Abstract
According to the National Institutes of Health, clear cell renal cell carcinoma (ccRCC) is the most common type of Renal Cell Carcinoma (RCC), making up approximately 75% of total renal carcinoma cases. Clear cell Renal Cell Carcinoma is characterized by a significant accumulation of lipids in the cytoplasm, which allows light from microscopes to pass through giving them a “clear” phenotype. Many of these lipids are in the form of fatty acids, both free and incorporated into lipid droplets. RCC is typically associated with a poor prognosis due to the lack of specific symptoms. Some symptoms include blood in urine, fever, lump on the side, weight loss, fatigue, to name a few; all of which can be associated with non-specific, non-cancerous, health conditions that contribute to difficult diagnosis. Treatment of RCC has typically been centered around radical nephrectomy as the standard of care, but due to the potentially small size of lesions and the possibility of causing surgically induced chronic kidney disease, treatments have shifted to more cautious, less invasive approaches. These approaches include active surveillance, nephron-sparing surgery, and other minimally invasive techniques like cryotherapy and renal ablation. Although these techniques have had the desired effect of reducing the number of surgeries, there is still considerable potential for renal impairment and the chance that tumors can grow out of control without surgery. With the difficulty that surrounds the treatment of ccRCC and its considerably high mortality rate amongst urological cancers, it is important to look for novel approaches to improve patient outcomes. This review looks at available literature and our data that suggests the lipogenic enzyme stearoyl-CoA desaturase may be more beneficial to patient survival than once thought. As our understanding of the importance of lipids in cell metabolism and longevity matures, it is important to present new perspectives that present a new understanding of ccRCC and the role of lipids in survival mechanisms engaged by transformed cells during cancer progression.
In this review, we provide evidence that pharmacological inhibition of lipid desaturation in renal cancer patients is not without risk, and that the presence of unsaturated fatty acids may be a beneficial factor in patient outcomes. Although more direct experimental evidence is needed to make definitive conclusions, it is clear that the work reviewed herein should challenge our current understanding of cancer biology and may inform novel approaches to the diagnosis and treatment of ccRCC.
Keywords: YB-1, ccRCC, SCD1, MUFA, cancer, lipid metabolism, TCGA, kidney
Introduction
Renal cell carcinoma (RCC) is not a single cancer type but a collection of various tumorous conditions of the kidney.1 The most common subtype of RCC is clear cell renal cell carcinoma (ccRCC), making up about 75% of all RCC cases. 2 Clear cell RCC consists of two subtypes; clear cell type A (ccA) and B (ccB), with ccA showing better prognosis. 3 The most distinct characteristic of ccRCC, is the high abundance of cytoplasmic lipid. For many years, lipid dysfunction has been an obvious target for therapeutics, however, there still has not been any substantial changes in patient outcomes to date. That is because ccRCC is an enigmatic disease with many aspects that are still incompletely understood, like how men are twice as likely to be diagnosed than women, or the lack of consistent biomarkers. 4,5 Further still, there are few genetic mutations associated with this disease (except for VHL), therefore early detection remains the best means to improve patient survival. 3 To improve our ability to detect ccRCC, several studies seek to identify specific genes overexpressed in ccRCC compared to other renal cancers or normal kidney. One study ranked overexpressed genes using hierarchal clustering from microarray analyses of numerous ccRCC, chromophobe RCC, and normal kidney samples. 6 When further verifying these genes, they found that only nicotinamide N-methyltransferase (NNMT) and adipose differentiation-related protein (ADFP) agreed with gene analysis studies done by others. ADFP is also known as perilipin 2 (PLIN2) and was of particular interest to our laboratory as it points to a role for lipogenic genes in the development of ccRCC. Additional experimental evidence from other labs showed that another lipogenic gene, stearoyl-CoA desaturase (SCD1), was overexpressed and critical to ccRCC cell viability. This has led to many studies looking for ways to block SCD1 activity to stop cancer growth. However, our laboratory has found that SCD1 is, in part, responsible for patient survival, and decreased SCD1 protein expression is correlated with a worse prognosis. 7 Furthermore, we showed that the expression of the SCD1 gene is inhibited by the oncogene Y-box protein 1 (YB-1). These findings demonstrate how complex the biology is within ccRCC and demands that we look more carefully at how these genes work together to affect patient survival. Here we will discuss the roles of specific genes related to lipid metabolism and how they influence ccRCC development, the potential interactions with oncogenic YB-1, and what that could mean for future studies in ccRCC.
Loss of VHL in ccRCC development.
