:1628–39. [DOI] [PubMed] [Google Scholar]
- [233].Luo K, Yang Y, Shao Z. Physically Crosslinked Biocompatible Silk-Fibroin-Based Hydrogels with High Mechanical Performance. Adv Funct Mater 2016;26:872–80. [Google Scholar]
- [234].Zhang J, Allardyce BJ, Rajkhowa R, Zhao Y, Dilley RJ, Redmond SL, et al. 3D Printing of Silk Particle-Reinforced Chitosan Hydrogel Structures and Their Properties. ACS Biomaterials Science & Engineering 2018;4:3036–46. [DOI] [PubMed] [Google Scholar]
- [235].Sun K, Li R, Li H, Li D, Jiang W. Comparison of three-dimensional printing for fabricating silk fibroin-blended scaffolds. Int J Polym Mater Polym Biomater 2018;67:480–6. [Google Scholar]
- [236].Dorishetty P, Balu R, Sreekumar A, de Campo L, Mata JP, Choudhury NR, et al. Robust and Tunable Hybrid Hydrogels from Photo-Cross-Linked Soy Protein Isolate and Regenerated Silk Fibroin. ACS Sustainable Chem Eng 2019;7:9257–71. [Google Scholar]
- [237].Kim SH, Yeon YK, Lee JM, Chao JR, Lee YJ, Seo YB, et al. Precisely printable and biocompatible silk fibroin bioink for digital light processing 3D printing. Nat Commun 2018;9:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Hong H, Seo YB, Kim DY, Lee JS, Lee YJ, Lee H, et al. Digital light processing 3D printed silk fibroin hydrogel for cartilage tissue engineering. Biomaterials 2020;232:119679. [DOI] [PubMed] [Google Scholar]
- [239].Cui X, Soliman BG, Alcala-Orozco CR, Li J, Vis MAM, Santos M, et al. Rapid Photocrosslinking of Silk Hydrogels with High Cell Density and Enhanced Shape Fidelity. Adv Healthcare Mater 2020;9:1901667. [DOI] [PubMed] [Google Scholar]
- [240].Jiang J, Zhang S, Qian Z, Qin N, Song W, Sun L, et al. Protein Bricks: 2D and 3D Bio - Nanostructures with Shape and Function on Demand. Adv Mater 2018;30:1705919. [DOI] [PubMed] [Google Scholar]
- [241].Applegate MB, Coburn J, Partlow BP, Moreau JE, Mondia JP, Marelli B, et al. Laser-based three-dimensional multiscale micropatterning of biocompatible hydrogels for customized tissue engineering scaffolds. Proc Natl Acad Sci 2015;112:12052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Kim CS, Yang YJ, Bahn SY, Cha HJ. A bioinspired dual-crosslinked tough silk protein hydrogel as a protective biocatalytic matrix for carbon sequestration. NPG Asia Mat 2017;9:e391–e. [Google Scholar]
- [243].Mehrotra S, de Melo BAG, Hirano M, Keung W, Li RA, Mandal BB, et al. Nonmulberry Silk Based Ink for Fabricating Mechanically Robust Cardiac Patches and Endothelialized Myocardium-on-a-Chip Application. Adv Funct Mater 2020;30:1907436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Liu J, Shao J, Zheng J. Radiation grafting/crosslinking of silk using electron - beam irradiation. J Appl Polym Sci 2004;91:2028–34. [Google Scholar]
- [245].Liu K, Jiang J, Tao TH. Fabrication of 3D Silk Microstructures by Direct Two-Photon Polymerization. 2019 IEEE 32nd International Conference on Micro Electro Mechanical Systems (MEMS): IEEE; 2019. p. 522–4. [Google Scholar]
- [246].Kim S, Marelli B, Brenckle MA, Mitropoulos AN, Gil E-S, Tsioris K, et al. All-water-based electron-beam lithography using silk as a resist. Nat Nanotechnol 2014;9:306. [DOI] [PubMed] [Google Scholar]
- [247].Gasperini L, Mano JF, Reis RL. Natural polymers for the microencapsulation of cells. J R Soc, Interface 2014;11:20140817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Weisel JW, Litvinov RI. Fibrin Formation, Structure and Properties. Sub-cellular biochemistry 2017;82:405–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Ahmed TA, Dare EV, Hincke M. Fibrin: a versatile scaffold for tissue engineering applications. Tissue Eng, Part B 2008;14:199–215. [DOI] [PubMed] [Google Scholar]
