Abstract
Background
IFITM3 is a viral restriction protein that enables sequestration of viral particles and subsequent trafficking to lysosomes. Recently, IFITM3 upregulation was found to induce gamma – secretase activity and the production of amyloid beta. The purpose of this study was to determine whether dysregulation of IFITM3-dependent pathways was present in neurons and peripheral immune cells donated by AD patients. As a secondary aim, we sought to determine whether these perturbations could be induced by viruses, including SARS-CoV-2.
Methods
Gene set enrichment analyses (GSEA) previously performed on publicly available transcriptomic data from tissues donated by AD patients were screened for enriched pathways containing IFITM3. Subsequently, signature containing IFITM3, derived from entorhinal cortex (EC) neurons containing neurofibrillary tangles (NFT) was screened for overlap with curated, publicly available, viral infection-induced gene signatures (including SARS-CoV-2).
Results
GSEA determined that IFITM3 gene networks are significantly enriched both in CNS sites (entorhinal and hippocampal cortices) and in peripheral blood mononuclear cells (PBMCs) donated by AD patients. Overlap screening revealed that IFITM3 signatures are induced by several viruses, including SARS-CoV, MERS-CoV, SARS-CoV-2 and HIV-1 (adjusted p-value <0.001; Enrichr Database).
Discussion
A data-driven analysis of AD tissues revealed IFITM3 gene signatures both in the CNS and in peripheral immune cells. GSEA revealed that an IFITM3 derived gene signature extracted from EC/NFT neurons overlapped with those extracted from publicly available viral infection datasets, including SARS-CoV-2. Our results are in line with currently emerging evidence on IFITM3’s role in AD, and SARS-CoV-2’s potential contribution in the setting of an expanded antimicrobial protection hypothesis.
Keywords: Alzheimer’s disease, IFITM3, Systems biology, Neuroimmunology, Epigenetics
Highlights
-
•
IFITM3 is a protein that sequesters viral particles upon cellular entry, redirecting them to the lysosome.
-
•
IFITM3 upregulation was recently found to promote beta amyloid production.
-
•
The study’s aim was to determine whether IFITM3 dysregulation in AD patients could be attributed to viral pathogens.
-
•
IFITM gene signatures were significantly enriched as perturbed pathways in the CNS and peripheral blood of AD patients.
1. Background
The antimicrobial protection hypothesis of Alzheimer’s disease (AD) describes a model of amyloidogenesis resulting from chronic activation of the innate immune cascades; this activation is not attributed to any single pathogen per se, but its progression to chronicity is considered the driver of beta-amyloid accumulation and its subsequent deposition (Moir et al., 2018).
Among potential crossroads between amyloidogenesis and immunity has been recently been found in interferon-induced transmembrane protein 3 (IFITM3) expression (Hur et al., 2020); Upregulation of IFITM3, a viral particle sequestration protein, was found to be a characteristic of neurons and astrocytes displaying higher γ-secretase, and consequently, increased amyloid beta (Aβ) generation (Hur et al., 2020).
In a recent study from our group, where we examined a data-driven, in silico model of tissue interaction in AD, several infection- and immune-related gene networks were found to be overlapping between the central nervous system and the peripheral immune system (Vavougios et al., 2020). We hypothesize that (a) IFITM3 is differentially expressed in both the CNS and the peripheral immune system in AD and that (b) viral infections, including SARS-CoV-2 can induce IFITM3’s expression.
2. Materials and methods
In a previous study, we had identified immune and infection-related pathways overlapping between the peripheral immune system and the CNS in a data driven manner (Vavougios et al., 2020). For the construction of our model, we had used gene expression data from peripheral blood mononuclear cells (PBMC), entorhinal cortex neurons containing neurofibrillary tangles (EC-NFT), and hippocampal cortex neurons, available from the National Institutes of Health (NIH) public repository, Gene Expression Omnibus (GEO). An overview of the datasets included in our analysis is presented on supplementary Table 1. Detailed descriptions of each study are found elsewhere (Blalock et al., 2004, Maes et al., 2007)(Dunckley et al., 2006).
For this study, we scrutinized these previously generated data for pathways containing IFITM3. Subsequently, we used an EC-NFT gene signature containing IFITM3 in order to determine whether IFITM3’s expression could be upregulated via viral infections, including SARS-CoV-2. For this purpose, we performed gene set enrichment analysis via the Enrichr web platform (Kuleshov et al., 2016), screening the “COVID-19 related gene sets”, “Virus Perturbations Up” modules Additionally, the DisGeNET (Piñero et al., 2017) and KEGG (Kanehisa et al., 2017) pathways subsets were also scrutinized, in order to determine significant enrichments with other disease and specific pathways, respectively.