It is not possible to discuss ccRCC without first mentioning the von Hippel-Lindau protein (pVHL). VHL is an E3 ubiquitin ligase that helps to promote protein degradation. VHL is largely considered a tumor suppressor gene because of its ability to negatively influence key transcriptional programs required for rapid cell proliferation. VHL is one of a few genes that are consistently mutated in ccRCC. However, there is still no clear evidence that the presence or absence of VHL, or mutation in the gene influences disease outcome. Studies have shown that the VHL gene (located on chromosome 3p) is mutated in 50% of early-stage and some advanced ccRCC tumors. 4,8 Additionally, mutations have been observed across all 3 of VHL’s exons, with varying degrees of association with ccRCC development. VHL gene can also be subjected to frameshift mutations (c.439del) which further implicates VHL in ccRCC. 9 Early-stage ccRCC tumors with loss of heterozygosity (LOH) in the VHL gene had no other obvious chromosomal deletions which led to the conclusion that VHL mutations must occur at the primary stage of ccRCC. 8 The most notable consequence of possessing deficient, or mutant, VHL proteins is the dysregulated expression of the oxygen-sensitive transcription factors HIF1α and HIF2 α, in conjunction with other transcription factors. This is because mutant VHL is unable to ubiquitylate HIF proteins, allowing abnormally high expression of HIF-target genes, which drive cell growth and proliferation in ccRCC. Furthermore, studies show that the expression of HIF transcription factors increases the expression of lipogenic genes, which in turn increases the ability of ccRCC cells to produce fat. 10 We suspect that high lipid production and storage may be in concert with the altered metabolic profile of renal cancer.
Metabolism in cancer
It is well established that cancer is as much a metabolic issue as it is a genetic one. A major metabolic shift that is often seen in cancer is abnormally high rates of aerobic glycolysis. This leads to an overall acceleration of the macromolecule biosynthesis required to support growth. High rates of glycolysis glutaminolysis in cancer cells give the opportunity of producing more adenosine triphosphate (ATP) which is needed to supply energy needed for cellular proliferation. 11–13 However, less appreciated is the fact that glycolysis cannot proceed to completion without sufficient activity in downstream metabolic pathways such as the tricarboxylic acid cycle and the pentose phosphate pathway. Not only is glycolysis a way for cells to generate energy in the form of ATP, but it is also a source of carbon. Carbon, in the form of glucose and glutamine, enters the cell and becomes part of newly synthesized amino acids, proteins, and fats. 13 For rapidly dividing cells, including cancer, all of these macromolecules are critical for survival. Lipids (fatty acids and sterols) are particularly needed for growing cells. De novo lipogenesis (DNL) is the pathway that converts excess carbohydrates that enter the cell via glucose into lipids. These lipids can be complexed together for storage, broken down through oxidation for energy, or immediately used to provide components for new membranes.14,15 DNL, under normal homeostatic conditions, occurs mostly in the liver and adipose tissue, with only a low level of lipogenesis in other tissues. 16–18 However, under certain conditions (obesity, infection, and cancer) lipogenesis can be rapidly increased in any tissues that demand it.
Lipid synthesis promotes cancer cell survival
Upregulation of fatty acid synthesis is essential for cancer cell survival because cells depend on this process for energy, membrane biosynthesis, and cell signaling. According to several studies, cancer cells can reprogram their lipid metabolism that will enable cell proliferation and metastasis. 19 In a study conducted by Arthur A. Spector, the main components found in the ascites fluid of the Ehrlich ascites tumor were palmitate, stearate, oleate, and linoleate which happen to be the most readily available lipids for use in the body. Fatty acids required by tumor cells are typically supplied by the host and most of the remnant is circulated to the tumor as a free fatty acid. 20
The primary product of glucose-derived fatty acid synthesis is typically saturated fatty acids (SFA). While this is common in cancer cells, there is also a notable increase in the production of monounsaturated fatty acids (MUFA). Given these observations, glycolysis and fatty acid production are suggested to be linked through the unsaturation of SFAs to MUFAs by the enzyme stearoyl-CoA desaturase (SCD-1). 19 In a study conducted by Peck et. al, functional genomics identified SCD-1 as an important enzyme for several cancer cell lines, such as breast and prostate cancer. 21 When the researchers inhibited SCD-1, a change in the lipid components that led to decreased cell viability was observed in the absence of exogenous lipid. Additionally, they found that the change in the cardio-lipid composition led to an absence of cytochrome C which eventually led to apoptosis. Further exploration revealed elevated levels of SCD-1 mRNA correlated with poor survival in far-progressed tumors. 21
SCD-1 role and its promise as a therapeutic target for metabolic diseases
As more evidence suggests that the ccRCC phenotype (and to a lesser extent its gene profile) is similar to adipocytes, the rationale for targeting the lipid synthetic pathway has taken root. There have been dozens of studies that have shown increased expression of lipogenic genes such as fatty acid synthase (FAS), acetyl-CoA carboxylase (ACC), and stearoyl-CoA desaturase (SCD1), and others.22–24 However, only SCD1 has been successfully targeted with minimal effect to non-malignant cells.25 Due to its role in metabolic disorders such as obesity, diabetes, and cancer, SCD1 inhibition has a broad potential impact on human disease. 18