- [250].Shpichka A, Osipova D, Efremov Y, Bikmulina P, Kosheleva N, Lipina M, et al. Fibrin-based Bioinks: New Tricks from an Old Dog. Int J Bioprint 2020;6:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Freeman S, Ramos R, Chando PA, Zhou L, Reeser K, Jin S, et al. A bioink blend for rotary 3D bioprinting tissue engineered small-diameter vascular constructs. Acta Biomater 2019;95:152–64. [DOI] [PubMed] [Google Scholar]
- [252].Zhang K, Fu Q, Yoo J, Chen X, Chandra P, Mo X, et al. 3D bioprinting of urethra with PCL/PLCL blend and dual autologous cells in fibrin hydrogel: An in vitro evaluation of biomimetic mechanical property and cell growth environment. Acta Biomater 2017;50:154–64. [DOI] [PubMed] [Google Scholar]
- [253].Cubo N, Garcia M, del Cañizo JF, Velasco D, Jorcano JL. 3D bioprinting of functional human skin: production and in vivo analysis. Biofabrication 2016;9:015006. [DOI] [PubMed] [Google Scholar]
- [254].England S, Rajaram A, Schreyer DJ, Chen X. Bioprinted fibrin-factor XIII-hyaluronate hydrogel scaffolds with encapsulated Schwann cells and their in vitro characterization for use in nerve regeneration. Bioprinting 2017;5:1–9. [Google Scholar]
- [255].Ahlfeld T, Cubo-Mateo N, Cometta S, Guduric V, Vater C, Bernhardt A, et al. A Novel Plasma-Based Bioink Stimulates Cell Proliferation and Differentiation in Bioprinted, Mineralized Constructs. ACS Appl Mater Interfaces 2020;12:12557–72. [DOI] [PubMed] [Google Scholar]
- [256].Homan KA, Kolesky DB, Skylar-Scott MA, Herrmann J, Obuobi H, Moisan A, et al. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci Rep 2016;6:34845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].de Melo BAG, Jodat YA, Mehrotra S, Calabrese MA, Kamperman T, Mandal BB, et al. 3D Printed Cartilage-Like Tissue Constructs with Spatially Controlled Mechanical Properties. Adv Funct Mater 2019;29:1906330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Lee C, Abelseth E, de la Vega L, Willerth S. Bioprinting a novel glioblastoma tumor model using a fibrin-based bioink for drug screening. Materials Today Chemistry 2019;12:78–84. [Google Scholar]
- [259].de Guzman RC, Merrill MR, Richter JR, Hamzi RI, Greengauz-Roberts OK, Van Dyke ME. Mechanical and biological properties of keratose biomaterials. Biomaterials 2011;32:8205–17. [DOI] [PubMed] [Google Scholar]
- [260].Vasconcelos A, Cavaco-Paulo A. The use of keratin in biomedical applications. Curr Drug Targets 2013;14:612–9. [DOI] [PubMed] [Google Scholar]
- [261].Wang B, Yang W, McKittrick J, Meyers MA. Keratin: Structure, mechanical properties, occurrence in biological organisms, and efforts at bioinspiration. Prog Mater Sci 2016;76:229–318. [Google Scholar]
- [262].Rouse JG, Van Dyke ME. A review of keratin-based biomaterials for biomedical applications. Materials 2010;3:999–1014. [Google Scholar]
- [263].Wang Y, Zhang W, Yuan J, Shen J. Differences in cytocompatibility between collagen, gelatin and keratin. Mater Sci Eng C 2016;59:30–4. [DOI] [PubMed] [Google Scholar]
- [264].Parry DAD, Smith TA, Rogers MA, Schweizer J. Human hair keratin-associated proteins: Sequence regularities and structural implications. J Struct Biol 2006;155:361–9. [DOI] [PubMed] [Google Scholar]
- [265].Cera L, Gonzalez GM, Liu Q, Choi S, Chantre CO, Lee J, et al. A bioinspired and hierarchically structured shape-memory material. Nat Mater 2020:1–8. [DOI] [PubMed] [Google Scholar]
- [266].Navarro J, Swayambunathan J, Santoro M, Fisher J. Assessment of the Effects of Energy Density in Crosslinking of Keratin-Based Photo-Sensitive Resin. 2018 IX International Seminar of Biomedical Engineering (SIB) 2018. p. 1–6. [Google Scholar]
