Abstract
Fragment based drug discovery (FBDD) by the aid of different modelling techniques have been emerged as a key drug discovery tool in the area of pharmaceutical science and technology. The merits of employing these methods, in place of other conventional molecular modelling techniques, endorsed clear detection of the possible structural fragments present in diverse set of investigated compounds and can create alternate possibilities of lead optimization in drug discovery. In this work, two fragment identification tools namely SARpy and Laplacian-corrected Bayesian analysis were used for previous SARS-CoV PLpro and 3CLpro inhibitors. A robust and predictive SARpy based fragments identification was performed which have been validated further by Laplacian-corrected Bayesian model. These comprehensive approaches have advantages since fragments are straight forward to interpret. Moreover, distinguishing the key molecular features (with respect to ECFP_6 fingerprint) revealed good or bad influences for the SARS-CoV protease inhibitory activities. Furthermore, the identified fragments could be implemented in the medicinal chemistry endeavors of COVID-19 drug discovery.
Keywords: SARS-CoV, PLpro, 3CLpro, SARpy, Bayesian model, Fragment based drug discovery
1. Introduction
World Health Organization (WHO) has declared COVID-19 disease as a global pandemic. To date over 120 million confirmed cases along with near 3 million COVID-19-related mortalities were reported worldwide [1,2]. Studies on COVID-19 revealed that beta-coronaviruses also known as SARS-CoV-2 usually produce several proteases during their life cycle [3]. Among them two key proteases essential for viral replications are papain-like protease (PLpro) and 3C like protease (3CLpro). The papain-like protease discharge several diUbLys48 products by cleaving ISG15, a two-domain Ub-like protein, and Lys48-linked polyUb chains [4], [5], [6]. Thus, they function by hijacking the host ubiquitin enzyme that is responsible for the host defence mechanism [7,8]. Whereas, in 3C like protease (3CLpro) also known as SARS-CoV-2 main protease (Mpro) encompasses of 306 amino acid long polypeptide chains and plays a significant role in enzymatic activity leading to its post-translational processing of replicase polyproteins. The 3CLpro monomer mainly consists of three domains [9]. Both PLpro and 3CLpro are equally important for viral life cycle and play a vital role in the transcription/replication during the infection. Hence, pinpointing these two proteases (i.e., PLpro and 3CLpro) may serve as important targets for designing of several antiviral drugs against the highly contagious COVID-19 [10,11].
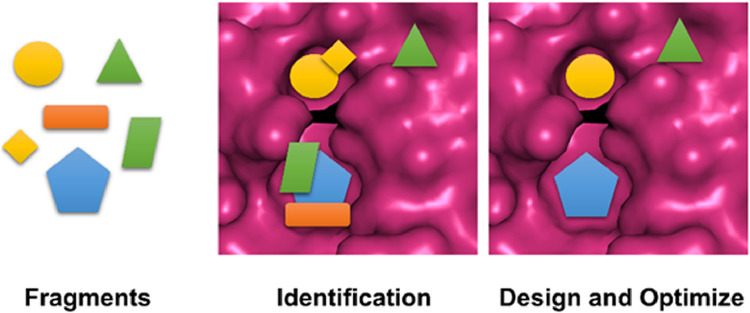
Discovering an effective inhibitor against the PLpro and 3CLpro are ongoing [12], [13], [14], [15], [16], [17]. The cost of bringing a single effective drug candidate to market not only requires billions of dollars but also requires huge time to accomplish [18]. In order to make this process faster, researchers around the globe has shifted their focus to the various emerging in silico lead discovery methods (Figure 1 ) [19,20]. Presently, among all the known computational techniques, the fragment based drug discovery (FBDD) proved to be very efficient method of drug design and discovery [21,22]. The identified important structural fragments will be helpful for efficient designing of lead molecule against SARS-CoV-2 main proteases - PLpro and 3CLpro. This way of lead designing process based on smaller fragments may remove the usual ADME associated problems with protease inhibitors based drug discovery [23].
Fig. 1.
Fragment based drug discovery process.
In the past 20 years, the concept of FBDD has recognized itself as a strategic approach for finding high-quality lead candidates [19]. It has a potential to address inflexible biological targets involving intracellular protein–protein interactions also. Today, more than ten FBDD based lead molecules targeting diverse protein families in different diseases have been progressed into clinical trials [20]. In FBDD the identification of very small molecules or so-called “fragments” which are responsible for binding to their specific target proteins are carried out [21]. Typically, screening of a small library of low molecular weight compounds for binding to a specific part of the target/receptor is the starting point of FBDD [22]. The structural evidence and thermodynamics of fragment binding proposed that they tends to bind to the crucial regions of the protein sites, mainly contribute to the enthalpy driven free energy change of ligand binding [24]. Thus, the fragments can contribute to the crucial part of a drug molecule.
In this study different fragment based computational tools like SARpy analysis and Bayesian classification was applied on to the dataset of previous SARS-CoV PLpro and 3CLpro inhibitors. The methods were successfully used previously to identify different fragments of various pharmacological and toxicological properties [25], [26], [27], [28], [29]. In these methods, the whole molecular structure was fragmented in a certain way into small parts and finally the contribution to increase or decrease the pharmacological activity/toxicological properties was identified. Thus, the identified fragments can give potential clues in the lead optimization process. Since the binding site of PLpro and 3CLpro enzymes were conserved between SARS-CoV and SARS-CoV-2 the fragments will be very useful for COVID-19 drug discovery [3,14,30,31]. The current study will offer an idea to in-depth qualification of fragment hits. This will stimulate further research by providing valuable guidance to the medicinal chemists for designing of novel PLpro and 3CLpro inhibitors against previous SARS as well as recent COVID-19 diseases.
