Abstract
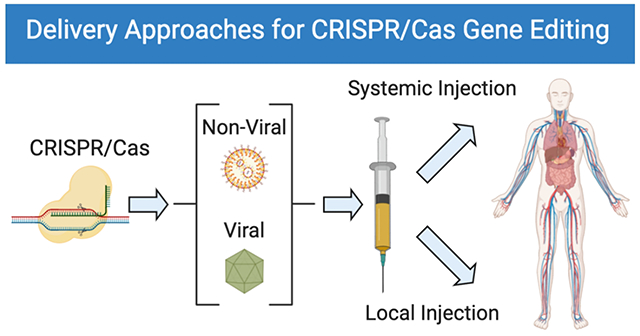
CRISPR/Cas9-based genome editing has quickly emerged as a powerful breakthrough technology for use in diverse settings across biomedical research and therapeutic development. Recent efforts toward understanding gene modification methods in vitro have led to substantial improvements in ex vivo genome editing efficiency. Because disease targets for genomic correction are often localized in specific organs, realization of the full potential of genomic medicines will require delivery of CRISPR/Cas9 systems targeting specific tissues and cells directly in vivo. In this Perspective, we focus on progress toward in vivo delivery of CRISPR/Cas components. Viral and nonviral delivery systems are both promising for gene editing in diverse tissues via local injection and systemic injection. We describe the various viral vectors and synthetic nonviral materials used for in vivo gene editing and applications to research and therapeutic models, and summarize opportunities and progress to date for both methods. We also discuss challenges for viral delivery, including overcoming limited packaging capacity, immunogenicity associated with multiple dosing, and the potential for off-target effects, and nonviral delivery, including efforts to increase efficacy and to expand utility of nonviral carriers for use in extrahepatic tissues and cancer. Looking ahead, additional advances in the safety and efficiency of viral and nonviral delivery systems for tissue- and cell-type-specific gene editing will be required to enable broad clinical translation. We provide a summary of current delivery systems used for in vivo genome editing, organized with respect to route of administration, and highlight immediate opportunities for biomedical research and applications. Furthermore, we discuss current challenges for in vivo delivery of CRISPR/Cas9 systems to guide the development of future therapies.
Keywords: CRISPR/Cas9, genome editing, viral delivery, non-viral delivery, nanoparticles, gene delivery, gene therapy, ribonucleoproteins (RNPs), homology-directed repair (HDR)
Graphical Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR) -associated protein (CRISPR/Cas) nuclease system has rapidly become a powerful toolbox for genome engineering with broad applications in biological research, including high-throughput screening applications, animal model creation, in vivo and ex vivo editing, and disease therapeutics.1–6 The canonical CRISPR/Cas9 system involves Cas9 protein and guide RNA structures formed by two complementary sequences consisting of the trans-activating tracrRNA and targeting crRNA. To simplify the system, the tracrRNA/crRNA duplex has been engineered into a single chimeric RNA, termed single guide RNA (sgRNA), making utility of gene editing easier for researchers across disciplines.1 Initially, Cas9 protein and sgRNA must first assemble into ribonucleoproteins (RNPs). After traversing the cell’s nuclear membrane and entering into the nucleus, the target sequence of sgRNA may then recognize a highly specific genomic DNA sequence flanked by a protospacer-adjacent motif (PAM). It is at this point that Cas9 protein creates a double-stranded break (DSB) cut in the genomic DNA. Following cleavage, these DSBs can generally be repaired via two distinct pathways: 1) non-homologous end joining (NHEJ), which introduces small deletions or insertions (indels) at random around the targeted gene locus; and 2) homology-directed repair (HDR), which incorporates a template strand of DNA into the DSB site, thereby enabling precise genome modification.7–11
A key component of efficient CRISPR/Cas9-mediated genome editing is the development of efficient delivery strategies that can effectively deliver CRISPR/Cas9 systems into targeted cells.6, 12–13 To date, multiple approaches have been reported for in vitro/ex vivo delivery of CRISPR/Cas9 machinery, including physical methods (microinjection, electroporation),14–15 viral delivery (lentiviral vectors, adenoviral vectors, and adeno-associated vectors),7, 13 and nonviral delivery (nanoparticle delivery systems).6, 12, 16 These methods are able to deliver CRISPR/Cas9 systems into cells efficiently and have been widely used for various applications, including editing mouse zygotes,17–18 stem cells,19–22 cultured cells,23–26 and organoids.27–30 Acknowledging that various reviews have been written on CRISPR/Cas summarizing gene editing approaches,6–7, 9–16 in this Perspective, we focus on how these approaches have been utilized to optimize various strategies for in vivo editing—ranging from viral to synthetic carriers as well as local to systemic routes of administration—and the advantages and challenges associated with each strategy. Therefore, we summarize current delivery strategies used for in vivo genome editing organized with respect to route of administration, and, looking forward, we elucidate current challenges impeding effective in vivo genome editing and propose potential avenues worth pursuing that may overcome these obstacles. Through continued development and optimization, CRISPR/Cas systems hold the potential to enable the realization of truly curative therapeutics that combat diseases at the DNA level, addressing the origin of the disorder rather than simply treating the symptoms.
Viral Delivery Systems for in Vivo Genome Editing
Viral vectors are a promising system employed for in vivo delivery of CRISPR/Cas9.13 Due to their high cellular uptake and editing efficiency, viral vectors have been widely used for gene therapy and genome editing. At present, the most commonly used viral vectors in vivo include lentiviral vectors (LVs), adenoviral vectors (AVs), and adeno-associated viruses (AAVs).31–32 These viral vectors have been used to deliver CRISPR/Cas9 systems successfully to multiple tissues/organs via a variety of administration methods (Figure 1).32
Figure 1.
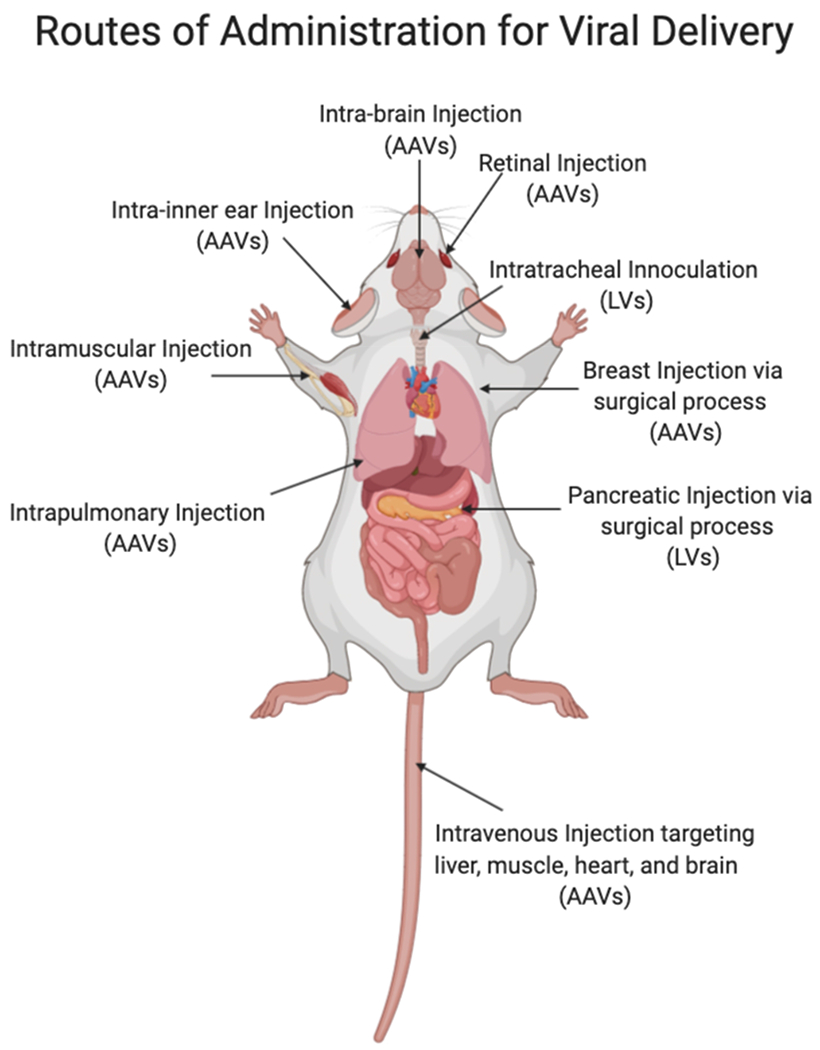
Routes of administration for viral-based delivery systems, including lentivirus (LV) and adeno-associated viruses (AAV), pertaining to in vivo genome editing.
Local Administration
Researchers have investigated LVs, AVs, and AAVs for the generation of various mouse models via local injection of gene-editing machinery directly into tissues. For example, LVs expressing both Cas9/sgAlk and Cas9/sgEml4 were delivered either by intratracheal inoculation or by direct intrapulmonary injection into lungs of adult mice, which successfully generated Eml4-Alk rearrangements and thereby induced tumor formation in mouse lungs for cancer model generation.33 Lentiviral vectors and AAVs have also been shown to deliver CRISPR systems into lung via intratracheal and intranasal inoculation. Similarly, these systems successfully knocked out tumor suppressor genes and were able to generate lung tumor mouse models.34 Furthermore, LVs containing CRISPR/Cas9 systems have been adapted for the generation of multiple other cancer models such as pancreatic cancer35 and invasive lobular breast cancer36 via local injections into the pancreas and breast tissue, respectively.
In addition to tumor modeling, viral vectors have been used to treat models of genetic diseases, such as Duchenne muscular dystrophy (DMD),37–39 Alzheimer’s disease (AD),40 and dominant progressive hearing loss.41 DMD is a fatal genetic disease caused by mutations in the dystrophin gene that result in the absence of this critical structural protein in muscle. Recently, AAVs were used to deliver CRISPR/Cas9 nucleases coupled with paired sgRNAs flanking mutated DMD exon23 in the mdx mouse model of DMD. Ultimately, the treatment was able to restore the DMD reading frame and partially recover muscle functional deficiency following intra-muscular injection.38 By deleting DMD exon 44 and exon 50 in DMD mice via AAV vector-mediated single-cut genome editing, the expression of dystrophin was rescued and muscle function was improved.42–43 It has been shown that a mutation in amyloid precursor protein (APP), termed APPswe (Swedish), leads to abnormally high levels of amyloid-β (Aβ) protein accumulation in the brain. In the interest of creating a mouse model of Alzheimer’s disease (AD) with this mutation, AAV vectors coding for sequences of both Cas9 protein and APPsw specific guide RNAs were co-injected into the hippocampus of adult mice. Combinatorial delivery of these AAVs successfully generated an AD model mice with site-specific indels at the mutated APPsw allele.40 Beethoven (Bth) mice, a mouse model for dominant progressive hearing loss, is related to a Bth point mutation in the Tmc1 allele. György et al. screened 14 Cas9/sgRNA combinations and identified one that selectively and efficiently disrupted the mutant Tmc1 allele, rather than the wild-type Tmc1/tmc1 allele, in Beethoven (Bth) mice.41 They found that AAV-mediated SaCas9-KKH delivery prevented deafness in Bth mice for up to 1 year post inner-ear injections. Overall, viral vectors have been able to induce gene editing in multiple tissues via local administration, including heart,44–45 lung,33,46–47 muscle,37–39,42–43 brain,40,48 breast,36 pancreas,35 inner ear,41 and retina.49–50
Systemic Administration
In addition to local administration, researchers have identified multiple AAV serotypes that have the capability to deliver CRISPR/Cas9 complexes into different tissues via systemic injection. Adeno-associated virus vectors are able to edit genes in liver tissues to treat diseases such as metabolic liver diseases,51–53 hemophilia B,54 and targets for control of serum cholesterol levels.55 For example, Yin et al. combined viral delivery (lipid nanoparticles carrying Cas9 mRNA) and nonviral delivery (AAV8 encoding a sgRNA and a HDR template), for the treatment of a mouse model of hereditary tyrosinemia type I (HT-1) via systemic injection. They demonstrated that this treatment successfully corrected a mutation in fumarylacetoacetate hydrolase (FAH) in hepatocytes and rescued disease symptoms, such as weight loss and liver damage.51 Ran et al. packaged SaCas9 and a sgRNA expression cassette into AAV8 and administered them via tail vein injection. The viruses then accumulated in the mouse liver and targeted the cholesterol regulatory gene PCSK9. At only 1 week post injection, they observed over 40% indel formation in liver, with similar levels maintained at 4 weeks post injection. Meanwhile, they detected significant reductions in serum PCSK9 (~95%) and total cholesterol levels (~40%) that were sustained throughout the whole study.55 Another tissue that has been targeted via viral vectors is muscle. Specifically, AAV6,56 AAV8,39 and AAV942–43 have been used to deliver CRISPR/Cas9 cassettes via systemic injection to restore the expression of dystrophin in muscles, resulting in improvements in muscle function in the mdx mouse model of DMD. Additionally, researchers have systematically targeted heart57 and brain58 using AAV9 vectors through the careful inclusion of tissue-specific promotors.
