Abstract
We applied whole genome sequencing to identify putative transmission clusters among clinical multidrug-resistant Escherichia coli sequence type 131-H30 isolates from 4 United States children’s hospitals. Of 126 isolates, 17 were involved in 8 putative transmission clusters; 4 clusters showed evidence of healthcare-associated epidemiologic linkages. Geographic clustering analyses showed weak geographic clustering.
Keywords: antimicrobial resistance, E coli infections, ST131
Extraintestinal pathogenic Escherichia coli (ExPEC) causes a wide range of nonintestinal illnesses, ranging from uncomplicated urinary tract infection to fatal bacteremia [1]. The widespread dissemination of antimicrobial-resistant lineages such as sequence type (ST) 131-H30 (also known as ST131 clade C), a dominant multidrug-resistant ExPEC lineage in both adults and children, has brought new interest to understanding the transmission dynamics of these common pathogens [2, 3]. While the majority of ST131-H30 infections in both adults and children are characterized as healthcare-associated according to history of healthcare exposure [3, 4], the frequency of healthcare acquisition vs community acquisition remains poorly defined.
The transmission dynamics of ST131-H30 (hereafter, H30) are challenging to study because, like other ExPEC lineages, long-term intestinal colonization likely results in numerous “silent” transmission events [5]. Additionally, the role of acquisition via contaminated common sources remains inadequately understood [6]. Whole genome sequencing (WGS) can shed light on pathogen transmission dynamics despite these complexities. Here, we used WGS to identify putative transmission clusters among passively collected clinical H30 isolates from 4 children’s hospitals across the United States (US). We also quantified genomic evidence of geographic clustering to characterize the spatiotemporal dynamics of H30 among children.
METHODS
Strain Collection and Whole Genome Sequencing
All isolates and clinical data came from a previously described multicenter case-control study [3, 7]. Between September 1, 2009 and September 30, 2013, 4 freestanding children’s hospitals—referred to as “West,” “Midwest 1,” “Midwest 2,” and “East”—collected clinical E coli from urine or other normally sterile sites from inpatient and outpatient individuals < 22 years old. All extended-spectrum cephalosporin-resistant and a subset of extended-spectrum cephalosporin-sensitive isolates were collected. The institutional review board at each hospital approved the study protocol. H30 isolates were identified using fumC/fimH genotyping [8]; only the first H30 isolate per individual was included.
All H30 isolates underwent WGS via Illumina NextSeq. Sequencing reads were quality filtered, mapped to an H30 reference genome (EC958), and single-nucleotide variants (SNVs) were called and filtered for quality and suspected recombination [9]. Filtered SNVs were used to construct a pairwise SNV distance matrix. See Supplementary Methods for more details. Sequence data are available from the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJNA578285 (see Supplementary Table 3 for sample metadata).
Identification and Characterization of Putative Clusters
Pairwise SNV distances within and between collection sites were visualized and the minimum SNV distance between 2 isolates from discordant sites was selected as a threshold for putative transmission clusters. Given the substantial geographic distance between the collection sites, we expect no recent person-to-person transmission events that span 2 collection sites. The transcluster package in R (version 3.5.1, R Core Team, 2018) was used to estimate transmission events separating isolates in putative clusters [10], assuming person-to-person transmission. See Supplementary Methods for more details.
Genomic Evaluation of Geographic Clustering
To examine the spatiotemporal dynamics of H30, we quantified genomic evidence of geographic clustering using an SNV-distance based approach that compared the median SNV distance within collection sites to that between collection sites (SNVwithin / SNVbetween) [11]. Permutation-based 95% interval estimates with 1000 permutations of collection site were used to assess statistical significance; an SNV distance ratio below the lower bound indicated evidence of clustering. Clustering analyses were executed on the full data and on temporally segregated sample sets in approximate 1-year increments to explore whether geographic signal interacted with sampling date.
Clustering by “geographically close” vs “geographically distant” sites was also examined. Pairs of isolates from Midwest 1 and Midwest 2 were classified as geographically close, while other discordant site pairs were classified as geographically distant. The same clustering methods were applied to the full data and to the temporally segregated sample sets.