- [267].Navarro J, Clohessy RM, Holder RC, Gabard AR, Herendeen GJ, Christy RJ, et al. In Vivo Evaluation of Three-Dimensional Printed, Keratin-Based Hydrogels in a Porcine Thermal Burn Model. Tissue Eng Part A 2020;25:265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Grigsby WJ, Scott SM, Plowman-Holmes MI, Middlewood PG, Recabar K. Combination and processing keratin with lignin as biocomposite materials for additive manufacturing technology. Acta Biomater 2020;104:95–103. [DOI] [PubMed] [Google Scholar]
- [269].Sun J, Zhou W, Huang D, Fuh JY, Hong GS. An overview of 3D printing technologies for food fabrication. Food Bioprocess Technol 2015;8:1605–15. [Google Scholar]
- [270].Lin H-H, Hsieh F-Y, Tseng C-S, Hsu S-h. Preparation and characterization of a biodegradable polyurethane hydrogel and the hybrid gel with soy protein for 3D cell-laden bioprinting. J Mater Chem B 2016;4:6694–705. [DOI] [PubMed] [Google Scholar]
- [271].Chen J, Mu T, Goffin D, Blecker C, Richard G, Richel A, et al. Application of soy protein isolate and hydrocolloids based mixtures as promising food material in 3D food printing. J Food Eng 2019;261:76–86. [Google Scholar]
- [272].Chien KB, Makridakis E, Shah RN. Three-dimensional printing of soy protein scaffolds for tissue regeneration. Tissue Eng, Part C 2013;19:417–26. [DOI] [PubMed] [Google Scholar]
- [273].Chien KB, Aguado BA, Bryce PJ, Shah RN. In vivo acute and humoral response to three-dimensional porous soy protein scaffolds. Acta Biomater 2013;9:8983–90. [DOI] [PubMed] [Google Scholar]
- [274].Liu Y, Liu D, Wei G, Ma Y, Bhandari B, Zhou P. 3D printed milk protein food simulant: Improving the printing performance of milk protein concentration by incorporating whey protein isolate. Innovative Food Sci Emerging Technol 2018;49:116–26. [Google Scholar]
- [275].Liu L, Meng Y, Dai X, Chen K, Zhu Y. 3D printing complex egg white protein objects: Properties and optimization. Food Bioprocess Technol 2019;12:267–79. [Google Scholar]
- [276].Liu L, Yang X, Bhandari B, Meng Y, Prakash S. Optimization of the formulation and properties of 3D-printed complex egg white protein objects. Foods 2020;9:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Anukiruthika T, Moses J, Anandharamakrishnan C. 3D printing of egg yolk and white with rice flour blends. J Food Eng 2020;265:109691. [Google Scholar]
- [278].Chang Q, He Y, Liu Y, Zhong W, Wang Q, Lu F, et al. Protein Gel’s Phase Transition: Toward Superiorly Transparent and Hysteresis-Free Wearable Electronics. Adv Funct Mater 2020;30:1910080/1–17. [Google Scholar]
- [279].Jalili-Firoozinezhad S, Rajabi-Zeleti S, Mohammadi P, Gaudiello E, Bonakdar S, Solati-Hashjin M, et al. Facile Fabrication of Egg White Macroporous Sponges for Tissue Regeneration. Adv Healthcare Mater 2015;4:2281–90. [DOI] [PubMed] [Google Scholar]
- [280].Kaipparettu BA, Kuiatse I, Chan BT-Y, Kaipparettu MB, Lee AV, Oesterreich S. Novel egg white–based 3-D cell culture system. BioTechniques 2008;45:165–71. [DOI] [PubMed] [Google Scholar]
- [281].Mousseau Y, Mollard S, Qiu H, Richard L, Cazal R, Nizou A, et al. In vitro 3D angiogenesis assay in egg white matrix: comparison to Matrigel, compatibility to various species, and suitability for drug testing. Lab Invest 2014;94:340–9. [DOI] [PubMed] [Google Scholar]
- [282].Ritzoulis C, Scoutaris N, Papademetriou K, Stavroulias S, Panayiotou C. Milk protein-based emulsion gels for bone tissue engineering. Food Hydrocolloids 2005;19:575–81. [Google Scholar]
- [283].Xu J, Fan Z, Duan L, Gao G. A tough, stretchable, and extensively sticky hydrogel driven by milk protein. Polym Chem 2018;9:2617–24. [Google Scholar]
- [284].Lee HJ, Kim YB, Ahn SH, Lee JS, Jang CH, Yoon H, et al. A new approach for fabricating collagen/ECM‐based bioinks using preosteoblasts and human adipose stem cells. Adv Healthcare Mater 2015;4:1359–68. [DOI] [PubMed] [Google Scholar]
