Abstract
The poultry industry in Colombia has implemented several changes and measures in chicken processing to improve sanitary operations and control pathogens’ prevalence. However, there is no official in-plant microbial profile reference data currently available throughout the processing value chains. Hence, this research aimed to study the microbial profiles and the antimicrobial resistance of Salmonella isolates in three plants. In total, 300 samples were collected in seven processing sites. Prevalence of Salmonella spp. and levels of Enterobacteriaceae were assessed. Additionally, whole-genome sequencing was conducted to characterize the isolated strains genotypically. Overall, the prevalence of Salmonella spp. in each establishment was 77%, 58% and 80% for plant A, B, and C. The mean levels of Enterobacteriaceae in the chicken rinsates were 5.03, 5.74, and 6.41 log CFU/mL for plant A, B, and C. Significant reductions were identified in the counts of post-chilling rinsate samples; however, increased levels were found in chicken parts. There were six distinct Salmonella spp. clusters with the predominant sequence types ST32 and ST28. The serotypes Infantis (54%) and Paratyphi B (25%) were the most commonly identified within the processing plants with a high abundance of antimicrobial resistance genes.
Keywords: microbial profile, Salmonella, chicken processing, antimicrobial resistance, Colombia
1. Introduction
The poultry industry in Colombia has grown steadily in the last ten years, favoring the investment in new processing technologies, genetic selection, good agricultural and manufacturing practices and harmonization with international standards. Additionally, Colombian producers have implemented biosecurity measures to prevent the dissemination of Newcastle and salmonellosis to meet the demands of national and global chains [1]. In 2014, the National Administrative Department of Statistics (DANE, Departamento Administrativo Nacional de Estadística) in collaboration with the National Federation of Poultry Producers of Colombia (FENAVI, Federación Nacional de Avicultores) reported that the valorization of the poultry sector in Colombia was estimated to be 14.8 billion Colombian pesos (4.79 million US Dollars), a record value in the poultry industry.
The Ministry of Agriculture and Rural Development, together with the Ministry of Health and Social Protection, established the Decree 1500 in 2007 to improve food safety programs in the meat and poultry sectors. This decree has been updated in recent years, and in 2016 the national government implemented the last phase of the sanitary laws (Decree 1500 of 2007, Decree 2270 of 2012, and Decree 1282 of 2016, resolution 240/2013), which target the reduction of foodborne illness and the implementation of food safety programs in meat and poultry productions. These updated decrees establish new microbiological verification programs for the conventional inspection system for meat and meat products.
In Colombia, chicken production is concentrated in five-Departments, Santander, Cundinamarca, Valle del Cauca, Cauca, and Antioquia, providing more than two-thirds of the country’s total production [2]. The poultry processing plants in Colombia are classified into two categories: industrial plants and special plants. The first produces more than 3000 birds/day, while the second presents a lower capacity. Industrial plants comprise 97% of the total production volume [3]. There are 118 authorized chicken slaughtering establishments in the country, and 397 more operate under provisional authorization based on the national sanitary standards [4]. In 2016, ten plants were closed because they did not meet the requirements of Decree 1500, while 49% (70/142) followed the general sanitary regulations.
The National Health Institute (INS, Instituto Nacional de Salud) of Colombia reported 11,502 foodborne illness cases related to 881 outbreaks in 2018 [5]. The Valle del Cauca presented the highest numbers of outbreaks (124/881), followed by Bogota D.C. (78/881) and Sucre (64/881) [5]. Salmonella spp. contamination of foods was identified as the attribution source for 29% (159/547) of the total number of outbreaks where the food or surface sample was available. The main food products linked with these outbreaks were cheese (19.4%), chicken meat (10.7%), mixed foods (9.6%), fast food (9.65%), fish and seafood (7.8%), beef and pork meat (6%), and fruit juices (2.6%) [5].
The INS initiated in 1997 a laboratory-based surveillance program to track Salmonella spp. clinical isolates around the country. In 2019, the institution reported 20 years of data from different health systems, with 12966 Salmonella spp. isolates from the 32 departments. From those isolates, more than 60% were recovered from the Antioquia department (26.4%; 3421/12966), Bogota D.C. (24.5%; 3182/12966), and Valle del Cauca (8.7%; 1127/12966). The top five serotypes identified were Typhimurium (27.6%), Enteritidis (27.1%), Typhi (11.4%), Dublin (3.3%), and Infantis (2.03%). Antimicrobial susceptibility testing of 5185 Salmonella spp. isolates was also included in the program from the period 2014–2018, where S. Enteritidis (24%; 1247/5195) and S. Typhimurium (21%; 1090/5195) presented the highest levels of resistance to ampicillin, cefotaxime, ceftazidime, ciprofloxacin, chloramphenicol, tetracycline and trimethoprim-sulfamethoxazole [6]. However, Salmonella spp. infection is an important concern for public health authorities in Colombia. There is no available data on illness source attribution to estimate common causes and determine opportunities to improve the health system.
Furthermore, there is no public document that collects together information on all the monitoring programs established in the processing plants. The National Institute of Food and Drug Surveillance (INVIMA, Instituto Nacional de Vigilancia de Medicamentos y Alimentos) and FENAVI initiated a national data collection of the prevalence of Salmonella spp. and Campylobacter spp. in chicken carcasses, and the common serovars found in processing environments. However, this information has not yet been published. Additionally, the INS and the Ministry of Health and Social Protection issued food safety risk profiles for Salmonella spp. [7], and Campylobacter spp. [8] associated with whole chicken carcasses and chicken parts, due to the increased number of foodborne illnesses in the country. These documents presented a detailed analysis of the risk factors associated with poultry production and processing in Colombia. They also identified the lack of microbial profiling, pathogens serotyping, and antimicrobial resistance surveillance in the design of science-based approaches to control and reduce foodborne pathogens in the poultry industry.
Hence, this study’s main objectives were to determine the microbial profile during chicken processing operations to assess pathogen prevalence, Enterobacteriaceae levels, and evaluate the genotypic characterization of antimicrobial resistant Salmonella spp. strains.
2. Materials and Methods
2.1. Characteristics of the Poultry Processing Plants
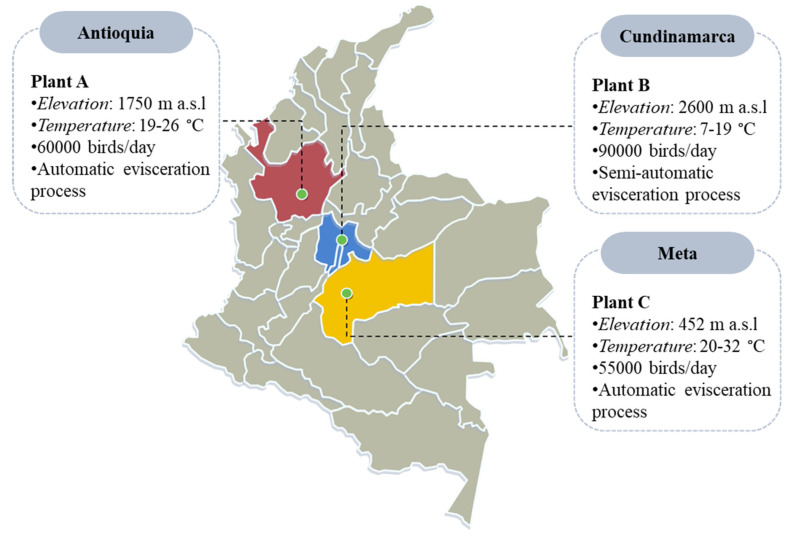
Three commercial poultry processing plants were sampled between July-October 2017. A summary of the locations and characteristics of each plant is presented in Figure 1. All three establishments utilize 10 ppm hypochlorous acid (HClO) as a chemical intervention in the immersion chiller tank, and tap water for washers and drench applications. They run two schedule shifts of 8 h/day for 5 days per week.
Figure 1.
Characteristics of the three poultry processing facilities included in this study.
The broiler chickens processed in the poultry establishments were fed with a diet based on maize and supplement with salinomycin and narasin as a coccidiostat. Subtherapeutic levels of antibiotics such as enramycin, avilamycin, bacitracin, and chlorhydroxy-quinoline were added to the poultry feed as a growth promoter. Therapeutic levels of chlortetracycline, tiamulin, and tilmicosin were also used to treat disease in chickens.