3. Results
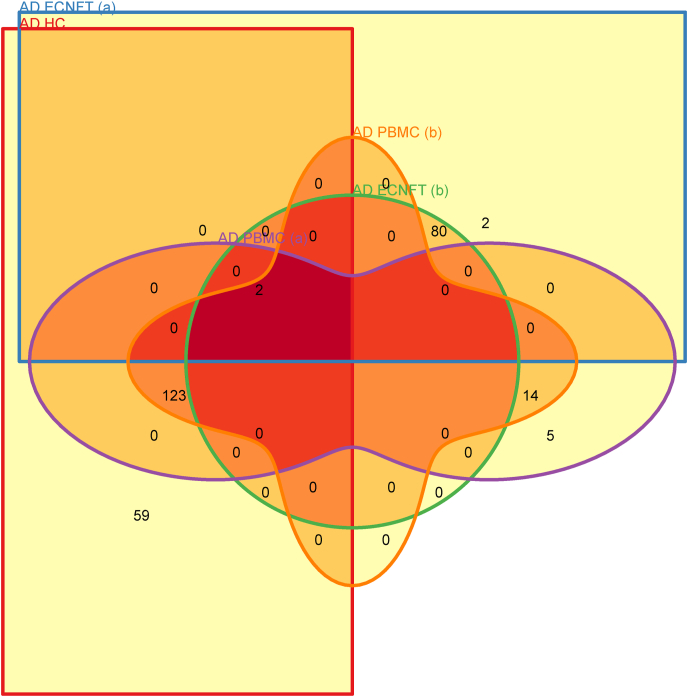
In our original data, IFITM3 which was part of several significantly enriched gene ontologies (GOs) in the PBMC dataset, the entorhinal cortex dataset and a hippocampal cortex dataset (Benjamini – Hochberg adjusted p-value <0.05; Table 1). These pathways were associated with interferon signaling and viral gene expression (Table 1); Several genes comprising these pathways overlapped between different tissues donated by AD patients (PBMC, HC, EC; See Venn Diagram (Khan and Mathelier, 2017), Fig. 1), as per the tissue interaction concept introduced by our study (Vavougios et al., 2020). In order to determine whether IFITM3 dysregulation could arise secondary to viral infection, we used an AD NFT-EC gene signature to determine whether it overlaps with upregulated genes from viral perturbation experiments in Gene Expression Omnibus (GEO), available via the Enrichr web service (under the GEO Virus Perturbations UP module) (Kuleshov et al., 2016). Enrichment analyses revealed that several predominantly respiratory pathogens, including SARS-CoV-2, could significantly upregulate IFITM3 (Table 2 – 5; See also Supplementary Materials 1). Aside from viral infections, DisGeNET enrichments included autoimmune diseases and neoplasms, with a notable enrichment of multiple sclerosis (Table 4 and Supplementary Materials 1) (see Fig. 2) (see Table 5).
Table 1.
Gene signatures containing IFITM3 in the Vavougios et al. study, 2020.
| Gene Ontology and corresponding dataset |
Reference |
Hits |
Adjusted P-value |
|---|---|---|---|
| AD EC – NFT (GDS2795) | |||
| response_to_type_I_interferon(5) | http://amigo.geneontology.org/amigo/term/GO:0034340 | 84 | 0.00200343 |
| cellular_response_to_type_I_interferon(6) | http://amigo.geneontology.org/amigo/term/GO:0071357 | 82 | 0.00229691 |
| type_I_interferon_signaling_pathway(7) | http://amigo.geneontology.org/amigo/term/GO:0060337 | 82 | 0.00229691 |
| AD PMBC (GDS2601) | |||
| multi_organism_metabolic_process(3) | http://amigo.geneontology.org/amigo/term/GO:0044033 | 144 | 0.000712085 |
| viral_gene_expression(4) | http://amigo.geneontology.org/amigo/term/GO:0019080 | 139 | 0.000712085 |
| viral_transcription(5) | http://amigo.geneontology.org/amigo/term/GO:0019083 | 130 | 0.0110638 |
| AD HC (GDS810) | |||
| viral_gene_expression(4) | http://amigo.geneontology.org/amigo/term/GO:0019080 | 184 | 0.0131733 |
GDSXXX or GDSXXXX format represents the Gene Expression Omnibus (GEO) dataset identifier used in these analyses. “Hits” refers to the number of genes comprising each signature.AD: Alzheimer’s Disease; PBMC: Peripheral Blood Mononuclear Cells; NEC: Normal Elderly Controls; EC: Entorhinal Cortex; NFT: Neurofibrillary tangles; FDR: False Discovery Rate; GO: Gene Ontology.
