Graphical abstract
Keywords: SARS-CoV2 evolution, Infectivity, Viral mutations, Lung organoid, Machine learning, Simulations of COVID-19
Abstract
The global pandemic caused by the SARS-CoV-2 virus continues to spread. Infection with SARS- CoV-2 causes COVID-19, a disease of variable severity. Mutation has already altered the SARS-CoV-2 genome from its original reported sequence and continued mutation is highly probable. These mutations can: (i) have no significant impact (they are silent), (ii) result in a complete loss or reduction of infectivity, or (iii) induce increase in infectivity. Physical generation, for research purposes, of viral mutations that could enhance infectivity are controversial and highly regulated. The primary purpose of this project was to evaluate the ability of the DeepNEU machine learning stem-cell simulation platform to enable rapid and efficient assessment of the potential impact of viral loss-of-function (LOF) and gain-of-function (GOF) mutations on SARS-CoV-2 infectivity. Our data suggest that SARS-CoV-2 infection can be simulated in human alveolar type lung cells. Simulation of infection in these lung cells can be used to model and assess the impact of LOF and GOF mutations in the SARS-CoV2 genome. We have also created a four- factor infectivity measure: the DeepNEU Case Fatality Rate (dnCFR). dnCFR can be used to assess infectivity based on the presence or absence of the key viral proteins (NSP3, Spike-RDB, N protein, and M protein). dnCFR was used in this study, not to only assess the impact of different mutations on SARS-CoV2 infectivity, but also to categorize the effects of mutations as loss of infectivity or gain of infectivity events.
1. Introduction
The continuing evolution of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome remains a major obstacle to developing effective antiviral and vaccine therapies [1], [2]. The potential to better understand and predict this evolution will assist in the early detection of drug-resistant strains and facilitate the development of effective antiviral drugs and vaccines [3].
One important focus in the field of virology is to develop a better understanding of the impact of genetic mutation on infectivity [4], [5]. Here we define infectivity as the ratio of individuals who become infected divided by the number who are exposed to the virus. Predicting viral evolution is a fundamental goal in virology and this is especially true for pathogenic viruses [6]. Genomic information about current viral pathogens like SARS-CoV-2 and their continued evolution will provide a better understanding of the dynamics of future virus evolution and shed light on effective strategies to contain future outbreaks [7], [8].
Even though RNA viruses have a limited genome and a relatively limited evolutionary capacity, they do present unique challenges to predicting the impact of evolutionary changes. RNA viruses are known for their high mutation rates (around 1 mutation in 1,000 bases) and frequent recombination that can produce novel genotypes from co-circulating strains [6]. RNA viruses additionally undergo frequent mutations as they circulate in the population as in response to host factors. Of note, a recent variant analysis of SARS-CoV-2 has shown that, out of 10 022 SARS CoV-2 genomes that were analyzed, there were 65,776 variants detected and 5775 of them were distinct variants. These 5775 distinct variants included 2969 missense mutations, and 1965 point-mutations [9].The feasibility of predicting viral evolution relies upon on the breadth and scale of well posed questions and calls for cautious optimism [10].
Machine learning-based predictions of the impact of genetic mutations has been efficiently utilized in the field of viral genetics for many years, and have been mostly focused on the prediction of viral mutations that are associated with drug resistance [10]. Given the ongoing SARS-CoV-2 global pandemic and the emergence of new variants of concern, it is highly desirable to have a fast, reliable and efficient machine learning platform for simulating viable mutations and studying their potential effects on the infectivity of SARS-CoV-2 as well as future viral pathogens for which we are not prepared.
An important goal of this study is to simulate the natural evolution of the post-pandemic strain of SARS-CoV-2 by systematically introducing both gain of function (GOF) and loss of function mutations (LOF) into the viral genome. The overarching objective of this study is to identify potential therapeutic targets and improve preparedness for future epidemic/pandemic outbreaks of new strains of SARS-CoV-2 and other viral pathogens. In this study we simulate the impact of predicted GOF and LOF mutations in the SARS-CoV-2 genome. We have also developed a measure of viral infectivity, DeepNEU case fatality rate (dnCFR) for estimating changes in the SARS-CoV-2 case CFR in response to predicted GOF and LOF mutations. Our literature validated deep machine learning platform, DeepNEU v5.0, has successfully identified the infectivity potential of SARS-CoV-2 mutations well ahead of when they could occur and be identified in nature. These discoveries will offer the possibility of improving viral pandemic preparedness and better targeting surveillance between and during epidemics/pandemics.
1.1. Methods
The DeepNEU stem cell simulation platform is a literature validated hybrid deep-machine learning system with elements of fully connected recurrent neural networks (RNN), cognitive maps (CM), support vector machines (SVM) and evolutionary systems (GA). The detailed methodology for simulation development and validation plus the description of the current database (DeepNEU v5.0) used in these experiments has been described in [11], [12], [13].
1.1.1. The DeepNEU simulations
The main goal of this project was to extend our previous DeepNEU based research into SARS-CoV-2 infection by evaluating the potential impact of simulated LOF and GOF mutations in the viral genome on viral infectivity. As described previously [13] we first created computer simulations (aiPSC) of human induced pluripotent stem cells (iPSC) and lung (aiLUNG) cells. Once validated, the aiLUNG simulations were exposed to simulated SARS-CoV-2 viremia by turning on extracellular Spike-RBP (Receptor Binding Domain) in the presence of active Transmembrane Serine Protease 2 (TMPRSS2) [13]. The simulated SARS-CoV-2 infection of AT1 and AT2 lung cells (aiLUNG-COVID-19) was confirmed using a profile of genotypic and phenotypic features from the published literature [14], [15]. Finally, the validated aiLUNG and aiLUNG-COVID-19 simulations were used to evaluate an inclusive set of factors derived from the published SARS-CoV-2 genome (Accession number: NC_045512.2; https://www.ncbi.nlm.nih.gov/sars-cov-2/) regarding their ability to affect an increase or decrease in infectivity. A summary of the fifteen SARS-CoV-2 gene/gene products evaluated in the current experiments are presented in Table 1.
Table 1.