2.2. Experimental Design
In total, 270 chicken rinsate and 30 fecal samples were collected in the three plants (100 samples from seven sites in each plant) during a normal processing day. The processing sites were selected based on the production line at the processing plants. Samples were collected at the arrival station (fecal), pre-scalding (after bleeding and before entering the scalding tank), post-scalding (immediately after exiting the scalder tank), post-IOBW (inside-outside body wash), pre-chiller (before entering the pre-chiller tank), post-chiller (immediately after exiting the immersion chill tank), and parts (reconstruct a chicken carcass with legs, breasts, and wings). Plant B did not have a chicken cut-up and deboning room installed. Hence, post-defeathering samples were collected instead of chicken parts. The prevalence of Salmonella spp. and the levels of Enterobacteriaceae in chicken rinsate were evaluated for all samples collected.
2.3. Chicken Carcass Rinsate Sample Collection
Whole chicken carcass and chicken part rinsates (one full chicken breast, two chicken wings, and two chicken legs) were collected at different processing sites in all three processing plants, according to the USDA-FSIS method MLG 4.09. Briefly, as described in Ramirez-Hernandez et al., [9], chicken carcasses and chicken parts were randomly selected and removed from the processing line and aseptically placed in individual sterile poultry rinse bags (Nasco, Fort Atkinson, WI, USA). Then, 400 mL of sterile buffered peptone water (BPW, BD, Detroit, MI, USA) was added to each bag and distributed by shaking for 1 min. After collection, samples were kept at 5 °C in a cooler and sent overnight to the Pontificia Universidad Javeriana in Bogota, D.C., Colombia.
2.4. Fecal Sample Collection
Sampled birds were scheduled to be processed as the first lots on the sampling day. For plant A, fecal samples were pooled by collecting aseptically 25 g from arriving cages (n = 10) into a sterile sampling bag (Whirl-Pak; Nasco, Fort Atkinson, WI, USA). For plants B and C, fecal samples were extracted aseptically from the cloaca of individual chickens (n = 10) into a sterile sampling bag. Samples were kept 5 °C in a cooler and shipped together with the rinsate samples to the Pontificia Universidad Javeriana in Bogota, D.C., Colombia.
2.5. Microbiological Analysis
The microbial analysis was conducted at the food microbiology laboratory in the Department of Microbiology at the Universidad Javeriana in Bogota D.C., Colombia.
2.5.1. Salmonella spp. Detection and Isolation
The prevalence of Salmonella spp. was evaluated using a molecular detection system (MDS100, 3MTM; St. Paul, MN, USA) with method AOAC 2013, following manufacturer’s instructions. Briefly, for Salmonella spp. detection (MDA2SAL96-3MTM molecular detection assay 2-Salmonella; St. Paul, MN, USA), 30 mL of rinsate sample were mixed with 30 mL of double strength BPW; fecal samples were diluted and homogenized thoroughly for 2 min and incubated at 37 ± 1 °C for 24 h. The enrichments were transferred to lysis tubes and heated at 100 ± 1 °C for 15 min. Lysates were transferred to a reagent tube, loaded into a molecular detection speed loader tray (3MTM), and analyzed using molecular detection software. Presumptive positive samples were then cultured based on the Microbiology Laboratory Guidebook MLG 4.09 (Salmonella). Briefly, an aliquot of 0.5 mL of each enriched sample was transferred to 10 mL of Tetrathionate (TT) broth (BD; Sparks, MD, USA), and 0.1 mL were transferred into 10 ml modified Rappaport-Vassiliadis (mRV) broth (BD; Sparks, MD, USA), and incubated at 42 ± 1 °C and 37 ± 1 °C for 24 h. After incubation, tubes were vortexed, and one loopful (10 µL) of the enrichment was streaked on Brilliant Green (BG) agar (Acumedia; Lasing, MI, USA) and Xylose Lysine TergitolTM 4 (XLT4) agar (EMD; Billerica, MA, USA), and incubated at 37 ± 1 °C for 24 h. Salmonella presumptive colonies were selected based on typical morphology and color. Subsequently, the screening slants media Triple Sugar Iron (TSI) agar (BD; Sparks, MD, USA) and Lysine Iron Agar (LIA) (BD; Sparks, MD, USA) were used by stabbing the butts and streaking the slants a single pick colony. Tubes were incubated at 37 ± 1 °C for 24 h, and typical reactions on TSI and LIA slants were evaluated.
2.5.2. Enumeration of Enterobacteriaceae
Dilutions prepared for Salmonella detection were used to quantify the levels of Enterobacteriaceae in the chicken carcass and chicken parts rinsates and fecal samples, following the MLG 3.02 protocol (Quantitative Analysis of Bacteria in Foods as Sanitary Indicator). Further, serial diluted (1:10) in BPW were done, and 1 ml of the corresponding dilution was transferred onto Enterobacteriaceae Petrifilm plates (3MTM, St. Paul, MN, USA), which were incubated at 35 °C for 24 h, following manufacturer’s instructions.
2.6. Whole Genome Sequencing (WGS)
The confirmed Salmonella spp. strains were sequenced following the methodology explained by Ramirez-Hernandez et al., [9,10]. WGS analysis was conducted as part of the GenomeTrakr Project: Texas Department of State Health Services. The genomic data of each sequenced strain have been deposited in National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/ (accessed on 24 February 2021)) under the Bioproject accession number PRJNA284276. Among the 85 Salmonella genome assemblies, there was a mean of 56x (min: 32, max: 103), 4.91 Mbp of genome size (min: 4.51, max: 5.10), and 120.60 of number of contigs (min: 24, max: 279).
2.7. Bioinformatic Analysis
Fastq files containing the sequence data were used to perform contig assemblies using Patric (http://www.patricbrc.org (accessed on 24 February 2021)). Briefly, assembled contig files were run into different pipelines available at the Center for Genomic Epidemiology website (http://www.genomicepidemiology.org/ (accessed on 24 February 2021)), including ResFinder 3.0, SeqSero 1.2, MLST 1.8, PlasmidFinder 1.3, SpeciesFinder 1.2 16S rRNA, and CSI phylogeny 1.4. Lastly, bacterial populations were defined based on the genotypic characteristics of the strains.
2.8. Statistical Analysis
Chi-square test was used to determine the statistical relationship of Salmonella spp. prevalence between the processing sites. Enterobacteriaceae counts were analyzed with a non-parametric test Kruskal-Wallis followed by pairwise multiple comparisons post hoc Conover’s tests to identify the significant variation in microbial level on fecal and rinsate samples collected in different sites. A p-value of 0.05 was selected for a significant difference in this study.
3. Results
3.1. Salmonella spp. Prevalence
Salmonella spp. were recovered from chicken carcass, chicken parts rinsates and fecal samples at different processing sites (Table 1). The overall prevalence of Salmonella spp. in each plant was: Plant A, 77% (77/100; CI, 68 to 84%), Plant B, 58% (58/100; CI, 48 to 67%), and Plant C, 80% (80/100; CI, 71 to 87%).
Table 1.
Salmonella spp. prevalence from chicken samples collected at different processing sites.
| Processing Site | Plant A | Plant B | Plant C | |||
|---|---|---|---|---|---|---|
| No. Positive (%) |
95% CI | No. Positive (%) |
95% CI | No. Positive (%) |
95% CI | |
| Fecal material | ||||||
| Arrival | 1/10 (10) | 0.5–46 | 0/1 (0) | 0–34 | 4/10 (40) | 14–73 |
| Chicken carcass rinse | ||||||
| Pre-scalding | 2/15 (13) a | 23–42 | 12/15 (80) a | 51–97 | 12/15 (80) a | 51–97 |
| Post-scalding | 14/15 (93) b | 66–99 | 8/15 (53) a | 27–78 | 15/15 (100) a | 75–100 |
| Post-defeathering | - | - | 13/15 (87) a | 58–98 | ||
| Post-IOBW (inside-outside body wash) | 15/15 (100) b | 75–100 | 6/15 (40) a | 17–67 | 11/15 (73) a | 45–91 |
| Pre-chiller | 15/15 (100) b | 75–100 | 12/15 (80) a | 51–97 | 13/15 (87) a | 58–98 |
| Post-chiller | 15/15 (100) b | 75–100 | 7/15 (47) a | 22–73 | 12/15 (80) a | 51–97 |
| Parts | 15/15 (100) b | 75–100 | 13/15 (87) a | 58–98 | - | |
a,b Prevalence followed by different superscripts is statistically different (Comparison between processing sites). 95% CI: 95% Confidence Interval.
Plant A: The prevalence of Salmonella spp. was unchanged from the post-IOBW processing sites, to 100% (CI, 75–100%) presumptive positive samples (p > 0.05). However, samples from the pre-scalding site exhibited a different prevalence than the subsequent processing sites (p = 0.001).