Challenges of Viral Delivery
Despite these achievements, there are several limitations and drawbacks to using viral vectors. First and foremost, viruses have a limited packaging capacity of approximately 5 kilobases (kb),9, 12–13 which severely hinders the size of the sequences that can be packaged into the vector. By using smaller Cas9 orthologues (e.g., StCas9, SaCas9, and cjCas9)9 or dual AAV vectors,59–60 some researchers have been able to circumvent the packaging issues, in part; however, achieving precise genome editing via HDR given these constraints remains a significant hurdle. The second cause for concern is the propensity of viruses to generate a negative and substantial immune response, which then eliminates the possibility of repeat dosing—an aspect that is critically important in cells with high turnover rates.12–13 Therefore, a single injection of viral vectors must yield a high enough permanent incidence of gene modification to reach levels that provide a therapeutic benefit, something that is difficult to accomplish in a single dose. A third point of consideration is that the construction process and resources necessary to create a virus with therapeutic potential—from both a financial as well as an industrial perspective—are complex, expensive, and time consuming. Finally, viruses used to achieve genome editing have been known to induce high frequencies of off-target events.6–7, 9–10, 12 The long term and/or continuous expression of CRISPR components—as is the case when using viruses—may significantly increase the risks of off-target events in vivo after viral delivery, some of which may indeed be deleterious to the host’s genome. Efforts to overcome these drawbacks are underway and may be addressed by new approaches. Due to high potency and tissue specificity, viral vectors are still the most widely used method at present for in vivo genome editing. Viral delivery systems for in vivo gene editing remain an attractive approach for difficult to target tissues, such as heart and brain. Due to their high editing efficiency, viral vectors have been investigated in several preclinical and clinical studies.32
Nonviral Delivery Systems for in Vivo Genome Editing
With the rapid development of synthetic vectors, nonviral delivery systems have already had a transformative impact on the genome editing field and they have the potential to drive advances further in a variety of widespread applications. Unlike viral vectors, which, as noted previously, have a limited packaging capacity, nonviral delivery systems are quite flexible with respect to loading capacity and can accommodate large nucleic acid and protein cargos such as Cas9 mRNA and RNPs (~160 kDa),23–24,61–63 in addition to sgRNAs64–65 and donor DNA.66 By encapsulating these cargos inside nanoparticles (NPs) rather than in viruses, immune detection, along with most immunogenicity can be minimized, thus allowing for repeat administration of NPs loaded with gene-editing cargo. Another benefit to using NPs as transfection agents is that when delivering Cas9 mRNA or RNPs, Cas9 expression is transient, and this transient expression of the nuclease greatly reduces the probability of insertional mutagenesis. Therefore, the threat of nuclease-induced off-target events is largely mitigated when compared to that of viral vectors.67–69 To date, a variety of synthetic vectors have been developed to deliver CRISPR/Cas9 systems in vivo. Here, we will introduce them as classified by method of administration (Figure 2).
Figure 2.
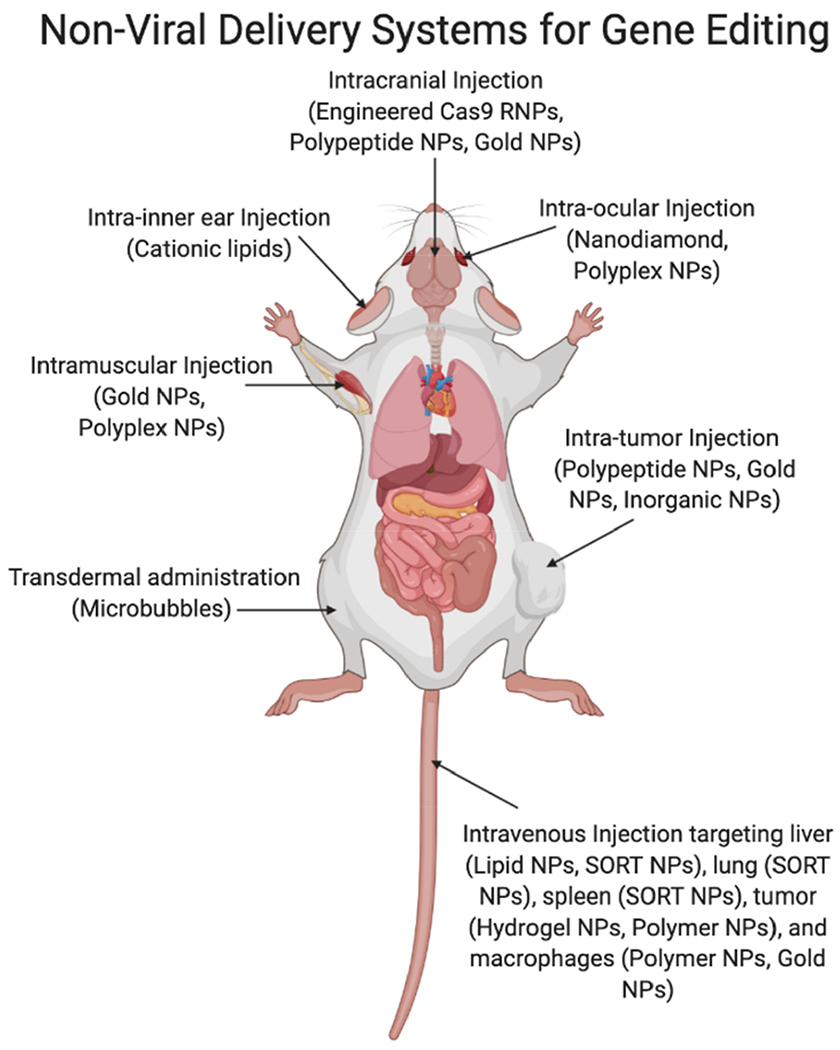
Different routes of nonviral-based delivery systems for in vivo genome editing. Nanoparticles, NPs; ribonucleoproteins, RNPs; selective organ targeting, SORT.
Local Administration
Intratumoral Injection
Multiple synthetic vectors have been developed that can successfully deliver CRISPR/Cas9 systems designed to knock out tumor-related genes and to suppress tumor growth following intratumor injection. As an example, Wang et al. developed PEGylated NPs based on α-helical polypeptide PPABLG (P-HNPs) that co-deliver Cas9 expression plasmids and sgRNAs targeting polo-like kinase 1 (Plk1), named P-HNPpcas9+sgPlk1.70 Following intratumor injection of P-HNPpcas9+sgPlk1 into HeLa xenograft tumor-bearing mice, the gene editing efficiency at the Plk1 locus reached 35%, which resulted in significant tumor suppression (higher than 71%) and improved the survival rate of tumor-bearing mice (60%) (Figure 3A). Wang et al. used TAT-peptide-coated gold nanoclusters (GNs) as a core to absorb Cas9 protein/sgPlk1 plasmid (they abbreviate the entire complex as GCP).71 After absorption, GCP was further encapsulated in a cationic lipid shell to form lipid-coated GCP (LGCP). Intratumoral injection of LGCP effectively inhibited tumor growth in a melanoma mice model by deleting the Plk1 gene in tumors and significantly down-regulating the Plk1 protein expression (Figure 3B). Most recently, the authors used these TAT-peptide-coated AuNPs (20 nm) to replace GNs for encapsulation of Cas9-sgPlk1 plasmids (CPs) and coated the outside in lipid to form lipid-encapsulated AuNPs-condensed CP (LACP). When irradiated with a 514 nm laser, the temperature generated by the localized surface plasmon resonances of AuNPs not only triggered release of the encapsulated cargo, but also provided a means of performing photothermal cancer therapy (Figure 3C). When injected directly into a tumor and irradiated, the tumor volume decreased to approximately 15% of that of the untreated group, indicating LACP-mediated tumor inhibition under irradiation.72 More recently, Pan et al. developed a near-infrared (NIR) upconversion-triggered CRISPR-Cas9 system (UCNPs-Cas9@PEI).73 The upconverting NPs (UCNPs) convert NIR light (980 nm) into ultraviolet light, which triggers the cleavage of photosensitive molecules, resulting in on-demand release of CRISPR-Cas9 machinery for gene editing. With NIR light irradiation for 20 min every other day, intratumor injection of UCNPs-Cas9@PEI delayed tumor progression by knocking-out the Plk1 gene in tumor tissues, with no detectable toxic side effects (Figure 3D).
Figure 3.
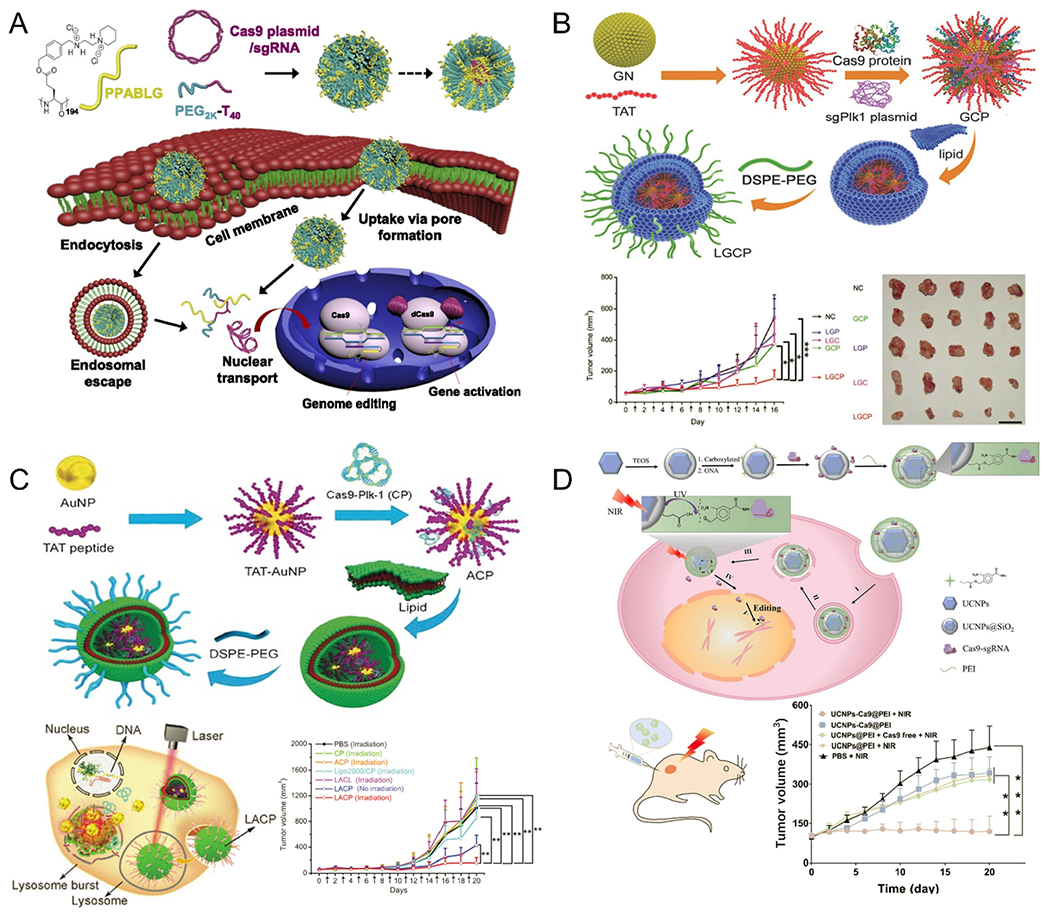
Nonviral, intratumor injection delivery systems for in vivo genome editing. (A) Scheme of formation of PEGylated nanoparticles based on α-helical polypeptide PPABLG (P-HNPs) and their intracellular delivery of Cas9 expression plasmid/single guide RNA for genome editing or gene activation. Reproduced with permission from ref 70. Copyright 2018 National Academy of Sciences. (B) Schematic illustration of synthesis process of nanoformulations (polyethylene glycol-lipid/Gold nanoclusters/Cas9 protein/sgPlk1 plasmid, LGCP). LGCP delivering Cas9 protein/sgPlk1 plasmid successfully inhibited tumor growth by knocking-out the Plk1 gene. Reproduced with permission from ref 71. Copyright 2017 John Wiley & Sons, Inc. (C) Schematic representation of synthesis process for lipid-encapsulated AuNPs-condensed Cas9-sgPlk1 plasmids (LACPs) and laser-enhanced knock-outs of targeted gene by LACPs, resulting in tumor inhibition in mouse melanoma models. Reproduced with permission from ref 72. Copyright 2018 John Wiley & Sons, Inc. (D) Design of upconverting nanoparticles (UCNPs)-based CRISPR-Cas9 delivery system for near-infrared (NIR) light–controlled gene editing. When exposed to NIR light, UCNPs-Cas9@PEI platform successfully inhibited tumor proliferation. Reproduced with permission from ref 73. Copyright 2019 American Association for the Advancement of Science.