Summary of evaluated LOF and GOF mutations in the SARS-CoV-2 genome (N = 15 × 2).
| SARS-CoV-2 Target | Loss of Function | Gain of Function |
|---|---|---|
| aiPSC-WT | N/A | N/A |
| aiLUNG (i.e. Uninfected) | N/A | N/A |
| aiLUNG + SARS-CoV-2 | N/A | N/A |
| Spike-RBD Mutation | −1, Locked OFF | +1, Locked ON |
| Furin Mutation | −1, Locked OFF | +1, Locked ON |
| NSP12 Mutation (RdRP) | −1, Locked OFF | +1, Locked ON |
| orf1ab Mutation | −1, Locked OFF | +1, Locked ON |
| orf10 Mutation | −1, Locked OFF | +1, Locked ON |
| (N)ucleoprotein Mutation | −1, Locked OFF | +1, Locked ON |
| (M)embrane Mutation | −1, Locked OFF | +1, Locked ON |
| NSP3 Mutation | −1, Locked OFF | +1, Locked ON |
| orf7a Mutation | −1, Locked OFF | +1, Locked ON |
| orf8 Mutation | −1, Locked OFF | +1, Locked ON |
| NSP5 Mutation | −1, Locked OFF | +1, Locked ON |
| (S)pike Mutation | −1, Locked OFF | +1, Locked ON |
| (E)nvelope Mutation | −1, Locked OFF | +1, Locked ON |
| NSP13 Mutation (Helicase) | −1, Locked OFF | +1, Locked ON |
| orf3a Mutation | −1, Locked OFF | +1, Locked ON |
NSP = Non-Structural Protein, RdRP = RNA-dependent RNA polymerase, orf = open reading frame, aiLUNG = Wild Type (uninfected), aiLUNG + SARS-CoV-2 = aiLUNG-COVID-19.
Prior to the application of simulated LOF and GOF mutations as described above, the predictions from the wild type aiPSC, aiLUNG-WT and aiLUNG-COVID-19 simulations regarding the expression or repression of genes and proteins and presence or absence of phenotypic features were validated directly against published data as outlined previously [13]. All experiments in this study were conducted in triplicate (n = 3) using different initial conditions in the form of initial state vectors.
1.2. DeepNEU platform statistical analysis
For these experiments we have used the unbiased binomial test to analyze aiPSC, aiLUNG and LUNG-COVID-19 simulation predictions versus the published literature . This test compensates for prediction bias and is most suitable for calculating the significance of differences when comparing outcomes that fall into just two categories (e.g., agree vs disagree) Analysis of the complete data set identified the pre-test probability of a positive outcome prediction is 0.661 and the pre-test probability of a negative prediction is therefore 0.339 . A p value < 0.05 is considered significant in that the predicted outcome is unlikely to have occurred by chance alone. To compare between group differences (e.g. LOF vs GOF) the non parametric Mann-Whitney u test was used to estimate significance [16]. We chose this non parametric test, because some of the data were not normally distributed.
For the purpose of this project it we created a simple method for estimating the impact of LOF and GOF mutations on infectivity based on a recent paper by [17]. These authors identified four gene products that were impacted in almost all the known mutations identified so far in the SARS-CoV-2 genome. These four gene products were Polyprotein-1ab (orf-1ab), Nucleocapsid protein (N), Spike protein (S) and Membrane protein (M). Refinement of the impact of these four proteins revealed that the non-structural protein cleavage products NSP3, NSP4 and NSP14 were largely responsible for mutations seen in the orf-1ab polyprotein, while the Spike-RBD protein appeared responsible for most of the variation in the Spike protein [17]. We therefore created an estimate of infectivity (dnCFR) by combining DeepNEU estimates of NSP3 derived from orf-1ab polyprotein (NSP4 and NSP14 are not implemented in DeepNEU (v5.0)), Nucleocapsid protein (N), Spike Receptor Binding Domain (S-RBD) and Membrane protein (M). The dnCFR measure was used to compare all imposed LOF and GOF mutations. In addition, the dnCFR measure was correlated with the calculated Angular Cosine Distance (ACD) a validated metric for evaluating the distance between real valued vectors with values between −1 and +1 [18], [19].
Validation of the dnCFR measure included calculation of Cosine Similarity (CS) for all LOF and GOF mutations to establish similarity to the wild type SARS-CoV-2 genome. Cosine Similarity is a commonly used measure for comparing the similarity of two or more real valued vectors with the same number of elements. In this study each SARS-CoV-2 genomic profile was represented as real valued vectors. As similarity between the genomic profile vectors increases, CS increases to + 1 or maximum similarity. As CS similarity decreases away from the reference vector and becomes increasingly dissimilar, CS decrease towards −1 or maximum dissimilarity [7]. We then used a simple mathematical transformation to derive Angular Cosine Distance (ACD) using the formula ACD = arccosine (CS)/Pi. The ACD metric was selected to evaluate the distance between wild type and mutated SARS-CoV-2 genomic vectors because [1] it conforms with all four properties of a valid distance metric, [2] sample sizes are relatively small (N < 20) minimizing any influence of the curse of dimensionality and [3] it is a widely used and well validated metric for comparing bounded real valued (−1 to + 1) vectors [18], [19].
Further validation of the dnCFR measure using publicly available CFR data
Following the initial validation as outlined above, the dnCFR was used to predict actual CFRs over a six-month period beginning August 1st, 2020 and ending on January 1st, 2021. The beginning and ending CFRs were obtained for continents, regions and countries around the globe. These data which are updated daily are available from Coronavirus Pandemic Data Explorer publicly available at SARS-CoV-2 worldwide CFR.
For this process, we chose to focus on the Spike-RBD component of dnCFR since this element contributed the most to the values generated for the global COVID-19 dnCFR (see below). During data generation, the M, N and NSP3 components were held constant while S-RND was varied between −1 (complete absence) and + 1 (maximum effect). This process produced a dataset of calculated dnCFR and actual CFRs that could be evaluated from continents, regions and major cities. Initially, predictions were compared to actual CFRs and plotted for visual analysis. Then the system predictions vs actual results were used to develop an optimal regression model for statistical analysis. Pearson r correlation coefficient, R2 and p value with n-2 degrees of freedom were used to assess the significance of results.
1.3. Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
2. Results
2.1. The aiPSC and wild type (uninfected) aiLUNG simulations
As reported previously both the unsupervised aiPSC simulations and the unsupervised aiLUNG simulations converged quickly (24 iterations) to a new system wide steady state without evidence of overtraining after 1000 iterations [13]. The aiPSC simulations expressed the same human hESC specific surface antigen and genomic profile as both undifferentiated human embryonic stem cells (hESC) and induced pluripotent stem cells (iPSC) [13]. The probability that all (N = 15) of these aiPSC-WT outcomes were correctly predicted by chance alone using the binomial test is 0.0021.
While the aiLUNG simulations reproduced several lung cell types including ATI and ATII precursors, Alveolar ATI and ATII cells, ATI and ATII Sacular cells plus epithelial Ciliated, Club, Goblet cells and pulmonary neuroendocrine cells (PNEC), we focused on the ATI and ATII alveolar cells because they are a primary target of SARS-CoV-2 respiratory infection. The aiLUNG simulations produced a similar genotypic and phenotypic expression profile when compared with the human wild type (ATI and ATII) lung cell specific factors taken from the literature [13]. The probability that all (N = 15) of these aiLUNG outcomes were correctly predicted by chance alone using the binomial test is 0.0021. Importantly, the data also indicate that the generation of aiLUNG cells from aiPSC produces a heterogenous population of alveolar cell precursors and more mature alveolar cells consistent with previous study [20].
