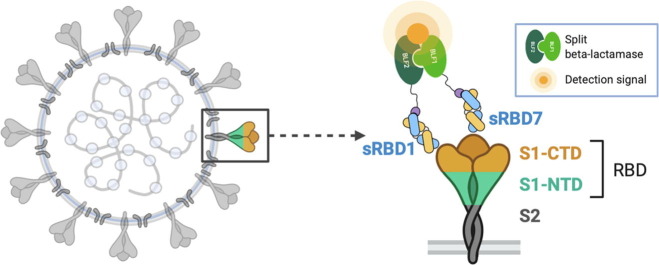
Graphical abstract
Keywords: point-of-care virus detection, Covid-19, antiviral therapeutics, protein engineering, split-enzyme complementation assay
Abstract
The Covid-19 pandemic is a centenarial global catastrophe. Similar events are likely to be recurring with more frequency in the future. The inability to control the virus’ impact is caused by many factors, but the lack of a technology infrastructure to detect and impede the virus at an early stage are principal shortcomings. Using phage display mutagenesis, we have generated a cohort of high performance antibody fragments (Fabs) that can be used in a sensitive point of care (POC) assay and are potent inhibitors (IC50-0.5 nM) to viral entry into cells. The POC assay is based on a split-enzyme (β-lactamase) complementation strategy that detects virus particles at low nM levels. We have shown that this assay is equally effective for detecting other viruses like Ebola and Zika. Importantly, its components can be freeze dried and stored, but becomes fully active when rehydrated.
Introduction
The COVID-19 (CoV-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become a global health and economic calamity. Unfortunately, at the onset of the pandemic, few methods and reagents were available to mount a timely response to the threat. The rate at which new viral infections like CoV-19 proliferate across populations makes it virtually impossible to respond effectively in real time without having specific infrastructure in place. The current standard for detection is through nucleic acid- based assays that involve utilization of polymerase chain reaction (PCR).1 While highly reliable, these have been shown to be impractical in rapid large scale systematic testing. Thus, there have been attempts to develop more rapid point of care (POC) approaches to detect low abundance viral antigens without the requirement of sophisticated instrumentation that the PCR based tests require. Clearly, what is needed is a set of detection assay formats that are well vetted by regulatory agencies so that when other pandemic threats are identified in their nascent state, there is a roadmap for how to assemble a rapid chain of responses.
With this in mind, we describe here the development of a sensitive and robust point of care (POC) viral detection assay. As with other protein antigen assays, the main component is a set of high performance antibody-based reagents used for antigen detection. Importantly, the same antibodies used for detection might also bind to epitopes that would lead to virus neutralization. It has been established that the viral entry of SARS-CoV-2 into cells is initiated by the receptor binding domain (RBD) of the trimeric spike protein of the virus binding to angiotensin-converting enzyme 2 (ACE2).2 Blocking this interaction is thought to be the most promising approach to neutralize the virus. For our applications, high affinity Fabs against the receptor binding domain (RBD) of the spike protein (S-protein) were generated by phage display mutagenesis technology. The Fabs are based on the Herceptin Fab scaffold that has been engineered for stability and expression.3 The principal epitope for Fab binding was found to be aligned with the ACE2 binding site on the RBD since it can be used as an effective viral inhibitor of cell entry. In this regard, using affinity maturation, we were able to generate a set of ultra high affinity binders that targeted this epitope with sub-nanomolar neutralization capabilities.
Most POC assays rely on some type of ELISA format for the detection readout.4 In contrast, the detection assay described here involves a split-enzyme complementation readout, which can have orders of magnitude better signal to noise discrimination than classical ELISA methods.4 The split-enzyme used in this complementation assay is based on the reconstitution of two fragments of β-lactamase to produce a fluorogenic signal. It is a variant of a traditional sandwich assay, the N and C-terminal fragments of β-lactamase are attached independently to different Fabs that recognize two distinct epitopes and thus, can bind simultaneously to the RBD target. Further, the complementation constructs containing the enzyme fragments are designed to be modular so that Fabs generated to other virus targets can be interchanged in a plug and play fashion. For instance, the same system detects Ebola and Zika antigens alongside SARS-CoV-2 antigen detection by simply adding the appropriate Fabs for the target of interest.3 Thus, the assay can be quickly recapitulated depending only on the step to generate Fabs to the desired target antigen. Other attributes built into our POC detection assay is that it can be packaged in a kit, which can be stored and rapidly activated without losing sensitivity.
Results
Generation of synthetic antibodies recognizing SARS-CoV-2 RBD
Our objective was to generate a cohort of high performance synthetic Fabs to the receptor binding domain (RBD) of SARS-CoV-2 that could be used for sensitive enzyme-complementation detection assays and for possible viral neutralization purposes. To generate Fabs that fill these criteria, we undertook a phage display mutagenesis campaign using the receptor binding domain (RBD) of SARS-CoV-2 as the target antigen. The RBD is highly glycosylated and thus was expressed in mammalian cells. Exploiting its high level of glycosylation, the protein was biotinylated through its carbohydrate groups with EZ-Link Hydrazide Biotins (Thermo) to facilitate immobilization onto streptavidin-coated paramagnetic beads during the phage display biopanning procedure. Five rounds of phage display selection were performed with a high diversity (1010) synthetic library5 using previously published protocols.6, 7 To obtain high affinity binders, the concentration of the RBD antigen was systematically reduced, starting from 500 nM during the first round and ending with 1 nM in round 5. To acquire binders with a slow dissociation rate, additional selection pressure with longer washes during biopanning was applied. Phage ELISA was performed on 192 clones resulting in identifying 5 unique high affinity variants: RBD1- RBD5 with RBD1 having the highest ELISA signal [Figure S1].
