Abstract
Novel twenty-three 3(2H)-pyridazinone derivatives were designed and synthesized based on the chemical requirements related to the anti-proliferative effects previously demonstrated within this scaffold. The introduction of a piperazinyl linker between the pyridazinone nucleus and the additional (un)substituted phenyl group led to some compounds endowed with a limited cytotoxicity against human gingival fibroblasts (HGFs) and good anti-proliferative effects against gastric adenocarcinoma cells (AGS) as evaluated by MTT and LDH assays, using doxorubicin as a positive control. Successive analyses revealed that the two most promising representative compounds (12 and 22) could exert their effects by inducing oxidative stress as demonstrated by the hydrogen peroxide release and the morphological changes (cell blebbing) revealed by light microscopy analysis after the haematoxylin-eosin staining. Moreover, to further assess the apoptotic process induced by compounds 12 and 22, Bax expression was measured by flow cytometry. These findings enlarged our knowledge of the structural requirements in this scaffold to display valuable biological effects against cancerous cell lines.
Keywords: 3(2H)-pyridazinone, hydrazone, fibroblasts, gastric cancer, piperazine, Bax, H2O2 release
1. Introduction
Gastric cancer (GC) is the third cause of cancer death in the world, despite a declining incidence due to improvements in screening methods and Helicobacter pylori eradication. Its symptomatology (early satiety, nausea, vomiting, asthenia, anaemia and weight loss) can be appreciated only when it has grown in size interfering with the nutritional process [1]. Complete surgical resection of the whole tumoral mass remains the first-line protocol for GC treatment, but being often diagnosed at an advanced stage, this malignant tumor can frequently be not operable [2]. Individualised treatments, involving radiotherapy and chemotherapy, only provide modest results especially in advanced or metastatic GC and due to the existence of four molecular subtypes of GC some chemotherapeutics may be unsuccessful. Moreover, trials with molecular-targeted therapy have provided contrasting results suggesting an urgent need to propose the development of different therapeutic protocols or new drugs for prolonging the survival of the fragile patients with GC [3]. Among the classical anti-GC drugs, 5-fluorouracil and doxorubicin display a potent anti-proliferative activity, while exerting important side effects and inducing the development of drug-resistant cancer cells [4].
With the aim of proposing an alternative therapeutic arsenal, our research group explored innovative chemical scaffolds [5,6], confirming how the introduction of nitrogen-containing heterocycles could be the driving force to achieve promising preliminary results [7]. Among the large plethora of nitrogenous heterocycles, pyridazinone compounds are of great interest to researchers because of their various biological effects [8,9]. These activities include anti-inflammatory, analgesic, anti-viral, anti-parasitic, anti-fungal, anti-bacterial anti-diabetic, anti-tubercular, anti-depressant, anti-hypertensive, anti-thrombotic, anti-feedant, and diuretic effects [10,11,12,13,14,15,16,17,18,19,20]. In addition, pyridazinone compounds have been also reported to have anticancer activity [21,22], and an interesting group of 3(2H)-pyridazinone derivatives have been synthesized and evaluated for their anti-proliferative effects against human colon carcinoma HCT116 cells [23]. These results suggest that pyridazinone compounds may be useful in cancer chemotherapy and that, according to the proposed SARs [23], pyridazinone derivatives bearing different substituents may exhibit varying degrees of cytotoxic effect.
In the present study, 23 new 3(2H)-pyridazinone-based compounds, which are thought to be effective against gastric adenocarcinoma (AGS cells) and have low toxicity, were synthesized and their biological activities were evaluated. In the design of these compounds, we kept into consideration the role exerted by the piperazine linker introduced between the pyridazinone core nucleus and the additional phenyl ring to display anti-proliferative effects against liver (HEP3B), colon (HCT116) and neuroblastoma (SHSY5Y) cell lines as previously reported [24,25].
These compounds were first submitted to a general screening for their toxicity against human gingival fibroblasts and AGS cells at established concentrations (10 and 50 µM) to select the most promising derivatives for further biological assessment of their possible antineoplastic activity evaluated by means of MTT test, lactate dehydrogenase (LDH) assay, light microscopy analysis, hydrogen peroxide (H2O2) release and Bax immunostaining by flow cytometry. All the results have been compared to those of doxorubicin as a positive control drug for gastric adenocarcinoma [26].
2. Results and Discussion
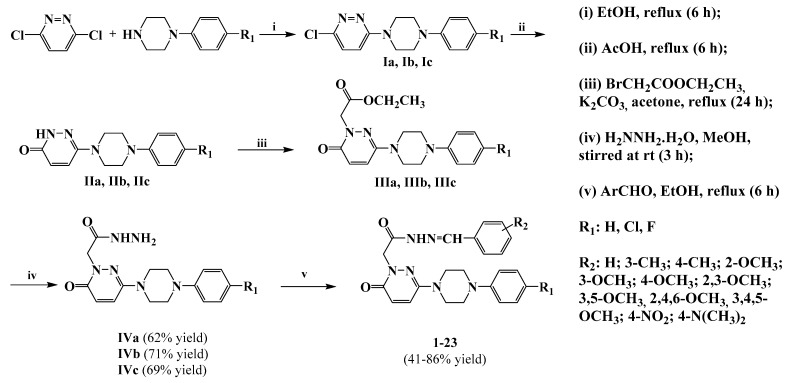
The novel compounds 1–23 were obtained by reacting different benzaldehyde derivatives with intermediates 4a–c, which were in turn prepared according to the literature [12,27,28], as shown in Scheme 1.
Scheme 1.
Synthesis of compounds 1–23.
As a result of the nucleophilic substitution reaction of 3,6-dichloropyridazine and the appropriate substituted phenylpiperazine, compounds Ia–c were obtained. Subsequently, by heating these compounds in glacial acetic acid, the pyridazine ring turned into a pyridazinone ring by hydrolysis. Compounds IIIa–c were synthesized by the reaction of ethyl bromoacetate with IIa–c in acetone in the presence of potassium carbonate. Substituted pyridazinone-2-yl acetohydrazide compounds IVa–c were obtained by the condensation reactions of hydrazine hydrate (99%) with IIIa–c. The new title compounds 1–23 were synthesized by the reaction of IVa–c with some benzaldehyde derivatives in ethanol and listed in Table 1. The reaction yields for the last step of the synthesis, after recrystallization from methanol/water, of the compounds have a wide range (41 to 86%). 1H-NMR, 13C-NMR, and mass spectra of compounds 1–23 are given as Supplementary Data.
Table 1.
Structures, yields and chemical-physical data of title compounds.

| Compd | R1 | R2 | Log P | Log S | tPSA | M.W. | Molecular Formula |
|---|---|---|---|---|---|---|---|
| 1 | H | H | 3.00 | −6.19 | 80.61 | 416.48 | C23H24N6O2 |
| 2 | H | 3002DCH3 | 3.49 | −6.54 | 80.61 | 430.50 | C24H26N6O2 |
| 3 | H | 4-CH3 | 3.49 | −6.55 | 80.61 | 430.50 | C24H26N6O2 |
| 4 | H | 4-OCH3 | 2.87 | −6.25 | 89.84 | 446.50 | C24H26N6O3 |
| 5 | H | 2,3-OCH3 | 2.75 | −6.07 | 99.07 | 476.53 | C25H28N6O4 |
| 6 | H | 3,5-OCH3 | 2.75 | −6.29 | 99.07 | 476.53 | C25H28N6O4 |
| 7 | H | 2,4,6-OCH3 | 2.62 | −6.34 | 108.30 | 506.55 | C26H30N6O5 |
| 8 | H | 3,4,5-OCH3 | 2.62 | −6.10 | 108.30 | 506.55 | C26H30N6O5 |
| 9 | H | 4-NO2 | 2.48 | −6.58 | 132.42 | 461.47 | C23H23N7O4 |
| 10 | Cl | 2-OCH3 | 3.43 | −6.95 | 89.84 | 480.95 | C24H25ClN6O3 |
| 11 | Cl | 3-OCH3 | 3.43 | −6.95 | 89.84 | 480.95 | C24H25ClN6O3 |
| 12 | Cl | 2,3-OCH3 | 3.31 | −6.77 | 99.07 | 510.97 | C25H27ClN6O4 |
| 13 | Cl | 2,4,6-OCH3 | 3.18 | −7.05 | 108.30 | 541.00 | C26H29ClN6O5 |
| 14 | Cl | 3,4,5-OCH3 | 3.18 | −6.80 | 108.30 | 541.00 | C26H29ClN6O5 |
| 15 | Cl | 4-NO2 | 3.10 | −7.29 | 132.42 | 495.92 | C23H22ClN7O4 |
| 16 | Cl | 4-N(CH3)2 | 3.84 | −7.29 | 83.85 | 493.99 | C25H28ClN7O2 |
| 17 | F | H | 3.16 | −6.46 | 80.61 | 434.47 | C23H23FN6O2 |
| 18 | F | 3-CH3 | 3.65 | −6.82 | 80.61 | 448.49 | C24H25FN6O2 |
| 19 | F | 3-OCH3 | 3.03 | −6.51 | 89.84 | 464.49 | C24H25FN6O3 |
| 20 | F | 4-OCH3 | 3.03 | −6.52 | 89.84 | 464.49 | C24H25FN6O3 |
| 21 | F | 2,4,6-OCH3 | 2.78 | −6.62 | 108.30 | 524.54 | C26H29FN6O5 |
| 22 | F | 4-NO2 | 2.62 | −6.86 | 132.42 | 479.46 | C23H22FN7O4 |
| 23 | F | 4-N(CH3)2 | 3.44 | −6.86 | 83.85 | 477.53 | C25H28FN7O2 |
Log P: partition coefficient; Log S: intrinsic solubility; tPSA: total polar surface area.
The capability of compounds 1–23 to affect cell viability was evaluated by means of the MTT test which measures the metabolic activity of viable cells. Firstly, the MTT test was carried out on primary HGFs, non-pathological cells, extracted from gingiva withdrawal of healthy donors. The effect of the compounds 1–23 was compared with the effect of doxorubicin, an antitumoral drug commonly used in the therapeutic protocols. Our control was represented by HGFs only treated with the vehicle DMSO. Compounds 1–23 were administered to HGFs up to 72 h at 10 µM evaluating the cell viability after both 48 and 72 h of treatment (Figure 1, upper and lower graphs). After 48 h of treatment only compound 2 and the doxorubicin are able to significantly reduce cell viability respect to DMSO-treated cells. Conversely, compounds 19 and 23 significantly increase this parameter with respect to DMSO-treated cells (upper graph). After 72 h of treatment only doxorubicin discloses a significant reduction in cell viability respect to control (lower graph).
Figure 1.
MTT test on HGFs treated with compounds 1–23 and with doxorubicin (DOXO) at 10 µM for 48 (upper graph) and 72 h (lower graph). *** p < 0.0002, ** p < 0.0005. DMSO: control vehicle.
