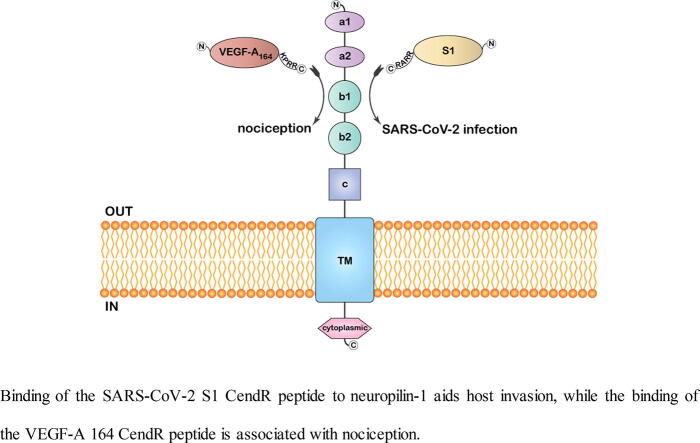
Graphical abstract
Binding of the SARS-CoV-2 S1 CendR peptide to neuropilin-1 aids host invasion, while the binding of the VEGF-A 164 CendR peptide is associated with nociception.
Keywords: Neuropilin, SARS-CoV-2, Spike protein, Host factor, VEGF-A, Nociception
Abstract
Viral internalization is aided by host cell surface receptors. In the case of SARS-CoV-2 and SARS-CoV, the primary host receptor is the angiotensin-converting enzyme 2 (ACE2). Considering the disparities in the transmission rate and viral tropism of the two coronaviruses, additional host factors were suspected. Recently, a novel host factor for SARS-CoV-2 entry, neuropilin-1 (NRP-1) has been identified. These receptors potentiate viral infection in the presence of other host factors like ACE2. Through its C-end rule (CendR) motif exposed following furin processing, the SARS-CoV-2 spike protein binds to the CendR pocket of NRP-1 and achieves cell entry through endocytosis. The binding of SARS-CoV-2 spike protein to the NRP-1 receptor interferes with the docking of its endogenous ligand VEGF-A, signaling that would otherwise promote nociception. This review looks at the function of neuropilins and how it contributes to SARS-CoV-2 infection and nociception.
1. Introduction
From a few cases of coronavirus disease 2019 (COVID-19) reported in late 2019, the viral outbreak rose to a pandemic level in a span of just a few months at a sharp rate of infectivity and mortality [1]. For many viruses, tissue tropism is dependent on the availability of receptors and entry cofactors on the surface of host cells since viral interaction with receptors on host cells is a pivotal early move for its invasion [2]. The RNA virus, ‘severe acute respiratory syndrome coronavirus 2′ (SARS-CoV-2), the causative agent of COVID-19 realizes target cell adherence and uptake through its viral spike (S) protein [3].
SARS-CoV-2 and the similar SARS-CoV, the causative agent of the previous outbreak in 2003, attach to angiotensin-converting enzyme 2 (ACE2), which is present primarily on the surface of cells in the lower respiratory epithelium, kidneys and the gastrointestinal tract. While it is quite probable that the principal entry gateway for the virus into the cell is the ACE2 receptor, given the reported dissimilarities in viral tropism, it has been proposed that other host factors may be implicated [2]. This could possibly be the justification behind the pandemic status of SARS-CoV-2 [4]. Additionally, SARS-CoV resulted in a far smaller epidemic, potentially owing to infection arising largely in the lower respiratory system; while SARS-CoV-2 propagates quickly via dynamic pharyngeal viral propagation [5].
It has been reported that SARS-CoV-2 affinity for ACE2 is ten to twenty-fold greater than that of SARS-CoV [6]. Another possible explanation for the higher transmission of SARS-CoV-2 is the bearing of a polybasic carboxyl-terminal sequence motif, Arg-Arg-Ala-Arg (RRAR) that serves as a furin cleavage site at the S1-S2 boundary in the SARS-CoV-2 S protein that is not found in SARS-CoV [7]. The occurrence of the polybasic furin-cleavage site in SARS-CoV-2, and not in SARS-CoV-2 causes greater infectivity by priming the integration process and could possibly generate other cell surface receptor attachment sites [8].