Fig. 1.
Venn diagram displaying common genes between signatures. Each distinct colored shape corresponds to a gene signature, with the numbers in their intersection representing the number of overlapping genes in each signature. AD NFT – EC (a): response_to_type_I_interferon(5); AD NFT – EC (b): cellular_response_to_type_I_interferon(6); AD NFT – EC (c): type_I_interferon_signaling_pathway(7); AD PBMC (a): multi_organism_metabolic_process(3); AD PBMC (b): viral_gene_expression(4) AD PBMC (c): viral_transcription(5); AD HC: viral_gene_expression(4). AD: Alzheimer’s Disease; PBMC: Peripheral Blood Mononuclear Cells; NEC: Normal Elderly Controls; EC: Entorhinal Cortex; NFT: Neurofibrillary tangles; FDR: False Discovery Rate; GO: Gene Ontology.
Table 2.
Gene set enrichment analysis, scrutinizing the “COVID-19 related gene sets” subset of the Enrichr database.
| Index | Name | P-value | Adjusted p-value |
|---|---|---|---|
| 1 | MERS-CoV Top 200 Predicted Genes from Geneshot GeneRIF via AutoRIF Co-Occurrence Gene Similarity | 6.993e-67 | 1.434e-64 |
| 2 | SARS-CoV perturbation Up Genes bronchial epithelial 2B4 from GSE17400:GPL570:6 | 3.746e-56 | 3.840e-54 |
| 3 | Up-regulated by SARS-CoV-2 in Calu-3 24hr from GSE148729 | 1.243e-46 | 8.495e-45 |
| 4 | Up-regulated by SARS-CoV-1 in Calu-3 from GSE148729 | 8.228e-42 | 4.217e-40 |
| 5 | SARS perturbation Up Genes airway epithelium (HAE) from GSE47961:GPL6480:4 | 5.631e-38 | 2.309e-36 |
| 6 | SARS perturbation Up Genes airway epithelium (HAE) from GSE47961:GPL6480:3 | 1.290e-37 | 4.407e-36 |
| 7 | Up-regulated by SARS-CoV-2 2 MOI in Calu-3 from GSE147507 | 4.427e-37 | 1.134e-35 |
| 8 | Up-regulated by SARS-COV-2 infection of Calu3 cells | 4.427e-37 | 1.296e-35 |
| 9 | SARS-CoV perturbation Up Genes bronchial epithelial 2B4 from GSE17400:GPL570:3 | 6.619e-37 | 1.508e-35 |
| 10 | SARS perturbation Up Genes airway epithelium (HAE) from GSE47961:GPL6480:6 | 1.825e-36 | 3.740e-35 |
Results have been truncated to the first 10 entries. For this analysis, the AD Entorhinal cortex gene signature designated as “response_to_type_I_interferon(5)” was supplied to Enrichr.
Table 4.
Gene set enrichment analysis, scrutinizing the “DisGeNET” subset of the Enrichr database.
| Name | P-value | Adjusted p-value | |
|---|---|---|---|
| 1 | Virus Diseases | 6.209e-39 | 1.421e-35 |
| 2 | Influenza | 5.146e-33 | 5.890e-30 |
| 3 | Hepatitis C | 6.045e-30 | 4.612e-27 |
| 4 | Infection | 3.631e-23 | 2.078e-20 |
| 5 | West Nile viral infection | 5.699e-22 | 2.609e-19 |
| 6 | Autoimmune Diseases | 8.919e-21 | 3.403e-18 |
| 7 | Hepatitis C, Chronic | 1.657e-20 | 5.418e-18 |
| 8 | Lupus Erythematosus, Systemic | 3.981e-20 | 1.139e-17 |
| 9 | Vesicular Stomatitis | 2.520e-19 | 6.408e-17 |
| 10 | Multiple Sclerosis | 3.907e-18 | 8.943e-16 |
Results have been truncated to the first 10 entries. For this analysis, the AD Entorhinal cortex gene signature designated as “response_to_type_I_interferon(5)” was supplied to Enrichr.