Intracranial Injection
In addition to viral delivery, there is great interest in developing nonviral vectors capable of editing genes in the brain for new disease therapies. Staahl et al. directly engineered Cas9 RNP complexes with multiple nuclear localization signals that enabled neuronal editing in the brain via intracranial injection.74 Park et al. constructed Cas9 nanocomplexes by mixing amphiphilic R7L10 peptide and Cas9/sgRNA RNPs to edit genes in post-mitotic neurons of the adult brain (Figure 4A).75 The Cas9 nanocomplex targeting beta-secretase 1 (Bace1) significantly reduced expression of amyloid beta (Aβ) peptides and suppressed Aβ-associated phenotypes and cognitive deficits in both five familial Alzheimer’s disease (5XFAD) and amyloid precursor protein (APP) knock-in Alzheimer’s disease mouse models.75 Beyond neuronal editing, the recently developed CRISPR-Gold was also able to induce gene editing in non-neuronal cells, including astrocytes and microglia, via delivery of Cas9 and Cpf1 RNPs. Upon intracranial injection, CRISPR-Gold inhibited 40–50% of Grm5 in the striatum and prompted behavioral improvements in a mouse model of fragile X syndrome (Figure 4B).76
Figure 4.
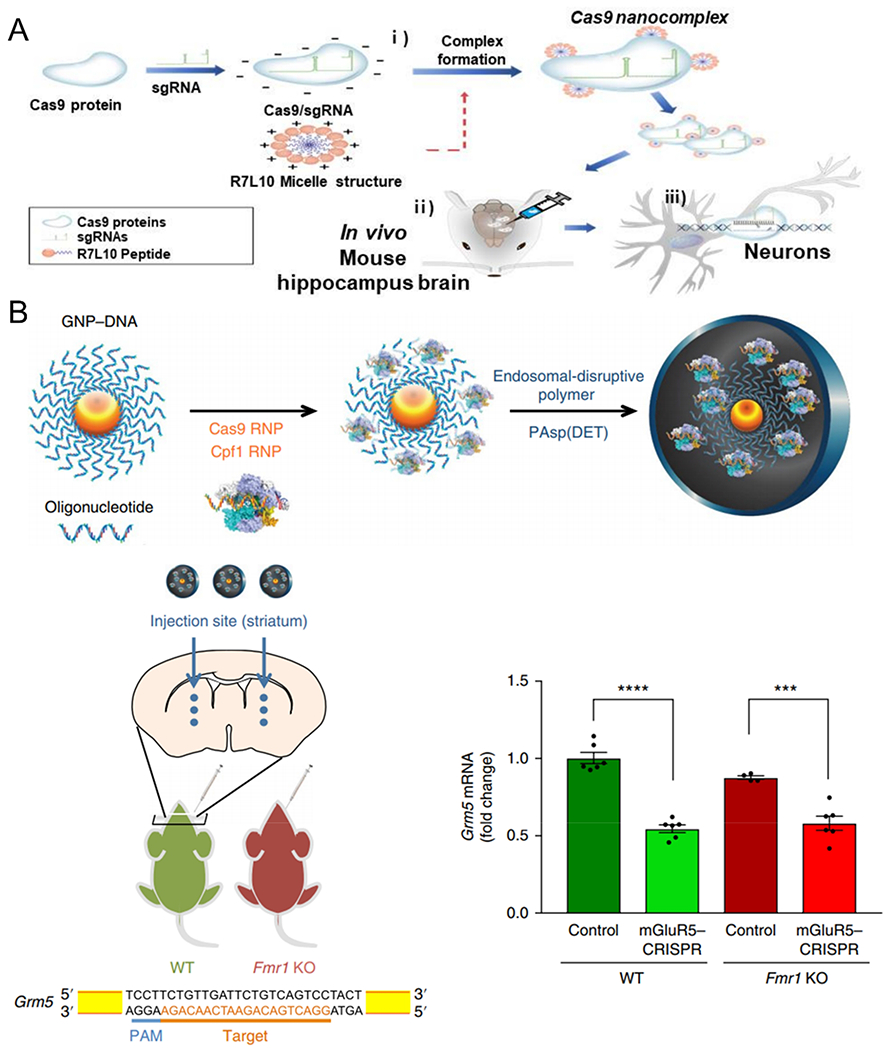
Nonviral delivery systems for in vivo genome editing via intracranial injection. (A) Schematic representation of the formation of CRISPR-Cas9 nanocomplex delivery system. Local administration of CRISPR-Cas9 nanocomplexes successfully edited target genes in post-mitotic neurons of the adult brain. Reproduced with permission from ref 75. Copyright 2019 Springer Nature. (B) Schematic representation of CRISPR-Gold synthesis. When injecting mGluR5–CRISPR into the striatum of wild-type (WT) and Fmr1 knockout (Fmr1 KO) mice, Grm5 mRNA levels were reduced by 40–50%. Reproduced with permission from ref 76. Copyright 2018 Springer Nature. Single-guide RNA, sgRNA; ribonucleoprotein, RNP.
Intramuscular Injection
There are more than 800 monogenic disorders that can cause dysfunction of skeletal muscles. Among them, DMD is the most severe and lethal monogenic disorder, resulting from mutations at the gene locus encoding dystrophin, a protein that maintains integrity of striated muscles.77 The development of genome-editing technology provides a possible means of permanently removing or repairing genetic mutations related to DMD, thereby restoring the expression of dystrophin protein and recovering muscle functions. For this purpose, CRISPR-Gold has been used to edit genes in muscles.78 Intramuscular injection of CRISPR-Gold into Ai9 mice resulted in deletion of a stop sequence in genomic DNA, which then induced expression of tdTomato, a fluorescent protein. Moreover, CRISPR-Gold produced correction of a point mutation in the dystrophin gene in the mdx model of DMD by using intramuscularly injected Cas9 RNPs along with donor DNA templates. Using this technique, researchers were able to achieve a local point mutation correction rate of 5.4% in mdx mice, leading to reduced muscle fibrosis and an increase in muscle function (Figure 5A). In addition, Chen et al. utilized in-situ polymerization to create nanocapsules (NCs) that are capable of encapsulating RNPs. Local injection of these RNP-loaded NCs into the tibialis anterior muscle of tdTomato mice efficiently induced the expression of tdTomato via gene editing (Figure 5B).79 Recently, Wei et al. reported a methodology that allows engineering of modified LNPs to deliver RNPs into muscle and other tissues.80 Optimized LNPs were able to deliver RNPs to restore 4.2% of dystrophin protein in ΔEx44 DMD mice.
Figure 5.
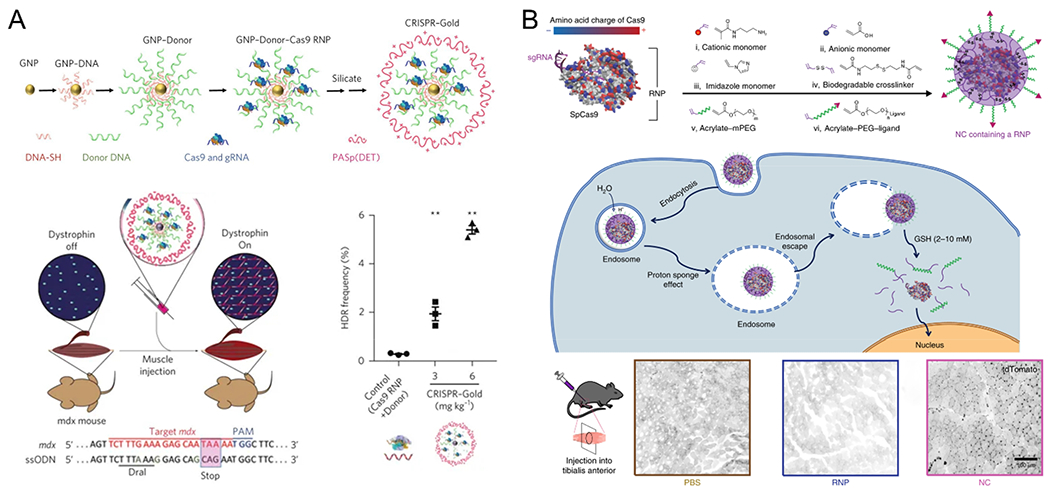
Nonviral delivery systems for in vivo genome editing via intramuscular injection. (A) Schematic representation of CRISPR-Gold synthesis. Intramuscular injection of CRISPR–Gold in mdx mice promoted homology-directed repair efficacy in the dystrophin gene, enhanced dystrophin protein expression, and reduced muscle fibrosis. Reproduced with permission from ref 78. Copyright 2017 Springer Nature. (B) A schematic illustration of the in situ free-radical polymerization of biodegradable nanocapsules (NCs) for the delivery of the Cas9 ribonucleoprotein (RNP) complex and its internalization and the subcellular release of the RNP. Local injections of NCs into the tibialis anterior muscle induced robust gene editing and strong tdTomato signals were observed. Reproduced with permission from ref 79. Copyright 2019 Springer Nature.
Intra-Inner Ear Injection
Nonviral delivery approaches have been used to treat genetic hearing loss disorders through local injection into the inner ear. Zuris et al. used the commercial cationic liposomal reagents Lipofectamine 2000 and RNAiMax to deliver Cas9 protein and EGFP sgRNA and then locally injected them into the cochlea of P2 Atoh1-GFP mice, where all hair cells express GFP.81 After 10 days, loss of GFP fluorescence in 20 ± 3% outer hair cells was observed in the Lipofectamine 2000 treatment group (Figure 6A). Given this high potential to treat genetic diseases of the inner ear, Zuris et al. then evaluated the ability of their system to treat autosomal-dominant hearing loss in a mouse model of a human genetic disease (Tmc1Bth/+ mice). After injection, they were able to knock out the mutated Tmc1Bth allele using Cas9-guideRNA-lipid complexes. This treatment effectively promoted hair cell survival and reduced progressive hearing loss in Tmc1Bth/+ mice (Figure 6B).82
Figure 6.
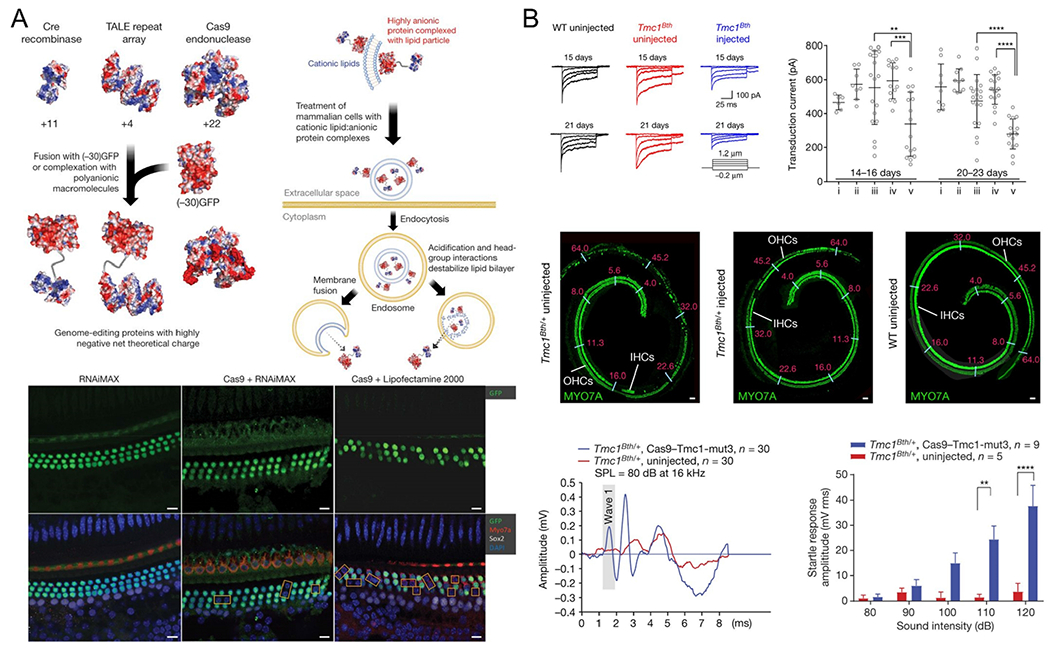
Nonviral delivery systems for in vivo genome editing via intra-inner ear injection. (A) Cationic lipids mediated delivery of proteins into mammalian cells through engineering proteins by fusion or noncovalent complexation with polyanionic macromolecules. Local injection of Cas9/sgRNA complexes delivered by RNAiMAX or Lipofectamine 2000 into the scala media successfully resulted in loss of green fluorescent protein (GFP) expression in hair cells. Reproduced with permission from ref 81. Copyright 2015 Springer Nature. (B) Scala media injection of Cas9/Tmc1-mut3 complexed with Lipofectamine 2000 (Cas9–Tmc1-mut3–lipid) promoted hair cell survival and reduced hearing loss in Tmc1Bth/+ mice. Reproduced with permission from ref 82. Copyright 2018 Springer Nature.