2. Materials and methods
2.1. Dataset
The dataset comprising of diverse SARS-CoV PLpro and 3CLpro inhibitors with their biological activities were retrieved from different literatures [32], [33], [34], [35], [36], [37], [38], [39], [40]. The duplicate compounds and the compounds with no inhibitory activities were removed from the dataset. Finally, 91 PLpro and 88 3CLpro inhibitors were selected for the determination of structural alerts using SARpy analysis [26], [27], [28]. The activity threshold was considered by taking the average of SARS-CoV PLpro pIC50 values (i.e., IC50 value of 6,200 nM), whereas in 3CLpro the activity threshold was set to the IC50 value of 10,000 nM. Thus, the molecules having the pIC50 values higher than the active threshold value were selected as actives compounds, whereas the molecules with pIC50 values lower than the active thresholds were considered as inactives. Finally, out of 91 PLpro inhibitors, 40 molecules were found to be active and 51 inactives Tables S1. Among 88 3CLpro inhibitors, 27 molecules were considered as actives and 61 inactives as depicted in the Table S2.
2.2. SARpy analysis
SARpy is a python based standalone software programme for automated QSAR model development [41], [42], [43], [44]. The software uses user-defined SMILEs notations for generating substructures in the set and tries to correlate between the particular molecular structures and their biological activity using three different steps. These are:
-
a
Fragmentation: In this step, recursive simple fragmentation algorithm is used to detect the chemical substructures present in the training set molecules. It iterates over every bond in the input structures and tries to generate possible pair of fragments.
-
b
Evaluation: Once all the substructures have been generated, their individual evaluation is done in order to detect the possible structural alerts (SAs) present in them.
-
c
Extraction: Finally, from the large collection of structural alerts that were generated, only the reduced sets of predicted rules were applied. In our current study, the rule sets were generated using two different settings they are: standard settings and modified settings. Further, the parameters selected in standard settings are minimum 2, maximum 18 atoms and occurring in a minimum 3 training set substances. Whereas, in modified settings minimum 3 and maximum 18 atoms and occurring in a minimum 5 training set substances was selected [45].
Additionally, each of the above SA settings was verified with two different single alert precision parameters they are: Auto MIN and Auto MAX. In the auto MIN settings the false negatives are minimized thus responsible for increasing the sensitivity values. Whereas, the auto MAX settings is considered for increasing the specificity values by minimizing the false positives in the training set. Thus, the aforementioned settings were first applied to split 1 to identify the settings generating the best overall result. Finally, the best optimal settings were then applied to both split 2 and split 3 for model development and evaluation [44], [45], [46], [47].
Evaluation of developed models is one of the key features for the prediction of model performances. From the three different splits, the number of true positives, true negatives, false positives and false negatives were determined. This information was further used for prediction of performances such as sensitivity, specificity, accuracy, Matthew's correlation coefficients (MCC), error rate (Table 1 ). The unpredicted rate was also calculated as prediction performance measure for different models by using the formula shown in Table 1.
Table 1.
The equations of statistical validation parameters.
| Entry | Parameter | Equation |
|---|---|---|
| 1 | Sensitivity | TP/(TP+FN) |
| 2 | Specificity | TN/(TN+FP) |
| 3 | Accuracy | (TP+TN)/(TP+FP+TN+FN) |
| 4 | MCC | (TP∗TN)−(FP∗FN)/√(TP+FP)(TP+FN)(TN+FP)(TN+FN) |
| 5 | Error rate | (FP+FN)/Total |
| 6 | Unpredicted rate | Number of unpredicted compounds / number of compounds in the dataset |
TP = True positive, TN = True negative, FP = False positive, FN = False negative.
2.3. Bayesian classification study
Bayesian classification study by the aid of Biovia Discovery Studio (DS) software [48] was conducted. Before conduct this Bayesian classification study, several fundamental molecular features namely, ALogP, Molecular weight (MW), Number of hydrogen bond donors (nHBD), Number of hydrogen bond acceptors (nHBA), Number of rotatable bonds (nRB), Number of rings (nRings), Number of aromatic rings (nAR), Molecular fractional polar surface area (MFPSA) of the dataset molecules have been calculated [48]. Alongside those molecular properties extended connectivity fingerprint of diameter 6 (ECFP_6), a topological fingerprint descriptor was also considered for this study [49]. The quality of this classification model was evaluated using the Receiver operating characteristics (ROC) [50]. Furthermore, the sensitivity, specificity and concordance were calculated for both the training and the test sets [25].
3. Results and discussions
3.1. Training Set and Test Set division
In this study 91 PLpro and 88 3CLpro inhibitors were selected as dataset in order to identify the fragments important for controlling the protease inhibition. The inhibitors were distributed into training set (80%) and test set (20%) randomly by using CORAL software. Again, the dataset was divided into training set A (80%) and test set A (20%) using the rational division algorithm (Kennard-Stone algorithm) [51], [52] by using the DatasetDivisionGUI 1.2 tool [53]. This algorithm selects the objects so they are divided evenly throughout the descriptor space of the original data set. The descriptors such as ALogP, Molecular weight (MW), Number of hydrogen bond donors (nHBD), Number of hydrogen bond acceptors (nHBA), Number of rotatable bonds (nRB), Number of rings (nRings), Number of aromatic rings (nAR), Molecular fractional polar surface area (MFPSA) and biological activity (active/inactive).
After the division of training and test set the key fragments from developed models were obtained by SARpy as well as Bayesian classification analysis and were interpreted for their importance in protease inhibition.
3.2. Model Development
3.2.1. SARpy analysis
In SARpy analysis, model building setting was considered as the initial step in order to build robust classification models. Different model buildings settings like: Standard-Auto MIN; Standard-Auto MAX; Modified-Auto MIN; Modified-Auto MAX was applied to the dataset. After model development, the three different statistical parameters like: error rate, unpredicted rate and number of structured matched along with the number of generated rules were considered for model evaluation for both training and test set. The performance of different model building settings for both PLpro and 3CLpro inhibitors are shown in Table 2 . Among these, the Modified-Auto MIN setting was found to have acceptable parameters such as: lowest unpredicted rate 0.04 and 0.22; highest structures matched 70 and 14 for both training and test set respectively in case of PLpro inhibitors, whereas, Standard-Auto MIN settings was found to be suitable for 3CLpro inhibitors. So, these settings were considered for further model development.
