Abstract
Triticum turgidum and T. timopheevii are two tetraploid wheat species sharing T. urartu as a common ancestor, and domesticated accessions from both of these allopolyploids exhibit nonbrittle rachis (i.e., nonshattering spikes). We previously described the loss-of-function mutations in the Brittle Rachis 1 genes BTR1-A and BTR1-B in the A and B subgenomes, respectively, that are responsible for this most visible domestication trait in T. turgidum. Resequencing of a large panel of wild and domesticated T. turgidum accessions subsequently led to the identification of the two progenitor haplotypes of the btr1-A and btr1-B domesticated alleles. Here, we extended the haplotype analysis to other T. turgidum subspecies and to the BTR1 homologues in the related T. timopheevii species. Our results showed that all the domesticated wheat subspecies within T. turgidum share common BTR1-A and BTR1-B haplotypes, confirming their common origin. In T. timopheevii, however, we identified a novel loss-of-function btr1-A allele underlying a partially brittle spike phenotype. This novel recessive allele appeared fixed within the pool of domesticated Timopheev’s wheat but was also carried by one wild timopheevii accession exhibiting partial brittleness. The promoter region for BTR1-B could not be amplified in any T. timopheevii accessions with any T. turgidum primer combination, exemplifying the gene-level distance between the two species. Altogether, our results support the concept of independent domestication processes for the two polyploid, wheat-related species.
Keywords: brittle rachis, domestication, haplotype analysis, progenitor, wild emmer wheat, Timopheev’s wheat
1. Introduction
Originating via hybridization among different progenitor species, allopolyploid wheat species are widespread due to their enhanced capacity for genetic adaptation relative to their diploid ancestors [1]. Triticum urartu [AA genome] underwent at least two independent spontaneous hybridizations with likely extinct forms of Aegilops speltoides Tausch (SS genome), one associated with the formation of T. turgidum ((2n = 4× = 28), BBAA genome) [2,3], and the other with the formation of T. timopheevii ((2n = 4× = 28), GGAtAt genome) [4,5,6], also known as Timopheev’s wheat.
As the common ancestor of all economically important domesticated wheat species, T. turgidum drew much scientific attention; the genetic and morphological characteristics of this polyploid lineage were intensely investigated in an effort to understand the process of wheat evolution and domestication. A breakthrough in this regard was the scientific discovery of “wild emmer wheat” (WEW), T. turgidum L. subsp. dicoccoides (Korn. Ex Asch. & Graebn. Thell.) by Aaronsohn more than a century ago [7]. The sustained cultivation of T. turgidum in the Fertile Crescent is linked to the subsequent emergence of “domesticated emmer wheat” (DEW), T. turgidum L. subsp. dicoccum (Schrank ex Schübl.) Thell., the domesticated form of wild emmer wheat. Possessing nonbrittle spikes but tough glumes, domesticated emmer served as an important evolutionary step toward the development of the various fully domesticated (free-threshing) tetraploid wheat subspecies: Durum wheat (DW—T. turgidum L. subsp. durum (Desf.) Husn.), rivet wheat (TRG—T. turgidum L. subsp. turgidum (Desf.) Husn.), Khorasan wheat (TRN—T. turanicum), Polish wheat (POL—T. polonicum), Persian wheat (CRT—T. carthlicum), and Georgian emmer (PLC—T. paleocolchicum). Whether they evolved from a single, common, domesticated emmer ancestor or emerged independently from separate wild or domesticated emmer accessions is still unknown [8]. However, as members of the T. turgidum lineage, they all share the same BBAA genomic composition.
In contrast to T. turgidum, T. timopheevii received far less scientific attention. Thought to be domesticated later than emmer [9], T. timopheevii (Zhuk.) Zhuk. subsp. timopheevii is the domesticated form (“domesticated Timopheev’s wheat”—DTW or domesticated timopheevii) of the wild T. timopheevii (Zhuk.) Zhuk. subsp. araraticum Jakubz (“wild Timopheev’s wheat—WTW or T. araraticum), both possessing an GGAtAt genomic composition. Geographically, T. araraticum grows from Armenia to Azerbaijan but also overlaps with wild emmer populations in southeastern Turkey, northern Iraq, and western Iran. In contrast, domesticated timopheevii is an endemic crop exclusively of western Georgia, to the north [9,10]. Using chloroplast DNA fingerprinting, the closest genetic similarity of T. araraticum accessions to domesticated timopheevii types was found among wild accessions collected from southern Turkey and northern Syria, implying T. timopheevii domestication might have taken place in these areas [9]. Archaeobotanical findings of early agrarian settlements discovered a different type of cultivated wheat other than domesticated emmer called “new glume wheat” (NGW), suggested to be related to T. timopheevii [11]. A recent study traced sequences from the G genome in NGW samples that were present across western Asia and Europe in the Neolithic and Bronze ages, indicating a wider spread than first thought and suggesting that domesticated timopheevii is of major importance to prehistoric Eurasian agriculture and not a just a minor crop restricted to western Georgia [12]. Supporting this, it was recently claimed that domestication of NGW took place in Transcaucasia and Anatolia in separate pathways [13], indicating the importance and widespread of the timopheevii species.