Secondly, the administration of compounds 1–23 and doxorubicin for 48 and 72 h was performed on a tumoral cell line of gastric adenocarcinoma (AGS). The newly synthetized molecules and doxorubicin were tested at 50 and 10 µM. When cells are treated with the higher dose of compounds 1–23 and doxorubicin (50 µM), most of them are able to significantly reduce cell viability, both after 48 and 72 h (Figure 2, upper and lower graphs).
Figure 2.
MTT test on AGS treated with compounds 1–23 and with doxorubicin (DOXO) at 50 µM for 48 (upper graph) and 72 h (lower graph). *** p < 0.0002, * p < 0.005. DMSO: control vehicle.
When AGS are treated with the lower dose (10 µM) after 48 h of treatment only the molecules 11, 17, 18 and 22 show a statistically significant decrease of AGS metabolic activity (Figure 3, upper graph), whereas, after 72 h of treatment several compounds (8, 14–19, 22, 23 and doxorubicin) show the capability to negatively affect the cell viability respect to DMSO treated AGS (Figure 3, lower graph).
Figure 3.
MTT test on AGS treated with compounds 1–23 and with doxorubicin (DOXO) at 10 µM for 48 (upper graph) and 72 h (lower graph). *** p < 0.0002, ** p < 0.0005 * p < 0.005. DMSO: control vehicle.
These preliminary data appear very promising considering that, except for a few of them and doxorubicin, they show an appreciable capability not to interfere with the proliferation of non-tumoral cells thus protecting them from any harmful effects. At the same time, compounds 17, 18 and 22 at 50 µM concentration revealed an appreciable capability to strongly counteract gastric adenocarcinoma cell proliferation after both 48 and 72 h of administration, whereas compounds 8 and 12 were effective only at 50 µM concentration after 72 h. Collectively, these results suggested that the aryl ring attached to the piperazine linker must be para-halogen (chloro or fluoro)-substituted, whereas the aryl ring on the hydrazido chain must have electron-donating or electron-withdrawing groups at the meta or para position, respectively, to display anti-proliferative effects against AGS cells.
Basing on these screening results, compounds 12 and 22 were chosen to perform specific assays with gastric adenocarcinoma cells (AGS) in order to better understand the molecular mechanisms underlying the biological effect of these molecules. In particular, 12 and 22 were selected because of their better ability to reduce the proliferation of AGS cells at 50 µM after 72 h, whereas they were safe and tolerated at 10 µM against both HGFs and AGS cells. Firstly, a cytotoxicity assay which measures the lactate dehydrogenase (LDH) released within the culture medium, was carried out after 48 and 72 h of treatment with 12 and 22 on AGS cells (Figure 4).
Figure 4.
Cytotoxicity assay of AGS gastric adenocarcinoma cells treated with molecules vehicle (DMSO), 12 and 22 for 48 and 72 h. LDH leakage is reported as the percentage of LDH leakage compared with vehicle treated cells (DMSO). Data are presented as the mean ± standard deviation of three separate experiments. ** p < 0.01, * p <0.05.
After 48 h of culture, both 12 and 22 treatments caused a statistically significant increase (p < 0.01) in LDH release compared to LDH release measured in DMSO sample. After 72 h of treatment this trend is maintained for AGS treated with 12 (p < 0.05), whereas a partial recovery for AGS treated with 22 could be hypothesized. Secondly, in order to evaluate the oxidative stress occurrence in AGS after administration of the two selected molecules, the H2O2 release within the culture medium was performed by an ELISA assay (Figure 5). The H2O2 release follows a similar trend to LDH release.
Figure 5.
Quantitative determination of the H2O2 released within the culture medium by AGS treated with vehicle (DMSO), 12 and 22, for 48 and 72 h. Values represent the means ± SD of three separate experiments. The bar graph shows the H2O2 release (ng/mL) in cell supernatants; * p < 0.05.
After 48 h of treatment a statistically significant increase in H2O2 levels of AGS treated with 12, compared to H2O2 levels of both vehicle-treated and 22-treated AGS, can be detected (p < 0.05). Again, after 72 h of administration, H2O2 release within the culture medium appears significantly augmented only when AGS are exposed to 12 (p < 0.05). Thirdly, a morphological analysis of AGS treated for 48 and 72 h with DMSO and with the two selected molecules, was carried out by means of haematoxylin-eosin staining (Figure 6 and Figure 7).
Figure 6.
Haematoxylin-eosin staining of AGS treated with molecules vehicle (DMSO), 12 and 22, for 48 h; upper line magnification 20×, lower line magnification 40×. Black arrows indicate mitotic cells, blue arrows indicate suffering or dead cells, red arrows indicate multinucleated cells.
Figure 7.
Haematoxylin-eosin staining of AGS treated with molecules vehicle (DMSO), 12 and 22, for 72 h; upper line magnification 20×, lower line magnification 40×. Black arrows indicate cells with modified chromatin, blue arrows indicate cells surrounded by little vesicles.
The morphological analysis put in evidence, after 48 h of treatment (Figure 6) in DMSO-treated AGS, an appreciable amount of cell mitosis (black arrows) along with a higher cell density respect to both 12 and 22-treated AGS. In 12-treated AGS and, to a lower extent, in 22-treated AGS, the occurrence of mitotic events appears negatively affected by the molecules administration as already evidenced by MTT test. Moreover, in 12 and 22-treated cells, several round shaped and darker cells, close to detachment and cell death are clearly detectable (blue arrows). In AGS treated with 12, some larger and multinucleated cells are also recognizable (red arrows).
After 72 h of treatment the previously described trend is maintained (Figure 7), however, in 12-treated sample, some cells with modified chromatin distribution (black arrows) start to appear along with several cells surrounded by numerous little vesicles (blue arrows), a morphological condition resembling to blebbing phenomenon.
Blebbing is often a typical feature of cells close to the apoptotic event [29,30]. Basing on this, to establish if 12 and 22 are able to trigger the apoptotic cascade thus explaining the rate of dead cells revealed by the viability test, Bax pro-apoptotic factor expression was evaluated through flow cytometry. Bax expression was measured in AGS cells after a 48 and 72 h of exposition to compounds 12 and 22 (Figure 8). The expression of the pro-apoptotic protein increases both at 48 and 72 h in the presence of compound 12 with respect to DMSO alone, while exposure to compound 22 is not effective as regard Bax levels of expression. In details, the percentage of Bax-positive cells significantly raises in a time-dependent manner, being assessed at 3.71% after 48 h (1.25% for DMSO alone) and 12.9% after 72 h (3.07% for DMSO alone).
Figure 8.
Bax expression in AGS cells in the presence of 12 and 22 after 48 and 72 h. (A) Bar graphs represent cell percentage of positive cells for Bax after 48 (upper histogram) and 72 h (lower histogram). (B) Representative peaks of treatment from the three independent experiments of the fluorescence emissions in the FL-2/PE channel. *** p < 0.0001 between DMSO and compounds.
In Silico Pharmacokinetic Parameters and Target Prediction of the Most Promising Compounds 12 and 22
In Table 1 and Table 2 we have reported the medicinal chemistry parameters obtained from the web tool SwissADME [31], describing some properties of the tested compounds that are crucial for the future drug development. Collectively, the two compounds fully comply with the limits of the Lipinski’s rule (no violation), supporting their feasible oral use and drug-likeness, as also further corroborated by their high passive absorption in the gastrointestinal tract. Finally, they were not categorized as Pan Assay Interference Compounds (PAINS substructures able to elicit promiscuous pharmacological behaviour.
Table 2.
In silico physical-chemical properties the most potent compounds 12 and 22.
| Compound | 12 | 22 |
|---|---|---|
| H-bond acceptors (HBA) | 5 | 6 |
| H-bond donators (HBD) | 1 | 1 |
| Consensus Log P | 2.59 | 2.41 |
| Lipinski violations | 0 | 0 |
| GI absorption | high | high |
| P-gp substrate | yes | yes |
| PAINS alerts | 0 | 0 |
The major drawback of these compounds, and likely of the entire scaffold proposed in this paper, is the propensity to be substrate of the permeability glycoprotein (P-gp), thus promoting the energy-dependent efflux out of the cytosol and reducing their antiproliferative efficacy. This behaviour is also the limiting drawback of doxorubicin.
Moreover, we also reported in Figure 9 the boiled-egg and bioavailability radar pictures as the two graphical outputs of the SwissADME tool, which considers several important parameters for determining the drug-likeness of a molecule. It allowed us to predict simultaneously two ADME characteristics: the passive absorption at the gastrointestinal tract (white area) and the ability to permeate the blood brain barrier (BBB, yellow area). Both the compounds are located in the yellow area implying that they are not likely able to cross the BBB, but can be passively absorbed at the GI level. Furthermore, the bioavailability radars provides the drug-likeness representation of the two compounds within the desirable pink area indicative for the optimal range of each physicochemical properties (lipophilicity, size, polarity, solubility, unsaturation, flexibility) for the oral administration.
Figure 9.
Representation of the boiled-egg graph and bioavailability radar calculated by SwissADME web-tool for compounds 12 and 22.
With the aim to identify the putative target(s) of our two compounds, we also applied the SwissTargetPrediction web-tool, performing the analysis on the two compounds. In Table 3 and Table 4 we reported the data concerning the most probable targets found, i.e., the ones exhibiting the highest values of probability to be a target. Unfortunately, both compounds did not display a main preference for one target, nor with a high probability value, despite some of those can be related to cancer cell growth and survival.
Table 3.
Protein target prediction for compound 12.
| Target | Common Name | Uniprot ID | ChEMBL ID | Target Class | Probability |
|---|---|---|---|---|---|
| Neuropeptide Y receptor type 5 | NPY5R | Q15761 | CHEMBL4561 | Family A G protein-coupled receptor | 0.12 |
| P2X purinoceptor 7 | P2RX7 | Q99572 | CHEMBL4805 | Ligand-gated ion channel | 0.12 |
| Adenosine A2a receptor | ADORA2A | P29274 | CHEMBL251 | Family A G protein-coupled receptor | 0.12 |
| Inhibitor of nuclear factor kappa B kinase beta subunit | IKBKB | O14920 | CHEMBL1991 | Kinase | 0.12 |
| Toll-like receptor 4 | TLR4 | O00206 | CHEMBL5255 | Toll-like and Il-1 receptors | 0.12 |
Table 4.