Table 2.
Performance of model building settings as obtained in case of PLpro and 3CLpro inhibitors.
| PLpro inhibitors | |||||||
| Settings | Rules (#) | Training set | Test set | ||||
| (Positive, Negative) | Error rate | Unpredicted rate | Structures matched | Error rate | Unpredicted rate | Structures matched | |
| Standard- Auto MIN |
14 (7, 7) | 0.19 | 0.03 | 71 | 0.06 | 0.39 | 11 |
| Standard- Auto MAX |
9 (3, 6) | 0.00 | 0.53 | 34 | 0.00 | 0.61 | 7 |
|
Modified- Auto MIN |
10 (5, 5) | 0.23 | 0.04 | 70 | 0.22 | 0.22 | 14 |
| Modified- Auto MAX |
5 (2, 3) | 0.00 | 0.62 | 28 | 0.22 | 0.22 | 0 |
| 3CLpro inhibitors | |||||||
| Standard- Auto MIN |
14 (4, 10) | 0.19 | 0.03 | 68 | 0.17 | 0.06 | 17 |
| Standard- Auto MAX |
7 (1, 6) | 0.00 | 0.41 | 41 | 0.39 | 0.00 | 7 |
|
Modified- Auto MIN |
8 (2, 6) | 0.11 | 0.11 | 62 | 0.17 | 0.00 | 17 |
| Modified- Auto MAX |
3 (1, 2) | 0.00 | 0.60 | 28 | 0.39 | 0.00 | 7 |
Further, to evaluate the capability of the selected methods the dataset was split again into training and test set two times (Split 2 and 3) separately and was used for the model development. The uniform distribution of the training and test set SARS-CoV PLpro and 3CLpro inhibitors in the PCA three dimensional plots referred a proper division of the training and the test sets from the split 2 and 3. After the division, by using Modified-Auto MIN settings for PLpro inhibitors and Standard-Auto MIN settings for 3CLpro inhibitors, total three different models from each setting were generated as shown in Table 3 .
Table 3.
Performance of prediction models of PLpro and 3CLpro inhibitors developed from 3 different splits.
| PLpro inhibitors | ||||||
| Training set | Test set | |||||
| split 1 | split 2 | split 3 | split 1 | split 2 | split 3a | |
| Sensitivity | 0.64 | 0.74 | 0.67 | 0.71 | 0.56 | 0.83 |
| Specificity | 1.00 | 0.89 | 0.93 | 0.71 | 1.00 | 0.75 |
| Accuracy | 0.76 | 0.80 | 0.79 | 0.71 | 0.71 | 0.79 |
| MCC | 0.61 | 0.62 | 0.61 | 0.43 | 0.56 | 0.58 |
| Error rate | 0.23 | 0.18 | 0.21 | 0.22 | 0.22 | 0.17 |
| Unpredicted rate | 0.04 | 0.11 | 0.04 | 0.22 | 0.22 | 0.22 |
| Structures matched (#) | 70 | 65 | 70 | 14 | 14 | 14 |
| 3CLpro inhibitors | ||||||
| Sensitivity | 0.83 | 0.81 | 0.70 | 0.50 | 0.66 | 1.00 |
| Specificity | 0.80 | 0.92 | 1.00 | 1.00 | 1.00 | 0.92 |
| Accuracy | 0.80 | 0.89 | 0.86 | 0.82 | 0.77 | 0.94 |
| MCC | 0.53 | 0.73 | 0.75 | 0.60 | 0.62 | 0.86 |
| Error rate | 0.19 | 0.10 | 0.13 | 0.17 | 0.17 | 0.06 |
| Unpredicted rate | 0.03 | 0.06 | 0.06 | 0.06 | 0.28 | 0.06 |
| Structures matched (#) | 68 | 66 | 66 | 17 | 13 | 17 |
The best model obtained from split 3 is marked in bold.
As expected, all the developed models were found to have significant statistical parameters. Among them, the model developed by using split 3 were found to have better statistically parameters in both the cases.
In case of PLpro inhibitors, the best model with sensitivity = 0.67, 0.83; specificity = 0.93, 0.75; and accuracy = 0.79, 0.79 were found for both training and test set respectively. Whereas, for 3Clpro inhibitors the training and test sets were found to have sensitivity = 0.73, 1.00; specificity = 1.00, 0.92; and accuracy = 0.86, 0.94, respectively. Additionally, the MCC value 0.61, 0.75 for training and 0.58, 0.86 for test in case of PLpro and 3CLpro respectively indicates that the classifier have good performance for the identification of structural fragments from the dataset. These fragments are high chemical reactivity molecular fragments responsible for modulating the protease inhibition. Therefore, the fragments obtained from compounds can be used to flag the potential chemical compounds responsible for protease inhibition. The fragments along with the SMARTS patters as obtained from the best model split 3 of both PLpro and 3CLpro inhibitors are shown below in Table 4 and 5 , respectively.
Table 4.
Important identified structural alerts of PLpro inhibitors as obtained from SARpy analysis.
 |
Table 5.
Important identified structural alerts of 3CLpro inhibitors as obtained from SARpy analysis.
 |
Furthermore, the outcomes from the Kennard-Stone rational division method yielded comparable results to the random division (Supplementary Table S3-S4). Similar fingerprints such as N1(CCC(CC1)C(=O)NCc1cc(ccc1)) and CC=C(CCC=C(C)C)C for PLpro inhibitors and N(C(=O)CSc1nccc(n1))c1ccccc1, S(=O)(=O)c1nc(c(c(c1))N(=O)=O) for 3CLpro inhibitors were also obtained from both Random division and Kennard-Stone rational division methods as can be seen from Supplementary Table S5-S6.