Although crosses between T. turgidum and T. timopheevii yielded F1 progeny, stable hybrid lines were not obtained, presumably due to failures in chromosome pairing, leading to infertility [4,14]. However, evidence of relatively good chromosome pairing in the F1 hybrids in some T. timopheevii × T. turgidum combinations were also reported [15]. Genome-wide variation between the two species was first demonstrated on the basis of large “species founder translocations” involving chromosomes 4A, 5A, 6A, 7B, 1G, and 4G [16,17]. Despite these significant distinctions between their genomes, T. timopheevii and T. turgidum share high homology with each other [18]. Indeed, cytogenetic examination suggests that genes can be transferred successfully between T. turgidum and T. timopheevii via direct crosses, though likely with a low success rate [19]. A more recent study, based on GBS data, supported the conclusion that T. turgidum and T. timopheevii are indeed distinct species [8], confirming the conclusions of earlier studies that the T. turgidum species evolved earlier [20].
Despite much progress in understanding the origin and evolution of T. timopheevii, the specific processes that shaped its domestication are still unknown. The hallmark trait of wheat domestication and arguably the most essential morphological change associated with this process is the transition from a brittle (shattering) to a nonbrittle (nonshattering) rachis [21]. During the domestication of T. turgidum, domesticated emmer acquired two recessive loss-of-function mutations in the A and B genome copies of the Brittle Rachis 1 (BTR1-A and BTR1-B), resulting in a nonshattering (i.e., intact, harvestable) spike [22]. The causative mutation in BTR1-A is a 2 bp deletion in the coding sequence which causes a loss-of-function frame shift. In BTR1-B, loss of function is due to a 4 kbp insertion, 50 bp upstream of the stop codon.
In a previous study, we resequenced the BTR1-A and BTR1-B regions in a wide panel of wild and domesticated tetraploid wheat accessions and identified wild emmer individuals that represent likely progenitors (“founder stocks”) of domesticated emmer, with its stacked, nonfunctional btr1-A and btr1-B haplotypes [23]. Since mutations in the BTR1 genes are associated with cereal domestication [21,22,24] we hypothesize that T. timopheevii was also domesticated through mutations in BTR1 genes, but should carry distinct, novel BTR1-A and BTR1-G mutated alleles. To shed light on the presumably analogous process of T. timopheevii domestication, we present here an examination of the sequence variation associated with spike shattering (i.e., the brittle rachis trait) in various wild and domesticated T. timopheevii and T. turgidum accessions. Our results showed that domesticated timopheevii carries a novel btr1-A allele whose causative mutation is different from that underlying domesticated T. turgidum accessions. Using T. turgidum primers, the BTR1-G promoter region could not be amplified in any T. timopheevii accession, further illustrating the completely different origin of the two species and specifically the domesticated alleles in these different genomes.
2. Materials and Methods
2.1. Plant Material and Phenotyping
We evaluated a total of 57 accessions investigated in this study, 34 from the T. timopheevi species and 23 from the T. turgidum species (Table S1). Within T. timopheevi, 32 accessions are wild (T. araraticum) and 2 are domesticated (DTW). Within the T. turgidum accessions were representatives from six subspecies, including domesticated emmer (6), T. carthlicum (5), T. turanicum (4), T. turgidum L. subsp. turgidum (4), T. polonicum (3), and T. paleocolchicum (1). In addition, the well-characterized wild emmer accession “Zavitan” [25] and durum wheat cv. “Svevo” were used as references. All accessions were grown in a greenhouse in four-liter pots at Tel Aviv University from January to May 2020. Based on five mature, senesced spikes from each accession, a visual assessment of brittle rachis phenotype was made according to three categories: (1) brittle—the spike completely shattered when harvested; (2) semi-brittle—only the upper half of the spike separated with a slight touch while the lower half remained intact; and (3) nonbrittle—the complete spike remained intact when harvested.