Protein target prediction for compound 22.
| Target | Common Name | Uniprot ID | ChEMBL ID | Target Class | Probability |
|---|---|---|---|---|---|
| Probable protein-cysteine N-palmitoyltransferase porcupine | PORCN | Q9H237 | CHEMBL1255163 | Enzyme | 0.12 |
| P2X purinoceptor 7 | P2RX7 | Q99572 | CHEMBL4805 | Ligand-gated ion channel | 0.12 |
| Dual specificity mitogen-activated protein kinase kinase 2 | MAP2K2 | P36507 | CHEMBL2964 | Kinase | 0.12 |
| Dual specificity mitogen-activated protein kinase kinase 1 | MAP2K1 | Q02750 | CHEMBL3587 | Kinase | 0.12 |
| Inhibitor of nuclear factor kappa B kinase beta subunit | IKBKB | O14920 | CHEMBL1991 | Kinase | 0.12 |
3. Material and Methods
3.1. Chemical Studies
All chemicals used in the synthesis of compounds are products supplied by Sigma Aldrich (Merck, Milan, Italy). The synthesis of compounds in the structure of N′-benzylidene-2-(substituted)pyridazine acetohydrazide, as seen in Scheme 1, consists of five steps and each synthesis was performed according to the literature [32,33]. Impurity monitoring at each stage of the syntheses was performed by TLC (Kieselgel F254 plates, Merck, Milan, Italy) method. Melting points of the compounds were measured with a IA9000 Melting Point Apparatus (Cole-Parmer, Staffordshire, UK) and checked with the literature data. IR spectra were taken by the ATR technique (Spectrum 3 FT-IR Spectrometer, Perkin Elmer, Milan, Italy). 1H-NMR and 13C-NMR spectra were obtained in DMSO-d6 at 600 MHz and 150 MHz, respectively. An Avance Ultrashield FT-NMR spectrometer (Bruker, Billerica, MA, USA) was used for these measurements. Mass spectra were recorded using an Agilent 6400. B.08.00 (B8023.0) system (Agilent Technology, Santa Clara, CA, USA). Elemental analyses of the compounds were performed with a CHNS932 ibstrument (Leco, St. Joseph, MI, USA). All analyses of the compounds were done at the Scientific and Technological Research Center of Inonu University.
3.1.1. Synthesis of Intermediate Compounds
Synthesis of 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazine-1-yl)-pyridazinesIa–c. 4-Chlorophenyl/4-fluorophenyl/phenyl piperazine (0.01 mol), 3,6-dichloropyridazine (0.01 mol) and ethanol (15 mL) were added to a flask. The mixture was stirred under reflux for 6 h. The reaction mixture was poured into ice water. The solid phase was separated by filtration and dried. For purification, the residue was crystallized from ethanol. The melting points of 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-pyridazines Ia–c were compatible with the literature data [34].
Synthesis of 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinonesIIa–c. Glacial acetic acid (30 mL) was added to the appropriate 6-substituted-3-chloropyridazine (0.05 mol). The mixture was stirred under reflux for 6 h. Glacial acetic acid was removed from the reaction medium with a rotary evaporator. The residue was extracted by adding water and chloroform. The organic phase was separated, sodium sulphate was added to dry, and then filtered. The chloroform phase was evaporated under reduced pressure. The residue was crystallized from ethanol. The melting points of 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinones IIa–c were in accordance with the literature [12,27].
Synthesis of ethyl 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinone-2-ylacetatesIIIa–c. 6-(4-(4-Chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinone (0.01 mol), potassium carbonate (0.02 mol), ethyl bromoacetate (0.02 mol), and acetone (40 mL) were added to a flask. The mixture was stirred under reflux overnight. The mixture was cooled and then filtered. Acetone was evaporated under reduced pressure. For purification, the residue was crystallized from methanol. The melting points of 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinone-2-ylacetates IIIa–c were compatible with the literature data [12,27,28].
Synthesis of ethyl 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinone-2-ylacetohydrazidesIVa–c. Ethyl 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)-3(2H)-pyridazinone-2-ylacetate (0.01 mol), hydrazine hydrate (3 mL, 99%) and methanol (25 mL) were added to a flask. The reaction mixture was stirred at 25 °C for 3 h. The product was filtered, washed with distilled water, dried, and crystallized from ethanol. The melting points of 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazine-1-yl)-3(2H)-pyridazinone-2-yl acetohydrazides (4a obtained in 62% yield, 4b in 71% yield, 4c in 69% yield) are 228-229, 231-232 and 251-252 °C, respectively. Characterization data of the compounds were compatible with the literature data [12,27,28].
3.1.2. General Synthesis of 6-(4-(4-Chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl)pyridazin-3(2H)-one-2-acetyl-2-(substituted/nonsubstituted benzal)hydrazones 1–23
A suitable benzaldehyde derivative (0.01 mol) and the appropriate 6-(4-(4-chlorophenyl/4-fluorophenyl/phenyl)piperazin-1-yl]-3(2H)-pyridazinone-2-yl acetohydrazide IVa–c (0.01 mol) were added to ethanol (15 mL). The mixture was stirred under reflux for 6 h. The reaction mixture was transferred into ice water, filtered and dried. The residue was crystallized from methanol:water mixtures. The structures of the compounds were determined by 1H-NMR, 13C-NMR, IR spectroscopy and elemental analysis. All spectral data of the compounds were in accordance with the assigned structures as given below.
N′-Benzylidene-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (1). White crystals (methanol/water); yield 83%; m.p. 236–237 °C; IR (ν cm−1, ATR): 3055 (C–H aromatic), 2947 (C–H aliphatic), 1652, 1629 (C=O), 1561 (C=N), 1231 (C–N). 1H-NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 5.07 (2H; s; NCH2CO), 6.92 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.94 (1H; d; J = 4.2 Hz, pyridazinone H4), 6.98–7.72 (10H; m; phenyl protons), 8.02 (1H; s; –N=CH–), 11.70 (1H; s; -NH–N); 13C–NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a + a’), 53.0 (1C; –N–CH2–C=O), 116.2 (1C; =CH), 119.7 (2C; phenyl C2,6), 127.6 (1C; phenyl C4), 129.4 (1C; =CH-phenyl C6), 130.4 (1C; =CH-phenyl C2), 130.6 (2C; phenyl C3,5), 131.1 (1C; =CH–phenyl C1), 134.4 (1C; =CH–phenyl C3), 144.3 (2C; =CH–phenyl C4,5), 147.4 (1C; pyridazinone C5), 149.1 (1C; phenyl C1), 151.3 (1C; pyridazinone C6), 158.2 (1C; pyridazinone C4), 163.8 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/API-ESMS m/z 439 [M + Na]+; Anal. Calcd. for C23H24N6O3.1/4 H2O: C, 65.62; H, 5.87; N, 19.96. Found: C, 65.96; H, 6.103; N, 20.12%.
N’-(3-Methylbenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)aceto-hydrazide (2). White crystals (methanol/water); yield 80%; m.p. 224–225 °C; IR (ν cm−1, ATR): 3052 (C–H aromatic), 2949 (C–H aliphatic), 1652 (C=O), 1581 (C=N), 1267 (C–N). 1H-NMR (DMSO-d6, 600 MHz): δ 2.34 (3H; s; CH3), 3.28 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 5.07 (2H; s; NCH2CO), 6.92 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.93 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.22–7.65 (9H; m; phenyl protons), 8.19 (1H; s; –N=CH–), 11.64 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 21.4 (1C; CH3), 39.6 (2C, CH2–N; b + b’), 39.7 (2C, CH2–N; a + a’), 53.5 (1C; –N–CH2–C=O), 116.2 (1C; =CH), 119.7 (2C; phenyl C2,6), 124.9 (1C; phenyl C4), 127.7 (1C; 3-methylphenyl C6), 129.4 (1C; 3-methylphenyl C2), 131.0 (2C; phenyl C3,5), 134.4 (1C; 3-methylphenyl C1), 134.6 (1C; 3-methylphenyl C3), 138.5 (2C; 3-methylphenyl C4,5), 138.5 (1C; pyridazinone C5), 144.4 (1C; phenyl C1), 149.0 (1C; pyridazinone C6), 149.1 (1C; pyridazinone C4), 163.7 (1C; CH2–N–C=O), 168.4 (1C; pyridazinone C3); LC/API-ESMS m/z 453 [M + Na]+; Anal. Calcd. for C24H26N6O2.1/3 H2O: C, 66.04; H, 6.16; N, 19.25. Found: C, 66.09; H, 6.10; N, 19.63%.
N’-(4-Methylbenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)aceto-hydrazide (3). White crystals (methanol/water); yield 74%; m.p. 210–212 °C; IR (ν cm−1, ATR): 3400 (N–H), 3052 (C–H aromatic), 2949 (C–H aliphatic), 1652 (C=O), 1581 (C=N), 1267 (C–N). 1H-NMR (DMSO-d6, 600 MHz): δ 2.34 (3H; s; CH3), 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 5.05 (2H; s; NCH2CO), 6.90 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.93 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.22–7.65 (9H; m; phenyl protons), 8.18 (1H; s; –N=CH–), 11.59 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 21.5 (1C; CH3), 46.4 (2C, CH2-N; b+b’), 48.4 (2C, CH2-N; a+a’), 53.1 (1C; -N-CH2-C=O), 116.2 (1C; =CH), 119.8 (2C; phenyl C2,6), 126.9 (1C; phenyl C4), 127.6 (1C; 4-methylphenyl C6), 129.8 (1C; 4-methylphenyl C2), 129.9 (2C; phenyl C3,5), 131.0 (1C; 4-methylphenyl C1), 131.9 (1C; 4-methylphenyl C3), 140.2 (2C; 4-methylphenyl C4,5), 140.4 (1C; pyridazinone C5), 144.3 (1C; phenyl C1), 144.4 (1C; pyridazinone C6), 149.1 (1C; pyridazinone C4), 158.2 (1C; CH2–N–C=O), 168.3 (1C; pyridazinone C3); LC/API-ESMS m/z 453 [M + Na]+; Anal. Calcd. for C24H26N6O2.2/3 H2O: C, 65.14; H, 6.23; N, 18.99. Found: C, 64.86; H, 5.99; N, 19.30%.
N’-(3-Methoxybenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (4). White crystals (methanol/water); yield 68%; m.p. 205–206 °C; IR (ν cm−1, ATR): 3389 (N–H), 3094 (C–H aromatic), 2971 (C–H aliphatic), 1681, 1652 (C=O), 1575 (C=N), 1243 (C–N), 1174 (C–O). 1H-NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 3.80 (3H; s; OCH3), 5.04 (2H; s; CH2CO), 6.93 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.98 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.01–7.96 (9H; m; phenyl protons), 8.16 (1H; s; –N=CH–), 11.51 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2-C=O), 55.8 (1C, O–CH3), 114.8 (1C; =CH), 116.2 (2C; phenyl C2,6), 127.1 (1C; phenyl C4), 128.9 (1C; 3-methoxyphenyl C5,6), 129.4 (1C; 3-methoxyphenyl C1), 131.0 (1C; phenyl C3,5), 131.1 (1C; 3-methoxyphenyl C4), 144.2 (1C; pyridazinone C4), 147.3 (1C; 3-methoxyphenyl C2), 149.0 (1C; pyridazinone C5), 151.3 (1C; phenyl C1), 158.1 (1C; pyridazinone C6), 158.2 (1C; 3-methoxyphenyl C3), 163.5 (1C; CH2–N–C=O), 168.2 (1C; pyridazinone C3); LC/API-ESMS m/z 469.2 [M + Na]+; Anal. Calcd. for C24H26N6O3.3/2 H2O: C, 60.87; H, 6.17; N, 18.01. Found: C, 61.70; H, 5.74; N, 18.01%.