Intraocular Injection
Nonviral delivery systems have also been assessed for delivery of CRISPR/Cas9 cargo into the eye via local injection. For example, Yang et al. functionalized nanodiamonds with mCherry protein and conjugated two linear DNA constructs onto the mCherry proteins (named cND-mC-C/C9).83 One of these DNA constructs encoded Cas9 protein and a GFP reporter protein, and the other encoded sgRNA and an HDR template targeting the Rs1 gene mutation, which is associated with X-linked retinoschisis (XLRS). Intravitreal injection of cND-mC-C/C9 NPs into mouse eye results in the presentation of pathological features of XLRS and, as such, may be used for the creation of an XLRS mouse model. Similarly, Chen et al. functionalized RNP-encapsulating nanocapsules (NCs) with a targeting ligand ATRA for retinal pigment epithelium (termed NC-ATRA), and found that subretinal injection of NC-ATRA into the eyes of transgenic Ai14 tdTom reporter mice induced higher gene editing efficiency than negative control groups.79
Transdermal Administration
Skin is also a targetable tissue for local treatment. Ryu et al. encapsulated RNP complexes into nanosized liposomes and conjugated them onto the surface of microbubbles.84 They then used this delivery system to edit the mouse SRD5A2 locus (mSRD5A2) for mouse androgenic alopecia therapy. They depilated the dorsal backs of C57BL/6 mice after an adaption, and topically applied testosterone daily for 7 weeks to maintain telogen retention as an androgenic alopecia model. Once activated by ultrasound, sonoporation generated by microbubble cavitation facilitated RNPs release and delivery into dermal papilla cells in hair follicles and resulted in mSRD5A2 gene knockout, leading to recovery of hair growth.
Systemic Administration
Targeting Liver
The liver is an attractive target for nonviral vector delivery because most NPs accumulate in the liver following systemic administration. To date, multiple lipid-based NPs have been identified for hepatocyte-specific delivery in liver via systemic injection.51, 64–65, 85–88 For example, Miller et al. developed zwitterionic amino lipid (ZAL) nanoparticles NPs (ZNPs) for delivery of Cas9 mRNA and sgLoxP targeting the stop cassette of a tdTomato reporter mouse model.64 Systemic injection of ZNPs enabled deletion of the stop cassette and induced tdTomato fluorescence in the liver, kidneys, and lungs of tdTomato mice (Figure 7A). Jiang et al. developed TT3 lipid-like nanoparticles (LLNs) that can efficiently deliver Cas9 mRNA and guide RNA to the liver, thereby disrupting HBV DNA and the PCSK9 gene for liver-related disease treatments.65 To increase gene editing efficiency in vivo further, end-modified sgRNA and fully modified sgRNA were synthesized. Upon single intravenous injection of biodegradable lipid nanoparticles (LNPs; LNP-INT01) co-delivering Cas9 mRNA and highly modified sgRNA targeting mouse transthyretin (Ttr) gene to CD-1 mice, ~70% of gene editing at Ttr locus and >97% knockdown of TTR serum protein were detected for at least 12 months (Figure 7B).85 Also, a single IV injection of CKK-E12 LNPs loaded with Cas9 mRNA and chemically modified sgRNA into mouse tail vein induced > 80% gene editing of the PCSK9 gene in the liver and resulted in an undetectable level of PCSK9 protein in the serum (Figure 7C).87 In addition to gene knock-out, researchers have also used LNPs for gene correction in the liver. Yin et al. used C12-200 formulated LNPs to encapsulate Cas9 mRNA and AAV vectors loaded with a cassette encoding a sgRNA and a donor template.51 The combined delivery of these components successfully repaired a Fah mutation in the Fahmut/mut mouse model of hepatorenal tyrosinemia type 1 (HT1) (0.81% of gene correction at Fah locus) and rescued the mice from weight loss and liver damage (Figure 7D).
Figure 7.
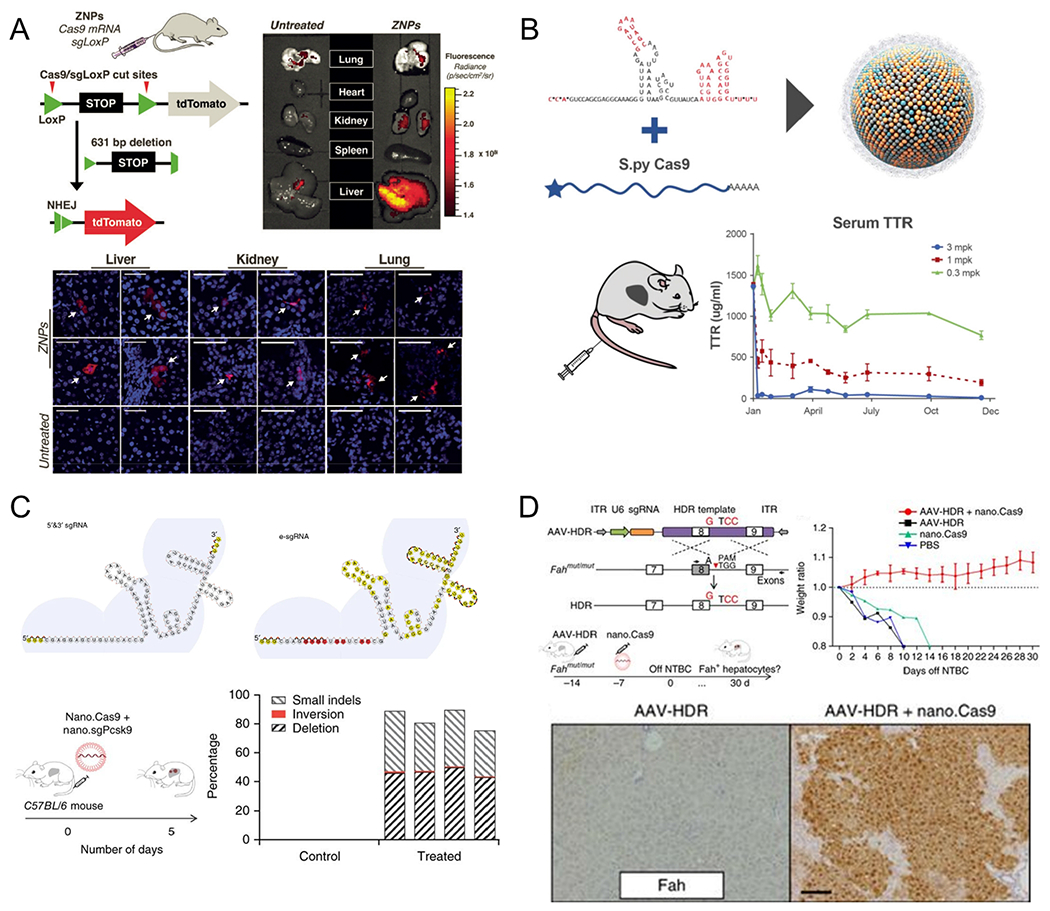
Nonviral delivery systems for liver-targeted genome editing via systemic administration. (A) zwitterionic amino lipid nanoparticles (ZNPs) co-delivering Cas9 mRNA and sgLoxP successfully deleted the stop cassette and activated expression of tdTomato protein mainly in the liver of tdTomato mouse model. Reproduced with permission from ref 64. Copyright 2017 John Wiley & Sons, Inc. (B) Single IV administration of a biodegradable lipid nanoparticle (LNP)-based delivery system (LNP-INT01) achieved and maintained ~70% of gene editing at Ttr locus and >97% knockdown of serum TTR levels for at least 12 months. Reproduced with permission from ref 85. Copyright 2018 Elsevier. (C) Structure illustration of the conventional 5′ and 3′ end modification (5′ and 3′ sgRNA) and the new e-sgRNA design. In vivo CKK-E12 LNP-based delivery of two e-sgRNAs targeting Pcsk9 and Cas9 mRNA mice induced > 80% gene editing of PCSK9 gene in the mouse liver. Reproduced with permission from ref 87. Copyright 2017 Springer Nature. (D) Combination of LNP-mediated delivery of Cas9 mRNA with adeno-associated viruses (AAVs) encoding a sgRNA and a repair template corrected the causative Fah-splicing mutation, generated Fah-positive hepatocytes, and rescued disease symptoms (including weight loss and liver damage) in type I tyrosinemia mice. Reproduced with permission from ref 51. Copyright 2016 Springer Nature.
Targeting Lungs and Spleen
As mentioned above, most systemic therapeutic efforts for gene editing using nonviral carriers have focused on targets in the liver. Delivering gene-editing components to extrahepatic tissues remains challenging. Solving this challenge represents a significant area of research for future expansion of gene editing because many important targets for gene correction are in tissues such as the lungs, spleen, kidney, bone marrow, and others. In the past few years, there have been reports of delivering siRNA and mRNA to extrahepatic tissues.64, 89–99 Although the mechanisms of targeting are not fully elucidated, these works provide a foundation upon which to develop tissue-targeted genomic medicines. For example, Sago et al. developed a high-throughput in vivo screening method, fast identification of nanoparticle delivery (FIND), to identify LNPs that can functionally deliver mRNA to different cell types.99 With this method, they identified two LNPs (7C2 and 7C3) that can deliver mRNA to endothelial cells, especially endothelial cells in the spleen. After IV injecting LSL-Tom mice twice with 7C3 encapsulating Cas9 mRNA:modified sgRNA (esgRNA), they observed gene editing in splenic endothelia cells (~20%) as efficiently as hepatocytes. As NPs developed for other drugs (e.g., siRNA) are reengineered for delivery of CRISPR/Cas, it is envisioned that more extrahepatic carriers will be discovered, especially as the mechanisms of targeting become clearer. In vivo screening methodologies, such as DNA barcoding, will play an important role in these efforts.
As an alternative to high-throughput screening, Cheng et al. recently developed a generalizable methodology, selective organ targeting (SORT), which enables researchers to reengineer LNPs for tissue-specific mRNA delivery targeting liver, lungs, and spleen.100 The approach involves the addition of SORT molecules into established NPs (including DLNP (Dendrimer-based lipid nanoparticles), Stable nucleic acid lipid particles (SNALPS), and Lipid-like nanoparticles (LLNPs)) to redirect delivery to extrahepatic tissues (Figure 8A). Liver-, lung-, and spleen-targeted SORT LNPs co-delivering Cas9 mRNA/sgTom selectively activated strong td-Tomato expression in liver, lung, and spleen, respectively, in tdTomato mice (Figure 8A). Lung-targeted SORT LNPs transfected 65% of all endothelial cells and 40% of all epithelial cells in the lungs of tdTomato mice following a single injection of 0.3 mg/kg Cre mRNA, suggesting potential for systemic correction of diseases that affect the lung epithelium. This approach also enabled the therapeutic editing of targets such as PCSK9, where SORT LNPs induced 60% editing at the PCSK9 locus and 100% PCSK9 reduction in both liver tissue and serum via co-delivery of Cas9 mRNA/sgPCSK9 (Figure 8A). In parallel, it was realized that 5-component SORT LNPs containing permanently cationic lipids (positively charged at neutral pH) could mediate self-assembly using neutral buffer solutions of RNPs to preserve protein structure and stability for effective genome editing.80 The developed 5A2-DOT-X LNPs could mediate tissue-specific, multiplexed editing of multiple genes following intravenous injection in mice. In addition to therapeutic applications, liver and lung tropic LNPs were used to create cancer models in situ through knockout of multiple tumor suppressor genes or by induction of programmed chromosomal rearrangement.80 With further development, the SORT strategy100 may enable editing in additional tissues in the future, thereby expanding the development of gene editing/correction therapeutics in difficult-to-access cells and organs.
Figure 8.