A key component of developing the split-enzyme complementation assay involves having a pair of high affinity Fabs that can bind simultaneously. An anticipated challenge was that since the SARS-CoV-2 RBD is heavily glycosylated, the available surface space to accommodate Fabs with non-overlapping epitopes would be reduced significantly. Having the high affinity RBD1 Fab in hand from the first selection, we sought to generate a second set of Fabs that bound to the RBD through independent epitopes. To obtain these Fabs, we employed an epitope masking strategy that involved adding the RBD1 Fab as a competitor during a second biopanning campaign. To do so, five rounds of selection with 1 μM RBD1 added as a competitor were performed starting with a 200 nM antigen concentration in round 1 and ending with an antigen concentration of 1 nM in round 5. After phage ELISA, positive clones were sequenced resulting in two unique sequences: sRBD6 and sRBD7, as well as the previously selected RBD5, indicating that this Fab from the original selection also had an independent epitope compared to RBD1 [Table S1]. From this set, we chose sRBD7 to pair with RBD1 from the first selection to carry forward for the development of the complementation assay.
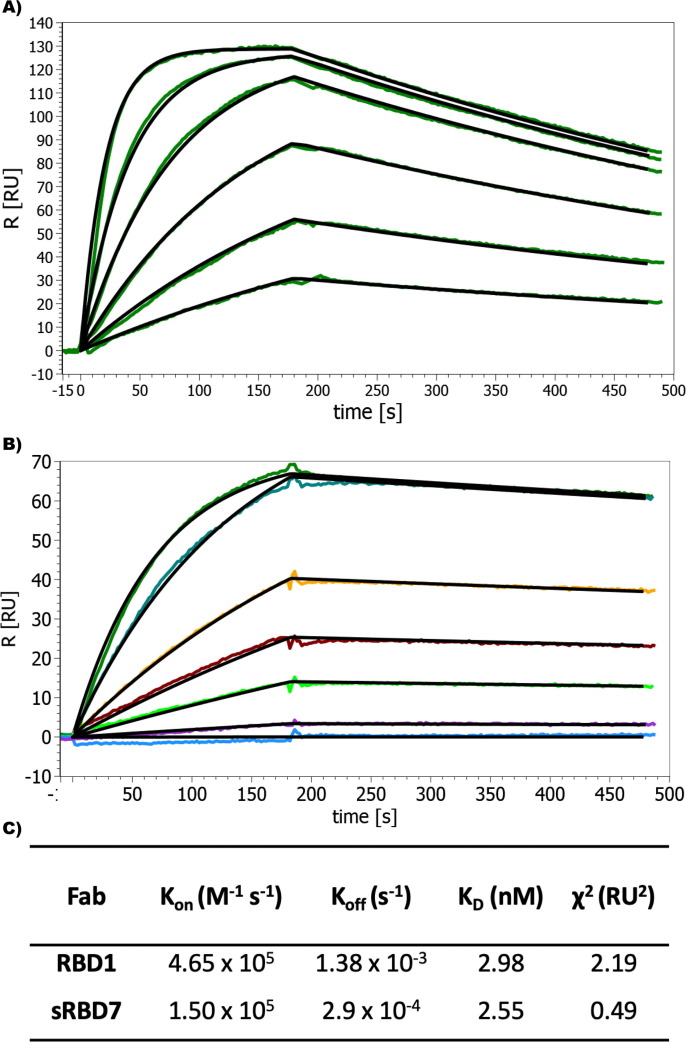
The binding affinities of RBD1 and sRBD7 were determinate by Surface Plasmon Resonance (SPR) showing they had similar binding constant (KD) of ~3 nM. However, their binding kinetics were quite different, where sRBD7 had almost an order of magnitude slower dissociation rate [Figure 1 ]. This reflects the success of the additional stringency placed on the biopanning conditions used for its selection.
Figure 1.
Characterization of Fab binding against RBD. (A) SPR sensogram showing the fast on-fast off kinetics between RBD1 and RBD. (B) Sensogram showing slower association and slower dissociation binding between RBD7 and RBD. (C) Kinetic parameters of binding. The concentration of Fabs were serially diluted two-fold for each run starting at 200 nM.
We have observed that epitope binning experiments based on protein ELISAs have to be interpreted with caution since Fabs that share a relatively small amount of epitope overlap can appear to be independent. To better quantify the independent epitopes, we performed epitope binning experiments by SPR. This confirmed that RBD1 and sRBD7 do bind to two distinct epitopes [Figure S2]. Further, although a full kinetic evaluation of RBD1 binding to the full SARS-CoV-2 spike protein could not be done due to a paucity of available protein, high affinity binding was confirmed based on a single injection SPR analysis (KD ~ 3 nM), which showed a similar binding profile to that observed for its binding to the RBD [Figure S3].
Affinity maturation of RBD1
Although the RBD1 has a high affinity (3 nM) and displays good kinetics, for the purposes of the complementation and neutralization assays, we decided to further improve its dissociation rate. Therefore, we initiated an affinity maturation campaign by designing and constructing phage display libraries to introduce higher diversity in CDR-H1. The choice of CDR-H1 was somewhat arbitrary since we did not have direct evidence to the extent of its engagement with the antigen; nevertheless, our experience has been that the heavy chain CDRs have a higher propensity to be involved antigen binding compared to light chain CDRs. We designed a combination of “tailored” and “hard” randomization strategies incorporated into six CDR-H1 positions. To that end, two sub-libraries were created, where in the first library mutated positions were bias to hydrophobic and aromatic amino acids, while the second library involved including all possible amino acids into the six positions. To ensure the improvement of Fab off-rates, additional selection pressure was applied during the biopanning. Three rounds of selection were performed with the RBD concentration being decreased accordingly, from 10 nM to 20 pM. Each round was additionally supplemented with four consecutives 30-min washing steps to strongly bias towards slow dissociation of the selected candidates.