Plant B: Different profiles were presented; Salmonella spp. were detected at various levels throughout the process. After pre-chilling, the prevalence was 47% (7/15; CI, 22 to 73%) and a higher level of contamination was detected in chicken parts with 80% (13/15; CI, 58 to 98%) of presumptive positive samples; however, there were no significant differences among the processing sites (p = 0.17).
Plant C: 80% (12/15; CI, 51–97) of Salmonella spp. positive samples were presented at the post-chilling site, with no statistical differences between processing sites (p = 0.29).
3.2. Enterobacteriaceae Levels
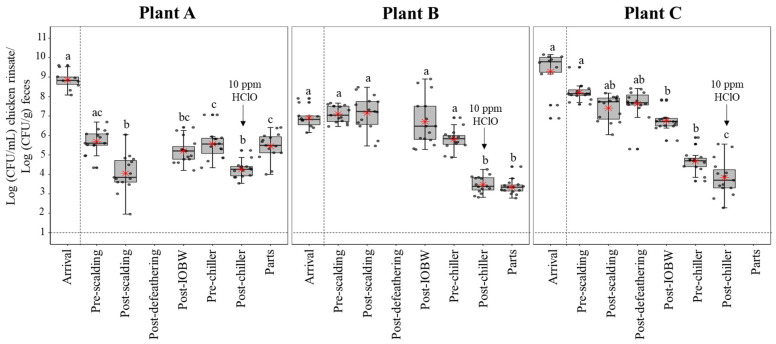
Enterobacteriaceae levels are presented in Figure 2.
Figure 2.
Enterobacteriaceae (Log CFU/mL for chicken rinsates; Log CFU/g for fecal samples) levels recovered from the chicken carcass, chicken parts, and fecal samples by each processing plant at the different processing sites. In each box plot, the heavy horizontal line crossing the box is the median, the bottom and top of the box are the lower and upper quartiles, and the whiskers are the maximum and minimum values of the date set. The red star inside the box represents the mean value. Boxes with different letters a, b, c, d are significantly different according to a non-parametric Kruskal-Wallis followed pairwise multiple comparison post hoc Conover’s test at p < 0.05. The grey points represent the actual data points. There are some that appears as darker points is because there are more than one data point with the same or close value.
Plant A: Fecal samples at the arrival location presented a mean of 8.85 Log CFU/g (CI, 8.51 to 9.19 Log CFU/g). From the first processing site evaluated (pre-scalding) to the final process (parts), counts of Enterobacteriaceae differed by 0.24 Log CFU/mL, but there were no statistically significant differences (p = 0.33). However, there was a considerable difference between Enterobacteriaceae levels in pre-chilling, compared to chicken parts (p = 0.003).
Plant B: Fecal samples at the arrival location presented a mean of 6.90 Log CFU/g (CI, 6.51 to 7.29 Log CFU/g). There was a significant reduction for chicken parts (p < 0.00001) of 3.76 Log CFU/ml from the pre-scalding. Likewise, the pre-chiller and post-chiller counts presented significant differences with a p = 0.017. Nevertheless, there was no statistically significant difference between pre-chiller and chicken parts (p = 0.29).
Plant C: Fecal samples at the arrival location presented a mean of 9.30 Log CFU/g (CI, 8.47 to 10.1 Log CFU/g). Similarly to plant B, there was a significant reduction in the Enterobacteriaceae counts from pre-scalding (8.21 Log CFU/mL) to chicken parts (3.83 Log CFU/mL). There was a significant difference between pre-chiller and post-chiller levels with a p = 0.01; however, no differences were found when comparing levels in post-chiller and chicken parts (p = 0.21).
3.3. WGS of Salmonella spp. Isolates
From a total of 215 Salmonella spp. positive isolates, only 85 isolates were fully characterized by whole genome sequence analysis.
Table 2 presents a summary of the Salmonella spp. serotypes identified from the processing sites in the three processing plants. In plant A, all 11 isolates were confirmed as S. Infantis. In plant B, the serotype Paratyphi B was the most common identified (54%, 14/26), followed by the serotypes Javiana (11.5%, 3/26), Typhimurium (11.5%, 3/26), Heidelberg (3.8%, 1/26), Infantis (3.8%, 1/26), and unrecognized serotypes ?:b:1,2 (3.8%, 1/26), 9:,z28:1,2 (3.8%, 1/26), and 9:-:1,2 (3.8%, 1/26), and an unknown (3.8%, 1/26). From the previous serotypes, they were identified with closely related strains. In the case of ?:b:1,2 which theO antigen was undetermined and the unknow serotype, they were both highly related to an identified S. Paratyphi strain with six single different nucleotide polymorphisms (SNPs); similarly, 9:,z28:1,2 with a S. Javiana strain differed in 6 SNPs, and the 9:-:1,2, which missed the phase 1 H antigen, was closely related with a S. Infantis strain with three different SNPs. In plant C, 48 isolates were sequenced where the serotype Infantis was confirmed in 73% of the samples (35/48), followed by Paratyphi B (12.5%, 6/48), Javiana (12.5%, 6/48) and 9:-:1,5 (2%, 1/48). The last serotype, missing the phase 1 H antigen was closely related to a S. Javiana strain differing in four SNPs. Clonal populations were characterized based on the combined analysis of phylogeny, serotype, multi-locus sequence type (MLST), plasmid incompatibility type, and genotype antimicrobial resistance (AMR) profiles. The sequence types identified among all Salmonella serotypes were ST32 (48/85), ST28 (25/85), ST24 (6/85), ST19 (3/85), ST1674 (2/85), and ST15 (1/85).
Table 2.
Salmonella spp. serotypes isolated from different processing sites (No. of isolates). SeqSero-1.2 pipeline from the Center of Genomic Epidemiology.
| Processing Site | Plant A | Plant B | Plant C |
|---|---|---|---|
| (n = 11) | (n = 26) | (n = 48) | |
| Arrival | Infantis (4) | ||
| Paratyphi B (2) | |||
| Pre-scalding | Typhimurium (3) | Infantis (7) | |
| Heidelberg (1) | Javiana (4) | ||
| Paratyphi B (2) | 9:-:1,5 * (1) | ||
| ?:b:1,2 * (1) | Paratyphi B (1) | ||
| Post-scalding | 9:-:1,2 * (1) | Infantis (10) | |
| Javiana (1) | |||
| ?:b:1,2 * (1) | |||
| Paratyphi B (2) | |||
| Post-defeathering | Infantis (10) | ||
| Post-IOBW | Infantis (1) | 9:I,z28:1,2 * (1) | Infantis (1) |
| Paratyphi B (2) | Javiana (1) | ||
| Pre-chiller | Infantis (4) | Paratyphi B (6) | Paratyphi B (3) |
| Unknown * (1) | |||
| Post-chiller | Infantis (2) | Infantis (2) | |
| Javiana (1) | |||
| Paratyphi B (1) | |||
| Parts | Infantis (4) | Infantis (1) | |
| Javiana (1) | |||
| Paratyphi B (2) |
* The predicted antigenic profile does not exist in the White-Kauffmann-Le Minor scheme.