Comparable sequences are present in the S proteins of various infectious human viruses, such as HIV-1, Ebola, and strongly pathogenic strains of avian influenza [9]. The high transmission feature of SARS-CoV-2 could also be attributed partly to specific structural characteristics of the S protein when compared to the earlier six human coronaviruses (HCoVs) that did not attain pandemic status [10]. These features include the strong binding association of the ‘receptor binding domain’ (RBD) to ACE2, the flat and non-submerged sialic acid-binding region, and the polybasic RRAR sequence motif.
2. Currently established SARS-CoV-2 host factors
A large body of work on SARS-CoV-2 uptake mainly focused on ACE2, though it exhibits minimal expression in the epithelia of respiratory and olfactory cells [11]. This indicated that other factors may be necessary to mediate virus-target cell contact in the event of low ACE2 expression. The low levels of ACE2 expression in a small subset of lung epithelial cells and negligible levels in endothelial cells allude that SARS-CoV-2 uptake and infection could potentially be through additional receptors, an integration of several receptors, or an enhancing cofactor. Though ACE2 expression has not been detected in the majority of neurons, there has been growing documentation of the prevalence of neurological manifestations among COVID-19 patients. Oddly enough, ACE2 expression levels drop with age, but higher COVID-19 gravity was observed in aged subjects, reinforcing the hypothesis that ACE2 alone cannot be the only pathway associated with SARS-CoV-2 uptake.
Interestingly, neuropilins (NRPs) have been noted to promote SARS-CoV-2 uptake [8], [12]. NRPs are transmembrane receptor glycoproteins associated initially with neurons. These receptors, NRP-1 and NRP-2, the two neuropilin isoforms in humans are abundantly present in both neuronal and endothelial cells. These multifaceted cell surface co-receptors recognize and associate with several membrane proteins and therefore mediate diverse biological activities such as semaphorin-based axon pathfinding, vascular endothelial growth factor VEGF-based angiogenesis, and vascular permeability. These receptors are also found to be implicated in tumor vascularization and are targeted in cancer-therapy [13]. Neuropilins typically exist as homodimers, but the two homologs can also form a heterodimer [14]. NRP-1 and NRP-2 exhibit 44% sequence identity and share identical domain organization [15]. Both NRP homologs engage their respective VEGF ligands through their core conserved b1 binding domain. Apart from the membrane-bound NRPs, soluble forms of NRP have also been identified. Soluble NRPs (sNRPs) are a product of alternative RNA splicing and ectodomain shedding; and carry an extracellular domain, with cytoplasmic and transmembrane domains lacking. Such non-membrane bound NRPs are promising candidates for novel biological markers and as synthetic pathway regulators. sNRPs can act as trap receptors to inhibit NRP-mediated activity, such as NRP-VEGF binding [13]. sNRP-1 inhibition of VEGF-A-NRP1 interaction was found to result in tumor cell death [16].
Single-cell RNA sequencing datasets of human lung tissue and olfactory epithelium showed extremely low levels of ACE2 expression. However, NRP-1 and NRP-2 were both enriched in nearly all olfactory and pulmonary cells, especially in endothelial cells. These findings were further validated through post-mortem analysis [17].
When expressed alone, ACE2 rendered cells become susceptible to infection. Even though NRP-1 alone fails to cause infection in HEK-293T cells, it promotes infection considerably when co-expressed along with ACE2 and transmembrane protease, serine 2 (TMPRSS2), indicating that NRP-1 can enhance SARS-CoV-2 infection in the presence of additional host factors [8]. This could be attributed to enhanced virus entry into the cell instead of mere binding to the cell surface [2].
SARS-CoV-2 S protein priming for viral entry is catalyzed by TMPRSS2; and the clinically approved protease inhibitor camostat mesylate has been shown to disrupt SARS-CoV-2 invasion in lung cells [18]. It was observed that along with TMPRSS2, the cathepsin B and L (CatB/L) protease could also prime SARS-CoV S protein, though TMPRSS2 is non-negotiable. The inhibition of CatB/L function was found to block SARS-CoV and SARS-CoV-2 uptake into TMPRSS2-deficient cells. Viral infection was blocked altogether when E64d, a CatB/L inhibitor and camostat mesylate were concomitantly introduced. This suggests that both TMPRSS2 and CatB/L are required for SARS-CoV-2 S protein priming.