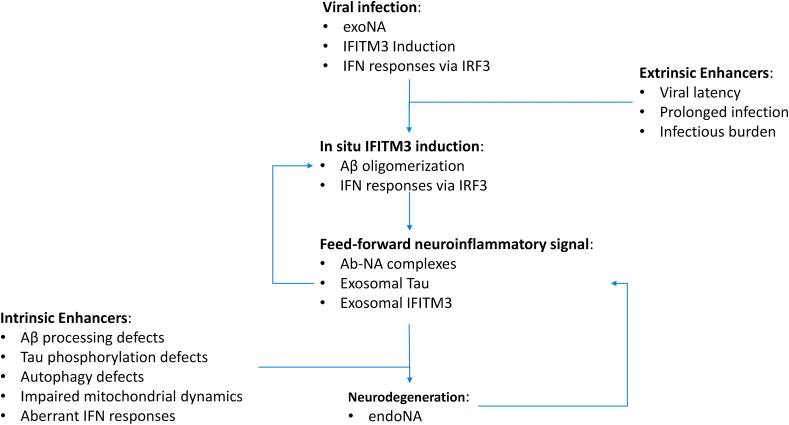
Fig. 2.
Schematic representation of the proposed IFITM3 feed forward mechanism in AD pathogenesis. Within this concept, viral infection initially upregulates IFITM3 (a) directly, via physical interactions between IFITM3 and viral particles (b) via promoting IRF3-mediated IFN cascades. IFITM3 in turn modulates γ-secretase towards increased production of Αβ oligomers, while other interactors such as FYN, which are also implicated in antiviral responses, promote Tau hyperphosporylation and aggregation. IFITM3 and Tau containing exosomes, in conjunction with Αβ oligomers would then propagate a neuroinflammatory signal from the primary infection site to other distal, non-infected sites via transsynaptic uptake. Aβ and nucleic acid (NA) complexes (Αβ-ΝΑ) would serve to stimulate microglia and trigger further inflammation. Both viral (exoNA) and endogenous (endoNA) nucleic acids could contribute to this process, either as a result of the viral lifecycle or as apoptotic debris. Prolonged infections, lifelong latency (i.e. HSV-1) or accumulating infectious burden on the primary infection site (i.e. the entorhinal cortex) could provide feed-forward neuroinflammatory stimulus even in the absence of an active pathogen. In a similar manner, intrinsic defects (i.e. pathogenic variants of the amyloid processing machinery, autophagic cascades and so forth) in either the neuroinflammatory induction and response aspect, autophagy or cellular bioenergetics would serve as enhancers, reinforcing the noxious biological effects of e.g. IFITM3 uptake by non-infected neurons.
Table 5.
Gene set enrichment analysis, scrutinizing the “KEGG Pathways 2019” subset of the Enrichr database.
| Index | Name | P-value | Adjusted p-value |
|---|---|---|---|
| 1 | Hepatitis C | 1.047e-48 | 8.457e-47 |
| 2 | Epstein-Barr virus infection | 1.859e-48 | 8.457e-47 |
| 3 | Influenza A | 3.364e-45 | 1.020e-43 |
| 4 | Measles | 2.016e-44 | 4.587e-43 |
| 5 | NOD-like receptor signaling pathway | 1.105e-42 | 2.011e-41 |
| 6 | Herpes simplex virus 1 infection | 1.925e-40 | 2.919e-39 |
| 7 | Kaposi sarcoma-associated herpesvirus infection | 3.494e-40 | 4.543e-39 |
| 8 | Human papillomavirus infection | 7.803e-39 | 8.876e-38 |
| 9 | JAK-STAT signaling pathway | 7.715e-33 | 7.801e-32 |
| 10 | Cytosolic DNA-sensing pathway | 4.126e-31 | 3.755e-30 |
Results have been truncated to the first 10 entries. For this analysis, the AD Entorhinal cortex gene signature designated as “response_to_type_I_interferon(5)” was supplied to Enrichr.
4. Discussion
SARS-CoV-2 impact on the CNS is increasingly recognized in the literature, and its potential specific relationship with AD is the subject of ongoing studies (Lempriere, 2020). Within the conceptual framework of the antimicrobial protection hypothesis, SARS-CoV-2’s interaction with IFITM3 is even more relevant.
4.1. A proposed IFITM3-driven feed-forward loop in AD pathogenesis
The IFITM family of proteins have been previously recognized as important regulators of MERS-CoV-2 and SARS-CoV-2 virulence, either by physiological restriction of their entry, or as infection enhancers, following loss-of-function mutations (Zhao et al., 2018). IFITM3 was found to be upregulated in SARS-CoV-2 -infected cells (Shi et al., 2020; Sardar et al., 2020); furthermore, specific polymorphisms, such as the rs12252-C have been identified as severity markers for COVID-19 (Zhang et al., 2020). A current concept for SARS-CoV-2’s immunoevasion of IFITM3 sequestration involves cleavage by TMPRSS2 (Zheng et al., 2020). Recently, yet another such host factor was identified in NRP1 (Cantuti-Castelvetri et al., 2020), indicating another CNS-specific interaction pathway. Interestingly, both IFITM3 and NRP1 have been identified as neuroinflammation-induced in a cellular model of AD (Correani et al., 2017).