N’-(2,3-Dimethoxybenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)aceto-hydrazide (5). White crystals (methanol/water); yield 86%; m.p. 249–250 °C; IR (ν cm−1, ATR): 3105 (C–H aromatic), 2966 (C–H aliphatic), 1695, 1659 (C=O), 1571 (C=N), 1232 (C–N), 1070 (C–O). 1H-NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 3.78 (3H; s; OCH3), 3.84 (3H; s; OCH3), 5.05 (2H; s; NCH2CO), 6.99 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.10 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.12–7.65 (8H; m; phenyl protons), 8.31 (1H; s; –N=CH–), 11.63 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2–C=O), 56.2 (1C; –O–CH3), 61.7 (1C; –O–CH3), 114.6 (1C; =CH), 116.2 (2C; phenyl C2,6), 119.7 (1C; phenyl C4), 124.8 (1C; 2,3-dimethoxyphenyl C6), 127.9 (1C; 2,3-dimethoxyphenyl C1), 130.9 (2C; phenyl C3,5), 131.1 (2C; 2,3-dimethoxyphenyl C4,5), 140.1 (2C; 2,3-dimethoxyphenyl C3), 142.9 (1C; 2,3-dimethoxyphenyl C2), 148.4 (1C; pyridazinone C5), 149.0 (1C; phenyl C1), 151.3 (1C; pyridazinone C6), 158.2 (1C; pyridazinone C4), 163.7 (1C; CH2–N–C=O), 168.4 (1C; pyridazinone C3); LC/API-ESMS m/z 496 [M]+; Anal. Calcd. for C25H28N6O4.1/3 H2O: C, 62.23; H, 5.99; N, 17.42. Found: C, 62.59; H, 5.77; N, 17.62%.
N’-(3,5-Dimethoxybenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)aceto-hydrazide (6). White crystals (methanol/water); yield 57%; m.p. 232–233 °C; IR (ν cm−1, ATR): 3094 (C–H aromatic), 2960 (C–H aliphatic), 1685, 1657 (C=O), 1574 (C=N), 1262 (C–N), 1150 (C–O). 1H–NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 3.78 (6H; s; OCH3), 5.10 (2H; s; NCH2CO), 6.87 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.91 (1H; d; J = 4.2 Hz, pyridazinone H4), 6.99–7.93 (8H; m; phenyl protons), 8.14 (1H; s; –N=CH–), 11.68 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a + a’), 53.0 (1C; –N–CH2–C=O), 55.8 (2C; –OCH3), 105.1 (1C; =CH), 116.2 (2C; phenyl C2,6), 119.7 (1C; phenyl C4), 127.1 (1C; 3,5-dimethoxyphenyl C6), 129.3 (1C; 3,5-dimethoxyphenyl C2), 131.0 (1C; phenyl C1), 136.5 (1C; 3,5-dimethoxyphenyl C1), 136.6 (1C; 3,5-dimethoxyphenyl C4), 144.1 (2C; 3,5- dimethoxyphenyl C3,5), 147.3 (1C; pyridazinone C5), 149.0 (1C; phenyl C1), 151.3 (1C; pyridazinone C6), 158.2 (1C; pyridazinone C4), 163.8 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/API-ESMS m/z 468.8 [M]+; Anal. Calcd. for C25H28N6O4.1/3 H2O: C, 62.23; H, 5.99; N, 17.42. Found: C, 62.35; H, 5.71; N, 17.57%.
N’-(2,4,6-Trimethoxybenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)aceto-hydrazide (7). White crystals (methanol/water); yield 82%; m.p. 254–255 °C; IR (ν cm−1, ATR): 3232 (N–H), 3086 (C–H aromatic), 2933 (C–H aliphatic), 1697, 1670 (C=O), 1595 (C=N), 1239 (C–N), 1126, 1057 (C–O). 1H-NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 3.81 (6H; s; OCH3), 3.83 (3H; s; OCH3), 4.94 (2H; s; NCH2CO), 6.29 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.94 (1H; d; J = 4.2 Hz, pyridazinone H4), 6.80–7.64 (8H; m; pyridazinone H4 and phenyl protons), 8.19 (1H; s; –N=CH–), 11.26 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a + a’), 53.4 (1C; –N–CH2–C=O), 55.89 (1C; –OCH3), 56.53 (2C; –OCH3), 104.3 (1C; =CH), 116.3 (2C; phenyl C2,6), 119.7 (1C; phenyl C4), 126.9 (1C; 2,4,6-trimethoxyphenyl C5), 129.4 (1C; 2,4,6-trimethoxyphenyl C3), 131.0 (2C; phenyl C3,5), 139.6 (1C; 2,4,6-trimethoxyphenyl C1), 143.2 (1C; 2,4,6-trimethoxyphenyl C4), 148.9 (2C; 2,4,6-trimethoxyphenyl C2,6), 151.3 (1C; pyridazinone C5), 158.2 (1C; phenyl C1), 158.3 (1C; pyridazinone C6), 160.3 (1C; pyridazinone C4), 162.7 (1C; CH2–N–C=O), 167.9 (1C; pyridazinone C3); LC/API-ESMS m/z 437 [M-2(OCH3)]+; Anal. Calcd. for C26H30N6O5: C, 61.65; H, 5.97; N, 16.59. Found: C, 61.28; H, 5.82; N, 16.67%.
N’-(3,4,5-Trimethoxybenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)aceto-hydrazide (8). White crystals (methanol/water); yield 83%; m.p. 238–239 °C; IR (ν cm−1, ATR): 3216 (N–H), 3065 (C–H aromatic), 2943 (C–H aliphatic), 1669 (C=O), 1578 (C=N), 1231 (C–N), 1124 (C–O). 1H-NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 3.83 (9H; s; OCH3), 5.08 (2H; s; NCH2CO), 6.82 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.92 (1H; d; J = 4.2 Hz, pyridazinone H4), 6.98–7.92 (7H; m; phenyl protons), 8.14 (1H; s; –N=CH–), 11.68 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a + a’), 53.5 (1C; –N–CH2–C=O), 56.4 (1C; –OCH3), 60.6 (2C; –OCH3), 104.6 (1C; =CH), 116.3 (2C; phenyl C2,6), 119.8 (1C; phenyl C4), 127.0 (1C; 3,4,5-trimethoxyphenyl C6), 129.4 (1C; 3,4,5-trimethoxyphenyl C2), 129.9 (2C; phenyl C3,5), 131.0 (1C; 3,4,5-trimethoxyphenyl C1), 139.5 (1C; 3,4,5-trimethoxyphenyl C3), 144.1 (2C; 3,4,5-trimethoxyphenyl C4,5), 147.4 (1C; pyridazinone C5), 149.1 (1C; phenyl C1), 151.3 (1C; pyridazinone C6), 153.6 (1C; pyridazinone C4), 163.7 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/API-ESMS m/z 529.2 [M + Na]+; Anal. Calcd. for C26H30N6O5.1/2 H2O: C, 60.57; H, 6.06; N, 16.30. Found: C, 60.92; H, 5.72; N, 16.55%.
N’-(4-Nitrobenzylidene)-2-(6-oxo-3-(4-phenylpiperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (9). White crystals (methanol/water); yield 54%; m.p. 239–240 °C; IR (ν cm−1, ATR): 3089 (C–H aromatic), 2967 (C–H aliphatic), 1692, 1660 (C=O), 1574 (C=N), 1339 (aromatic-NO2), 1235 (C–N). 1H-NMR (DMSO-d6, 600 MHz): δ 3.23 (4H; t; CH2N; Hb + Hb’), 3.39 (4H; t; CH2N; Ha + Ha’), 5.11 (2H; s; NCH2CO), 6.82 (1H; d; J = 4.0 Hz, pyridazinone H5), 6.92 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.22–8.33 (9H; m; phenyl protons), 8.30 (1H; s; –N=CH–), 11.96 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C, CH2–N; a+a’), 53.0 (1C; –N–CH2–C=O), 105.1 (1C; =CH), 116.2 (2C; phenyl C2,6), 124.4 (1C; phenyl C4), 124.5 (1C; 4-nitrophenyl C6), 128.3 (1C; 4-nitrophenyl C2), 128.4 (2C; phenyl C3,5), 129.4 (1C; 4-nitrophenyl C1), 140.8 (2C; 4-nitrophenyl C3,5), 141.9 (2C; =CH–phenyl C4), 142.1 (1C; pyridazinone C5), 148.2 (1C; phenyl C1), 149.1 (1C; pyridazinone C6), 151.3 (1C; pyridazinone C4), 158.2 (1C; CH2–N–C=O), 168.9 (1C; pyridazinone C3); LC/API-ESMS m/z 413 [M - NO2]+; Anal. Calcd. for C23H23N7O4.3/2 H2O: C, 56.55; H, 5.36; N, 20.07. Found: C, 56.71; H, 4.99; N, 19.97%.
N’-(2-Methoxybenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)-acetohydrazide (10). White crystals (methanol/water); yield 80%; m.p. 241–242 °C; IR (ν cm−1, ATR): 3236 (N–H), 3074 (C–H aromatic), 2970 (C–H aliphatic), 1683, 1668 (C=O), 1573 (C=N), 1231 (C–N), 1122 (C–O), 837 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 3.23 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 3.86 (3H; s; OCH3), 5.04 (2H; s; CH2CO), 7.24 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.25 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.26–7.86 (8H; m; phenyl protons), 8.36 (1H; s; –N=CH–), 11.61 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 600 MHz): δ 46.3 (2C, CH2–N; b + b’), 48.2 (2C, CH2–N; a + a’), 53.0 (1C; –N–CH2–C=O), 56.1 (1C; –OCH3), 112.3 (1C; =CH), 117.7 (2C; 4-chlorophenyl C2,6), 121.2 (1C; 2-methoxyphenyl C6), 122.4 (1C; 2-methoxyphenyl C5), 123.2 (2C; 4-chlorophenyl C3,5), 125.9 (2C; 2-methoxyphenyl C3,4), 127.1 (1C; pyridazinone C5), 129.1 (1C; 4-chlorophenyl C1), 131.1 (1C; 2-methoxyphenyl C1), 139.9 (1C; 2-methoxyphenyl C2), 142.9 (1C; 4-chlorophenyl C4), 148.9 (1C; pyridazinone C4), 150.1 (1C; pyridazinone C6), 163.6 (1C; CH2–N–C=O), 166.3 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 503 [M + Na]+; Anal. Calcd for C24H25ClN6O3.1/5H2O: C, 59.49; H, 5.28; N, 17.34. Found: C, 59.13; H, 5.21; N, 17.93%.