Wang and colleagues previously demonstrated that CD147, a transmembrane glycoprotein receptor which also goes by the name basigin or EMMPRIN, imparts functional significance in mediating SARS-CoV infection through a novel entry pathway. Specifically, they identified an interaction between CD147 and the SARS-CoV-2 S protein and found that the deletion of CD147 or its inhibition by meplazumab, an ‘anti-CD147 antibody’, blocks SARS-CoV-2 propagation [19]. Additionally, the drug apilimod was found to promote the inhibition of endosomal PIKfyve kinase, an effect that suppresses SARS-CoV-2 and Zaire Ebolavirus infection by blocking the secretion of the viral components from endosomes [20].
3. SARS-CoV-2 spike glycoprotein
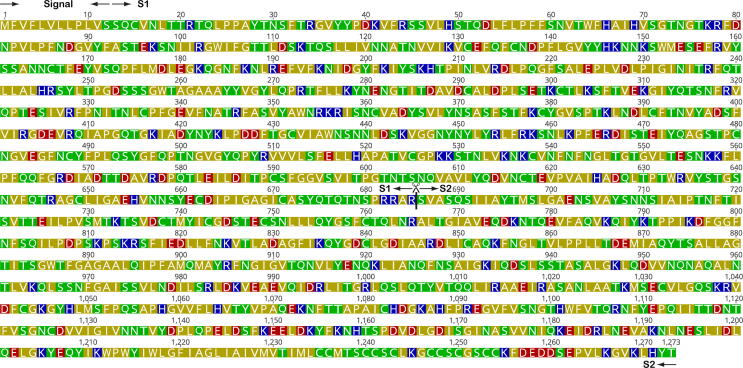
The 10 nm long S glycoprotein precursor of SARS-CoV-2, consisting of 1273 amino acids is cleaved by the protease furin present in endocytic compartments into S1 and S2, which remain associated non-covalently. The former is sub-divided into the C-terminal domain (CTD) and N-terminal domain (NTD) [10]. The cytoplasmic tail at the C-terminal is believed to facilitate signaling and promote receptor-guided endocytosis [21]. Proteolytic furin-cleavage at the S1-S2 boundary occurs either during viral synthesis or host cell entry and generates the polybasic carboxyl-terminal sequence motif (682RRAR685) on S1, and is necessary for invasion [22] (Fig. 1). This sequence motif corresponds to a [R/K]XX[R/K] motif, which is denoted as the C-end rule (CendR). In this motif, R represents arginine and X could be any amino acid; R can be replaced by K (lysine). Peptides with this motif can achieve cell entry and infection via binding to NRP-1 and NRP-2 on the cell surface [23].
Fig. 1.
Sequence of the SARS-CoV-2 spike protein (Uniprot accession: P0DTC2). Amino acids are colored based on their physiochemical property (red – acidic; blue – basic; green – polar; olive – hydrophobic). The furin cleavage site that exposes the CendR motif (682RRAR685) is marked with a  . Signal, S1 and S2 regions are also shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
. Signal, S1 and S2 regions are also shown. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Protein glycosylation imparts great significance in viral infectivity. Similar to the S proteins of other coronaviruses, the SARS-CoV-2 S protein undergoes abundant glycosylation, which enables it to bypass the host immune reaction by masking set antigenic determinants from antibody counteraction [6]. Such masking has been noted in influenza hemagglutinin and HIV-1 envelope protein. In the SARS-CoV-2 S protein, each protomer holds twenty-two N-linked glycosylation regions. Other than masking, molecular dynamics simulations indicate that N165 and N234 glycosylation may add to the structural integrity of the receptor-binding pocket in its detection by ACE2.
4. Interaction of spike S1 and neuropilin
Using X-ray crystallography and biochemical methods, the direct interaction of the S1 CendR motif with NRP-1 was demonstrated [12]. S1 CendR motif was observed to interact with Tyr297, Trp301, Thr316, Asp320, Ser346, Thr349 and Tyr353 of NRP-1. Similar binding was also reported with NRP-2, which exhibits 44% sequence identity with NRP-1. In fact, the residues involved in binding the CendR motif are conserved in both NRP-1 and NRP-2 b1 domains, which share approximately 52% sequence identity (Fig. 2). Surprisingly, both instances involve residual interaction with a ΔRRAR mutant (a mutant with the CendR RRAR sequence deleted), hinting at a supporting CendR-independent binding between S1 protein and neuropilins.
Fig. 2.