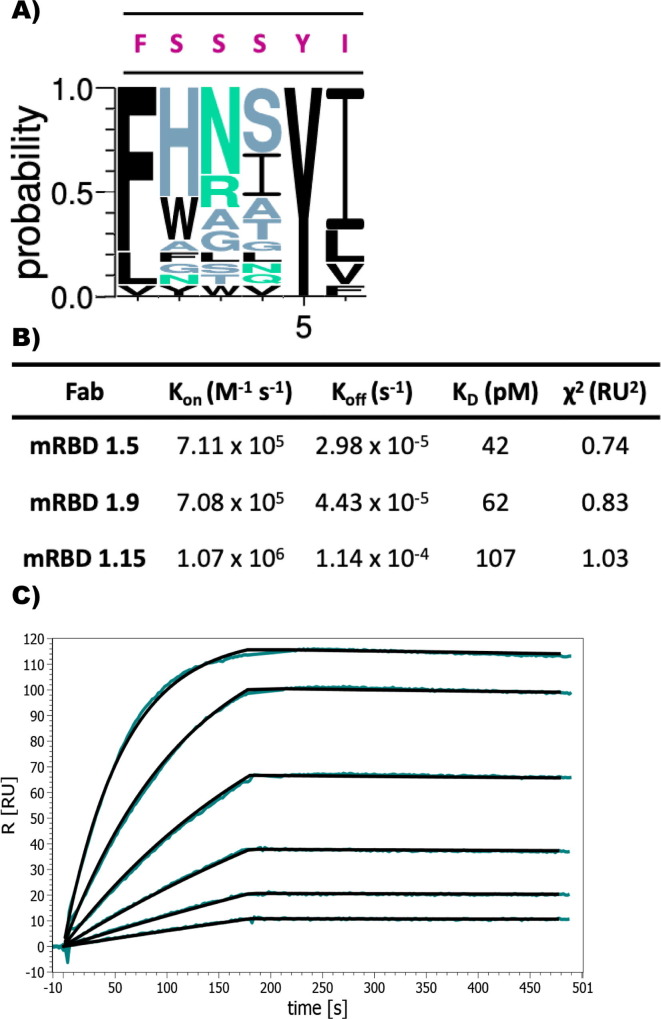
After round 3, 96 clones were picked and tested in phage ELISA leading to the identification of 19 sequence unique Fabs. Sequence alignment of these unique CDR-H1 variants showed no randomization at position 1 and a very limited randomization at position 5 and 6 compared to original RBD1 CDR-H1. High diversity at position 2, 3 and 4, with a preference for His at position 2 was observed [Figure 2 (A)]. The binding analysis showed the 10 to100-fold affinity improvement of matured CDR-H1 variants compared to the parent RBD1 Fab [Table S2]. Based on the overall binding kinetics, the affinity matured RBD1.5 variant (designated as mRBD1.5) had an affinity (KD) of 42 pM, where both the association and dissociation rates were improved by two orders of magnitude in comparison to WT RBD1 [Figure 2(B) and (C)].
Figure 2.
Affinity maturation of sRBD1. (A) WebLogo plot showing the sequence variants of the affinity matured CDR-H1 variants. Original sequence is presented in red. (B) Kinetic parameters of selected affinity maturated Fabs binding to RBD. Table of all variants is found in supplementary material. (C) mRBD 1.5 SPR sensogram showing the improved slow-dissociation rate. The concentration of Fab was serially diluted 2-fold starting at 25 nM.
Detection of SARS-CoV-2 with pGA1-FABLRT complementation assay
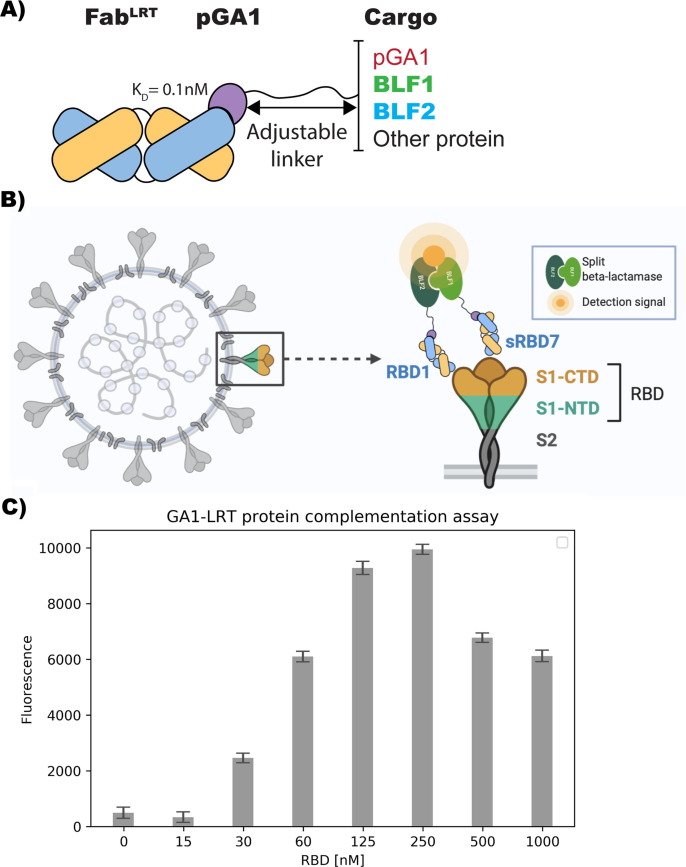
Previously we developed a sensitive protein complementation assay that was successfully applied for the detection of Ebola and Zika virus proteins.3 The system involves the use of the well established proximity-driven activity reconstitution of a TEM1 -lactamase (BL) split-enzyme system.8 In this assay, the two separate fragments, BFL1 (residues 26–196) and BFL2 (residues 198–220), are attached respectively to the two target molecules whose proximity is being evaluated. The assay system employed here does not involve linking the BFL fragments directly to the target molecule, but attaches these fragments to Fabs that are themselves directed against two distinct epitopes of the target, in this case the RBD of SARS-CoV-2. However, we have found that direct fusion constructs containing Fabs with linkers comprising the BLF fragments are difficult to express and tend to aggregate.3 Therefore, to circumvent these problems, we developed an alternative approach that exploits the high affinity (100 pM) between engineered forms of protein-G (pG) and a Fab’s constant domain [Figure 4(A)]. These engineered components are referred to as pGA1 (for protein G) and FabLRT (for the Fab). In this assay, the two separate fragments of the BL (BLF1 and BLF2) are attached through a linker to engineered protein pGA1.9 Thus, using the ultra-high affinity of FabLRT scaffold3 to protein pGA1, we were able to generate the pair of RBD1-BLF2 and sRBD7-BLF1 by premixing them separately to a final concentration of 250 nM. Conceptually, this non-wash assay requires simultaneous binding of the two FabLRT pairs to the target RBD protein [Figure 3 (B)]. Without knowledge of the RBD1 and sRBD7 epitope positions on the RBD, we decided to use a Gly-Ser linker of 15 residues between the protein pGA1-BLF (1 and 2) constructs. Titrating with increasing RBD concentration resulted in a detectable fluorescent signal at 30 nM, which increased linearly over the range from 30 to 125 nM with a distinct maximum at 250 nM [Figure 3(C)]. A reduction of the signal is observed at RBD excess, presumably caused by a breakdown of the stoichiometry at high antigen concentration due to the hook effect. To establish how the assay is influenced by the affinity levels of the Fab, we replaced the RBD1 Fab (KD ~ 3 nM) with the mRBD1.5 Fab (KD ~ 42 pM) coupled to BLF2, while keeping sRBD7 attached to BLF1. This resulted in about ~ 4-fold improvement in detection (Figure S4]. Control experiments involving attaching the two BLF fragments to either Fab RBD1 or sRBD7 resulted in no detectable signal when added in the presence of 250 nM RBD. This indicates that productive enzyme complementation requires the two Fabs to bind simultaneously to the same RBD molecule [Figure S5].