As a viral restriction protein, endocytic vesicle IFITM3 tags viral particles for lysosomal fusion (Spence et al., 2019). Viral particle restriction via IFITM3 trafficking is a second line of defense active against a variety of pathogens including HIV-1, Dengue, Ebola, Influenza A (IAV) and CMV, among others (Wellington et al., 2019).
Several components of antiviral immunity have recently been recognized as per their potential contribution in the pathogenesis of AD and neuroinflammation. Type I interferon (IFNA) responses, while pivotal in restricting viruses from propagating within the CNS (Roy et al., 2020), display dose-dependent neurotoxicity via both IFNAR- and NMDAR-initiated cascades (Kessing and Tyor, 2015). This paradigm has been studied in the HIV-associated neurocognitive disorder (HAND) spectrum, where cognitive impairment is considered to progress via accumulating exposure of the CNS to IFNAR-signaling (Thaney and Kaul, 2019).
In the setting of AD, IFN responses were recently shown to be elicited by Aβ-Νucleic Acid (NA) complexes, with IFITM3 among the IFN-gene signatures detected (Roy et al., 2020). The immunogenicity and of these Aβ-ΝΑ complexes and the resulting microglial activation appears to be independent of the source of NA (i.e. whether or not they are xenobiotic). This concept, along with its close connection with IFN responses is another finding of both our current analyses (Cytosolic DNA-sensing pathway, adj. p-value = 3.755e−30; Table 3) and our previous study (Vavougios et al., 2020).
Table 3.
Gene set enrichment analysis, scrutinizing the “GEO Virus Perturbations UP” subset of the Enrichr database.
| Index | Name | P-value | Adjusted p-value |
|---|---|---|---|
| 1 | A-CA-04-2009(H1N1) 12Hour GSE47960 | 1.345e-38 | 4.345e-36 |
| 2 | SARS-BatSRBD 72Hour GSE47960 | 6.268e-37 | 1.012e-34 |
| 3 | SARS-CoV 96Hour GSE47961 | 1.719e-30 | 1.388e-28 |
| 4 | A-CA-04-2009(H1N1) 6Hour GSE47961 | 1.719e-30 | 1.851e-28 |
| 5 | SARS-ddORF6 72Hour GSE47961 | 6.086e-29 | 2.457e-27 |
| 6 | RSV 48Hour GSE32138 | 6.086e-29 | 2.808e-27 |
| 7 | A-CA-04-2009(H1N1) 18Hour GSE37571 | 6.086e-29 | 3.276e-27 |
| 8 | A-CA-04-2009(H1N1) 12Hour GSE47961 | 6.086e-29 | 3.932e-27 |
| 9 | hMPV 24Hour GSE8961 | 2.035e-27 | 6.573e-26 |
| 10 | SARS-dORF6 72Hour GSE47960 | 2.035e-27 | 7.304e-26 |
Results have been truncated to the first 10 entries. For this analysis, the AD Entorhinal cortex gene signature designated as “response_to_type_I_interferon(5)” was supplied to Enrichr.
In this setting, Αβ-ΝΑ complexes may act as the immunogenic substrate of neuroinflammation propagating across the CNS in AD, a hypothesis supported by a correlation between IFN signaling upregulation (including IFITM3) and Braak staging in the study of Roy et al. (2020). This observation would further outline a positive feedback loop, where Aβ-ΝΑ induces IFITM3 (Roy et al., 2020), and IFITM3 furthermore induces Aβ production2∗. Notably, this model can readily account for the increased AD risk conferred by HSV-1 neuroinfection (Cairns et al., 2020), and may be further incorporate lysosomal/autophagosomal dysfunction noted in AD. Specifically, HSV-1 induced IFITM3 upregulation (Nicholl et al., 2000) in the brain during neuroinfection (Cairns et al., 2020), would not lead to viral clearance in the case of perturbed IFITM3, i.e. unsuccessful lysosomal fusion (Orr and Oddo, 2013). In this sense, the discovery of HSV-1 genomes entangled in amyloid-β plaques in the CNS (Bearer, 2012) would reflect the debris of a defunct IFITM3 shuttling system upregulated by the positive feedback of unresolved viral infection, and acting as an Aβ-NA immunogenic template, inducing neuroinflammation.