SCD-1 is the primary enzyme involved in the biosynthesis of monounsaturated fatty acids (MUFAs) through the desaturation of saturated fatty acids (SFAs). 26,27 During the desaturation process, SCD-1 introduces a cis-9 double bond on the SFA involved in the process. 27 Most commonly, stearic acid is converted to oleic acid, which is regularly abundant and readily available for the body to use. 28 SCD-1 in its active state has been recognized as the biochemical hallmark of cancer cells due to the overproduction of fatty acids. When SCD-1 activity is significantly increased, the metabolic balance between SFAs and MUFAs is tilted and leans more towards greater lipogenesis and lower lipid oxidation. High levels of SCD-1 activity is shown to induce cancer cell proliferation, survival, and its ability to potentially invade other major areas of the tissue/body.19
Through the action of the master regulator of fatty acid synthesis, transcription factor sterol regulatory element-binding protein-1 (SREBP-1), all de novo synthesis of SFAs occurs. In a mouse model, it was demonstrated that a high SFA diet induces lipogenic genes including SCD-1. Theses result indicated SCD-1-mediated desaturation of SFAs was essential to lipogenesis. Inhibition of the action of SCD-1 led to SFA-induced obesity in mice. This suggested that SCD-1 served as a molecular switch that prevents lipid-induced disorders by the consumption of excess SFAs. 29
SCD-1 appears to have a connection to tumor-promoting pathways
SCD-1 function has been suggested to have major roles in tumor-promoting pathways such as Wnt, AKT, NF-kappa B, EMT, and other metabolic reprogramming pathways. 26 This has been demonstrated by the function of SCD-1 being linked to inducing cancer cell proliferation. 30 Specifically looking at its role in tumor growth and cell viability, in vitro, and in vivo, SCD1 has the potential to be an effective molecular target for the treatment of ccRCC. Experimentally this was demonstrated when SCD1 inhibitors significantly reduced viability in ccRCC cell lines compared to normal kidney epithelial cells. 25 Few studies have investigated SCD1 in ccRCC patient populations, but those that we were able to access agreed with von Roemeling et al. in stating that SCD1 correlates with a worse prognosis and is likely a good target for treatment. However, this is in contrast to our data that looks at proteomic data from over 450 ccRCC patient samples deposited in the Cancer Genome Atlas (TCGA). From our analysis, we found that patients with high levels of SCD1 protein have better overall survival. 7 Although it is not uncommon for studies to have opposing conclusions, this case requires an explanation as to how the same molecule can be both the hero and the villain in two different patient cohorts. Later in this review, we will provide our explanations as to why that is the case.
Overexpression of perilipin2 in ccRCC
ccRCC is comprised of cells that take on an almost adipocyte appearance, thus, it would be expected that many genes related to adipogenesis and lipid trafficking are overexpressed. However, that is not necessarily the case and only select lipogenic genes show significantly altered expression across studies. One consistently overexpressed gene is perilipin 2 or adipose differentiation-related protein (ADFP). Importantly, the upregulation of perilipin 2/ADFP appears to be associated with a better prognosis. 10,31,32 Perilipin 2 functions in the generation and stabilization of lipid droplets in epithelial cells. In the context of malignancy, increased expression of preilipin2 has been observed in low grade/stage 1 and 2 ccRCC samples. 32These findings indicate that the handling and storage of abundant lipid stores in ccRCC are likely an early adaptation and not necessarily a causal factor. So similar to our results involving SCD1, perilipin 2 may also promote better overall survival in patients through an incompletely described mechanism that may involve the VHL/HIF pathway and stabilization of the endoplasmic reticulum. 10,33
Clearly defining the biological pathways that lead to cellular differentiation seen in ccRCC continues to be an active area of research. For example, one study showed the presence of distinct biological alterations present in ccRCC development which included loss of normal renal function, downregulation of metabolism, and the activation of immune pathways.34 As CcRCC cells lose normal functions, they undergo adipogenic differentiation due to the downregulation of key transcription factors (GATA3, TFCP2L1, TFAP2B, DMRT2). A major aspect of ccRCC behavior relies heavily on becoming more capable of taking on the characteristics of a fat cell. A question that remains unanswered is how the presence of accumulated lipids contributes to the disease progression and patient outcomes.
YB-1 as a multifunctional protein
Recent work from our laboratory showed that the expression of lipogenic protein SCD-1 is negatively influenced by Y-box binding protein 1 (YB-1).7 This places the role of a known oncogene in direct opposition to a key molecule in the lipogenic pathway. YB-1, encoded by the gene YBX1, is a multifunctional protein capable of binding to DNA and RNA and is involved in the regulation of transcription, translation, DNA repair, and even multidrug resistance. To our knowledge, this is the first time a relationship between YB-1 and SCD-1 or any lipogenic proteins has been described. Therefore, it is important to provide background on YB-1 to underscore the importance of its relationship to ccRCC.