2.2. Amplification and Sequencing of the BTR1-A and BTR1-B Gene Regions
Using the methods described by [23], PCR amplification of the BTR1-A gene region was carried out using the forward primer 5′-TTGCTGTTGACAAAGGCCAG-3′ (located 93 bp upstream of the start codon) and the reverse primer 5′-TTTTCTCGTTCGCTACCACAC-3′ (located 912 bp downstream from the stop codon). Due to low sequencing quality at the 3′ end of the resulting 1596 bp amplicon, we developed and used an alternative reverse primer, 5′-TCGGGAGCTCATTTGACCTT-3′, located only 734 bp downstream from the stop codon. Amplification of the BTR1-B and BTR1-G gene regions was attempted with the same primers and PCR conditions were described by [23], encompassing a 2447 bp region starting 1856 bp upstream of the BTR1-B start codon and including 483 bp of the BTR1-B coding sequence. Despite designing 25 different primers specific to the promoter and the coding sequence of BTR1-B (Table S2), only the T. turgidum accessions (BTR1-B) were successfully amplified under these conditions.
2.3. BTR1-A/btr1-A Marker Development
To discriminate between wild and domesticated T. timopheevii alleles of BTR1-A, we developed a cleaved amplified polymorphic sequence (CAPS) marker using the forward primer 5′-GTCCGGTTCATGCTTCACAG-3′ and the reverse primer 5′-TGCCAATGTACGTTGCAAGT-3′. The 456 bp amplicon was digested with AciI restriction enzyme (New England Biolabs, Ipswich, MA, USA) and the resulting fragments were imaged via electrophoresis (2% agarose gel) to distinguish between the wild (456 bp) and domesticated (148 + 307 bp) T. timopheevii alleles.
2.4. Sequencing, Phylogenetic, Haplotypic, and Functional Analyses
Sequencing of the target regions was obtained using ABI 3500xl Genetic analyzer (Applied Biosystems, Foster City, CA, USA). This method of sequencing utilizes the BigDye Terminator Cycle Sequencing Kit (Thermo Fisher Scientific, Waltham, MA, USA) with fluorescent dye attached to each of the dideoxy terminators.
Amplified sequences of the two targeted regions were imported into Sequencher v5.4 (www.genecodes.com) and low-quality reads were manually removed. Sequence alignments were generated using ClustalW within MEGA 6 [26] and haplotypes were defined using DnaSP 5.10.01 [27]. Orthologous sequences from the T. urartu reference genome [28] were used as outgroups. Phylogenetic trees were constructed based on the neighbor-joining method using MEGA 6 [26], assuming uniform rates among sites. A bootstrap analysis (1000 replicates) was performed to provide confidence estimates for branch nodes. Translation to peptides based on the coding sequence of each accession was done with EMBOSS Transeq © EMBL 2020 (https://www.ebi.ac.uk/Tools/st/emboss_transeq/ (accessed on 31 January 2021)).
3. Results
3.1. BTR1-A Haplotype Analysis
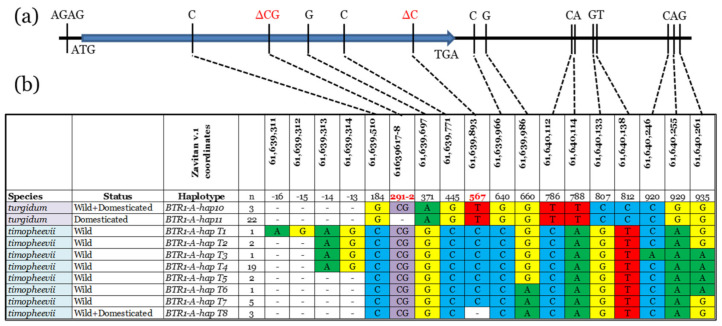
High-quality sequence data of the BTR1-A region were obtained from 1244 bp amplicons, starting 27 bp upstream of the start codon and ending 627 bp downstream from the stop codon (chr. 3A 61,639,300–61,640,544, [22]), a region that overlaps with the one described by [23]. In addition to the previously described 2 bp frameshift deletion within the coding sequence (position 291–292), which separates wild from domesticated wheat [22], our analysis identified three other variants within the BTR1-A coding region, another 1 bp frameshift deletion (position 567), as well as nine SNPs located downstream from the coding region (Figure 1a).