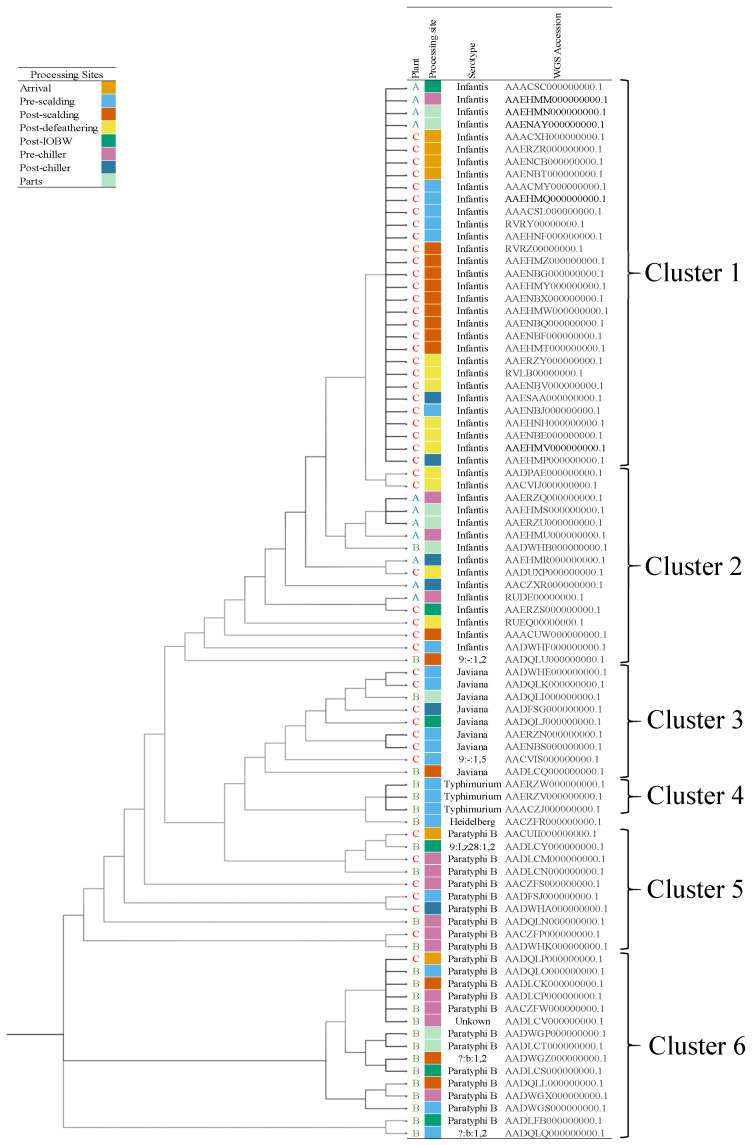
Figure 3 presents the phylogenetic trees of all Salmonella spp. strains recovered from the processing plants, created based on the concatenated high-quality single nucleotide polymorphisms, performed using CSI phylogeny 1.4 pipeline (Center of Genomic Epidemiology website). Figure 4 presents the distribution of AMR genes and plasmids of all Salmonella spp. strains recovered from the plants at the differing sites.
Figure 3.
Phylogenetic tree of Salmonella spp. strains isolated from the poultry processing plants defined by the concatenated alignment of the high-quality single nucleotide polymorphisms (SNPs). The description in the column represents the plant name, processing site, serotype, and the genome accession number from the NCBI (GenomeTrakr Project: Texas Department of States Health Services, SRA. study: SRP059203, Bioproject: PRJNA284276).
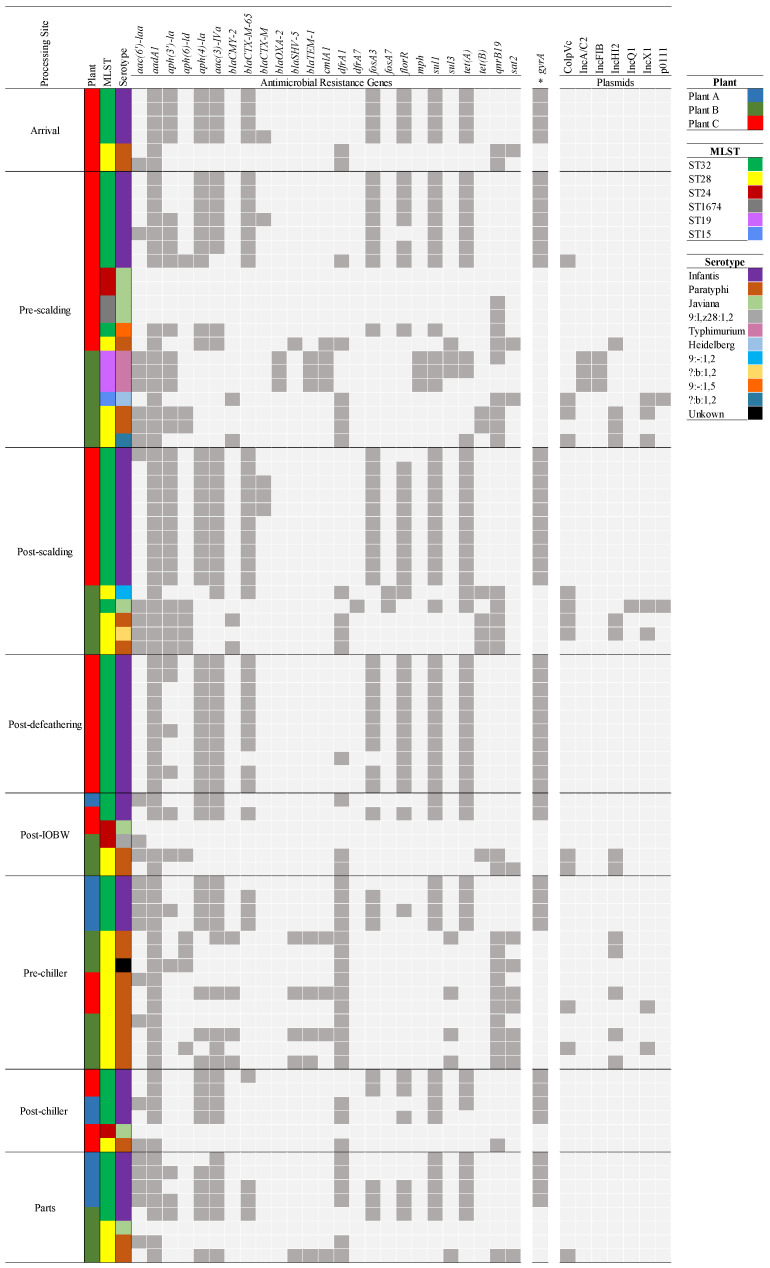
Figure 4.
Distribution of antimicrobial resistance (AMR) genes and plasmids of Salmonella spp. strains recovered from the poultry processing plants at the differing sites. Colors indicate processing plant, serotype, and MLST. The asterisk represents a chromosomal point mutation within the target genes. Dark grey and light grey show the presence and ab-sence of genes and plasmids in the bacterial genome.
Plant A: Two clusters attributed to the serotype Infantis were identified among the Salmonella spp. isolates. Although all presented the same sequence type ST32, they differed in the AMR genotypic profile. Strains from the Infantis cluster 1 (4/11) harbor multiple AMR genes aac(6′)-laa, aadA1, aph(4)-la, aac(3)-IVa, dfrA1, fosA, sul1, and tet(A). The Infantis cluster 2 (7/11) presented the same eight AMR genes profile as cluster 1, but four (4/6) of the clonal population harbored additional genes, blaCTX-M-65, fosA, and aac(3)-laa, and an extra florR gene in two (2/4). All strains presented aminoglycosides and aminoglycoside transferase genes (aac(6′)-laa, aadA1, aph(4)-la, aac(3)-IVa) which confer resistance to streptomycin and spectinomycin. Moreover, dfrA1 gene conferred resistance to trimethoprim (integron-encoded dihydrofolate reductase), fosA3 is identified as a Fosfomycin resistance gene, sul1 confers sulfonamide resistance, and tet(A) is associated with the resistance to tetracycline. The blaCTX-M-65 gene is correlated with resistance to beta-lactam antibiotics, and florR gene is typically associated with intrinsic resistance to phenicol antibiotic class. A chromosomal point mutation was present in all isolates with gyrA gene, which is the primary cause of quinolone resistance (nalidixic acid and ciprofloxacin). No plasmids were found on the bacterial genome of the isolates from this processing site.