3.2.2. Bayesian classification analysis
The processes of Laplacian-corrected Bayesian classification model development was done on training set and externally validated on test set compounds. The selection of the training and test sets was considered from the best model generated from SARpy model. The results of Laplacian-corrected Bayesian classification model analysis derived from training set compounds are summarized in Table 6 . It showed that the Bayesian classification model with sensitivity, specificity and concordance values of 0.767, 0.930 and 0.863, respectively, was obtained for the training set of SARS-CoV PLpro inhibitors. Likewise, the Bayesian classification model for the training set of SARS-CoV 3CLpro inhibitors displays sensitivity, specificity and concordance values of 1.000, 0.729 and 0.814, respectively.
Table 6.
Statistics of training and test set of SARS-CoV PLpro and 3CLpro inhibitors.
| Target | Set | ROC | TP | FN | FP | TN | Sensitivity | Specificity | Concordance |
|---|---|---|---|---|---|---|---|---|---|
| PLpro | Training# | 0.751 | 23 | 7 | 3 | 40 | 0.767 | 0.930 | 0.863 |
| Test | 0.662 | 8 | 2 | 3 | 5 | 0.800 | 0.625 | 0.722 | |
|
3CLpro |
Training# | 0.840 | 22 | 0 | 13 | 35 | 1.000 | 0.729 | 0.814 |
| Test | 0.908 | 5 | 0 | 7 | 6 | 1.000 | 0.462 | 0.611 |
Receiver operating characteristics (ROC); #a 5-fold cross validation is performed for the training set to calculate the statistics.
The ROC of 0.751, 0.840 and 0.662, 0.908 were revealed for the training, test set of SARS-CoV PLpro and SARS-CoV 3CLpro inhibitors, respectively. These results demonstrate that the both models were robust and also provide a good predictive value.
3.3. Mining of the fragments those modulate biological activity
In order to identify the potential SARS-CoV protease inhibitor activity regulators, particular focus is give on the molecular modelling techniques like SARpy and Bayesian classification analysis to identify the fragments controlling the protease inhibition. The fragments can increase or reduce the protease inhibitions are classified as good or bad fragments, respectively. For this rational division method and their corresponding QSAR methodologies, random selection provides essentially equivalent results.
3.3.1. Interpretation of fragments for SARS-CoV PLpro inhibitors
The important fragments for the PLpro inhibitors were identified as shown in Table 4. The possible role of these fragments controlling the PLpro inhibition is shown in Figure 2 .
Fig. 2.
Important structural alerts for PLpro inhibitors.
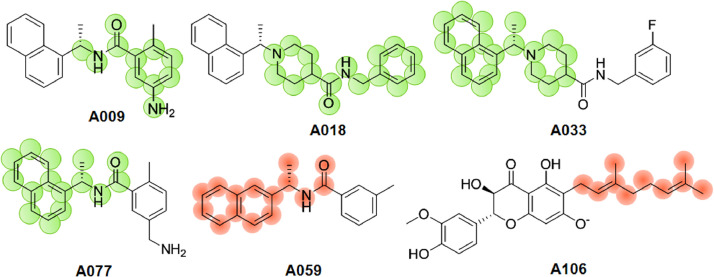
The molecular fragment Cc1cc(ccc1)C(=O)NC signifies the presence of any benzamide moiety found in compounds A009 (SARS-CoV PLpro IC50 = 0.6 µM) is responsible for inducing activity. Hence, this feature is responsible for modulating activities of SARS-CoV PLpro inhibitors. This fingerprint is marked in the Figure 2. Not surprisingly, this observation is in agreement with fingerprints G9, G10, G12, G14 (Figure S1) predicted by the Bayesian classification model on SARS-CoV PLpro inhibitors. The inhibitor A009 (GRL0617) and SARS-CoV PLpro interaction in Discovery Studio (DS) [48] suggests a pair of hydrogen bonds and some hydrophobic interactions which stabilized the complex (PDB: 3E9S). Notably, the amide group of benzamide moiety of GRL0617 helps in the interaction with the side chain of D165 and the backbone nitrogen of Q270 to form hydrogen bonds (Figure 3 A).
Fig. 3.
3D interaction plots of naphthyl based PLpro inhibitors with SARS-CoV PLpro active site amino acid residues (A) PDB: 3E9S and (B) PDB: 3MJ5.
Next, fragment N1(CCC(CC1))C(C)c1cccc2ccccc12 found in compound A018, A033 and others (Figure 2), A022-A025, A027, A030, A032, A033, A040-A042, A047, A048, A053, A054 signify the presence of 1-(1-(naphthalen-1-yl)ethyl)piperidine group is also answerable for SARS-CoV PLpro inhibitory activities. Meanwhile, we have analyzed the ligand-receptor interaction in Discovery Studio (DS) [48] where the piperidine ring engages in π-sigma interaction with the Y265 while the naphthalene ring forms several interactions with amino acids P248, P249, Y269 at the solvent exposed site of the enzyme (PDB: 3MJ5), as illustrated in Figure 3 B. Therefore, this indicates that naphthalene and piperidine rings are pivotal for SARS-CoV PLpro inhibition which also supports Bayesian modelling result in which G1-G7 fingerprints predicted these features as positive regulator of the PLpro inhibitory activities. Recent study by Freitas and co-workers suggested that 1-naphthalene based derivatives are also capable of inhibiting SARS-CoV-2 PLpro up to IC50 value of 2.4 µM [54].