4.2. A hypothesis on the potential consequences of SARS-CoV-2-mediated induction of IFITM3 signaling in the CNS
SARS-CoV-2’s modus operandi as a neuroinvasive pathogen provides further insight as to how viruses may trigger or sustain the proposed IFITM3 feedback loop. Even indirectly, SARS-CoV-2 may trigger IFITM3 by eliciting delayed and IFN responses (Meinhardt et al., 2021). In the scenario of transcribial neuroinvasion (Lei et al., 2020), SARS-CoV-2 would gain access to the entorhinal cortex. SARS-CoV-2 neuroinvasion has been shown to induce neurodegeneration in 3D brain organoids, pathologically associated with altered subcellular localization of hyperphosphorylated tau (Ramani et al., 2020). Interestingly, SARS-CoV-2 cellular entry and host-virus interactions further extent from IFITM3 to regulators of tau aggregation and phosphorylation, such as FYN (Briner et al., 2020; Glebov, 2020).
Direct and indirect induction of IFITM3 by neuroinvasion and IFN responses correspondingly, further contributed by Aβ oligomerization and Aβ-ΝΑ (both viral and i.e. fragments from neuronal apoptosis or genotoxic stress) would finally serve to evolve an in situ tauopathy to a distal Aβ-synaptopathy. Up to this point, our proposed model reiterates previously described mechanisms of HSV-1 modus operandi in β-amyloidogenesis (Bearer, 2012), with several important differences however.
On its own right, IFITM3 induction is not restricted to intracellular cascades. IFITM3 has been previously shown to be a mutant-tau exclusive cargo in AD exosomes (Podvin et al., 2020), whereas IFITM3-loaded exosome trafficking from infected to non-infected cells has been shown to confer resistance to DENV infection (Zhu et al., 2015). On these premises, IFITM3, exosomal hyperphosphorylated tau (Wang et al., 2017) and Αβ-ΝΑ may constitute a feed-forward signal expanding from a primary site of neuroinfection (i.e. the entorhinal cortex) via afferent projections (i.e. the hippocampi). Interestingly, while the primary site would be infected, the signal itself would propagate AD pathology even in the absence of infection: i. directly via Aβ oligomers, Aβ-ΝΑ and Tau and ii. indirectly, via IFITM3 uptake and the subsequent boosting in IFN and Aβ-oligomer production. The latter hypothesis could account for previously observed correlations between IFITM3 expression and Braak staging (Roy et al., 2020). In this setting, both the reactivation of a latent virus or de novo infection would reinforce the IFITM3 feed-forward loop.
4.3. Limitations and strengths
The proposed model of infection-induced, perturbed IFITM3 signaling in the pathogenesis of AD, as well as its specific iteration with SARS-CoV-2, should be interpreted within their respective limitations. Viral infection could have been a perturbator to any number of samples or studies included in our analyses. While the original studies report strict and stem from established brain banks (Supplementary Table 1), the possibility cannot be excluded and should be taken into consideration as a potential confounder on the results we report herein. Studying a significantly enriched signature that overlaps between different tissues (and by extent, studies) furthermore decreases but does not nullify this possibility. In a similar manner, as there is no detailed infection history on any of the original studies’ participants, we cannot account for its impact on our findings.
Another important limitation of our study is that our results indicate overlapping mechanisms, rather than a mechanistic effect. In this sense, we detect pathways and gene signatures that are perturbed in a similar manner in both AD and viral infections; we do not however determine a causal relationship directly. While a possible explanation for this overlap in AD would be neuroinvasion, our study by design does not provide mechanistic evidence of such. Furthermore, The overlapping IFITM3 signatures we report on are significantly enriched both in neuropathologically determined AD and viral infections, including SARS-CoV-2. Using IFITM3 signatures from AD EC-NFT, we reconstruct multiple viral infection-induced signatures containing IFITM3, indicating both candidate targets and the potential pathways by which said infections may either induce or contribute to AD pathobiology. Notably, while IFITM3 was focused on due to its discovery as a γ-secretase modulator, our approach identifies IFN responses, viral latency and innate immunity as common mechanisms enriched by IFITM3 networks.