Plant B: Salmonella isolates were classified between clusters 2–6 (Figure 3), except the serotype Heidelberg, of which just one isolate was identified from a pre-scalding rinsate sample. Eleven Paratyphi B strains were categorized in cluster 6, and the most frequent AMR genes among all were qnrB19, aadA1, and dfrA1, which confer resistance to quinolone, aminoglycoside, and trimethoprim. Five strains presented the blaCMY-2 gene associated with the beta-lactam resistance; moreover, two ?:b:1,2 (para-typhic B variant, lacking the O-4 antigen) strains isolated from pre- and post-scalding sites were in cluster 6, and harbor five AMR genes, with an additional blaCMY-2 gene in the strain isolated from the post-scalding tank. Three Paratyphi B strains were assigned to cluster 5, two of the strains were isolated from the pre-chilling tank and carry four AMR genes (aadA1, aph(6)-laa, dfrA1, and qnrB19), and one strain isolated from the post-IOBW presented eight genes more (aac(3)-IV, aadA2, aph(4)-la, blaCMY-2, blaSHV-5, cmlA1, and sul3). In addition, one 9:I,z28:1,2 strain isolated from the post-IOBW was also classified in cluster 5 and presented multiple AMR genes (blaCMY-2, qnrB19, tet(B), aadA1, aph(6)-ld, aph(3″)-lb, and dfrA1). Three S. Typhimurium strains isolated from the pre-scalding site were grouped in cluster 4; they harbor between 17 to19 AMR genes, among them four aminoglycoside resistance genes, aph(3″)-la, aac(3)-lld, aadA1, and aadA2, three sulfonamide resistance genes, sul1, sul2, and sul3, two beta-lactam resistance genes blaOXA-2, and blaTEM-1-B, two tetracycline-resistant genes, tet(A) and tet(M), two trimethoprim resistance genes, dfrA29 and dfrA12, two macrolide resistance gene mph and Inu(F), a chloramphenicol resistance gene cmlA1, and a quinolone resistance gene, qnrB19. Next, two S. Javiana were in cluster 3, one of them isolated from chicken parts rinsate which did not have AMR gene in its genome; on the other hand, the second Javiana strain isolated from the post-scalding site carried ten AMR genes, including a beta-lactam resistance gene blaTEM-1 and qnrB19 previously described. Finally, one S. Infantis and one 9:-:1,2 (monophasic variant, lacking the expression of flagellar phase 1) strains were identified in cluster 2, both presenting ten AMR genes in common that conferred resistance to aminoglycosides, beta-lactams, trimethoprim, phenicol, sulfonamides, Fosfomycin, and tetracycline; but the strain 9:-:1,2, harbors the qnrB19 gene, and extra aph(3″)-lb, aph(6)-lb, aac(6′)-laa, and tet(B) genes.
Plasmid incompatibility types were identified in 20 out of the 26 Salmonella spp. strains; the most common plasmids among the S. Parathypi B and the variant ?:b:1,2 were IncHI2 (13/20) and ColpVC (10/20); S. Heidelberg carry the same plasmids with the addition of IncX1. All S. Typhimurium carry the IncFIB and IncA/C2 plasmids. The S. Javiana strain isolated from the post-scalding tank has IncQ1, IncX1, ColpVC, and p0111 plasmids.
Plant C: Twenty-seven S. Infantis were classified in cluster 1, strains were isolated from different processing sites (arrival, pre- and post-scalding, post-defeathering, and post-chiller), the common resistance genes in all strains were aadA1, aph(4)-la, aac(3)-IV, sul1, and tet(A). Furthermore, 26 out of 27 strains harbored the fosA3 gene, and 25 of them carry the blaCTX-M-65 and florR genes. Besides, five S. Infantis were categorized in cluster 2; they were characterized to have the aadA1, aph(4)-la, aac(3)-IVa, blaCTX-M-65, fosA3, florR1, sul1, and tet(A) genes, and four had an extra aph(3’)-la gene. A chromosomal point mutation (gyrA gene) was presented in all Salmonella Infantis strains. Furthermore, seven S. Javiana strains were classified in cluster 3, and five of these were isolated from the pre-scalding, one from post-IOBW, and one from the post-chiller tank. Only two carry AMR genes associated with quinolone resistance (qnrB19). Five S. Paratyphi B strains were categorized in cluster 5, all carrying the dfrA1, aadA1, and qnrB19 genes, and two strains isolated from the pre-scalding site harbor seven genes more, aac(3)-IV, aadA2, aph(4)-la, blaSHV-5, blaTEM-1, cmlA1, and sul3. One S. Paratyphi B strain isolated from the arrival site was grouped in cluster 6, and had aac(6′)-laa, aadA1, qnrB19, and dfrA1 genes.
ColpVC plasmid was found in one out of the 34 S. Infantis strains isolated from the post-scalding tank. S. Paratyphi B strains (2/8) carry the plasmids ColpVC and IncX1 and one (1/8) also carries IncH12 and Incl1 plasmids.
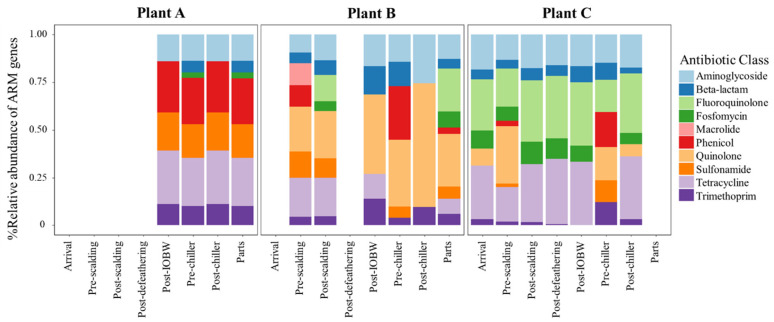
3.4. The Relative Abundance of AMR Genes within Processing Sites
Figure 5 shows the relative abundance percentage of the most-abundant AMR genes grouped by the antibiotic class they provide resistance to.
Figure 5.
The relative abundance percentage of antimicrobial resistance genes classed into antibiotic classes among the processing sites.
Plant A: AMR genes were grouped in the following antimicrobial classes: aminoglycoside, beta-lactam, Fosfomycin, phenicol, sulfonamide, tetracycline, and trimethoprim. There were no statistically significant differences (p > 0.05) in the abundance of AMR genes classified in the antimicrobial families within the processing sites (post-IOBW, pre-chiller, post-chiller, and parts) where the strains were isolated.
Plant B: The abundance of AMR genes changed throughout the stages of processing. There was a statistically significant difference in the abundance of aminoglycoside resistance genes in the processing sites post-scalding (p = 0.0247), pre-chiller (p = 0.0217), and pre-scalding (p = 0.0126) compared with chicken parts. Moreover, there were differences in the relative abundance of beta-lactam AMR genes from the pre-chiller and parts (p = 0.0134), pre-chiller, and post-chiller (p = 0.0076) sites. The abundance of macrolide resistance genes was characterized only in five strains isolated from the pre-scalding site; hence, there were statistical differences with the following sites. Similarly, variation in the abundance of sulfonamide resistant genes was identified from pre-scalding compared with pre-chiller (p = 0.0073), post-chiller (p = 0.0036), and chicken parts (p = 0.002). The abundance of tetracycline resistance genes presented significant differences from the pre-scalding site contrasted with the abundance in chicken parts (p = 0.0015), pre-chiller (p = 0.0007), and post-chiller (p = 0.0063). There was no statistically significant difference in the abundance of trimethoprim resistance genes within the processing sites.
Plant C: The isolates recovered from this processing plant presented the highest variability and abundance of AMR genes. There were statistically significant differences in the abundance of aminoglycoside resistance genes in the post-scalding site compare with the arrival (p = 0.0011), post-chiller (p = 0.0001), and post-IOBW (p = 0.0063); the pre-chiller site also had the lower abundance of resistance genes. There were considerable differences within post-defeathering (p = 0.009), post-scalding (p = 0.0005), and pre-scalding (p = 0.0007).
Genes associated with Fosfomycin resistance presented significant differences in the pre-chiller and post-chiller sites compared with post-defeathering (p = 0.0232 and 0.0232) and post-scalding (p = 0.0098 and 0.0098). There was also a difference between the abundance during pre-scalding and post-scalding (p = 0.0168). The abundance of fluoroquinolone was significantly lower in arrival (p = 0.0061), pre-chiller (p = 0.0052), post-chiller (p = 0.0026), and post-IOBW (p = 0.00148) compared with post-scalding site; besides, there was statistical significance in post-defeathering and post-chiller (p = 0.0224). Phenicol resistance genes were present only in the pre-scalding and pre-chiller sites, and there was a significant difference (p = 0.0023). There were considerable differences between the abundance of tetracycline resistance genes from post-scalding site compared with arrival (p = 0.0057), pre-scalding, (p = 0.0007), post-IOBW, (p = 0.0034), pre-chiller (p = 0.0040), and post-chiller (p = 0.0304). In addition, the relative abundance in post-defeathering differed from pre-scalding (p = 0.0102), pre-chiller (p = 0.0024), and post-chiller (p = 0.0204). Trimethoprim resistance genes were present in all processing sites except in pre-chiller, and there were no statistically significant differences among the other sites. Genes associated with quinolone resistance were statistically different in pre-chiller contrasted with post-scalding (p = 0.0043) and post-defeathering (p = 0.0043). There was no statistically significant difference in the abundance of beta-lactam and sulfonamide resistance genes within the processing sites.