N’-(3-Methoxybenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)-acetohydrazide (11). White crystals (methanol/water); yield 82%; m.p. 214–215 °C; IR (ν cm−1, ATR): 3065 (C–H aromatic), 2943 (C–H aliphatic), 1696, 1656 (C=O), 1567 (C=N), 1231 (C–N), 1124 (C–O), 836 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 3.23 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 3.80 (3H; s; OCH3), 5.06 (2H; s; CH2CO), 6.99 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.00–7.98 (9H; m; phenyl protons and pyridazinone H4), 8.18 (1H; s; –N=CH–), 11.67 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.3 (2C, CH2–N; b + b’), 48.1 (2C, CH2–N; a + a’), 53.0 (1C; –N–CH2–C=O), 55.6 (1C; –OCH3), 111.6 (1C; =CH), 116.6 (2C; 4-chlorophenyl C2,6), 117.7 (1C; 3-methoxyphenyl C6), 123.2 (1C; 3-methoxyphenyl C2), 127.1 (2C; 4-chlorophenyl C3,5), 129.1 (2C; 3-methoxyphenyl C4,5), 130.4 (1C; pyridazinone C5), 131.0 (1C; 4-chlorophenyl C1), 135.9 (1C; 3-methoxyphenyl C1), 144.1 (1C; 3-methoxyphenyl C3), 148.9 (1C; 4-chlorophenyl C4), 150.1 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 160.0 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 414 [M-(Cl + OCH3)]+; Anal. Calcd for C24H25ClN6O3.1/9H2O: C, 59.69; H, 5.26; N, 17.40. Found: C, 59.48; H, 5.12; N, 18.23%.
N’-(2,3-Dimethoxybenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (12). White crystals (methanol/water); yield 80%; m.p. 252–253 °C; IR (ν cm−1, ATR): 3196 (N–H), 3045 (C–H aromatic), 2968 (C–H aliphatic), 1690, 1667 (C=O), 1561 (C=N), 1230 (C–N), 1065 (C–O), 843 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 3.23 (4H; t; CH2N; Hb + Hb’), 3.37 (4H; t; CH2N; Ha + Ha’), 3.78 (3H; s; OCH3), 3.84 (3H; s; OCH3), 5.04 (2H; s; NCH2CO), 6.92 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.99 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.10–7.65 (7H; m; phenyl protons), 8.30 (1H; s; –N=CH–), 11.62 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.3 (2C, CH2–N; b + b’), 48.1 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2–C=O), 56.2 (1C; –OCH3), 61.7 (1C; –OCH3), 114.6 (1C; =CH), 117.4 (2C; 4-chlorophenyl C2,6), 123.2 (2C; 4-chlorophenyl C3,5), 127.9 (1C; 2,3-dimethoxyphenyl C6), 129.1 (1C; 2,3-dimethoxyphenyl C1), 131.0 (1C; 4-chlorophenyl C1), 140.1 (2C; 2,3-dimethoxyphenyl C4,5), 148.3 (1C; 2,3-dimethoxyphenyl C2), 148.4 (1C; 2,3-dimethoxyphenyl C3), 148.9 (1C; pyridazinone C5), 150.1 (1C; 4-chlorophenyl C4), 153.1 (1C; pyridazinone C6), 158.2 (1C; pyridazinone C4), 163.7 (1C; CH2–N–C=O), 168.4 (1C; pyridazinone C3); LC/API-ESMS m/z 533 [M + Na]+; Anal. Calcd. for C25H27ClN6O4.1/9 H2O: C, 58.53; H, 5.35; N, 16.38. Found: C, 58.15; H, 5.16; N, 17.30%.
N’-(2,4,6-Trimethoxybenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (13). White crystals (methanol/water); yield 86%; m.p. 281–282 °C; IR (ν cm−1, ATR): 3231 (N–H), 3086 (C–H aromatic), 2933 (C–H aliphatic), 1697, 1668 (C=O), 1592 (C=N), 1236 (C–N), 1156 (C–O), 851 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 3.22 (4H; t; CH2N; Hb + Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 3.81 (6H; s; OCH3), 3.83 (3H; s; OCH3), 4.93 (2H; s; NCH2CO), 6.29 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.89 (1H; d; J = 4.2 Hz, pyridazinone H4), 6.91–7.64 (6H; m; phenyl protons), 8.18 (1H; s; –N=CH–), 11.25 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.4 (2C, CH2–N; b + b’), 48.1 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2–C=O), 55.9 (1C; –OCH3), 56.5 (2C; –OCH3), 104.3 (1C; =CH), 117.7 (2C; 4-chlorophenyl C2,6), 123.2 (2C; 2,4,6-trimethoxyphenyl C3,5), 126.9 (1C; 4-chlorophenyl C3), 129.1 (1C; 4-chlorophenyl C5), 131.0 (1C; 4-chlorophenyl C1), 139.6 (1C; 2,4,6-trimethoxyphenyl C1), 143.2 (1C; 2,4,6-trimethoxyphenyl C2,4), 148.9 (2C; 2,4,6-trimethoxyphenyl C6), 150.1 (1C; pyridazinone C5), 158.3 (1C; 4-chlorophenyl C4), 160.3 (1C; pyridazinone C6), 160.4 (1C; pyridazinone C4), 162.7 (1C; CH2–N–C=O), 167.9 (1C; pyridazinone C3); LC/API-ESMS m/z 563 [M + Na]+; Anal. Calcd. for C26H29ClN6O5.1/6 H2O: C, 57.40; H, 5.43; N, 15.45. Found: C, 57.01; H, 5.23; N, 16.47%.
N’-(3,4,5-Trimethoxybenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (14). White crystals (methanol/water); yield 77%; m.p. 285–286 °C; IR (ν cm−1, ATR): 3213 (N–H), 3062 (C–H aromatic), 2938 (C–H aliphatic), 1681, 1663 (C=O), 1595 (C=N), 1232 (C–N), 1157 (C–O), 851 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 3.23 (4H; t; CH2N; Hb + Hb’), 3.36 (4H; t; CH2N; Ha + Ha’), 3.70 (6H; s; OCH3), 3.83 (3H; s; OCH3), 5.07 (2H; s; NCH2CO), 6.91 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.93-7.92 (7H; m; phenyl protons and pyridazinone H4), 8.14 (1H; s; –N=CH–), 11.67 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.4 (2C, CH2–N; b + b’), 48.1 (2C, CH2–N; a + a’), 53.1 (1C; –N–CH2–C=O), 56.5 (1C; –OCH3), 60.5 (2C; –OCH3), 104.6 (1C; =CH), 117.7 (2C; 4-chlorophenyl C2,6), 119.8 (2C; 2,4,6-trimethoxyphenyl C2,6), 123.2 (1C; 4-chlorophenyl C3), 129.1 (1C; 4-chlorophenyl C5), 129.9 (1C; 4-chlorophenyl C1), 131.1 (1C; 2,4,6-trimethoxyphenyl C1), 139.5 (1C; 2,4,6-trimethoxyphenyl C3,5), 144.0 (2C; 2,4,6-trimethoxyphenyl C4), 148.9 (1C; pyridazinone C5), 150.1 (1C; 4-chlorophenyl C4), 153.6 (1C; pyridazinone C6), 158.2 (1C; pyridazinone C4), 163.7 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/API-ESMS m/z 560 [M]+; Anal. Calcd. for C26H29ClN6O5.1/4 H2O: C, 57.25; H, 5.45; N, 15.41. Found: C, 56.86; H, 5.20; N, 16.63%.
N’-(4-Nitrobenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (15). White crystals (methanol/water); yield 86%; m.p. 238–239 °C; IR (ν cm−1, ATR): 3077 (C–H aromatic), 2964 (C–H aliphatic), 1689, 1654 (C=O), 1580 (C=N), 1340 (aromatic NO2), 1236 (C–N), 836 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 3.23 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 5.10 (2H; s; CH2CO), 6.92 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.99 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.24–8.29 (8H; m; phenyl protons), 8.33 (1H; s; –N=CH–), 11.96 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.3 (2C, CH2–N; b + b’), 48.1 (2C, CH2-N; a + a’), 53.0 (1C; –N–CH2–C=O), 105.1 (1C; =CH), 117.7 (2C; 4-chlorophenyl C2,6), 124.5 (1C; 4-nitrophenyl C6), 127.1 (1C; 4-nitrophenyl C2), 128.3 (2C; 4-chlorophenyl C3,5), 129.1 (2C; 4-nitrophenyl C3,5), 131.0 (1C; pyridazinone C5), 141.9 (1C; 4-chlorophenyl C1), 145.0 (1C; 4-nitrophenyl C1), 148.2 (1C; 4-nitrophenyl C4), 149.0 (1C; 4-chlorophenyl C4), 150.1 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 164.3 (1C; CH2–N–C=O), 168.9 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 414 [M-(Cl + NO2)]+; Anal. Calcd for C23H22ClN7O4.1/9H2O: C, 55.48; H, 4.50; N, 19.69. Found: C, 55.19; H, 4.42; N, 20.76%.
N’-(4-Dimethylaminobenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (16). White crystals (methanol/water); yield 68%; m.p. 243–244 °C; IR (ν cm−1, ATR): 3082 (C–H aromatic), 2962 (C–H aliphatic), 1678, 1665 (C=O), 1589 (C=N), 1227 (C–N), 836 (C–Cl). 1H-NMR (DMSO-d6, 600 MHz): δ 2.96 (6H; s; –N(CH3)2), 3.23 (4H; t; CH2N; b + b’), 3.37 (4H; t; CH2N; a + a’), 5.01 (2H; s; CH2CO), 6.72 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.74 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.91–7.88 (8H; m; phenyl protons), 8.06 (1H; s; –N=CH–), 11.36 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.4 (2C, CH2–N; b + b’), 46.5 (2C; –N(CH3)2), 48.1 (2C, CH2–N; a+a’), 52.9 (1C; –N–CH2–C=O), 112.3 (1C; =CH), 117.7 (2C; 4-chlorophenyl C2,6), 121.8 (1C; 4-dimethylaminophenyl C6), 127.1 (1C; 4-dimethylaminophenyl C2), 128.9 (2C; 4-chlorophenyl C3,5), 129.1 (2C; 4-dimethylaminophenyl C3,5), 131.1 (1C; pyridazinone C5), 145.1 (1C; 4-chlorophenyl C1), 148.2 (1C; 4-dimethylphenyl C1), 149.0 (1C; 4-dimethylphenyl C4), 150.1 (1C; 4-chlorophenyl C4), 151.9 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 163.1 (1C; CH2–N–C=O), 167.8 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 516 [M + Na]+; Anal. Calcd for C25H28ClN7O2.1/6 H2O: C, 60.42; H, 5.75; N, 19.73. Found: C, 60.03; H, 5.55; N, 20.48%.
N’-Benzylidene-2-(6-oxo-3-(4-(4-fluorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (17). White crystals (methanol/water); yield 47%; m.p. 207–208 °C; IR (ν cm−1, ATR): 3178 (N–H), 3056 (C–H aromatic), 2960 (C–H aliphatic), 1652 (C=O), 1585 (C=N), 1222 (C–N), 837 (C–F). 1H-NMR (DMSO-d6, 600 MHz): δ 3.17 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 5.06 (2H; s; CH2CO), 7.01 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.02 (1H; d; J = 4.2 Hz, pyridazinone H4), 7.06-7.72 (9H; m; phenyl protons), 8.26 (1H; s; –N=CH–), 11.66 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 49.1 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2-C=O), 115.7 (1C; =CH), 118.1 (2C; phenyl C4), 127.2 (1C; phenyl C3,5), 129.3 (1C; phenyl C2,6), 130.4 (2C; 4-fluorophenyl C3,5), 131.0 (2C; 4-fluorophenyl C2,6), 134.4 (1C; 4-fluorophenyl C3,5), 144.3 (1C; 4-fluorophenyl C1), 147.4 (1C; phenyl C1), 148.2 (1C; pyridazinone C5), 149.1 (1C; 4-fluorophenyl C4), 155.9 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 163.8 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 457 [M + Na]+; Anal. Calcd for C23H23FN6O2.1/2H2O: C, 62.29; H, 5.45; N, 18.95. Found: C, 62.21; H, 5.32; N, 18.81%.