Protein sequence alignment of the b1 domains of NRP-1 (Uniprot accession: O14786) and NRP-2 (Uniprot accession: O60462). Conserved residues are highlighted in green. Residues that interact with the CendR motif of SARS-CoV-2 spike protein are marked with a solid triangle below it. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
S1 constitutes the “head” of the glycoprotein and mediates the ACE2 interaction, while S2 remains fixed on the virus membrane and induces membrane fusion. Similar to fusion proteins of HIV-1 and influenza virus, S2 attaches a fusion peptide present at its amino end into the host cell membrane and then bends back to fuse the virus and host membranes [24]. The S2 protein requires an additional proteolytic activity which is mediated by TMPRSS2 or alternative proteases, to “liberate” its fusion peptide [18]. A cryo–electron micrograph of the SARS-CoV-2 S protein structure illustrates that the S1-S2 boundary is found at a solvent-exposed loop, making it free for receptor binding [22]. Monoclonal antibody directed against the extracellular b1b2 region of NRP-1 inhibits the substantial augmentation of SARS-CoV-2 pathogenicity. Antibody binding with the b1b2 region of wild-type (WT) NRP-1 was detected, unlike the case with the triple-mutant - S346A, E348A, and T349A- CendR binding site in the b1b2 domain [8].
NRP-2, a receptor for the Lujo virus does not exhibit b1 domain interaction like in SARS-CoV-2 [25]. Though the S1 protein can also engage NRP-2, its function in SARS-CoV-2 infection remains unclear [12]. This implies that a broad range of viruses engage NRPs in the process of infection, though more work is required to obtain a deeper understanding of the molecular mechanism involved. The NRP-1 specific inhibitor EG00229 binds to the b1 domain of NRP-1, inhibiting the direct interaction of the S1 CendR peptide with the b1 domain. Inhibiting S protein-NRP-1 interaction through RNA interference or EG00229 lowered SARS-CoV-2 load and invasion in cell culture. Hence, SARS-CoV-2 could drastically heighten its pathogenicity through NRP-1 binding, which augments its uptake and infection. Interestingly, no binding was observed following the mutation of the C terminal arginine to alanine ‘(679NSPRRAA685)’ of the S1 CendR peptide sequence. Furthermore, viral entry reduced by 50% in NRP-1-depleted cells relative to control cells following 0.5 h of incorporation. Therefore, NRP-1 represents a host agent for SARS-CoV-2 invasion and may possibly present a therapeutic target for COVID-19.
5. Expression of NRP-1 in COVID-19
Notably, ‘omic’ studies reported an appreciable overexpression of NRP-1 in biological specimens obtained from COVID-19 subjects relative to disease-free controls [11]. Additionally, post-mortem examination of olfactory epithelium collected from human COVID-19 patients demonstrated that SARS-CoV-2 invaded NRP-1-carrying cells opposite the nasal cavity [17]. These findings are interesting considering that various COVID-19 subjects lose their perception of odor [2].
Gene expression analysis showed a remarkable up-regulation of NRP-1 and NRP-2 in lung tissue samples from COVID-19 patients [12]. It has been observed in hamster models that deleted S1/S2 furin cleavage site in the SARS-CoV-2 virus resulted in weakened infection. Thus, NRP-1 interaction with the CendR peptide in S1 could possibly be implicated in the higher pathogenicity of SARS-CoV-2 relative to SARS-CoV.
Since some COVID-19 subjects reported neurologic symptoms, the probability of SARS-CoV-2 penetrating the CNS similar to SARS-CoV was proposed [26]. Shortly after, evidence of SARS-CoV-2 invading the CNS emerged [27]. Human-related coronaviruses can trigger neurological effects such as seizures, visual disturbances, challenges in motor function, and various other neurological manifestations [28].
6. Inhibition of NRP-1 and S1 interaction
Intriguingly, it has been reported that NRP-1 mediates cellular uptake of other viruses, including the Epstein-Barr virus (EBV) and Human T-Cell Lymphotropic Virus Type 1 (HTLV-1) [29]. NRP-1 exhibits direct interaction with glycoprotein B of EBV, and its knockdown blocks EBV infection. In the case of HTLV-1, NRP-1 interacts with the CendR motif KPXR.
Therefore, the S protein–NRP-1 binding is a possible antiviral target, and the capacity to aim for this particular interaction may offer a pathway for COVID-19 treatment.