Figure 1.
(a) Schematic showing the locations of the sequence polymorphisms used in the BTR1-A haplotype analysis. Highlighted in red font are two frameshift deletions in the BTR1-A coding sequence. (b) Among the materials included in this study, a total of 10 BTR1-A haplotypes were identified, two within the T. turgidum species and eight within the T. timopheevii species. In T. timopheevii, BTR1-A-hapT7 is the closest wild haplotype to the domesticated haplotype BTR1-A-hap T8, differing only in the 1 bp frameshift deletion at position 567. The “Status” column refers to the historical domestication status according to the relevant seed source. Colors help to detect the differences.
This set of polymorphisms divided the studied accessions into ten haplotypic groups, two belonging to T. turgidum accessions and eight representing T. timopheevii accessions (Figure 1b). The Zavitan and Svevo sequences were identical to their corresponding reference genomes [22,29], and we used the same haplotype terminology as in Nave et al. [23], namely, BTR1-A-hap10 and BTR1-A-hap11, to describe the Zavitan (wild emmer) and Svevo (durum wheat) haplotypes, respectively. Nearly all of the domesticated T. turgidum accessions carried the BTR1-A-hap11 haplotype, except for DEW–10489 and DEW–10516, two accessions found to carry the wild-type BTR1-A-hap10 haplotype (in combination with the loss-of-function btr1-B allele). Accession no. DEW–10489 originated from Jordan and exhibits some wild emmer characteristics (semi-brittle spike, wild emmer spike shape, and spikelet hairiness), calling into question its original classification by the genebank as domesticated emmer. Originating from Yemen, accession DEW–10516 possesses the typical domesticated emmer morphology, including a nonbrittle spike.
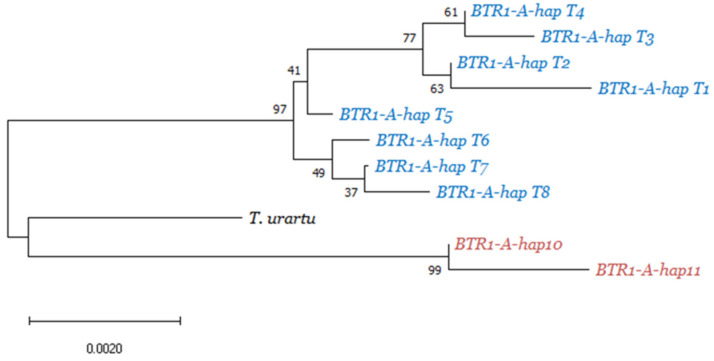
We identified eight distinct haplotypes among the T. timopheevii accessions (BTR1-A-hapT1 to T8), with 31 of the 32 T. araraticum accessions belonging to the first seven (BTR1-A-hapT1 to T7). Among the accessions included in this study, the largest T. araraticum haplotypic group is BTR1-A-hapT4, comprised of 19 accessions of broad geographic provenance, spanning Armenia, Azerbaijan, Iraq, and Iran (Table S1). Haplotypes BTR1-A-hapT1, T2, T3, T5, and T6 are each represented by only one or two T. araraticum accessions, while BTR1-A-hapT7 is carried by five T. araraticum accessions, all originating from Iraq. All 31 T. araraticum accessions from the first seven haplotypic groups exhibit a clear brittle rachis phenotype, as expected from wild plants. In contrast, the single T. araraticum from Iraq (WTW–102) carrying the BTR1-A-hapT8 haplotype exhibits a semi-brittle rachis phenotype. Also in this haplotypic group are the two domesticated timopheevii accessions, with their nonbrittle rachis spikes. The BTR1-A-hapT8 haplotype is distinguished by a cytosine deletion at position 567 of the coding sequence, 22 bp upstream of the stop codon. This single bp frameshift mutation is the only polymorphism that differentiates BTR1-A-hapT8 from its most closely related haplotype BTR1-A-hapT7 (Figure 1b), and we verified the deletion using a CAPS marker. A phylogenetic tree based on these results (Figure 2) showed a clear separation of the T. turgidum and T. timopheevi species, as well as two separate branches within each.
Figure 2.