- [285].Visser J, Melchels FP, Jeon JE, Van Bussel EM, Kimpton LS, Byrne HM, et al. Reinforcement of hydrogels using three-dimensionally printed microfibres. Nat Commun 2015;6:6933. [DOI] [PubMed] [Google Scholar]
- [286].Anil Kumar S, Alonzo M, Allen SC, Abelseth L, Thakur V, Akimoto J, et al. A visible light-cross-linkable, fibrin–gelatin-based bioprinted construct with human cardiomyocytes and fibroblasts. ACS Biomater Sci Eng 2019;5:4551–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Włodarczyk-Biegun MK, del Campo A. 3D bioprinting of structural proteins. Biomaterials 2017;134:180–201. [DOI] [PubMed] [Google Scholar]
- [288].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat Biotechnol 2016;34:312–9. [DOI] [PubMed] [Google Scholar]
- [289].Latza V, Guerette PA, Ding D, Amini S, Kumar A, Schmidt I, et al. Multi-scale thermal stability of a hard thermoplastic protein-based material. Nat Commun 2015;6:8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Chaunier L, Leroy E, Valle GD, Dalgalarrondo M, Bakan B, Marion D, et al. 3D printing of maize protein by fused deposition modeling. AIP Conference Proceedings: AIP Publishing LLC; 2017. p. 190003/1–5. [Google Scholar]
- [291].Tavares-Negrete JA, Aceves-Colin AE, Rivera-Flores DC, Diaz-Armas GG, Mertgen A-S, Trinidad-Calderon PA, et al. Biofabrication using maize protein: 3D printing using zein formulations. bioRxiv 2020. [Google Scholar]
- [292].Zein I, Hutmacher DW, Tan KC, Teoh SH. Fused deposition modeling of novel scaffold architectures for tissue engineering applications. Biomaterials 2002;23:1169–85. [DOI] [PubMed] [Google Scholar]
- [293].Kotikian A, Truby RL, Boley JW, White TJ, Lewis JA. 3D printing of liquid crystal elastomeric actuators with spatially programed nematic order. Adv Mater 2018;30:1706164. [DOI] [PubMed] [Google Scholar]
- [294].Levato R, Jungst T, Scheuring RG, Blunk T, Groll J, Malda J. From shape to function: the next step in bioprinting. Adv Mater 2020;32:1906423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Wang Y, Huang W, Wang Y, Mu X, Ling S, Yu H, et al. Stimuli-responsive composite biopolymer actuators with selective spatial deformation behavior. Proc Natl Acad Sci 2020;117:14602–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Khoury LR, Slawinski M, Collison DR, Popa I. Cation-induced shape programming and morphing in protein-based hydrogels. Sci Adv 2020;6:eaba6112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Urciuolo A, Poli I, Brandolino L, Raffa P, Scattolini V, Laterza C, et al. Intravital three-dimensional bioprinting. Nat Biomed Eng 2020;4:901–15. [DOI] [PubMed] [Google Scholar]
- [298].Chen Y, Zhang J, Liu X, Wang S, Tao J, Huang Y, et al. Noninvasive in vivo 3D bioprinting. Sci Adv 2020;6:eaba7406.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Ho CMB, Ng SH, Li KHH, Yoon Y-J. 3D printed microfluidics for biological applications. Lab Chip 2015;15:3627–37. [DOI] [PubMed] [Google Scholar]
- [300].Torculas M, Medina J, Xue W, Hu X. Protein-based bioelectronics. ACS Biomater Sci Eng 2016;2:1211–23. [DOI] [PubMed] [Google Scholar]
- [301].Zhu B, Wang H, Leow WR, Cai Y, Loh XJ, Han MY, et al. Silk fibroin for flexible electronic devices. Adv Mater 2016;28:4250–65. [DOI] [PubMed] [Google Scholar]
- [302].Valentine AD, Busbee TA, Boley JW, Raney JR, Chortos A, Kotikian A, et al. Hybrid 3D printing of soft electronics. Adv Mater 2017;29:1703817. [DOI] [PubMed] [Google Scholar]
- [303].Baumgartner M, Hartmann F, Drack M, Preninger D, Wirthl D, Gerstmayr R, et al. Resilient yet entirely degradable gelatin-based biogels for soft robots and electronics. Nat Mater 2020;19:1102–9. [DOI] [PubMed] [Google Scholar]