YB-1 and Renal Cell Carcinoma
YB-1 is a gene that is increased in RCC in several studies. In primary renal cancer, YB-1 can be found in the nucleus at significantly higher levels compared with para-carcinoma normal tissues. Similarly, YB-1 overexpression was associated with decreased patient survival. YB-1 coordinates cellular transactivation in RCC cells via TGF-β signaling. 35 Furthermore, YB-1 can be used to help with the staging of RCC, which makes it useful as a potential biomarker for diagnosis. 35,36
YB-1 Structure
The YB-1 protein is made up of 324 amino acids. It is composed of a variable N-terminal domain, a cold-shock domain (CSD), and a C-terminal domain (CTD) (Figure 1). 37 The N-terminal domain contains a rich content of the amino acids, alanine and proline, and the C-terminal domain consists of alternating clusters of positively and negatively charged amino acids. 38 The most prevalent amino acids in the YB-1 structure are arginine (11.7%), glycine (12%), proline (11%), and glutamate (8.3%). Although the actual molecular weight of YB-1 is 35.9 kDa, in gel electrophoresis is migrates around 50 kDa due to aberrant mobility. 39
Figure 1.
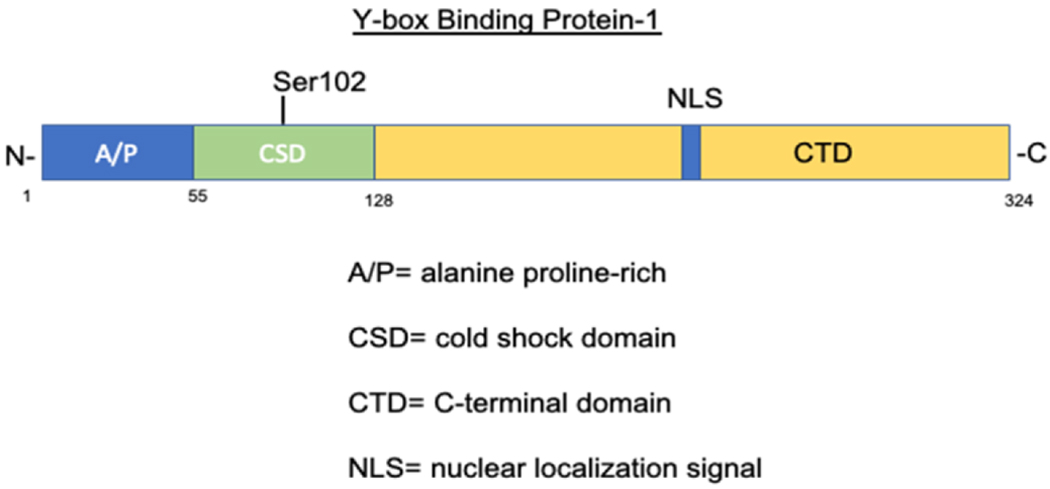
Schematic of YB-1 protein structure and major functional domains.
In vitro, YB-1 has been found to form oligomers, depending upon protein concentration as well as ionic strength. 40 At high ionic strength, YB-1 takes on an unfolded conformation. At low concentration, neutral pH with moderate ionic strength, YB-1 forms a more monomeric, and more globular/compact structure. YB-1 was shown to be partially ordered, but highly compact, and the lack of a rigid structure is believed to be an advantage when it comes to its capability to interact with multiple molecules. Chromatographic and thermodynamic data suggest that the high compactness of YB-1 may be due to interactions between A/P, CSD, and its C-terminal domain. 40
The CSD contains RNP1 and RNP2-like consensus motifs that may be involved in the binding of nucleic acids. The CSD has 5 β-strands that form a barrel and has a partiality towards binding G-rich sequences with more affinity towards binding RNA than DNA. 41 YB-1 CSD forms a homodimer in crystal and in solution and dimerization is important for RNA binding and splicing regulation. In the same study done by Yang et. al, they found that YB-1 CSD preferentially binds ssRNA oligomers and the hydrophobic/basic surface promotes RNA binding. 42 Induced mutation of the YB-1 CSD showed that there was a reduction in RNA binding which further suggests that dimerization is important for CSD interaction with RNA.