Phylogram based on BTR1-A variants showing clear differentiation between the T. turgidum (red) and T. timopheevii (blue) species, relative to the diploid T. urartu paraphyletic outgroup. The phylogram illustrates the close relationship between T. araraticum haplotype BTR1-A-hapT7 and domesticated timopheevii haplotype BTR1-A-hapT8. Local bootstrap values after 1000 replicates are indicated at the nodes.
From analysis of the translated coding sequences of the BTR1-A haplotypes, we identified three amino acid polymorphisms (positions 62, 124 and 149) between the T. timopheevii accessions and the wild emmer Zavitan reference. For BTR1-A-hapT8, carrying the unique cysteine deletion, all seven amino acids (positions 190–197) downstream of the frameshift were different, compared with other haplotypes.
3.2. BTR1-B Haplotype Analysis
We used a similar approach to study the haplotypic variation of the Btr1-B and Btr1-G genes. In agreement with previous analyses, all domesticated T. turgidum accessions were found to carry BTR1-B-hap8, the domesticated haplotype described by Nave et al. [23]. Unlike the BTR1-A case, none of the T. araraticum or domesticated timopheevii accessions could be amplified for the BTR1-G promoter region using any primer combinations based on available T. turgidum references (Table S2). However, most of the coding sequence region was successfully amplified from T. timopheevi accessions using the forward primer 5′-CGCAATGGAAGAAGATGTACCA-3′ (located 14 bp upstream of the start codon in T. turgidum) and various reverse primers (Table S2). Since the Btr1-B region in T. turgidum is a complex of gene duplication clusters (with four Btr1-B copies) [22,29]; given the partial sequence data obtained for the T. timopheevi accessions, misidentifying the correct Btr1-B ortholog was possible. Therefore, in this study, the haplotype analysis was exclusively focused on the BTR1-A gene for the T. timopheevi accessions.
4. Discussion
Haplotype analyses of the BTR1-A gene region reconfirmed the clear separation between the T. turgidum and the T. timopheevii at the gene-level, in agreement with previous observations of the large genetic distance between them [5,8,9,20]. Consistent with this observation, the lack of consistent amplification for the BTR1 locus from T. timopheevii G subgenome likewise supported the significant genetic distance between T. turgidum and T. timopheevii. New insight into the comparative histories of domestication within these lineages was afforded by the BTR1-A haplotype analysis, however. Specifically, the causative mutation in BTR1-A that underlies the domestication syndrome of emmer was discovered to be different from that carried by domesticated accessions of Timopheev’s wheat, a result which suggests independent domestication pathways for the two species.
4.1. The Triticum Turgidum Lineage
Previous haplotype analyses [22,23] suggested that the recessive, loss-of-function btr1-A and btr1-B alleles are fixed in all domesticated emmer accessions. Here, however, we discovered two domesticated emmer accessions (DEW–10489 and DEW–10516) carrying the wild-type Btr1-A allele (BTR1-A-hap10) in combination with the domesticated btr1-B allele. BTR1-A-hap10 found in these two accessions and shared by some wild emmer accessions was identified to be the closest haplotype to BTR1-A-hap11, the haplotype carried by all other domesticated emmer lines studied to date. BTR1-A-hap10 is the basal haplotype present in most wild emmer populations from both Northern and Southern Levant [23]; therefore, it is possible that accessions DEW–10489 and DEW–10516 represent intermediate accessions that acquired only the loss-of-function allele of the BTR1-B gene (btr1-B), but not the causative mutation of BTR1-A (btr1-A).
Such accessions might thus be viewed as “missing links” in emmer wheat domestication, exhibiting phenotypes that fall intermediately along the domestication spectrum depending on their overall, genome-wide loads of domesticated alleles. Understood in this way, the intermediate brittle-rachis phenotype of DEW–10489 and the apparently nonbrittle phenotype of DEW–10516 may represent different stages of nonbrittle rachis allele fixation during the process of domestication. One alternative explanation is that these accessions could instead be progeny from spontaneous crosses between wild and domesticated accessions. Under this scenario, we hypothesize that accession DEW–10489 may represent a progeny of such a cross that was not (spontaneously) backcrossed to domesticated wheat, or lost many domestication-related alleles, hence retaining some characteristics of wild emmer. In contrast, accession DEW–10516 may represent a hybrid progeny that somehow was backcrossed to domesticated emmer (once or more), hence the typical domesticated phenotype. Further study is needed to clarify why the effect of the active Btr1-A allele on rachis brittleness was not detected in DEW–10516.