4. Discussion
In this study, the prevalence of the primary poultry-associated pathogen was assessed during processing. Salmonella spp. are commonly detected at high rates throughout processing, including after the chilling operation with a prevalence of 100%, 47%, and 87%, respectively, for plants A, B, and C. It could be inferred that the application of 10 ppm of HClO is not enough to reduce the microbial load in the chicken carcasses. Besides, for chloride to be efficient, the pH of the water must remain in a range of 5.8 to 6.8 [11], and increasing of water pH and organic load will decrease its efficacy [12,13]. The poultry processing plants sampled were characterized by measuring the pH and chloride concentration in the first shift of the processing day, moreover, having a counterflow immersion chiller that moves toward the chicken carcasses to the cleanest water. However, during the day, organic load accumulation is likely to occur in the chiller tank, causing either direct contamination or cross-contamination with Salmonella spp. [14].
On the other hand, the levels of Enterobacteriaceae were quantified throughout the processing. Overall, there were significant reductions in the counts of Enterobacteriaceae from the pre-scalding to the post-chilling sites. Nevertheless, in plant A, samples from chicken parts exhibited higher Enterobacteriaceae levels than in previous stages (post-chiller). In the process of cutting the chicken carcass into chicken parts, which usually requires the intervention of experienced workers, handling and manipulation increased the chances of cross-contamination. Bacterial contamination can occur from equipment surfaces, water, and animal microbiota; microorganisms from the environment and air can contaminate chicken parts [15,16].
In Colombia, few studies have investigated the prevalence of foodborne pathogens throughout processing. Most researchers have focused on retail market scenarios [17,18,19]. Nonetheless, Ramirez-Hernandez et al., [20] reported the prevalence of Salmonella in three poultry processing establishments, with a maximum overall rate of 21.2% (51/240 samples) in an establishment located in the Department of Valle; additionally, the numbers of presumptive Salmonella-positive samples after chilling were lower (<18%) compared to the results obtained in this study. Differences in food safety programs and manufacturing practices implemented in the processing plants, and other variables such as geolocation and seasonality, could explain the different levels of indicator microorganism and Salmonella rates in the chicken carcasses.
Decree 1500 established Salmonella spp. as a microorganism when verifying performance standards in processing facilities. Generic E. coli is also considered an indicator microorganism to check the control process and cleaning and sanitation procedures. These measures are conducted only in the post chilling operation, and there is no monitoring before and after this site. The cut-up and deboning processing room have been shown to include the most critical steps for cross-contamination of chicken meat. Because chicken parts, comminuted chicken, and seasoned chicken parts have become an emerging product category in Colombia, it is crucial to assess the levels and prevalence of indicator microorganisms and pathogens to design appropriate strategies to ensure product safety before packing.
AMR has become a global public health concern for both human and animal health [21]. Improper use of antibiotics in animal production and human medicine has been shown to be the leading cause of the emergence of AMR bacteria [22,23,24]. Antimicrobial resistance of poultry-associated pathogens in Colombia is a new field of study that has gained more attention from associated government institutions and research groups in universities. Donado-Godoy et al., [25] are among the pioneers in the Colombian Integrated Surveillance Program for Antimicrobial Resistance (COIPARS) pilot project established in 2013. They reported that 23% (139/600) of the carcass rinsates collected at the post-chilling sites in different processing plants in Colombia were positive for Salmonella. Among these isolates, high levels of resistance were presented to ampicillin (64%, 84/132), cefotaxime (57%, 75/132), ceftiofur (58%, 72/125) and ciprofloxacin (85%, 111/131). In the same way, Campylobacter spp. was detected in 36% (215/600) of the samples. Isolates exhibited resistance to ciprofloxacin (92%, 70/76) and tetracycline (93%, 71/76).
Results obtained in this work on the genotypic characterization of Salmonella spp. indicate high rates of AMR profiles among isolates. It is considered that the presence of AMR genes can represent the phenotypical resistance of antimicrobial agents. However, there are several mechanisms of antimicrobial resistance in bacteria, and there is not always an association with a specific gene. Moreover, such resistance mechanisms can naturally occur or acquire transferable genetic elements (i.e., plasmids or resistance gene encoding integrons). Cross-resistance to antimicrobials can also occur with resistance to different group members of chemical-related components, and/or with the same or similar mechanism of action [26,27]. The presence of qnrB19 gene in the Salmonella genomes (36%, 31/85) of the isolates collected in this study is associated with quinolone resistance and could be attributed to the extensive use of chlohydroxy-quinoline (quinolone class) in the feed to promote the healthy growth of the chickens.
More extensively, research work has been conducted on antimicrobial resistant bacteria from retailed chicken meat in Colombia. Donado-Godoy et al., [18] reported high levels of resistance to tilmicosin (100%, 51/51) and nalidixic acid (66%, 34/51) among Salmonella spp. isolates recovered from retailed chicken carcasses in Bogota. Another study conducted by the same group [28] reported the AMR profiles of 378 Salmonella spp. isolates from chicken carcasses available in wet markets, supermarkets, and independent markets, collected in six different Departments. Overall, 94% (354/378) of the isolates were resistant to at least one antimicrobial. High levels of resistance were identified for nalidixic acid (70%) tetracycline (57%), streptomycin (67%), and trimethoprim-sulfamethoxazole (54%). Furthermore, 59% presented a multi-drug resistant (MDR) phenotype with resistance to 6 to 15 antimicrobial agents. Additionally, a recent study identified Salmonella Heidelberg (3%, 15/540) strains recovered from a poultry processing plant in Santander. Phenotypical characterization indicated Salmonella MDR profiles, including resistance to the antibiotic classes of quinolones, fluoroquinolones, cephalosporins, beta-lactams, aminoglycosides, and tetracyclines [29].
The serotype Infantis was the most commonly isolated among the poultry processing plants. S. Infantis has emerged as one of the most common serovars causing human salmonellosis in Europe [30] and the United States [31]. An outbreak of MDR S. Infantis strain in the United States linked to raw chicken products infected 129 people in 32 states; 21 were hospitalized, and one death has been reported [32]. The outbreak strain was identified in samples from raw chicken products from 76 slaughter and processing establishments, and from live chickens [32]. The increasing prevalence of S. Infantis has been characterized by MDR profiles and the harboring of MDR genes, such as the extended-spectrum β-lactamases (ESBLs) [33]. Of the ESBL enzymes, the CTX-family is the most widely reported. Before 2014, the blaCTX-M-65 gene had only been described in E. coli isolates from a patient in the United States. In Colombia, the INS reported results of extended-spectrum cephalosporin-resistance markers from Salmonella spp. clinical isolates recovered from 1997–2018. The most frequent ESBL genotype identified was the CTX-M (79%; 164/208), especially from the serotypes Typhimurium (40%; 65/164) and Infantis (29%; 48/164) (18). The closely related MDR of Infantis was disseminated among the broiler population and associated with animal production environments, eventually spreading into the food chain and potentially into humans [30,34].
The WGS analysis using ResFinder confirmed the presence of the blaCTX-M-65 in 31 (31/40; 77%) S. Infantis isolated from the poultry processing plants in Colombia. All the blaCTX-M-65- positive isolates carried other resistance genes to aminoglycoside, Fosfomycin, phenicol, sulfonamide, tetracyclines, and trimethoprim. Our results of the genotypic characterization of S. Infantis correlate with the profiles of strains isolated from chicken broilers and human cases in Switzerland [30], Great Britain [35], Italy [36], United States [33], and Ecuador [37]. Additionally, Castellanos et al. [38] reported blaCMY-2 and blaCTX-M-65 genes which encode resistance associated with AmpC β-lactamases and ESBLs in both S. Paratyphi B (ST28) and S. Heidelberg (ST15) from poultry-associated isolates in Colombia. The blaCMY-2 gene was present in nine MDR S. Paratyphi B, and S. Heidelberg isolates from plant B and C. Similarly, Castro-Vargas et al., [29] showed the presence of blaCMY-2 gene and blaCTX-M in S. Heidelberg isolates recovered from broilers’ cloacal swabs in poultry farms in Colombia.
Furthermore, S. Paratyphi B was the second most frequently isolated serotype among all Salmonella isolates, and were distributed throughout the different processing sites. Previously published work reported that S. Paratyphi B dT+ and S. Heidelberg were the most prevalent serotypes isolated from farms and retail meat samples in Colombia during 2008 and 2009 [28]. There were also three S. Typhimurium ST19 strains isolated from plant B with a high abundance of AMR genes. This strain ST19 has been recently reported in a study from Salmonella spp. clinical isolates (87%; 182/209) in Colombia from 1999 to 2017. These strains were resistant to nalidixic acid and ciprofloxacin, and they are associated with the presence of qnrB19 genes [39], also identified in our strains. The high rate of AMR genes in the Salmonella spp. isolates was associated with multiclass resistance antibiotics. The AMR gene abundance of ten antimicrobial classes were distinct when comparing processing plants, and within processing sites. Understanding the AMR gene profiles of foodborne pathogens could help to predict the risk associated with poultry products’ consumption, target intervention, and limit the dissemination of AMR genes through the food chain.