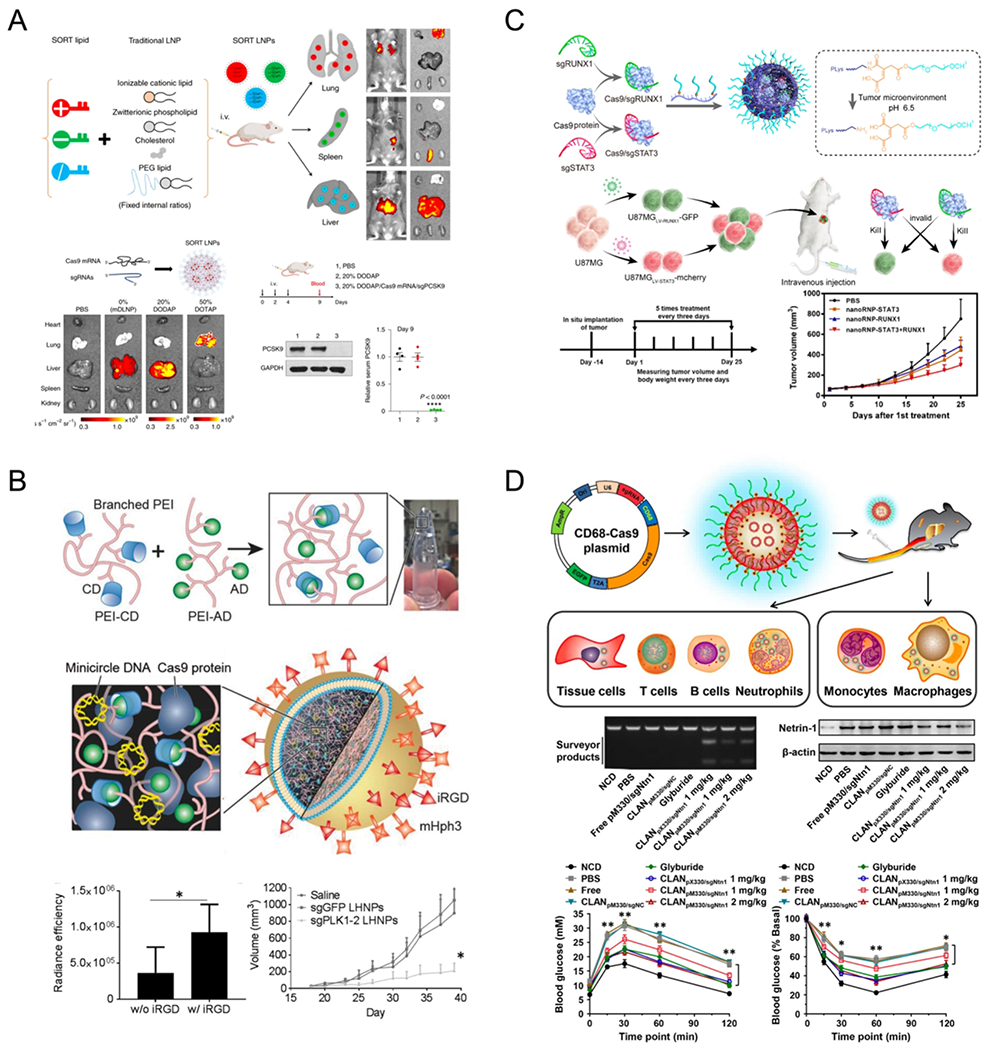
Nonviral delivery systems for genome editing targeting extrahepatic tissues via systemic administration. (A) Selective organ targeting (SORT) enables lipid nanoparticles (LNPs) to be systematically and predictably engineered to deliver mRNA into lung and spleen, in addition to liver. mRNA dendrimer-based lipid nanoparticles (mDLNPs) and SORT LNPs (20% DODAP) induced tdTom fluorescence specifically in the liver and SORT LNPs (50% DOTAP) selectively edited the lung. Co-delivery of Cas9 mRNA and modified sgPCSK9 was achieved with 20% DODAP SORT LNPs in C57BL/6 mice via 3 IV injections (days 0, 2, 4). After 9 days, ~60% indel at the PCSK9 locus of liver tissue was detected, leading to ~100% PCSK9 protein reduction in mice serum. Reproduced with permission from ref 100. Copyright 2020 Springer Nature. (B) Liposome-templated hydrogel nanoparticles (LHNPs) were core–shell structures, where the core and shell were formed by DOTAP liposomes and PEI hydrogel, respectively. Systemic injection of iRGD conjugated LHNPs targeting polo-like kinase 1 (Plk1) significantly suppressed tumor proliferation in mice bearing U87 flank tumors. Reproduced from ref 101. Copyright 2017 John Wiley & Sons, Inc. (C) Tumor environment responsive copolymer (PLys100-CA-mPEG77) were mixed with Cas9 ribonucleoproteins and formed stable nanostructures, named as nanoRNP. Intravenous injection of nanoRNP targeting STAT3 and RUNX1 gene loci effectively inhibited tumor growth in a heterogeneous tumor model. Reproduced from ref 102. Copyright 2019 American Chemical Society. (D) Cationic lipid-assisted PEG-b-PLGA nanoparticles (CLANs) encapsulating CD68 promoter-driven CRISPR/Cas9 plasmids could be internalized into diverse cells post intravenous injection, but only allowing Cas9 expressed in monocytes and macrophages. Intravenous injection of CLANpM330/sgNtn1 successfully knocked-out at the Ntn1 gene locus, specifically in the macrophages and monocytes, inhibited the expression of netrin-1 in macrophages and finally improved type 2 diabetes symptoms in the mouse model. Reproduced with permission from ref 103. Copyright 2018 American Chemical Society.
Targeting Tumors
Nonviral delivery systems have been widely explored for antitumor therapy. With the development of genome-editing technology, knocking out tumor-related genes using CRISPR/Cas9 systems becomes a promising strategy to inhibit tumor proliferation. Chen et al. constructed iRGD conjugated liposome-templated hydrogel nanoparticles (LHNPs) as a targeted CRISPR/Cas9 delivery system with the ability to encapsulate Cas9 protein and mini circle DNA encoding sgRNA.101 Systemic treatment with iRGD-LHNPs targeting Plk1 gene suppressed tumor proliferation in mice bearing U87 flank tumors. Furthermore, they encapsulated Lexiscan, a small drug known to improve blood-brain barrier permeability, into iRGD-LHNPs. Intravenous administration of these engineered LHNPs increased survival rates for mice bearing intracranial U87 tumors (Figure 8B). Recently, Liu et al. designed a tumor-environment-responsive copolymer (PLys100-CA-mPEG77) that was able to form nanostructures by mixing with Cas9 ribonucleoproteins, named nanoRNP.102 Upon intravenous injection, nanoRNP carrying two sgRNA sequences targeting STAT3 and RUNX1 gene loci effectively inhibited tumor growth in a heterogeneous tumor model (Figure 8C).
Targeting Macrophages
Nonviral vectors deliver CRISPR/Cas9 systems into macrophages via systemic administration. Wang et al. constructed Cas9 expression plasmids driven by a macrophage-specific promoter (human CD68 promoter).103 Cationic lipid-assisted PEG-b-PLGA nanoparticles (CLANs) encapsulating these plasmids were shown to express Cas9 protein in macrophages and monocytes in vivo. By adding a guide RNA sequence targeting Ntn1 into these plasmids, CLANs encapsulating these plasmids efficiently accomplished three things: 1) knock-out of the Ntn1 gene, specifically in the macrophages and monocytes in vivo after intravenous injection; 2) inhibition of netrin-1 expression in macrophages; and 3) improvement of type 2 diabetes symptoms in the mouse model (Figure 8D). In addition to CRISPR/Cas9 plasmids, the authors also screened and found an optimized CLAN that co-delivered Cas9 mRNA and sgRNA into macrophages in vivo for the treatment of inflammatory diseases.104 Lee et al. reported the discovery of AuNP-based nanocomposites that were generated through the co-engineering of AuNPs and Cas9 proteins including a 20-glutamic acid tag (Cas9 E20). Systemic injection of this complex into mice led to 8% and 4% of gene editing at the PTEN locus in the macrophages of the liver and spleen. 105
Challenges for Nonviral Delivery
Efficiency
Of paramount importance will be the ability of nonviral vectors to achieve gene editing at levels that are high enough to result in therapeutic benefits to patients in the clinic. Great progress has been made for high editing efficiency in the liver, and this efficiency will next need to be achieved in other tissues and cell types. As discussed previously, nonviral delivery methods enable multiple doses to be administered sequentially, which inherently increases the total cumulative editing levels that are achievable. Advantages of synthetic carriers, such as minimized immunogenicity, a reduction in the number of off-target effects, and the ability to administer repeat doses, may overcome some issues related to viral delivery. An additional concern is the fact that many genetic diseases are not able to be treated effectively via knockout of specific genes and/or gene sequences. Rather, many of these genetic diseases require correction of a mutated genetic sequence or knock-in of a specific gene. Currently, the frequency of HDR, which would enable these types of corrections, is much lower than that of indels. Therefore, the elucidation of methods that increase the efficiency of HDR over NHEJ is of exceptional importance and yet also challenging, as the exact mechanisms underlying these processes are, at this time, not well understood. Some recent efforts have been focused on increasing HDR. For example, covalently linking a DNA repair template to Cas9/sgRNA RNPs has been reported to improve HDR efficiency.106–107 Additionally, Cpf1 (or Cas12a) with staggered cutting was demonstrated to prefer HDR more than traditional Cas9 protein, which generates blunt-end cutting.20 Also, a fusion nickase named hRad51-Cas9(D10A) was able to mediate HDR without generating double-stranded DNA breaks, which again favors a higher HDR:indel ratio and low off-target events.108 Thus, developments in Cas protein design may further aid advancement of HDR correction in combination with improved synthetic carriers.
Specificity
Developing nonviral vectors that can preferentially deliver CRISPR components into targeted tissues and targeted cell types will expand the potential targets of genome-editing therapies. Currently, LNPs targeting hepatocytes in the liver via systemic delivery are the most advanced,109 but a number of studies have described LNP delivery to nonhepatic tissues such as the endothelium80, 99–100, 110 and immune cells.80, 100, 110 One obstacle to broad application of nonviral vector-based gene editing is the lack of nanovectors that can target specific tissues or cell types for precise gene modification. That being said, the SORT technology described above is a promising advance that may overcome some of these problems due to its predictable nature and tissue selectivity.100
Off-Target
In addition to high on-target efficiency and specificity, the risk of off-target effects, which may potentially knockout other genes and lead to fatal diseases, should also be seriously considered. Many efforts have been made to minimize off-target events. Several bioinformatic online tools have been developed to help design sgRNAs with an explanation of all possible off-target sites, so that sgRNA sequences can be selected with minimized numbers of off-target sites for in vivo gene editing. The choice of Cas9 protein and sgRNAs is also essential, as high-fidelity Cas9 proteins were identified to reduce off-target mutation in vivo.111 Chemically or structurally modified sgRNAs also demonstrate a reduction in off-target cleavage compared to unmodified sgRNAs.112 Off-target edits may also be related to the delivery strategies of the CRISPR/Cas9 system. Viral delivery and plasmid delivery show significantly higher off-target effects than do mRNA delivery and RNP delivery.113 Even though they are targeting the correct gene loci, gene cutting at unwanted tissues or cells (i.e., low specificity) may also be considered off-target effects. Therefore, developing nonviral vectors that can target specific tissues will largely reduce off-target events.
Immunogenicity
Components of CRISPR/Cas9 systems (Cas9 proteins, Cas9 mRNAs, or sgRNAs) delivered by nonviral vectors may trigger host immune responses. Anti-Cas9 antibodies have been reported to exist in human populations.114 RNA cargos delivered may also activate innate immune responses in vivo.115–116 In addition, some nonviral vectors may also be recognized by host immune systems. The activation of the immune system may not only significantly decrease editing efficiency in vivo but may also result in severe safety concerns in patients. Encapsulating CRISPR components inside nanovectors partially shields their recognition by the host immune system and largely reduces the generation of immune responses in comparison to viral systems. In sum, the potential of nonviral vectors to trigger an immune response should be carefully considered when designing nonviral CRISPR delivery systems.
Conclusions and Prospects
The CRISPR/Cas9 platform is an unprecedented technological leap in genome editing that is quickly reaching all aspects of biomedical research and therapeutic discovery. In the short time since CRISPR/Cas was first reported, numerous biotechnology companies have been founded to develop the therapeutic potential of this revolutionary technology further. Large pharma has also become involved, positioning CRISPR to affect many therapeutic modalities and areas. Currently, most of the ongoing clinical trials employing CRISPR/Cas-mediated gene editing are relating to chimeric antigen receptor T cells (CAR-T) therapies, which are generated using ex vivo editing approaches (Table 1). These approaches may largely avoid current challenges associated with in vivo techniques (e.g., specificity, off-targets, and immunogenicity), but ex vivo approaches are still challenging and could induce innate negative immune responses. For the clinical translation of direct in vivo genome editing, challenges remain ahead. Encouragingly, many pharmaceutical companies are developing gene therapies employing CRISPR/Cas9 systems for genetic disease treatments. Vertex Pharmaceuticals recently acquired Exonics Therapeutics, a company focusing on DMD therapies using in vivo CRISPR/Cas9 technology to repair exon mutations and restore dystrophin expression. CRISPR Therapeutics and Editas Medicine are also developing DMD therapies via in vivo CRISPR-based gene editing. Likewise, Vertex and Editas are also actively developing CRISPR/Cas9-based gene therapies for cystic fibrosis (CF) treatments. In conjunction with DMD and CF, local delivery of CRISPR/Cas9 systems targeting other genetic diseases such as Leber congenital amaurosis 10 and alpha-I antitrypsin deficiency, are also under development, according to these companies’ pipelines and clinical trial registries. Going forward, developing delivery strategies that combine high tissue specificity, high editing efficiency, and minimized toxicity are critical to accelerate the clinical translation of in vivo CRISPR/Cas9 based therapies. Given the rapid progress of CRISPR/Cas9 therapeutics in just a few short years, there is optimism that advances in viral and nonviral delivery approaches will someday yield the creation of safe and effective cell-specific therapies to treat human disease.
Table 1.