NRP-1 has long been a focus for drug design, given its involvement in cancer. For over two decades, exploratory attempts were aimed at the progress of NRP-1 antibody therapeutics, such as a ‘dual-specificity’ antibody against NRP-1 and the angiogenic cytokine vascular endothelial growth factor-A (VEGF-A) 164. VEGF-A 164 is a physiological ligand of NRP-1 and binds to the CendR binding pocket through its CendR motif [21]. Notable NRP-1 inhibitors, EG00229 and EG01377 carry an end Arg-like component and carboxyl group found to be critical in NRP-1 binding [30] similar to the novel EG01377-derived fluorescent molecule [31]. Other reported molecules carry arginine-based moieties and various chemotypes including bis-guanidines acylthioureas, aryl benzylethers, or benzamidosulfonamides. A virtual screen of approximately half a million molecules targeting the NRP-1 CendR region proposed 9 chemical strings from both natural and synthetic sources, carrying a lead- or drug-like physico-chemical features and offer a pharmacophore prototype for directing future molecular design [21]. Additionally, six compounds that disrupt VEGF-A interaction with a higher efficacy relative to the well-documented NRP-1 blocker EG00229, and three that block VEGF-A binding and the furin-processed S1 pocket were presented. The identified compounds engage NRP-1 in a SARS-CoV-2 S protein-like fashion, and could potentially disrupt the role of NRP-1 in SARS-CoV-2 viral infection. Interestingly, a couple of the compounds blocked SARS-CoV-2 viral action and may hold promise for further advances, though further investigation is required to elucidate their underlying mechanism. At a minimum, the identified compounds share a 2(1H)-pyridone core, and those with ‘pyridone and pyrimidone cores’ inhibited VEGF-A-NRP-1 binding with higher efficiency than EG00229; and as efficient as the SARS-CoV-2 S glycoprotein association with NRP-1. Interestingly, the introduction of sNRP-1 was also found to disrupt NRP-1-mediated viral infection [23]. Given the implication of numerous proteases, the suggested anti-viral application of protease blockers with the potential to suppress the proteolytic furin-processing of the S glycoprotein is to be expected [6].
7. S1-NRP-1 interaction and pain relief
The interaction of NRP-1 with the S1 CendR peptide bears a high level of semblance with its endogenous partner VEGF-A 164. The core residues of NRP-1 involved in the interaction with the C-terminal R685 of the CendR peptide - Tyr297, Trp301, Thr316, Asp320, Ser346, Thr349 and Tyr353 - are nearly identical in both complexes [12]. Jointly, the Arg682 and Arg685 side chains engage Tyr297 and Tyr353 side chains in NRP-1 through stacked cation-π interactions. Site-specific mutagenesis of the S1 Arg685 to aspartic acid drastically lowered GFP-S1493-685 immunoprecipitation, emphasizing the importance of the C-end arginine. Mutating Thr316 to arginine within the b1 domain of NRP-1 also decreased the interaction with GFP-S1493-685, in line with its suppressive effect on VEGF-A164 interaction.
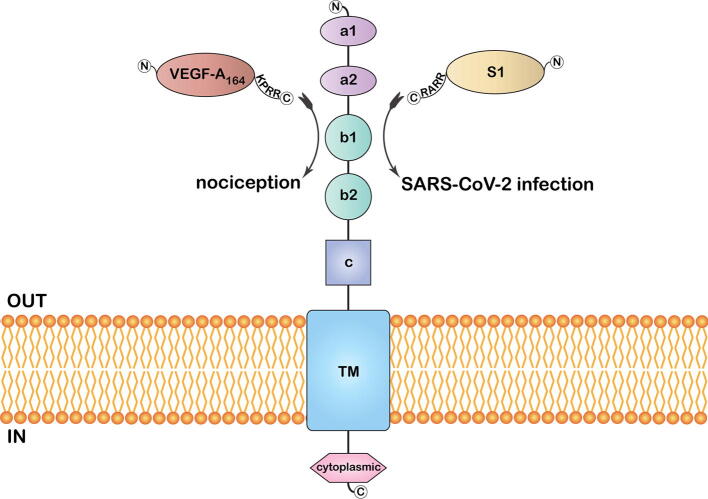
Given that both S protein and VEGF-A - a pro-nociceptive and angiogenesis agent - engage NRP-1 at a common binding domain, the potential of S protein to inhibit VEGF-A/NRP-1 signaling is worth examining (Fig. 3). Considering the higher VEGF-A levels observed in COVID-19 patients, pain-related complaints are expected [11]. Instead, findings hint that the S protein ceases NRP-1 signaling to alleviate VEGF-A driven pain. VEGF-A-induced neuronal activity could be inhibited by NRP-1 inhibitor EG00229. It was also found that S protein blocks VEGF-A/NRP-1 signaling, which otherwise promotes nociception. Such ‘silencing’ of pain perception, a timely warning of COVID-19 through blockage of VEGF-A/NRP-1 signaling may be behind the higher disease circulation in asymptomatic individuals, and such implicit transmission of the virus challenges the control of the outbreak. The findings do not rule out the possibility that other components of the S protein or alternative viral proteins may promote pro-nociception.