The national poultry sector in Colombia faces a variety of challenges motivated by the emergence of fast and dynamic markets that demand better quality, safety, and diversified products. The abundance of AMR Salmonella spp. strains circulating in chicken meat represents a public health concern for potential foodborne outbreaks in the country. Hence, data collected at different stages throughout the chicken processing value-chain can help support the implementation of science-based risk management options focused on proven mitigation stages for pathogen control, while ensuring microbial safety in chicken meat products.
5. Conclusions
The results of our study identified that Enterobacteriaceae levels decreased throughout the processing sites; however, counts ranging from 3 to 5 Log/CFU/mL rinsate were found after chilling. There was a high prevalence of Salmonella spp. in the processing sites and a high rate of AMR genotype among Salmonella spp. isolates recovered from the three plants. Serotypes Infantis and Paratyphi B were the most common isolates from the chicken rinsates, and they exhibited different genotypic characteristics. The diversity of AMR genes on the Salmonella genomes from the processing sites suggests that changes in their abundance could be attributed to the site conditions, resulting in variations in the natural resistance genes or the acquisition of new AMR genes by mutation or plasmids transfer. Nevertheless, further studies are necessary for identifying emerging or reemerging Salmonella serotypes and the routes of contamination in chickens to improve prevention and control methods in poultry processing operations.
Acknowledgments
The authors are very thankful to Francisco Garay, a professor from the Veterinary and Medicine Department at CES University in Medellin, for his collaboration and support in the plants’ sampling. We also thank 3MTM Food Safety for the kind donation of the laboratory supplies required to conduct this study.
Author Contributions
Conceptualization, A.R.-H., A.K.C.-C. and M.X.S.-P.; Methodology, A.R.-H., A.V.-G. and A.K.C.-C.; Validation, A.R.-H. and M.X.S.-P.; Formal analysis, A.R.-H.; Investigation, A.R.-H., A.V.-G. and A.K.C.-C.; Resources, M.M.B.; Data curation, A.R.-H., A.V.-G., A.K.C.-C. and M.X.S.-P.; Writing—original draft preparation, A.R.-H.; Writing—review and editing, A.R.-H. and M.X.S.-P.; Supervision, M.X.S.-P.; funding acquisition, M.X.S.-P. and M.M.B. All authors have read and agreed to the published version of the manuscript.
Funding
We would also like to express our gratitude to the International Cultural Center at Texas Tech University for the awarded International Research and Development Seed Grant that supports this research in Colombia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the image of the poultry processing plants.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Procolombia Colombian Poultry Sector: Features of the Colombia Poultry Sectore. [(accessed on 15 October 2018)];2016 Available online: https://compradores.procolombia.co/en/explore-business-opportunities/colombian-poultry-sector.
- 2.Instituto Colombiano Agropecuario (ICA)/Colombian Agricultural Institute Censo Pecuario Nacional año 2019/National Livestock Census Year. [(accessed on 26 September 2018)];2019 Available online: https://www.ica.gov.co/areas/pecuaria/servicios/epidemiologia-veterinaria/censos-2016/censo-2018.
- 3.Díaz M.A. Determinantes del Desarrollo en la Avicultura en Colombia: Instituciones, Organizaciones y Tecnologías. Documento de Trabajo Sobre Economía Regional/Determinants Poultry Farming in Colombia Development: Institutions, Organizations and Technologies. White Paper on Regional Economy. [(accessed on 25 September 2018)];2014 Available online: http://www.banrep.gov.co/sites/default/files/publicaciones/archivos/dtser_214.pdf.
- 4.Instituto Nacional de Vigilancia de Medicamentos y Alimentos (INVIMA)/National Institute of Food and Drug Surveillance Plantas de Beneficio, Desposte, Desprese y Acondicionadores que Cumplen con los Requisitos Sanitarios Exigidos en el Decreto 1500 de 2007, decreto 2270 de 2012 y Resoluciones Reglamentarias/Processing Plants, Slaughtering Process, Cutting and Deboning to Comply with the Sanitary Requirements of Decree 1500 of 2007, Decree 2270 of 2012 and Regulatory Resolutions. [(accessed on 14 March 2020)];2019 Updated 20 August 2019. Available online: https://www.invima.gov.co/documents/20143/426809/PLANTAS-DE-BENEFICIO-DESPOSTE-Y-DESPRESE-AUTORIZADAS-POR-EL-INVIMA-PARA-SU-FUNCIONAMIENTO-A-JULIO-DE-2019-as-asp.pdf.
- 5.Instituto Nacional de Salud (INS)/National Health Institute Boletín Epidemiológico Semanal, las Enfermedades Transmitidas por Alimentos-ETA. Semana Epidemiológica 52/Weekly Epidemiological Bulletin, Foodborne Diseases. Epidemiological Week 52. [(accessed on 14 March 2020)];2018 Available online: https://www.ins.gov.co/buscador-eventos/BoletinEpidemiologico/2018%20Bolet%C3%ADn%20epidemiol%C3%B3gico%20semana%2052.pdf.
- 6.Instituto Nacional de Salud (INS)/National Health Institute Informe de Vigilancia por Laboratorio de Salmonella spp.: “Colombia 1997–2017”/Laboratory Surveillance for Salmonella spp.: “Colombia 1997–2017”. [(accessed on 18 November 2019)];2019 Available online: https://www.ins.gov.co/buscador-eventos/Informacin%20de%20laboratorio/Informe%20de%20vigilancia%20por%20laboratorio%20Salmonella%20spp%20Colombia%201997%202018.pdf.
- 7.Ministerio de Salud y Protección Social, Instituto Nacional de Salud, Unidad de Evaluación de Riesgos Para la Inocuidad de Los alimentos/Ministry of Health and Social Protection, National Health Institute, Risk Assessment Unit for Food Safety Perfil de riesgo Salmonella spp. (No Tifoideas) en Pollo Entero y en Piezas/Risk profile of Salmonella spp. in Broilers and Chicken Parts. [(accessed on 15 October 2018)];2011 Available online: https://www.minsalud.gov.co/sites/rid/Lists/BibliotecaDigital/RIDE/IA/INS/perfil-salmonella-spp.pdf.
- 8.Ministerio de Salud y Protección Social, Instituto Nacional de Salud/Ministry of Health and Social Protection, National Health Institute Perfil del Riesgo Campylobacter spp. en Pollos de Engorde/Risk Profile of Campylobacter spp. In Broilers. [(accessed on 9 October 2019)];2013 Available online: https://www.minsalud.gov.co/sites/rid/Lists/BibliotecaDigital/RIDE/IA/INS/Perfil-campylobacter-en-pollos.pdf.