Ongoing clinical trials of CRISPR/Cas9 based gene therapies.
All clinical trials listed in this table are based on ClinicalTrials.gov.117 AAVS, adeno-associated viruses; hHSPCs, human hematopoietic stem and progenitor cells; EBV-CTLs, Epstein-Barr virus-specific cytotoxic T lymphocytes; CAR-T, chimeric antigen receptor (CAR) T-cell; iHSCs, induced hematopoietic stem cells; MSCs, mesenchymal stem cells; CISH, cytokine-induced SH2 protein; TIL, tumor infiltrating lymphocytes.
| Study Title | Conditions/Disease | Target Gene | Type | Delivery Systems | Phase | Status | ClinicalTrials.gov identifier |
|---|---|---|---|---|---|---|---|
| Single Ascending Dose Study in Participants With LCA10 | Blindness; Leber Congenital Amaurosis 10 Vision Disorders Eye Diseases Retinal Disease | CEP290 (Correction) | Virus (AAV5) | Adeno-associated virus | Phase 1/2 | Recruiting | NCT03872479 |
| A Safety and Efficacy Study Evaluating CTX001 in Subjects With Transfusion-Dependent β-Thalassemia | Beta-Thalassemia Thalassemia Genetic Diseases, Inborn Hematologic Diseases Hemoglobinopathies | BCL11A | CD34+ hHSPCs | Ex vivo | Phase 1/2 | Recruiting | NCT03655678 |
| A Safety and Efficacy Study Evaluating CTX001 in Subjects With Severe Sickle Cell Disease | Sickle Cell Disease Hematological Diseases Hemoglobinopathies | BCL11A | CD34+ hHSPCs | Ex vivo | Phase 1/2 | Recruiting | NCT03745287 |
| PD-1 Knockout EBV-CTLs for Advanced Stage Epstein-Barr Virus (EBV) Associated Malignancies | Stage IV Gastric Carcinoma Stage IV Nasopharyngeal Carcinoma T-Cell Lymphoma Stage IV Stage IV Adult Hodgkin Lymphoma Stage IV Diffuse Large B-Cell Lymphoma | PD-1 | EBV-CTLs | Ex vivo | Phase 1/2 | Recruiting | NCT03044743 |
| A Study of Metastatic Gastrointestinal Cancers Treated With Tumor Infiltrating Lymphocytes in Which the Gene Encoding the Intracellular Immune Checkpoint CISH Is Inhibited Using CRISPR Genetic Engineering | Gastrointestinal Epithelial Cancer Gastrointestinal Neoplasms Cancer of Gastrointestinal Tract Cancer, Gastrointestinal Gastrointestinal Cancer Colo-rectal Cancer Pancreatic Cancer Gall Bladder Cancer Colon Cancer Esophageal Cancer Stomach Cancer | CISH | TIL | Ex vivo | Phase 1/2 | Recruiting | NCT04426669 |
| A Safety and Efficacy Study Evaluating CTX110 in Subjects With Relapsed or Refractory B-Cell Malignancies | B-cell Malignancy Non-Hodgkin Lymphoma B-cell Lymphoma | CD19 | CAR-T | Ex vivo | Phase 1/2 | Recruiting | NCT04035434 |
| A Feasibility and Safety Study of Universal Dual Specificity CD19 and CD20 or CD22 CAR-T Cell Immunotherapy for Relapsed or Refractory Leukemia and Lymphoma | B Cell Leukemia B Cell Lymphoma | CD19 and CD20 or CD22 | CAR-T | Ex vivo | Phase 1/2 | Recruiting | NCT03398967 |
| A Study Evaluating UCART019 in Patients With Relapsed or Refractory CD19+ Leukemia and Lymphoma | B Cell Leukemia B Cell Lymphoma | CD19 | CAR-T | Ex vivo | Phase 1/2 | Recruiting | NCT03166878 |
| A Safety and Efficacy Study Evaluating CTX120 in Subjects With Relapsed or Refractory Multiple Myeloma | Multiple Myeloma | BCMA | CAR-T | Ex vivo | Phase 1 | Recruiting | NCT04244656 |
| Study of PD-1 Gene-knocked Out Mesothelin-directed CAR-T Cells With the Conditioning of PC in Mesothelin Positive Multiple Solid Tumors | Solid Tumor, Adult | PD-1 | CAR-T | Ex vivo | Phase 1 | Recruiting | NCT03747965 |
| Cell Therapy for High Risk T-Cell Malignancies Using CD7-Specific CAR Expressed On Autologous T Cells | T-cell Acute Lymphoblastic Leukemia T-cell Acute Lymphoblastic Lymphoma T-non-Hodgkin Lymphoma | CD-7 | CAR-T | Ex vivo | Phase 1 | Not yet recruiting | NCT03690011 |
| Study of CRISPR-Cas9 Mediated PD-1 and TCR Gene-knocked Out Mesothelin-directed CAR-T Cells in Patients With Mesothelin Positive Multiple Solid Tumors. | Solid Tumor, Adult | PD-1/TCR | CAR-T | Ex vivo | Phase 1 | Recruiting | NCT03545815 |
| CRISPR (HPK1) Edited CD19-specific CAR-T Cells (XYF19 CAR-T Cells) for CD 19+ Leukemia or Lymphoma | Leukemia Lymphocytic Acute (ALL) in Relapse Leukemia Lymphocytic Acute (All) Refractory Lymphoma, B-Cell CD 19 Positive | CD19 | CAR-T | Ex vivo | Phase 1 | Recruiting | NCT04037566 |
| A Safety and Efficacy Study Evaluating CTX130 in Subjects With Relapsed or Refractory Renal Cell Carcinoma | Renal Cell Carcinoma | CD70 | CAR-T | Ex vivo | Phase 1 | Recruiting | NCT04438083 |
| TACE Combined With PD-1 Knockout Engineered T Cell in Advanced Hepatocellular Carcinoma | Advanced Hepatocellular Carcinoma | PD-1 | T cells | Ex vivo | Phase 1 | Recruiting | NCT04417764 |
| PD-1 Knockout Engineered T Cells for Metastatic Non-small Cell Lung Cancer | Metastatic Non-small Cell Lung Cancer | PD-1 | T cells | Ex vivo | Phase 1 | Active, not recruiting | NCT02793856 |
| A Safety and Efficacy Study of TALEN and CRISPR/Cas9 in the Treatment of HPV-related Cervical Intraepithelial Neoplasial | Human Papillomavirus-Related Malignant Neoplasm | E6/E7 | Plasmid | Gel | Phase 1 | Unknown | NCT03057912 |
| Cell Therapy for High Risk T-Cell Malignancies Using CD7-Specific CAR Expressed On Autologous T Cells | T-cell Acute Lymphoblastic Leukemia T-cell Acute Lymphoblastic Lymphoma T-non-Hodgkin Lymphoma | CD7 | CAR-T | Ex vivo | Phase 1 | Not yet recruiting | NCT03690011 |
| iHSCs With the Gene Correction of HBB Intervent Subjests With β-thalassemia Mutations | Thalassemia | HBB (Correction) | iHSCs | Ex vivo | Early Phase 1 | Not yet recruiting | NCT03728322 |
Acknowledgments
R.L. acknowledges support from the National Institutes of Health (NIH) (EB000244). D.J.S. acknowledges support from the Cystic Fibrosis Foundation (CFF) (SIEGWA18XX0), NIH National Institute of Biomedical Imaging and Bioengineering (NIBIB) (R01 EB025192-01A1), American Cancer Society (ACS) (RSG-17-012-01), and the Robert A. Welch Foundation (I-1855). T.W. acknowledges support from the Cancer Prevention and Research Institute of Texas (CPRIT) Training Grant (RP160157).
Footnotes
The authors declare competing financial interests. D.G.A. is a founder of CRISPR Tx and a consultant with Translate Bio. For a list of entities with which R.L. is involved, compensated, or uncompensated, see www.dropbox.com/s/yc3xqb5s8s94v7x/Rev%20Langer%20COI.pdf?dl=0. DJ.S. is a co-founder of, and consultant to, ReCode Therapeutics.
References
- (1).Jinek M; Chylinski K; Fonfara I; Hauer M; Doudna JA; Charpentier E, A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337(6096), 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cong L; Ran FA; Cox D; Lin SL; Barretto R; Habib N; Hsu PD; Wu XB; Jiang WY; Marraffini LA; Zhang F, Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339 (6121), 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Mali P; Yang LH; Esvelt KM; Aach J; Guell M; DiCarlo JE; Norville JE; Church GM, RNA-guided human genome engineering via Cas9. Science 2013, 339 (6121), 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Doudna JA; Charpentier E, Genome editing: The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346 (6213), 1258096. [DOI] [PubMed] [Google Scholar]
- (5).Sander JD; Joung JK, CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol 2014, 32 (4), 347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Wang HX; Li M; Lee CM; Chakraborty S; Kim HW; Bao G; Leong KW, CRISPR/Cas9-based genome editing for disease modeling and therapy: Challenges and opportunities for nonviral delivery. Chem. Rev 2017,117 (15), 9874–9906. [DOI] [PubMed] [Google Scholar]
- (7).Yin H; Xue W; Anderson DG, CRISPR-Cas: A tool for cancer research and therapeutics. Nat. Rev. Clin. Oncol 2019,16 (5), 281–295. [DOI] [PubMed] [Google Scholar]
- (8).Nelson CE; Robinson-Hamm JN; Gersbach CA, Genome engineering: a new approach to gene therapy for neuromuscular disorders. Nat. Rev. Neurol 2017, 13 (11), 647–661. [DOI] [PubMed] [Google Scholar]
- (9).Pickar-Oliver A; Gersbach CA, The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20 (8), 490–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sanchez-Rivera FJ; Jacks T, Applications of the CRISPR-Cas9 system in cancer biology. Nat. Rev. Cancer 2015,15 (7), 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wu WY; Lebbink JHG; Kanaar R; Geijsen N; van der Oost J, Genome editing by natural and engineered CRISPR-associated nucleases. Nat. Chem. Bio. 2018,14 (7), 642–651. [DOI] [PubMed] [Google Scholar]
- (12).Wilbie D; Walther J; Mastrobattista E, Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc. Chem. Res 2019, 52 (6), 1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Yin H; Kauffman KJ; Anderson DG, Delivery technologies for genome editing. Nat. Rev. Drug Discov 2017,16 (6), 387–399. [DOI] [PubMed] [Google Scholar]
- (14).Lino CA; Harper JC; Carney JP; Timlin JA, Delivering CRISPR: a review of the challenges and approaches. Drug Deliv. 2018, 25 (1), 1234–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Wang L; Zheng W; Liu S; Li B; Jiang X, Delivery of CRISPR/Cas9 by novel strategies for gene therapy. Chembiochem 2019, 20 (5), 634–643. [DOI] [PubMed] [Google Scholar]
- (16).Li L; Hu S; Chen X, Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018,171, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Qin W; Dion SL; Kutny PM; Zhang Y; Cheng AW; Jillette NL; Malhotra A; Geurts AM; Chen YG; Wang H, Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics 2015, 200 (2), 423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Raveux A; Vandormael-Pournin S; Cohen-Tannoudji M, Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote. Sci. Rep 2017, 7, 42661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wu Y; Zeng J; Roscoe BP; Liu P; Yao Q; Lazzarotto CR; Clement K; Cole MA; Luk K; Baricordi C; Shen AH; Ren C; Esrick EB; Manis JP; Dorfman DM; Williams DA; Biffi A; Brugnara C; Biasco L; Brendel C; Pinello L; Tsai SQ; Wolfe SA; Bauer DE, Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat. Med. 2019, 25 (5), 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Shahbazi R; Sghia-Hughes G; Reid JL; Kubek S; Haworth KG; Humbert O; Kiem H-P; Adair JE, Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat. Mater 2019,18 (10), 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Patmanathan SN; Gnanasegaran N; Lim MN; Husaini R; Fakiruddin KS; Zakaria Z, CRISPR/Cas9 in stem cell research: Current application and future perspective. Curr. Stem Cell Res. Ther 2018,13 (8), 632–644. [DOI] [PubMed] [Google Scholar]
- (22).Hsu MN; Chang YH; Truong VA; Lai PL; Nguyen TKN; Hu YC, CRISPR technologies for stem cell engineering and regenerative medicine. Biotechnol Adv 2019, 107447. [DOI] [PubMed] [Google Scholar]
- (23).Wang M; Zuris JA; Meng FT; Rees H; Sun S; Deng P; Han Y; Gao X; Pouli D; Wu Q; Georgakoudi I; Liu DR; Xu QB, Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A 2016,113 (11), 2868–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yin J; Wang Q; Hou S; Bao LC; Yao WB; Gao XD, Potent protein delivery into mammalian cells via a supercharged polypeptide. J. Am. Chem. Soc 2018,140 (49), 17234–17240. [DOI] [PubMed] [Google Scholar]
- (25).Zhou W; Cui H; Ying L; Yu XF, Enhanced cytosolic delivery and release of CRISPR/Cas9 by black phosphorus nanosheets for genome editing. Angew. Chem. Int. Ed. 2018, 57 (32), 10268–10272. [DOI] [PubMed] [Google Scholar]
- (26).Yue HH; Zhou XM; Cheng M; Xing D, Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale 2018,10 (3), 1063–1071. [DOI] [PubMed] [Google Scholar]
- (27).Schwank G; Koo BK; Sasselli V; Dekkers JF; Heo I; Demircan T; Sasaki N; Boymans S; Cuppen E; van der Ent CK; Nieuwenhuis EE; Beekman JM; Clevers H, Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13 (6), 653–8. [DOI] [PubMed] [Google Scholar]