N’-(3-M-ethylbenzylidene)-2-(6-oxo-3-(4-(4-fluorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (18). White crystals (methanol/water); yield 51%; m.p. 201–202 °C; IR (ν cm−1, ATR): 3170 (N–H), 3045 (C–H aromatic), 2988 (C–H aliphatic), 1652 (C=O), 1576 (C=N), 1264 (C–N). 1H-NMR (DMSO-d6, 600 MHz): δ 2.51 (3H; s; CH3), 3.17 (4H; t; CH2N; Hb+Hb’), 3.38 (4H; t; CH2N; Ha + Ha’), 5.07 (2H; s; NCH2CO), 7.00 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.02–7.98 (9H; m; phenyl protons and pyridazinone H5), 8.19 (1H; s; –N=CH–), 11.67 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 21.4 (1C; CH3), 46.5 (2C, CH2–N; b + b’), 49.1 (2C, CH2–N; a + a’), 55.6 (1C; –N–CH2–C=O), 111.6 (1C; =CH), 115.9 (2C; 4-fluorophenyl C2,6), 118.0 (2C; 4-fluorophenyl C3,5), 120.2 (1C; 3-methylphenyl C6), 130.4 (1C; 3-methylphenyl C2), 131.0 (1C; 4-fluorophenyl C1), 135.9 (1C; 3-methylphenyl C1), 144.1 (1C; 3-methylphenyl C3), 147.3 (2C; 3-methylphenyl C4,5), 148.2 (1C; pyridazinone C5), 149.0 (1C; 4-fluorophenyl C4), 155.9 (1C; pyridazinone C6), 158.2 (1C; pyridazinone C4), 163.8 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/API-ESMS m/z 414 [M-(F + CH3)]+; Anal. Calcd. for C24H25FN6O2.4/5 H2O: C, 61.20; H, 5.80; N, 17.84. Found: C, 60.85; H, 5.40; N, 17.84%.
N’-(3-Methoxybenzylidene)-2-(6-oxo-3-(4-(4-fluorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (19). White crystals (methanol/water); yield 48%; m.p. 202–203 °C; IR (ν cm−1, ATR): 3069 (C–H aromatic), 2960 (C–H aliphatic), 1698, 1656 (C=O), 1569 (C=N), 1234 (C–N), 1158 (C–O), 836 (C–F). 1H-NMR (DMSO-d6, 600 MHz): δ 3.17 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 3.80 (3H; s; OCH3), 5.06 (2H; s; CH2CO), 7.00 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.01–7.98 (9H; m; phenyl protons and pyridazinone H4), 8.19 (1H; s; –N=CH–), 11.67 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 49.1 (2C, CH2–N; a + a’), 53.0 (1C; –N–CH2–C=O), 55.6 (1C; –OCH3), 111.6 (1C; =CH), 115.6 (2C; 4-fluorophenyl C2,6), 118.1 (1C; 3-methoxyphenyl C6), 120.2 (1C; 3-methoxyphenyl C2), 130.4 (2C; 4-fluorophenyl C3,5), 131.0 (2C; 3-methoxyphenyl C4,5), 135.9 (1C; pyridazinone C5), 144.1 (1C; 4-fluorophenyl C1), 147.3 (1C; 3-methoxyphenyl C1), 149.0 (1C; 3-methoxyphenyl C3), 155.9 (1C; 4-fluorophenyl C4), 157.5 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 160.0 (1C; CH2–N–C=O), 168.5 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 487 [M + Na]+; Anal. Calcd for C24H25FN6O3.2/3H2O: C, 60.49; H, 5.57; N, 17.64. Found: C, 60.06; H, 5.13; N, 17.43%.
N’-(4-Methoxybenzylidene)-2-(6-oxo-3-(4-(4-chlorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (20). White crystals (methanol/water); yield 41%; m.p. 247–248 °C; IR (ν cm−1, ATR): 3065 (C–H aromatic), 2966 (C–H aliphatic), 1686 (C=O), 1570 (C=N), 1247 (C–N), 1024 (C–O), 836 (C–F). 1H-NMR (DMSO-d6, 600 MHz): δ 3.17 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 3.80 (3H; s; OCH3), 5.04 (2H; s; CH2CO), 6.91 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.92-7.96 (9H; m; phenyl protons and pyridazinone H4), 8.16 (1H; s; –N=CH–), 11.52 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 49.1 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2–C=O), 55.8 (1C; –OCH3), 114.8 (1C; =CH), 115.9 (2C; 4-fluorophenyl C2,6), 118.1 (1C; 4-methoxyphenyl C6), 127.0 (1C; 4-methoxyphenyl C2), 128.9 (2C; 4-fluorophenyl C3,5), 129.2 (2C; 4-methoxyphenyl C3,5), 131.0 (1C; pyridazinone C5), 144.1 (1C; 4-fluorophenyl C1), 147.3 (1C; 4-methoxyphenyl C1), 148.2 (1C; 4-methoxyphenyl C4), 149.1 (1C; 4-fluorophenyl C4), 155.9 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 163.5 (1C; CH2–N–C=O), 168.2 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 487 [M + Na]+; Anal. Calcd for C24H25FN6O3.2/3H2O: C, 60.49; H, 5.57; N, 17.64. Found: C, 60.05; H, 5.24; N, 17.50%.
N’-(2,4,6-Trimethoxybenzylidene)-2-(6-oxo-3-(4-(4-fluorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (21). White crystals (methanol/water); yield 44%; m.p. 275–276 °C; IR (ν cm−1, ATR): 3231 (N–H), 3085 (C–H aromatic), 2963 (C–H aliphatic), 1673, 1668 (C=O), 1595 (C=N), 1240 (C–N), 1128 (C–O), 851 (C–F). 1H-NMR (DMSO-d6, 600 MHz): δ 3.18 (4H; t; CH2N; Hb + Hb’), 3.37 (4H; t; CH2N; Ha + Ha’), 3.82 (6H; s; OCH3), 3.83 (3H; s; OCH3), 4.94 (2H; s; NCH2CO), 6.29 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.90 (1H; d; J = 4.2 Hz, pyridazinone H4), 6.91–7.64 (6H; m; phenyl protons), 8.19 (1H; s; –N=CH–), 11.26 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 49.2 (2C, CH2–N; a + a’), 52.9 (1C; –N–CH2–C=O), 55.9 (2C; –OCH3), 56.5 (2C; –OCH3), 104.2 (1C; =CH), 115.9 (2C; 4-fluorophenyl C2,6), 118.1 (2C; 2,4,6-trimethoxyphenyl C3,5), 126.9 (1C; 4-fluorophenyl C3), 127.0 (1C; 4-fluorophenyl C5), 131.0 (1C; 4-fluorophenyl C1), 139.6 (1C; 2,4,6-trimethoxyphenyl C1), 148.2 (3C; 2,4,6-trimethoxyphenyl C2,4,6), 148.9 (1C; pyridazinone C5), 155.9(1C; 4-fluorophenyl C4), 158.3 (1C; pyridazinone C6), 160.4 (1C; pyridazinone C4), 162.6 (1C; CH2–N–C=O) and 167.9 (1C; pyridazinone C3); LC/API-ESMS m/z 524 [M]+; Anal. Calcd. for C26H29FN6O5.H2O: C, 57.56; H, 5.76; N, 15.49. Found: C, 57.75; H, 5.30; N, 15.64%.
N’-(4-Nitrobenzylidene)-2-(6-oxo-3-(4-(4-fluorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (22). White crystals (methanol/water); yield 49%; m.p. 243–244 °C; IR (ν cm−1, ATR): 3077 (C–H aromatic), 2963 (C–H aliphatic), 1690, 1659 (C=O), 1580 (C=N), 1339 (aromatic NO2), 1236 (C–N), 836 (C–F). 1H-NMR (DMSO-d6, 600 MHz): δ 3.17 (4H; t; CH2N; b + b’), 3.39 (4H; t; CH2N; a + a’), 5.11 (2H; s; NCH2CO), 6.94 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.99 (1H; d; J = 4.2 Hz, pyridazinone H5), 7.00-8.39 (8H; m; phenyl protons), 8.88 (1H; s; –N=CH–), 11.96 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 49.1 (2C, CH2–N; a+a’), 53.0 (1C; –N–CH2–C=O), 115.9 (1C; =CH), 118.0 (2C; 4-fluorophenyl C2,6), 124.5 (1C; 4-nitrophenyl C6), 127.1 (1C; 4-nitrophenyl C2), 128.4 (2C; 4-fluorophenyl C3,5), 128.5 (2C; 4-nitrophenyl C3,5), 131.0 (1C; pyridazinone C5), 140.8 (1C; 4-fluorophenyl C1), 141.9 (1C; 4-nitrophenyl C1), 148.2 (1C; 4-nitrophenyl C4), 149.1 (1C; 4-fluorophenyl C4), 155.9 (1C; pyridazinone C4), 157.5 (1C; pyridazinone C6), 158.2 (1C; CH2–N–C=O), 168.9 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 414 [M-(F + NO2)]+; Anal. Calcd for C23H22FN7O4.2/3H2O: C, 56.21; H, 4.79; N, 19.95. Found: C, 56.07; H, 4.89; N, 19.63%.
N’-(4-dimethylaminobenzylidene)-2-(6-oxo-3-(4-(4-fluorophenyl)piperazin-1-yl)pyridazin-1(6H)-yl)acetohydrazide (23). White crystals (methanol/water); yield 45%; m.p. 248–249 °C; IR (ν cm−1, ATR): 3046 (C–H aromatic), 2994 (C–H aliphatic), 1646, 1612 (C=O), 1584 (C=N), 1265 (C–N), 846 (C–F). 1H-NMR (DMSO-d6, 600 MHz): δ 2.97 (6H; s; –N(CH3)2), 3.17 (4H; t; CH2N; b + b’), 3.38 (4H; t; CH2N; a + a’), 5.01 (2H; s; CH2CO), 6.75 (1H; d; J = 4.1 Hz, pyridazinone H5), 6.77 (1H; d; J = 4.2 Hz, pyridazinone H5), 6.91–7.88 (8H; m; phenyl protons), 8.06 (1H; s; –N=CH–), 11.35 (1H; s; –NH–N); 13C-NMR (DMSO-d6, 150 MHz): δ 46.5 (2C, CH2–N; b + b’), 48.4 (2C; –N(CH3)2), 49.1 (2C, CH2–N; a+a’), 52.9 (1C; –N–CH2–C=O), 112.3 (1C; =CH), 115.9 (2C; 4-fluorophenyl C2,6), 118.1 (1C; 4-dimethylaminophenyl C6), 121.8 (1C; 4-dimethylaminophenyl C2), 126.9 (2C; 4-fluorophenyl C3,5), 128.9 (2C; 4-dimethylaminophenyl C3,5), 129.9 (1C; pyridazinone C5), 145.1 (1C; 4-fluorophenyl C1), 148.2 (1C; 4-dimethylphenyl C1), 151.9 (1C; 4-dimethylphenyl C4), 155.9 (1C; 4-fluorophenyl C4), 157.5 (1C; pyridazinone C4), 158.2 (1C; pyridazinone C6), 163.1 (1C; CH2–N–C=O), 167.8 (1C; pyridazinone C3); LC/MSMS (ESI+) m/z 501 [M + Na]+; Anal. Calcd for C25H28FN7O2.1/3 H2O: C, 62.10; H, 5.98; N, 20.28. Found: C, 62.24; H, 5.80; N, 20.37%.