- 9.Ramirez-Hernandez A., Bugarel M., Kumar S., Thippareddi H., Brashears M.M., Sanchez-Plata M.X. Phenotypic and genotypic characterization of antimicrobial resistance in Salmonella strains isolated from chicken carcasses and parts collected at different stages during processing. J. Food Protect. 2019;82:1793–1801. doi: 10.4315/0362-028X.JFP-19-056. [DOI] [PubMed] [Google Scholar]
- 10.Ramirez-Hernandez A. Ph.D. Thesis. Texas Tech University; Lubbock, TX, USA: 2019. Assessing the Effect of Interventions on Pathogens and the Microbial Ecology of the Poultry Processing Chain by Microbial Profiling and the Phenotypic and Genotypic Characterization of Antimicrobial Resistance. [Google Scholar]
- 11.Keener K.M., Bashor M.P., Curtis P.A., Sheldon B.W., Kathariou S. Campylobacter and Poultry Processing. Compr. Rev. Food Sci. Safe. 2004;3:105–116. doi: 10.1111/j.1541-4337.2004.tb00060.x. [DOI] [PubMed] [Google Scholar]
- 12.Byrd J.A., Rand McKee S.R. Improving Slaughter and Processing Technologies. In: Mead G.C., editor. Food Safety Control in the Poultry Industry. CRC Press L.L.C.; Boca Raton, FL, USA: 2005. pp. 310–327. [Google Scholar]
- 13.Nagel G.M., Bauermeister L.J., Bratcher C.L., Singh M., McKee S.R. Salmonella and Campylobacter reduction and quality characteristics of poultry carcasses treated with various antimicrobials in a post-chill immersion tank. Int. J. Food Microbiol. 2013;165:281–286. doi: 10.1016/j.ijfoodmicro.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Smith D.P., Northcutt J.K., Cason J.A., Hinton A., Jr., Buhr R.I., Ingram K.D. Effect of external or internal fecal contamination on numbers of bacteria on prechilled broiler carcasses. Poultry Sci. 2003;86:1241–1244. doi: 10.1093/ps/86.6.1241. [DOI] [PubMed] [Google Scholar]
- 15.Rouger A., Tresse O., Zagorec M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms. 2017;5:50. doi: 10.3390/microorganisms5030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vihavainen E.J., Björkroth J. Handbook of Poultry Science and Technology, Secondary Processing. John Wiley & Sons, Inc; New Jersey, NY, USA: 2010. Microbial Ecology and Spoilage of Poultry Meat and Poultry Meat Products; pp. 485–493. [Google Scholar]
- 17.Donado-Godoy P., Clavijo V., León M., Tafur M.A., Gonzales S., Hume M., Alali W., Walls I., Lo Fo Wong D.M.A., Doyle M.P. Prevalence of Salmonella on retail broiler chicken meat carcasses in Colombia. J. Food Protect. 2012;75:1134–1138. doi: 10.4315/0362-028X.JFP-11-513. [DOI] [PubMed] [Google Scholar]
- 18.Donado-Godoy P., Byrne B.A., Leon M., Castellanos R., Vanegas C., Coral A., Arévalo A., Clavijo V., Vargas M., Romero Zuñiga J.J., et al. Prevalence, resistance patterns, and risk factors for antimicrobial resistance in bacteria from retail chicken meat in Colombia. J. Food Protect. 2015;78:751–759. doi: 10.4315/0362-028X.JFP-14-349. [DOI] [PubMed] [Google Scholar]
- 19.Karczmarczyk M., Martins M., McCusker M., Mattar S., Amaral L., Leonard N., Aarestrup F.N., Fanning S. Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: Identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett. 2010;313:10–19. doi: 10.1111/j.1574-6968.2010.02119.x. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez-Hernandez A., Varón-García A., Sanchez-Plata M.X. Microbiological profile of three commercial poultry processing plants in Colombia. J. Food Protect. 2017;80:1980–1986. doi: 10.4315/0362-028X.JFP-17-028. [DOI] [PubMed] [Google Scholar]
- 21.Ciorba V., Odone A., Veronesi L., Pasquarella C., Signorelli P. Antibiotic resistance as a major public health concern: Epidemiology and economic impact. Ann. Igiene Med. Prev. Comunita. 2015;27:562–579. doi: 10.7416/ai.2015.2048. [DOI] [PubMed] [Google Scholar]
- 22.Dutil L., Irwin R., Finley R., Ng L.K., Avery B., Boerlin P., Bourgault A., Cole L., Daignault D., Desruisseau A., et al. Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans Canada. Emerg. Infect. Dis. 2010;16:48–54. doi: 10.3201/eid1601.090729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laxminarayan R., Matsoso P., Pant S., Brower C., Røttingen J.A., Klugman K., Davies S. Access to effective antimicrobials: A worldwide challenge. Lancet. 2016;387:168–175. doi: 10.1016/S0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- 24.McEwen S.A., Fedorka-Cray P.J. Antimicrobial use and resistance in animals and human beings. Clin. Infect. Dis. 2002;34:S93–S106. doi: 10.1086/340246. [DOI] [PubMed] [Google Scholar]
- 25.Donado-Godoy P., Castellanos R., León M., Arévalo A., Clavijo V., Bernal J., León D., Tafur M.A., Byrne B.A., Smith W.A., et al. The establishment of the Colombian integrated program for antimicrobial resistance surveillance (C.O.I.P.A.R.S.): A pilot project on poultry farms, slaughterhouses and retail market. Zoonoses Public Health. 2015;62:58–69. doi: 10.1111/zph.12192. [DOI] [PubMed] [Google Scholar]
- 26.Sefton A.M. Mechanisms of antimicrobial resistance: Their clinical relevance in the new millennium. Drugs. 2002;62:557–566. doi: 10.2165/00003495-200262040-00001. [DOI] [PubMed] [Google Scholar]
- 27.Sosa A.D.J., Byarugaba D.K., Amabile-Cuevas C.F., Hsueh P.R., Kariuki S., Okeke I.N. Antimicrobial Resistance Mechanism in Developing Countries. Springer; New York, NY, USA: 2010. [Google Scholar]
- 28.Li Y., Pulford C.V., Díaz P., Perez-Sepulveda B.M., Duarte C., Predeus A.V., Wiesner M., Heavens D., Low R., Shudoma C., et al. Genomic and phylogenetic analysis of Salmonella Typhimurium and its monophasic variants responsible for invasive endemic infections in Colombia. BioRxiv. 2019 doi: 10.1101/588608. [DOI] [Google Scholar]
- 29.Castro-Vargas R., Fandiño de Rubio L.C., Vega A., Rondón-Barragán I. Phenotypic and genotypic resistance of Salmonella Heidelberg isolated from one of the largest poultry production region from Colombia. Int. J. Poult. Sci. 2019;18:610–617. doi: 10.3923/ijps.2019.610.617. [DOI] [Google Scholar]
- 30.Hindermann D., Gopinath G., Chase H., Negrete F., Althaus D., Zurfluh K., Tall B.D., Stephan R., Nüesch-Inderbinen M. Salmonella enterica serovar Infantis from Food and Human Infections, Switzerland, 2010–2015: Poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front. Microbiol. 2017;8:1322. doi: 10.3389/fmicb.2017.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marder M.E., Griffin P., Cieslak P., Dunn J., Hurd S., Jervis R., Lathrop S., Muse A., Ryan P., Smith K., et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne diseases active surveillance network, 10 U.S. sites, 2006-2017. MMWR-Morbid. Mortal Wkly. Rep. 2018;23:324–328. doi: 10.15585/mmwr.mm6711a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Centers for Disease Control and Prevention (CDC) Outbreak of Multidrug-Resistant Salmonella Infections liNked to Raw Chicken Products. [(accessed on 15 March 2020)];2019 Available online: https://www.cdc.gov/salmonella/infantis-10-18/index.html.
- 33.Tate H., Folster J.P., Hsu C.H., Chen J., Hoffmann M., Li C., Morales C., Tyson G.H., Mukherjee S., Brown A.C., et al. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nógrády N., Király M., Davies B., Nagy B. Multidrug resistant clones of Salmonella Infantis of broiler origin in Europe. Int. J. Food Microbiol. 2012;157:108–112. doi: 10.1016/j.ijfoodmicro.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Burke L., Hopkins K.L., Meunier D., De Pinna E., Fitzgerald-Hughes D., Humphreys H., Woodford N. Resistance to third-generation cephalosporins in human non-typhoidal Salmonella enterica isolates from England and Wales, 2010–2012. J. Antimicrob. Chemother. 2014;69:977–981. doi: 10.1093/jac/dkt469. [DOI] [PubMed] [Google Scholar]
- 36.Franco A., Leekitcharoenphon P., Feltrin F., Alba P., Cordaro G., Iurescia M., Tolli R., D’Incau M., Staffolani M., Di Giannatale E., et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS ONE. 2015;10:e0144802. doi: 10.1371/journal.pone.0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cartelle-Gestal M., Zurita J., Paz-Mino A., Ortega-Paredes D., Alcocer I. Characterization of a small outbreak of Salmonella enterica serovar Infantis that harbor CTX-M-65 in Ecuador. Braz. J. Infect. Dis. 2005;2:406–407. doi: 10.1016/j.bjid.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellanos L.R., van der Graaf-van Bloois L., Donado-Godoy P., León M., Clavijo V., Arévalo A., Bernal J.F., Mevius D.J., Wagenaar J.A., Zomer A., et al. Genomic characterization of extended-spectrum cephalosporin-resistant Salmonella enterica in the Colombian poultry chain. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.02431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donado-Godoy P., Clavijo V., Leon M., Arevalo A., Castellanos R., Bernal J., Tafur M.A., Ovalle M.V., Alali M.Q., Hume M., et al. Counts, serovars, and antimicrobial resistance phenotypes of Salmonella on raw chicken meat at retail in Colombia. J. Food Protect. 2014;77:227–235. doi: 10.4315/0362-028X.JFP-13-276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to their containing information that could compromise the image of the poultry processing plants.