2.2. Simulation of SARS-CoV-2-infected aiLUNG cells (aiLUNG-COVID-19)
The next step in the experiments was to expose the aiLUNG cells to simulated SARS-CoV-2 virus. For this simulated infection, the concept of SARS-CoV-2 viremia was activated (turned on). The viremia activates the viral life cycle beginning with the interaction of the viral Spike protein with its receptor protein Angiotensin-converting enzyme 2 (ACE2) and ending with exocytosis of new viral particles which completes the cycle by contributing new viral particles to the ongoing viremia [21]. The SARS-CoV-2 genome consists of four structural genes, at least six non-structural genes and produces at least ten proteins. As described previously, the seventeen gene/protein expression profile was compared with the uninfected aiLUNG simulations to assess the validity of simulated COVID-19. All genes and proteins studied were expressed in the aiLUNG-COVID-19, but not aiLUNG simulations. The probability that all (N = 17) of these aiLUNG-COVID-19 simulation outcomes were correctly predicted by chance alone using the binomial test is 0.0009 [13].
A phenotypic profile of aiLUNG-COVID-19 was also developed based on the published literature and has been described previously [13]. These phenotypic features (N = 8) include: New Extracellular Virus release, Spike-ACE2 Interface, Spike-RBD, TMPRSS2, Virus Clearance, Virus Intracellular RNA release, Virus Internalization and Virus Replication. Strictly speaking, TMPRSS2 is a transmembrane host factor with protease activity but was included in the disease phenotype because its’ protease activity is required for [1] priming S-protein, [2] activating ACE2 through cleaving its C terminal and [3] it may be impacted by mutational pressure from S-protein variations [22]. The presence of these phenotypic features of COVID-19 was correctly predicted by the aiLUNG-COVID-19 simulations when compared with the aiLUNG simulations. The probability that all (N = 8) of these aiLUNG-COVID-19 outcomes were predicted correctly by chance alone using the binomial test is 0.0364.
When we combined the genotypic and phenotypic profiles, the probability that all (N = 25) features of simulated aiLUNG-COVID-19 were accurately predicted by chance alone using the binomial test is 0.00003.
2.3. Evaluation of the validated aiLUNG-COVID-19 simulations for estimating the impact of LOF and GOF mutations on SARS-CoV-2 infectivity.
2.3.1. LOF mutations:
The LOF mutations (N = 15), representing fifteen genes and proteins of the SARS-CoV-2 genome listed above, were simulated by setting the gene/gene product concepts to −1 and locking them off during system development. This is the computational analogue to creating a gene deletion and therefore an absent gene product that is propagated from each iteration to the next until system convergence is achieved. All unsupervised aiLUNG-COVID-19 and aiLUNG-COVID-19 with LOF simulations converged quickly to a new system wide steady state without evidence of overtraining after 1000 iterations.
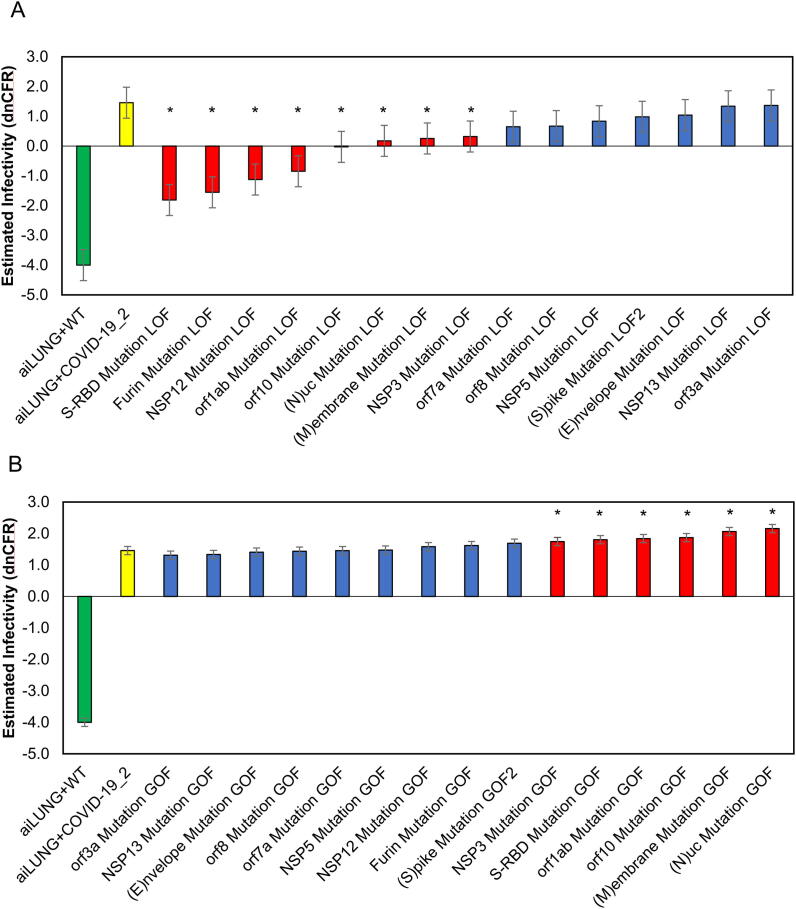
The dnCFR measure for the LOF mutations ranged from a value of −4.000 for the aiLUNG-WT (uninfected) simulations to a maximum of 1.365 (95% CI ± 0.521) with a theoretical maximum of + 4.000. The mean dnCFR for LOF mutations was (0.152 ± 0.521). Analysis of system outputs using the dnCFR identified eight LOF mutations that significantly decreased SARS-CoV-2 infectivity (p < 0.05) when compared with the wild type SARS-CoV-2 genome. The most impactful LOF mutation was in Spike-RBD (-1.812 ± 0.521) followed closely by LOF mutation in the S protein cleavage enzyme Furin (-1.553 ± 0.521). The remainder of the LOF mutations did not significantly alter infectivity as estimated by the dnCFR (p > 0.05)(Fig. 1A and Fig. 4).