RESULTS
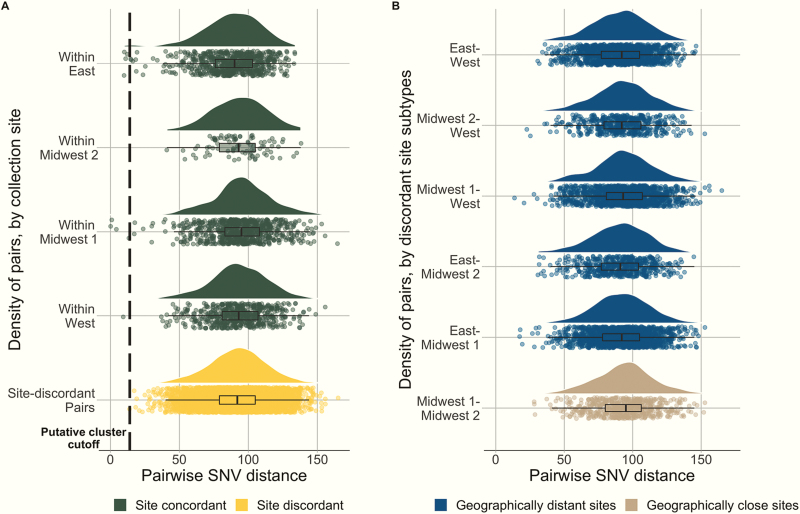
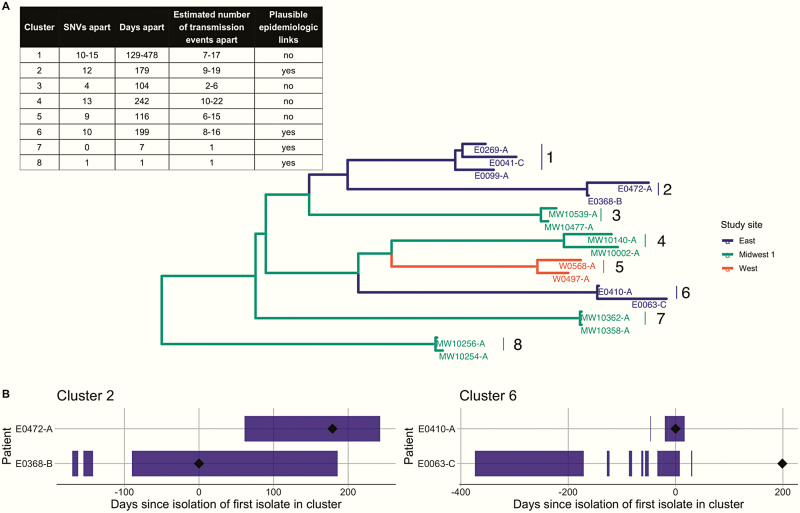
One hundred twenty-six H30 isolates were identified during the 4-year study from 126 unique children. There were 7875 pairwise comparisons made; the pairwise SNV distance ranged from 0 to 165 SNVs. The minimum SNV distance between isolates from discordant collection sites was 14 SNVs, and pairs of isolates separated by ≤ 14 SNVs were included in putative transmission clusters (Figure 1A). Using this threshold, 8 putative clusters were identified involving 17 isolates, 7 clusters containing 2 isolates, and 1 cluster containing 3 isolates (Figure 2A, Supplementary Figure 1). The putative cluster with 3 isolates (Cluster 1) consisted of 1 pair separated by 15 SNVs, but because the other 2 pairs were separated by ≤ 14 SNVs, all 3 isolates were included in further analyses.
Figure 1.
Distribution of pairwise single-nucleotide variant (SNV) distances between H30 clinical isolates from 4 children’s hospitals in the United States. Each dot represents 1 pair of isolates; the boxplot illustrates the median, interquartile range, and range of pairwise SNV distances; and the curve represents the density of pairs over the distribution of pairwise SNV distances. A, Pairs are stratified by concordant collection site vs discordant collection site. The dashed line indicates the selected threshold for identifying putative transmission clusters. B, Pairs are stratified by geographically distant discordant collection sites vs geographically close discordant collection sites.
Figure 2.
A, Maximum-likelihood phylogeny of 8 identified putative transmission clusters of H30 identified from 4 children’s hospitals in the United States colored by study site, with the number of days separating their collection; the number of single-nucleotide variants (SNVs) separating them after quality filtering; a range of estimated number of transmission events separating them as calculated by the R package transcluster; and an indicator of documented plausible epidemiologic links. The range of estimated transmission events reflect a range of reasonable values for substitution and transmission rates, but do not account for potential intrahost evolution or diversity. B, Temporal depiction of the overlapping hospitalizations of individuals in Cluster 2 and Cluster 6. The black diamond indicates the date of isolate collection and the purple bars represent time hospitalized. Time is measured in days since the isolation of the first isolate in each cluster.