3.2. Cell Culture
3.2.1. Human Gingival Fibroblasts (HGFs) Culture
A total of 10 donors, periodontally and systemically healthy, subjected to the extraction of the third molar, signed the informed consent according to the Italian Legislation and in accordance with the code of Ethical Principles for Medical Research comprising Human Subjects of the World Medical Association (Declaration of Helsinki). The project obtained the approval of the Local Ethical Committee of the University of Chieti (Chieti, Italy; approval number. 1173, approved on 31 March 2016). The tissue samples were immediately placed in Dulbecco’s modified Eagle’s medium (DMEM) then rinsed in phosphate-buffered saline buffer (PBS), cut into smaller fragments and cultured in DMEM, supplemented with 10% foetal bovine serum (FBS), 1% penicillin/ streptomycin and 1% fungizone (all purchased from EuroClone, Milan, Italy). Cells were kept at 37 °C in a humidified atmosphere of 5% (v/v) CO2. After 10 days of culture fungizone was removed and cells used after 7–14 passages, as previously reported [35].
3.2.2. AGS Culture
AGS human gastric adenocarcinoma cell line (ECACC 89090402, Sigma Aldrich, Milan, Italy) was cultured in Ham’s F12 medium supplemented with 10% of FBS, 1% of penicillin/streptomycin, and 1% of L-glutamine (all purchased from EuroClone). Cell culture was kept at 37 °C in a humidified atmosphere with 5% CO2.
3.3. HGF and AGS Treatment
For each tested compound a stock solution 0.1 M in DMSO was prepared. The stock solution was then diluted in DMEM or Ham’s F12 medium (for HGFs and AGS, respectively) to obtain final solutions of 10 µM for HGFs and of 10 µM and 50 µM for AGS cells. Doxorubicin was used as a reference drug. To exclude DMSO cytotoxicity, in all the incubation media, the final concentration of DMSO was maintained at 0.2%.
The HGFs and AGS cells were seeded at 8000 cells/well in 96 well plate and the day after cell seeding the medium (DMEM and Ham’s for HGFs and AGS, respectively) was replaced by a fresh one containing compounds 1–23 at 10 µM for HGFs and at 10 µM and 50 µM for AGS. Cells were incubated up to 72 h in a humidified atmosphere of 5% (v/v) CO2 at 37 °C.
3.4. MTT Metabolic Activity Assay
After 48 and 72 h of culture of HGFs and AGS in presence of compounds 1–23 and of doxorubicin, an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) test was carried out. MTT test. The MTT test is based on the viable cells capability to reduce MTT into a violet formazan salt. At the established experimental times, the medium was replaced by a fresh one containing 10% of MTT (Sigma Aldrich, Milan, Italy) and probed with cells at 37 °C for 5 h. The plate was then incubated in DMSO for 30 min at 37 °C to allow formazan salts dissolution. The final violet solution was read at 540 nm by means of a microplate reader (Multiskan GO, Thermo Scientific, Waltham, MA, USA). Data obtained without cells were established as background. Viability level was normalized with values derived from cells treated with DMSO.
3.5. Cytotoxicity Assay (LDH Assay)
To assess membrane integrity of AGS cells, lactate dehydrogenase (LDH) leakage into the culture medium was measured by means of “CytoTox 96 nonradioactive cytotoxicity assay” (Promega, Madison, WI, USA), following the manufacturer’s instructions, after 48 and 72 of culture in presence of compounds 12 and 22. In each well, the measured LDH leakage in the supernatant was normalized to the MTT optical density values obtained from MTT test.
3.6. Light Microscopy Analysis
AGS cells cultured on Thermanox were fixed in 4% paraformaldehyde for 50 min, washed in PBS and counterstained with haematoxylin-eosin solution. Slides were washed in PBS, dehydrated through the alcohol ascending series and mounted with a permanent xylene-based balsam. The coloured slides were examined by means of Leica DM 4000 light microscope (Leica Cambridge Ltd., Cambridge, UK) equipped with a Leica DFC 320 camera (Leica Cambridge Ltd., Cambridge, UK). Computerized images were acquired with LASX image software (Leica Cambridge Ltd., Cambridge, UK).
3.7. Hydrogen Peroxide (H2O2) Release
At the established experimental times cell supernatants were collected. The quantitative measurement of H2O2 released in supernatant was carried out through the Hydrogen Peroxide colorimetric detection kit (Enzo Life Sciences, Inc., Farmingdale, NY, USA). For each sample 50 µL of supernatant/well were loaded, in duplicate, in a Half-Area Microtiter Plate, then 100 µL of Color Reagent were added in each well. The multiwell plate was shacked for 10 s and then incubated for 30 min at room temperature. The optical density (OD) was measured at 550 nm by means of a spectrophotometer (Multiskan GO; Thermo Fisher Scientific, Inc., Waltham, MA USA). The results were calculated by subtracting the average blank OD from the average OD for each sample.
3.8. Bax Immunostaining by Flow Cytometry
At the established exposure times, the incubation medium was discarded and cells were incubated with the Accutase® solution (Sigma-Aldrich, St. Louis, MO, USA) for 5 min at 37 °C and 5% CO2 for cell detachment. After that, cells were collected, centrifuged at 1200 rpm for 10 min and the obtained pellets were afterwards washed with PBS without calcium and magnesium. Next, supernatants were discarded and cell pellets were fixed for 15 min at room temperature with the PerFix-nc Buffer 1 from the commercial PerFix-nc Kit (no-centrifuge assay) (Beckman Coulter, Indianapolis, IN, USA). Fixed sample were gently vortexed after having added 100 µL/sample of the permeabilizing reagent PerFix-nc Buffer 2 (Beckman Coulter) containing 5% of FBS, and incubated for additional 20 min at room temperature. After the permeabilization, the mouse anti-Bax monoclonal antibody (sc-7480, Santa Cruz Biotechnology, Santa Cruz, CA, USA) was added in a dilution of 1:50 and samples were incubated for 1 h at 4 °C. Next, cells were centrifuged for 10 min at 4 °C, the supernatant was discarded and the secondary phycoerythrin (PE) horse anti-mouse IgG (H+L) antibody (Vector Laboratories, Inc., Burlingame, CA, USA) was added (1:50) and incubated at 4 °C in the dark for 45 min. Secondary antibody overage was removed by centrifugation and cells were suspended in 300 µL of PerFix-nc Buffer 3 1X (Beckman Coulter). Finally, 10,000 cells for each sample were run in a CytoFlex Flow cytometer (Beckman Coulter) equipped with a 488 nm laser using the FL-2 (PE) channel in a linear mode. Relative fluorescence emissions of gated cells by means of their forward and side scatter properties (FSC/SSC) were analyzed with the CytExpert Software (Beckman Coulter) and they were expressed as the percentage of positive cells for Bax-PE conjugated. Individual values obtained from three independent experiments (n = 3) were summarized as means and standard deviations.
3.9. Statistical Analysis
Statistical analysis was performed using the GraphPad 7 software (GraphPad Software, San Diego, CA, USA) by means of t-test and Ordinary One-Way ANOVA followed by post-hoc Tukey’s multiple comparisons tests. Values of p < 0.05 were considered statistically significant.
4. Conclusions
We previously investigated the anti-proliferative effects of a large number of pyridazinones against HCT116 cells [36]. To further expand the knowledge of the SARs within this scaffold, we introduced a piperazinyl linker between the pyridazinone core nucleus and the additional phenyl ring aiming at potentiating the biological activity and reducing the cytotoxicity against a non-cancerous cell line. The results of the MTT tests have been compared to those of doxorubicin as a reference drug. We selected the two best-in-class compounds 12 and 22 and better explored their mechanism of action in terms of pro-apoptotic ability. These compounds induced a change in the cell morphology, a release of oxidant hydrogen peroxide and the expression of Bax, thus confirming their anti-proliferative role against AGS cells. On the basis of these results, we were also able to update the SAR studies within this scaffold as reported in Figure 10.
Figure 10.
Update of the SARs within the pyridazinone scaffold for the design of biologically active compounds. The structural information gained in this work have been highlighted as red text.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-8247/14/3/183/s1, 1H-NMR, 13C-NMR, and mass spectra for each compound.