Fig. 1.
aiLUNG simulations of the effect of LOF and GOF mutations in the SARS-CoV-2 genome on viral infectivity. A, LOF simulations and their virulence estimation. Red bars represent mutations in SARS-CoV2 that result in a significant decrease in infectivity. B, GOF simulations their infectivity estimation. Red bars represent mutations in SARS-CoV2 that result in a significant increase in infectivity. A, B, Green bar represents aiLUNG-WT; Yellow bar represents aiLUNG-COVID-19 (unmutated original SARS-CoV2 genome) simulations; blue bars represent mutations that has no significant effect on infectivity. These data represent the average from 3 experiments ± the 95% confidence interval around the average. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
Heat map representation of summary DeepNEU simulations data of the effects of individual LOF mutations. A, effect of LOF on the SARS-CoV-2 genome and B, effect of LOF on phenotypic profile of simulated ATI and ATII cells. Data are the average of 3 separate experiments. Data represented as dnCFR measure +/- the 95% CI around the estimates is presented. dnCFR is the DeepNEU measure of SAR-CoV-2 infectivity, where (-4) represents the maximum reduction in viral infectivity and (+4) represents the maximum increase in infectivity.
2.3.2. GOF mutations:
The GOF mutations (N = 15) were simulated by setting the gene/gene product concepts to + 1 and locking them on during system development. This is the computational analogue to creating a maximum increase in gene function and therefore a maximum gene product that is propagated from each iteration to the next until system convergence is achieved. All unsupervised aiLUNG-COVID-19 and aiLUNG-COVID-19 with GOF simulations converged quickly to a new system wide steady state without evidence of overtraining after 1000 iterations.
The dnCFR measure for the GOF mutations ranged from a value of −4.000 for the aiLUNG-WT simulations to a maximum of 2.156 (95% CI ± 0.131) with a theoretical maximum of + 4.000. The mean dnCFR for GOF mutations was (1.652 ± 0.131). Analysis of system outputs using the dnCFR measure identified six GOF mutations that significantly increased infectivity (p < 0.05) when compared with aiLUNG-COVID-19 without mutations. The most impactful GOF mutation was in the N protein (2.156 ± 0.131) followed closely by GOF mutation in the M protein (2.063 ± 0.131). A significant increase in SARS-CoV-2 infectivity was also predicted to result from GOF mutations in ORF10, ORF1ab and Spike protein RNA binding domain (Spike-RBD). The remainder of the GOF mutations did not significantly alter infectivity (p > 0.05) (Fig. 1B and Fig. 5).
Fig. 5.
Heat map representation of summary DeepNEU simulations data of the effects of individual GOF mutations. A, effect of GOF on the SARS-CoV-2 genome and B, effect of GOF on phenotypic profile of simulated ATI and ATII cells. Data are the average of 3 separate experiments. Data represented as dnCFR measure +/- the 95% CI around the estimates.
2.4. Using the dnCFR measure to compare LOF and GOF mutations
The first step in assessing the dnCFR measure was to evaluate the ability of each of the four individual genomic components of the measure (NSP3, S-RBP, M, N proteins) to distinguish LOF from GOF mutations. Based on the 2 tailed Mann-Whitney u test, each component of the dnCFR measure easily distinguished LOF and GOF mutations from each other (all exact p values ≤ 0.000044) (Fig. 2 and Fig. 6).
Fig. 2.
DeepNEU summary analysis summary and statistics A, DeepNEU summary results comparing the impact of LOF and GOF mutations on SARS-CoV-2 individual genotypic features. Data from 3 separate experiments. E = Envelope, M = Membrane, N = Nucleocapsid, S-RBD = Spike-Receptor Binding Domain. B, Summary results from DeepNEU simulations of the individual mutated components and composite dnCFR measure effects on SAR-CoV-2 infectivity. Results are the average of 3 experiments ± the 95% CI.
Fig. 6.
DeepNEU simulations of the calculated 4 component dnCFR as a measure of SARS-CoV-2 infectivity. A, calculated 4 component dnCFR measure for LOF mutations. B, calculated 4 component dnCFR measure for GOF mutations. dnCFR is the DeepNEU measure of SAR-CoV-2 infectivity, where (-4) represents the maximum reduction in viral infectivity and (+4) represents the maximum increase in infectivity. Results are from 3 separate experiments ± 95% CI.
Next, we explored the ability of the dnCFR measure to distinguish between LOF and GOF mutations. The mean dnCFR (±95%CI) for LOF mutations was 0.152 ± 0.521 and 1.652 ± 0.131 for GOF mutations. The exact Mann-Whitney u test p value for the direct comparison was 1.55E-07 suggesting that the dnCFR measure was better at distinguishing between LOF and GOF mutations than any single element of the measure (Fig. 2A and Fig. 6).
We also compared the dnCFR measure using a widely used and validated distance metric, the Angular Cosine Distance (ACD). The ACD was used to estimate the distance between genomic profiles. As we described previously, we first calculated the Cosine Similarity (CS) measure for each LOF and GOF mutation profile. This value was then converted to the ACD using the equation ACD = arccosine (ACD)/Pi. ACD values were calculated for each LOF and GOF mutation. Like the dnCFR measure, the ACD metric also easily distinguished between the LOF and GOF mutations (Mann-Whitney u test p = 3.87E-07). We next compared the ACD metric directly with the dnCFR measure using the Spearman rank correlation coefficient. The Spearman coefficient for the LOF mutations was 0.973 (critical value (N = 15) is 0.779, p < 0.001) (Fig. 3A). The Spearman coefficient for the GOF mutations was 0.903 (critical value (N = 15) is 0.779, p < 0.001) (Fig. 3B). These data indicate that the dnCFR measure and ACD metric are both able to accurately distinguish between LOF and GOF mutations and are strongly positively correlated with each other.
Fig. 3.
DeepNEU simulations of the effect of LOF and GOF mutations on SARS-CoV2 infectivity. A, Correlation between ACD metric and dnCFR measure regarding there ability to identify LOF mutations in the SARS-CoV-2 genome. Data from from 3 experiments ± the 95% confidence around the estimates is presented. Spearman r = 0.973, n = 15, p < 0.001. B, Correlation between ACD metric and dnCFR measure regarding there ability to identify GOF mutations in the SARS-CoV-2 genome. Data from from 3 experiments plus the 95% confidence around the estimates is presented. Spearman r = 0.903, n = 15, p < 0.001.
2.5. The dnCFR estimates vs actual CFR
A total of 46 (df = 44) predictions for dnCFR were generated for this analysis. The initial analysis revealed a weak linear correlation between calculated dnCFR and actual CFR (Pearson r = 0.514). However, when we evaluated the log of the CFR (LogCFR) and the dnCFR, a much stronger correlation was revealed (Pearson r = 0.982, df = 44, critical value ~ 0.490, p < 0.001). The derived simple log-linear regression equation is Log(CFR) = 0.5091*dnCFR-2.1925, (R2 = 0.9635). The predicted Log(CFR) can be converted to the corresponding CFR using the transform CFR = 10(LogCFR). The data are summarized graphically in Fig. 7 and numerically in Table 2.
Fig. 7.