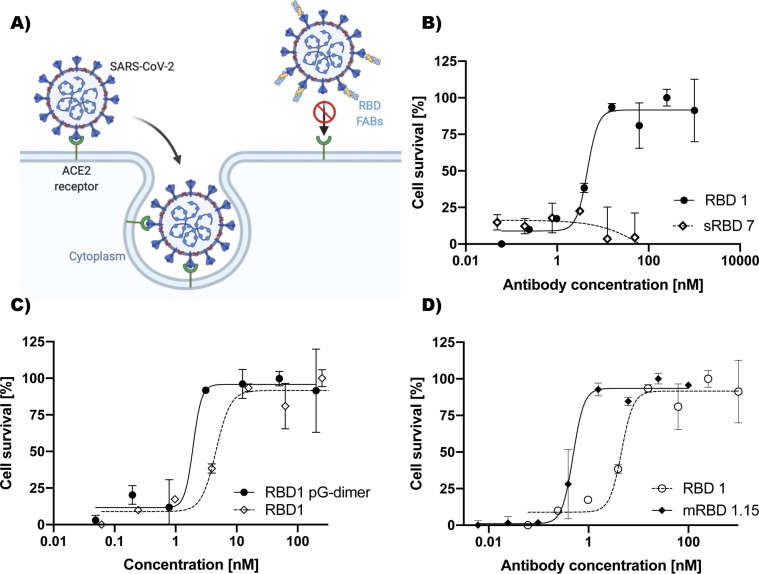
Figure 4.
Antibody mediated SARS-CoV-2 neutralization. (A) Neutralization mechanism model. Anti-RBD Fabs block the interaction with human ACE2 receptor on the cell surface. (B) Protection from cell death achieved by different concentrations of the RBD1 and RBD7 Fabs. Neutralization IC50 value for RBD1 is 4.8 nM. No efficient neutralization with RBD7 was observed. (C) Virus neutralization improved over 2.5-fold by RBD1 dimerization. IC50 neutralization for the dimerized RBD1 was 1.9 nM. (D) Improvement in SARS-CoV-2 neutralization. IC50 of mRBD 1.15 was 10-fold improved with a value of 0.48 nM. Results were normalized. All Experiments were done in triplicate and the error bars represent SD value from the mean.
Figure 3.
GA1-FABLRT protein complementation assay. (A) Model of FabLRT-pGA1 modular platform. (B) Model of SARS-CoV-2 detection using pGA1-FABLRT protein complementation assay. Two separate fragments of BL enzyme are attached to two different Fabs via protein pGA1-LRT interaction. BL fragments can associate to form an active enzyme state when Fabs bind to the Receptor Binding Domain (RBD). (C) Detection of SARS-CoV-2 at different concentrations using complementary parts: pGA1-BLF1/sRBD7 and BLF2-pGA1/RBD1. Detectable signal was observed from 15 to 250 nM. At higher concentration, the hook effect is observed. Reaction was incubated for 20 min at RT; Results were normalized. Experiment was done in triplicate and the error bars display SD value from the mean.
Next, we tested if the complementation system could be used for detection of full trimeric S-protein, since the RBD Fab epitopes are unknown and might not be accessible in a context of a full-length S-protein. Notably, the S-protein was readily detected with a higher sensitivity with a detectable concentration starting at 7 nM with a distinct maximum at 125 nM [Figure S6]. A similar control experiment to the one above, where each Fab was premixed with both components (i.e. RBD1- BLF1, RBD1-BLF2) showed that measurable signal was not produced in this format [Figure S7]. Thus, as was the case for the RBD domain alone, triggering a signal requires the Fabs to bind to two independent epitopes on the same trimeric S-protein configuration.
Post lyophilization stability of pGA1-FABLRT complementation assay
One of the key features we wished to instill in our point-of-care assay for ease of storage was the ability to preserve the functionality of the components upon reconstitution from a freeze-dried form. To that end, single Fabs, RBD1-BLF2 and sRBD7-BLF1 at 500 nM were lyophilized overnight. Upon reconstitution no aggregation was observed and the Fabs’ binding properties were unchanged [Figure S8(A)]. Moreover, the pGA1-FABLRT complementation assay performed equally well as the control (not lyophilized) when the sample was supplemented with a protein carrier (BSA) as part of the freeze-drying procedure. Lack of BSA supplementation led to 40% signal reduction [Figure S8(B)].