Author Contributions
Conceptualization, S.C., B.M., and Z.Ö.; methodology, M.A.A., M.U., A.R. and M.G.; in silico investigation, S.C.; writing and original draft preparation, S.Z., M.G., S.C. and B.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financed by intramural grants by Ministero Italiano dell’Università e della Ricerca (MIUR) FAR2019 (ex 60%), held by Simone Carradori.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ajani J.A., Lee J., Sano T., Janjigian Y.Y., Fan D., Song S. Gastric adenocarcinoma. Nat. Rev. Dis. Primers. 2017;3:17036. doi: 10.1038/nrdp.2017.36. [DOI] [PubMed] [Google Scholar]
- 2.Ilson D.H. Advances in the treatment of gastric cancer. Curr. Opin. Gastroenterol. 2018;34:465–468. doi: 10.1097/MOG.0000000000000475. [DOI] [PubMed] [Google Scholar]
- 3.Song H., Zhu J., Lu D. Molecular-targeted first-line therapy for advanced gastric cancer. Cochrane Database Syst. Rev. 2016;7:CD011461. doi: 10.1002/14651858.CD011461.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alsina M., Miquel J.M., Diez M., Castro S., Tabernero J. How I treat gastric adenocarcinoma. ESMO Open. 2019;4(Suppl. S2):e000521. doi: 10.1136/esmoopen-2019-000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sisto F., Carradori S., Guglielmi P., Traversi C.B., Spano M., Sobolev A.P., Secci D., Di Marcantonio M.C., Haloci E., Grande R., et al. Synthesis and biological evaluation of carvacrol-based derivatives as dual inhibitors of H. pylori strains and AGS cell proliferation. Pharmaceuticals. 2020;13:405. doi: 10.3390/ph13110405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marconi G.D., Carradori S., Ricci A., Guglielmi P., Cataldi A., Zara S. Kinesin Eg5 targeting inhibitors as a new strategy for gastric adenocarcinoma treatment. Molecules. 2019;24:3948. doi: 10.3390/molecules24213948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lang D.K., Kaur R., Arora R., Saini B., Arora S. Nitrogen-containing heterocycles as anticancer agents: An overview. Anticancer Agents Med. Chem. 2020;20:2150–2168. doi: 10.2174/1871520620666200705214917. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee P.S. Various Biological Activities of Pyridazinone Ring Derivatives. Asian J. Chem. 2011;23:1905–1910. [Google Scholar]
- 9.Akhtar W., Shaquiquzzaman M., Akhter M., Verma G., Khan M.F., Alam M.M. The therapeutic journey of pyridazinone. Eur. J. Med. Chem. 2016;123:256–281. doi: 10.1016/j.ejmech.2016.07.061. [DOI] [PubMed] [Google Scholar]
- 10.Yamali C., Ozan G.H., Kahya B., Çobanoğlu S., Şüküroğlu M.K., Doğruer D.S. Synthesis of some 3(2H)-pyridazinone and 1(2H)-phthalazinone derivatives incorporating aminothiazole moiety and investigation of their antioxidant, acetylcholinesterase, and butyrylcholinesterase inhibitory activities. Med. Chem. Res. 2015;24:1210–1217. doi: 10.1007/s00044-014-1205-8. [DOI] [Google Scholar]
- 11.Utku S., Gökçe M., Aslan G., Bayram G., Ülger M., Emekdaş G., Şahin M.F. Synthesis and in vitro antimycobacterial activities of novel 6-substituted-3(2H)-pyridazinone-2-acetyl-2-(substituted/nonsubstituted acetophenone)hydrazone. Turk. J. Chem. 2011;35:331–339. [Google Scholar]
- 12.Şahin M.F., Badıçoğlu B., Gökçe M., Küpeli E., Yesilada E. Synthesis and analgesic and antiinflammatory activity of methyl [6-substituted-3(2H)-pyridazinone-2-yl]acetate derivatives. Arch. Pharm. Pharm. Med. 2004;33:445–452. doi: 10.1002/ardp.200400896. [DOI] [PubMed] [Google Scholar]
- 13.Siddiqui A.A., Mishra R., Shaharyar M., Husain A., Rashid M., Pal P. Triazole incorporated pyridazinones as a new class of antihypertensive agents: Design, synthesis and in vivo screening. Bioorg. Med. Chem. Lett. 2011;21:1023–1026. doi: 10.1016/j.bmcl.2010.12.028. [DOI] [PubMed] [Google Scholar]
- 14.Siddiqui A.A., Mishra R., Shaharyar M. Synthesis, characterization and antihypertensive activity of pyridazinone derivatives. Eur. J. Med. Chem. 2010;45:2283–2290. doi: 10.1016/j.ejmech.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Mogilski S., Kubacka M., Redzicka A., Kazek G., Dudek M., Malinka W., Filipek B. Antinociceptive, anti-inflammatory and smooth muscle relaxant activities of the pyrrolo[3,4-d]pyridazinone derivatives: Possible mechanisms of action. Pharmacol. Biochem. Behav. 2015;133:99–110. doi: 10.1016/j.pbb.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 16.Saini M., Mehta D.K., Das R., Saini G. Recent advances in anti-inflammatory potential of pyridazinone derivatives. Mini-Rev. Med. Chem. 2016;16:996–1012. doi: 10.2174/1389557516666160611015815. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y.J., Lu D., Xu Y.B., Xing W.Q., Tong X.K., Wang G.F., Feng C.-L., He P.-L., Yang L., Tang W., et al. A novel pyridazinone derivative inhibits hepatitis B virus replication by inducing genome-free capsid formation. Antimicrob Agents Chemother. 2015;59:7061–7072. doi: 10.1128/AAC.01558-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacomini A.P., Silva M.J.V., Silva R.G.M., Goncalves D.S., Volpato H., Basso E.A., Paula F.R., Nakamura C.V., Sarragiotto M.H., Rosa F.A. Synthesis and evaluation against Leishmania amazonensis of novel pyrazolo[3,4-d]pyridazinone-N-acylhydrazone-(bi)thiophene hybrids. Eur. J. Med. Chem. 2016;124:340–349. doi: 10.1016/j.ejmech.2016.08.048. [DOI] [PubMed] [Google Scholar]
- 19.Rathish I.G., Javed K., Bano S., Ahmad S., Alam M.S., Pillai K.K. Synthesis and blood glucose lowering effect of novel pyridazinone substituted benzenesulfonylurea derivatives. Eur. J. Med. Chem. 2009;44:2673–2678. doi: 10.1016/j.ejmech.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Nagle P., Pawar Y., Sonawane A., Bhosale S., More D. Docking simulation, synthesis and biological evaluation of novel pyridazinone containing thymol as potential antimicrobial agents. Med. Chem. Res. 2013 doi: 10.1007/s00044-013-0685-2. [DOI] [Google Scholar]
- 21.Bruel A., Logé C., Tauzia M.L., Ravache M., Le Guevel R., Guillouzo C., Lohier J.F., Oliveira Santos J.S., Lozach O., Meijer L., et al. Synthesis and biological evaluation of new 5-benzylated 4-oxo-3,4-dihydro-5H-pyridazino[4,5-b]indoles as PI3Kα inhibitors. Eur. J. Med. Chem. 2012;57:225–233. doi: 10.1016/j.ejmech.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 22.El-Ghaffar N.F.A., Mohamed M.K., Kadah M.S., Radwan A.M., Said G.H., Abd el Al S.N. Synthesis and anti-tumor activities of some new pyridazinones containing the 2-phenyl-1H-indolyl moiety. J. Chem. Pharm. Res. 2011;3:248–259. [Google Scholar]
- 23.Özdemir Z., Utku S., Mathew B., Carradori S., Orlando G., di Simone S., Alagöz M.A., Özçelik A.B., Uysal M., Ferrante C. Synthesis and biological evaluation of new 3(2H)-pyridazinone derivatives as non-toxic anti-proliferative compounds against human colon carcinoma HCT116 cells. J. Enzym. Inhib. Med. Chem. 2020;35:1100–1109. doi: 10.1080/14756366.2020.1755670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ciftci O., Ozdemir Z., Acar C., Sözen M., Başak-Türkmen N., Ayhan İ., Gözükara H. The novel synthesized pyridazinone derivates had the antiproliferative and apoptotic effects in SHSY5Y and HEP3B cancer cell line. Lett. Org. Chem. 2018;15:323–331. doi: 10.2174/1570178614666170707154210. [DOI] [Google Scholar]
- 25.Özdemir Z., Başak-Türkmen N., Ayhan İ., Çiftçi O., Uysal M. Synthesis of new 6-[4-(2-fluorophenylpiperazine-1-yl)]-3(2H)-pyridazinone2-acethyl-2-(substitutedbenzal)hydrazone derivatives and evulation of their cytotoxic effects in liver and colon cancer cell line. Pharm. Chem. J. 2019;52:923–929. doi: 10.1007/s11094-019-01927-y. [DOI] [Google Scholar]
- 26.Firouzi Amoodizaj F., Baghaeifar S., Taheri E., Farhoudi Sefidan Jadid M., Safi M., Seyyed Sani N., Hajazimian S., Isazadeh A., Shanehbandi D. Enhanced anticancer potency of doxorubicin in combination with curcumin in gastric adenocarcinoma. J. Biochem. Mol. Toxicol. 2020;34:e22486. doi: 10.1002/jbt.22486. [DOI] [PubMed] [Google Scholar]
- 27.Utku S., Gökçe M., Aslan G., Bayram G., Ülger M., Emekdaş G., Şahin M.F. Synthesis of novel 6-substituted-3(2H)-pyridazinone-2-acetyl-2-(substituted/-nonsubstituted benzal)hydrazone derivatives and acetylcholinesterase and butyrylcholinesterase inhibitory activities in vitro. Arzneim. Forsch. 2011;61:1–7. doi: 10.1055/s-0031-1296161. [DOI] [PubMed] [Google Scholar]
- 28.Bozbey İ., Özdemir Z., Uslu H., Özçelik A.B., Şenol F.S., Orhan-Erdoğan İ., Uysal M. A Series of New Hydrazone Derivatives: Synthesis, Molecular Docking and Anticholinesterase Activity Studies. Min. Rev. Med. Chem. 2020;20:1042–1060. doi: 10.2174/1389557519666191010154444. [DOI] [PubMed] [Google Scholar]
- 29.Khairul W.M., Hashim F., Mohammed M., Shah N.S.M.N., Johari S.A.T.T., Rahamathullah R., Daud A.I., Ma N.L. Synthesis, Molecular Docking and Biological Activity Evaluation of Alkoxy Substituted Chalcone Derivatives: Potential Apoptosis Inducing Agent on MCF-7 Cells. Anticancer Agents Med. Chem. 2020 doi: 10.2174/1871520620999201110190709. [DOI] [PubMed] [Google Scholar]
- 30.Kaushal R., Kumar N., Thakur A., Nehra K., Awasthi P., Kaushal R., Arora S. Synthesis, Spectral Characterization, Antibacterial and Anticancer Activity of some Titanium Complexes. Anticancer Agents Med. Chem. 2018;18:739–746. doi: 10.2174/1871520618666171219122431. [DOI] [PubMed] [Google Scholar]
- 31.Daina A., Michielin O., Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Önkol T., Gökçe M., Orhan İ., Kaynak F. Design, synthesis and evaluation of some novel 3(2H)-pyridazinone-2-yl acetohydrazides as acetylcholinesterase and butyrylcholnesterase inhibitors. Org. Commun. 2013;6:55–67. [Google Scholar]
- 33.Özdemir Z., Yılmaz H., Sarı S., Karakurt A., Şenol F.S., Uysal M. Design, synthesis, and molecular modeling of new 3(2H)-pyridazinone derivatives as acetylcholinesterase/butyrylcholinesterase inhibitors. Med. Chem. Res. 2017;26:2293–2308. doi: 10.1007/s00044-017-1930-x. [DOI] [Google Scholar]
- 34.Stokbroekx R.A., Van der Aa M.J.M., Willems J.J.M., Luyckx M.G.M. Anti-virally active pyridazinamines. Eur. Pat. Appl. 1985 EP 156433 A2 19851002. [Google Scholar]
- 35.De Colli M., Tortorella P., Marconi G.D., Agamennone M., Campestre C., Tauro M., Cataldi A., Zara S. In vitro comparison of new bisphosphonic acids and zoledronate effects on human gingival fibroblasts viability, inflammation and matrix turnover. Clin. Oral Invest. 2016;20:2013–2021. doi: 10.1007/s00784-015-1690-2. [DOI] [PubMed] [Google Scholar]
- 36.Kumar R., Harilal S., Carradori S., Mathew B. A Comprehensive Overview of Colon Cancer- A Grim Reaper of the 21st Century. Curr. Med. Chem. 2020 doi: 10.2174/0929867327666201026143757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article.