Linear Regression analysis between DeepNEU estimated dnCFR vs Actual CFR. Results from experiments applying the equation Log(CFR) = 0.5091*dnCFR-0.2915 to publicly available CFR data. Three of the dnCFR measure components were held constant at their baseline values while the S-RBD varied from a minimum of −1 to a maximum of + 1. Over this range of values for S-RBD activity, predicted dnCFR ranged between −0.118 to + 1.88. After computing the value for Log(CFR), the predicted CFR was recovered by using the transform CFR = 10dnCFR .
Table 2.
Correlation between predicted dnCFR and actual CFR of global SARS-CoV2 variants.
| Continent | Region | M protein | N protein | NSP3 | S-RBD | dnCFR | CFR | Log(CFR) | |
|---|---|---|---|---|---|---|---|---|---|
| Best case (WT) | COVID-19 | −1 | −1 | −1 | −1 | −4.0000 | 0.000 | −4.000 | |
| Global CFR | COVID-19 | 0.202 | 0.202 | 0.483 | 0.571 | 1.4580 | 0.035 | −1.456 | |
| Africa | S_Africa | 0.202 | 0.202 | 0.483 | 0.02 | 0.9070 | 0.016 | −1.796 | |
| Africa | S_Africa _6m | 0.202 | 0.202 | 0.483 | 0.495 | 1.3820 | 0.027 | −1.569 | |
| Africa | Africa | 0.202 | 0.202 | 0.483 | 0.148 | 1.0350 | 0.021 | −1.678 | |
| Africa | Africa + 6 m | 0.202 | 0.202 | 0.483 | 0.259 | 1.1460 | 0.024 | −1.620 | |
| Asia | China | 0.202 | 0.202 | 0.483 | 1 | 1.8870 | 0.053 | −1.276 | |
| Asia | China + 6 m | 0.202 | 0.202 | 0.483 | 0.999 | 1.8860 | 0.050 | −1.301 | |
| Asia | Asia | 0.202 | 0.202 | 0.483 | 0.571 | 1.4580 | 0.035 | −1.456 | |
| Asia | Asia + 6 m | 0.202 | 0.202 | 0.483 | −0.041 | 0.8460 | 0.017 | −1.770 | |
| Asia | Russia | 0.202 | 0.202 | 0.483 | −1 | −0.1130 | 0.005 | −2.301 | |
| Asia | Russia + 6 m | 0.202 | 0.202 | 0.483 | 0.01 | 0.8970 | 0.018 | −1.745 | |
| Europe | UK | 0.202 | 0.202 | 0.483 | 1 | 1.8870 | 0.135 | −0.870 | |
| Europe | UK + 6 m | 0.202 | 0.202 | 0.483 | 0.56 | 1.4470 | 0.029 | −1.538 | |
| Europe | UK_B1.1.7* | 0.202 | 0.202 | 0.483 | 0.784 | 1.6710 | 0.044 | −1.357 | |
| Europe | Italy | 0.202 | 0.202 | 0.483 | 1 | 1.8870 | 0.142 | −0.848 | |
| Europe | Italy + 6 m | 0.202 | 0.202 | 0.483 | 0.571 | 1.4580 | 0.035 | −1.456 | |
| Europe | Denmark | 0.202 | 0.202 | 0.483 | 0.936 | 1.8230 | 0.044 | −1.357 | |
| Europe | Denmark + 6 m | 0.202 | 0.202 | 0.483 | −0.603 | 0.2840 | 0.008 | −2.097 | |
| Europe | EU | 0.202 | 0.202 | 0.483 | −0.041 | 0.8460 | 0.017 | −1.770 | |
| Europe | EU + 6 m | 0.202 | 0.202 | 0.483 | 0.259 | 1.1460 | 0.024 | −1.620 | |
| Europe | France | 0.202 | 0.202 | 0.483 | −0.15 | 0.7370 | 0.015 | −1.824 | |
| Europe | France + 6 m | 0.202 | 0.202 | 0.483 | 0.259 | 1.1460 | 0.024 | −1.620 | |
| Europe | Europe | 0.202 | 0.202 | 0.483 | 1 | 1.8870 | 0.078 | −1.108 | |
| Europe | Europe + 6 m | 0.202 | 0.202 | 0.483 | 0.224 | 1.1110 | 0.023 | −1.638 | |
| Europe | Europe + B1.1.7* | 0.202 | 0.202 | 0.483 | 0.314 | 1.2010 | 0.026 | −1.593 | |
| N_America | USA | 0.202 | 0.202 | 0.483 | 0.703 | 1.5900 | 0.034 | −1.469 | |
| N_America | USA + 6 m | 0.202 | 0.202 | 0.483 | −0.041 | 0.8460 | 0.017 | −1.770 | |
| N_America | USA_B1.1.7* | 0.202 | 0.202 | 0.483 | −0.025 | 0.8620 | 0.017 | −1.762 | |
| N_America | N_America | 0.202 | 0.202 | 0.483 | 1 | 1.8870 | 0.064 | −1.194 | |
| N_America | N_America + 6 m | 0.202 | 0.202 | 0.483 | 0.183 | 1.0700 | 0.022 | −1.658 | |
| N_America | Canada | 0.202 | 0.202 | 0.483 | −0.593 | 0.2940 | 0.009 | −2.046 | |
| N_America | Canada + 6 m | 0.202 | 0.202 | 0.483 | 0.362 | 1.2490 | 0.027 | −1.569 | |
| Oceania | Australia | 0.202 | 0.202 | 0.483 | −0.237 | 0.6500 | 0.012 | −1.921 | |
| Oceania | Austrakia + 6 m | 0.202 | 0.202 | 0.483 | 0.649 | 1.5360 | 0.032 | −1.495 | |
| Oceania | New Zealand | 0.202 | 0.202 | 0.483 | −0.21 | 0.6770 | 0.014 | −1.854 | |
| Oceania | New Zealand + 6 m | 0.202 | 0.202 | 0.483 | −0.343 | 0.5440 | 0.012 | −1.921 | |
| Oceania | Oceania | 0.202 | 0.202 | 0.483 | −0.274 | 0.6130 | 0.013 | −1.886 | |
| Oceania | Oceania + 6 m | 0.202 | 0.202 | 0.483 | 0.454 | 1.3410 | 0.030 | −1.523 | |
| S_America | Brazil | 0.202 | 0.202 | 0.483 | 0.571 | 1.4580 | 0.035 | −1.456 | |
| S_America | Brazil + 6 m | 0.202 | 0.202 | 0.483 | 0.48 | 1.3670 | 0.025 | −1.602 | |
| S_America | SAmerica | 0.202 | 0.202 | 0.483 | −0.5 | 0.3870 | 0.010 | −2.000 | |
| S_America | SAmerica + 6 m | 0.202 | 0.202 | 0.483 | 0.363 | 1.2500 | 0.027 | −1.569 | |
| S-RBD GOF | Other | 0.202 | 0.202 | 0.483 | 1 | 1.8870 | 0.057 | −1.244 | |
| S-RBD LOF | Other | 0.202 | 0.202 | 0.483 | −1 | −0.1130 | 0.004 | −2.398 | |
| Worst case (GOF) | COVID-19 | 1 | 1 | 1 | 1 | 4.0000 | 1.000 | 0.000 | |
| N = 46 | df = 44 | Log-Linear Regression = | Log(CFR) = 0.5091*(dnCFR)−2.1915, R2 = 0.9635 | ||||||
| Pearson r = | 0.514 for dnCFR vs actual CFR | * | Calculations for B1.1.7 mutation, ~40% increase in S-RBD function | ||||||
| 0.982 for dnCFR vs Log (actual CFR), p < 0.001 | |||||||||
3. Discussion
Recently, we evaluated the capability of the DeepNEU (v5.0) machine learning platform to simulate SARS-CoV-2 infection in simulated Type 1 (AT1) and Type 2 (AT2) alveolar lung cells (aiLUNG-COVID-19) [13]. In our most recent research, we reported the ability of the DeepNEU platform to enable the rapid identification of therapeutic targets and drug repurposing for treating COVID-19 [13](While we have used the same approach that we reported in our previous research [13],),the primary purpose of this project was to extend our previous work by evaluating the ability of the DeepNEU platform to enable rapid and efficient assessment of the impact of LOF and GOF mutations in the SARS-CoV-2 genome. As of this writing and with a few exceptions, the diversity and impact of known mutations in the SARS-CoV-2 genome are relatively unknown and this is particularly true for GOF mutations that have increased potential to amplify SARS-CoV-2 infectivity.