Fig. 3.
An illustration of the membrane-embedded full-length neuropilin-1 with its distinct domains. Binding of the S1 CendR peptide aids host invasion, while the binding of the VEGF-A 164 CendR peptide is associated with nociception.
VEGF-A isoforms also end in a polybasic sequence terminating with an arginine residue, conforming to the CendR rule. The Arg164 residue at the C-terminal of the 164 amino acid long VEGF-A isoform binds to the NRP-1 b1 domain binding site; the guanidine engages in a bidentate salt bridge with the conserved Asp320 of NRP-1 and the carboxylate group interacts with the conserved Ser346, Thr349, and Tyr353 residues through hydrogen bonding [11].
In spite of the structural relatedness between NRP-1 and NRP-2, they mediate different signalling pathways through their ligands; semaphorins and corresponding players of the VEGF family [32]. NRP-1 mainly facilitates VEGF-A-based angiogenesis while NRP-2 mostly mediates VEGF-C-based lymphangiogenesis. Similarly, despite high structural homology between NRP-1 and NRP-2 b1 domains, there are variations in certain regions, particularly in the residues of the electronegative L1 loop present exclusively in NRP-2. The Thr-299 within the L1 loop of NRP-1 is substituted by Asp-301 in the L1 loop of NRP-2. This substitution consequently presents an electrostatic repulsion between NRP-2 and the electronegative exon 7 residues of the VEGF-A heparin binding domain (HBD), specifically Glu154. This is not the case with NRP-1, where the strong interaction between NRP-1 and VEGF-A is ensured by Glu154 and results in strong and selective binding of VEGF-A to NRP-1. Therefore, heparin considerably strengthens the selectivity of VEGF-A for NRP-1, but not NRP-2. Exon 8 residues within the HBD determines strong ‘affinity’ interaction, while exon 7 residues mainly direct ‘selectivity’. Hence, though VEGF-A interacts with both NRP-1 and NRP-2, it exhibits a fifty-fold greater affinity for NRP-1.
8. Dissimilarities between NRP-1 and NRP-2 interactions
Molecular factors of neuropilin ligand specificity remain unclear. Site-specific mutagenesis studies looking at sequence variations in NRP-1 and NRP-2 binding pockets suggested that electrostatic repulsion mechanism explains the molecular underpinning for ligand specificity [33]. Additionally, it is likely that other determinants such as structural disparities in other regions of the proteins, entropic shifts owing to solvent or side chain reorganization present on the ligand binding regions, and energy of the other hydrogen bonds within the NRP-ligand complexes could explain the observed dissimilarities in the ligand partialities of the two neuropilins. Furthermore, other factors at the molecular level, such as ‘post-translational modifications’ of NRPs or VEGF ligands and glycosylation could contribute to ligand identification. Tsai et al. also reported a Zn2+ binding site on the conserved NRP-2 b1 domain that fits three zinc ions, present in proximity to the b1-b2 domain junction in the ectopic site. This ion binding site is distant from the VEGF binding site on the NRP-2 b1 domain and is improbable to impact VEGF ligand binding. Since this binding region is not found in NRP-1, it could possibly be implicated in regulating NRP-2 ligand binding. The affinity of NRP-2 for zinc implies that the zinc-binding pocket may be of regulatory significance, and not structural, and comes into play in the event of shifting zinc concentrations. NRP-2 heat resistance drops with the introduction of zinc. Heparin offers resistance from zinc-triggered protein instability. Previous studies documented that a string of heparin molecules promote dimerization of NRP-1 through binding to its b1-b2 domains.