- (28).Matano M; Date S; Shimokawa M; Takano A; Fujii M; Ohta Y; Watanabe T; Kanai T; Sato T, Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med 2015, 21 (3), 256–262. [DOI] [PubMed] [Google Scholar]
- (29).Fujii M; Matano M; Nanki K; Sato T, Efficient genetic engineering of human intestinal organoids using electroporation. Nat. Protoc 2015,10 (10), 1474–1485. [DOI] [PubMed] [Google Scholar]
- (30).Roper J; Yilmaz OH, Breakthrough moments: Genome editing and organoids. Cell Stem Cell 2019, 24 (6), 841–842. [DOI] [PubMed] [Google Scholar]
- (31).Xu CL; Ruan MZC; Mahajan VB; Tsang SH, Viral Delivery Systems for CRISPR. Viruses 2019,11 (1), E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Lau CH; Suh Y, In vivo genome editing in animals using AAV-CRISPR system: applications to translational research of human disease. F1000Res. 2017, 6, 2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Blasco RB; Karaca E; Ambrogio C; Cheong TC; Karayol E; Minero VG; Voena C; Chiarle R, Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep. 2014, 9 (4), 1219–1227. [DOI] [PubMed] [Google Scholar]
- (34).Platt RJ; Chen SD; Zhou Y; Yim MJ; Swiech L; Kempton HR; Dahlman JE; Parnas O; Eisenhaure TM; Jovanovic M; Graham DB; Jhunjhunwala S; Heidenreich M; Xavier RJ; Langer R; Anderson DG; Hacohen N; Regev A; Feng GP; Sharp PA; Zhang F, CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014,159 (2), 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Chiou SH; Winters IP; Wang J; Naranjo S; Dudgeon C; Tamburini FB; Brady JJ; Yang DA; Gruner BM; Chuang CH; Caswell DR; Zeng H; Chu P; Kim GE; Carpizo DR; Kim SK; Winslow MM, Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Gene Dev. 2015, 29 (14), 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Annunziato S; Kas SM; Nethe M; Yucel H; Del Bravo J; Pritchard C; Bin Ali R; van Gerwen B; Siteur B; Drenth AP; Schut E; van de Ven M; Boelens MC; Klarenbeek S; Huijbers IJ; van Miltenburg MH; Jonkers J, Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Gene Dev. 2016, 30 (12), 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Tabebordbar M; Zhu KX; Cheng JKW; Chew WL; Widrick JJ; Yan WX; Maesner C; Wu EY; Xiao R; Ran FA; Cong L; Zhang F; Vandenberghe LH; Church GM; Wagers AJ, In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351 (6271), 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nelson CE; Hakim CH; Ousterout DG; Thakore PI; Moreb EA; Rivera RMC; Madhavan S; Pan XF; Ran FA; Yan WX; Asokan A; Zhang F; Duan DS; Gersbach CA, In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351 (6271), 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Nelson CE; Wu Y; Gemberling MP; Oliver ML; Waller MA; Bohning JD; Robinson-Hamm JN; Bulaklak K; Castellanos Rivera RM; Collier JH; Asokan A; Gersbach CA, Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019, 25 (3), 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Gyorgy B; Loov C; Zaborowski MP; Takeda S; Kleinstiver BP; Commins C; Kastanenka K; Mu DK; Volak A; Giedraitis V; Lannfelt L; Maguire CA; Joung JK; Hyman BT; Breakefield XO; Ingelsson M, CRISPR/Cas9 mediated disruption of the Swedish APP allele as a therapeutic approach for early-onset Alzheimer’s Disease. Mol. Ther. - Nucl. Acids 2018,11, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Gyorgy B; Nist-Lund C; Pan BF; Asai Y; Karavitaki KD; Kleinstiver BP; Garcia SP; Zaborowski MP; Solanes P; Spataro S; Schneider BL; Joung JK; Geleoc GSG; Holt JR; Corey DP, Allele-specific gene editing prevents deafness in a model of dominant progressive hearing loss. Nat. Med. 2019, 25 (7), 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Min YL; Li H; Rodriguez-Caycedo C; Mireault AA; Huang J; Shelton JM; McAnally JR; Amoasii L; Mammen PPA; Bassel-Duby R; Olson EN, CRISPR-Cas9 corrects Duchenne muscular dystrophy exon 44 deletion mutations in mice and human cells. Science Adv. 2019, 5 (3), eaav4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Amoasii L; Long C; Li H; Mireault AA; Shelton JM; Sanchez-Ortiz E; McAnally JR; Bhattacharyya S; Schmidt F; Grimm D; Hauschka SD; Bassel-Duby R; Olson EN, Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 2017, 9 (418), eaan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zincarelli C; Soltys S; Rengo G; Koch WJ; Rabinowitz JE, Comparative cardiac gene delivery of Adeno-Associated Virus serotypes 1–9 reveals that AAV6 mediates the most efficient transduction in mouse heart. Clin. Transl. Sci. 2010, 3 (3), 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Long CZ; Li H; Tiburcy M; Rodriguez-Caycedo C; Kyrychenko V; Zhou HY; Zhang Y; Min YL; Shelton JM; Mammen PPA; Liaw NY; Zimmermann WH; Bassel-Duby R; Schneider JW; Olson EN, Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Science Adv. 2018, 4 (1), eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Rogers ZN; McFarland CD; Winters IP; Naranjo S; Chuang CH; Petrov D; Winslow MM, A quantitative and multiplexed approach to uncover the fitness Landscape of tumor suppression in vivo. Nat. Methods 2017,14 (7), 737–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Sanchez-Rivera FJ; Papagiannakopoulos T; Romero R; Tammela T; Bauer MR; Bhutkar A; Joshi NS; Subbaraj L; Bronson RT; Xue W; Jacks T, Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014, 516 (7531), 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Nishiyama J; Mikuni T; Yasuda R, Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron 2017, 96 (4), 755–768 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Yu WH; Mookherjee S; Chaitankar V; Hiriyanna S; Kim JW; Brooks M; Ataeijannati Y; Sun X; Dong LJ; Li TS; Swaroop A; Wu ZJ, Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun 2017, 8, 14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Kim J; Jo D; Kim J, Long-term effects of in vivo genome editing in the mouse retina using Campylobacter jejuni Cas9 expressed via adeno-associated virus. Brit. J. Pharmacol. 2019, 176 (16), 2984–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Yin H; Song C-Q; Dorkin JR; Zhu LJ; Li Y; Wu Q; Park A; Yang J; Suresh S; Bizhanova A; Gupta A; Bolukbasi MF; Walsh S; Bogorad RL; Gao G; Weng Z; Dong Y; Koteliansky V; Wolfe SA; Langer R; Xue W; Anderson DG, Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016, 34 (3), 328–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Jarrett KE; Lee CM; Yeh YH; Hsu RH; Gupta R; Zhang M; Rodriguez PJ; Lee CS; Gillard BK; Bissig KD; Pownall HJ; Martin JF; Bao G; Lagor WR, Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci. Rep 2017, 7, 44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).De Caneva A; Porro F; Bortolussi G; Sola R; Lisjak M; Barzel A; Giacca M; Kay MA; Vlahovicek K; Zentilin L; Muro AF, Coupling AAV-mediated promoterless gene targeting to SaCas9 nuclease to efficiently correct liver metabolic diseases. JCI Insight 2019, 5, 128863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Ohmori T; Nagao Y; Mizukami H; Sakata A; Muramatsu SI; Ozawa K; Tominaga SI; Hanazono Y; Nishimura S; Nureki O; Sakata Y, CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci. Rep. 2017, 7 (1), 4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Ran FA; Cong L; Yan WX; Scott DA; Gootenberg JS; Kriz AJ; Zetsche B; Shalem O; Wu XB; Makarova KS; Koonin EV; Sharp PA; Zhang F, In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520 (7546), 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Bengtsson NE; Hall JK; Odom GL; Phelps MP; Andrus CR; Hawkins RD; Hauschka SD; Chamberlain JR; Chamberlain JS, Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun 2017, 8, 14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Xie C; Zhang YP; Song L; Luo J; Qi W; Hu J; Lu D; Yang Z; Zhang J; Xiao J; Zhou B; Du JL; Jing N; Liu Y; Wang Y; Li BL; Song BL; Yan Y, Genome editing with CRISPR/Cas9 in postnatal mice corrects PRKAG2 cardiac syndrome. Cell Res. 2016,26 (10), 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Gaj T; Ojala DS; Ekman FK; Byrne LC; Limsirichai P; Schaffer DV, In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Science Adv. 2017, 3 (12), eaar3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).McClements ME; MacLaren RE, Adeno-associated Virus (AAV) Dual Vector Strategies for Gene Therapy Encoding Large Transgenes. Yale J. Biol. Med 2017, 90 (4), 611–623. [PMC free article] [PubMed] [Google Scholar]
- (60).Chamberlain K; Riyad JM; Weber T, Expressing transgenes that exceed the packaging capacity of Adeno-Associated Virus capsids. Hum. Gene Ther. Method 2016, 27 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Alsaiari SK; Patil S; Alyami M; Alamoudi KO; Aleisa FA; Merzaban JS; Li M; Khashab NM, Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework. J. Am. Chem. Soc. 2018,140 (1), 143–146. [DOI] [PubMed] [Google Scholar]
- (62).Mout R; Ray M; Yesilbag Tonga G; Lee YW; Tay T; Sasaki K; Rotello VM, Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano 2017,11 (3), 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Sun WJ; Ji WY; Hall JM; Hu QY; Wang C; Beisel CL; Gu Z, Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed 2015, 54 (41), 12029–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Miller JB; Zhang S; Kos P; Xiong H; Zhou K; Perelman SS; Zhu H; Siegwart DJ, Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed 2017, 56 (4), 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Jiang C; Mei M; Li B; Zhu XR; Zu WH; Tian YJ; Wang QN; Guo Y; Dong YZ; Tan X, A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and PCSK9 in vivo. Cell Res. 2017, 27 (3), 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Nguyen DN; Roth TL; Li PJ; Chen PA; Apathy R; Mamedov MR; Vo LT; Tobin VR; Goodman D; Shifrut E; Bluestone JA; Puck JM; Szoka FC; Marson A, Polymer-stabilized Cas9 nanoparticles and modified repair templates increase genome editing efficiency. Nat. Biotechnol. 2020, 38 (1), 44–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Mout R; Ray M; Lee YW; Scaletti F; Rotello VM, In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: Progress and challenges. Bioconjugate Chem. 2017, 28 (4), 880–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Liu C; Zhang L; Liu H; Cheng K, Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications. J. Controlled Release 2017, 266, 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Chen F; Alphonse M; Liu Q, Strategies for nonviral nanoparticle-based delivery of CRISPR/Cas9 therapeutics. WIRES Nanomed. Nanobi. 2020,12 (3), e1609. [DOI] [PubMed] [Google Scholar]
- (70).Wang HX; Song Z; Lao YH; Xu X; Gong J; Cheng D; Chakraborty S; Park JS; Li M; Huang D; Yin L; Cheng J; Leong KW, Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. U.S.A 2018,115 (19), 4903–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Wang P; Zhang L; Xie Y; Wang N; Tang R; Zheng W; Jiang X, Genome editing for cancer therapy: Delivery of Cas9 protein/sgRNA plasmid via a gold nanocluster/lipid core-shell nanocarrier. Adv. Sci 2017, 4 (11), 1700175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Wang P; Zhang L; Zheng W; Cong L; Guo Z; Xie Y; Wang L; Tang R; Feng Q; Hamada Y; Gonda K; Hu Z; Wu X; Jiang X, Thermo-triggered release of CRISPR-Cas9 system by lipid-encapsulated gold nanoparticles for tumor therapy. Angew. Chem. Int. Ed 2018, 57 (6), 1491–1496. [DOI] [PubMed] [Google Scholar]