5. Concluding remarks
In the case of SARS-CoV-2, independent studies have shown that the novel coronavirus is capable of transcribial neuroinvasion and gain access to the entorhinal cortex (Meinhardt et al., 2021), is associated with AD-like dyscognitive phenotypes (Woo et al., 2020) correlated with hippocampal atrophy (Lu et al., 2020). Furthermore, SARS-CoV-2 both induces and evades IFN responses during its latency (Lei et al., 2020), indicating another plausible explanation for the transcriptomic overlap we outline. The latter finding may account for the noted deterioration of AD symptoms in patients during infection (Takeda et al., 2014), as well as post-infection risk of dementia(Muzambi et al., 2020). This hypothesis however, would require a singular experiment where all these independent observations, as well as our own findings, are replicated.
The interaction concept introduced in our study (Vavougios et al., 2020) with the global (PMBCs, Hippocampi, Entorhinal Cortex) presence of IFITM3 networks furthermore supports an “outside – in” trigger for neurodegeneration. Finally, taking into account that multiple viruses could upregulate IFITM3 based on overlap between our recent work (Vavougios et al., 2020), Hur et al.‘s study (Hur et al., 2020) and GEO experimental data, the putative viral trigger for AD may not need to be any single viral pathogen, but rather a process of viral infection induced, positive/feed-forward feedback of the IFITM3 trafficking system. Future experiments should explore a mechanistic model of IFITM3’s perturbations introduced by viral infections, and their effects on β-amyloidogenesis and tau oligomerization.
Declaration of competing interest
None declared.
Acknowledgements
To Dr Athanasia Kefala, for her assistance with mathematical modeling and Edwards’ Venn diagrams.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2021.100243.
Author contributions
GDV conceived the study, performed the original analyses and wrote the first draft. GS oversaw the analyses and provided data quality assessments. CN, HSP and TD contributed with interpretations of the results in the clinical concept. KIG, SGZ contributed with interpretations infection-related pathways. TD, HSP, KIG, GS and SGZ contributed equally in the paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Bearer E.L. HSV, axonal transport and Alzheimer’s disease: in vitro and in vivo evidence for causal relationships. Future Virol. 2012;7:885–899. doi: 10.2217/fvl.12.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blalock E.M., Geddes J.W., Chen K.C., Porter N.M., Markesbery W.R., Landfield P.W. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc. Natl. Acad. Sci. U. S. A. 2004;101(7):2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briner A., Götz J., Polanco J.C. Fyn kinase Controls tau aggregation in vivo. Cell Rep. 2020;32(7):108045. doi: 10.1016/j.celrep.2020.108045. [DOI] [PubMed] [Google Scholar]
- Cairns D.M., Rouleau N., Parker R.N., Walsh K.G., Gehrke L., Kaplan D.L. A 3D human brain–like tissue model of herpes-induced Alzheimer’s disease. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aay8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020 doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correani V., Di Francesco L., Mignogna G. Plasma membrane protein profiling in beta-amyloid-treated microglia cell line. Proteomics. 2017;17:1600439. doi: 10.1002/pmic.201600439. [DOI] [PubMed] [Google Scholar]
- Dunckley T., Beach T.G., Ramsey K.E. Gene expression correlates of neurofibrillary tangles in Alzheimer’s disease. Neurobiol. Aging. 2006;27(10):1359–1371. doi: 10.1016/j.neurobiolaging.2005.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020;287(17):3664–3671. doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur J.-Y., Frost G.R., Wu X. The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer’s disease. Nature. 2020 doi: 10.1038/s41586-020-2681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessing C.F., Tyor W.R. Interferon-alpha induces neurotoxicity through activation of the type I receptor and the GluN2A subunit of the NMDA receptor. J. Interferon Cytokine Res. 2015;35(4):317–324. doi: 10.1089/jir.2014.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinf. 2017;18:287. doi: 10.1186/s12859-017-1708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11(1):3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempriere S. SARS-CoV-2 and the brain to be studied long-term. Nat. Rev. Neurol. 2020;16:522. doi: 10.1038/s41582-020-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Li X., Geng D., Mei N., Wu P.Y., Huang C.C. Cerebral micro-structural changes in COVID-19 patients - an MRI-based 3-month follow-up study. EClin. Med. 2020;25:100484. doi: 10.1016/j.eclinm.2020.100484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes O.C., Xu S., Yu B., Chertkow H.M., Wang E., Schipper H.M. Transcriptional profiling of Alzheimer blood mononuclear cells by microarray. Neurobiol. Aging. 2007;28(12):1795–1809. doi: 10.1016/j.neurobiolaging.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Meinhardt J., Radke J., Dittmayer C., Franz J., Thomas C., Mothes R. Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 2021;24(2):168–175. doi: 10.1038/s41593-020-00758-5. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Lathe R., Tanzi R.E. The antimicrobial protection hypothesis of Alzheimer’s disease. Alzheimers Dement. 2018;14:1602–1614. doi: 10.1016/j.jalz.2018.06.3040. [DOI] [PubMed] [Google Scholar]