Considering that the interaction of S1 CendR peptide with NRP-1 mimics that of its endogenous partner VEGF-A 164, the possible basis of differential binding of VEGF-A to NRP-1 and NRP-2 could be relevant to that of S1 to the two NRP isoforms.
9. Future perspectives
In the case of SARS-CoV-2, NRP-1 was found to be up-regulated in the epithelia of olfactory and respiratory cells where it heightens SARS-CoV-2 infection [1]. Though NRP-1 is considerably expressed in immune cells, in situ or in vivo attempts will be required to validate whether NRP-1 is implicated in SARS-CoV-2 invasion of immune cells [34]. NRP-1 was also shown to augment EBV uptake through the epithelia of nasopharyngeal cells, while NRP-2 was found to exhibit an opposing outcome in inhibiting EBV infection [29]. While the precise basis of the opposing effects of NRP-1 and NRP-2 in terms of EBV infection is subject to further study, the discrepancy could be credited to the various physiological ligands they engage, and the varying signalling cascades they mediate. NRP-1 also mediates human T-cell lymphotropic virus type 1 (HTVL-1) infection which enters host cells through engagement with the glucose transporter GLUT1.
Through binding with NRP-1, the S protein interferes with the docking of VEGF-A on its receptor, signalling that would otherwise promote nociception [11]. Therefore, NRP-1 constitutes a subject for the management of neuropathic pain. More so, NRP-1 offers a strategy to block viral access into cells to lower viral burden. The potential pain silencing aspect of S1 protein is also worth further investigation. Finally, the basis of residual S1 interaction with both NRP-1 and NRP-2 in the case of the ΔRRAR mutant needs to be elucidated as it alludes to a supporting CendR peptide-independent interaction between NRPs and the S1 glycoprotein.
Funding
This work received no funding.
CRediT authorship contribution statement
Amie Jobe: Writing - original draft, Writing - review & editing. Ranjit Vijayan: Conceptualization, Writing - original draft, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Davies J., Randeva H., Chatha K., Hall M., Spandidos D., Karteris E. Neuropilin–1 as a new potential SARS–CoV–2 infection mediator implicated in the neurologic features and central nervous system involvement of COVID–19. Mol Med Rep. 2020;22(5):4221–4226. doi: 10.3892/mmr.2020.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kielian M. Enhancing host cell infection by SARS-CoV-2. Science. 2020;370(6518):765–766. doi: 10.1126/science.abf0732. [DOI] [PubMed] [Google Scholar]
- 3.Tang Tiffany, Bidon Miya, Jaimes Javier A., Whittaker Gary R., Daniel Susan. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiv Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bittmann S., Weissenstein A., Moschüring-Alieva E., Villalon G., Luchter E. Neuropilin-1 in transmission process of COVID-19. J Regen Biol Med. 2020;2(4):1–2. [Google Scholar]
- 5.Wölfel Roman, Corman Victor M., Guggemos Wolfgang, Seilmaier Michael, Zange Sabine, Müller Marcel A. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 6.Papageorgiou A.C., Mohsin I. The SARS-CoV-2 spike glycoprotein as a drug and vaccine target: structural insights into its complexes with ACE2 and antibodies. Cells. 2020;9(11):2343. doi: 10.3390/cells9112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiv Res. 2020;176:104742. doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tse L.V., Hamilton A.M., Friling T., Whittaker G.R. A novel activation mechanism of avian influenza virus H9N2 by furin. J Virol. 2014;88(3):1673–1683. doi: 10.1128/JVI.02648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seyran M, Takayama K, Uversky VN, Lundstrom K, Palù G, Sherchan SP, Attrish D, Rezaei N, Aljabali AAA, Ghosh S, et al. 2020. The structural basis of accelerated host cell entry by SARS‐CoV‐2. FEBS J Dec 2. [DOI] [PMC free article] [PubMed]