Meanwhile, it should be noted that the 1-naphthyl containing PLpro inhibitors are more effective than the corresponding 2-naphthyl analogue (A003: SARS-CoV PLpro IC50 = 14.8 µM). Bad fragment B2 (Figure S2) predicted by the Bayesian classification model on SARS-CoV PLpro inhibitors illustrated the negative contribution of 2-naphthyl analogues. In addition, fragment SA N1(CCC(CC1)C(=O)NCc1cc(ccc1)) representing the N-benzylpiperidine-4-carboxamide group found in several active compounds (A022-A025, A027, A030, A032, A033, etc.) and C(=O)NC(c1cc2c(cc1)cccc2)C representing the N-(1-(naphthalen-2-yl)ethyl)formamide group found in compounds (example A077 in Figure 2) are the potential fragment that are liable for increasing the PLpro inhibitory activities of these compounds. Interestingly, this information is aligned with the experimental results by means of X-ray structural analyses of inhibitor bound SARS-CoV PLpro enzyme (PDB: 4OW0) [32]. The carboxyamide nitrogen of the inhibitor (compound A033) engages in the formation of a 3 Å hydrogen bond with the backbone carbonyl of Y269 at the active site (PDB: 4OW0) as depicted in Figure 4 drawn by PyMOL tool [55].
Fig. 4.
Binding mode of compound A033 with SARS-CoV PLpro active site amino acid residues (PDB: 4OW0).
Further, fragment like 2,6-dimethylocta-2,6-dieneas found in compounds A106-A112 is represented by the molecular fragment CC=C(CCC=C(C)C)C (Figure 2). Likewise, bad fingerprints B8, B14 and B15 (as depicted in Figure S2) also supported the negatively contribution of 2,6-dimethylocta-2,6-dienefunction (compounds A106-A112) to SARS-CoV PLpro inhibitory activities. Additionally, the study was also performed with Kennard-Stone rational dataset division method, similar types of good fingerprints like G5, G7, G8, G9, G10, G20 etc. (Figure S5) and bad fingerprints like B8, B13, B14 etc. (Figure S6) were obtained.
3.3.2. Interpretation of fragments for 3CLpro inhibitors
The fragments crucial for inducing activity to 3CLpro inhibitors were predicted and were further used for interpretation as shown in Table 5. The structural alert C(=O)Oc1cnccc1 defines the presence of pyridin-3-yl formate group in compounds B001-B003 and B005-B011 is responsible for inducing biological activities against SARS-CoV 3CLpro (Figure 5 ).
Fig. 5.
Important structural alerts for SARS-CoV 3CLpro inhibitors.
This indicates that pyridin-3-yl formate group is favoured for SARS-CoV 3CLpro inhibition which is also supported by G3, G4, G5 and G6 good fingerprints (Figure S3) as derived from the ECFP_6 fingerprint. In contrast, the presence of pyridine nucleus having branching with NO2 and SO2 group represented by the fragment S(=O)(=O)c1nc(c(c(c1))N(=O)=O) is also considered as negative fragment in compounds B032 and B033 (Figure 5). This indicates that pyridine nucleus having branching with NO2 and SO2 group is unfavoured which also support previously published results [56]. The fragments like CN1C(=O)C(=O)c2cc(I)ccc12 defines the presence of 5-iodo indoline-2,3-dione group is accountable for SARS-CoV 3CLpro inhibitory activity. Active compounds B013, B026 bearing the fragment are illustrated in Figure 5. Further, the fragments like CC(C)NC(=O)CNC(=O) and CN1C(=O)C(=O)c2ccccc12 define the presence of 2-formamido-N-isopropylacetamide and 1-methylindoline-2,3-dione groups, respectively are also accountable for SARS-CoV 3CLpro inhibitory activity. These observations can be justified by observing the compounds B007, B004, B017 for fragment CC(C)NC(=O)CNC(=O) and B013, B014, B015, B019, B024 for SA CN1C(=O)C(=O)c2ccccc12. Notably, the CC(C)NC(=O)CNC(=O) feature of compound B007 formed several interactions with SARS-CoV 3CLpro active site amino acid residues (PDB: 3ATW) as illustrated in Figure 6 .
Fig. 6.
3D interaction plot of compound B007 with SARS-CoV 3CLpro active site amino acid residues (PDB: 3ATW).
By contrast, compounds B052, B057 bearing fragment CC(C)NC(=O)CNC(=O) possess lower SARS-CoV 3CLpro inhibitory activities. Likewise, the fragment, N(C(=O)CSc1nccc(n1))c1ccccc1 found in compounds B063-B068 and B070-B077 represent the N-phenyl-2-(pyrimidin-2-ylthio)acetamide group is a significant negative fragment for protease inhibition. This inspection is in agreement with bad fingerprints (Figure S4) as constructed from the ECFP_6 fingerprint. This observation is also verified by the experimental results such as the activity of compound compounds B063-B068 and B070-B077 bearing such fragment exhibited poor SARS-CoV 3CLpro inhibitory activities. Hence, this sub-structural feature is responsible for lowering the proteolytic activity against SARS-CoV Mpro (Figure 7 ). Further, with Kennard-Stone rational dataset division method, comparable good fingerprints including G1, G8, G9 etc. (Figure S7) and bad fingerprints like: B3, B7, B8 etc. (Figure S8) were also obtained. Thus, these fingerprints will be important in optimizing the activity of the 3CLpro inhibitors in future.
Fig. 7.
Structures of some inactive inhibitors containing bad Bayesian fragments.
Further, the fragment c1ccc(cc1C)CCCC represents the presence of 1-butyl benzene. This function should also consider as potential modulator of protease inhibition and is responsible for hindrance of activity in compounds. Not surprisingly, the Bayesian modelling based Good fragments G7, G18, G19 and G20 clearly indicated the importance of furan ring against SARS-CoV 3CLpro inhibition. This observation is in agreement with our pervious observation [56] in which compounds bearing both furan and pyridine exhibited effective proteolytic activities (IC50 in between 50 to 63 nM). In addition, compound B018 having a furan ring in its structure exhibits promising SARS-CoV 3CLpro inhibitory activities. In a structure-based analysis, Jacobs et al [57] found that the furan oxygen atom of compound B018 forms hydrogen bonding interaction the backbone NH of G143. At the S1´ site, the catalytic C145 resides beneath the furan oxygen atom at a short distance (Figure 8 ). Further, we have analyzed the ligand (B018)-receptor (SARS-CoV 3CLpro) interaction in DS [48] where a π-donor hydrogen bond is noticed between the furan ring and the catalytic C145 [57] (Figure S9).