Except for accessions DEW–10489 and DEW–10516, all tested T. turgidum accessions from the six domesticated subspecies (DEW, CRT, TRN, TRG, POL, and PLC) were found to carry the domesticated haplotypes in both the A and B subgenomes (BTR1-A-hap11/BTR1-B-hap8) that contain the loss-of-function alleles of the BTR1-A and BTR1-B genes. These results strengthen the conclusion that domesticated accessions within the T. turgidum lineage have a monophyletic origin [22,23].
4.2. Triticum Timopheevii Domestication
Haplotype analysis of wheat domestication gene BTR1-A revealed seven T. araraticum haplotypes (BTR1-A-hapT1 to T7) and one haplotype (BTR1-A-hapT8) carried by all domesticated accessions and one wild accession (WTW–102). The single bp frameshift mutation found exclusively in the coding sequence of the BTR1-A-hapT8 haplotype is responsible for changing seven amino acids in the C-terminal end of the protein. Such a modification is expected to alter the function of the protein [30], potentially contributing to the nonbrittle rachis phenotype exhibited by the domesticated timopheevii accessions in this study. The existence of two independent, functional mutations in the BTR1-A genes within the T. turgidum and T. timopheevii species supports that the turgidum and timopheevii domestication processes were distinct from one another and occurred post polyploidization.
Observed in 19 out of 32 T. araraticum accessions, BTR1-A-hapT4 is suggested by its abundance and wide geographic distribution to be the most ancestral (basal) T. araraticum haplotype. A majority of the T. araraticum accessions in this work were collected from Iraq (21 out of 32), and most of the haplotypes contained Iraqi representatives (six out of seven), the latter point suggesting that this region may be the center of diversity for T. timopheevii. Moreover, BTR1-A-hapT7 (Figure 2), the closest wild haplotype to the domesticated BTR1-A-hapT8 haplotype, consists only of T. araraticum accessions of Iraqi provenance. These results suggest that the T. araraticum founder stock carried the BTR1-A-hapT7 haplotype and pinpoints Iraq as the likely place of T. timopheevii domestication, conflicting with previous results that point to northern Syria and southern Turkey as the domestication regions [9], but supporting relatively new evidence of a larger geographical distribution of domesticated timopheevii than previously thought [12,13].
We failed to amplify the BTR1-G promoter region from any T. timopheevi accession, suggesting that this gene is highly polymorphic between the T. turgidum and the T. timopheevii species. Aegilops speltoides is the G-genome donor for the formation of T. timopheevii, and this wild species carries an intact Btr1 homolog [31], implying that Btr1 may have mutated in T. timopheevii after polyploidization. Because a reference genome of at least one T. timopheevii accession is likely required to resolve this point, we focused here only on the BTR1-A gene.
In addition to the strongly determinant BTR1-A and BTR1-B genes, the nonbrittle rachis phenotype in T. turgidum lineage is associated with several other smaller effect loci (QTLs) as well [22], something that is also likely the case in the T. timopheevii lineage. Accordingly, the nonbrittle phenotypes of DTW–2729 and DTW–2804 may be explained by the combination of the domesticated BTR1-A-hapT8 haplotype with as-yet unidentified mutation(s) in the At and/or G subgenomes of domesticated timopheevii. We suggest that the semi-brittle accession WTW–102 from Iraq, also carrying the BTR1-A-hapT8 haplotype, may represent a missing link in T. timopheevii domestication, analogous to DEW–10489 and DEW–10516 within the T. turgidum lineage.
5. Conclusions
This is the first report of the likely nonfunctional btr1-A allele that contributes to the domesticated nonbrittle phenotype in T. timopheevii, noteworthy due to its difference from the domesticated T. turgidum loss-of-function btr1-A allele. While extended haplotype analysis confirmed the clear distinction between the T. timopheevii and T. turgidum lineages, it is the unique single bp frameshift in domesticated timopheevii that suggests an independent domestication process for Timopheev’s wheat. In seeking to understand the domestication processes for these two lineages, we presented the concept of “missing link” accessions that carry only one loss-of-function allele and exhibit intermediate brittle rachis phenotypes. Our results point to Iraq as an important center of divergence for T. timopheevii and the region where domestication probably occurred. We also found that all six domesticated T. turgidum subspecies carry the same BTR1-A and BTR1-B haplotypes, suggesting a shared domestication pathway.