Of the 8 putative clusters, documented epidemiologic data associated with 4 clusters (Clusters 2, 6, 7, and 8) was consistent with possible nosocomial acquisition (Figure 2A, Supplementary Table 2). Clusters 2 and 6 involved individuals with documented overlapping hospitalizations at the same hospital within the 6 months preceding their infection (Figure 2B). The genomic evidence for direct transmission within these clusters is less clear: they were separated by 12 and 10 SNVs, respectively, and the transcluster method estimated between 8 and 19 transmission events separating the isolates in these clusters (Figure 2A), assuming person-to-person transmission. Additionally, only 1 of the 2 isolates in Cluster 2 was phenotypically nonsusceptible to trimethoprim-sulfamethoxazole and had trimethoprim-sulfamethoxazole resistance-associated genetic determinants (Supplementary Figure 2). However, the within-cluster difference in collection dates was 179 and 199 days, so long-term colonization and within-host evolution may have inflated the estimated number of transmission events and resulted in lost resistance determinants. Clusters 7 and 8 consisted of isolates that differed by 0–1 SNVs and were collected between 1 and 7 days apart, with the transcluster method estimating direct transmission (Figure 2A). While there were no documented overlapping hospitalizations, both individuals within Cluster 7 had surgical site infections associated with neurological procedures, while both individuals within Cluster 8 were paraplegic. These connections are consistent with plausible epidemiological links in inpatient or outpatient care, although conclusively establishing such links is outside the scope of these data. These data also cannot distinguish between plausible links associated with person-to-person transmission or transmission via common sources such as surfaces or equipment, and do not rule out the possibility of nosocomial acquisition among the clusters without documented epidemiologic links, particularly due to a lack of data about outpatient exposures.
Genomic clustering analyses demonstrated minimal evidence of geographic clustering (Supplementary Figure 1). The median SNV distance within pairs across concordant sites was not significantly different from that within pairs across discordant sites. (SNVwithin / SNVbetween = 1.01 [95% interval estimate, 0.99–1.01]; Figure 1A). Similarly, the median SNV distance within pairs across geographically close discordant sites was not significantly lower than that within pairs across geographically distant sites (SNVwithin / SNVbetween = 1.03 [95% interval estimate, 0.99–1.02]; Figure 1B). Results for temporally segregated sample sets were similar, with most measures not supporting evidence of clustering (Supplementary Table 1 and Supplementary Figures 3 and 4). Additionally, there was no evidence of temporal clustering independent of geography (Supplementary Figure 5).
Discussion
We applied WGS to clinical isolates collected from 4 freestanding US children’s hospitals over 4 years to identify putative transmission clusters and investigate the spatiotemporal dynamics of E coli ST131-H30. We identified 8 putative transmission clusters of H30, including 2 clusters with documented overlapping hospitalizations and 2 clusters with other plausible healthcare-associated epidemiologic links, although these data cannot conclusively establish that these epidemiologic links represent nosocomial acquisition. Genomic spatiotemporal analyses demonstrated little evidence of geographic clustering of H30 more broadly.
To our knowledge, there are no available data about transmission of H30 between children within healthcare settings. Evidence of the frequency of within-healthcare transmission of H30 in adults is mixed and varies by population: some long-term care facilities and rehabilitation wards have reported high carriage rates and substantial transmission, whereas studies conducted in university hospitals and among cancer patients have reported very minimal transmission [12–15]. Our observation of limited plausible within-hospital transmission is consistent with a framework where within-hospital transmission is not a dominant contributor to the propagation of H30 among children. Interesting recent work identifying the important role of community factors in children’s risk of multidrug-resistant ExPEC infections also supports this framework [16]. However, the identification of some plausible nosocomial transmission highlights the utility of WGS of clinical isolates to uncover potential silent acquisition events.
There are also no data, to our knowledge, describing the spatiotemporal dynamics of H30 within the US using geographically diverse isolates. Our observation of limited geographic clustering was unexpected; we anticipated a genomic signature associated with sustained local circulation at the various geographic sites. These findings may reflect the rapid and recent dissemination of H30 at the time of this data collection—perhaps insufficient time had passed since the emergence of H30 in this population to allow for the establishment of noticeable genomic differences. The observed lack of clustering is also consistent with multiple introductions via common sources such as food, although recent studies do not support a major role of food sources in the dissemination of H30 [6, 17]. Whether these patterns remain the same today, almost 2 decades after H30 is believed to have disseminated globally, is worthy of further study [2].
The results of this study should be interpreted in the context of multiple limitations. First, epidemiologic data were limited and all plausible acquisition events should be interpreted cautiously. Second, applying an SNV threshold for defining putative clusters has limitations [10], and defining the SNV threshold using geographic criteria does not account for certain transmission mechanisms, such as food, that might span greater geographical distances. However, as noted above, current literature supports the human fecal-oral route of transmission as the dominant mode of transmission for H30, with little evidence of H30 found in animals or food [6, 17]. Finally, the local epidemiology of H30 may have changed since the collection of these isolates. This study also had several strengths, including a multicenter design, a large collection of H30 isolates from children, and the use of WGS to identify clusters. Taken together, our findings of minimal evidence of transmission clusters or broader geographic clustering are consistent with the prevailing conceptualization of H30 as a globally and recently disseminated strain that may be more frequently transmitted in the community than in healthcare [2, 14]. Future studies should consider focusing on community-based exposures when investigating the transmission dynamics of H30.