- Muzambi R., Bhaskaran K., Brayne C., Davidson J.A., Smeeth L., Warren-Gash C. Common bacterial infections and risk of dementia or cognitive decline: a systematic review. J Alzheimers Dis. 2020;76(4):1609–1626. doi: 10.3233/JAD-200303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholl M.J., Robinson L.H., Preston C.M. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 2000;81:2215–2218. doi: 10.1099/0022-1317-81-9-2215. [DOI] [PubMed] [Google Scholar]
- Orr M.E., Oddo S. Autophagic/lysosomal dysfunction in Alzheimer’s disease. Alzheimer’s Res. Ther. 2013;5:53. doi: 10.1186/alzrt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñero J., À Bravo, Queralt-Rosinach N. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017;45(D1):D833–D839. doi: 10.1093/nar/gkw943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podvin S., Jones A., Liu Q., Aulston B., Ransom L., Ames J. Dysregulation of exosome cargo by mutant tau expressed in human-induced pluripotent stem cell (iPSC) neurons revealed by proteomics analyses. Mol. Cell. Proteomics. 2020;19(6):1017–1034. doi: 10.1074/mcp.RA120.002079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani A., Muller L., Ostermann P.N., Gabriel E., Abida-Islam P., Muller-Schiffmann A. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020;39(20) doi: 10.15252/embj.2020106230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy E.R., Wang B., Wan Y.W., Chiu G., Cole A., Yin Z. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J. Clin. Invest. 2020;130(4):1912–1930. doi: 10.1172/JCI133737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardar R., Satish D., Birla S., Gupta D. Integrative analyses of SARS-CoV-2 genomes from different geographical locations reveal unique features potentially consequential to host-virus interaction, pathogenesis and clues for novel therapies. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e04658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., Kenney A.D., Kudryashova E. Opposing activities of IFITM proteins in SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.08.11.246678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.S., He R., Hoffmann H.-H. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019;15:259–268. doi: 10.1038/s41589-018-0213-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Sato N., Morishita R. Systemic inflammation, blood-brain barrier vulnerability and cognitive/non-cognitive symptoms in Alzheimer disease: relevance to pathogenesis and therapy. Front. Aging Neurosci. 2014;6:171. doi: 10.3389/fnagi.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaney V.E., Kaul M. Type I interferons in NeuroHIV. Viral Immunol. 2019;32(1):7–14. doi: 10.1089/vim.2018.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavougios G.D., Nday C., Pelidou S.H. Double hit viral parasitism, polymicrobial CNS residency and perturbed proteostasis in Alzheimer’s disease: a data driven, in silico analysis of gene expression data. Mol. Immunol. 2020;127:124–135. doi: 10.1016/j.molimm.2020.08.021. [DOI] [PubMed] [Google Scholar]
- Wang Y., Balaji V., Kaniyappan S., Krüger L., Irsen S., Tepper K. The release and trans-synaptic transmission of Tau via exosomes. Mol. Neurodegener. 2017;12(1):5. doi: 10.1186/s13024-016-0143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington D., Laurenson-Schafer H., Abdel-Haq A., Dong T. IFITM3: how genetics influence influenza infection demographically. Biomed. J. 2019;42:19–26. doi: 10.1016/j.bj.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo M.S., Malsy J., Pottgen J., Seddiq Zai S., Ufer F., Hadjilaou A. Frequent neurocognitive deficits after recovery from mild COVID-19. Brain Commun. 2020;2(2) doi: 10.1093/braincomms/fcaa205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qin L., Zhao Y. Interferon-induced transmembrane protein 3 genetic variant rs12252-C associated with disease severity in coronavirus disease 2019. J. Infect. Dis. 2020;222:34–37. doi: 10.1093/infdis/jiaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Sehgal M., Hou Z. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J. Virol. 2018;92 doi: 10.1128/JVI.01535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Zhao X., Zheng S. Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion. Emerg. Microb. Infect. 2020;9:1567–1579. doi: 10.1080/22221751.2020.1787797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., He Z., Yuan J., Wen W., Huang X., Hu Y. IFITM3-containing exosome as a novel mediator for anti-viral response in dengue virus infection. Cell Microbiol. 2015;17(1):105–118. doi: 10.1111/cmi.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.