Acknowledgments
We would like to thank Valentina Portnoy for her excellent technical assistance with the experiments.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/12/3/338/s1: Table S1: Distribution and phenotypic assessment of the 57 genotypes used for haplotype analysis; Table S2: List of primers designed to amplify the promoter and the coding sequence of BTR1-B.
Author Contributions
Conceptualization: H.O., J.P., T.K. and A.D.; formal analysis, M.N., M.T., J.R., V.K.T. and A.D.; funding acquisition, J.P., I.H. and A.D.; project administration, A.D.; supervision, A.D.; writing—original draft, M.N., I.H. and A.D.; writing—review and editing, all authors. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the United States—Israel Binational Science Foundation (BSF grant 2015409), the US-Israel Binational Agricultural Research and Development Fund (BARD project No. IS-5188-19), and the Israel Science Foundation (ISF grant 1137/17).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or supplementary material.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dubcovsky J., Dvorák J. Genome plasticity a key factor in the success of polyploid wheat under domestication. Science. 2007;316:1862–1866. doi: 10.1126/science.1143986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ogihara Y., Tsunewaki K. Diversity and evolution of chloroplast DNA in Triticum and Aegilops as revealed by restriction fragment analysis. Theor. Appl. Genet. 1988;76:321–332. doi: 10.1007/BF00265331. [DOI] [PubMed] [Google Scholar]
- 3.Dvořák J., di Terlizzi P., Zhang H.B., Resta P. The evolution of polyploid wheats: Identification of the A genome donor species. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- 4.Wagenaar E.B. Studies on the genome constitution of Triticum timopheevi zhuk. ii. The T. timopheevi complex and its origin. Evolution. 1966;20:150–164. doi: 10.1111/j.1558-5646.1966.tb03351.x. [DOI] [PubMed] [Google Scholar]
- 5.Matsuoka Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011;52:750–764. doi: 10.1093/pcp/pcr018. [DOI] [PubMed] [Google Scholar]
- 6.Mosulishvili M., Bedoshvili D., Maisaia I. A consolidated list of Triticum species and varieties of Georgia to promote repatriation of local diversity from foreign genebanks. Ann. Agrar. Sci. 2017;15:61–70. doi: 10.1016/j.aasci.2017.02.006. [DOI] [Google Scholar]
- 7.Aaronsohn A. Agricultural and Botanical Explorations in Palestine. US Government Printing Office; Washington, DC, USA: 1910. pp. 1–63. Bureau Plant Industry Bulletin, U.S.D.A. [Google Scholar]
- 8.Oliveira H.R., Jacocks L., Czajkowska B.I., Kennedy S.L., Brown T.A. Multiregional origins of the domesticated tetraploid wheats. PLoS ONE. 2020;15:e0227148. doi: 10.1371/journal.pone.0227148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori N., Kondo Y., Ishii T., Kawahara T., Valkoun J., Nakamura C. Genetic diversity and origin of timopheevi wheat inferred by chloroplast DNA fingerprinting. Breed. Sci. 2009;59:571–578. doi: 10.1270/jsbbs.59.571. [DOI] [Google Scholar]
- 10.Zohary D., Hopf M. Domestication of Plants in the Old World. 3rd ed. Oxford University Press; New York, NY, USA: 2000. p. 316. [Google Scholar]
- 11.Jones G., Valamoti S., Charles M. Early crop diversity: A “new” glume wheat from northern Greece. Veget. Hist. Archaebot. 2000;9:133–146. doi: 10.1007/BF01299798. [DOI] [Google Scholar]
- 12.Czajkowska B.I., Bogaard A., Charles M., Jones G., Kohler-Schneider M., Muller-Bieniek A., Brown T.A. Ancient DNA typing indicates that the “new” glume wheat of early Eurasian agriculture is a cultivated member of the Triticum timopheevii group. J. Archaeol. Sci. 2020;123:105258. doi: 10.1016/j.jas.2020.105258. [DOI] [Google Scholar]
- 13.Ulaş B., Fiorentino G. Recent attestations of “new” glume wheat in Turkey: A reassessment of its role in the reconstruction of Neolithic agriculture. Veget. Hist. Archaeobot. 2020:1–17. doi: 10.1007/s00334-020-00807-w. [DOI] [Google Scholar]