The A/P domain of the N-terminus may be involved in its transcriptional activity and, in a study done by Okamoto et. al, has been shown to interact with p53 through this domain to regulate transcription. 43 The localization of the YB-1 protein is typically in the cytosol and is shuttled between the cytoplasm and the nucleus. Translocation into the nucleus results from different stresses including PI3K-Akt signaling, viral infection, and even hyperthermia. 44–46
YB-1 Function
YB-1 is a molecule that has many interacting partners. YB-1 can bind to the promoters of various genes to either activate or repress transcription. Under cellular stress conditions (cold shock or mechanical stress), YB-1 is phosphorylated and translocated into the nucleus where it contributes to transcriptional regulation. YB-1 has been shown to bind directly to the promoter of oncogenic PIK3CA driving its transcription. 47 YB-1 has also been characterized as a transcriptional repressor. One study demonstrated that YB-1 was a component of the COL1A2 transrepression complex. 48 Another study illustrated how Y-box repressor proteins (dpbA and dbpB/YB-1), once phosphorylated by ERK2 and GSK3β in vitro, bound to VEGF HRR DNA and repress the promoter. 49
YB-1 as an oncogene
YB-1 is considered to be an oncogene because it is overexpressed in cancers and is highly associated with increased metastasis and mortality. Much of its oncogenic capacity is found in the transcriptional regulation of genes in numerous cancer models. Many studies have shown that YB-1 is involved in the regulation of cell proliferation, migration, invasion, and apoptosis. 47,50–56 Through activation of the EGFR/AKT pathway, YB-1 has been demonstrated to promote tumor formation and progression in chordoma. 57 In another study, YB-1 was expressed in immature and anaplastic multiple myeloma (MM) cells and the subsequent knockdown of YB-1, by siRNA, leading to growth arrest and induction of apoptosis in MM cell lines. 50 Further results showed that the mechanism was by cellular resistance against chemotherapeutic agents, like the cytotoxic drug, doxorubicin. Similar findings were observed in breast cancer cells where YB-1 was linked to intrinsic multidrug resistance . 58. This trend carries over into ccRCC models where drug resistance was observed in cells that had higher expression of YB-1 and ABCB-1 proteins. 59
The role of YB-1 in tumor invasiveness is also well documented in the literature. In invasive breast cancer cells YB-1 levels correlated with reduced expression of the cell-cell adhesion molecule, E-cadherin, which promoted epithelial-mesenchymal transition (EMT). Overexpression of YB-1 in pre-malignant breast epithelial grafts in mice lead to decreased E-cadherin expression and increased dissemination into the fat pads. 51
Consistently, higher expression of YB-1 has been associated with increased proliferation markers and cell abundance. YB-1 has also been associated with cell survival through selective inhibition of p53 preventing the activation of proapoptotic genes. In a study done by Homer et. al, increased YB-1 expression was shown to decrease p53 in ubiquitin-dependent manner, or by direct blockade of the transactivator domain. 60
In addition to YB-1’s role in transcription, it also contributes to the regulation of translation and mRNA stability. It is known that YB-1 is a component of messenger ribonucleoprotein particles (mRNPs) and it has different effects on translation. 61 The consequence of YB-1-dependent translational regulation is metastasis, and a more invasive cancer phenotype. This occurs through the induction of many mRNAs associated with growth and differentiation genes, like Snail 1, or the protection of mRNAs from degradation. 51 YB-1 binding to RNA also has implications for RNA processes such as pre-mRNA splicing. For example YB-1 was shown to bind to ACE elements on CD44 RNA sequence and promote splicing of an alternative exon 4. 62 It has also been shown to interact with other proteins of the serine/arginine (SR)-rich proteins that work in concert to process pre-mRNAs through alternative splicing. 63
The wide variety of interactions is what initially led our research group to investigate YB-1 and its role in regulating genes in ccRCC. A protein as dynamic as YB-1 should be understood as completely as possible so that scientist and clinicians develop ways to overcome its oncogenic function, thus improving the treatment landscape for ccRCC and other related diseases.
Discussion
Further understanding the connection between the YB-1 protein and lipogenesis in ccRCC could potentially identify more effective ways to diagnose this disease. Recent work by Jeffords et al. has shown that a previously unknown relationship exists between YB-1 and the lipogenic enzyme SCD-1. This finding points to a balance between oncogenic potential of YB-1 and cellular and organismal survival. For many years SCD-1 expression in RCC has been considered an oncogene. The data supporting this thinking were compelling as it was reiterated in several other models of cancer including breast and lung cancers. However, there has been little evidence that demonstrates that SCD-1 expression is altered significantly in early-stage disease. Similarly, there is no known mutation in the SCD1 gene, which indicates that SCD-1 is not a driver of tumorigenesis in ccRCC. In this review, we have proposed a different perspective of SCD-1 in ccRCC. This alternative view suggests that SCD-1 is the consequence of some upstream driver mutation. As such, SCD-1 appears to be playing a role in a survival mechanism that is amplified as cell division increases. This viewpoint is supported not only by our in vitro data, but also experimental evidence in model organisms that show increased longevity when SCD-1 orthologs are overexpressed. In humans, similar evidence can be gleaned from the 7-countries dietary study. 64 During the multi-site dietary intervention study, it was concluded that individuals with diets high in oleic acid-rich Olive oil (similar to unsaturated fatty acids produced by SCD-1), had longer life spans, and fewer life-threatening health conditions.
Our observation that the oncogene YB-1 correlates with poor prognosis in nearly every study (including our own), and our unique finding of SCD-1’s effect on survival indicates a difference in either patient cohorts, samples type, or analysis methods as compared previous studies. However, with access to additional large patient datasets, it may be possible to identify the specific conditions present across all studies involving proteomic analysis on tumor samples.