Supplementary Data
Supplementary materials are available at the Journal of The Pediatric Infectious Diseases Society online (http://jpids.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank Carey-Ann Burnham, Alexis Elward, Jason Newland, Rangaraj Selvarangan, Kaede Sullivan, Theoklis Zaoutis, and Xuan Qin for provision of bacterial isolates and associated clinical data; Brad Cookson for helpful advice; Jeff Myers and Huxley Smart for assistance with molecular typing of isolates; the Northwest Genomics Center for execution of whole genome sequencing; and members of the Snitkin Lab at the University of Michigan for their critical review of the manuscript. A portion of this work was presented at the American Society for Microbiology Conference on Rapid Applied Microbial Next-Generation Sequencing and Bioinformatics Pipelines, September 2018, Tysons, Virginia, USA.
Financial support. This work was supported by the Seattle Children’s Research Institute’s Center for Clinical and Translational Research Pediatric Pilot Fund program, and the National Institutes of Health via the National Institute of Allergy and Infectious Diseases (grant number R01AI083413) and the National Center for Advancing Translational Sciences (grant number TL1TR000422).
Potential conflicts of interest. All authors: No reported conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 2003; 5:449–56. [DOI] [PubMed] [Google Scholar]
- 2. Johnson JR, Tchesnokova V, Johnston B, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J Infect Dis 2013; 207:919–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miles-Jay A, Weissman SJ, Adler AL, et al. Epidemiology and antimicrobial resistance characteristics of the sequence type 131-H30 subclone among extraintestinal Escherichia coli collected from US children. Clin Infect Dis 2018; 66:411–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Banerjee R, Johnston B, Lohse C, et al. The clonal distribution and diversity of extraintestinal Escherichia coli isolates vary according to patient characteristics. Antimicrob Agents Chemother 2013; 57:5912–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Logan LK, Hujer AM, Marshall SH, et al. Analysis of β-lactamase resistance determinants in Enterobacteriaceae from Chicago children: a multicenter survey. Antimicrob Agents Chemother 2016; 60:3462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lazarus B, Paterson DL, Mollinger JL, Rogers BA. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis 2015; 60:439–52. [DOI] [PubMed] [Google Scholar]
- 7. Zerr DM, Miles-Jay A, Kronman MP, et al. Previous antibiotic exposure increases risk of infection with extended-spectrum-β-lactamase- and AmpC-producing Escherichia coli and Klebsiella pneumoniae in pediatric patients. Antimicrob Agents Chemother 2016; 60:4237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weissman SJ, Johnson JR, Tchesnokova V, et al. High-resolution two-locus clonal typing of extraintestinal pathogenic Escherichia coli. Appl Environ Microbiol 2012; 78:1353–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forde BM, Ben Zakour NL, Stanton-Cook M, et al. The complete genome sequence of Escherichia coli EC958: a high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS One 2014; 9:e104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stimson J, Gardy J, Mathema B, et al. Beyond the SNP threshold: identifying outbreak clusters using inferred transmissions. Mol Biol Evol 2019; 36:587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eyre DW, Davies KA, Davis G, et al. EUCLID Study Group . Two distinct patterns of Clostridium difficile diversity across Europe indicating contrasting routes of spread. Clin Infect Dis 2018; 67:1035–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burgess MJ, Johnson JR, Porter SB, et al. Long-term care facilities are reservoirs for antimicrobial-resistant sequence type 131 Escherichia coli. Open Forum Infect Dis 2015; 2:ofv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adler A, Gniadkowski M, Baraniak A, et al. MOSAR WP5 and WP2 Study Groups . Transmission dynamics of ESBL-producing Escherichia coli clones in rehabilitation wards at a tertiary care centre. Clin Microbiol Infect 2012; 18:E497–505. [DOI] [PubMed] [Google Scholar]
- 14. Hilty M, Betsch BY, Bögli-Stuber K, et al. Transmission dynamics of extended-spectrum β-lactamase-producing Enterobacteriaceae in the tertiary care hospital and the household setting. Clin Infect Dis 2012; 55:967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vehreschild MJ, Hamprecht A, Peterson L, et al. A multicentre cohort study on colonization and infection with ESBL-producing Enterobacteriaceae in high-risk patients with haematological malignancies. J Antimicrob Chemother 2014; 69:3387–92. [DOI] [PubMed] [Google Scholar]
- 16. Logan LK, Medernach RL, Rispens JR, et al. Community origins and regional differences highlight risk of plasmid-mediated fluoroquinolone resistant Enterobacteriaceae infections in children. Pediatr Infect Dis J 2019; 38:595–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Day MJ, Hopkins KL, Wareham DW, et al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis 2019; 19:1325–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.