While there is some variation in the definition of infectivity, we have defined it here to mean the ratio of individuals who become infected with SARS-CoV-2 divided by the number who are exposed to the virus. In addition, the SARS-CoV-2 associated case fatality rate (CFR) is the proportion of people who die from documented COVID-19. For the purposes of this project we have created a new measure called dnCFR. This new measure represents a logical extension of the insights into mutations in the SARS-CoV-2 genome provided in [17]. These authors identified four gene products (proteins) that result from the most common mutations in the SARS-CoV-2 genome. These mutated proteins are [1] Non-Structural Protein 3 (NSP3), [2] Spike-Receptor Binding Domain (S-RBD), [3] Membrane (M) protein and [4] Nucleocapsid (N) protein. All four of these proteins were implemented in DeepNEU (v5.0) and could be evaluated individually and in combination to constitute the dnCFR measure. Individually each of the four mutated proteins could easily distinguish LOF from GOF mutations (p < 4.4E-05). Importantly, when combined the dnCFR measure appeared to be about 75 times better at distinguishing between LOF and GOF mutations (p = 1.55E-07). The dnCFR measure also performed well when compared with a validated and widely used metric, the Angular Cosine Distance (ACD) which we used to measure the distance between two real valued genomic vectors. Specifically, we wanted to measure the distance between the un-mutated SARS-CoV-2 genome and the mutated genomes using the ACD. The calculated Spearman correlation coefficient was>0.900 (p < 0.001) for all comparisons supporting the existence of a strong positive correlation between the two.
To provide further validation, we explored the relationship between the calculated dnCFR and actual CFR as an estimate of infectivity. By varying the function of the Spike-RBD between a complete loss of function (S-RBD = -1) and a qualitative maximum gain of function (S-RBD =+1), we generated a dataset of 46 dnCFR predictions. On the basis of these data, we found a weak correlation between actual CFR and calculated dnCFR but a strongly positive correlation between the Log(CFR) and the dnCFR. This relationship allowed us to derive a simple log-linear regression equation in the form of Log(CFR) = 0.5091*dnCFR-2.1925, (R2 = 0.9635, p < 0.001, df = 44). The predicted Log(CFR) was then converted to the corresponding CFR using the transform CFR = 10(LogCFR).
Given that the dnCFR has four components and three of them were held constant for this analysis, it is not surprising that varying Spike-RBD between −1 and + 1 had a significant impact on CFR over a limited range. For example, a Spike-RBD set to −1 produced a minimum dnCFR value of −0.113 and a CFR of 0.004 or 0.4% while a maximum S-RBD set at + 1 produced dnCFR of 1.887 and a CFR of ~ 5%. As a result, actual CFR values > 5% will always return the same dnCFR of 1.887. The same limitation applies to dnCFR < -0.113 but would be expected to have little or no impact on actual CFR since the current low dnCFR in already close to zero. It should be noted that the validation derived from these data only applies to Spike-RBD and over the range of dnCFR from −0.113 to + 1.887. This type of analysis could easily be repeated for the remaining three predictors (i.e. M, N and NSP3 proteins) of the dnCFR measure.
As part of our analysis, we also examined the impact of a simulated B1.1.7 GOF mutation(s) in the S-RBD region of the S-protein in 3 areas; the USA, the UK and the continent of Europe. This mutation is thought to be responsible for a 30%-50% increase in receptor binding and viral transmissibility. This simulated GOF mutation was applied at the end of the study period (i.e. January 1, 2021). The derived linear equation linking Log(CFR) to the calculated dnCFR was used to predict the potential impact by multiplying the S-RBD value by a factor of 1.4 to simulate a 40% increase in function. A small increase in CFR was predicted for both the USA, 0.3% from 1.7% to ~ 2.0% and from 2.3% to ~ 2.6% for Europe. The largest change in the CFR was in the UK where it was predicted to increase from 2.9% to ~ 4.4% over the next 6 months if current policies and behaviors continue.
The dnCFR measure has a few important advantages namely its simplicity and ease of use. It is easily calculated by adding four values whereas the ACD calculation requires a two-step calculation involving a trigonometric transformation. The dnCFR can also be directly linked to the global, regional or local CFR estimate. As of this writing the number of global SARS-CoV-2 infections is 22,200,000 with 783,000 deaths producing a CFR of 0.035. This global CFR of 3.5% or 35,270 deaths per million is associated with a dnCFR estimate of 1.457 for the infectivity of the wild type SARS-CoV-2 genome. For example, a GOF mutation that produces a dnCFR of 2 or a 1.373 increase in infectivity would result in a CFR of 0.048 or 48,044 per million people infected or 12,774 excess deaths per million. Perhaps most importantly the dnCFR measure can be modified for any viral genome for which there are validated insights into the genome mutational landscape. While the dnCFR is based on sound logic and mathematical principles there are a few drawbacks. These potential drawbacks of the current version of the dnCFR measure are related to its newness and lack of additional independent validation and widespread use.