- (73).Pan YC; Yang JJ; Luan XW; Liu XL; Li XQ; Yang J; Huang T; Sun L; Wang YZ; Lin YH; Song YJ, Near-infrared upconversion-activated CRISPR-Cas9 system: A remote-controlled gene editing platform. Science Adv. 2019, 5 (4), eaav7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Staahl BT; Benekareddy M; Coulon-Bainier C; Banfal AA; Floor SN; Sabo JK; Urnes C; Munares GA; Ghosh A; Doudna JA, Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol 2017, 35 (5), 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Park H; Oh J; Shim G; Cho B; Chang Y; Kim S; Baek S; Kim H; Shin J; Choi H; Yoo J; Kim J; Jun W; Lee M; Lengner CJ; Oh YK; Kim J, In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat. Neurosci 2019, 22 (4), 524–528. [DOI] [PubMed] [Google Scholar]
- (76).Lee B; Lee K; Panda S; Gonzales-Rojas R; Chong A; Bugay V; Park HM; Brenner R; Murthy N; Lee HY, Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2 (7), 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Min Y-L; Bassel-Duby R; Olson EN, CRISPR correction of Duchenne muscular dystrophy. Annu. Rev. Med. 2019, 70, 239–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Lee K; Conboy M; Park HM; Jiang F; Kim HJ; Dewitt MA; Mackley VA; Chang K; Rao A; Skinner C; Shobha T; Mehdipour M; Liu H; Huang WC; Lan F; Bray NL; Li S; Corn JE; Kataoka K; Doudna JA; Conboy I; Murthy N, Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017,1, 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Chen G; Abdeen AA; Wang Y; Shahi PK; Robertson S; Xie R; Suzuki M; Pattnaik BR; Saha K; Gong S, A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 2019,14 (10), 974–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Wei T; Cheng Q; Min YL; Olson EN; Siegwart DJ, Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 2020, 11 (1), 3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zuris JA; Thompson DB; Shu Y; Guilinger JP; Bessen JL; Hu JH; Maeder ML; Joung JK; Chen ZY; Liu DR, Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015, 33 (1), 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Gao X; Tao Y; Lamas V; Huang M; Yeh WH; Pan B; Hu YJ; Hu JH; Thompson DB; Shu Y; Li Y; Wang H; Yang S; Xu Q; Polley DB; Liberman MC; Kong WJ; Holt JR; Chen ZY; Liu DR, Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 2018, 553 (7687), 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Yang TC; Chang CY; Yarmishyn AA; Mao YS; Yang YP; Wang ML; Hsu CC; Yang HY; Hwang DK; Chen SJ; Tsai ML; Lai YH; Tzeng YH; Chang CC; Chiou SH, Carboxylated nanodiamond-mediated CRISPR-Cas9 delivery of human retinoschisis mutation into human iPSCs and mouse retina. Acta Biomater. 2020,101, 484–494. [DOI] [PubMed] [Google Scholar]
- (84).Ryu J-Y; Won E-J; Lee HAR; Kim JH; Hui E; Kim HP; Yoon T-J, Ultrasound-activated particles as CRISPR/Cas9 delivery system for androgenic alopecia therapy. Biomaterials 2020, 232, 119736. [DOI] [PubMed] [Google Scholar]
- (85).Finn JD; Smith AR; Patel MC; Shaw L; Youniss MR; van Heteren J; Dirstine T; Ciullo C; Lescarbeau R; Seitzer J; Shah RR; Shah A; Ling D; Growe J; Pink M; Rohde E; Wood KM; Salomon WE; Harrington WF; Dombrowski C; Strapps WR; Chang Y; Morrissey DV, A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018, 22 (9), 2227–2235. [DOI] [PubMed] [Google Scholar]
- (86).Liu J; Chang J; Jiang Y; Meng X; Sun T; Mao L; Xu Q; Wang M, Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 2019, 31 (33), 1902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Yin H; Song CQ; Suresh S; Wu Q; Walsh S; Rhym LH; Mintzer E; Bolukbasi MF; Zhu LJ; Kauffman K; Mou H; Oberholzer A; Ding J; Kwan SY; Bogorad RL; Zatsepin T; Koteliansky V; Wolfe SA; Xue W; Langer R; Anderson DG, Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 2017, 35 (12), 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Zhang L; Wang L; Xie Y; Wang P; Deng S; Qin A; Zhang J; Yu X; Zheng W; Jiang X, Triple-targeting delivery of CRISPR/Cas9 to reduce the risk of cardiovascular diseases. Angew. Chem. Int. Ed. 2019, 58 (36), 12404–12408. [DOI] [PubMed] [Google Scholar]
- (89).Kranz LM; Diken M; Haas H; Kreiter S; Loquai C; Reuter KC; Meng M; Fritz D; Vascotto F; Hefesha H; Grunwitz C; Vormehr M; Husemann Y; Selmi A; Kuhn AN; Buck J; Derhovanessian E; Rae R; Attig S; Diekmann J; Jabulowsky RA; Heesch S; Hassel J; Langguth P; Grabbe S; Huber C; Tureci O; Sahin U, Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534 (7607), 396–401. [DOI] [PubMed] [Google Scholar]
- (90).Miller JB; Kos P; Tieu V; Zhou K; Siegwart DJ, Development of cationic quaternary ammonium sulfonamide amino lipids for nucleic acid delivery. ACS Appl. Mater. Inter. 2018, 10 (3), 2302–2311. [DOI] [PubMed] [Google Scholar]
- (91).Fehring V; Schaeper U; Ahrens K; Santel A; Keil O; Eisermann M; Giese K; Kaufmann J, Delivery of therapeutic siRNA to the lung endothelium via novel lipoplex formulation DACC. Mol. Ther. 2014, 22 (4), 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Dahlman JE; Barnes C; Khan OF; Thiriot A; Jhunjunwala S; Shaw TE; Xing YP; Sager HB; Sahay G; Speciner L; Bader A; Bogorad RL; Yin H; Racie T; Dong YZ; Jiang S; Seedorf D; Dave A; Sandhu KS; Webber MJ; Novobrantseva T; Ruda VM; Lytton-Jean AKR; Levins CG; Kalish B; Mudge DK; Perez M; Abezgauz L; Dutta P; Smith L; Charisse K; Kieran MW; Fitzgerald K; Nahrendorf M; Danino D; Tuder RM; von Andrian UH; Akinc A; Panigrahy D; Schroeder A; Koteliansky V; Langer R; Anderson DG, In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 2014, 9 (8), 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Kaczmarek JC; Patel AK; Kauffman KJ; Fenton OS; Webber MJ; Heartlein MW; DeRosa F; Anderson DG, Polymer-lipid nanoparticles for systemic delivery of mRNA to the lungs. Angew. Chem. Int. Ed. 2016, 55 (44), 13808–13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Fenton OS; Kauffman KJ; Kaczmarek JC; McClellan RL; Jhunjhunwala S; Tibbitt MW; Zeng MD; Appel EA; Dorkin JR; Mir FF; Yang JH; Oberli MA; Heartlein MW; DeRosa F; Langer R; Anderson DG, Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. 2017, 29 (33), 1606944. [DOI] [PubMed] [Google Scholar]
- (95).Yan Y; Xiong H; Zhang X; Cheng Q; Siegwart DJ, Systemic mRNA delivery to the lungs by functional polyester-based carriers. Biomacromolecules 2017,18 (12), 4307–4315. [DOI] [PubMed] [Google Scholar]
- (96).Kowalski PS; Capasso Palmiero U; Huang Y; Rudra A; Langer R; Anderson DG, Ionizable amino-polyesters synthesized via ring opening polymerization of tertiary amino-alcohols for tissue selective mRNA delivery. Adv. Mater. 2018, 1801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Lokugamage MP; Sago CD; Gan Z; Krupczak BR; Dahlman JE, Constrained nanoparticles deliver siRNA and sgRNA to T cells in vivo without targeting ligands. Adv. Mater. 2019, 31 (41), el902251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Sago CD; Lokugamage MP; Islam FZ; Krupczak BR; Sato M; Dahlman JE, Nanoparticles that deliver RNA to bone marrow identified by in vivo directed evolution. J. Am. Chem. Soc. 2018,140 (49), 17095–17105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Sago CD; Lokugamage MP; Paunovska K; Vanover DA; Monaco CM; Shah NN; Gamboa Castro M; Anderson SE; Rudoltz TG; Lando GN; Munnilal Tiwari P; Kirschman JL; Willett N; Jang YC; Santangelo PJ; Bryksin AV; Dahlman JE, High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. U.S.A. 2018,115 (42), E9944–E9952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Cheng Q; Wei T; Farbiak L; Johnson LT; Dilliard SA; Siegwart DJ, Selective ORgan Targeting (SORT) nanoparticles for tissue specific mRNA delivery and CRISPR/Cas gene editing. Nat. Nanotechnol. 2020,15 (4), 313–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Chen Z; Liu F; Chen Y; Liu J; Wang X; Chen AT; Deng G; Zhang H; Liu J; Hong Z; Zhou J, Targeted delivery of CRISPR/Cas9-mediated cancer gene therapy via liposome-templated hydrogel nanoparticles. Adv. Fund. Mater 2017, 27 (46), 1703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Liu Q; Cai J; Zheng Y; Tan Y; Wang Y; Zhang Z; Zheng C; Zhao Y; Liu C; An Y; Jiang C; Shi L; Kang C; Liu Y, NanoRNP overcomes tumor heterogeneity in cancer treatment. Nano Lett. 2019, 19 (11), 7662–7672. [DOI] [PubMed] [Google Scholar]
- (103).Luo YL; Xu CF; Li HJ; Cao ZT; Liu J; Wang JL; Du XJ; Yang XZ; Gu Z; Wang J, Macrophage-specific in vivo gene editing using cationic lipid-assisted polymeric nanoparticles. ACS Nano 2018,12 (2), 994–1005. [DOI] [PubMed] [Google Scholar]
- (104).Xu C; Lu Z; Luo Y; Liu Y; Cao Z; Shen S; Li H; Liu J; Chen K; Chen Z; Yang X; Gu Z; Wang J, Targeting of NLRP3 inflammasome with gene editing for the amelioration of inflammatory diseases. Nat. Commun 2018, 9 (1), 4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Lee YW; Mout R; Luther DC; Liu Y; Castellanos-García L; Burnside AS; Ray M; Tonga GY; Hardie J; Nagaraj H, In vivo editing of macrophages through systemic delivery of CRISPR-Cas9-ribonucleoprotein-nanoparticle nanoassemblies. Adv. Ther 2019, 2(10), 1900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Savic N; Ringnalda FC; Lindsay H; Berk C; Bargsten K; Li Y; Neri D; Robinson MD; Ciaudo C; Hall J; Jinek M; Schwank G, Covalent linkage of the DNA repair template to the CRISPR-Cas9 nuclease enhances homology-directed repair. eLife 2018, 7, e33761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Aird EJ; Lovendahl KN; St Martin A; Harris RS; Gordon WR, Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun. Biol 2018, 7, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Rees HA; Yeh WH; Liu DR, Development of hRad51-Cas9 nickase fusions that mediate HDR without double-stranded breaks. Nat. Commun 2019, 10 (1), 2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Cheng Q; Wei T; Jia Y; Farbiak L; Zhou K; Zhang S; Wei Y; Zhu H; Siegwart DJ, Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater 2018, 30 (52), e1805308. [DOI] [PubMed] [Google Scholar]
- (110).Kaczmarek JC; Kauffman KJ; Fenton OS; Sadtler K; Patel AK; Heartlein MW; DeRosa F; Anderson DG, Optimization of a degradable polymer-lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 2018, 18 (10), 6449–6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Kleinstiver BP; Pattanayak V; Prew MS; Tsai SQ; Nguyen NT; Zheng ZL; Joung JK, High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529 (7587), 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Yin H; Song CQ; Suresh S; Kwan SY; Wu Q; Walsh S; Ding J; Bogorad RL; Zhu LJ; Wolfe SA; Koteliansky V; Xue W; Langer R; Anderson DG, Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat. Chem. Bio 2018,14 (3), 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Lattanzi A; Meneghini V; Pavani G; Amor F; Ramadier S; Felix T; Antoniani C; Masson C; Alibeu O; Lee C; Porteus MH; Bao G; Amendola M; Mavilio F; Miccio A, Optimization of CRISPR/Cas9 delivery to human hematopoietic stem and progenitor cells for therapeutic genomic rearrangements. Mol. Ther 2019, 27(1), 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Charlesworth CT; Deshpande PS; Dever DP; Camarena J; Lemgart VT; Cromer MK; Vakulskas CA; Collingwood MA; Zhang L; Bode NM; Behlke MA; Dejene B; Cieniewicz B; Romano R; Lesch BJ; Gomez-Ospina N; Mantri S; Pavel-Dinu M; Weinberg KI; Porteus MH, Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med 2019, 25 (2), 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Kauffman KJ; Mir FF; Jhunjhunwala S; Kaczmarek JC; Hurtado JE; Yang JH; Webber MJ; Kowalski PS; Heartlein MW; DeRosa F; Anderson DG, Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 2016,109, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (116).Guo S; Li H; Ma M; Fu J; Dong Y; Guo P, Size, shape, and sequence-dependent immunogenicity of RNA nanoparticles. Mol. Ther. - Nucl. Acids 2017, 9, 399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (117).ClinicalTrials.gov is a database of privately and publicly funded clinical studies conducted around the world. https://clinicaltrials.gov/. [Google Scholar]