Antibody mediated SARS-CoV-2 neutralization
To determine the potential level of virus neutralization, the Fabs were tested in a plaque reduction assay that measures the cell survival afforded by the Fabs. This is a stringent assay that is the gold standard currently in use.10 RBD1 showed a dose-dependent SARS-CoV-2 infection neutralization with an IC50 of 4.8 nM [Figure 4 (B)]. Conversely, as expected, sRBD7 showed no virus neutralization capacity [Figure 4(B)], since it recognizes a different epitope than RBD1. Additionally, we anticipated that neutralization could be further increased by converting the RBD1 Fab into a bivalent IgG type format. As an alternative approach to introducing bivalency, we tested the Protein pGA1 dimer with 52 Gly-Ser linker with two LRT grafted3 RBD1 Fabs. This resulted in 2.5-times improvement in viral neutralization (IC50 = 1.9 nM) [Figure 4(C)]. Additionally, the affinity matured Fab, mRBD1.15, had over 10-times improved neutralization capabilities (IC50 = 0.48 nM) in a monovalent form [Figure 4(D)]. These findings suggest a potential for the development of anti-viral drugs based on Fabs that bind tightly to the RBD.
Discussion
We have described the generation and validation of a cohort of Fabs produced by phage display mutagenesis (Table S1) that have proven to be potent blocking inhibitors to viral entry into host cells and can be utilized in a sensitive point of care split-enzyme complementation assay to detect the presence of SARS-CoV-2 viral particles. In both applications, we targeted the Fabs to the receptor binding domain (RBD) of the S1 subunit of the spike protein (S-protein). The S-protein protrudes from the virus surface as a homo-trimeric assembly making it a potentially effective target for antibody engagement. Cryo-EM studies revealed the existence of conformational heterogeneity in the trimeric organization of the RBD within the S-protein; one of the RBD domains being in an open position and accessible for binding to the ACE2 receptor, while the other two were in an inaccessible “closed position.11 While there may be several targetable epitopes on the RBD that bind agnostically to either the open or closed states, any antibody that binds to the RBD- ACE2 receptor binding interface most probably targets only the single open conformation of the RBD domain in the S-protein.
An additional challenge for generating antibodies to the RBD is that the S-protein is heavily glycosylated, presumably to help the virus evade the immune system.12 In phage display selections, it is rarely observed that the glycosylation sites are part of the binding epitope for myriad reasons. Thus, the actual available epitope space depends on the amount and organization of the glycosylation, both on the RBD itself and other spatially proximal segments of the S-protein. Recent cryo-EM structures of the S-protein show that glycosylation does indeed shield a significant portion of the RBD, but there are patches of unimpeded surface that we presumed could be targeted.13
Phage display selection produced four Fab binders (RBD1-4), which shared the same epitope. Of these binders, RBD1, had superior binding properties, as judged by ELISA and SPR (KD- 3 nM). A competition assay with soluble ACE2 showed that this epitope is associated with the ACE2-RBD interface [Figure S9]. To assess the ability of RBD1 to neutralize the viral activity of SARS-CoV-2, a plaque reduction assay was performed.10 This is a stringent assay that measures the percentage of cell survival as a function of Fab concentration. Serial additions of the RBD1 resulted in a clear dose dependent cell survival response with an IC50 of 4.8 nM [Figure 4(B)]. Notably, using the monovalent RBD1 Fab alone displayed comparable neutralization potency with other reported IgG antibodies having bivalent formats.14, 15, 16 We next decided to evaluate whether further improvement could be achieved by converting RBD1 into a format mimicking IgG bivalency. This was done by employing an engineered construct we had previously developed that effectively couples two Fabs together through a linker of adjustable length3 [Figure 3(A)]. An important consideration for the linker length was to ensure the linked Fabs could bind on different S-proteins on the viral coat since there is only a single accessible RBD available for Fab binding per spike. As a compromise between limiting the linker length, while ensuring that it was long enough to facilitate the two RBD1 Fabs to bind across spikes (~160 Å apart),17 a 52 residue Gly-Ser linker was employed. The viral neutralization potential of this bivalent construct was evaluated in the plaque reduction assay and showed a 2.5- fold improvement (IC50- 1.9 nM) in cell protection over the monovalent RBD1 Fab [Figure 4(C)]. Because these assays have to be run at a special containment facility and consequently are low throughput, we were not able to systematically test different linker lengths to further optimize the bivalent effects of the construct.
While the RBD1 Fab had potent neutralization properties, we sought to further improve it through an affinity maturation process. From the affinity maturation phage display selection, 19 unique clones were isolated and reformatted into protein format. The affinities and kinetics of these Fabs showed marked reductions in KD and dissociation rates; for instance, affinity matured Fab, mRBD1.5 had a 100-fold improved affinity and off-rate compared to the parent RBD1; a number of the other affinity matured Fabs showed affinity increases in the range of 50-fold [Table S2]. Notably, a number of these Fabs exhibited extremely slow off-rates which were at the limits of what can be measured by SPR accurately, so their affinities may be underestimated.
To establish whether these higher affinities would translate into more effective inhibitors of viral entry, the plaque reduction assay was performed using mRBD1.15 (KD- 100 pM). Indeed, this Fab displayed a 10-fold greater neutralization capacity (EC50- 0.48 nM) than the RBD1 parent (EC50- 4.8 nM). Although there were several affinity matured Fabs with higher affinities (mRBD1.5–42 pM, mRBD1.9–62 pM), these were not tested because at affinities below 1 nM, the dynamic range of the assay is significantly reduced. Likewise, improved neutralization that may have been achieved by converting these Fabs into a bivalent format could not be evaluated for similar reasons.