Conclusion
Clear cell Renal Cell Carcinoma is difficult to treat for a variety of reasons. Therefore, it is important to identify novel approaches to diagnose and treat patients. For nearly a decade, the research described SCD-1 as an oncogenic factor in renal cancer cells. However, no clinical trials with SCD-1 inhibitors have shown evidence of increased patient survival, to date. This may be due to off-target effects that make SCD-1 inhibition toxic to other tissues, through uncharacterized alterations in key cellular mechanisms. This is plausible because SCD-1 is not simply an oncogene, it is also responsible for preventing cellular lipotoxicity and it provides the components of several downstream lipid-containing molecules necessary for normal physiology. Although blocking SCD-1 function may appear an effective way to kill growing cancer cells, in many cases, SCD-1 inhibition can damage normal cells as well. Without targeted therapy directed at the tumor, the off-target effects of shutting down SCD-1 are likely going to lead to deleterious outcomes in the patient.
Vigorous in vitro and in vivo experimentation must be carried out to evaluate how lipids are uniquely utilized in ccRCC compared to normal kidney epithelia. Only then will we be able to understand where the true vulnerabilities are. To accomplish this, it will certainly take an interdisciplinary approach using key resources from fields such as proteomics, bioinformatics, molecular biology, and biochemistry.
References:
- 1.Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906. [DOI] [PubMed] [Google Scholar]
- 2.Zhang P, Ro JY. Renal cell carcinoma. Annals of nrologic oncology. 2018;1(1): 1–18. [Google Scholar]
- 3.Cairns P Renal cell carcinoma. Cancer Biomark. 2010;9(1-6):461–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669): 1119–1132. [DOI] [PubMed] [Google Scholar]
- 5.Brannon AR, Reddy A, Seiler M, et al. Molecular Stratification of Clear Cell Renal Cell Carcinoma by Consensus Clustering Reveals Distinct Subtypes and Survival Patterns. Genes Cancer. 2010;1(2): 152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao M, Tabuchi H, Nagashima Y, et al. Gene expression analysis of renal carcinoma: adipose differentiation-related protein as a potential diagnostic and prognostic biomarker for clear-cell renal carcinoma. J Pathol. 2005;205(3):377–387. [DOI] [PubMed] [Google Scholar]
- 7.Samuel Jeffords EF; Breanna Cole; Kate Root; Thierry Chekouo; Melvin Richard G.; Lynne Bemis; Simmons Glenn E. Jr . Y-box binding protein 1 acts as a negative regulator of stearoyl CoA desaturase 1 in clear cell renal cell carcinoma. Oncology Letters 2020;20(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet. 1994;7(1):85–90. [DOI] [PubMed] [Google Scholar]
- 9.Kuroda N, Hosokawa T, Michal M, et al. Clear cell renal cell carcinoma with focal renal angiomyoadenomatous tumor-like area. Ann Diagn Pathol. 2011;15(3):202–206. [DOI] [PubMed] [Google Scholar]
- 10.Qiu B, Ackerman D, Sanchez DJ, et al. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov. 2015;5(6):652–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008; 105(48): 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem Sci. 2010;35(8):427–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeNicola GM, Cantley LC. Cancer’s Fuel Choice: New Flavors for a Picky Eater. Mol Cell 2015;60(4):514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Kang Y. Lipid Metabolism Fuels Cancer’s Spread. Cell Metab. 2017;25(2):228–230. [DOI] [PubMed] [Google Scholar]
- 15.Braig S Chemical genetics in tumor lipogenesis. Biotechnol Adv. 2018;36(6): 1724–1729. [DOI] [PubMed] [Google Scholar]
- 16.Aljohani A, Khan MI, Bonneville A, et al. Hepatic stearoyl CoA desaturase 1 deficiency increases glucose uptake in adipose tissue partially through the PGC-lalpha-FGF21 axis in mice. J Biol Chem. 2019;294(51): 19475–19485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ameer F, Scandiuzzi L, Hasnain S, Kalbacher H, Zaidi N. De novo lipogenesis in health and disease. Metabolism. 2014;63(7):895–902. [DOI] [PubMed] [Google Scholar]
- 18.Iida T, Ubukata M, Mitani I, et al. Discovery of potent liver-selective stearoyl-CoA desaturase-1 (SCD1) inhibitors, thiazole-4-acetic acid derivatives, for the treatment of diabetes, hepatic steatosis, and obesity. Ear J Med Chem. 2018;158:832–852. [DOI] [PubMed] [Google Scholar]
- 19.Igal RA. Stearoyl-CoA desaturase-1: a novel key player in the mechanisms of cell proliferation, programmed cell death and transformation to cancer. Carcinogenesis. 2010;31(9):1509–1515. [DOI] [PubMed] [Google Scholar]