3.1. dnCFR and LOF mutations
When we applied the dnCFR measure to each of the fifteen LOF mutations evaluated, eight significant mutations were identified. All these mutations produced a significant decrease in SARS-CoV-2 infectivity compared with the wild type genome. The most significant LOF mutation was in Spike-RBD with a dnCFR of −1.812 ± 0.521 representing a 224.37% decrease in infectivity. This complete LOF mutation leads to a profound loss of infectivity as evidenced by an estimated CFR of 0.00 deaths per million people infected and a decrease of 35,270 deaths per million. There are three other LOF mutations that produce a negative dnCFR. These other mutated proteins are Furin (-1.553 ± 0.521), NSP12/RdRP (-1.124 ± 0.521) and orf1ab (-0.846 ± 0.521) suggesting that SARS-CoV-2 infectivity is dependent to some degree, on each of these mutations. The least, but still significant LOF mutation was in the NSP3 protein with a dnCFR of 0.321 ± 0.521 representing a 17.9% decrease in infectivity. This LOF mutation results in a decrease in infectivity and is associated with a CFR of 2.90% or 28,949 per million people infected and an expected decrease of 6,322 deaths per million.
These data have important implications for future research focusing on SARS-CoV-2 pandemic preparedness. Importantly, the rapid identification of drug and drug combinations, monoclonal antibodies or vaccines that target one or more of these LOF mutations would be expected to produce a LOF or LOFs situation that could reduce CFR by a minimum of 6,322 deaths per million infections.
3.2. dnCFR and GOF mutations
When the dnCFR measure was applied to the each of the fifteen GOF mutations evaluated, six significant mutations were identified. All these mutations produced a significant increase in SARS-CoV-2 infectivity compared with the wild type genome. The most significant GOF mutation was in the N protein with a dnCFR of 2.156 ± 0.131 representing a 47.98% increase in infectivity. This GOF mutation produces an increase in infectivity associated with a CFR of 5.18% or 52,191 deaths per million people infected and an increase of 16,921 deaths per million. The second most significant GOF mutation was in the M protein with a dnCFR of 2.063 ± 0.131. In addition, GOF mutations in ORF10, ORF1ab and Spike protein RNA binding domain (Spike-RBD) also produced significant increases in SARS-CoV-2 infectivity. The least, but still significant GOF mutation was in the NSP3 protein with a dnCFR of 1.743 ± 0.131 representing a 19.63% increase in infectivity. This GOF mutation produces an increase in infectivity associated with a CFR of 42,732 deaths per million people infected and an expected increase of 7,012 deaths per million.
These data also have important implications for future research focusing on SARS-CoV-2 pandemic preparedness. For example, these data suggest that GOF mutations other than in the S-protein and particularly in N and M gene/proteins may have even greater impact on infectivity and CFR of evolving SARS-CoV-2. The implication is that going forward SARS-CoV-2 research should be refocused to assign greater importance to potential N and M mutations. Importantly, the rapid identification of drug and drug combinations, monoclonal antibodies or vaccines that target one or more of these GOF mutations would be expected to produce a GOF or GOFs situation that could reduce CFR by at least 7,012 and perhaps as much as 17,000 deaths per million infections. Early application of this literature validated technology could have even greater beneficial effects on future viral pandemics for which we remain unprepared.
Interestingly, when we combined all mutations, six of the mutated proteins, both LOF and GOF, significantly impacted the infectivity of SARS-CoV-2 as estimated by the dnCFR. These six proteins were N, M, S-RBD, Orf1ab, Orf10 and NSP3.
3.3. Evolution of the SARS-CoV-2 genome so far
The evolution of the SARS-CoV-2 genome is ongoing and so far, it appears to have evolved into at least six clades defined based on a common ancestor. These currently identified clades are labelled as G, GH. GR, S, V and L plus an O clade representing Other. These clades have different geographic representation as well as mutational profiles. Worldwide the most common clades are G, GH and GR accounting for ~ 74% of identified mutations. Importantly, clades GH and GR are believed to be derived from the G clade. The G glade has NSP3, RdRP (NSP12) and Spike (S) mutations. The GH clade has the same mutations as G plus an ORF3a mutation and similarly, the GR clade has the same mutations as G plus a Nucleocapsid (N) mutation [23]. All these individual mutations have been evaluated by the DeepNEU platform.
Beginning with Africa, the most common clade is G followed by GH, GR, and O. In Asia the largest clade is O followed by GH, S, GR and G. The most common European clade is GR followed G, V and GH. In North America the dominant clade is GH followed by S and G. The most common clade in South America is GR followed by GH and G. Finally, in Oceania GH is the most common clade followed by O, G, V and GR. The G, GH and GR clades are variably but substantially represented in all regions of the globe discussed [9], [23], [24], [25], [26], [27].
The DeepNEU platform can be used to assess the impact of regionally specific SARS-CoV-2 clades by combining LOF and/or GOF mutations. For example, globally the most common clade is GR [18] and the worse-case scenario can be simulated by combining NSP3 + RdRP (NSP12) + Spike (S) + Nucleocapsid (N) GOF mutations. Of note, the current version of DeepNEU could easily handle an almost unlimited number of LOF and/or GOF mutations. The cumulative mutational impact on the GR clade CFR can be estimated by the dnCFR as outlined above. For example, GOF mutations of all four proteins of the GR clade would result in a dnCFR of 3.052 which equates to a worst case, CFR of 7.333% or 73,891 deaths per million and an expected increase in CFR of 38,621 deaths per million. Given that the average number of mutations in the SARS-CoV-2 genome so far has been > 7, this scenario is unlikely but not impossible.
3.4. Future viral pandemic preparedness: We are not prepared!
Finally, we must act now and fortunately we can begin with the world health organization (WHO) list of top 10 pathogens for which we are not now prepared. Importantly, all the pathogens on this list have been recognized longer than SARS-CoV-2(2019) has. For example, the Rift Valley fever virus was officially recognized in 1931, the Zika virus was recognized in 1947, Crimean-Congo Fever in 1967, Lassa Fever in 1969, Ebola in 1976, Nipah virus in 1998 and the MERS virus in 2012. Although none of these pathogens have effective therapies, all of them have a considerable body of knowledge regarding their genome and changes over time. So far, this is the only absolute requirement for implementing other analyses like that for SARS-CoV-2. In other words, an approach that combines the DeepNEU platform and a dnCFR measure modified for a specific viral genome can be used with the other members of the WHO list of top 10 pathogens for which, midway through 2020, we are not prepared.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors wish to thank Dr. Jim Koropatnick, Professor Emeritus, Departments of Oncology, Physiology and Pharmacology, and Pathology and Laboratory Medicine, Adjunct Professor, Department of Microbiology and Immunology. The University of Western Ontario, Cancer Research Laboratory Program, London Canada for his constructive review and suggested edits that materially improved the final manuscript.