The second goal of the project was to develop a sensitive high throughput point-of-care (POC) detection system to identify virus infection. To accomplish this, we employed an assay built upon our previously developed and deployed split-enzyme complementation assay for the detection of Ebola and Zika virus based on the split β-lactamase system.3 This complementation format is a variation of a classical sandwich assay and has superior target specificity and sensitivity compared to traditional ELISA assays.4 Having established this proof of concept, we endeavored to repurpose the detection platform for detecting SARS-CoV-2.
For the complementation assay to detect the RBD, we used RBD1 from the first selection and sRBD7, which was obtained using an epitope masking strategy ensuring it bound to an orthogonal epitope to RBD1. Connecting these Fabs to the β-lactamase fragments using the pGA1- FabLRT constructs resulted in detection with excellent signal-to-noise of the RBD domain at a concentration of 30 nM [Figure 3(C)]. We next tested the sensitivity in the context of the full S-protein. In this format, possible interference due to glycosylation or other steric shielding involving the trimeric nature of the S-protein could be assessed. For instance, in the full trimeric S-protein, the RBD would be displayed in two conformational states, one copy in the open position making it accessible for ACE2 binding and two copies in the down position, which presumably covers this binding site. We anticipated that RBD1 would bind to the open state since its mode of neutralization involves direct epitope competition with ACE2. Notably, the epitope for sRBD7 is also available since adding the two Fabs with their associated β-lactamase fragments leads to enzyme reconstitution and activity. Importantly, the detection level is more sensitive in the context of the full S-protein leading to appreciable signal over noise discrimination at 7 nM concentration of the S-protein [Figure S6]. Thus, we postulate that using the affinity matured mRBD1.5 version would lead to a further increase in detection sensitivity.
The linker lengths on the BLFs are too short to accommodate complementation between Fab- BLF constructs binding across two adjacent spike assemblies on the virus surface. However, it was an open question as to whether the signaling induced by the complementation in the format described here requires the two Fabs to bind in cis (to the same RBD domain) or could also work in trans (different RBDs on the same S-protein). Control experiments involving different combinations of the RBD1 and sRBD7 Fabs coupled to the β-lactamase fragments were tested by pairing each of the two Fabs coupled to both fragments: i.e. RBD1-BLF1, RBD1-BLF2 or sRBD7-BLF1, sRBD7-BLF2. In these configurations no signal was observed. This would be expected in the case of the RBD1- BLF1, RBD1- BLF2 pair since the Fab can only bind to the single RBD in the open state conformation per spike. However, it was not clear whether sRBD7 could bind to the RBD in both the open and closed conformations. If that were true, however, it would be expected that sRBD7 constructs paired with both BLF fragments could complement successfully on one trimeric S-protein. The observation that this configuration produced no signal suggests that the mechanism of complementation that is in play in our system involves RBD1 and sRBD7 binding on the same RBD unit, with both RBD1 and sRBD7 binding to its open conformation state.
An additional enhancement is the assay’s portability that provides for the POC aspects of its utility. The objective was to establish a kit where all the reagents could be packaged and stored on the shelf, to be reconstituted when needed upon rehydration. To accomplish this, we tested whether the Fab and pGA1 linkers with the associated enzyme fragments could be lyophilized and reconstituted with no loss in functionality. Figure S8 shows that these components are not affected by lyophilization and are as active after reconstitution as their counterparts which were not lyophilized. Thus, it will be possible to package an assay like this into a kit that is cheap, storable and readily useable in the field.
To put this work into context, we undertook this project with the full appreciation that the tools and reagents that we produced would not make an immediate impact during the current pandemic. However, the strategies devised and applied here provide a blueprint for quickly responding to future challenges, which will unfortunately occur. As in the case of many efforts to respond to the Cov-19 pandemic in real time, this work was accomplished during a lab shutdown with minimal research material and staff. Nevertheless, exploiting our phage display mutagenesis pipeline, we generated a set of high performance inhibitors to viral entry. Further, repurposing an assay platform that was previously developed for detecting Ebola and Zika infections, we were able to quickly incorporate a set of high performance Fab binders into a detection platform that could be utilized for sensitive virus detection in an easily used kit format.
Material and Methods
Protein cloning, expression and purification
The sequences of the constructs used are provided in Table S3. The RBD protein was produced in mammalian cells (gifted by Wilson and Hubbell labs from the University of Chicago). Proteins for the split enzyme proximity assay were cloned into pHFT2 vector18 with the strategy described previously.3 Selected Fabs were cloned from phage into Sph1 sites of pSFV4 expression vector using an Infusion HD cloning kit (Takara Bio) according to the protocol. All selected Fab CDR sequences are provided in Table S1. The FabLRT scaffold was grafted into Fab light chain at aa positions 123–127 (SQLKS -> LRT) using quick change site-directed mutagenesis.
Fabs and Fab-GA1 fusions were expressed in the periplasm of E. coli BL21 cells for 4 hours at 37 °C post induction with 1 mM IPTG at OD600 = 0.8–1. The cells were harvested by sonication in Protein G-wash buffer9 (50 mM Phosphate buffer, 500 mM NaCl, pH 7.4). After centrifugation the supernatant was applied on the protein G-F affinity column created in the lab (Slezak et al. manuscript in preparation) using SulfoLink Coupling Resin (Thermo Scientific). Proteins were eluted from the column with 0.1 M glycine, pH 2.6, and neutralized with 1 M Tris-HCl, pH 8.5. Fabs and Fab-GA1 fusions were dialyzed overnight into HBS.
For the proximity assay, pGA1-BLF fusions were expressed and purified as described previously.3 Briefly, proteins were expressed in BL21 (DE3) overnight in 20 °C post induction with 1 mM IPTG at OD600 = 0.6. Cells were sonicated in buffer A containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl and 10% glycerol. The insoluble His-tagged pGA1-BLF fusions were extracted from the cell pellets by 6 M Gua-HCl in buffer A with 0.3 mM TCEP and purified on TALON resin (Takara Bio) using a denaturation-condition protocol and on-column renaturations reached by six washes of the column with serially diluted 6 M Gua-HCl, followed by the final wash in buffer A alone. Proteins were eluted from the column with 100 mM imidazole in buffer A. Fusion proteins were stored on ice and never frozen.