Fig. 8.
Binding mode of compound B018 with SARS-CoV 3CLpro active site amino acid residues as derived from the crystal structure (PDB: 3V3M) [57].
In summary, SARpy analysis based structural fragments for SARS-CoV 3CLpro inhibitory activity suggested that heterocyclic rings such as pyridine, pyrimidine andindoline modulate the biological properties. Moreover, the results of Bayesian modelling study are closely associated with the SARpy analysis. Additionally, the Bayesian modelling study predicted the positive influence of furan ring in the SARS-CoV 3CLpro inhibitory property. Therefore, to achieve an attractive potency level, several heterocyclic rings controlled the 3CLpro inhibitory activity with respect to their polarity, size and hydrogen bonding capabilities.
4. Implications for COVID-19 Drug Discovery
The medicinal chemistry outlook of novel coronavirus disease 2019 (COVID-19), caused by SARS-CoV-2, is just few months old. Molecular modelling driven anti-viral study can generate several possibilities in drug discovery against COVID-19. The fragments studied and extensively interpreted in this current study surely leaving much room for follow-up investigations against SARS-CoV-2. Notably, the fragments identified by the SARpy analysis and Bayesian classification study may be an effective approach to accelerate drug design against SARS-CoV-2.
Since the binding site of PLpro and 3CLpro enzymes were conserved between SARS-CoV and SARS-CoV-2 the fragments will be very useful for COVID-19 drug discovery. Here, the identified fragments can also found in recently reported compounds (Figures 9 and 10 ) [7,9,17,54]. The current study will offer an idea to explore the important existing structural data and an in-depth qualification of fragment hits. This will stimulate further research by providing valuable guidance to the medicinal chemists for designing of novel PLpro and 3CLpro inhibitors against previous SARS as well as recent COVID-19 diseases.
Fig. 9.
Structures of two active SARS-CoV-2 PLpro inhibitors highlighting important fragments. The activity values were taken from literature [54].
Fig. 10.
Structures of some active SARS-CoV-2 3CLpro inhibitors highlighting important fragments. The activity values were taken from literatures [9,17].
Meanwhile, looking the exact crystal binding position of an inhibitor and changing/replacing the different fragment(s) may offers the possibility of modifying the lead compounds, hence, their inhibitory potential against the proteases. Besides, potential protease inhibition activities are also accompanied by further increases in scaffold diversity. However, care should be taken to design effective protease inhibitors by keeping ADME properties in mind. Nevertheless, this study may offer a bigger direction in broad spectrum anti-viral drug design and discovery.
5. Conclusion
Here, classification-based QSAR models for diverse set of SARS-CoV PLpro and 3CLpro were developed and validated. All models were successfully used to hunt promising fragments. The fragments can be further utilised for the virtual screening of chemical libraries or FDA approved drugs to identify effective protease inhibitors.
The closely related fragment concepts are experiencing increasing interest in medicinal chemistry. In our analysis, we have refined the assessment of fragments by exploring structure activity relationships (SARs). The fragments encode an attractive knowledge base for compound design as well as utilized for lead optimization.
Since the size of the database of SARS-CoV protease inhibitor continues to grow, collectively our modeling studies emphasize that our approach could be used to aid in the process of lead optimization against the proteases. In diseases like COVID-19 with the lack of potential drug and large-scale inhibitors, it is quite difficult to connect the dots to quaerere significant therapeutics. Reciprocally, one can apply these identified fragments to increase the protease inhibition of weak hits resulting from QSAR/VS studies etc. against targeted proteases.
To summarize, we have developed robust and validated models which may be applicable to drug design and lead optimization, opening up opportunities to design small molecules targeting the coronavirus proteases.
CRediT author statement
Kalyan Ghosh: Data curation, Methodology, Software, Investigation, Writing- Original draft preparation.
Sk. Abdul Amin: Conceptualization, Methodology, Software, Investigation, Visualization, Writing- Original draft preparation.
Shovanlal Gayen: Conceptualization, Writing- Reviewing and Editing.
Tarun Jha: Writing- Reviewing and Editing, Supervision.
Declaration of Competing Interest
The authors have no conflict of interests.
Acknowledgment
Sk. Abdul Amin is thankful to the Council of Scientific and Industrial Research (CSIR), New Delhi, India for providing financial assistance in form of a Fellowship [FILE NO.: 09/096(0967)/2019-EMR-I, Dated: 01-04-2019]. Tarun Jha is thankful for the financial support from RUSA 2.0 of UGC, New Delhi, India to Jadavpur University, Kolkata, India. We are very much thankful to the Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India and Department of Pharmaceutical Sciences, Dr. Harisingh Gour University, India for providing the research facilities.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.130366.
Appendix. Supplementary materials
References
- 1.World Health Organization . WHO Director-General's opening remarks at the media briefing on COVID-19-11 March 2020. 2000. https://www.who.int/dg/speeches/detail/who-director-general-s-openingremarks-at-the-media-briefing-on-covid-19 (Date of Access: Aug 18, 2020) [Google Scholar]
- 2.WHO data https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (Date of Access: March 30, 2021).