- 14.Maan S.S. Cytoplasmic and cytogenetic relationships among tetraploid Triticum species. Euphytica. 1973;22:287–300. doi: 10.1007/BF00022637. [DOI] [Google Scholar]
- 15.Maestra B., Naranjo T. Structural chromosome differentiation between Triticum timopheevii and T. turgidum and T. aestivum. Theor. Appl. Genet. 1999;98:744–750. doi: 10.1007/s001220051130. [DOI] [Google Scholar]
- 16.Naranjo T. Chromosome structure of durum wheat. Theor. Appl. Genet. 1990;79:397–400. doi: 10.1007/BF01186085. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J., Gill B.S. Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploid wheats. Chromosome Res. 1994;2:59–64. doi: 10.1007/BF01539455. [DOI] [PubMed] [Google Scholar]
- 18.Gill B.S., Chen P.D. Role of cytoplasm-specific introgression in the evolution of the polyploid wheats. Proc. Natl. Acad. Sci. USA. 1987;84:6800–6804. doi: 10.1073/pnas.84.19.6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gill B.S., Friebe B. Cytogenetics, Phylogeny and Evolution of Cultivated Wheats. FAO; Rome, Italy: 2002. [(accessed on 22 January 2021)]. Available online: http://www.fao.org/3/y4011e/y4011e07.htm. [Google Scholar]
- 20.Wang G.Z., Miyashua N.T., Tsunewaki K. Plasmon analyses of Triticum (wheat) and Aegilops: PCR-single-strand conformational polymorphism (PCR-SSCP) analyses of organellar DNAs. Proc. Natl. Acad. Sci. USA. 1997;94:14570–14577. doi: 10.1073/pnas.94.26.14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pourkheirandish M., Dai F., Sakuma S., Kanamori H., Distelfeld A., Willcox G., Kawahara T., Matsumoto T., Kilian B., Komatsuda T. On the Origin of the Non-brittle Rachis Trait of Domesticated Einkorn Wheat. Front. Plant Sci. 2018;8:2031. doi: 10.3389/fpls.2017.02031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avni R., Nave M., Barad O., Baruch K., Twardziok S.O., Gundlach H., Hale I., Mascher M., Spannagl M., Wiebe K., et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017;357:93–97. doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- 23.Nave M., Avni R., Çakır E., Portnoy V., Sela H., Pourkheirandish M., Ozkan H., Iago H., Komatsuda T., Dvorak J., et al. Wheat domestication in light of haplotype analyses of the Brittle rachis 1 genes (BTR1-A and BTR1-B) Plant Sci. 2019;285:193–199. doi: 10.1016/j.plantsci.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 24.Pourkheirandish M., Hensel G., Kilian B., Senthil N., Chen G., Sameri M., Azhaguvel P., Sakuma S., Dhanagond S., Sharma R., et al. Evolution of the Grain Dispersal System in Barley. Cell. 2015;162:527–539. doi: 10.1016/j.cell.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Avni R., Nave M., Eilam T., Sela H., Alekperov C., Peleg Z., Dvorak J., Korol A., Distelfeld A. Ultra-dense genetic map of durum wheat × wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol. Breed. 2014;34:1549–1562. doi: 10.1007/s11032-014-0176-2. [DOI] [Google Scholar]
- 26.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Librado P., Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 28.Ling H.Q., Ma B., Shi X., Liu H., Dong L., Sun H., Cao Y., Gao Q., Zheng S., Li Y., et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature. 2018;557:424–428. doi: 10.1038/s41586-018-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maccaferri M., Harris N.S., Twardziok S.O., Pasam R.K., Gundlach H., Spannagl M., Ormanbekova D., Lux T., Prade V.M., Milner S.G., et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat. Genet. 2019;51:885–895. doi: 10.1038/s41588-019-0381-3. [DOI] [PubMed] [Google Scholar]
- 30.Sharma S., Schiller M.R. The carboxy-terminus, a key regulator of protein function. Crit. Rev. Biochem. Mol. Biol. 2019;54:85–102. doi: 10.1080/10409238.2019.1586828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng X., Mishina K., Jia J., Distelfeld A., Maughan P.J., Kikuchi S., Sassa H., Komatsuda T. The brittle rachis trait in species belonging to the Triticeae and its controlling genes Btr1 and Btr2. Front. Plant Sci. 2020;11:1000. doi: 10.3389/fpls.2020.01000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article or supplementary material.