Phage display selection protocol
To obtain high affinity binders, five rounds of selection were performed using phage display selection protocol.6, 19 RBD protein was biotinylated via glycoproteins with EZ-Link Hydrazide Biotin (Thermo Scientific) as recommended in order to immobilize onto streptavidin-coated paramagnetic beads (Promega and Dynabeads M0270, Invitrogen). In the first round of selection, 500 nM of RBD was immobilized on 200 l SA magnetic beads (Promega) and was incubated with 1 mL phage library (1011 cfu) for 1 hour at room temperature with gentle shaking. The beads were washed three times to remove nonspecific phage and added to log phase E. coli XL-1 blue cells (Stratagene) and incubated for 20 min at room temperature. Then, media containing 100 g/mL ampicillin and 109p.f.u./mL of M13K07 helper phage (NEB) was added for overnight phage amplification at 37 °C. For all subsequent rounds, the amplified phage was precipitated in 20% PEG/2.5 M NaCl for 20 min on ice. Before each round, the phage pool was negatively selected against empty paramagnetic beads for 30 min with shaking to eliminate nonspecific binders. The final concentration of antigen was dropped systematically from 500 to 1 nM from the first to the fifth round (2nd round: 200 nM, 3rd round: 50 nM, 4th round 10 nM and 5th round 1 nM). After phage binding, the beads were subjected to five washing rounds. Last two washes steps were 20 and 25 min respectively. This additional selection pressure was supposed to remove all low affinity binders. The bound phages were eluted using 0.1 M glycine, pH 2.6 and neutralized with TRIS-HCl, pH 8. Then, the phage eluate was used for E. coli infection and phage amplification as described above. Additional selection to generate non-overlapping epitope with sRBD1 was performed. Selection protocol was as described above with addition of 1 of sRBD1 in every step to ensure binding to a different epitope. The final concentration of antigen was dropped gradually from 200 to 1 nM from the first to the fifth round (2nd round: 50 nM, 3rd round: 20 nM, 4th round 10 nM and 5th round 1 nM). After round 4th and 5th phages were plated on ampicillin plates and 96 single colonies were picked for single-point phage ELISA assays. The promising clones demonstrating high ELISA signal and low non-specific binding were sequenced and reformatted into a pSFV4 expressing vector as described in Protein expression and purification.
Enzyme-Linked immunoabsorbent assays (ELISA)
The ELISA protocol was described previously.20 Briefly, 50 nM of RBD was directly immobilized on high binding experimental wells (Greiner Bio) and BSA was immobilized in control wells, followed by extensive blocking with BSA. After 15 min incubation with phage, the wells were extensively washed three times and incubated with Protein L- HRP (Thermo Scientific, 1:5000 dilution in HBST) for 20 min. The plates were again washed and developed with TMB substrate (Thermo Scientific) and quenched with 10% H3PO4, followed by the absorbance at A450 determination.
Affinity maturation of RBD1
Phage libraries of RBD1 for affinity maturation were created using the strategy previously published.21 To that end, a stop codon was introduced in CDR-H1 with quick-change mutagenesis. Two phage libraries with “hard” and “tailored” randomization strategies were created with the phosphorylated oligos. ssDNA containing stop codon introduced in the middle of CDR-H1 was isolated from phage (using QIAprep Spin M13 Kit, Qiagen) and used in a Kunkel mutagenesis protocol.22 The Kunkel reaction was purified (with Wizard SV Gel and PCR Clean-Up System, Promega), electroporated into TG1 cells (Lucigen) and after 1-hour recovery in 37 °C with shaking, 40 mL of 2xYT media (supplemented with 100 g/mL ampicillin and helper phage) was added to initiate phage production. The next day, the libraries were precipitated as described above. Three rounds of biopanning were performed with different target concentration (1st round: 10 nM, 2nd round: 1 nM, 3rd round: 20 pM). To ensure the improvement in dissociation constant, selection process included 4 consecutive 30 min washing steps. Affinity improvement of the selected clones was tested by SPR.
Phosphorylated oligos used for libraries design:
H1: CAGCTTCTGGCTTCAACNTTNNCNNSNWWNNTNTTCACTGGGTGCGTCAGGCC,
H2: GCTTCTGGCTTCAACTTCNNKNNKNNKNNKATACACTGGGTGCGTCAG
(N standing for all 4 bases, S standing for G or C, W standing for A or T).
Surface plasmon resonance analysis
All Surface plasmon resonance (SPR) analyses were performed on a MASS-1 (Bruker). All targets were immobilized via a 6x His-tag to a Ni-NTA sensor chip. Fabs in twofold dilutions were run as analytes at 30 l/min flow rate at 20 °C. Sensograms were corrected through double referencing and 1:1 binding model fit was done using Sierra Analyzer (Bruker). For epitope binning experiments, after injection of a saturating concentration of the RBD1 Fab, an equal molar mixture of sRBD7 fab was injected and increase in Response Unit (RU) was observed.
pGA1-FABLRT protein complementation assay
Viral detection protocol using non-wash pGA1-FABLRT protein complementation assay was previously described.3 Briefly, 250 nM of each pGA1-BLF fusion, premixed with RBD1 or RBD7 Fabs, were combined in a black FluoroNunc 96-well plate (Nunc). Different concentrations of viral proteins and 2 M fluorogenic BL substrate (Fluorocillin Green 495/525, Life Technologies) was added to the final volume of 100 l. Fluorescent signal was monitored at room temperature using Safire2 Tecan Plate Reader (483 nm excitation, 525 nm emission). The results were reproduced at least three times. Results were normalized by subtraction of a substrate background fluorescence.