- 20.Spector AA. The importance of free fatty acid in tumor nutrition. Cancer Res. 1967;27(9):1580–1586. [PubMed] [Google Scholar]
- 21.Peck B, Schug ZT, Zhang Q, et al. Inhibition of fatty acid desaturation is detrimental to cancer cell survival in metabolically compromised environments. Cancer and Metabolism. 2016;4(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi M, Rhodes DR, Furge KA, et al. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci U S A. 2001;98(17):9754–9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saito K, Arai E, Maekawa K, et al. Lipidomic Signatures and Associated Transcriptomic Profiles of Clear Cell Renal Cell Carcinoma. Sci Rep. 2016;6:28932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Z, Liu Y, Liu Q, et al. The mRNA Expression Signature and Prognostic Analysis of Multiple Fatty Acid Metabolic Enzymes in Clear Cell Renal Cell Carcinoma. J Cancer. 2019;10(26):6599–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Roemeling CA, Marlow LA, Wei JJ, et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin Cancer Res. 2013;19(9):2368–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kikuchi K, Tsukamoto H. Stearoyl-CoA desaturase and tumorigenesis. Chemico-Biological Interactions. 2020;316:108917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koeberle A, Loser K, Thurmer M. Stearoyl-CoA desaturase-1 and adaptive stress signaling. Biochim Biophys Acta. 2016;1861(11):1719–1726. [DOI] [PubMed] [Google Scholar]
- 28.Paton CM, Ntambi JM. Biochemical and physiological function of stearoyl-CoA desaturase. Am J Physiol Endocrinol Metab. 2009;297(1):E28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampath H, Miyazaki M, Dobrzyn A, Ntambi JM. Stearoyl-CoA desaturase-1 mediates the pro-lipogenic effects of dietary saturated fat. J Biol Chem. 2007;282(4):2483–2493. [DOI] [PubMed] [Google Scholar]
- 30.Igal RA. Stearoyl CoA desaturase-1: New insights into a central regulator of cancer metabolism. Biochim Biophys Acta. 2016;1861(12 Pt A):1865–1880. [DOI] [PubMed] [Google Scholar]
- 31.Song Y, Zhong L, Zhou J, et al. Data-Independent Acquisition-Based Quantitative Proteomic Analysis Reveals Potential Biomarkers of Kidney Cancer. Proteomics Clin Appl. 2017;11(11–12). [DOI] [PubMed] [Google Scholar]
- 32.Cao Q, Ruan H, Wang K, et al. Overexpression of PLIN2 is a prognostic marker and attenuates tumor progression in clear cell renal cell carcinoma. Int J Oncol. 2018;53(1):137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yao M, Huang Y, Shioi K, et al. Expression of adipose differentiation-related protein: a predictor of cancer-specific survival in clear cell renal carcinoma. Clin Cancer Res. 2007;13(1):152–160. [DOI] [PubMed] [Google Scholar]
- 34.Tun HW, Marlow LA, von Roemeling CA, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PLoS One. 2010;5(5):e10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Su J, Fu D, et al. The Role of YB1 in Renal Cell Carcinoma Cell Adhesion. Int J Med Sci. 2018;15(12):1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Chen Y, Geng H, Qi C, Liu Y, Yue D. Overexpression of YB1 and EZH2 are associated with cancer metastasis and poor prognosis in renal cell carcinomas. Tumour Biol. 2015;36(9):7159–7166. [DOI] [PubMed] [Google Scholar]
- 37.Cohen SB, Ma W, Valova VA, et al. Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene. 2010;29(3):403–410. [DOI] [PubMed] [Google Scholar]
- 38.Yadav BS, Singh S, Shaw AK, Mani A. Structure prediction and docking-based molecular insights of human YB-1 and nucleic acid interaction. J Biomol Struct Dyn. 2016;34(12):2561–2580. [DOI] [PubMed] [Google Scholar]
- 39.Kang S, Lee TA, Ra EA, et al. Differential control of interleukin-6 mRNA levels by cellular distribution of YB-1. PLoS One. 2014;9(11):e112754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guryanov SG, Filimonov VV, Timchenko A A, et al. The major mRNP protein YB-1: structural and association properties in solution. Biochim Biophys Acta. 2013;1834(2):559–567. [DOI] [PubMed] [Google Scholar]
- 41.Kljashtorny V, Nikonov S, Ovchinnikov L, et al. The Cold Shock Domain of YB-1 Segregates RNA from DNA by Non-Bonded Interactions. PLoS One. 2015; 10(7):e0130318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X-J, Zhu H, Mu S-R, et al. Crystal structure of a Y-box binding protein 1 (YB-1)-RNA complex reveals key features and residues interacting with RNA. J Biol Chem. 2019;294(28): 10998–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okamoto T, Izumi H, Imamura T, et al. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene. 2000; 19(54):6194–6202. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland BW, Kucab J, Wu J, et al. Akt phosphorylates the Y-box binding protein 1 at Seri02 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24(26):4281–4292. [DOI] [PubMed] [Google Scholar]