The authors also wish to thank Dr. Mark Poznansky (past president and CEO, Ontario Genomics Institute) for his constructive review and suggested edits that materially improved the final manuscript.
Author contributions
SE and WD conceptualized, and analyzed the experimental work, wrote the manuscript, and prepared the figures. WD performed all computational simulations of COVID-19 disease and SARS-CoV2 GOF and LOF mutations modeling.
Transparency statement regarding methodology.
123Genetix aims to be transparent and accountable to researchers, stakeholders and other
Service- users. We aim to make information easy to find for other interested parties such as partners, regulators, and members of the scientific communities. Please see appendix 1 for detailed methodology.
References
- 1.Acter, T., Uddin, N., Das, J., Akhter, A., Choudhury, T. R., and Kim, S. Evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as coronavirus disease 2019 (COVID-19) pandemic: A global health emergency. Sci Total Environ, 2020; 138996. [DOI] [PMC free article] [PubMed]
- 2.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18(1) doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanjuán R., Domingo-Calap P. Mechanisms of viral mutation. Cell Mol Life Sci. 2016;73(23):4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geoghegan J.L., Holmes E.C. The phylogenomics of evolving virus virulence. Nat Rev Genet. 2018;19(12):756–769. doi: 10.1038/s41576-018-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolan P.T., Whitfield Z.J., Andino R. Mapping the evolutionary potential of RNA viruses. Cell Host Microbe. 2018;23(4):435–446. doi: 10.1016/j.chom.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull World Health Organ. 2020;98(7):495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, H., Liu, S.-M., Yu, X.-H., Tang, S.-L., and Tang, C.-K. Coronavirus disease 2019 (COVID-19): current status and future perspective. Int J Antimicrob Agent; 2020. 105951. [DOI] [PMC free article] [PubMed]
- 9.Zheng J. SARS-CoV-2: an emerging coronavirus that causes a global threat. Int J Biol Sci. 2020;16(10):1678–1685. doi: 10.7150/ijbs.45053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, J., Zhang, S., Li, B., Hu, Y., Kang, X.-P., Wu, X.-Y., Huang, M.-T., Li, Y.-C., Zhao, Z.-P., Qin, C.-F. Machine Learning Methods for Predicting Human-Adaptive Influenza A Viruses Based on Viral Nucleotide Compositions. Mol Biol Evolut; 2020. 37, 1224-1236. [DOI] [PMC free article] [PubMed]
- 11.Esmail S., Danter W.R. DeepNEU: artificially induced stem cell (aiPSC) and differentiated skeletal muscle cell (aiSkMC) simulations of infantile onset POMPE disease (IOPD) for potential biomarker identification and drug discovery. Front Cell Dev Biol. 2019;7:325. doi: 10.3389/fcell.2019.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danter W.R. DeepNEU: cellular reprogramming comes of age–a machine learning platform with application to rare diseases research. Orphanet J Rare Dis. 2019;14:13. doi: 10.1186/s13023-018-0983-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esmail S., Danter R.W. Viral Pandemic Preparedness: a pluripotent stem cell-based Machine Learning platform for simulating COVID-19 infection to enable Drug Discovery and Repurposing. Stem Cells Transl Med. 2020;10:239–250. doi: 10.1002/sctm.20-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamò L., Hibaoui Y., Kallol S., Alves M.P., Albrecht C., Hostettler K.E. Generation of an alveolar epithelial type II cell line from induced pluripotent stem cells. Am J Physiol-lung Cell Mol Physiol. 2018;315(6):L921–L932. doi: 10.1152/ajplung.00357.2017. [DOI] [PubMed] [Google Scholar]
- 15.Jacob A., Morley M., Hawkins F., McCauley K.B., Jean J.C., Heins H. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell. 2017;21(4):472–488.e10. doi: 10.1016/j.stem.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Addinsoft. Addinsoft Long Island; New York: 2019. XLSTAT statistical and data analysis solution. [Google Scholar]
- 17.Gussow Ayal B., Auslander Noam, Faure Guilhem, Wolf Yuri I., Zhang Feng, Koonin Eugene V. Genomic determinants of pathogenicity in SARS-CoV-2 and other human coronaviruses. Proc Natl Acad Sci. 2020;117(26):15193–15199. doi: 10.1073/pnas.2008176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanwimalueang Theerasak, Mandic Danilo. Cosine similarity entropy: Self-correlation-based complexity analysis of dynamical systems. Entropy. 2017;19(12):652. doi: 10.3390/e19120652. [DOI] [Google Scholar]
- 19.Cai Stanley, Georgakilas Georgios K., Johnson John L., Vahedi Golnaz. A cosine similarity-based method to infer variability of chromatin accessibility at the single-cell level. Front Genet. 2018;9 doi: 10.3389/fgene.2018.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Changgong, Smith Susan M, Peinado Neil, Gao Feng, Li Wei, Lee Matt K. WNT5a-ROR Signaling Is Essential for Alveologenesis. Cells. 2020;9(2):384. doi: 10.3390/cells9020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J Adv Res. 2020 doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calcagnile M., Forgez P., Alifano P., Alifano M. The lethal triad: SARS-CoV-2 Spike, ACE2 and TMPRSS2. Mutations in host and pathogen may affect the course of pandemic. bioRxiv. 2021 doi: 10.1101/2021.01.12.426365. [DOI] [Google Scholar]
- 23.Mercatelli, D., Giorgi, F. M. (2020) Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. Doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed]
- 24.Kim Jun-Sub, Jang Jun-Hyeong, Kim Jeong-Min, Chung Yoon-Seok, Yoo Cheon-Kwon, Han Myung-Guk. Genome-Wide Identification and Characterization of Point Mutations in the SARS-CoV-2 Genome. Osong Public Health and Research Perspectives. 2020;11(3):101–111. doi: 10.24171/j.phrp.2020.11.3.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laha Sayantan, Chakraborty Joyeeta, Das Shantanab, Manna Soumen Kanti, Biswas Sampa, Chatterjee Raghunath. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect, Genet Evolut. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., Li H., Wu X., Zhong Y., Zhang K., Zhang Y.-P. Moderate mutation rate in the SARS coronavirus genome and its implications. BMC Evol Biol. 2004;4:21. doi: 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nasir Abdullahi, I., Uchenna Emeribe, A., Abimbola Ajayi, O., Soji Oderinde, B., Ohinoyi Amadu, D., Iherue Osuji, A. Implications of SARS-CoV-2 genetic diversity and mutations on pathogenicity of the COVID-19 and biomedical interventions; 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.