Plaque reduction neutralization assay
Vero E6 cells (ATCC) were infected under biosafety level 3 conditions with SARS-CoV-2 (nCoV/Washington/1/2020, kindly provided by the National Biocontainment Laboratory, Galveston, TX). The neutralization assay was performed as previously described23 with some modifications. Briefly, the Fabs were serially diluted 4-fold and mixed with 400 PFU of SARS-CoV-2 for one hour at 37 °C, then used to infect Vero E6 cells for three days. Cells were fixed with 3.7% formalin and stained with 0.25% crystal violent. Crystal violet-stained cells were then quantified by absorbance at (595 nm) with a Tecan m200 microplate reader. The 50% neutralization titer was then calculated using GraphPad Prism.
CRediT authorship contribution statement
Tomasz Slezak: Conceptualization, Methodology, Investigation, Formal analysis, Writing - original draft, Writing - review & editing. Anthony A. Kossiakoff: Conceptualization, Supervision, Writing - original draft, Writing - review & editing, Funding acquisition.
Acknowledgments
Acknowledgments
We thank the Wilson and Hubbell labs from the University of Chicago for providing the RBD protein and Wells lab from the UCSF for ACE2 protein. We acknowledge the assistance and advice of S. Mukherjee, E. Davydova and A. Prominski. SARS-CoV-2 infections were performed under BSL-3 containment by the SARS-CoV-2 Research Core at the Howard T. Ricketts Laboratory.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Edited by Sachdev Sidhu
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2021.166956.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Udugama B., Kadhiresan P., Kozlowski H.N., Malekjahani A., Osborne M., Li V.Y.C., Chen H., Mubareka S., et al. Diagnosing covid-19: The disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 2.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of sars-cov-2 by full-length human ace2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slezak T., Bailey L.J., Jaskolowski M., Nahotko D.A., Filippova E.V., Davydova E.K., Kossiakoff A.A. An engineered ultra-high affinity fab-protein g pair enables a modular antibody platform with multifunctional capability. Protein Sci. 2020;29(1):141–156. doi: 10.1002/pro.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hart R.W., Mauk M.G., Liu C., Qiu X., Thompson J.A., Chen D., Malamud D., Abrams W.R., et al. Point-of-care oral-based diagnostics. Oral Dis. 2011;17(8):745–752. doi: 10.1111/j.1601-0825.2011.01808.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller K.R., Koide A., Leung B., Fitzsimmons J., Yoder B., Yuan H., Jay M., Sidhu S.S., et al. T cell receptor-like recognition of tumor in vivo by synthetic antibody fragment. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0043746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paduch M., Koide A., Uysal S., Rizk S.S., Koide S., Kossiakoff A.A. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods. 2013;60(1):3–14. doi: 10.1016/j.ymeth.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellouse F.A., Esaki K., Birtalan S., Raptis D., Cancasci V.J., Koide A., Jhurani P., Vasser M., et al. High-throughput generation of synthetic antibodies from highly functional minimalist phage-displayed libraries. J. Mol. Biol. 2007;373(4):924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 8.Galarneau A., Primeau M., Trudeau L.E., Michnick S.W. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nature Biotechnol. 2002;20(6):619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 9.Bailey L.J., Sheehy K.M., Hoey R.J., Schaefer Z.P., Ura M., Kossiakoff A.A. Applications for an engineered protein-g variant with a ph controllable affinity to antibody fragments. J. Immunol. Methods. 2014;415:24–30. doi: 10.1016/j.jim.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muruato A.E., Fontes-Garfias C.R., Ren P., Garcia-Blanco M.A., Menachery V.D., Xie X., Shi P.Y. A high-throughput neutralizing antibody assay for covid-19 diagnosis and vaccine evaluation. Nature Commun. 2020;11(1):4059. doi: 10.1038/s41467-020-17892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-em structure of the 2019-ncov spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the sars-cov-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sikora M., von Bülow S., Blanc F.E.C., Gecht M., Covino R., Hummer G. Map of sars-cov-2 spike epitopes not shielded by glycans. bioRxiv. 2020 doi: 10.1371/journal.pcbi.1008790. 2020.2007.2003.186825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., et al. A noncompeting pair of human neutralizing antibodies block covid-19 virus binding to its receptor ace2. Science. 2020;368(6496):1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., et al. Potent neutralizing antibodies against sars-cov-2 identified by high-throughput single-cell sequencing of convalescent patients' b cells. Cell. 2020;182(1):73–84 e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., et al. Structures of human antibodies bound to sars-cov-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182(4):828–842 e816. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turonova B., Sikora M., Schurmann C., Hagen W.J.H., Welsch S., Blanc F.E.C., von Bulow S., Gecht M., Bagola K., Horner C., et al. In situ structural analysis of sars-cov-2 spike reveals flexibility mediated by three hinges. Science. 2020;370(6513):203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J., Koide A., Makabe K., Koide S. Design of protein function leaps by directed domain interface evolution. Proc. Natl. Acad. Sci. USA. 2008;105(18):6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizk S.S., Kouadio J.L., Szymborska A., Duguid E.M., Mukherjee S., Zheng J., Clevenger C.V., Kossiakoff A.A. Engineering synthetic antibody binders for allosteric inhibition of prolactin receptor signaling. Cell Commun. Signal. 2015;13:1. doi: 10.1186/s12964-014-0080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paduch M., Kossiakoff A.A. Generating conformation and complex-specific synthetic antibodies. Methods Mol. Biol. 2017;1575:93–119. doi: 10.1007/978-1-4939-6857-2_6. [DOI] [PubMed] [Google Scholar]
- 21.Sidhu S.S., Li B., Chen Y., Fellouse F.A., Eigenbrot C., Fuh G. Phage-displayed antibody libraries of synthetic heavy chain complementarity determining regions. J. Mol. Biol. 2004;338(2):299–310. doi: 10.1016/j.jmb.2004.02.050. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA. 1985;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Madrid A.T., Porterfield J.S. A simple micro-culture method for the study of group b arboviruses. Bull. World Health Organ. 1969;40(1):113–121. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.