- 3.Amin S.A., Jha T. Fight against novel coronavirus: A perspective of medicinal chemists. Eur. J. Med. Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adhikari N., Baidya S.K., Saha A., Jha T. In: Viral Proteases and their inhibitors. Gupta S.P., editor. Academic Press; USA: 2017. Structural insight into the viral 3Clike protease inhibitors: Comparative SAR/QSAR approaches; pp. 317–402. Chapter 11. [Google Scholar]
- 5.Chen Y., Liu Q., Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., Ge J., Zheng L., Zhang Y., Wang H., Zhu Y., Zhu C., Hu T., Hua T., Zhang B., Yang X., Li J., Yang H., Liu Z., Xu W., Guddat W.L., Wang Q., Lou Z., Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai W., Zhang B., Su H., Li J., Zhao Y., Xie X., Jin Z., Liu F., Li C., Li Y., Bai F., Wang H., Cheng X., Cen X., Hu S., Yang X., Wang J., Liu X., Xiao G., Jiang H., Liu H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368:1331–1335. doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goyal B., Goyal D. Targeting the Dimerization of the Main Protease of Coronaviruses: A Potential Broad-Spectrum Therapeutic Strategy. ACS Comb. Sci. 2020 doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keng C.T., Åkerström S., Leung C.S., Poon L.L., Peiris J.M., Mirazimi A., Tan Y.J. SARS coronavirus 8b reduces viral replication by down-regulating E via an ubiquitinin dependent proteasome pathway. Microbesinfect. 2011;13:179–188. doi: 10.1016/j.micinf.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 12.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngo S.T., Quynh Anh Pham N., Thi Le L., Pham D.H., Vu V.V. Computational determination of potential inhibitors of SARS-CoV-2 main protease. J. Chem. Inf. Model. 2020 doi: 10.1021/acs.jcim.0c00491. [DOI] [PubMed] [Google Scholar]
- 16.Wang J. Fast Identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J. Chem. Inf. Model. 2020 doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. BioRxiv. 2020 doi: 10.1101/2020.04.20.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna I. Drug discovery in pharmaceutical industry: productivity challenges and trends. Drug Discov. Today. 2012;17:1088–1102. doi: 10.1016/j.drudis.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Murray C.W., Rees D.C. The rise of fragment-based drug discovery. Nat. Chem. 2009;1:187–192. doi: 10.1038/nchem.217. [DOI] [PubMed] [Google Scholar]
- 20.Erlanson D.A., Fesik S.W., Hubbard R.E., Jahnke W., Jhoti H. Twenty years on: the impact of fragments on drug discovery. Nat. Rev. Drug Discov. 2016;15:605–619. doi: 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- 21.Chen H., Zhou X., Wang A., Zheng Y., Gao Y., Zhou J. Evolutions in fragment-based drug design: The deconstruction–reconstruction approach. Drug Discov. Today. 2014;20:105–113. doi: 10.1016/j.drudis.2014.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erlanson D.A., Jahnke W. Wiley-VCH; Weinheim, Germany: 2016. Fragment-based Drug Discovery. Lessons and Outlook. ISBN 9783527337750. [Google Scholar]
- 23.Drag M., Salvesen G.S. Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferenczy G.G., Keserű G.M. Thermodynamics of fragment binding. J. Chem. Inf. Model. 2012;52:1039–1045. doi: 10.1021/ci200608b. [DOI] [PubMed] [Google Scholar]
- 25.Amin S.A., Adhikari N., Jha T. Diverse classes of HDAC8 inhibitors: in search of molecular fingerprints that regulate activity. Future Med. Chem. 2018;10:1589–1602. doi: 10.4155/fmc-2018-0005. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari T., Gini G., Bakhtyari N.G., Benfenati E. Mining toxicity structural alerts from SMILES: A new way to derive Structure Activity Relationships. IEEE; 2000. pp. 120–127. In 2011 IEEE Symposium on Computational Intelligence and Data Mining (CIDM) 2011 Apr 11. [Google Scholar]
- 27.Lombardo A., Pizzo F., Benfenati E., Manganaro A., Ferrari T., Gini G. A new in silico classification model for ready biodegradability, based on molecular fragments. Chemosphere. 2014;108:10–16. doi: 10.1016/j.chemosphere.2014.02.073. [DOI] [PubMed] [Google Scholar]
- 28.Golbamaki A., Benfenati E., Golbamaki N., Manganaro A., Merdivan E., Roncaglioni A., Gini G. New clues on carcinogenicity-related substructures derived from mining two large datasets of chemical compounds. J. Environ. Sci. Health C. 2016;34:97–113. doi: 10.1080/10590501.2016.1166879. [DOI] [PubMed] [Google Scholar]
- 29.Gaikwad R., Ghorai S., Amin S.A., Adhikari N., Patel T., Das K., Jha T., Gayen S. Monte Carlo based modelling approach for designing and predicting cytotoxicity of 2-phenylindole derivatives against breast cancer cell line MCF7. Toxicol. In Vitro. 2018;52:23–32. doi: 10.1016/j.tiv.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem. 2020 doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bobrowski T., Alves V.M., Melo-Filho C.C., Korn D., Auerbach S., Schmitt C., Muratov EN., Tropsha A. Computational models identify several FDA approved or 1 experimental drugs as putative agents against SARS-CoV-2. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12153594.v1. [DOI] [Google Scholar]