- 45.Holm PS, Bergmann S, Jurchott K, et al. YB-1 relocates to the nucleus in adenovirus-infected cells and facilitates viral replication by inducing E2 gene expression through the E2 late promoter. J Biol Chem. 2002;277(12): 10427–10434. [DOI] [PubMed] [Google Scholar]
- 46.Stein U, Jürchott K, Walther W, Bergmann S, Schlag PM, Royer HD. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J Biol Chem. 2001;276(30):28562–28569. [DOI] [PubMed] [Google Scholar]
- 47.Astanehe A, Finkbeiner M, To K, Dunn SE. The Transcription Factor Y-Box Binding Protein-1 (Yb-1) Induces Expression of the Pik3ca Oncogene Leading to Increased Invasion Ofbasal-Like Breast Carcinoma Cells. Clinical & Investigative Medicine. 2008;31(4):3. [Google Scholar]
- 48.Higashi K, Inagaki Y, Suzuki N, et al. Y-box-binding Protein YB-1 Mediates Transcriptional Repression of Human α2(I) Collagen Gene Expression by Interferon-γ. Journal of Biological Chemistry. 2003;278(7):5156–5162. [DOI] [PubMed] [Google Scholar]
- 49.Coles LS, Lambrusco L, Burrows J, et al. Phosphorylation of cold shock domain/Y-box proteins by ERK2 and GSK3beta and repression of the human VEGF promoter. FEBS Lett. 2005;579(24):5372–5378. [DOI] [PubMed] [Google Scholar]
- 50.Chatteijee M, Rancso C, Stiihmer T, et al. The Y-box binding protein YB-1 is associated with progressive disease and mediates survival and drug resistance in multiple myeloma. Blood. 2008; 111(7):3714–3722. [DOI] [PubMed] [Google Scholar]
- 51.Evdokimova V, Tognon C, Ng T, et al. Translational activation of snail 1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15(5):402–415. [DOI] [PubMed] [Google Scholar]
- 52.Ha B, Lee EB, Cui J, Kim Y, Jang HH. YB-1 overexpression promotes a TGF-beta1-induced epithelial-mesenchymal transition via Akt activation. Biochem Biophys Res Commun. 2015;458(2):347–351. [DOI] [PubMed] [Google Scholar]
- 53.Harada M, Kotake Y, Ohhata T, et al. YB-1 promotes transcription of cyclin D1 in human non-small-cell lung cancers. Genes Cells. 2014;19(6):504–516. [DOI] [PubMed] [Google Scholar]
- 54.Sutherland BW, Kucab J, Wu J, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24(26):4281–4292. [DOI] [PubMed] [Google Scholar]
- 55.Somasekharan SP, El-Naggar A, Leprivier G, et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208(7):913–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee C, Dhillon J, Wang MYC, et al. Targeting YB-1 in HER-2 overexpressing breast cancer cells induces apoptosis via the mTOR/STAT3 pathway and suppresses tumor growth in mice. Cancer Res. 2008;68(21):8661–8666. [DOI] [PubMed] [Google Scholar]
- 57.Liang C, Ma Y, Yong L, et al. Y-box binding protein-1 promotes tumorigenesis and progression via the epidermal growth factor receptor/AKT pathway in spinal chordoma. Cancer Sci. 2019;110(1):166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bargou RC, Jürchott K, Wagener C, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3(4):447–450. [DOI] [PubMed] [Google Scholar]
- 59.D’Costa NM, Lowerison MR, Raven PA, et al. Y-box binding protein-1 is crucial in acquired drug resistance development in metastatic clear-cell renal cell carcinoma. J Exp Clin Cancer Res. 2020;39(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Homer C, Knight DA, Hananeia L, et al. Y-box factor YB1 controls p53 apoptotic function. Oncogene. 2005;24(56):8314–8325. [DOI] [PubMed] [Google Scholar]
- 61.Evdokimova VM, Ovchinnikov LP. Translational regulation by Y-box transcription factor: involvement of the major mRNA-associated protein, p50. Int J Biochem Cell Biol. 1999;31(1):139–149. [DOI] [PubMed] [Google Scholar]
- 62.Stickeler E, Fraser SD, Honig A, Chen AL, Berget SM, Cooper TA. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20(14):3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li J, Hawkins IC, Harvey CD, Jennings JL, Link AJ, Patton JG. Regulation of alternative splicing by SRrp86 and its interacting proteins. Mol Cell Biol. 2003;23(21):7437–7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kromhout D, Menotti A, Alberti-Fidanza A, et al. Comparative ecologic relationships of saturated fat, sucrose, food groups, and a Mediterranean food pattern score to 50-year coronary heart disease mortality rates among 16 cohorts of the Seven Countries Study. Eur J Clin Nutr. 2018;72(8):1103–1110. [DOI] [PubMed] [Google Scholar]