- 11.Moutal A., Martin L.F., Boinon L., Gomez K., Ran D., Zhou Y. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain. 2021;162(1):243–252. doi: 10.1097/j.pain.0000000000002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H.-F., Vander Kooi C.W. Neuropilin functions as an essential cell surface receptor. J Biol Chem. 2015;290(49):29120–29126. doi: 10.1074/jbc.R115.687327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzog B., Pellet-Many C., Britton G., Hartzoulakis B., Zachary I.C., Heldin C.-H. VEGF binding to NRP1 is essential for VEGF stimulation of endothelial cell migration, complex formation between NRP1 and VEGFR2, and signaling via FAK Tyr407 phosphorylation. Mol Biol Cell. 2011;22(15):2766–2776. doi: 10.1091/mbc.E09-12-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plein Alice, Fantin Alessandro, Ruhrberg Christiana. Neuropilin regulation of angiogenesis, arteriogenesis, and vascular permeability. Microcirculation. 2014;21(4):315–323. doi: 10.1111/micc.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prud’homme Gérald J., Glinka Yelena. Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget. 2012;3(9):921–939. doi: 10.18632/oncotarget.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amraei R, Yin W, Napoleon MA, Suder EL, Berrigan J, Zhao Q, Olejnik J, Chandler KB, Xia C, Feldman J, et al. 2020. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. bioRxiv [accessed 2021 Jan 28]. https://doi.org/10.1101/2020.06.22.165803
- 18.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Ke, Chen Wei, Zhang Zheng, Deng Yongqiang, Lian Jian-Qi, Du Peng. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther. 2020;5(1) doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang Y.-L., Chou Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc Natl Acad Sci. 2020;117(34):20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perez-Miller S, Patek M, Moutal A, Cabel CR, Thorne CA, Campos SK, Khanna R. 2020. In silico identification and validation of inhibitors of the interaction between neuropilin receptor 1 and SARS-CoV-2 Spike protein. bioRxiv. [accessed 2021 Jan 28]. https://doi.org/10.1101/2020.09.22.308783
- 22.Wrapp Daniel, Wang Nianshuang, Corbett Kizzmekia S., Goldsmith Jory A., Hsieh Ching-Lin, Abiona Olubukola. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teesalu T., Sugahara K.N., Kotamraju V.R., Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci. 2009;106(38):16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison S.C. Viral membrane fusion. Virology. 2015;479-480:498–507. doi: 10.1016/j.virol.2015.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raaben M, Jae LT, Herbert AS, Kuehne AI, Stubbs SH, Chou Y, Blomen VA, Kirchhausen T, Dye JM, Brummelkamp TR, Whelan SP. 2017. NRP2 and CD63 Are host factors for lujo virus cell entry. Cell Host Microbe 22(5):688–96.e5. [DOI] [PMC free article] [PubMed]
- 26.Li Y.-C., Bai W.-Z., Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020;92(6):552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Xue Q., Xu X. Involvement of the nervous system in SARS-CoV-2 infection. Neurotox Res. 2020;38(1):1–7. doi: 10.1007/s12640-020-00219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arabi Y.M., Harthi A., Hussein J., Bouchama A., Johani S., Hajeer A.H. Severe neurologic syndrome associated with Middle East respiratory syndrome corona virus (MERS-CoV) Infection. 2015;43(4):495–501. doi: 10.1007/s15010-015-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Hong-Bo, Zhang Hua, Zhang Jing-Ping, Li Yan, Zhao Bo, Feng Guo-Kai. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat Commun. 2015;6(1) doi: 10.1038/ncomms7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarvis A., Allerston C.K., Jia H., Herzog B., Garza-Garcia A., Winfield N. Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction. J Med Chem. 2010;53(5):2215–2226. doi: 10.1021/jm901755g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conole D., Chou Y.-T., Patsiarika A., Nwabo V., Dimitriou E., Soudy C. Discovery of a novel fluorescent chemical probe suitable for evaluation of neuropilin-1 binding of small molecules. Drug Dev Res. 2020;81(4):491–500. doi: 10.1002/ddr.21641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker M.W., Xu P., Li X., Vander Kooi C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J Biol Chem. 2012;287(14):11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai Y.-C.I., Fotinou C., Rana R., Yelland T., Frankel P., Zachary I. Structural studies of neuropilin-2 reveal a zinc ion binding site remote from the vascular endothelial growth factor binding pocket. FEBS J. 2016;283(10):1921–1934. doi: 10.1111/febs.13711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar A., Narayan R., Kumar S., Kumari C., Pareek V., Prasoon P. Expression of SARS-CoV-2 host cell entry factors in immune system components of healthy individuals and its relevance for COVID-19 immunopathology. Authorea. 2020 doi: 10.22541/au.159656128.87988725. [DOI] [PubMed] [Google Scholar]