- 32.Báez-Santos Y.M., Barraza S.J., Wilson M.W., Agius M.P., Mielech A.M., Davis N.M., Baker S.C., Larsen S.D., Mesecar A.D. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J. Med. Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng K.W., Cheng S.C., Chen W.Y., Lin M.H., Chuang S.J., Cheng I.H., Sun C.Y., Chou C.Y. Thiopurineanalogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antiviral Res. 2015;115:9–16. doi: 10.1016/j.antiviral.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chou C.Y., Chein C.H., Han Y.S., Prebanda M.T., Hsieh H.P., Turk B., Chang G.G., Chen X. Thiopurine analogues inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biochem. Pharmacol. 2008;75:1601–1609. doi: 10.1016/j.bcp.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frieman M., Basu D., Matthews K., Taylor J., Jones G., Pickles R., Baric R., Engel D.A. Yeast based small molecule screen for inhibitors of SARS-CoV. PLoS One. 2011;6:e28479. doi: 10.1371/journal.pone.0028479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh A.K., Takayama J., Rao K.V., Ratia K., Chaudhuri R., Mulhearn D.C., Lee H., Nichols D.B., Baliji S., Baker S.C., Johnson M.E. Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein− ligand X-ray structure and biological evaluation. J. Med. Chem. 2010;53:4968–4979. doi: 10.1021/jm1004489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghosh A.K., Takayama J., Aubin Y., Ratia K., Chaudhuri R., Baez Y., Sleeman K., Coughlin M., Nichols D.B., Mulhearn D.C., Prabhakar B.S. Structure-Based Design, Synthesis, and Biological Evaluation of a Series of Novel and Reversible Inhibitors for the Severe Acute Respiratory Syndrome− Coronavirus Papain-Like Protease. J. Med. Chem. 2009;52:5228–5240. doi: 10.1021/jm900611t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J.Y., Kim J.H., Kim Y.M., Jeong H.J., Kim D.W., Park K.H., Kwon H.J., Park S.J., Lee W.S., Ryu Y.B. Tanshinones as selective and slow-binding inhibitors for SARS-CoV cysteine proteases. Bioorg. Med. Chem. 2012;20:5928–5935. doi: 10.1016/j.bmc.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratia K., Saikatendu K.S., Santarsiero B.D., Barretto N., Baker S.C., Stevens R.C., Mesecar A.D. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Nat. Acad. Sci. 2006;103:5717–5722. doi: 10.1073/pnas.0510851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.https://chemrxiv.org/articles/Computational_Models_Identify_Several_FDA_Approved_or_Experimental_Drugs_as_Putative_Agents_Against_SARS-CoV-2/12153594/1
- 41.Toropova A.P., Toropov A.A., Benfenati E. A quasi-QSPR modelling for the photo catalytic decolourization rate constants and cellular viability (CV%) of nanoparticles by CORAL. SAR QSAR Environ. Res. 2015;26:29–40. doi: 10.1080/1062936X.2014.984327. [DOI] [PubMed] [Google Scholar]
- 42.Toropov A.A., Toropova A.P., Puzyn T., Benfenati E., Gini G., Leszczynska D., Leszczynski J. QSAR as a random event: modeling of nanoparticles uptake in PaCa2 cancer cells. Chemosphere. 2013;92:31–37. doi: 10.1016/j.chemosphere.2013.03.012. [DOI] [PubMed] [Google Scholar]
- 43.Van Bossuyt M., Raitano G., Honma M., Van Hoeck E., Vanhaecke T., Rogiers V., Mertens B., Benfenati E. New QSAR models to predict chromosome damaging potential based on the in vivo micronucleus test. Toxicol.Lett. 2020 doi: 10.1016/j.toxlet.2020.04.016. [DOI] [PubMed] [Google Scholar]
- 44.Ferrari T., Cattaneo D., Gini G., Golbamaki Bakhtyari N., Manganaro A., Benfenati E. Automatic knowledge extraction from chemical structures: the case of mutagenicity prediction. SAR QSAREnviron. Res. 2013;24:365–383. doi: 10.1080/1062936X.2013.773376. [DOI] [PubMed] [Google Scholar]
- 45.Yang H., Li J., Wu Z., Li W., Liu G., Tang Y. Evaluation of different methods for identification of structural alerts using chemical ames mutagenicity data set as a benchmark. Chem. Res. Toxicol. 2017;30:1355–1364. doi: 10.1021/acs.chemrestox.7b00083. [DOI] [PubMed] [Google Scholar]
- 46.Baderna D., Gadaleta D., Lostaglio E., Selvestrel G., Raitano G., Golbamaki A., Lombardo A., Benfenati E. New in silico models to predict in vitro micronucleus induction as marker of genotoxicity. J. Hazard. Mater. 2020;385 doi: 10.1016/j.jhazmat.2019.121638. [DOI] [PubMed] [Google Scholar]
- 47.Hemmerich J., Troger F., Füzi B.; F., Ecker G. Using machine learning methods and structural alerts for prediction of mitochondrial toxicity. Mol. Inform. 2020;39 doi: 10.1002/minf.202000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biovia D.S. Discovery Studio 2016. 2000. San Diego, CA, USA. [Google Scholar]
- 49.David R., Mathew H. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 50.Fawcett T. An introduction to ROC analysis. Pattern Recog. Lett. 2006;27:861–874. [Google Scholar]
- 51.Martin T.M., Harten P., Young D.M., Muratov E.N., Golbraikh A., Zhu H., Tropsha A. Does rational selection of training and test sets improve the outcome of QSAR modeling? J. Chem. Inf. Model. 2012;52:2570–2578. doi: 10.1021/ci300338w. [DOI] [PubMed] [Google Scholar]
- 52.Golbraikh A., Shen M., Xiao Z., Xiao Y.D., Lee K.H., Tropsha A. Rational selection of training and test sets for the development of validated QSAR models. J. Comput. Aided Mol. Des. 2003;17:241–253. doi: 10.1023/a:1025386326946. [DOI] [PubMed] [Google Scholar]
- 53.http://teqip.jdvu.ac.in/QSAR_Tools/ (as accessed on 17th Feb, 2021)
- 54.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020 doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 55.PyMOL is a molecular visualization system on an open-source foundation. 2000. [Google Scholar]
- 56.Ghosh K., Amin S.A., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J. Mol. Struct. 2020;1224 doi: 10.1016/j.molstruc.2020.129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacobs J., Grum-Tokars V., Zhou Y., Turlington M., Saldanha S.A., Chase P., Eggler A., Dawson E.S., Baez-Santos Y.M., Tomar S., Mielech A.M. Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease. J. Med. Chem. 2013;56:534–546. doi: 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.